Abstract
MicroRNAs (miRNAs) are small, non-coding RNAs that play essential roles in plant growth, development, and stress response. Populus euphratica is a typical abiotic stress-resistant woody species. This study presents an efficient method for genome-wide discovery of new drought stress responsive miRNAs in P. euphratica. High-throughput sequencing of P. euphratica leaves found 197 conserved miRNAs between P. euphratica and Populus trichocarpa. Meanwhile, 58 new miRNAs belonging to 38 families were identified, an increase in the number of P. euphratica miRNAs. Twenty-six new and 21 conserved miRNA targets were verified by degradome sequencing, and target annotation showed that these targets were involved in multiple biological processes, including transcriptional regulation and response to stimulus. Furthermore, comparison of high-throughput sequencing with miRNA microarray profiling data indicated that 104 miRNA sequences were up-regulated, whereas 27 were down-regulated under drought stress. This preliminary characterization provides a framework for future analysis of miRNA genes and their roles in key poplar traits such as stress resistance, and could be useful for plant breeding and environmental protection
Keywords: Drought, miRNA, mirtron, Populus euphratica
Introduction
Forests play an important role in fixing carbon and protecting the environment; however, most fast-growing tree species, such as poplars (Populus spp.), require large amounts of water for development. Thus, enhancing water use efficiency (WUE) and drought resistance of such trees is pressing and challenging work. Populus euphratica is the only arboreal species that can be established in the world's largest shifting-sand desert, the Taklimakan Desert, which is characterized by a wide temperature range as well as salinity, aridity, and especially drought stress (Gries, 2003). Thus, P. euphratica is widely considered an ideal model system for researching into abiotic stress resistance of woody plants (Ottow et al., 2005). Studies on this species will further understanding of the resistance mechanisms of woody plants to drought stress and provide the possibility of increasing plant WUE.
MicroRNAs (miRNAs) are endogenous non-coding small RNAs (sRNAs), typically ∼21 nucleotides (nt) in length, playing negative regulatory functions at post-transcription level by repressing gene translation or degrading target mRNAs. In plants, after transcription by Pol II or Pol III enzyme into primary miRNA (pri-miRNA), the miRNA gene is processed by Dicer-like (DCL) into a stem–loop miRNA::miRNA* duplex (Kurihara and Watanabe, 2004), called an miRNA precursor (pre-miRNA). After that, the miRNA::miRNA* duplex is cleaved from the pre-miRNA and transported from the nucleus into the cytoplasm (Bartel, 2004). This miRNA::miRNA* duplex then joins with Argonaute (AGO) forming the RNA-induced silencing complex (RISC) (Baumberger and Baulcombe, 2005). Finally, the silencing complex down-regulates targets by either cleaving target mRNAs or repressing the translation process (Bartel, 2004). Increasing amounts of published research has shown that miRNAs are involved in multiple crucial developmental and metabolic pathways in plants, including development-phase change (Aukerman and Sakai, 2003), signal transduction (Jones-Rhoades et al., 2006), mechanical stress responses (Lu et al., 2005, 2008), cold, and dehydration stress responses (Jones-Rhoades and Bartel, 2004; Li et al., 2009).
To identify miRNAs that are responsive to drought stress and high WUE, P. euphratica plants were exposed to four levels of relative soil moisture content (RSMC). Leaves of samples at 35–40% and 70–75% RSMC were used for high-throughput sequencing experiments. The sequencing data showed 58 new P. euphratica miRNAs belonging to 38 families and 197 conserved P. trichocarpa miRNAs. Meanwhile, a putative mirtron was also identified along with 14 miRNA*s of new P. euphratica miRNA and 127 miRNA*s of conserved Populus trichocarpa miRNAs. Furthermore, all the known plant miRNA sequences and new P. euphratica miRNAs were used as probes for miRNA microarray analysis. Comparison between high-throughput sequencing and microarray results indicated that the expression of 104 up- and 27 down-regulated miRNAs was consistent in these two experiments under drought stress. The method of combining high-throughput sequencing and microarray technologies allowed the successful discovery of new and stress responsive miRNAs and will serve as a basis for future comparative functional genomic analyses using syntenic orthologues.
Materials and methods
Plant materials and total RNA extraction
One-year-old seedlings of P. euphratica, obtained from the Xinjiang Uygur Autonomous Region of China, were planted in individual pots (15 l) containing loam soil and placed in a greenhouse at Beijing Forestry University. Each pot contained three individuals. Potted plants were well irrigated according to evaporation demand and watered with 1 l of full-strength Hoagland nutrient solution every 2 weeks for 2 months before drought stress treatment. The temperature in the greenhouse was 20–25 °C with a 16-h photoperiod (04:00am–08:00pm). In the drought stress treatment, P. euphratica plants were submitted to soil water deficiency at four RSMC levels for 2 months according to previous research (Hasio, 1973). They were Group A with RSMC 70–75%; Group B with RSMC 50–55%; Group C with RSMC 35–40%; and Group D with RSMC 15–20%. Soil with sufficient irrigation every day kept RSMC at 70–75% because of transpiration, so Group A was used as the control sample. Leaf water potential (WP) was measured by PsyPro WP data logger (Wescor). Net photosynthetic rate and transpiration rate were measured by Li-6400 Photosynthesis System (Li-Cor). All data were statistically analysed by one-way ANOVA using SPSS (SPSS statistical package 10.1; SPSS, Chicago, IL, USA). For material harvest, mature leaves from the same position on eight different plants in each treatment were mixed and ground immediately in liquid nitrogen. Total RNA was extracted from mixed leave tissues by the standard CTAB method for plants (Chang et al., 1993). Then these total RNAs were used for high-throughput sequencing and microarray profiling.
