Abstract
Research efforts spanning the past two decades have established a clear link between nuclear receptor function, regulation of the circadian clock and lipid homeostasis. As such, this family of receptors represents an important area of research. Recent advances in the field have identified two nuclear receptor subfamilies, the REV-ERBs and the ‘retinoic acid receptor-related orphan receptors’ (RORs), as critical regulators of the circadian clock with significant roles in lipid homeostasis. In this review, the latest information garnered from cutting-edge research on these two nuclear receptor subfamilies will be discussed. Through direct targeting of the REV-ERBs and RORs with synthetic ligands, generation of novel tools aimed at characterizing their function in vivo have been developed, which may lead to novel therapeutics for the treatment of metabolic disorders.
Nuclear receptors: ligand-activated transcription factors
Nuclear receptors (NRs) are a family of highly conserved transcription factors that regulate gene transcription in response to various environmental stimuli. There are 48 NRs that exist in humans, but some organisms such as worms contain nearly 300 members of the family [1]. NRs share considerable amino acid sequence homology with highly conserved domain structures, including a variable amino-terminal A/B domain, a central, highly conserved DNA-binding domain containing two zinc fingers, a hinge region and a carboxy-terminal ligand-binding domain (LBD, or E region) [2]. Several NRs contain an additional C-terminal F domain, the function of which is poorly understood. Examples of classical and well-characterized NRs include the estrogen receptor (ER), progesterone receptor, glucocorticoid receptor and thyroid receptor.
Nuclear receptors work in concert with other proteins to regulate the expression of genes important for an organism’s development, homeostasis and metabolism. NRs bind directly to specific DNA response elements in the regulatory region of their target genes and regulate the rate of transcription in a ligand-dependent manner. Most NRs bind to DNA as dimers: either homodimers, as is the case for steroid hormones, or heterodimers with another member of the NR superfamily (the retinoid X receptors [RXRs]) [3,4]. A small number of NRs have the ability to bind to DNA as monomers. Ligand binding induces a conformational change within the LBD, exposing a surface for specific cofactor interactions, which results in the recruitment of additional proteins that subsequently modify the chromatin and contact the basal transcriptional machinary to alter gene transcription. NR ligands are small, hydrophobic molecules, such as steroid hormones, fatty acids, lipophilic vitamin derivatives, dietary metabolites, as well as antibiotics and synthetic drugs. Ligands are classified according to their ability to regulate a NR’s transcriptional activity. Agonists bind to the LBD and induce a conformational change that results in increased recruitment of cofactor proteins that result in alterations in target gene transcription eliciting a ‘maximal’ response. An antagonist does not provoke a response of the receptor, rather it blocks the ability of an agonist to bind and generate a response. If a particular receptor has ligand-independent ‘basal’ activity, inverse agonists may also be identified. An inverse agonist binds to the receptor but inhibits the constitutive activity of the receptor. This class of NR ligands can be observed when a particular receptor has a basal conformation that permits interaction with a cofactor protein (coactivator or corepressor) leading to basal activity. An inverse agonist induces a conformational change within the receptor that decreases the affinity of the receptor for the cofactor protein. Partial agonists bind and activate a receptor, but have only partial efficacy at the receptor relative to a maximally efficacious ‘full’ agonist. In addition, analogous to partial agonists, partial inverse agonists display only limited ability to block the basal activity of a receptor. Thus, there are many different types of ligands that can modulate the transcriptional activity of a NR, the commonality being their ability to bind a NR. Approximately half of the NRs found in humans have identified endogenous ligands [1]. The remaining NRs are considered ‘orphan’ receptors, as they have no known, or generally agreed upon, ligands.
Orphan NRs represent an active area of research due to the potential for identification of ligands that may be used to modulate these receptors and may lead to treatments for various diseases. Recently, heme was identified as the ligand for the orphan receptors REV-ERBα/β, the well-known regulators of the circadian clock and lipid metabolism [5,6]. Another orphan subclass, the ‘retinoic acid receptor-related orphan receptors’ (RORs), recognize the same DNA binding sites as the REV-ERBs and are often coexpressed in the same tissues as the REVERBs [2,7]. The RORs are also well known regulators of the circadian clock and recent evidence has associated their activity with metabolic processes, including lipid homeostasis. De-orphanizing these receptors will further serve to advance our knowledge of the two NRs, their dynamic interplay and ultimately their roles in an organism’s physiology. Furthermore, the information garnered from these studies will help increase the opportunities for therapeutic intervention of disorders caused by aberrant ROR and REV-ERB activity.
The RORs & REV-ERBs
The first member of the ROR subfamily, RORα, was identified in the early 1990s based on sequence similarities to the retinoic acid receptors (RARs) and the RXRs [7,8]. The highly similar receptors, RORβ and RORγ, were identified soon thereafter [9,10].
The three RORs display significant sequence similarities. Each gene generates multiple isoforms based on alternative promoter usage and exon splicing, with all of the isoforms varying only in the amino-terminal region of the receptor [11]. In humans, four isoforms of RORα exist (α1–α4), while only α1 and α4 are present in mice. Tissue expression of RORα includes the liver, skeletal muscle, skin, lungs, adipose tissue, kidney, thymus and brain. Two isoforms of RORβ are found in mice (β1 and β2) whereas only β1 is present in humans. The expression of RORβ is limited to the CNS. Finally, two isoforms of RORγ are found in both humans and mice (γ1 and γ2). RORγ2 is often regarded as RORγt and found primarily in the immune system. RORγ is most highly expressed in the thymus (mainly RORγt) but is also expressed in the liver, skeletal muscle, adipose tissue and kidney [11]. RORγt has been the focus of considerable attention due to its role in TH17 cell development and its exclusive expression in key cells in the immune system. The three RORs and their various isoforms display distinct patterns of tissue expression and are involved in the regulation of physiological processes utilizing specific target genes. However, owing to significant sequence similarities, in cells where RORs are coexpressed, the different RORs may exhibit functional overlap [11]. RORs exhibit a typical NR domain structure (Figure 1A), as discussed in the introduction. All RORs recognize and bind as monomers to specific sequences of DNA, termed ROR response elements (ROREs), typically consisting of an AGGTCA ‘half site’ with a 5′ AT-rich extension in the regulatory region of the target gene [8–10]. When bound to this element, the RORs recruit coactivators resulting in constitutive activation of transcription of their target genes [11,12].
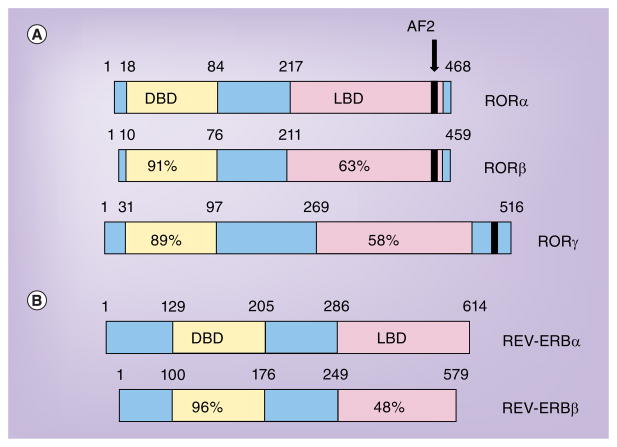
Figure 1. Comparison of human retinoic acid receptor-related orphan receptors and REV-ERB nuclear hormone receptors.
(A) The numbers indicate amino acid identity relative to RORα. RORα4, RORβ1 and RORγ1 are shown. (B) The numbers indicate amino acid identity relative to REV-ERBα. AF: Activation-function; DBD: DNA-binding domain; LBD: Ligand-binding domain; ROR: Retinoic acid receptor-related orphan receptor.
Another NR subclass, the REV-ERBs (REVERBα and REV-ERBβ), display significant crosstalk with the RORs. The REV-ERBs were originally identified as orphan receptors based on homology to the canonical NR domain structure (Figure 1B). REV-ERBα and REV-ERBβ have overlapping expression patterns and are found in adipose tissue, skeletal muscle, the brain and liver. However, the receptors display different relative tissue-specific expression levels; for example, REV-ERBα is broadly expressed at the same level in many different tissues, whereas REV-ERBβ is very highly expressed in certain tissues, showing high expression in parts of the brain (pineal and prefrontal cortex), thyroid, uterus and pituitary.
REV-ERBs regulate the expression of many target genes in a circadian manner. However, in contrast to the RORs, the REV-ERBs are transcriptional repressors (Figure 2). This is in part because REV-ERBs lack the activation-function 2 (AF-2) region, which is involved in coactivator binding [13]. Thus, the REV-ERBs bind corepressor proteins such as the nuclear receptor corepressor (NCoR) [14,15]. REV-ERBs bind to the same RORE DNA response elements as the RORs [16]. REV-ERBs can bind as a monomer to an AGGTCA ‘half site’ with a 5′AT-rich extension. Although this form has been shown to be transcriptionally inactive, REVERB monomers function as transcriptional silencers [17]. REV-ERBs can also bind as a homodimer to a RORE direct repeat 2. They are highly stable homodimers that function as transcriptional repressors [17].
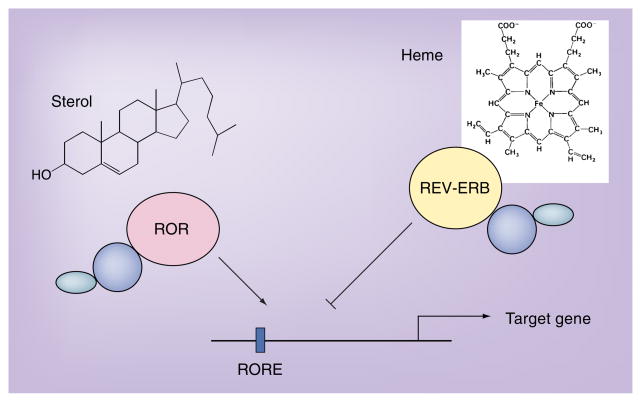
Figure 2. Model illustrating the regulation of a target gene by both retinoic acid receptor-related orphan receptors and REV-ERB.
