Abstract
Phosphatidylcholine-hydrolysing phospholipase C, also known as non-specific phospholipase C (NPC), is a new member of the plant phospholipase family that reacts to environmental stresses such as phosphate deficiency and aluminium toxicity, and has a role in root development and brassinolide signalling. Expression of NPC4, one of the six NPC genes in Arabidopsis, was highly induced by NaCl. Maximum expression was observed from 3 h to 6 h after the salt treatment and was dependent on salt concentration. Results of histochemical analysis of PNPC4:GUS plants showed the localization of salt-induced expression in root tips. On the biochemical level, increased NPC enzyme activity, indicated by accumulation of diacylglycerol, was observed as early as after 30 min of salt treatment of Arabidopsis seedlings. Phenotype analysis of NPC4 knockout plants showed increased sensitivity to salinity as compared with wild-type plants. Under salt stress npc4 plants had shorter roots, lower fresh weight, and reduced seed germination. Expression levels of abscisic acid-related genes ABI1, ABI2, RAB18, PP2CA, and SOT12 were substantially reduced in salt-treated npc4 plants. These observations demonstrate a role for NPC4 in the response of Arabidopsis to salt stress.
Keywords: Arabidopsis thaliana, diacylglycerol, phospholipase C, salt stress
Introduction
Phospholipases are key components of the plant phospholipid signalling network. Besides their metabolic functions they play a key role in signal transduction mechanisms in plant cells. This phospholipid signalling network includes in particular phosphoinositide-specific phospholipase C (PI-PLC), phospholipase D (PLD), and phospholipases A1 and A2 (PLA1 and PLA2). Stimulation of the signalling network is involved in many responses of plants to both biotic and abiotic adverse environmental factors such as wounding, fungal and bacterial attack, drought, cold, and salt stress (Li et al., 2009; Xue et al., 2009; Munnik, 2010).
A link between water stress (salt, osmotic, and drought stress) and the phospholipid signalling network, particularly PLC and PLD, has been shown many times. Recently Munnik and Vermeer (2010) described osmotic stress-induced PI-PLC signalling in detail, whereas Hong et al. (2010) presented data on PLD and phosphatidic acid (PA) signalling in response to drought and salinity. Rapid accumulation of both the PI-PLC substrate phosphatidylinositol 4,5-bisphosphate (PIP2) and the product inositol 1,4,5-trisphosphate (IP3) as a response to water stress was described in, for example, Arabidopsis, tobacco, and rice. The importance of PI-PLC in water stress-related processes was confirmed by PI-PLC gene manipulation (Wang et al., 2008; Georges et al., 2009). The involvement of PLDα1, α3, δ, and ϵ in salt, osmotic, and drought stress was also demonstrated (Mane et al., 2007; Hong et al., 2008a, b; Bargmann et al., 2009).
Besides PI-PLC, phospholipase C hydrolysing phosphatidylcholine (PC-PLC), also termed non-specific phospholipase C (NPC), has been described. This enzyme that generates diacylglycerol (DAG) through glycerophospholipid hydrolysis [mainly phosphatidylcholine (PC)] has been characterized in animals (Exton, 1994) and is well known and characterized in bacteria (Titball, 1993). Based on the amino acid sequence similarity with the bacterial Gram-negative (non-haemolytic) PC-PLC family, Nakamura et al. (2005) identified six putative PC-PLC genes in Arabidopsis designated NPC1–NPC6. In that work, NPC4 was expressed in Escherichia coli, and NPC4 protein revealed Ca2+-independent phospholipase activity that preferred PC over phosphatidylethanolamine (PE) as the substrate. NPC4 showed very low activity for PIP2 as a substrate. To date, PC-PLC activity was reported to have a metabolic function in DAG exchange from phospholipid to galactolipids in plants (Andersson et al., 2005; Gaude et al., 2008; Tjellström et al., 2008). Down-regulation of PC-PLC in response to elicitor signalling has been described previously. A rapid decrease of DAG levels in tobacco VBI-0 cells was found after treatment with the elicitor cryptogein from Phytophthora cryptogea (Scherer et al., 2002). Similarly, inhibition of PC-PLC activity was observed after treatment of both tobacco Bright Yellow 2 (BY-2) cells and pollen tubes with aluminium (Pejchar et al., 2010). The role of NPC3 and NPC4 in root development and brassinolide signalling was also shown in Arabidopsis (Wimalasekera et al., 2010). Recently Reddy et al. (2010) expressed NPC3 in E. coli and showed that purified NPC3 protein has lysophosphatidic acid phosphatase activity.
The aim of this work was to investigate the function of Arabidopsis NPCs further in relation to salt stress. Here it is shown that NPC4 is a component of the salt stress response in Arabidopsis.
Materials and methods
Plant material
Arabidopsis thaliana Columbia (Col-0) seeds were obtained from Lehle seeds and used as wild-type (WT) controls. Two T-DNA insertion lines were used in experiments: npc4-1 (SALK_046713) from the Salk Institute Genome Analysis Laboratory (SIGNAL) collection (Alonso, 2003) and npc4-2 (GK-571E10) from the GABI-KAT collection (Rosso et al., 2003). For the first characterization of these mutants, see Wimalasekera et al. (2010).
Salt treatment
T-DNA mutants and WT Arabidopsis plants were grown on agar plates containing 4.4 g l−1 MS (Murashige–Skoog) basal salts, sucrose (10 g l−1), MES (0.5 g l−1), inositol (0.1 g l−1), 1% (w/v) agar (pH 5.8) supplemented with 50 mM or 100 mM NaCl. Seeds were surface sterilized using 30% (v/v) bleach solution for 10 min and rinsed five times with sterile water. After planting seeds on agar (45 seeds per plate for weighing and 13 seeds per plate for root growth analysis), the plates were transferred for 4 d to the dark at 4 °C in order to synchronize germination. The plants were grown in a horizontal (weight) or vertical (root growth) position in a growth chamber at 22 °C under long day conditions (16 h/8 h light/dark cycle) and weighted or measured after 14 d of cultivation. Documentation was done by scanning (Canon CanoScan 8800F). Root measurements were done using JMicroVision 1.2.7 software.
Germination
The same basal medium as in the growth experiment with 45 seeds per plate and four replicates and with 150 mM NaCl was used in the germination test. The growth conditions were continuous light at 23 °C. Germinated seeds were counted at 24, 30, 36, and 42 h after transferring seeds from 4 °C.
Hydroponic cultivation
The seeds were surface sterilized and stratified as described above and sown onto rollers cut from Grodan® Master slab saturated with nutrient solution. Plants were cultivated in containers of 2.5 l in modified half-strength Hoagland's solution (Hoagland and Arnon, 1950). The nutrient concentrations were as follows: 0.5 mM NH4H2PO4, 3 mM KNO3, 2 mM Ca(NO3)2·4H2O, 1 mM MgSO4·7H2O, 24.5 μM ferric citrate, 0.45 μM KI, 4.85 μM H3BO3, 5.92 μM MnSO4·4H2O, 0.7 μM ZnSO4.7H2O, 0.1 μM Na2MoO4·2H2O, 0.01 μM CuSO4·5H2O, 0.01 μM CoCl2·6H2O, 10.02 μM Na2EDTA, 10 μM FeSO4·7H2O, 55.51 μM myo-inositol, 0.81 μM nicotinic acid, 0.49 μM pyridoxin, 2.97 μM thiamin. The solution was replaced for the first time when plants were 2 weeks old, and thereafter once a week. Aeration of the solution was carried out for 15 min every 3 h using an aquarium air pump. A 10 h/14 h light/dark cycle at 75% relative humidity and day/night temperatures of 22 °C and 20 °C, respectively, were used.