P. euphratica high-throughput sequencing and miRNA identification
The high-throughput sequencing followed the Illumina protocol based on a small RNA kit and Illumina Genome Analyzer. Sequencing reads were first aligned against the P. trichocarpa genome (JGI P. trichocarpa genome V 1.1) by SOAP software (Li et al., 2008). Sequences with a perfect match were retained for further analysis. By comparing the existing sRNA database, all sRNAs were annotated in the order of the following categories: (i) rRNAetc: rRNA, tRNA, snRNA, scRNA, and snoRNA deposited at GenBank (http://www.ncbi.nih.gov/GenBank/) and Rfam (http://rfam.sanger.ac.uk/) databases. In this category, RNA sequences based on structural conservation were also considered. GtRNAdb (http://lowelab.ucsc.edu/GtRNAdb/Ptric/popTri2-tRNAs.fa), a high-confidence level Populus tRNA database predicted by tRNAscan based on structure (Schattner et al., 2005), was also used to exclude tRNA. (ii) Known miRNA: previously discovered miRNAs in miRBase13.0; (iii) exon_sense/antisense: genomic exon sequences in sense/antisense direction; (iv) intron_sense/antisense: genomic intron sequences in sense/antisense direction. Both of the exon_sense/antisense and intron_sense/antisense were classified by the Populus genome data from P. trichocarpa genome V 1.1 (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html). (v) Unknown sRNA.
To further analyse the RNA secondary structure comprising genome-matched sequencing reads, 100 nt of the genomic sequences flanking each side of these sequences were extracted, the secondary structures were predicted using RNAfold (http://www.tbi.univie.ac.at/%7Eivo/RNA/RNAfold.html), and analysed by Mireap (http://sourceforge.net/projects/mireap/). Mireap is software that can be used to identify miRNAs from sRNA high-throughput sequencing data. The consideration of sequencing read abundance, pre-miRNA hairpin energy, and the secondary structure of the miRNA::miRNA* complex confirmed Mireap as reliable software for discovering new miRNAs. In this work, Mireap parameters were adjusted to meet the demands of plant miRNA identification as follows: (i) the length range of the miRNA sequence should be 20–23 bp; (ii) the maximal free energy allowed for an miRNA precursor should be –18 kcal/mol; (iii) the minimal common base pairs between miRNA and miRNA* should be 16, with no more than four bulges; and (iv) the maximal asymmetry of miRNA::miRNA* duplex should be four bases.
P. euphratica degradome sequencing
New P. euphratica miRNA targets were predicted as described (Edwards et al., 2005). Predicted targets of conserved miRNA for P. trichocarpa and P. euphratica were already available at the PopGenIE ftp site (ftp://aspnas.fysbot.umu.se/v1_archive/miRNA/). Both conserved and new miRNA targets were experimentally verified by P. euphratica mRNA degradome sequencing following the previously published Parallel Analysis of RNA Ends (PARE) protocol (German et al., 2009). The leaf total RNA from the control sample that was used for degradome sequencing library construction was also used for miRNA target identification. Illumina Genome Analysis II data of PARE were then analysed by the CleaveLand pipeline (Addo-Quaye et al., 2009), using P. trichocarpa annotated transcripts of Jamboree gene model v1.1.
MiRNA microarrays
Microarray assays were performed using a service provider (LC Sciences). Total RNAs extracted from pooled samples of four drought treatment levels were used. This experiment was based on technical replicate, which was carried out using three replicates for every miRNA probe in each chip. Group B and Group C were profiled in the same chip, while Group A along with Group D was in another. The assay started using 2–5 μg of total RNA, which was size fractionated using a YM-100 Microcon centrifugal filter (Millipore) and the sRNAs (<300 nt) isolated were 3' extended with a poly(A) tail using poly(A) polymerase. An oligonucleotide tag was then ligated to the poly(A) tail for later fluorescent dye staining; two different tags were used for the two RNA samples in dual-sample experiments. Hybridization was performed overnight on a μParaflo™ microfluidic chip using a micro-circulation pump (Atactic Technologies). On the microfluidic chip, each detection probe consisted of a chemically modified nucleotide coding segment complementary to the target miRNA (from miRBase 13.0, http://microrna.sanger.ac.uk/sequences/) or newly identified P. euphratica miRNA or candidates and a spacer segment of polyethylene glycol to extend the coding segment away from the substrate. The hybridization melting temperatures were balanced by chemical modifications of the detection probes. Hybridization used 100 μl of 6×SSPE buffer (0.90 M NaCl, 60 mM Na2HPO4, and 6 mM EDTA at pH 6.8) containing 25% formamide at 34 °C. After hybridization, detection used fluorescence labelling and tag-specific Cy3 and Cy5 dyes. Hybridization images were collected using a laser scanner (GenePix 4000B; Molecular Device) and digitized using Array-Pro image analysis software (Media Cybernetics).
High-throughput sequencing abundance profile analysis
The high-throughput sequencing abundance profile analysis was based on the sequence reads of each library for the drought treatment and control. The first step was to normalize the miRNA sequence reads in the drought treatment and control to tags per million. The calculation of the P-value for comparing miRNA expression between the two libraries was based on previously established methods (Audic and Claverie, 1997; Man et al., 2000). Specifically, the log2 ratio formula was: log2 ratio=log2 (miRNA reads in drought treatment/miRNA reads in control).
P-value formulas were:
 |
where N1 is the total number of reads in the sequencing library of control, N2 is the total number of reads in the sequencing library of drought treatment, x is the number of reads for an miRNA in the control library, y is the number of reads for an miRNA in the drought treatment library.
All calculations were performed on a BGI Bio-Cloud Computing platform (http://cloud.genomics.org.cn). MiRNA tags per million of <1 were filtered in both libraries.
MiRNA microarray abundance profile analyses
Hierarchical clustering of miRNA expression patterns involved normalization, data adjustment, t-test, and clustering. Normalization was carried out using a cyclic LOWESS (locally weighted regression) method (Bolstad et al., 2003). The normalization was to remove system-related variations, such as sample amount variations, different labelling dyes, and signal gain differences of scanners so that biological variations can be faithfully revealed.
Data adjustment included data filtering, log2 transformation, and gene centring and normalization. The data filtering removed genes (or miRNAs) with (normalized) intensity values below a threshold value of 32 across all samples. The log2 transformation converted intensity values into log2 scale. Gene centring and normalization transform the log2 value using the mean and the standard deviation of individual genes across all samples.