REV-ERB functions as a receptor for heme, which is required for its activity as a repressor. Several sterols have been suggested as ligands for the RORs. In this cartoon, sterols regulate the constitutive activity of the RORs.
ROR: Retinoic acid receptor-related orphan receptor; RORE: ROR response element.
The roles of RORs & REV-ERBs in lipid homeostasis, metabolism & circadian rhythms
Circadian rhythms are daily cycles of biochemical, behavioral and physiological processes in living organisms regulated by endogenous ‘clocks’. Regulation of the circadian clock is important for an organism’s physiological function and behavior since it relies on it to anticipate and adapt to predictable daily changes such as day/night and activity/rest cycles, metabolism, hormone secretion, insulin sensitivity, blood pressure and feeding time. Abnormalities in circadian rhythm have been implicated in a number of diseases including sleep and mood disorders, diabetes, obesity and cancer [2,18].
Numerous studies have established links between expression of a certain number of NRs driven by the circadian clock, NR function and participation in the circadian control of various metabolic processes. In particular, RORα and REV-ERBα are part of the core clock machinery. In mammals, the master circadian clock is located in the suprachiasmatic nucleus (SCN) of the hypothalamus. Semi-autonomous clocks in the periphery can be entrained to signals from the SCN as well as other cues including nutrient status [18]. There are several interlocked transcriptional and post-translational auto-regulatory feedback loops controlling the circadian cycle. The Per, Arnt and Single-minded (PAS) domain basic helix-loop-helix transcription factor circadian locomotor output cycles kaput (CLOCK), brain and muscle aryl hydrocarbon receptor nuclear translocator (ARNT)-like protein 1 (BMAL1), and the CLOCK paralog neuronal PAS domain protein 2 (NPAS2), form a heterodimer and control the positive limb of the circadian cycle by activating transcription of the Period (PER) 1, PER2, and Cryptochrome (CRY) 1 and CRY2 genes through their E-box DNA elements in their promoters [19,20]. Once the PER/CRY heterodimer concentration reaches a critical level, the PER/CRY heterodimers enter the nucleus and repress BMAL1/CLOCK transactivation as well as their own transcription. This results in relief of BMAL1/CLOCK inhibition and allows a new cycle to start. This feedback loop results in the oscillations in expression of BMAL1/CLOCK and CRY/PER that follow a circadian pattern [21].
RORα and REV-ERBα are major regulators of the cyclic expression of BMAL1, the secondary feedback loop in the circadian cycle [22,23]. REV-ERBα transcription is activated by the BMAL1/CLOCK heterodimer and transrepressed by CRY/PER, resulting in circadian oscillations of REV-ERBα. In turn, REV-ERBα represses BMAL1 and CLOCK transcription. REV-ERBβ expression also oscillates in a circadian fashion and can repress BMAL1 transcription [22]. RORα competes with REV-ERBα for binding of their shared DNA binding elements, the RORE, in the BMAL1 promoter leading to BMAL1 expression being repressed by REVERBα and activated by RORα. The oscillating expression of RORα and REV-ERBα in the SCN leads to the circadian pattern of BMAL1 expression [23,24]. This REV-ERBα/RORα feedback loop interconnects the positive and negative limbs of the core circadian clock.
NPAS2, like CLOCK, forms heterodimers with BMAL1 and effectively functions in the regulation of mammalian circadian rhythms. As BMAL1 forms obligate heterodimers with NPAS2 or CLOCK, it was unclear as to how the expression of these two proteins was coordinated with BMAL1 expression. We recently demonstrated that NPAS2, like BMAL1, is a RORα and REV-ERBα target gene. We discovered two functional ROREs within the NPAS2 promoter and demonstrated that both RORα and REVERBα regulate the expression of NPAS2, thus suggesting a mechanism by which RORα and REV-ERBα coordinately regulate the positive limb of the circadian clock [25]. The importance of these proteins in circadian rhythms has been established through genetic knock-out experiments. Rev-erbα-deficient mice display altered circadian rhythms characterized by an increase in Bmal1 and Clock expression and shorter period lengths when compared with wild-type mice [26]. Rorα– and Rorβ-deficient mice display aberrant circadian behavior, while no abnormalities are apparent in Rorγ-deficient mice [23,24,27–29].
An organism’s energy homeostasis depends upon a coordinated control of key metabolic processes. Organs with high metabolic activity, including liver and adipose tissue, display circadian rhythm in the expression of genes involved in key metabolic pathways [30,31]. Growing evidence suggests that RORs play a critical role in the regulation of several metabolic pathways. Staggerer mice (RORαsg/sg), a natural mouse mutant in which RORα is rendered inactive due to a deletion within the RORα gene, has shed some light on the role of RORα in metabolism [32]. When fed a normal diet, staggerer mice display hypo-α-lipoproteinemia and have lower levels of total plasma cholesterol, the high-density lipoprotein, Apoa1, the major constituent of high-density lipoprotein, ApoCIII, Apoa2 and triglycerides, compared with wild-type mice [33–35]. In addition, expression of the reverse cholesterol transporters Abca1 and Abca8/G1 are decreased in the liver and intestine of staggerer mice [36]. These mice are less susceptible to hepatic steatosis and despite their higher food consumption, have reduced body fat [36]. Fat cells in brown and white adipose tissue are smaller and liver triglyceride levels lower in these mice than their wild-type counterparts. When fed a high fat diet (HFD), staggerer mice are resistant to HFD-induced obesity and hepatic steatosis [36]. However, despite their resistance to weight gain, these mice develop severe atherosclerosis when fed a HFD, suggesting that RORα may play an atheroprotective role [36]. Rorγ−/− mice display normal cholesterol and triglyceride levels, however, their blood glucose levels are slightly lower than wild-type mice [37]. Mice deficient in both Rorα and Rorγ (double knock-out) exhibit similar changes in cholesterol, triglyceride and blood glucose levels as the single knock-out mice. Gene expression analysis from livers of double knock-out mice suggests a degree of functional redundancy between the two RORs, most likely due to their similarities in RORE binding affinities [37].
The effect on cholesterol and triglyceride homeostasis in the Ror knock-out mice is most likely due to RORα’s regulation on a number of genes involved in the control of lipogenesis and fatty acid oxidation. For instance, sterol regulatory element-binding protein 1, isoform c (Srebp-1c) is reduced in the livers and muscle of staggerer mice as is fatty acid synthase (Fas) [36,38]. Expression of the coactivators PGC1α and β, proteins involved in the regulation of oxidative metabolism and gluconeogenesis, is increased in staggerer mice [29]. Finally, RORs have been demonstrated to regulate the oxysterol 7α-hydroxylase gene (Cyp7b1), the gene product is an enzyme that plays a crucial role in the alternative pathway of cholesterol metabolism. Expression of Cyp7b1 is reduced in staggerer mice and to a lesser degree in Rorγ−/− mice [37–39]. One intriguing notion is that the regulated conversion of cholesterol into bile acids, yielding hydroxycholesterols as intermediates, may regulate the circadian rhythm via modulation of ROR activity. Circadian control of cholesterol biosynthesis has been well documented and it is believable that the levels of hydroxycholesterols may also follow a circadian pattern [40,41]. Therefore, expression of Cyb7a1 and Cyp7b1 may regulate themselves through a dynamic feedback loop involving the RORs and REV-ERBs and ultimately be another mechanism of control of the circadian rhythm.
Recent evidence has established a role for RORα in glucose metabolism. Chopra et al. found that loss of the NR coactivator steroid receptor coactivator 2 (SRC-2) resulted in a murine phenotype similar to von Gierke’s disease. This disease is associated with severe hypoglycemia and abnormal accumulation of glucose in the liver [42]. Furthermore, our group demonstrated that RORα regulates both the expression and secretion of FGF21, a hepatic hormone that regulates peripheral glucose tolerance, torpor and hepatic lipid metabolism [39,43,44].
REV-ERBs play a central role in regulating clock genes, as discussed earlier, which in turn affects many processes coordinated by the circadian clock. Although currently there have been no specific reports detailing the precise role of REV-ERBs in clock-associated diseases, dysfunction of the circadian lock (perhaps via REVERB dysfunction) can have profound effects on energy homeostasis and metabolic disorders. In this sense, Rev-erbα−/− mice display a 0.5 h shorter period and are more sensitive to phase shifts induced by light pulses [26,45].
Several studies have shown that REV-ERBα plays an important role in lipid metabolism. Rev-erbα−/− mice display elevated very low-density lipoprotein triglyceride levels, which correlates with elevated levels of Apoc3 mRNA in the serum and liver [46,47]. Apoc3 is a key player in serum triglyceride metabolism, and in combination with data from RORαsg/sg staggerer mice, suggests that REV-ERBs and RORs are both important in the regulation of Apoc3 expression and lipid metabolism [46,48]. REV-ERBα is also a regulator of other genes involved in metabolism, including Apoa1 (in certain species; mentioned above for ROR), as well as Elov3 (a very long-chain fatty acid elongase) and Pai-1 (regulator of the fibrinolytic system and modulator of inflammation and atherosclerosis) [49–51].