Quantitative RT-PCR
Hydroponically grown 5-week-old plants at the stage of leaf rosettes were used for expression analysis. For the determination of the basal expression of the studied genes, the root and leaf samples were collected immediately prior to the exchange of the nutrient solution. The original nutrient solution was replaced with a fresh solution with or without salt. NaCl concentrations were 25, 50, and 100 mM. Root and leaf samples were collected 1, 2, 3, 6, 12, and 36 h after changing the solution and instantly frozen in liquid nitrogen. RNA was isolated using a Spectrum Plant Total RNA Kit (Sigma-Aldrich), a Turbo DNA-free Kit (Applied Biosystems) was used for DNA removal, and a Transcriptor High Fidelity cDNA Synthesis Kit (Roche) was used for cDNA synthesis. The reverse transcription reaction was primed with anchored-oligo(DT)18 primer. Quantitative PCR was performed with a LightCycler® 480 system (Roche) using the LightCycler® Probes Master with the corresponding hydrolysis probe (UPL Roche) to detect the expression of NPC isoforms and reference genes, and LightCycler® 480 SYBR Green I Master (Roche) for the other genes. Actin2 and UBQ10 were used as reference genes for the normalization of target gene expression. Fold change in expression of the target gene was calculated using the equation (Pfaffl, 2001):
Primers and the probes are described in Supplementary Tables S1 and S2 available at JXB online.
Histochemical β-glucuronidase (GUS) staining
Construction of promoter:GUS plants was described previously (Wimalasekera et al., 2010). The histochemical GUS assay (Jefferson et al., 1987) was carried out on seedlings and 5-week-old plants. T2 seeds of PNPC3:GUS and PNPC4:GUS were grown on agar plates under the same conditions as described in ‘Salt treatment’ (see above). Ten-day-old seedlings were transferred to a 12-well plate containing 1 ml of half-strength Hoagland's solution with 2% (w/v) sucrose with or without 100 mM NaCl. After 24 h incubation, the plants were immersed in X-Gluc buffer [2 mM X-Gluc, 50 mM NaPO4 pH 7.0, 0.5% (v/v) Triton-X, 0.5 mM K-ferricyanide] for 16 h at 37 °C. Chlorophyll of the green parts was removed by repeated washing in 80% (v/v) ethanol. To determine NaCl-mediated expression of NPC4 in adult plants, PNPC3:GUS and PNPC4:GUS were grown hydroponically for 5 weeks and exposed to 100 mM NaCl for 4 h. The staining procedure was the same as in the case of seedlings. Observations were done on a Nikon SMZ 1500 zoom stereoscopic microscope coupled to a Nikon DS-5M digital camera.
PC-PLC activity in salt-treated Arabidopsis seedlings
Seven-day-old Arabidopsis seedlings (five seedlings for each sample) were transferred from agar plates to 900 μl of water and labelled with 0.66 μg ml−1 of fluorescent phosphatidylcholine (bodipy-PC, D-3771, Invitrogen, USA) for 10 min. Then, 100 μl of NaCl solution was added to obtain final concentrations of 10–100 mM and seedlings were incubated on an orbital shaker at 23 °C in the dark for different times. Lipids were extracted by the modified method of Bligh and Dyer (1959) by addition of 4 ml of methanol/chloroform 2/1 (v/v) and 2 ml of 0.1 M KCl 30 min later. Samples were centrifuged for 15 min at 420 g. The lower phase was evaporated to dryness by a vacuum evaporator and redissolved in ethanol. Samples were applied on HP-TLC silica gel-60 plates (Merck KGaA, Darmstadt, Germany) by an ATS4 sampler (Camag, Muttenz, Switzerland). After 10 min of saturation, plates were developed in the horizontal developing chamber (Camag) in a mobile phase of acetone/chloroform 1/1 (v/v). Plates were dried, scanned using a Fuji FLA-7000 fluorescence scanner (Fujifilm, Tokyo, Japan), and analysed by Kodak ds 1D software. Identification of the spot corresponding to bodipy-DAG was based on comparison with the bodipy-DAG standard prepared as described previously (Pejchar et al., 2010).
Results
Expression of the NPC gene family in response to salt
Arabidopsis plants were grown hydroponically for 5 weeks and afterwards were treated with Hoagland medium supplemented with 100 mM NaCl for 1, 2, 3, 6, 12, and 36 h. Expression of all members of the NPC gene family was measured by quantitative RT-PCR in both roots and leaves of control non-treated plants and salt-treated plants. A slight increase in NPC6 gene expression in roots was observed after 6 h of treatment (Fig. 1). Similarly, gene expression of NPC2 was slightly increased after 12 h of salt treatment (data not shown). However, the most meaningful rise in gene expression was shown by NPC4. Quantification of expression of NPC5 was not possible because the transcript level in both non-treated and treated plants was too low in roots and leaves. This is consistent with published observations (Nakamura et al., 2005; Wimalasekera et al., 2010) where NPC5 transcript was found only in flowers.
Fig. 1.
Expression pattern of the NPC genes in NaCl-treated plants. The transcript levels of NPC genes were measured by quantitative real-time PCR in leaves and roots of non-treated plants and in plants treated with 100 mM NaCl for 6 h. Actin2 and UBQ10 were used as internal controls. The expression of each gene in non-treated controls was set to 1. Data represent the means ±SE, n=3 discrete samples from one biological experiment. This experiment was repeated three times with similar results. NPC, non-specific phospholipase C; nd, not determined.
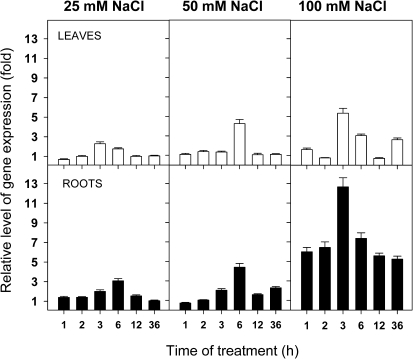
In order to investigate the expression pattern of NPC4 further, Arabidopsis plants were also subjected to lower NaCl concentrations (25 mM and 50 mM) for identical amounts of time to those used in previous experiments. The results showed very distinct time and concentration dependence (Fig. 2). In both leaves and roots there was a transient increase in expression observed after 3–6 h of treatment. Expression in roots was about twice as high as that in leaves. In roots treated with 25 mM NaCl for 6 h the expression of NPC4 was three times higher than NPC4 expression in control plants. Treatment with 50 mM NaCl for 6 h led to a 5-fold increase in the expression level of NPC4 in roots. Treatment with 100 mM NaCl induced an increase in expression in roots as early as after 1 h of the treatment, and the expression remained high for all sampled times. A rapid and transient increase of NPC4 expression may indicate a signalling function for NPC4.
Fig. 2.