A t-test was performed between ‘control’ and ‘drought’ sample groups (Pan, 2002). t-values were calculated for each miRNA, and P-values were computed from the theoretical t-distribution. Only miRNAs with P<0.01 were selected for cluster analysis. Hirearchical clustering was performed based on average linkage Euclidean distance metrics (Eisen et al., 1998).
All data processes, except the clustering plot, were carried out using computer programs developed in house. The clustering plot was generated using TIGR MeV (http://www.tm4.org/) software from the Institute for Genomic Research.
Accession number
Sequencing data obtained in this work have been submitted to the Gene Expression Omnibusunder the accession number GSE25747.
Results
P. euphratica under soil water deficiency
P. euphratica plants were submitted to soil water deficiency at four RSMC levels according to previous research (Hasio, 1973). They were Group A with RSMC 70–75%, Group B with RSMC 50–55%, Group C with RSMC 35–40%, and Group D with RSMC 15–20%. The leaf WP was first detected as a measure of the ability of plants to absorb water. There was an obvious decrease (P<0.01) in leaf WP from –1.12 to –2.98 MPa between Group B and Group C (Fig. 1A), suggesting a significant gene expression change between these two groups. Although these two groups showed leaf net photosynthetic rates of 9.01 and 8.55 μmol CO2 m−2 s−1 at the same difference level (P<0.01), the transpiration rate between Group B and Group C was significantly reduced from 4.85 to 2.83 mmol H2O m−2 s−1 (Fig. 1B, C). Lastly, the WUE was calculated, which equals photosynthetic rate divided by transpiration rate, to indicate the prospect of drought-resistant breeding and woody plant productivity with limited water (Fig. 1D). WUE was significantly reduced from 1.86 to 3.02 μmol CO2 per mmol H2O between Group B and C. Finally, Group C was chosen for sRNA high-throughput sequencing; it was significantly lower in leaf WP and significantly higher in WUE than Group B. Leaves of Group A (RSMC 70–75%) were used as control samples because soil with sufficient irrigation every day could keep the RSMC at 70–75% because of transpiration. All differentiation was observed at a significance level of P<0.01.
Fig. 1.
P. euphratica under soil water deficiency. Leaf WP (A), leaf net photosynthetic rates (B), transpiration rate (C), and WUE (D) of P. euphratica under soil water deficiency at four RSMC levels. The values with different capital letters and ** were significantly different at the P<0.01 level.
Overview of P. euphratica RNA high-throughput sequencing
P. euphratica RNA high-throughput sequencing acquired 7,035,135 sequences in Group C and 8,186,600 sequences in the control, sRNAs 20–22 nt in length accounted for 54.77% and 78.71% of these, respectively, representing the major components of sRNA (Fig. 2A). By comparing the sRNA databases at GenBank (http://www.ncbi.nih.gov/Genbank/), Rfam (http://rfam.sanger.ac.uk/), miRBase 13.0 (http://microrna.sanger.ac.uk/sequences), and poplar genome (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html), high-throughput sequencing data were annotated and classified into seven categories: rRNAetc (rRNA, tRNA, snRNA, scRNA, and snoRNA deposited in the GenBank and Rfam databases), known miRNA (P. trichocarpa miRNAs in miRBase 13.0), exon_sense, exon_antisense, intron_sense, intron_antisense, and unknown sRNAs (Fig. 2B, C). Results showed that the proportion of rRNAetc was obviously increased from 11.32% to 35.8% under drought stress, and so were exon sequences from 5.39% to 12.29%, suggesting the expression of many functional genes. The unknown sRNAs also increased from 6.98% to 13.34%, implying that unknown drought responsive sRNAs remain to be discovered, such as miRNA, siRNA, or piRNA. Although the sRNAs of two samples were sequenced to the same depth, the distribution of the reads showed that drought stress significantly reduced the percentage of overall miRNA counts from 75.67% to 37.79% between control and drought-stressed plants (Fig. 2B).
Fig. 2.
P. euphratica high-throughput sequencing overview. The length distribution of the P. euphratica sRNA high-throughput sequencing. (A) Distribution of different sRNA annotation categories. (B) Distribution of different sRNA annotation categories. (C) Distribution of unique (remove redundancies) sRNAs reads. For the purpose of each sRNA has unique annotation, all sRNAs were annotated in the order of rRNAetc, known as miRNA, exon_sense, exon_antisense, intron sense, intron_antisense, and unknown sRNAs (see Materials and methods).
Generally, the reads of different sRNA categories varied significantly between two leaf samples as analysed above, while after removing redundant sequence reads, percentages of unique sequence reads in the same categories were comparatively consistent in these two sequenced samples, with the greatest differentiation being 2.71% between the category exon_sense in two samples (Fig. 2C). It was proposed that the distribution of the total reads of different sRNA categories could represent their expression situations, while distribution of the unique sequence reads could represent the proportions of sRNA categories in the P. euphratica genome. This result also indicated that high-throughput sequencing generated sufficient data that the sequencing depth was sufficient to cover most of the sRNA sequences expressed from the P. euphratica genome.
Conserved miRNAs between P. euphratica and P. trichocarpa
To detect conserved miRNAs between P. euphratica and P. trichocarpa, all the high-throughput sequencing data that could map completely to the P. trichocarpa genome (http://genome.jgi-psf.org/Poptr1_1/Poptr1_1.home.html) were aligned with the known P. trichocarpa pre-miRNA sequences in the miRBase 13.0 (http://www.mirbase.org/) (Griffiths-Jones et al., 2008). It was found that of 234 previously known P. trichocarpa miRNAs, 197 were expressed in at least one of the P. euphratica sRNA libraries, corresponding to 193 and 177 miRNA genes in control and Group C, respectively (Table 1, Supplementary Table S1, available at JXB online). These results showed that under drought stress P. euphratica employed most of the known P. trichocarpa miRNAs. Furthermore, 127 miRNA* sequences of the 197 conserved miRNA genes in P. euphratica were identified, a larger increase in number than previously reported. The detection of miRNA* from the opposite arm of the miRNA in the pre-miRNA sequences illustrated the sensitivity of the high-throughput sequencing approach in finding miRNA. In the classical plant miRNA generation pathway, an miRNA::miRNA* complex is generated from the stem of the pre-miRNA hairpin structure by an RNase III-like enzyme (DCL), as previously published (Bartel, 2004). Comparing with a single RNA strand, this double-stranded RNA complex is generally short lived. In most cases, miRNA* is more easily degraded when exposed in the nucleus, while miRNA is protected for combining with the RISC (Baumberger and Baulcombe, 2005). The choice of whether miRNA or miRNA* loads to the RISC seems to be determined by their stability and whether their 5' ends are less tightly paired (Bartel, 2004). Typically, miRNA*s from the opposing arm in the cloned miRNA libraries are found at much lower frequency than miRNAs, while high-throughput sequencing technology can detect a large number of them.