REV-ERBα has also been shown to be a direct regulator of Fgf21 expression. Overexpression of the receptor results in decreased Fgf21 mRNA levels in primary mouse hepatocytes in a heme-dependent manner, and is dependent on Alas1 expression levels [52]. Alas1, a PGC-1α target gene, produces a product that functions as a rate-limiting enzyme in the synthesis of heme, the natural ligand for REV-ERBs. PGC-1α regulates the expression of Alas1, thus linking the roles for both REV-ERB and PGC-1α in Fgf21 expression [53]. Rev-erbβ knock-out mice have not yet been reported, but additional studies have provided insights into its role in metabolism. Expression of a dominant negative form of REV-ERBβ lacking the LBD results in decreased expression of several genes involved in lipid metabolism. These loss-of-function studies identified Fatty acid translocase (Fat/CD36), Fabp3, Fabp4 Ucp3, Srebp-1c and Scd-1 as REV-ERBβ target genes [54].
Discovery of endogenous ligands
The LBDs of NRs are multifunctional. They play a role not only in ligand binding, but also nuclear localization, receptor dimerization and they provide an interface for coactivator and corepressor binding. Ligand binding induces a conformational change in the receptor resulting in helix 12 in the LBD (also referred to as the AF-2 region) making contact with the ligand. For agonist ligands, this results in the dissociation of corepressor molecules to be replaced by coactivator molecules [55,56]. In the case of most NRs, coactivator/corepressor interaction is ligand-dependent. In addition to the typical 12 canonical α-helices, the RORs contain two extra helices, H2′ and H11′ [11]. However, the RORs have intrinsic transcriptional activity, where ligand binding actually represses the activity of the receptor, meaning that the RORs are constitutively active and that coactivators bind to the AF-2 surface in the absence of ligand.
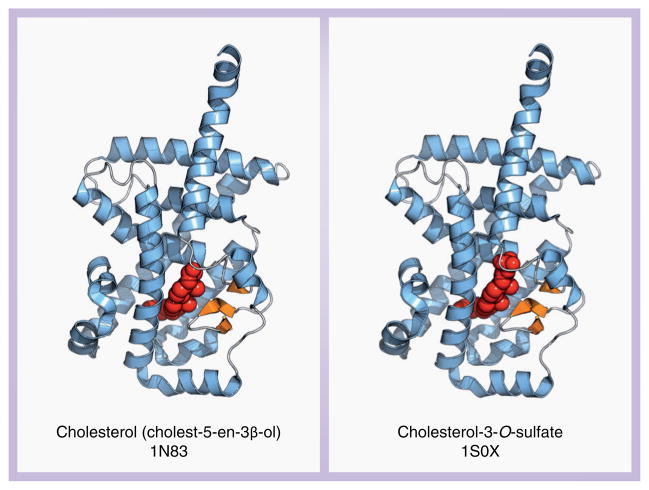
Identification of endogenous ligands for the RORs has been controversial (Figure 3A). In the mid 1990s, two putative ligands for RORα, melatonin and thiazolidinedione, were suggested [57]. However, these data have yet to be confirmed. Later, x-ray crystallographic studies provided insights into the structure, size and potential ligands for RORα’s LBD (Figure 4). When the RORα LBD protein was expressed in Sf-9 insect cells, crystallographic analysis revealed that RORα co-purified with cholesterol, which was bound in the ligand-binding pocket, thus suggesting a potential endogenous ligand for RORs [58]. In addition to cholesterol, 7-dehydrocholesterol, cholesterol sulfate and 25-hydroxycholesterol were also identified as RORα agonists [58,59]. All ligands were found to bind RORα in a reversible manner and enhanced RORE-dependent transcriptional activation by RORα. These findings suggested that RORα may function as a lipid sensor, thus implicating RORα in the regulation of lipid metabolism. In fact, recent reports indicate that RORα regulates the expression of several genes involved in lipid metabolism [35–37,58]. Regardless, these data suggest that RORα may bind to a number of sterols leaving open the question as to which ligands are physiologically relevant.
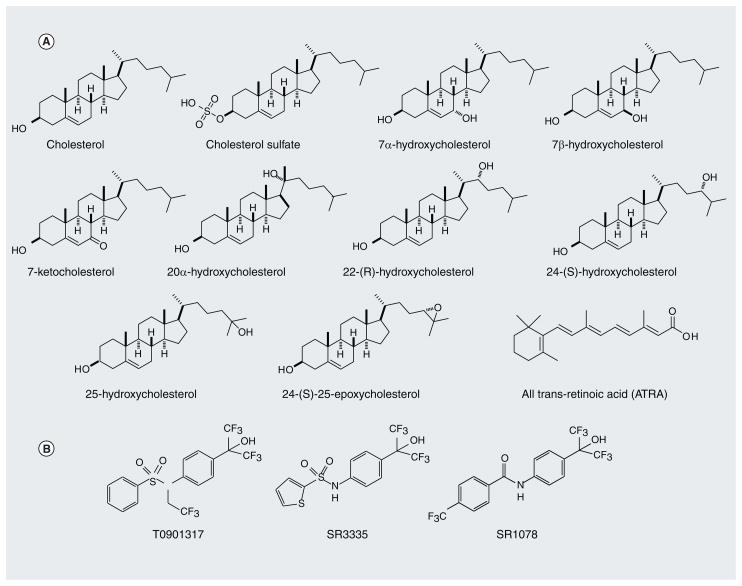
Figure 3. Examples of retinoic acid receptor-related orphan receptor ligands.
(A) Endogenous ROR ligands that have been proposed. (B) Synthetic ROR ligands that have been identified.
ROR: Retinoic acid receptor-related orphan receptor.
Figure 4. Crystal structures of retinoic acid receptor-related orphan receptor α with two proposed endogenous ligands within the ligand-binding pocket.
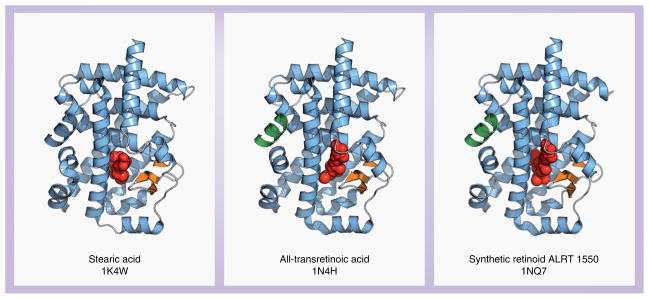
Putative ligands for RORβ have been revealed through studies involving x-ray crystallography (Figure 5). Similar to cholesterol for RORα, stearic acid co-purified with recombinantly expressed RORβ LBD protein [60]. Crystallographic analysis revealed that the stearic acid molecule bound to the ligand-binding pocket in RORβ. However, functional experiments suggest that stearic acid likely acts more as a ‘filler’ than a functional ligand [60]. Several retinoids, including all-trans retinoic acid, and the synthetic retinoid ALRT 1550 (Allergan Ligand Retinoid Therapeutics, Inc.), were identified as functional inverse agonists as they reversibly bound RORβ in the ligand-binding pocket with high affinity and altered RORβ-mediated transcriptional activation. These retinoids were also able to bind to RORγ not RORα, and inhibit RORγ-mediated transactivation [61]. Further analysis of these ligands is needed, since it is still unclear whether they act as genuine physiological ligands for RORβ and RORγ and can regulate ROR target genes in a ligand-dependent manner.
Figure 5. Crystal structures of retinoic acid receptor-related orphan receptor β with three proposed ligands.
Despite the reports describing cholesterol and cholesterol sulfate as ligands for RORα, it was still unclear as to whether these two sterols were physiologically relevant ligands. Recently, we demonstrated that the 7-oxygenated sterols (Figure 3A) function as high-affinity ligands for both RORα and RORγ, demonstrating Ki values in the range of 10–20 nM as determined by radioligand-binding assays. Our data indicate that the 7-oxygenated sterols (7α-OHC, 7β-OHC and 7-ketocholesterol) bind to RORα and RORγ with higher affinity than cholesterol sulfate, with cholesterol binding barely detectable. We found that RORα and RORγ, when produced in Escherichia coli, were devoid of any endogenous sterols yet displayed constitutive activity as evidenced by their ability to bind to coactivator peptides and were transcriptionally active in the absence of a ligand. The 7-oxygenated sterols functioned as inverse agonists to both receptors while cholesterol and cholesterol sulfate failed to modulate the activity of RORα and RORγ in cell based assays. Finally, the 7-oxygenated sterols modulated the expression of RORα/γ-dependent target genes in a receptor-dependent manner [12]. Our data suggest that while cholesterol and cholesterol sulfate bind RORα and RORγ, these sterols lack the ability to induce the conformational change necessary to alter cofactor binding and transcriptional activity.
We also discovered that 24S-hydroxycholestrol (24S-OHC; Figure 3A) is a high-affinity ligand for RORα and RORγ, binding to these receptors with Ki values of approximately 25 nM [62]. 24S-OHC acts as an inverse agonist for both RORα and RORγ, as it dose-dependently reduces the constitutive activity of these receptors in Gal4 cotransfection assays, as well as Bmal::luc and Glucose-6-Phosphatase (G6Pase)::luc reporter assays. In addition, 24S-OHC reduces the expression of BMAL1 and REV-ERBα mRNA levels in a RORα-dependent manner, as shown by siRNA knock-down of RORα, via decreased SRC-2 recruitment to promoter regions, as shown in a BMAL1 ChIP-reChIP assay. In this report, we also showed that 24S,25-epoxycholesterol (24,25-epoC) and 24R-hydroxycholesterol (24R-OHC) (Figure 3A) selectively bind and regulate the activity of RORγ, with similar Ki values (20 and 102 nM, respectively) and IC50 values in a Gal4 dose titration assay (280 and 90 nM, respectively) [62].