Detailed analysis of NPC4 expression in NaCl-treated plants. The transcript levels of the NPC4 gene were measured by quantitative real-time PCR in leaves and roots of non-treated plants and in plants treated with different concentrations of NaCl (25–100 mM) for different times (1–36 h). Actin2 and UBQ10 were used as internal controls. The expression of NPC4 in non-treated controls at the respective times was set to 1. Data represent the means ±SE, n=2 discrete samples from one biological experiment. This experiment was repeated twice with similar results.
Activity of NPC in salt-treated Arabidopsis seedlings
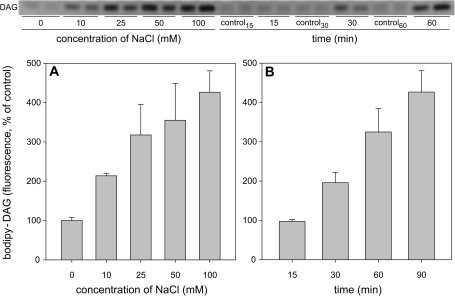
A method based on using fluorescently labelled PC (bodipy-PC) to determine the activity of NPC in tobacco cells was recently reported (Pejchar et al., 2010; Wimalasekera et al., 2010). A similar approach was employed to determine the activity of NPC in Arabidopsis seedlings. In order to find out if the observed increase in NPC4 expression is recognizable at the level of enzyme activity, 1-week-old Arabidopsis seedlings were labelled with bodipy-PC and then treated with different NaCl concentrations (10–100 mM NaCl) for 90 min. The quantity of bodipy-DAG, the product of NPC activity, doubled after 90 min of 10 mM NaCl treatment and increased 4-fold after 100 mM NaCl treatment. (Fig. 3A). Arabidopsis seedlings were also treated with 100 mM NaCl for different times (15, 30, 60, and 90 min). Changes in NPC activity were already detectable after 30 min of NaCl treatment. Thus, it is obvious that salt-stressed Arabidopsis seedlings respond by the rapid alteration of the NPC activity and that this effect is caused even by very mild salt stress (10 mM NaCl). Rapid changes in expression of NPC4 in 7-day-old seedlings were also determined after 4 h of 100 mM salt treatment and 2.5-fold (±0.3) increase found.
Fig. 3.
Effect of NaCl on DAG production in Arabidopsis seedlings. Arabidopsis seedlings were grown on agar plates for 7 d. Prior to treatment seedlings were removed from agar and incubated with bodipy-PC for 10 min in water. (A) Seedlings were treated for 90 min with 0–100 mM NaCl. Lipids were extracted, separated by high-performance thin-layer chromatography (HP-TLC) and quantified. The quantity of bodipy-DAG in control non-treated seedlings was set to 100%. Data represent means ±SE from independently analysed parallel samples. This experiment was repeated three times with similar results. (B) Seedlings were treated with 100 mM NaCl. Lipids were extracted at the time intervals indicated, separated by HP-TLC, and quantified. The quantity of bodipy-DAG in control non-treated seedlings was set to 100%. Data represent the means ±SE from independently analysed parallel samples. This experiment was repeated twice with similar results. DAG, diacylglycerol.
Expression of NPC4 is localized in root tips
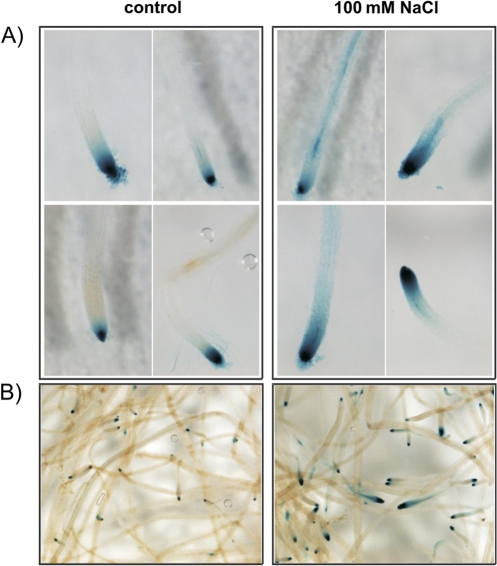
To gain insights into the localization of expression of NPC4 in roots after salt treatment, the expression pattern was investigated using promoter:GUS plants. Histochemical analysis of PNPC4:GUS plants confirmed that the expression pattern under salt stress was changed only in the roots (Fig. 4; data from other plant organs are not shown). In 10-day-old seedlings the expression was located predominantly in the root tips of the main and lateral roots of both control and treated plants (Fig. 4A). The highest intensity of GUS staining in control plants was found in the apical meristem and partly in the region of elongation, whereas after incubation in 100 mM NaCl the staining had spread to the region of mature cells. The highest expression outside the root tips was observed in epidermal cells, vascular tissues, and lateral root primordium.
Fig. 4.
Histochemical analysis of PNPC4:GUS expression under salt stress in Arabidopsis plants. (A) Effect of salt on the PNPC4:GUS expression pattern in the main root of 10-day-old seedlings. The plants were grown on agar and transferred to liquid nutrient solution with or without 100 mM NaCl for 24 h. (B) Effect of salt on the PNPC4:GUS expression pattern in roots of 5-week-old Arabidopsis plants. The plants were grown hydroponically in modified half-strength Hoagland's solution and exposed to 100 mM NaCl for 4 h.
Hydroponically grown 5-week-old plants were analysed to see if a similar expression pattern would occur in adult plants of the same age used to observed NPC expression measured by quantitative PCR. Despite the differences in the age of plants, the GUS expression pattern remained the same (Fig. 4B).
Sensitivity of NPC4 knockout Arabidopsis lines to salt
Two lines of homozygous Arabidopsis T-DNA insertion mutants of NPC4 were used as described in Wimalasekera et al. (2010): npc4-1 (SALK_046713) and npc4-2 (GK_571E10) (Supplementary Fig. S2 at JXB online). There is no obvious phenotype of either of these lines when grown in normal conditions in soil either in the greenhouse or in the growth chamber.
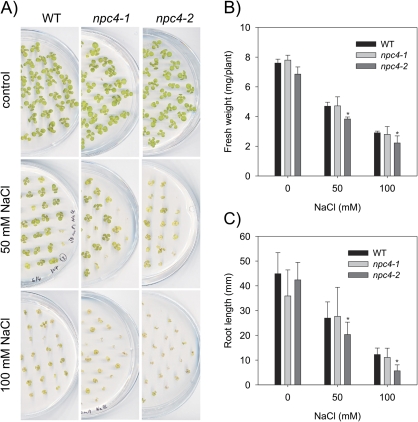
However, when mutant plants were grown in Petri dishes on agar MS medium supplemented with MES, inositol, and sucrose (see Materials and methods), there were slight but not significant differences in npc4-1, npc4-2, and the WT in fresh weight and root length. At the same time, significant decreases (t-test P <0.05) in both fresh weight and root length were revealed under salt stress conditions (Fig. 5A–C). The fresh weight and main root length phenotype of npc4-2 was dose dependent. The fresh weight of npc4-2 seedlings was reduced by 20% at 50 mM NaCl compared with the WT and by almost 25% at 100 mM NaCl (Fig. 5B). Similarly the main root length of npc4-2 was reduced by 25% at 50 mM NaCl compared with the WT and by ∼55% at 100 mM NaCl compared with the WT (Fig. 5C). Even though the fresh weight and the main root length of npc4-1 were also reduced at 100 mM compared with the WT (Fig. 5B, C), these reductions were not significant (t-test P <0.05).