Table 1.
Number of miRNAs identified in P. euphratica by high-throughput sequencing
| Sample | Categories | miRNA | Difference | miRNA* | miRNA*≥miRNA |
| Control | Conserved miRNAs | 193 | 76 | 114 | 33 |
| New miRNA candidates | 112 | – | 15 | – | |
| Group C | Conserved miRNAs | 177 | 71 | 115 | 40 |
| New miRNA candidates | 109 | – | 11 | – | |
| Control and Group C | Conserved miRNAs | 173 | 31 | 102 | 24 |
| New miRNAs | 46 | – | 6 | – | |
| Control or Group C | Conserved miRNAs | 197 | 99 | 127 | 42 |
| New miRNAs | 58 | – | 14 | – |
‘Conserved miRNAs’, conserved miRNAs between P. euphratica and P. trichocarpa; ‘New miRNAs’, new miRNAs in P. euphratica; ‘Control and Group C’, number of sequences identified in both control and Group C leaf libraries; ‘Control or Group C’, number of sequences identified in control or Group C libraries; ‘Difference’, the most sequenced sRNA in the pri-miRNA was not the mature miRNA registered in miRBase 13.0; ‘miRNA*≥miRNA’, the number of sequenced miRNA* reads larger than or equal to miRBase 13.0 registered mature miRNA.
If miRNA and miRNA* have equivalent thermodynamic stabilities and tightly paired 5' ends, sequencing counts will show similar frequencies of miRNA and miRNA*. This means that both strands of the miRNA or miRNA* enter the RISC with equal probability and may have biological functions. Thus degradation of miRNA* is less probable than of typical miRNA genes. A few of this kind of miRNA genes have been predicted and validated in vertebrates and insects but few have been found in plants (Lagos-Quintana et al., 2002; Schwarz et al., 2003; Zhu et al., 2008). However, nine miRNAs (ptc-miR160f, ptc-miR169b, ptc-miR169l, ptc-miR171h, ptc-miR171m, ptc-miR172h, ptc-miR393a, ptc-miR393b, and ptc-miR403c) of the 197 conserved miRNAs in both of the two libraries have been identified reported here that demonstrated a nearly equal number of sequence reads from both arms of the stem–loop precursor (Supplementary Table S1 at JXB online). This suggested that either end of their pre-miRNA sequences could generate mature miRNA. It was also found 24 miRNA genes that had more sequence reads in the opposite arm than the miRNA13.0 annotated miRNA in at least one of the sRNA libraries. Moreover, 15 of these miRNAs showed a reversal in the ratios of the 5'- and 3'-derived sequence reads across the two RNA libraries (Supplementary Table S1 at JXB online). These results exhibited the alternative use of the pre-miRNA 5' and 3' arms as well as the complexity of the mature miRNA-generating process. Heterogeneity at the 5' or 3' end indicated a bias towards using different arms of the pre-miRNA between P. euphratica and P. trichocarpa.
Meanwhile, 76 and 71 conserved miRNAs in control and Group C, respectively, 31 miRNA in both libraries, had more reads of other sequences in their stem–loop than the miRNA sequence registered in miRBase 13.0; these read count dominant sequences were usually one or two nucleotides away from their registered miRNA sequences (Table 1, Supplementary Table S1 at JXB online). A similar phenomenon was discovered in many previous studies in animals and rice (Landgraf et al., 2007; Morin et al., 2008; Zhu et al., 2008). The most frequently sequenced miRNA isoforms could be utilized to refine miRBase annotations of poplar miRNAs.
Analysis of all conserved miRNA sequence reads showed that the number varied significantly, from hundreds of thousands for the most abundant miRNAs to zero for the 37 previously discovered P. trichocarpa miRNAs that were not detected. The six most abundant ones (ptc-miR156h, ptc-miR156g, ptc-miR156i, ptc-miR156j, ptc-miR156c, ptc-miR156a) represented ∼90% of the total conserved miRNA reads in both the libraries (Supplementary Table S1 at JXB online). Similar phenomena have been observed in studies of chickens (Glazov et al., 2008).
New P. euphratica miRNAs
Besides discovering conserved miRNAs, the high-throughput sequencing data also provided possibilities of finding new miRNA genes. Mireap software (Kwak et al., 2009) was used to identify new P. euphratica miRNAs with adjusted parameters, which were a better fit for plant miRNA identification (Kwak et al., 2009). In total, 142 unique sequences were identified as potential novel poplar miRNAs or miRNA*s. They were classified into 120 families, including 189 candidate miRNAs (189 genomic loci) that appeared in at least one of the libraries (Table 1, Supplementary Table S2 at JXB online).
A recently published article proposed precise and strict new miRNA annotation criteria (Meyers et al., 2008). Besides the primary criteria used by Mireap, two elementary requirements are demanded in high-throughput sequencing data analysis: (i) high-throughput sequencing data should represent both the miRNA and miRNA*; and (ii) in miRNA*-deficient cases, isolation and sequencing of the candidate miRNA should come from multiple and independent libraries. Among the 189 new miRNA candidates, 58 miRNAs from 38 families were categorized as highly confident according to these precise criteria and named as new P. euphratica miRNAs (Supplementary Table S2 at JXB online). The remaining 131 miRNA candidates remain designated as potential P. euphratica miRNAs and could provide the reference for further miRNA identification. Thus these 131 miRNA candidates together with the 58 new miRNAs were processed by the additional miRNA expression and miRNA target analysis reported here.