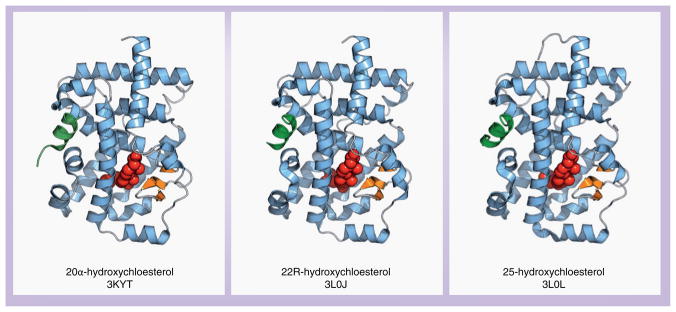
Alongside these discoveries, another group discovered that 20α-hydroxycholesterol (20α-OHC), 22R-hydroxycholestrol (22R-OHC) and 25-hydroxycholestrol (25-OHC) (Figure 3A) are ligands for RORγ [63]. These ligands dose-dependently increased the recruitment of coactivator peptides in an AlphaScreen biochemical assay. Crystal structures of RORγ bound to these ligands were determined (Figure 6), which revealed that all three ligands bound to RORγ in a similar manner. Mutagenesis studies validated the role of several key residues involved in the interaction with the hydroxycholestrol ligands, including one mutation (Ile397Asn) that created a new hydrogen bond with 22R-OHC; for this mutation, all ligands except 22R-OHC lost activity [64].
Figure 6. Crystal structures of retinoic acid receptor-related orphan receptor γ with three proposed endogenous ligands within the ligand-binding pocket.
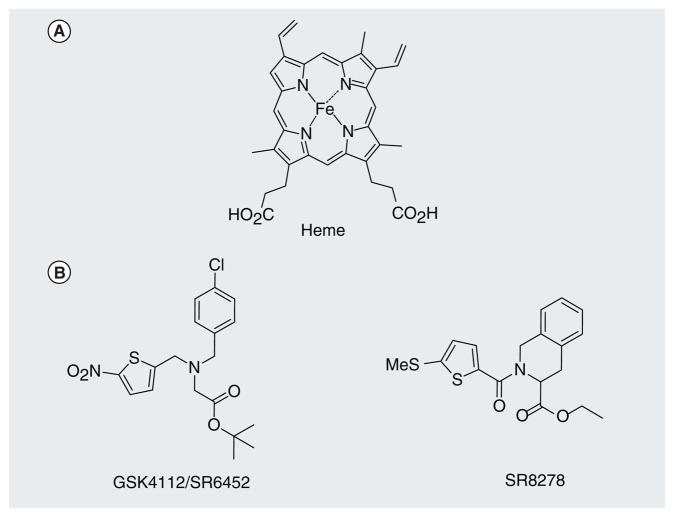
For many years, REV-ERBs were thought to function in a ligand-independent manner, much like the RORs. Their function as constitutive transcriptional repressors was attributed to their lack of anAF-2 region, or helix 12 in the LBD. However, in 2007, our laboratory, as well as the Lazar laboratory, discovered that REV-ERBs are indeed ligand-regulated and identified the porphyrin heme as a physiological ligand (Figure 7) [5,6]. We showed that heme binds to REV-ERBs with a Kd of 2–3 μM and thereby induced a conformational change in the receptor, increasing recruitment of the corepressor NCoR and leading to suppression of the expression of REV-ERB target genes. Additional data from the Lazar laboratory showed that heme regulates the expression of additional REV-ERB target genes, including the gluconeogenic genes G6Pase and PEPCK, as well as interaction with the NCoR–HDAC3 corepressor complex.
Figure 7. Examples of REV-ERB ligands.
(A) Heme, the identified endogenous ligand for REV-ERB. (B) Two recently characterized REV-ERB-specific synthetic ligands.
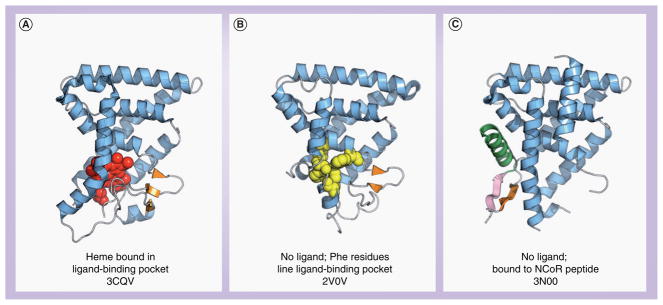
Crystal structures of REV-ERBα and REVERBβ provided a structural validation of heme as a ligand for REV-ERBs (Figure 8), including some molecular details concerning the repression-only function of these receptors. A structural study of REV-ERBβ bound to heme demonstrated one heme molecule hexa-coordinated within the ligand-binding pocket, making contacts to the side chains of Cys-384 and His-568 (Figure 8A) [65]. In the absence of heme (Figure 8B), side chains of aromatic residues line the REV-ERBβ ligand-binding pocket (Phe-405, Phe-409, Phe-443 and Phe-450), which stabilizes the ligand-free form of the receptor [66], supported by circular dichroism thermal melt plots showing only a slight increase in the thermal stability of REV-ERBs upon binding heme [5].
Figure 8. Crystal structures of REV-ERB ligand-binding domain.
(A) Heme bound in the ligand-binding pocket of REV-ERBβ, (B) REV-ERBβ in the absence of a ligand. (C) NCoR bound to REV-ERBα in the absence of heme.
NCoR: Nuclear receptor corepressor.
More recently, the structure of REV-ERBα bound to the NCoR interaction domain 1 (ID1) peptide was reported in the absence of heme (Figure 8C) [67]. The NCoR ID1 domain forms an antiparallel β-sheet interface with the C-terminal ‘Y domain’ of REV-ERBα. This type of antiparallel β-sheet interface has only been observed in one other structural study (RAR bound to the NCoR ID1 peptide) and may be a general structural mechanism for corepressor binding. However, an interesting point to note is that this interaction is facilitated in the absence of REV-ERB’s physiological ligand, heme. The authors suggest that the NCoR ID1 region may enable REV-ERB to perpetuate a basal level of repression in the absence of heme. In agreement, we previously noted that heme displaced the NCoR ID1 peptide in a FRET-based biochemical assay [5] – suggesting that the ID1 region may prefer to bind to heme-free receptor. The authors also suggest that heme binding may induce a conformational change resulting in the recruitment of the NCoR ID2 region, perhaps by displacing the ID1 region.
Discovery of synthetic ligands
The wide range of physiological processes regulated by NRs and pathological consequences resulting from aberrant NR activity have made this family of proteins a popular area of research as potential targets for drug development. The NRs contain naturally occurring hydrophobic, ligand-binding pockets that when bound by small hydrophobic molecules can be turned ‘on’ or ‘off’. The most effective drugs are small hydrophobic compounds that can cross the plasma membrane. The steroid receptors, including the mineralcorticoid, estrogen, androgen, glucocorticoid and progesterone receptors have been successful targets for a range of diseases including cancer, inflammation and cardiovascular disease. Thyroid receptor, peroxisome proliferator-activated receptors, RARs and RXRs have been successful targets for the development of drugs for the treatment of metabolic and skin disorders. Accordingly, a tremendous amount of effort has been generated in order to elucidate and determine the function of human NR ligands.
Recently, we identified the first high-affinity synthetic ligand for both RORα and RORγ. Using a NR-specificity screen containing all 48 human NRs, we identified the benzene-sulfonamide liver X receptor (LXR) agonist, T0901317 (Figure 3B), as a RORα/γ inverse agonist. We determined that T0901317 bound with high affinity to both RORα and RORγ (Ki = 132 and 52 nM, respectively) resulting in the modulation of the transcriptional activity of each receptor and their ability to interact with cofactor peptides. Furthermore, T0901317 repressed the expression of ROR-dependent target genes, including the gluconeogenic enzyme G6Pase [68]. While we were the first to discover that T0901317 modulates ROR activity, it had already been well characterized as a LXRα and LXRβ agonist. Furthermore, T0901317 acts as a potent agonist to the farnesoid X receptor and pregnane X receptor [69,70]. Although the promiscuity of T0901317 makes it difficult to use this compound as a selective probe, it lays the groundwork for the development of ROR-selective modulators.
Using the benzenesulfonamide as a scaffold, we synthesized an array of compounds and assessed their activity on RORα, RORγ, farnesoid X receptor, LXRα and LXRβ. Using this approach, we discovered one compound, the amide SR1078 (Figure 3B), to be a selective RORα/γ modulator as it displayed no activity on the other NRs tested. Further examination of SR1078 revealed that it is a RORα/γ agonist as it stimulated the expression of two ROR target genes in the liver, G6Pase and FGF21 [71]. Additional work from our group has elucidated a selective RORα synthetic ligand, SR3335 (Figure 3B). Similar to T091317, treatment with SR3335 suppressed the expression of G6Pase and PhosPhoenolpyruvate Carboxykinase (PEPCK), RORα target genes involved in hepatic gluconeogenesis. Furthermore, in a diet-induced obesity mouse model, SR3335 treatment led to reduced plasma glucose levels following a pyruvate tolerance test, and indicator of gluconeogenesis [72]. These studies prove the feasibility of developing ROR-selective modulators as tools to probe the function of these receptors in vitro and in vivo. Currently, the design and synthesis of compounds with improved physicochemical properties for the individual ROR family members is underway and would further increase their utility as therapeutics in the treatment of ROR-mediated metabolic disorders, including Type II diabetes.
The first synthetic REV-ERB compound, GSK4112 (Figure 7B), was identified in 2008 in a FRET-based biochemical assay as a ligand that dose-dependently increased the interaction of a peptide derived from the NCoR corepressor with REV-ERBα [73]. This study also showed that treatment of Rat-1 cells with GSK4112 reduces the expression of the REV-ERB target gene Pai-1. GSK4112 treatment also resets the circadian expression of a Per2:Luc reporter in Rat-1 cells and primary lung fibroblasts.