Fig. 5.
Effect of NaCl on fresh weight and root length of npc4-1 and npc4-2 knockouts. (A) Arabidopsis thaliana Col wild-type (WT), npc4-1, and npc4-2 were grown on agar plates supplemented with 0, 50, and 100 mM NaCl for 14 d. (B) Fresh weight of 14-day-old WT, npc4-1, and npc4-2 seedlings. Forty-five seedlings from one agar plate were pooled and weighed. Data represent the means ±SE, n=4. This experiment was repeated three times with similar results. (C) Root length of 14-day-old WT, npc4-1, and npc4-2 seedlings. Data represent the means ±SE, n=52. This experiment was repeated three times with similar results. Asterisks indicate a statistically significant (t-test P <0.05) difference in comparison with the WT.
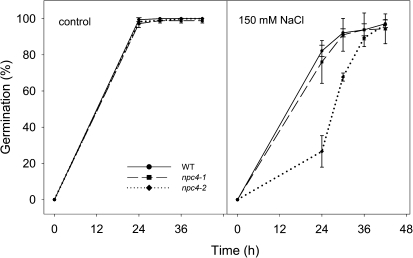
It is well known that salt inhibits seed germination. Therefore, germination of npc4 seeds on control medium and on medium supplemented with 150 mM NaCl was determined. Germination of seeds of both npc4 lines was indistinguishable from germination of WT seeds on control medium, and nearly 100% of WT and npc4 seeds germinated within 24 h. However, on the medium with 150 mM NaCl, 82% of WT seeds, 76% of npc4-1, and only 27% of npc4-2 seeds germinated after 24 h. Still nearly all seeds of the WT and npc4 germinated within 42 h. Thus it was shown that the germination rate of npc4 seeds decreased in comparison with WT seeds, but 150 mM NaCl does not inhibit germination completely (Fig. 6).
Fig. 6.
Effect of NaCl on the seed germination phenotype of npc4-1 and npc4-2 knockouts. Forty-five seeds of the WT, npc4-1, or npc4-2 were germinated on an agar plate with or without 150 mM NaCl. Germinated seeds were counted at 24, 30, 36, and 42 h after transferring seeds from 4 °C. Data represent the means ±SE, n=4. This experiment was repeated three times with similar results.
These results show a higher sensitivity of npc4 knockout lines to NaCl and demonstrate that NPC4 plays a role in response of Arabidopsis to salt stress.
Transcription pattern of salt and phospholipid signalling-related genes
The results mentioned above demonstrate that NPC4 is involved in the response of Arabidopsis plants to salt stress. Genes which are known to be related to salt stress as well as those related to phospholipid signalling were selected and their expression pattern was tested for alterations in npc4-1, npc4-2, and the WT under salt stress conditions. Expression of the genes was monitored in roots and leaves of 5-week-old hydroponically grown Arabidopsis plants treated with 100 mM NaCl. Samples were taken 3, 6, 9, and 12 h after addition of NaCl to the medium.
Differences between the gene expression pattern of salt-treated WT and knockout plants were found only in roots, and data from leaves are, therefore, not shown.
Expression of salt overly sensitive (SOS) signal transduction pathway genes:
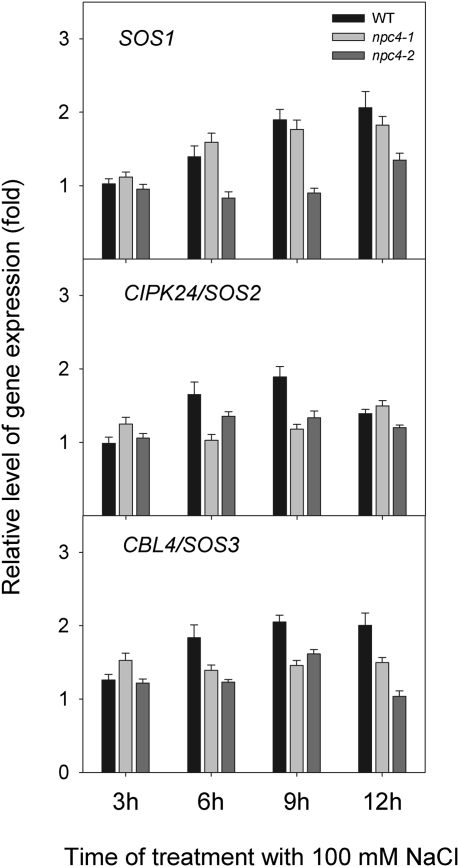
The SOS signalling pathway is probably the best characterized salt-specific signal transduction pathway (Zhu, 2002, 2003; Munns and Tester, 2008). The calcineurin B-like protein (CBL4/SOS3) interacts with CBL-interacting protein kinase (CIPK24/SOS2) (Gong et al., 2004) and the CBL4–CIPK24 complex then activates the Na+/H+ antiporter SOS1 located at the plasma membrane (Qiu et al., 2002). In roots of salt-treated WT plants, expression of all SOS genes doubled during 12 h. Expression of both SOS2 and SOS3 genes preceded expression of the SOS1 gene. This increase was distinctly smaller in npc4 knockouts (Fig. 7). Such results might indicate a link between the SOS signalling pathway and NPC4.
Fig. 7.
Relative level of gene expression of SOS signal transduction genes in the WT, npc4-1, and npc4-2 under 100 mM NaCl treatment. The transcript levels of SOS1, CIPK24/SOS2, and CBL4/SOS3 were measured by quantitative real-time PCR in roots of non-treated plants and in plants treated with 100 mM NaCl for 3, 6, 9, and 12 h. Actin2 and UBQ10 were used as internal controls. The expression of the genes in non-treated controls at the respective times was set to 1. Data represent the means ±SE, n=2 discrete samples from one biological experiment. This experiment was repeated three times with similar results.
Expression of ion transporters:
During salt stress, Na+ ions enter mainly passively across the plasma membrane into the cytoplasm of root cells. A low concentration of Na+ in the cytoplasm has to be maintained and, therefore, vacuolar Na+/H+ antiporter (NHX) is activated during salt stress and Na+ is sequestered into vacuoles (Pardo et al., 2006; Munns and Tester, 2008). Expression of NHX1 and NHX8 was monitored. NHX1 is one of the predominant isoforms in Arabidopsis and is an important member of the salt tolerance machinery (Rodríguez-Rosales et al., 2009). NHX8 also belongs to the monovalent cation:proton antiporter-1 family; however, it is more specific as a plasma membrane Li+/H+ antiporter (An et al., 2007). No clear tendency in the NHX expression pattern in WT and npc4 plants under salt stress was observed (Supplementary Fig. S3A at JXB online).