Similar to the discovery of miRNA* of the conserved miRNA above, 14 miRNA*s of the 58 P. euphratica miRNAs were found in at least one of the sRNA libraries; only six (peu-miR6*, peu-miR50*, peu-miR102*, peu-miR106*, peu-miR129*, and peu-miR71*) of the 14 miRNA*s appeared in both drought-stressed and control sRNA libraries. Moreover, it was found that sequences of LG_V:16747064:16747196:+ and LG_V:16751220:16751352:+ in the Populus genome (http://genome.jgi-psf.org/poplar) can generate miRNA sequences at different ends (5' or 3') under different living conditions; peu-miR30bb and peu-miR30bc at the 3' arm under normal conditions (control), and peu-miR71b and peu-miR71c at the 5' arm under drought stress (Group C). Because there were other miRNA family members in both the peu-miR30 and peu-miR70 families, they were not mutually named as miRNA and miRNA*. This observation demonstrated the alternative choice of the pre-miRNA arms in expressing mature miRNA sequences in different growing conditions.
Sequence comparisons between new P. euphratica miRNA or miRNA candidates and other plant miRNA hairpin sequences registered in miRBase 13.0 revealed that nine (peu-miR101a, peu-miR101b, peu-miR106*, peu-miR131, peu-miR132, peu-miR28, peu-miR32, peu-miR49, and peu-miR50*) sequences were orthologues of miRNAs identified in other plant species (with two base pair mismatches). Comparison with previous research on new miRNA identification in Populus balsamifera (Abdelali, 2007) has shown that peu-miR102 and peu-miR129 were conserved in this species. To investigate evolutionary conservation of the 142 new P. euphratica miRNA or miRNA candidate sequences, highly similar DNA sequences in the genome assemblies of Arabidopsis thaliana, Brachypodium distachyon, Carica papaya, Glycine max, Lotus japonicus, Manihot esculenta, Medicago truncatula, Oryza sativa, Solanum lycopersicum, Sorghum bicolor, Vitis vinifera, and Zea mays were sought. It was found that 22 of the 142 sequences were conserved (perfect match) in at least one of these genomes (Supplementary Table S2 at JXB online).
The nucleotide bias at each position of the 142 newly identified miRNA or miRNA candidates (Supplementary Fig. S1 at JXB online) showed that the first nucleotide of the new miRNA genes tended to be (U) in general. As expected, miRNAs are loaded to the RISC assisted by AGO. Research has shown that AGO proteins have more affinities with uracil in the 5' terminus of miRNA, thus resulting in cloned miRNA sequences with uracil nucleotide bias in the first position (Mi et al., 2008). The newly identified miRNAs in P. euphratica also followed this trend.
Degradome sequencing analysis of new and conserved miRNA targets in P. euphratica
New P. euphratica miRNA targets were predicted as described before (Edwards et al., 2005). In total, the 58 new P. euphratica miRNA sequences were predicted to match 129 targets (Supplementary Table S3 at JXB online). The targets of conserved miRNAs from P. trichocarpa are already predicted and available at the PopGenIE ftp site. As for experimentally verifying the predicted miRNA targets, a PARE was performed for the P. euphratica mRNA degradome sequencing following a previously published method (German et al., 2009). Specifically, after extracting the 3'- polyadenylated mRNA from total P. euphratica RNA, only miRNA-cleaved mRNA and other degraded mRNA could be ligated by a 5' RNA adapter because the 5'- phosphate and intact mRNAs were 5' protected by the 5' cap. Accordingly, the 5'-adapter-ligated RNA could be used for high-throughput sequencing library construction. The PARE acquired 18,980,835 20-nt sequences in total and 1,055,227 sequences after removing redundancy; 11,105,167 reads (435,488 distinct) could be matched to the P. trichocarpa genome V 1.1 without mismatch (Supplementary Table S4 at JXB online). The degradome sequencing data were further analysed by the CleaveLand pipeline (Addo-Quaye et al., 2009). The cleaved target transcripts were categorized into three classes as was previously reported for Arabidopsis (Addo-Quaye et al., 2009).
In total, 26 new P. euphratica miRNA pairs and 22 conserved miRNAs and their target genes were qualified by PARE (Table 2; Supplementary Table S5 at JXB online). Among the 26 new miRNA pairs and their targets (Fig. 3; Supplementary Fig. S2 at JXB online), 13 belonged to category I, meaning that the expected cleavage site had the most abundant sequencing reads in the target mRNA. Furthermore, four of the category I miRNA and target pairs had >60% of the cleavage at the expected site. With respect to the PARE-qualified conserved miRNA targets, the 21 pairs of conserved miRNAs and their targets included 11 miRNAs and 15 genes. Eleven of the 21 pairs belonged to category I and five pairs had >60% of the cleavage at the expected site (Supplementary Fig. S3; Supplementary Table S5 at JXB online). All of these results indicated that both P. euphratica sRNA and degradome sequencing generated reliable results in new miRNA identification and miRNA target verification. The annotation of the PARE-verified targets was found to be diverse and included not only transcription factors but also signal transduction factors and other proteins involved in various biological processes (Table 2; Supplemenary Table S4 at JXB online).
Table 2.