Both ROR and REV-ERB have been shown to play an important role in regulation of adipogenesis [74–78]. We tested the pharmacology of GSK4112 within the context of a REV-ERB responsive gene, Bmal1, in a recent paper characterizing the ability of GSK4112 to regulate adipogenesis in 3T3-L1 cells in a REV-ERB-dependent manner [79]. First, we biochemically verified the ability of GSK4112 to dose-dependently increase the interaction of the receptor with a NCoR corepressor peptide revealing that GSK4112 is a dual REV-ERBα/β agonist. GSK4112 decreases the activity of a luciferase reporter under the control of the Bmal1 reporter in a cotransfection assay with a REVERBα expression vector in both HepG2 and HEK293 cells. Furthermore, GSK4112 treatment of HepG2 cells decreases the expression of Bmal1 in a dose-dependent manner [80,81]. In addition, ChIP analysis reveals that GSK4112 treatment of HepG2 cells increases the occupancy of NCoR on the Bmal1 promoter. These data reveal that GSK4112 acts as a REV-ERB agonist, regulating the expression of REVERB responsive target genes and increases the recruitment of the NCoR corepressor complex in a manner similar to REV-ERB’s physiologically ligand, heme [5]. In this study, we also showed that GSK4112 induces adipogenesis in 3T3-L1 cells in a synergistic manner with the peroxisome proliferator-activated receptor γ agonist rosiglitazone.
Later, GSK4112 was further characterized and suggested that it could be used as a chemical probe to define the function of REVERB in vitro [80]. In this study, data were presented showing that heme and GSK4112 compete for binding in FRET-based biochemical assay with a NCoR peptide. In addition, the authors present some information regarding the structure–activity relationship in the discovery of GSK4112. Data are also presented, validating our prior study, showing that GSK4112 decreases the expression of the REV-ERB target gene Bmal1 in a REV-ERBα-dependent manner, and further show it decreases the expression of other REV-ERB target genes, including G6Pase, PEPCK and PGC1α. GSK4112 also increases the occupancy of HDAC3 to the G6Pase promoter. Furthermore, GSK4112 treatment represses glucose output in primary mouse hepatocytes, as well as decreases the expression of Bmal1 and G6Pase. In addition, GSK4112 represses the circadian expression of Bmal1, Cry1 and Pgc1α in primary mouse hepatocytes.
Our unpublished studies and the findings of other groups reveal that GSK4112 displays poor pharmacokinetic properties [80]. In particular, high clearance and rapid metabolism lead to low plasma drug levels. Thus, the poor pharmaco-kinetic properties of this compound preclude it from use as a probe of REV-ERB function in vivo. However, these studies reveal that small-molecule compounds can indeed be used to define the function of REV-ERB, providing a basis for future REV-ERB-based medicinal chemistry efforts with significantly better in vivo pharmacokinetic/pharmacodynamic properties.
We recently identified a compound, SR8278, which functions as a REV-ERB antagonist (Figure 7B) [81]. In a Gal4 cotransfection assay, where the LBDs of REV-ERBα and REV-ERBβ are fused to the Gal4 DNA-binding domain, the GSK4112 agonist causes repression, whereas the SR8278 antagonist stimulates transcription. We further verified the antagonist activity of SR8278 using luciferase reporters containing the promoter sequences for the REV-ERB target genes BMAL1, G6Pase and PEPCK. Similar to the observation in the Gal4 assay, SR8278 stimulates transcription for the REV-ERB responsive promoters and does so in a dose-dependent manner that competes with GSK4112 and heme, the endogenous agonist. We further characterized the antagonist activity of SR8278 by treating HepG2 cells with the compounds. Whereas GSK4112 decreased the expression of PEPCK and G6Pase, SR8278 stimulated the expression of these genes. Unfortunately, SR8278 also displays poor pharmacokinetic properties that will limit its use to in vitro and biochemical studies. However, this discovery of the first antagonist of REV-ERB activity, and future structure–activity relationship studies to find compounds with favorable pharmacokinetic properties, will allow the field to better define the function of REV-ERBs in vivo.
Metabolic disorders associated with RORs and REV-ERBs
The circadian rhythm plays an essential role in the regulation of metabolism. Several lines of evidence suggest that the incidence of cardiovascular disease and metabolic disturbances is higher in individuals who alter their normal sleep–wake pattern for shift work [82–84]. Additional studies have uncovered the prevalence of metabolic syndrome, increased body-mass index and cardiovascular events in shift workers, raising the possibility that disrupted cycles of rest and activity and fasting and feeding may contribute to the initiation and progression of metabolic syndrome [85,86]. Circadian control of glucose metabolism is a well-recognized aspect of clinical diabetes management. Individuals diagnosed with Type I diabetes mellitus are acutely aware of this pattern as patients must adjust their daily insulin requirements to accommodate the fluctuations of insulin in accordance with the light–dark cycle and timing of meals [87]. One hallmark of Type II diabetes is alterations in the normal patterns of glucose regulation/tolerance [88]. Further understanding of the physiological and molecular mechanisms regulating circadian control of glucose tolerance may lead to better strategies for diabetes management.
Insight into RORs regulation of glucose metabolism comes from studies of patients with Von Gierke’s disease, a genetic disorder resulting from a deficiency in the enzyme glucose-6-phosphatase. Deficiency of this enzyme leads to the inability of the liver to produce free glucose from glycogen and from gluconeogenesis. Therefore, reduced glycogen breakdown results in increased glycogen storage in both the liver and kidneys, causing enlargement of both organs and severe hypoglycemia. Both organs function normally in childhood but are susceptible to a variety of problems in adulthood. Mice deficient in the NR coactivator, SRC-2, develop symptoms similar to those with von Gierke’s disease. Chopra et al. demonstrated that RORα, along with SRC-2, was required to regulate G6Pase in a normal manner [42].
The 7-oxygenated sterols (7α-hydroxycholesterol [7α-OHC], 7β-hydroxycholesterol [7β-OHC], and 7-ketocholeseterol [7-KC]) function as high-affinity ligands for both RORα and RORγ, behaving as inverse agonists, thus suppressing the constitutive activity of the receptors [89]. The 7-oxygenated sterols play a very important role in both bile acid biosynthesis and in the pathology of atherosclerosis. 7β-OHC and 7-KC are the two most enriched oxysterols found in atherosclerotic plaques following 27-OHC. 7-KC is the most enriched oxysterol in oxidized low density lipoprotein and in the macrophage foam cells [90,91], and has been shown to exert an atherogenic activity by inhibiting sterol efflux from these cells [92]. Plasma levels of 7β-OHC are elevated in patients with cardiovascular disease [93]. Clearly modulators of RORs would prove beneficial for elucidation of the function of RORs in vivo and provide the basis for therapeutic intervention of ROR-mediated disorders.
To date, no human diseases or disorders have been exclusively attributed to aberrant REVERB activity. However, empirical evidence links REV-ERB function with the regulation of lipid homeostasis and metabolism. Rev-erbα−/− mice display a dyslipidemic phenotype with elevated very low-density lipoprotein triglyceride levels along with increased liver and serum ApoCIII expression [34,47]. In addition, Rev-erbα−/− mice exhibit decreased levels of expression of cholesterol 7α-hydroxylase (Cyp7a1), the enzyme that catalyzes the rate-limiting step of bile acid biosynthesis [94,95]. Furthermore, REV-ERB has been shown to suppress the expression of G6Pase and PEPCK and hepatocyte glucose output. Clearly, modulators of this NR could also serve as useful treatments for metabolic disorders.
Future perspective: targeting RORs & REV-ERBs
As more information surrounding the biology of the RORs and REV-ERBs comes to light, it becomes more apparent that these two NR subfamilies play important roles in numerous physiological processes, specifically the regulation of circadian rhythms, lipid homeostasis and glucose homeostasis. Several studies have revealed that the activity of these NRs can be regulated by both endogenous and synthetic ligands. Recent breakthroughs in the development of synthetic ligands selective to the RORs and REV-ERBs promises to provide us with both chemical tools to probe the function of the receptors as well as develop therapeutics towards the treatments of disorders in which the RORs and REV-ERBs play significant roles.
One major challenge in the development of selective synthetic ligands for the treatment of ROR-mediated disorders is our limited knowledge of the function of each subfamily member. Genetic evidence suggests that RORα and RORγ, may have overlapping roles in the regulation of metabolism. However, exactly how much these two NRs overlap in terms of function has yet to be established. While the generation of dual RORα/γ agonists and inverse agonists will significantly aid in elucidating the function of the RORs in a biological context, development and optimization of synthetic ligands, both agonists and inverse agonists, for RORα as well as RORγ will help define the specific roles that each member plays in metabolic processes. Application of the synthetic ligands as chemical tools will prove useful. For example, in vitro use will enhance our knowledge of the function of the receptors at the molecular and cellular level. Furthermore, the physiochemical properties of the ligands must be optimized, including their pharmacokinetics, drug solubility and plasma protein binding. Once optimized, administration of the compounds in vivo will be more telling of the function of the RORs in an organism’s physiology.
While there has been successful development of synthetic ligands for RORα and RORγ, little is still known about RORβ. Genetic experiments have provided some insight into the function of this third ROR family member, however, ligands specific to RORβ will aid in our understanding of this NR. Considering the rapid development of synthetic compounds for RORα and RORγ, it is conceivable that the next few years will see the development of compounds specific to RORβ.
Similarly, our recent advances in developing synthetic REV-ERB ligands will allow us to probe the physiological function of these receptors as well as to evaluate the clinical utility of these compounds as potential drugs. Currently, there are only two synthetic ligands available (the agonist GSK4112 and the antagonist SR8278) and both of these ligands have poor pharmacokinetic properties limiting their use in vivo. In addition, both demonstrate activity at REV-ERBα and REV-ERBβ. Current efforts in our laboratory are focused on developing REVERB ligands with greater potency/efficacy and improved pharmacokinetic properties as well as receptor subtype selectivity.