Expression of phospholipid signalling-related genes:
The role of DAG, the product of PC-PLC activity, in plants is not very clear. DAG is likely to act as a signalling molecule at least in some systems such as tobacco pollen tubes (Helling et al., 2006). However, it is generally assumed that the most probable mechanism of DAG action is the conversion of DAG to PA by diacylglycerol kinase (DGK). It was shown that the PI-PLC/DGK pathway is activated during water stress. Therefore, expression of DGK1, the isoform which is induced by salt and drought according to the Genevestigator database (https://www.genevestigator.com) (Hruz et al., 2008), was monitored. PA is a well documented signalling molecule that is also involved in responses to salt stress. One of the genes which is known to be activated by PA is TOR (Fang et al., 2001). TOR is important in cell growth and development and also in hyperosmotic stress (Menand et al., 2002; Mahfouz et al., 2006). Thus, expression of TOR was monitored in WT and npc4 plants. Expression of DGK1 was somewhat higher after salt treatment at later times; however, there were not very clear differences between WT and npc plants. Expression of TOR in WT and knockouts remained unchanged after salt treatment (Supplementary Fig. S3B at JXB online).
Expression of ABA-related genes:
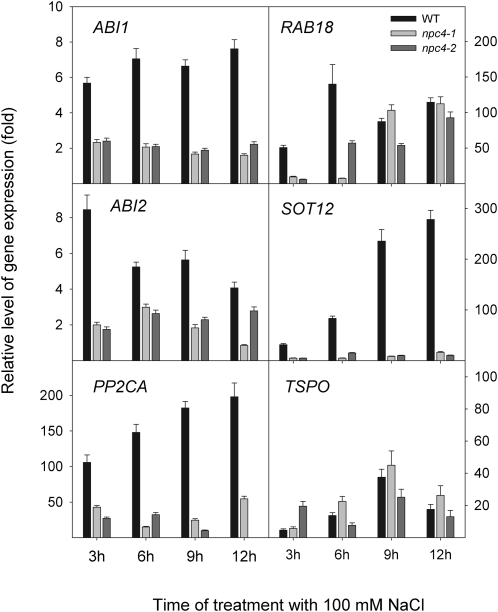
Abscisic acid (ABA) is a hormone whose concentration is elevated during salt stress, and ABA-related genes are induced afterwards (Knight et al., 1997; Kreps et al., 2002; Zhu, 2002; Christmann et al., 2006). Among them, expression of ABI1, ABI2, RAB18, PP2CA, SOT12, and TSPO was monitored. Additionally, expression of the stress-responsive gene RD29/LTI78/COR78 that is supposed to be functional in both ABA-dependent and ABA-independent signalling pathways was also monitored (Agarwal and Jha, 2010). Expression of ABI1, ABI2, PP2CA, and SOT12 in roots of salt-treated npc4-1 and npc4-2 plants clearly decreased in comparison with expression of these genes in salt-treated WT plants (Fig. 8). The decrease was detected at all monitored time points. However, the expression of RAB18 in npc4-1 and npc4-2 roots was decreased only at early time points (3 h and 6 h). The expression pattern of TSPO (Fig. 8) as well as of RD29/LTI78/COR78 (Supplementary Fig. S3C) did not differ distinctly in WT and npc4 plants.
Fig. 8.
Relative level of gene expression of ABA-related genes in the WT, npc4-1, and npc4-2 under 100 mM NaCl treatment. The transcript levels of ABI1, ABI2, RAB18, PP2CA, and SOT12 were measured by quantitative real-time PCR in roots of non-treated plants and in plants treated with 100 mM NaCl for 3, 6, 9, and 12 h. Actin2 and UBQ10 were used as internal controls. The expression of the genes in non-treated controls at the respective times was set to 1. Data represent the means ±SE, n=2 discrete samples from one biological experiment. This experiment was repeated three times with similar results.
Discussion
The level of NPC4 transcript and enzyme activity increased rapidly in response to salt treatment
Results of quantitative RT-PCR analysis demonstrated that salt stress induced expression of genes coding for PC-PLC, NPC2, NPC4, and NPC6. Among them, NPC4 was the most highly induced gene. The expression was rapid (within 1 h), transient, and induced by a relatively low salt concentration (25 mM NaCl). A rapid reaction to low salt concentration was also observed at the level of NPC activity. The activity of NPC doubled during 30 min treatment with 100 mM NaCl, and even 10 mM NaCl treatment led to a 2-fold increase in activity within 90 min.
Enzymatic activity measurements were done using seedlings (Fig. 3) because lipid labelling can only be done with seedlings, whereas the time course of expression of NPC genes was carried out on 5-week-old plants (Fig. 2). However, in seedlings, an increase in promoter activity was found after 24 h NaCl treatment (Fig. 4) and when expression of NPC4 was checked in experimental conditions similar to those used for enzymatic activity measurements a 2.5-fold increase of NPC4 transcript was found in the WT seedlings. All these results suggest a signalling role for NPC4.
NPC4 expression response to salt stress was localized to roots
Quantitative RT-PCR and PNPC4:GUS results localize salt-induced expression of NPC4 to roots, more precisely to root tips. On the basis of previous results it was hypothesized that NPC4 expression correlates with auxin abundance (Wimalasekera et al., 2010). The present results support this assumption because the changes in the distribution of auxin under salt stress detected by Wang et al. (2009) using the DR5:GUS system and the changes observed in the localization of PNPC4:GUS showed a similar pattern. Expression of PNPC3:GUS after salt treatment was also analysed because it was shown earlier (Wimalasekera et al., 2010) that NPC3 and NPC4 expression patterns are similar. Both control plants and plants under salt stress showed no change in PNPC3:GUS expression (Supplementary Fig. S1 at JXB online). These results indicate a specific role for NPC4 in salt stress response.
Expression of SOS signalling and ABA response genes was down-regulated in NPC4 knockout plants
SOS1 together with SOS2 and SOS3 plays an important role in the initial phases of salt stress. These proteins in concert play a role in maintenance of cell sodium homeostasis in high salt conditions. Similarly to NPC4 (this work), expression of SOS1 and SOS2 under salt stress was localized to roots or root tips, respectively (Shi et al., 2000, 2002). In the npc4 mutants, the expression of SOS genes was diminished. A possible explanation can be found in the reports of SOS1 regulation by PA via mitogen-activated protein kinase 6 (MPK6). Yu et al. (2010) showed direct PA stimulation of MPK6 phosphorylation of SOS1 under salt stress where PA was derived from PC by PLDα activity. However, the role of PA originating from DAG by DGK activity is not excluded. Rapid conversion of DAG to PA has already been reported (Bargmann et al., 2009).
Expression of ABI1, ABI2, and PP2CA together with SOT12 was strongly decreased in salt-treated npc4 plants. ABI1, ABI2, and PP2CA are key players in ABA signalling, and products of these genes function as negative regulators of ABA response (Gosti et al., 1999; Merlot et al., 2001; Kuhn et al., 2006). These proteins control various ABA responses such as stomatal closure, seed dormancy, or plant growth (Leube et al., 1998). ABI1, ABI2, and PP2CA belong to the large family of Mg2+- and Mn2+-dependent serine/threonine phosphatases type 2C (PP2Cs) and they interact with the ABA receptor RCARs/PYR1/PYLs (Raghavendra et al., 2010).