Targets of new P. euphratica miRNA verified by degradome sequencing
| miRNA | Target gene | Category | Cleavage site | Percentage of cleavage at the expected site (%) | Reads at cleavage site (tpb) | Position penalty score | MFE ratio (%) | Target annotation |
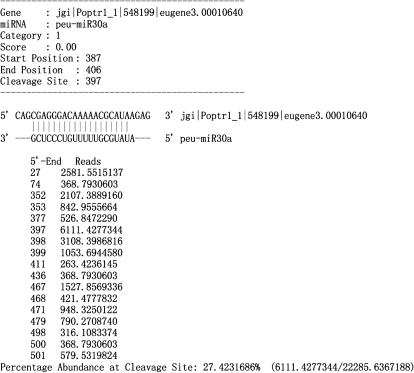
| peu-miR30a | eugene3.00010640 | 1 | 397 | 27.42 | 6111.43 | 1 | 97.92 | Electron carrier activity |
| peu-miR30a | eugene3.105640001 | 1 | 340 | 15.89 | 553.19 | 1 | 97.92 | Electron carrier activity |
| peu-miR30a | fgenesh4_pg.C_scaffold_263000013 | 1 | 310 | 15.89 | 553.19 | 1 | 97.92 | Electron carrier activity |
| peu-miR30b | eugene3.00010640 | 2 | 399 | 4.73 | 1053.69 | 0.5 | 97.33 | Electron carrier activity |
| peu-miR71* | eugene3.00010640 | 1 | 398 | 13.95 | 3108.4 | 1 | 86.89 | Electron carrier activity |
| peu-miR71* | grail3.0008024501 | 3 | 230 | 2.12 | 1527.9 | 3 | 97.92 | Electron carrier activity |
| peu-miR71* | eugene3.105640001 | 1 | 341 | 21.94 | 763.93 | 2 | 86.89 | Electron carrier activity |
| peu-miR71* | fgenesh4_pg.C_scaffold_263000013 | 1 | 311 | 21.94 | 763.93 | 2 | 86.89 | Electron carrier activity |
| peu-miR77 | eugene3.00002056 | 2 | 1053 | 8.43 | 553.19 | 4 | 71.54 | Electron carrier activity |
| peu-miR77 | estExt_Genewise1_v1.C_LG_XIV3469 | 3 | 1275 | 8.43 | 553.19 | 4 | 71.54 | Electron carrier activity |
| peu-miR84* | fgenesh4_pm.C_LG_XIII000061 | 3 | 344 | 27.27 | 474.16 | 4.5 | 65.09 | Electron carrier activity |
| peu-miR101a | gw1.I.9350.1 | 1 | 298 | 68.11 | 3319.14 | 1 | 87.26 | Transcription factor |
| peu-miR131 | eugene3.00120942 | 1 | 1088 | 69.44 | 658.56 | 5 | 76.15 | Electron carrier activity |
| peu-miR131 | fgenesh4_pg.C_LG_X001404 | 1 | 1186 | 60.00 | 316.11 | 3.5 | 83.46 | DNA binding |
| peu-miR131 | estExt_Genewise1_v1.C_LG_XV2187 | 2 | 1230 | 21.93 | 658.56 | 4 | 82.69 | Electron carrier activity |
| peu-miR131 | fgenesh4_pg.C_scaffold_9189000001 | 1 | 97 | 59.57 | 1475.17 | 4 | 82.69 | Electron carrier activity |
| peu-miR131 | fgenesh4_pg.C_LG_II001303 | 1 | 820 | 75.00 | 316.11 | 4 | 82.69 | DNA binding |
| peu-miR58 | estExt_Genewise1_v1.C_LG_XV2187 | 2 | 1229 | 4.39 | 131.71 | 5 | 65.45 | Transcription factor, SBP-box |
| peu-miR58 | fgenesh4_pg.C_LG_X001404 | 1 | 1185 | 40.00 | 210.74 | 5 | 65.45 | DNA binding |
| Peu-miR67* | gw1.VIII.1137.1 | 2 | 326 | 7.88 | 1001.01 | 4.5 | 64.08 | Function unknown |
| Peu-miR67* | eugene3.00031501 | 1 | 518 | 11.91 | 1475.17 | 4.5 | 64.08 | Vesicle transport v-SNARE |
| peu-miR93a | grail3.0010018301 | 2 | 172 | 11.41 | 895.64 | 4.5 | 74.10 | Function unknown |
| peu-miR93a | estExt_Genewise1_v1.C_LG_IV3721 | 3 | 1236 | 0.86 | 421.48 | 4.5 | 70.82 | NADH-ubiquinone oxidoreductase |
| peu-miR93b | grail3.0010018301 | 2 | 173 | 10.07 | 790.27 | 4.5 | 71.91 | Function unknown |
| Peu-miR106* | estExt_fgenesh4_pg.C_17020003 | 3 | 157 | 1.55 | 158.05 | 5 | 72.09 | Cytochrome c oxidase biogenesis protein |
| Peu-miR106* | estExt_fgenesh4_pm.C_1230037 | 3 | 67 | 2.12 | 158.05 | 5 | 72.09 | Function unknown |
| peu-miR115aa | gw1.57.264.1 | 3 | 373 | 0.12 | 158.05 | 4 | 61.32 | Function unknown |
| peu-miR123aa | estExt_fgenesh4_pg.C_LG_III1182 | 1 | 1225 | 0.95 | 842.96 | 4 | 74.26 | Development/cell death domain |
‘Cleavage site’, the cleavage site location at the gene model sequence; ‘Percentage of cleavage at the expected site’, percentage of sequence reads at cleavage site divided by all the cleavage reads in the same gene model sequence; ‘Position penalty score’, the same penalty score as the prediction of new miRNA targets; ‘MFE ratio’, minimum free energy percentage of the miRNA bound to its target divided by their perfect complement without mismatch; tbp, tags per billion; a miRNA candidates.
Fig. 3.
Cleavage site distribution of peu-miR30a verified by degradome sequencing. The standard CleaveLand output results for the degradome sequencing analysis of peu-miR30a. ‘Score’, the position penalty score used as the prediction of miRNA targets. ‘Start Position’ and ‘End position’, the start and end location at the miRNA pairing with the target gene sequence. ‘Cleavage site’, the verified cleavage site of the miRNA in the target gene sequence. Below the sequence of miRNA and target, the left column numbers represented all the cleavage sites discovered by degradome sequencing, the right column numbers represent the corresponding sequence reads in tags per million.
Putative mirtron in P. euphratica
For typical animal miRNA genes, after being transcribed from the genome, the formed pri-miRNA is first cleaved by the RNase III enzyme Drosha in the nucleus, which changes the pri-miRNA into a pre-miRNA hairpin structure. Subsequently, the pre-miRNA is transported into the cytoplasm and continually cleaved by Dicer (another RNase III) to generate the miRNA::miRNA* complex. In plants, DCL plays the role of Drosha and may also have the function of Dicer, and both the pri-miRNA and pre-miRNA are cleaved in the nucleus (Bartel, 2004). Alternatively, many recent studies have described a new mechanism to generate miRNA by a nuclear pathway that appears to bypass Drosha or DCL, but instead involves intron splicing to generate the pre-miRNA. This kind of miRNA was named mirtron miRNA; it uses short (∼200 bp) spliced introns as the pre-miRNA, and is further cleaved by Dicer or DCL to generate the miRNA::miRNA* duplex (Okamura et al., 2007). Several mirtrons have been reported in animals but few such miRNA-generating introns have been found in plants (Ruby et al., 2007; Glazov et al., 2008; Zhu et al., 2008).