In conclusion, the data generated from the in vitro characterization of the ROR and REVERB synthetic ligands are extremely promising. However, how these ligands affect the physiology of the organism in vivo has yet to be extensively studied. While studies in knockout animals are extremely telling regarding the roles these receptors play in the development and an organism’s overall physiology, the ultimate roles of these two opposing NRs in disease progression and resolution can only be determined through in vivo use of specific synthetic analogs. Efforts in our laboratory to characterize ROR and REV-ERB compounds in vivo are currently underway and are yielding promising, exciting and unexpected results, making the study of the RORs and REV-ERBs even more stimulating.
Executive summary.
Nuclear receptors (NRs) are ligand-dependent transcription factors that regulate the expression of genes important in development, homeostasis and metabolism.
Ligands to NRs are classified according to their ability to regulate a NRs transcriptional activity. For example, they can be agonists, antagonists, inverse agonists and partial agonists.
Two subfamilies of NRs, the retinoic acid receptor-related orphan receptors (RORs) and REV-ERBs, have been demonstrated to play critical roles in the regulation of circadian rhythms and lipid homeostasis.
Identification of endogenous ligands for the RORs has proven controversial and includes various sterols, including cholesterol and its derivatives, as well as several retinoids.
Recently, heme was identified as an endogenous ligand for the REV-ERBs, thus de-orphanizing this NR.
Discovery of synthetic compounds to the RORs and REV-ERBs (e.g., T0901317 and GSK4112, respectively) has led to an expansion of structure–activity relationship studies aimed at the identification of novel compounds for both the RORs and REV-ERBs.
An array of new selective synthetic ligands for ROR and REV-ERBs have recently been discovered creating an opportunity to evaluate these receptors as drug targets for metabolic diseases.
Key Term
- REV-ERBs
Initially, those in the field of the nuclear receptor research thought that REV-ERBs had no endogenous ligands and were ligand-independent constitutive transcriptional repressors. However, recent studies revealed that REV-ERBs respond to an endogenous ligand, heme, by repressing the transcription of their target genes. Without heme binding, the REV-ERBs are transcriptionally inactive, thus there is no basal transcription activity. All cells have some intracellular heme present, thus leading to the appearance that REV-ERBs have constitutive repressor activity
Footnotes
For reprint orders, please contact reprints@future-science.com
Financial & competing interests disclosure
This work was supported by NIH grants to TPB (DK080201, DK089984 and MH092769) and LAS (DK088499). The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.Sladek FM. What are nuclear receptor ligands? Mol Cell Endocrinol. 2010;334(1–2):3–13. doi: 10.1016/j.mce.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burris TP. Nuclear hormone receptors for heme: REV-ERBα and REV-ERBβ are ligand-regulated components of the mammalian clock. Mol Endocrinol. 2008;22(7):1509–1520. doi: 10.1210/me.2007-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangelsdorf DJ, Thummel C, Beato M, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83(6):835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mangelsdorf DJ, Evans RM. The RXR heterodimers and orphan receptors. Cell. 1995;83(6):841–850. doi: 10.1016/0092-8674(95)90200-7. [DOI] [PubMed] [Google Scholar]
- 5▪▪.Raghuram S, Stayrook KR, Huang P, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBα and REV-ERBβ. Nat Struct Mol Biol. 2007;14(12):1207–1213. doi: 10.1038/nsmb1344. In conjunction with [6], these studies identified heme as an endogenous physiological ligand for the orphan nuclear receptors REV-ERBα and β. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6▪▪.Yin L, Wu N, Curtin JC, et al. REV-ERBα, a heme sensor that coordinates metabolic and circadian pathways. Science. 2007;318(5857):1786–1789. doi: 10.1126/science.1150179. In conjunction with [5], these studies identified heme as an endogenous physiological ligand for the orphan nuclear receptors REV-ERBα and β. [DOI] [PubMed] [Google Scholar]
- 7.Becker-Andre M, Andre E, DeLamarter JF. Identification of nuclear receptor mRNAs by RT-PCR amplification of conserved zinc-finger motif sequences. Biochem Biophys Res Commun. 1993;194(3):1371–1379. doi: 10.1006/bbrc.1993.1976. [DOI] [PubMed] [Google Scholar]
- 8.Giguere V, Tini M, Flock G, et al. Isoform-specific amino-terminal domains dictate DNA-binding properties of RORα, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994;8(5):538–553. doi: 10.1101/gad.8.5.538. [DOI] [PubMed] [Google Scholar]
- 9.Carlberg C, Hooft van Huijsduijnen R, Staple JK, DeLamarter JF, Becker-André M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol Endocrinol. 1994;8(6):757–770. doi: 10.1210/mend.8.6.7935491. [DOI] [PubMed] [Google Scholar]
- 10.Hirose T, Smith RJ, Jetten AM. RORγ: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem Biophys Res Commun. 1994;205(3):1976–1983. doi: 10.1006/bbrc.1994.2902. [DOI] [PubMed] [Google Scholar]
- 11.Jetten AM. Retinoid-related orphan receptors (RORs): critical roles in development, immunity, circadian rhythm, and cellular metabolism. Nucl Recept Signal. 2009;7:e003. doi: 10.1621/nrs.07003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Kumar N, Solt LA, et al. Modulation of retinoic acid receptor-related orphan receptor α and γ activity by 7-oxygenated sterol ligands. J Biol Chem. 2010;285(7):5013–5025. doi: 10.1074/jbc.M109.080614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burke L, Downes M, Carozzi A, Giguère V, Muscat GE. Transcriptional repression by the orphan steroid receptor RVR/REV-ERBβ is dependent on the signature motif and helix 5 in the E region: functional evidence for a biological role of RVR in myogenesis. Nucleic Acids Res. 1996;24(18):3481–3489. doi: 10.1093/nar/24.18.3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Downes M, Burke LJ, Bailey PJ, Muscat GE. Two receptor interaction domains in the corepressor, N-CoR/RIP13, are required for an efficient interaction with REV-ERBAα and RVR: physical association is dependent on the E region of the orphan receptors. Nucleic Acids Res. 1996;24(22):4379–4386. doi: 10.1093/nar/24.22.4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamir I, Harding HP, Atkins GB, et al. A nuclear hormone receptor corepressor mediates transcriptional silencing by receptors with distinct repression domains. Mol Cell Biol. 1996;16(10):5458–5465. doi: 10.1128/mcb.16.10.5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harding HP, Lazar MA. The orphan receptor REV-ERBAα activates transcription via a novel response element. Mol Cell Biol. 1993;13(5):3113–3121. doi: 10.1128/mcb.13.5.3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harding HP, Lazar MA. The monomer-binding orphan receptor REV-ERB represses transcription as a dimer on a novel direct repeat. Mol Cell Biol. 1995;15(9):4791–4802. doi: 10.1128/mcb.15.9.4791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134(5):728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10(5):543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gachon F, Nagoshi E, Brown SA, Ripperger J, Schibler U. The mammalian circadian timing system: from gene expression to physiology. Chromosoma. 2004;113(3):103–112. doi: 10.1007/s00412-004-0296-2. [DOI] [PubMed] [Google Scholar]
- 21.Duez H, Pavlic M, Lewis GF. Mechanism of intestinal lipoprotein overproduction in insulin resistant humans. Atheroscler Suppl. 2008;9(2):33–38. doi: 10.1016/j.atherosclerosissup.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 22.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20(5):391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 23▪.Sato TK, Panda S, Miraglia LJ, et al. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43(4):527–537. doi: 10.1016/j.neuron.2004.07.018. Study suggesting that the opposing activities of the nuclear receptors RORa and REV-ERBa are important in the maintenance of the core mammalian circadian clock. [DOI] [PubMed] [Google Scholar]
- 24▪▪.Akashi M, Takumi T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat Struct Mol Biol. 2005;12(5):441–448. doi: 10.1038/nsmb925. Study identified RORα as a functional component of the cell-autonomous core circadian clock. [DOI] [PubMed] [Google Scholar]
- 25.Crumbley C, Wang Y, Kojetin DJ, Burris TP. Characterization of the core mammalian clock component, NPAS2, as a REV-ERB{α}/ROR{α} target gene. J Biol Chem. 2010;285:35386–35392. doi: 10.1074/jbc.M110.129288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26▪▪.Preitner N, Damiola F, Molina L, et al. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110(2):251–260. doi: 10.1016/s0092-8674(02)00825-5. Study identified REV-ERBα as being an integral part of the regulation of the circadian clock. [DOI] [PubMed] [Google Scholar]