The expression data presented here allow the hypothesis that NPC4 interacts with the RCARs/PP2Cs signalling pathway. This, together with rapid activation of NPC4 enzyme activity (Fig. 3), makes NPC4 a good candidate for a salt signalling protein. The signalling mechanism of NPC4 might be based on rapid conversion of DAG, the product of NPC4 activity, to PA by DGK. Similarly, it has been shown (Munnik, 2001; Meijer and Munnik, 2003; Bargmann et al., 2009) that DAG as a product of PI-PLC hydrolysis of PIP2 was rapidly phosphorylated by DGK to PA. PA is the product of PLD, and both molecules, PA and PLD, were shown to participate in ABA signalling. PLDα1 plays a role in ABA-mediated stomatal closure (Zhang et al., 2004; Mishra et al., 2006). ABI1 is a well characterized plant PA target. PA binding decreases the phosphatase activity of ABI1 and thus promotes ABA signalling (Zhang et al., 2004). The role of PA–MPK6 interaction during salt stress was discussed earlier. PA also plays an important role in ABA signalling during seed germination (Katagiri et al., 2005). However, further investigation is required to reveal whether the mode of action of NPC4 in ABA signalling is PA mediated.
RAB18 is a drought-, salt-, ABA-, sugar-, and phosphate starvation-inducible gene (Knight et al., 1997; Ciereszko and Kleczkowski, 2002). The induction of its expression requires Ca2+ influx via specific plasmalemma channels (Ghelis et al., 2000). Changes in the expression pattern of both RAB18 and ABI1 were similar in plants with overexpressed PLDα1 (Peng et al., 2010), which is in agreement with the interpretations of the present results. Hallouin et al. (2002) showed that stimulation of PLD and not PI-PLC activity is necessary for ABA-induced RAB18 expression. The present observation suggests that NPC4 may be essential for salt-induced RAB18 expression. Whether signalling steps of salt induction and ABA induction leading to RAB18 expression are the same or different is not clear.
SOT12 is a member of sulphotransferase protein family that seems to play an important role in plant growth, development, and adaptation to stress (Klein and Papenbrock, 2004). It was shown that SOT12 in Arabidopsis was highly expressed under salt and osmotic stress and that the sot12 mutant was hypersensitive to salt stress and ABA in seed germination (Kreps et al., 2002; Baek et al., 2010). Taking into account the observation that expression of SOT12 was greatly decreased in npc4 plants, it is possible to conclude that PP2C-mediated ABA signalling and SOT12 are on the identical branch of responses to salt stress. TSPO is a membrane-bound ABA-regulated protein that is proposed to amplify ABA signalling (Guillaumot et al., 2009). In contrast to the above-mentioned genes, regulation by ABA of this gene is nearly identical in magnitude and time course in the mutants and WT plants.
Conclusion
In conclusion, the NPC4 gene, the member of the novel PLC gene family hydrolysing PC, is highly and specifically expressed in roots in salt-treated Arabidopsis. NPC4 loss-of-function mutant plants revealed higher sensitivity to salt stress when compared with WT plants, which may be explained by putative partial disruption of the ABA signalling network. Expression of SOS signalling and ABA response genes in npc4 plants suggests a positive regulatory function for NPC4 in ABA salt signalling processes. Further experiments are now required to determine precisely the location of NPC4 in this network and to clarify the molecular mechanism of NPC4 function in response to salt stress.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Histochemical analysis of PNPC3:GUS expression under salt stress in Arabidopsis plants.
Figure S2. The position of T-DNA insertions in two Arabidopsis npc4 mutant lines.
Figure S3. Relative level of gene expression of ion transporter genes, phospholipid signalling-related genes, and of RD29/LTI78/COR78 in WT, npc4-1, and npc4-2 under 100 mM NaCl treatment.
Table S1. List of UPL (Universal Probe Library) probe numbers and corresponding quantitative RT-PCR primers
Table S2. List of quantitative RT-PCR primers
Acknowledgments
The authors thank Hana Fialová for her technical assistance. This work was supported by the Czech Science Foundation (grant No. 522/07/1614) and by the Ministry of Education, Youth, and Sports (grant Nos. LC 06034 and MSM 6046137305).
Glossary
Abbreviations
- ABA
abscisic acid
- ABI
ABA-insensitive
- bodipy
boron-dipyrromethene (4,4-difluoro-4-bora-3a,4a-diaza-s-indacene)
- BY-2
Bright Yellow 2
- CBL
calcineurin B-like
- CIPK24
CBL-interacting protein kinase 24
- COR78
cold-regulated 78
- DAG
diacylglycerol
- DGK
diacylglycerol kinase
- DR5
direct repeat5
- GUS
β-glucuronidase
- IP3
inositol 1,4,5-trisphosphate
- LTI78
low-temperature-induced 78
- MS
Murashige–Skoog, NHX, Na+/H+ exchanger
- NPC
non-specific phospholipase C
- PA
phosphatidic acid
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PLA
phospholipase A
- PC-PLC
phosphatidylcholine-hydrolysing phospholipase C
- PIP2
phosphatidylinositol 4,5-bisphosphate
- PI-PLC
phosphoinositide-specific phospholipase C
- PLD
phospholipase D
- PP2CA
protein phosphatase 2CA
- RAB18
responsive to ABA 18
- RD29
desiccation-responsive 29
- SIGNAL
Salk Institute Genome Analysis Laboratory
- SOS
salt overly sensitive
- SOT
sulphotransferase
- TOR
target of rapamycin
- TSPO
tryptophan-rich sensory protein
- UBQ10
ubiquitin 10
- UPL
Universal Probe Library
- VBI-0
Virginia Bright Italia 0
- WT
wild-type
- X-Gluc
5-bromo-4-chloro-3-indolyl glucuronide
References
- Agarwal PK, Jha B. Transcription factors in plants and ABA dependent and independent abiotic stress signalling. Biologia Plantarum. 2010;54:201–212. [Google Scholar]
- Alonso JM. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- An R, Chen QJ, Chai MF, Lu PL, Su Z, Qin ZX, Chen J, Wang XC. AtNHX8, a member of the monovalent cation:proton antiporter-1 family in Arabidopsis thaliana, encodes a putative Li+/H+ antiporter. The Plant Journal. 2007;49:718–728. doi: 10.1111/j.1365-313X.2006.02990.x. [DOI] [PubMed] [Google Scholar]
- Andersson MX, Larsson KE, Tjellström H, Liljenberg C, Sandelius AS. Phosphate-limited oat. The plasma membrane and the tonoplast as major targets for phospholipid-to-glycolipid replacement and stimulation of phospholipases in the plasma membrane. Journal of Biological Chemistry. 2005;280:27578–27586. doi: 10.1074/jbc.M503273200. [DOI] [PubMed] [Google Scholar]
- Baek D, Pathange P, Chung J-S, Jiang J, Gao L, Oikawa A, Hirai MY, Saito K, Pare PW, Shi H. A stress-inducible sulphotransferase sulphonates salicylic acid and confers pathogen resistance in Arabidopsis. Plant, Cell and Environment. 2010;33:1383–1392. doi: 10.1111/j.1365-3040.2010.02156.x. [DOI] [PubMed] [Google Scholar]