The sequencing data obtained showed that one of the newly identified miRNA candidates (peu-miR11) was located at the 3' end of an intron in the estExt_Genewise1_v1.C_LG_I7094 gene (Fig. 4A). This intron was predicted to form a pre-miRNA-like hairpin structure by RNAfold software (Fig. 4B) and further met all the Mireap criteria for a new P. euphratica miRNA candidate. Consequently, this new P. euphratica miRNA was identified as a new putative mirtron; it might represent a new mechanism of miRNA generation in plants. Comparing nucleotide bias with 19 mammalian and 19 invertebrate mirtrons reported previously (Berezikov et al., 2007), it was concluded that the P. euphratica putative mirtron has the same typical conserved sequences in the 5' (GAAGU) and 3' (UAG) ends as invertebrates. These conserved sequences were previously reported as a characteristic of animal mirtron genes (Okamura et al., 2007) (Fig. 4C). This conservation increased the possibility that this miRNA candidate could be identified as a mirtron and showed that this putative mirtron was conserved with invertebrates.
Fig. 4.
P. euphratica putative mirtron. (A) The gene structure of estExt_Genewise1_v1.C_LG_I7094 in the genome. (B) Predicted secondary structure of the putative peu-miR11 mirtron. Mature sequence in a red line, miRNA* sequence in a blue line, potential hnRNP binding site in a green line, and arrows pointing to the DCL cleavage site are shown. (C) Conservation between peu-miR11 and sequence logos of other 5' and 3' mirtron products. Data represent 19 primate/mammalian mirtrons (Berezikov et al., 2007), and 19 invertebrate (15 fly and 4 worm) mirtrons (Ruby et al., 2007). Peu-miR11 has the same conserved sequence at the 5' (GAAGU) and 3' (UAG) ends as animals and is more conserved to invertebrate mirtrons.
MiRNAs and miRNA family expression profiling
To analyse miRNA expression under drought stress, miRNA expression profiling of leaf samples between drought-stressed (Group C) and control (Group A) P. euphratica plants was established. Specifically, the expression amount of a specific miRNA was represented by the sequence reads of the most numerous miRNA sequence in the pre-miRNA plus its ±2 nt adjacent sequence reads. Then the expression amounts were normalized for the purpose of calculating P-values and log2 ratios between drought-stressed and normal growth plants. Because the possibility remained that miRNA candidates were true new miRNAs, miRNA expression analysis reported here also included these candidates. The results will be referenced in future research on identification of new or drought response plant miRNAs. Results showed that 92 known sequenced P. trichocarpa miRNAs and 34 newly discovered P. euphratica miRNAs or candidates were up-regulated, whereas 36 conserved and 6 newly discovered leaf miRNA or candidates were down-regulated under drought stress (Fig. 5A, Supplementary Table S6 at JXB online).
Fig. 5.
Expression of the miRNAs and miRNA families in P. euphratica. (A) MiRNA expression scatter plot of high-throughput sequencing between drought-treated and normal growth P. euphratica. For each miRNA, sequence reads were divided by the total sequence number then multiplied to 1,000,000 (reads per million). (B) Venn diagrams of the tags detected by microarray profiling and high-throughput sequencing. The number in the middle of the microarray and high-throughput sequencing circle represented miRNAs that had the same expression pattern during drought stress in the two experimental results. The upper Venn diagram is the result without consideration of the significance level and the lower Venn diagram is the result under the condition of P<0.01 in both experiments.
Because different miRNA genes belonging to the same family may have the same mature sequence, the expression of the mature miRNA sequences as they represent the expression of whole or part of different miRNA families is of interest, in addition to the expression of miRNA genes analysed above. For this purpose, the concept of miRNA sequence tags (‘tags’ for short) was introduced, which was defined as the unique (remove redundancy) sequences of all the mature plant miRNA sequences registered in miRBase 13.0 and all the 142 new P. euphratica miRNA or miRNA candidate sequences. In total, 1014 plant miRNA and 142 new P. euphratica miRNA or candidate sequence tags could be extracted. The 1014 plant miRNA tags were named according to the miRNA name first appearing in miRBase 13.0, except that all 114 P. trichocarpa tags were named according to the name that first appeared in P. trichocarpa (Supplementary Table S7 at JXB online). Then the high-throughput sequencing read counts of each miRNA sequence tags were further analysed.
In the high-throughput sequencing data, 260 tags were found, which included 173 up-regulated and 47 down-regulated tags. Only expression significant at P<0.01 and a log2 ratio >1 were calculated. Furthermore, all 1014 miRBase 13.0 and 142 new P. euphratica miRNA or candidate sequence tags were used as probes in the miRNA expression microarray. Among the 260 tags identified by high-throughput sequencing, 104 up- and 27 down-regulated tags could be verified by microarray profiling (Supplementary Table S7 at JXB online). At a significance level of P<0.01, only 21 up- and 2 down-regulated tags had the same expression pattern between the drought treatment and control samples in the results of sequencing and microarray (Fig. 5B; Table 3).
Table 3.