- 27.Masana MI, Sumaya IC, Becker-Andre M, Dubocovich ML. Behavioral characterization and modulation of circadian rhythms by light and melatonin in C3H/HeN mice homozygous for the RORβ knockout. Am J Physiol Regul Integr Comp Physiol. 2007;292(6):R2357–R2367. doi: 10.1152/ajpregu.00687.2006. [DOI] [PubMed] [Google Scholar]
- 28.Schaeren-Wiemers N, André E, Kapfhammer JP, Becker-André M. The ExDression pattern of the orphan nuclear receptor RORβ in the developing and adult rat nervous system suggests a role in the processing of sensory information and in circadian rhythm. Eur J Neurosci. 1997;9(12):2687–2701. doi: 10.1111/j.1460-9568.1997.tb01698.x. [DOI] [PubMed] [Google Scholar]
- 29.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBα and β and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4(2):e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ando H, Yanagihara H, Hayashi Y, et al. Rhythmic messenger ribonucleic acid expression of clock genes and adipocytokines in mouse visceral adipose tissue. Endocrinology. 2005;146(12):5631–5636. doi: 10.1210/en.2005-0771. [DOI] [PubMed] [Google Scholar]
- 31.Panda S, Antoch MP, Miller BH, et al. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109(3):307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 32.Hamilton BA, Frankel WN, Kerrebrock AW, et al. Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature. 1996;379(6567):736–739. doi: 10.1038/379736a0. [DOI] [PubMed] [Google Scholar]
- 33.Vu-Dac N, Gervois P, Grötzinger T, et al. Transcriptional regulation of apolipoprotein A-I gene expression by the nuclear receptor RORα. J Biol Chem. 1997;272(36):22401–22404. doi: 10.1074/jbc.272.36.22401. [DOI] [PubMed] [Google Scholar]
- 34.Raspe E, Duez H, Gervois P, et al. Transcriptional regulation of apolipoprotein C-III gene expression by the orphan nuclear receptor RORα. J Biol Chem. 2001;276(4):2865–2871. doi: 10.1074/jbc.M004982200. [DOI] [PubMed] [Google Scholar]
- 35.Mamontova A, Séguret-Macé S, Esposito B, et al. Severe atherosclerosis and hypoαlipoproteinemia in the staggerer mouse, a mutant of the nuclear receptor RORα. Circulation. 1998;98(24):2738–2743. doi: 10.1161/01.cir.98.24.2738. [DOI] [PubMed] [Google Scholar]
- 36.Lau P, Fitzsimmons RL, Raichur S, Wang SC, Lechtken A, Muscat GE. The orphan nuclear receptor, RORα, regulates gene expression that controls lipid metabolism: staggerer (SG/SG) mice are resistant to diet-induced obesity. J Biol Chem. 2008;283(26):18411–18421. doi: 10.1074/jbc.M710526200. [DOI] [PubMed] [Google Scholar]
- 37.Kang HS, Angers M, Beak JY, et al. Gene expression profiling reveals a regulatory role for RORα and RORγ in Phase I and Phase II metabolism. Physiol Genomics. 2007;31(2):281–294. doi: 10.1152/physiolgenomics.00098.2007. [DOI] [PubMed] [Google Scholar]
- 38.Wada T, Kang HS, Angers M, et al. Identification of oxysterol 7α-hydroxylase (Cyp7b1) as a novel retinoid-related orphan receptor α (RORα) (NR1F1) target gene and a functional cross-talk between RORα and liver X receptor (NR1H3) Mol Pharmacol. 2008;73(3):891–899. doi: 10.1124/mol.107.040741. [DOI] [PubMed] [Google Scholar]
- 39.Kharitonenkov A, Shiyanova TL, Koester A, et al. FGF-21 as a novel metabolic regulator. J Clin Invest. 2005;115(6):1627–1635. doi: 10.1172/JCI23606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mayer D. The circadian rhythm of synthesis and catabolism of cholesterol. Arch Toxicol. 1976;36(3–4):267–276. doi: 10.1007/BF00340534. [DOI] [PubMed] [Google Scholar]
- 41.Edwards PA, Muroya H, Gould RG. In vivo demonstration of the circadian thythm of cholesterol biosynthesis in the liver and intestine of the rat. J Lipid Res. 1972;13(3):396–401. [PubMed] [Google Scholar]
- 42▪.Chopra AR, Louet JF, Saha P, et al. Absence of the SRC-2 coactivator results in a glycogenopathy resembling Von Gierke’s disease. Science. 2008;322(5906):1395–1399. doi: 10.1126/science.1164847. Identified G6Pase as an ROR target gene, and linked ROR transcriptional activity with glucose metabolism. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E. Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab. 2007;5(6):426–437. doi: 10.1016/j.cmet.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 44.Inagaki T, Dutchak P, Zhao G, et al. Endocrine regulation of the fasting response by PPARα-mediated induction of fibroblast growth factor 21. Cell Metab. 2007;5(6):415–425. doi: 10.1016/j.cmet.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Duez H, Staels B. REV-ERBα: an integrator of circadian rhythms and metabolism. J Appl Physiol. 2009;107(6):1972–1980. doi: 10.1152/japplphysiol.00570.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chomez P, Neveu I, Mansén A, et al. Increased cell death and delayed development in the cerebellum of mice lacking the REV-ERB(α) orphan receptor. Development. 2000;127(7):1489–1498. doi: 10.1242/dev.127.7.1489. [DOI] [PubMed] [Google Scholar]
- 47.Raspe E, Duez H, Mansén A, et al. Identification of REV-ERBα as a physiological repressor of apoC-III gene transcription. J Lipid Res. 2002;43(12):2172–2179. doi: 10.1194/jlr.m200386-jlr200. [DOI] [PubMed] [Google Scholar]
- 48.Shachter NS. Apolipoproteins C-I and C-III as important modulators of lipoprotein metabolism. Curr Opin Lipidol. 2001;12(3):297–304. doi: 10.1097/00041433-200106000-00009. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Yin L, Lazar MA. The orphan nuclear receptor REV-ERBα regulates circadian expression of plasminogen activator inhibitor type 1. J Biol Chem. 2006;281(45):33842–33848. doi: 10.1074/jbc.M607873200. [DOI] [PubMed] [Google Scholar]
- 50.Anzulovich A, Mir A, Brewer M, Ferreyra G, Vinson C, Baler R. Elovl3: a model gene to dissect homeostatic links between the circadian clock and nutritional status. J Lipid Res. 2006;47(12):2690–2700. doi: 10.1194/jlr.M600230-JLR200. [DOI] [PubMed] [Google Scholar]
- 51.Vu-Dac N, Chopin-Delannoy S, Gervois P, et al. The nuclear receptors peroxisome proliferator-activated receptor α and REV-ERBα mediate the species-specific regulation of apolipoprotein A-I expression by fibrates. J Biol Chem. 1998;273(40):25713–25720. doi: 10.1074/jbc.273.40.25713. [DOI] [PubMed] [Google Scholar]
- 52.Estall JL, Ruas JL, Choi CS, et al. PGC-1α negatively regulates hepatic FGF21 expression by modulating the heme/REV-ERB(α) axis. Proc Natl Acad Sci USA. 2009;106(52):22510–22515. doi: 10.1073/pnas.0912533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng J, Shan Y, Lambrecht RW, Donohue SE, Bonkovsky HL. Differential regulation of human ALAS1 mRNA and protein levels by heme and cobalt protoporphyrin. Mol Cell Biochem. 2008;319(1–2):153–161. doi: 10.1007/s11010-008-9888-0. [DOI] [PubMed] [Google Scholar]
- 54.Ramakrishnan SN, Lau P, Burke LJ, Muscat GE. REV-ERBβ regulates the expression of genes involved in lipid absorption in skeletal muscle cells: evidence for cross-talk between orphan nuclear receptors and myokines. J Biol Chem. 2005;280(10):8651–8659. doi: 10.1074/jbc.M413949200. [DOI] [PubMed] [Google Scholar]
- 55.Gronemeyer H, Gustafsson JA, Laudet V. Principles for modulation of the nuclear receptor superfamily. Nat Rev Drug Discov. 2004;3(11):950–964. doi: 10.1038/nrd1551. [DOI] [PubMed] [Google Scholar]
- 56.Rosenfeld MG, Lunyak VV, Glass CK. Sensors and signals: a coactivator/corepressor/ epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 2006;20(11):1405–1428. doi: 10.1101/gad.1424806. [DOI] [PubMed] [Google Scholar]
- 57.Wiesenberg I, Missbach M, Kahlen JP, Schräder M, Carlberg C. Transcriptional activation of the nuclear receptor RZRα by the pineal gland hormone melatonin and identification of CGP 52608 as a synthetic ligand. Nucleic Acids Res. 1995;23(3):327–333. doi: 10.1093/nar/23.3.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58▪▪.Kallen JA, Schlaeppi JM, Bitsch F, et al. X-ray structure of the hRORα LBD at 1.63 Å: structural and functional data that cholesterol or a cholesterol derivative is the natural ligand of RORα. Structure. 2002;10(12):1697–1707. doi: 10.1016/s0969-2126(02)00912-7. Solved the first crystal structure of RORa’s ligand-binding domain (LBD) revealing cholesterol bound within the ligand-binding pocket. [DOI] [PubMed] [Google Scholar]
- 59▪.Kallen JA, Schlaeppi JM, Bitsch F, Delhon I, Fournier B. Crystal structure of the human RORα ligand binding domain in complex with cholesterol sulfate at 2.2 Å. J Biol Chem. 2004;279(14):14033–14038. doi: 10.1074/jbc.M400302200. Through x-ray crystallography, this group identified cholesterol sulfate within the ligand-binding domain of RORα, and characterized it as another endogenous ligand for ROR. [DOI] [PubMed] [Google Scholar]
- 60.Stehlin C, Wurtz JM, Steinmetz A, et al. X-ray structure of the orphan nuclear receptor RORβ ligand-binding domain in the active conformation. EMBO J. 2001;20(21):5822–5831. doi: 10.1093/emboj/20.21.5822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stehlin-Gaon C, Willmann D, Zeyer D, et al. All-trans retinoic acid is a ligand for the orphan nuclear receptor RORβ. Nat Struct Biol. 2003;10(10):820–825. doi: 10.1038/nsb979. [DOI] [PubMed] [Google Scholar]