- Bargmann BOR, Laxalt AM, ter Riet B, van Schooten B, Merquiol E, Testerink C, Haring MA, Bartels D, Munnik T. Multiple PLDs required for high salinity and water deficit tolerance in plants. Plant and Cell Physiology. 2009;50:78–89. doi: 10.1093/pcp/pcn173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bligh EG, Dyer WJ. A rapid method of total lipid extraction and purification. Canadian Journal of Biochemistry and Physiology. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- Christmann A, Moes D, Himmelbach A, Yang Y, Tang Y, Grill E. Integration of abscisic acid signalling into plant responses. Plant Biology. 2006;8:314–325. doi: 10.1055/s-2006-924120. [DOI] [PubMed] [Google Scholar]
- Ciereszko I, Kleczkowski LA. Effects of phosphate deficiency and sugars on expression of rab18 in Arabidopsis: hexokinase-dependent and okadaic acid-sensitive transduction of the sugar signal. Biochimica et Biophysica Acta. 2002;1579:43–49. doi: 10.1016/s0167-4781(02)00502-x. [DOI] [PubMed] [Google Scholar]
- Exton JH. Phosphatidylcholine breakdown and signal transduction. Biochimica et Biophysica Acta. 1994;1212:26–42. doi: 10.1016/0005-2760(94)90186-4. [DOI] [PubMed] [Google Scholar]
- Fang YM, Vilella-Bach M, Bachmann R, Flanigan A, Chen J. Phosphatidic acid-mediated mitogenic activation of mTOR signaling. Science. 2001;294:1942–1945. doi: 10.1126/science.1066015. [DOI] [PubMed] [Google Scholar]
- Gaude N, Nakamura Y, Scheible WR, Ohta H, Dormann P. Phospholipase C5 (NPC5) is involved in galactolipid accumulation during phosphate limitation in leaves of Arabidopsis. The Plant Journal. 2008;56:28–39. doi: 10.1111/j.1365-313X.2008.03582.x. [DOI] [PubMed] [Google Scholar]
- Georges F, Das S, Ray H, Bock C, Nokhrina K, Kolla VA, Keller W. Over-expression of Brassica napus phosphatidylinositol-phospholipase C2 in canola induces significant changes in gene expression and phytohormone distribution patterns, enhances drought tolerance and promotes early flowering and maturation. Plant, Cell and Environment. 2009;32:1664–1681. doi: 10.1111/j.1365-3040.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- Ghelis T, Dellis O, Jeannette E, Bardat F, Miginiac E, Sotta B. Abscisic acid plasmalemma perception triggers a calcium influx essential for RAB18 gene expression in Arabidopsis thaliana suspension cells. FEBS Letters. 2000;483:67–70. doi: 10.1016/s0014-5793(00)02088-3. [DOI] [PubMed] [Google Scholar]
- Gong D, Guo Y, Schumaker KS, Zhu J- K. The SOS3 family of calcium sensors and SOS2 family of protein kinases in Arabidopsis. Plant Physiology. 2004;134:919–926. doi: 10.1104/pp.103.037440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti F, Beaudoin N, Serizet C, Webb AAR, Vartanian N, Giraudat J. ABI1 protein phosphatase 2C is a negative regulator of abscisic acid signaling. The Plant Cell. 1999;11:1897–1909. doi: 10.1105/tpc.11.10.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillaumot D, Guillon S, Déplanque T, Vanhee C, Gumy C, Masquelier D, Morsomme P, Batoko H. The Arabidopsis TSPO-related protein is a stress and abscisic acid-regulated, endoplasmic reticulum–Golgi-localized membrane protein. The Plant Journal. 2009;60:242–256. doi: 10.1111/j.1365-313X.2009.03950.x. [DOI] [PubMed] [Google Scholar]
- Hallouin M, Ghelis T, Brault M, Bardat F, Cornel D, Miginiac E, Rona JP, Sotta B, Jeannette E. Plasmalemma abscisic acid perception leads to RAB18 expression via phospholipase D activation in Arabidopsis suspension cells. Plant Physiology. 2002;130:265–272. doi: 10.1104/pp.004168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helling D, Possart A, Cottier S, Klahre U, Kost B. Pollen tube tip growth depends on plasma membrane polarization mediated by tobacco PLC3 activity and endocytic membrane recycling. The Plant Cell. 2006;18:3519–3534. doi: 10.1105/tpc.106.047373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI. The water-culture method for growing plants without soil. California Agricultural Experiment Station Circular. 1950;347 [Google Scholar]
- Hong Y, Pan X, Welti R, Wang X. Phospholipase Dα3 is involved in the hyperosmotic response in Arabidopsis. The Plant Cell. 2008a;20:803–816. doi: 10.1105/tpc.107.056390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong YY, Zhang WH, Wang XM. Phospholipase D and phosphatidic acid signalling in plant response to drought and salinity. Plant, Cell and Environment. 2010;33:627–635. doi: 10.1111/j.1365-3040.2009.02087.x. [DOI] [PubMed] [Google Scholar]
- Hong Y, Zheng S, Wang X. b. Dual functions of phospholipase Dα1 in plant response to drought. Molecular Plant. 2008;1:262–269. doi: 10.1093/mp/ssm025. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. Genevestigator V3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics. 2008;2008:420747. doi: 10.1155/2008/420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katagiri T, Ishiyama K, Kato T, Tabata S, Kobayashi M, Shinozaki K. An important role of phosphatidic acid in ABA signaling during germination in Arabidopsis thaliana. The Plant Journal. 2005;43:107–117. doi: 10.1111/j.1365-313X.2005.02431.x. [DOI] [PubMed] [Google Scholar]
- Klein M, Papenbrock J. The multi-protein family of Arabidopsis sulphotransferases and their relatives in other plant species. Journal of Experimental Botany. 2004;55:1809–1820. doi: 10.1093/jxb/erh183. [DOI] [PubMed] [Google Scholar]
- Knight H, Trewavas AJ, Knight MR. Calcium signalling in Arabidopsis thaliana responding to drought and salinity. The Plant Journal. 1997;12:1067–1078. doi: 10.1046/j.1365-313x.1997.12051067.x. [DOI] [PubMed] [Google Scholar]
- Kreps JA, Wu YJ, Chang HS, Zhu T, Wang X, Harper JF. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn JM, Boisson-Dernier A, Dizon MB, Maktabi MH, Schroeder JI. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in Arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiology. 2006;140:127–139. doi: 10.1104/pp.105.070318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leube MP, Grill E, Amrhein N. ABI1 of Arabidopsis is a protein serine/threonine phosphatase highly regulated by the proton and magnesium ion concentration. FEBS Letters. 1998;424:100–104. doi: 10.1016/s0014-5793(98)00149-5. [DOI] [PubMed] [Google Scholar]
- Li M, Hong Y, Wang X. Phospholipase D- and phosphatidic acid-mediated signaling in plants. Biochimica et Biophysica Acta. 2009;1791:927–935. doi: 10.1016/j.bbalip.2009.02.017. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Kim S, Delauney AJ, Verma DPS. Arabidopsis TARGET OF RAPAMYCIN interacts with RAPTOR, which regulates the activity of S6 kinase in response to osmotic stress signals. The Plant Cell. 2006;18:477–490. doi: 10.1105/tpc.105.035931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mane SP, Vasquez-Robinet C, Sioson AA, Heath LS, Grene R. Early PLDα-mediated events in response to progressive drought stress in Arabidopsis: a transcriptome analysis. Journal of Experimental Botany. 2007;58:241–252. doi: 10.1093/jxb/erl262. [DOI] [PubMed] [Google Scholar]