- 62.Wang Y, Kumar N, Crumbley C, Griffin PR, Burris TP. A second class of nuclear receptors for oxysterols: regulation of RORα and RORγ activity by 24S-hydroxycholesterol (cerebrosterol) Biochim Biophys Acta. 2010;1801(8):917–923. doi: 10.1016/j.bbalip.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63▪▪.Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORγ. Mol Endocrinol. 2010;24(5):923–929. doi: 10.1210/me.2009-0507. Identified several hydroxycholesterols as endogenous ligands for RORγ through crystallographic methods. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adare A, Afanasiev S, Aidala C, et al. Enhanced production of direct photons in Au + Au collisions at square root(S(NN)) = 200 GeV and implications for the initial temperature. Phys Rev Lett. 2010;104(13):132301. doi: 10.1103/PhysRevLett.104.132301. [DOI] [PubMed] [Google Scholar]
- 65▪.Pardee KI, Xu X, Reinking J, et al. The structural basis of gas-responsive transcription by the human nuclear hormone receptor REV-ERBβ. PLoS Biol. 2009;7(2):e43. doi: 10.1371/journal.pbio.1000043. Provided the crystal structure of REV-ERBβ when bound to heme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Woo EJ, Jeong DG, Lim MY, et al. Structural insight into the constitutive repression function of the nuclear receptor REV-ERBβ. J Mol Biol. 2007;373(3):735–744. doi: 10.1016/j.jmb.2007.08.037. [DOI] [PubMed] [Google Scholar]
- 67▪.Phelan CA, Gampe RT, Jr, Lambert MH, et al. Structure of REV-ERBα bound to N-CoR reveals a unique mechanism of nuclear receptor-co-repressor interaction. Nat Struct Mol Biol. 2010;17(7):808–814. doi: 10.1038/nsmb.1860. Provides the structure of the LBD of REV-ERBα when bound to the corepressor NCoR and suggests that REV-ERBα may be active in the absence of heme. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68▪▪.Kumar N, Solt LA, Conkright JJ, et al. The benzenesulfoamide T0901317 [N-(2,2,2-trifluoroethyl)-N-[4-[2,2,2-trifluoro-1-hydroxy-1-(trifluorometh yl)ethyl] phenyl]-benzenesulfonamide] is a novel retinoic acid receptor-related orphan receptor-α/γ inverse agonist. Mol Pharmacol. 2010;77(2):228–236. doi: 10.1124/mol.109.060905. Identifies the first synthetic ligand that modulates both RORα and RORγ activity through direct binding of the receptors, resulting in the modulation of the receptor’s ability to interact with transcriptional cofactor proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Houck KA, Borchert KM, Hepler CD, et al. T0901317 is a dual LXR/FXR agonist. Mol Genet Metab. 2004;83(1–2):184–187. doi: 10.1016/j.ymgme.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 70.Mitro N, Vargas L, Romeo R, Koder A, Saez E. T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS Lett. 2007;581(9):1721–6172. doi: 10.1016/j.febslet.2007.03.047. [DOI] [PubMed] [Google Scholar]
- 71▪▪.Wang Y, Kumar N, Nuhant P, et al. Identification of SR1078, a synthetic agonist for the orphan nuclear receptors RORα and RORγ. ACS Chem Biol. 2010;5(11):1029–1034. doi: 10.1021/cb100223d. Identified and characterized the first selective RORα/γ synthetic agonist. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kumar N, Kojetin DJ, Solt LA, et al. Identification of ML176 (SR3335): a synthetic RORα selective inverse agonist. ACS Chem Biol. 2010 doi: 10.1021/cb1002762. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73▪▪.Meng QJ, McMaster A, Beesley S, et al. Ligand modulation of REV-ERBα function resets the peripheral circadian clock in a phasic manner. J Cell Sci. 2008;121(Pt. 21):3629–3635. doi: 10.1242/jcs.035048. Identified and characterized the first selective REV-ERBα synthetic agonist, GSK4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fu M, Sun T, Bookout AL, et al. A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol Endocrinol. 2005;19(10):2437–2450. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- 75.Fontaine C, Rigamonti E, Pourcet B, et al. The orphan nuclear receptor REV-ERBα is a peroxisome proliferator-activated receptor (PPAR) γ target gene and promotes PPARγ-induced adipocyte differentiation. J Biol Chem. 2003;278(39):37672–37680. doi: 10.1074/jbc.M304664200. [DOI] [PubMed] [Google Scholar]
- 76.Chawla A, Lazar MA. Induction of REV-ERBAα, an orphan receptor encoded on the opposite strand of the α-thyroid hormone receptor gene, during adipocyte differentiation. J Biol Chem. 1993;268(22):16265–16269. [PubMed] [Google Scholar]
- 77.Duez H, Duhem C, Laitinen S, et al. Inhibition of adipocyte differentiation by RORα. FEBS Lett. 2009;583(12):2031–2036. doi: 10.1016/j.febslet.2009.05.019. [DOI] [PubMed] [Google Scholar]
- 78.Austin S, Medvedev A, Yan ZH, Adachi H, Hirose T, Jetten AM. Induction of the nuclear orphan receptor RORγ during adipocyte differentiation of D1 and 3T3-L1 cells. Cell Growth Differ. 1998;9(3):267–276. [PubMed] [Google Scholar]
- 79▪.Kumar N, Solt LA, Wang Y, et al. Regulation of adipogenesis by natural and synthetic REV-ERB ligands. Endocrinology. 2010;151(7):3015–3025. doi: 10.1210/en.2009-0800. Series of studies suggesting that heme, functioning as a REV-ERB ligand, is an important regulator of adipogenesis and that synthetic ligands can modulate adipogenesis and may be useful therapeutics for the treatment of metabolic diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grant D, Yin L, Collins JL, et al. GSK4112, a small-molecule chemical probe for the cell biology of the nuclear heme receptor REV-analogα. ACS Chem Biol. 2010;5(10):925–932. doi: 10.1021/cb100141y. [DOI] [PubMed] [Google Scholar]
- 81▪▪.Kojetin D, Wang Y, Kamenecka TM, Burris TP. Identification of SR8278, a synthetic antagonist of the nuclear heme receptor REV-ERB. ACS Chem Biol. 2011;6(2):131–134. doi: 10.1021/cb1002575. Study that identified and characterized the first synthetic REV-ERB antagonist, SR8278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lund J, Arendt J, Hampton SM, English J, Morgan LM. Postprandial hormone and metabolic responses amongst shift workers in Antarctica. J Endocrinol. 2001;171(3):557–564. doi: 10.1677/joe.0.1710557. [DOI] [PubMed] [Google Scholar]
- 83.Kawachi I, Colditz GA, Stampfer MJ, et al. Prospective study of shift work and risk of coronary heart disease in women. Circulation. 1995;92(11):3178–3182. doi: 10.1161/01.cir.92.11.3178. [DOI] [PubMed] [Google Scholar]
- 84.Knutsson A, Hallquist J, Reuterwall C, Theorell T, Akerstedt T. Shiftwork and myocardial infarction: a case-control study. Occup Environ Med. 1999;56(1):46–50. doi: 10.1136/oem.56.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Ellingsen T, Bener A, Gehani AA. Study of shift work and risk of coronary events. J R Soc Promot Health. 2007;127(6):265–267. doi: 10.1177/1466424007083702. [DOI] [PubMed] [Google Scholar]
- 86.Hermansson J, Gillander Gådin K, Karlsson B, Lindahl B, Stegmayr B, Knutsson A. Ischemic stroke and shift work. Scand J Work Environ Health. 2007;33(6):435–943. doi: 10.5271/sjweh.1165. [DOI] [PubMed] [Google Scholar]
- 87.Van Cauter E, Polonsky KS, Scheen AJ. Roles of circadian rhythmicity and sleep in human glucose regulation. Endocr Rev. 2007;18(5):716–738. doi: 10.1210/edrv.18.5.0317. [DOI] [PubMed] [Google Scholar]
- 88.Holterhus PM, Odendahl R, Oesingmann S, et al. Classification of distinct baseline insulin infusion patterns in children and adolescents with Type I diabetes on continuous subcutaneous insulin infusion therapy. Diabetes Care. 2007;30(3):568–573. doi: 10.2337/dc06-2105. [DOI] [PubMed] [Google Scholar]
- 89▪▪.Wang Y, Kumar N, Solt LA, et al. Modulation of ROR{α} and ROR{γ} activity by 7-oxygenated sterol ligands. J Biol Chem. 2009;285(1):5013–5025. doi: 10.1074/jbc.M109.080614. Study demonstrating that 7-oxygenated sterols function as high-affinity ligands for both RORα and RORγ leading to suppression of the transcriptional activity of the receptors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Brown AJ, Jessup W. Oxysterols and atherosclerosis. Atherosclerosis. 1999;142(1):1–28. doi: 10.1016/s0021-9150(98)00196-8. [DOI] [PubMed] [Google Scholar]
- 91.Brown AJ, Leong SL, Dean RT, Jessup W. 7-hydroperoxycholesterol and its products in oxidized low-density lipoprotein and human atherosclerotic plaque. J Lipid Res. 1997;38(9):1730–1745. [PubMed] [Google Scholar]
- 92.Gelissen IC, Brown AJ, Mander EL, Kritharides L, Dean RT, Jessup W. Sterol efflux is impaired from macrophage foam cells selectively enriched with 7-ketocholesterol. J Biol Chem. 1996;271(30):17852–17860. doi: 10.1074/jbc.271.30.17852. [DOI] [PubMed] [Google Scholar]
- 93.Zieden B, Kaminskas A, Kristenson M, et al. Increased plasma 7β-hydroxycholesterol concentrations in a population with a high risk for cardiovascular disease. Arterioscler Thromb Vasc Biol. 1999;19(4):967–971. doi: 10.1161/01.atv.19.4.967. [DOI] [PubMed] [Google Scholar]
- 94.Duez H, van der Veen JN, Duhem C, et al. Regulation of bile acid synthesis by the nuclear receptor REV-ERBα. Gastroenterology. 2008;135(2):689–698. doi: 10.1053/j.gastro.2008.05.035. [DOI] [PubMed] [Google Scholar]
- 95.Le Martelot G, Claudel T, Gatfield D, et al. REV-ERBα participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7(9):e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]