- Meijer HJG, Munnik T. Phospholipid-based signaling in plants. Annual Review of Plant Biology. 2003;54:265–306. doi: 10.1146/annurev.arplant.54.031902.134748. [DOI] [PubMed] [Google Scholar]
- Menand B, Desnos T, Nussaume L, Berger F, Bouchez D, Meyer C, Robaglia C. Expression and disruption of the Arabidopsis TOR (target of rapamycin) gene. Proceedings of the National Academy of Sciences, USA. 2002;99:6422–6427. doi: 10.1073/pnas.092141899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlot S, Gosti F, Guerrier D, Vavasseur A, Giraudat J. The ABI1 and ABI2 protein phosphatases 2C act in a negative feedback regulatory loop of the abscisic acid signalling pathway. The Plant Journal. 2001;25:295–303. doi: 10.1046/j.1365-313x.2001.00965.x. [DOI] [PubMed] [Google Scholar]
- Mishra G, Zhang WH, Deng F, Zhao J, Wang XM. A bifurcating pathway directs abscisic acid effects on stomatal closure and opening in Arabidopsis. Science. 2006;312:264–266. doi: 10.1126/science.1123769. [DOI] [PubMed] [Google Scholar]
- Munnik T. Phosphatidic acid: an emerging plant lipid second messenger. Trends in Plant Science. 2001;6:227–233. doi: 10.1016/s1360-1385(01)01918-5. [DOI] [PubMed] [Google Scholar]
- Munnik T, editor. Lipid signaling in plants. Berlin: Springer; 2010. [Google Scholar]
- Munnik T, Vermeer JEM. Osmotic stress-induced phosphoinositide and inositol phosphate signalling in plants. Plant, Cell and Environment. 2010;33:655–669. doi: 10.1111/j.1365-3040.2009.02097.x. [DOI] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Awai K, Masuda T, Yoshioka Y, Takamiya K, Ohta H. A novel phosphatidylcholine-hydrolyzing phospholipase C induced by phosphate starvation in Arabidopsis. Journal of Biological Chemistry. 2005;280:7469–7476. doi: 10.1074/jbc.M408799200. [DOI] [PubMed] [Google Scholar]
- Pardo JM, Cubero B, Leidi EO, Quintero FJ. Alkali cation exchangers: roles in cellular homeostasis and stress tolerance. Journal of Experimental Botany. 2006;57:1181–1199. doi: 10.1093/jxb/erj114. [DOI] [PubMed] [Google Scholar]
- Pejchar P, Potocký M, Novotná Z, Veselková Š, Kocourková D, Valentová O, Schwarzerová K, Martinec J. Aluminium ions inhibit the formation of diacylglycerol generated by phosphatidylcholine-hydrolysing phospholipase C in tobacco cells. New Phytologist. 2010;188:150–160. doi: 10.1111/j.1469-8137.2010.03349.x. [DOI] [PubMed] [Google Scholar]
- Peng Y, Zhang J, Cao G, Xie Y, Liu X, Lu M, Wang G. Overexpression of a PLDα1 gene from Setaria italica enhances the sensitivity of Arabidopsis to abscisic acid and improves its drought tolerance. Plant Cell Reports. 2010;29:793–802. doi: 10.1007/s00299-010-0865-1. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Research. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Q-S, Guo Y, Dietrich MA, Schumaker KS, Zhu J- K. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proceedings of the National Academy of Sciences, USA. 2002;99:8436–8441. doi: 10.1073/pnas.122224699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavendra AS, Gonugunta VK, Christmann A, Grill E. ABA perception and signalling. Trends in Plant Science. 2010;15:395–401. doi: 10.1016/j.tplants.2010.04.006. [DOI] [PubMed] [Google Scholar]
- Reddy VS, Rao DKV, Rajasekharan R. Functional characterization of lysophosphatidic acid phosphatase from Arabidopsis thaliana. Biochimica et Biophysica Acta. 2010;1801:455–461. doi: 10.1016/j.bbalip.2009.12.005. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Rosales MP, Gálvez FJ, Huertas R, Aranda MN, Baghour M, Cagnac O, Venema K. Plant NHX cation/proton antiporters. Plant Signaling and Behavior. 2009;4:265–276. doi: 10.4161/psb.4.4.7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Scherer GFE, Paul RU, Holk A, Martinec J. Down-regulation by elicitors of phosphatidylcholine-hydrolyzing phospholipase C and up-regulation of phospholipase A in plant cells. Biochemical and Biophysical Research Communications. 2002;293:766–770. doi: 10.1016/S0006-291X(02)00292-9. [DOI] [PubMed] [Google Scholar]
- Shi HZ, Ishitani M, Kim CS, Zhu JK. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proceedings of the National Academy of Sciences, USA. 2000;97:6896–6901. doi: 10.1073/pnas.120170197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Quintero FJ, Pardo JM, Zhu J- K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. The Plant Cell. 2002;14:465–477. doi: 10.1105/tpc.010371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titball RW. Bacterial phospholipases C. Microbiological Reviews. 1993;57:347–366. doi: 10.1128/mr.57.2.347-366.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjellström H, Andersson MX, Larsson KL, Sandelius AS. Membrane phospholipids as a phosphate reserve: the dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant, Cell and Environment. 2008;31:1388–1398. doi: 10.1111/j.1365-3040.2008.01851.x. [DOI] [PubMed] [Google Scholar]
- Wang CR, Yang AF, Yue GD, Gao Q, Yin HY, Zhang JR. Enhanced expression of phospholipase C 1 (ZmPLC1) improves drought tolerance in transgenic maize. Planta. 2008;227:1127–1140. doi: 10.1007/s00425-007-0686-9. [DOI] [PubMed] [Google Scholar]
- Wang YN, Li KX, Li X. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. Journal of Plant Physiology. 2009;166:1637–1645. doi: 10.1016/j.jplph.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Wimalasekera R, Pejchar P, Holk A, Martinec J, Scherer GFE. Plant phosphatidylcholine-hydrolyzing phospholipases C NPC3 and NPC4 with roles in root development and brassinolide signalling in Arabidopsis thaliana. Molecular Plant. 2010;3:610–625. doi: 10.1093/mp/ssq005. [DOI] [PubMed] [Google Scholar]
- Xue HW, Chen X, Mei Y. Function and regulation of phospholipid signalling in plants. Biochemical Journal. 2009;421:145–156. doi: 10.1042/BJ20090300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Nie J, Cao C, Jin Y, Yan M, Wang F, Liu J, Xiao Y, Liang Y, Zhang W. Phosphatidic acid mediates salt stress response by regulation of MPK6 in Arabidopsis thaliana. New Phytologist. 2010;188:762–773. doi: 10.1111/j.1469-8137.2010.03422.x. [DOI] [PubMed] [Google Scholar]
- Zhang WH, Qin CB, Zhao J, Wang XM. Phospholipase Dα1-derived phosphatidic acid interacts with ABI1 phosphatase 2C and regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, USA. 2004;101:9508–9513. doi: 10.1073/pnas.0402112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. Regulation of ion homeostasis under salt stress. Current Opinion in Plant Biology. 2003;6:441–445. doi: 10.1016/s1369-5266(03)00085-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.