Expression pattern consistent miRNA tags in microarray profiling and high-throughput sequencing under drought stress
| Tag name | Sequence (5'–3') | Microarray |
High-throughput sequencing |
||||||
| Group C median signal | Control median signal | log2 (Group C/control) | Control expressed | Group C expressed | Control normalized | Group C normalized | log2 (Group C/control) | ||
| ath-miR156g | CGACAGAAGAGAGUGAGCAC | 6106.99 | 4801.66 | 0.35 | 106. | 72 | 22.07 | 40.18 | 0.86 |
| ath-miR319a | UUGGACUGAAGGGAGCUCCCU | 9658.96 | 6994.93 | 0.41 | 19 | 35 | 3.96 | 19.53 | 2.30 |
| bna-miR156a | UGACAGAAGAGAGUGAGCACA | 4760.84 | 3484.23 | 0.47 | 7229 | 9584 | 1505.19 | 5348.95 | 1.83 |
| ghr-miR156c | UGUCAGAAGAGAGUGAGCAC | 4302.29 | 3028.75 | 0.59 | 15 | 23 | 3.12 | 12.84 | 2.04 |
| peu-miR102 | UCUUUCCGAGUCCUCCCAUACC | 3937.86 | 3270.72 | 0.27 | 73 | 72 | 15.20 | 40.18 | 1.40 |
| peu-miR102* | UAUGGGAGAGGCGGGAAUGACU | 665.89 | 310.08 | 1.10 | 8 | 15 | 1.67 | 8.37 | 2.33 |
| peu-miR123a | UGUCGCAGGAGAGAUGGCGCU | 302.07 | 109.51 | 1.59 | 515 | 387 | 107.23 | 215.99 | 1.01 |
| peu-miR123b | UGUCGCAGGAGAGAUGGCGCUA | 263.11 | 88.76 | 1.60 | 588 | 344 | 122.43 | 191.99 | 0.65 |
| peu-miR129 | UUCAUUCCUCUUCCUAAAAUGG | 248.30 | 64.90 | 1.88 | 89 | 87 | 18.53 | 48.56 | 1.39 |
| pta-miR319 | UUGGACUGAAGGGAGCUCC | 8945.88 | 6395.55 | 0.49 | 4 | 12 | 1.00 | 6.70 | 2.74 |
| ptc-miR156a | UGACAGAAGAGAGUGAGCAC | 6205.64 | 4932.89 | 0.41 | 122451 | 109321 | 25496.16 | 61013.36 | 1.26 |
| ptc-miR156k | UGACAGAAGAGAGGGAGCAC | 3549.59 | 2359.96 | 0.68 | 439 | 248 | 91.41 | 138.41 | 0.60 |
| ptc-miR162a | UCGAUAAACCUCUGCAUCCAG | 9416.06 | 6869.24 | 0.45 | 3671 | 1859 | 764.36 | 1037.53 | 0.44 |
| ptc-miR167f | UGAAGCUGCCAGCAUGAUCUU | 8097.06 | 6035.68 | 0.41 | 3202 | 1616 | 666.71 | 901.91 | 0.44 |
| ptc-miR319a | UUGGACUGAAGGGAGCUCCC | 12057.19 | 10260.72 | 0.23 | 67 | 183 | 13.95 | 102.13 | 2.87 |
| ptc-miR396a | UUCCACAGCUUUCUUGAACUG | 4357.86 | 1427.49 | 1.68 | 186 | 352 | 38.73 | 196.46 | 2.34 |
| ptc-miR396c | UUCCACAGCUUUCUUGAACUU | 542.90 | 83.90 | 2.66 | 92 | 110 | 19.16 | 61.39 | 1.68 |
| ptc-miR473a | ACUCUCCCUCAAGGCUUCCA | 2335.32 | 1671.64 | 0.52 | 1153 | 2216 | 240.07 | 1236.78 | 2.37 |
| sbi-miR156e | UGACAGAAGAGAGCGAGCAC | 4269.41 | 3034.67 | 0.53 | 118 | 81 | 24.57 | 45.21 | 0.88 |
| vvi-miR156e | UGACAGAGGAGAGUGAGCAC | 3387.98 | 2394.35 | 0.51 | 52 | 47 | 10.83 | 26.23 | 1.28 |
| vvi-miR167c | UGAAGCUGCCAGCAUGAUCUC | 4819.80 | 2951.66 | 0.77 | 27 | 23 | 5.62 | 12.84 | 1.19 |
| ptc-miR166n | UCGGACCAGGCUUCAUUCCUU | 2540.95 | 4532.28 | –0.89 | 799 | 33 | 166.36 | 18.42 | –3.18 |
| ptc-miR169z | CAGCCAAGAAUGAUUUGCCGG | 8.87 | 69.20 | –2.96 | 320 | 59 | 66.63 | 32.93 | –1.02 |
‘Group C/control median signal’, the median signal for each of the three miRNA tags probe in the microarray. ‘Group C/control expressed’, the high-throughput sequencing reads for each miRNA tags; ‘Group C/control normalized’, the normalized miRNA tag expression in tags per million; ‘log2 (GroupC/control)’, log2 ratio of the median signal in the microarray or the normalized expression in the high-throughput sequencing between drought treatment (Group C) and control samples.
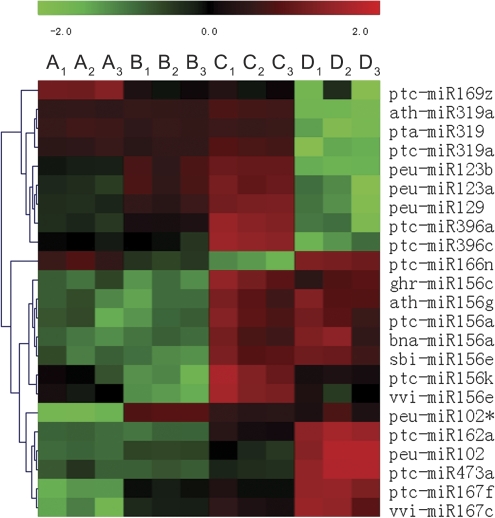
In addition to Group C and control (Group A), miRNA tag expression was also profiled in other drought treatments (Groups B and D) using microarray. After analysing and filtering data, all 23 sequence tags that had consistent expression between the high-throughput sequencing and microarray at the P<0.01 level were cluster analysed. They were approximately divided into three groups (Fig. 6). (i) ghr-miR156c, ath-miR156g, ptc-miR156a, bna-miR156a, sbi-miR156e, ptc-miR156k, vvi-miR156e, ptc-miR162a, peu-miR102, ptc-miR473a, ptc-miR167f, and vvi-miR167c were down-regulated in the treatment groups growing well (control and Group B) and up-regulated in all drought-influenced conditions (Groups C and D). (ii) ath-319a, pta-miR319, ptc-miR319a, peu-miR123b, peu-miR123a, peu-miR129, ptc-miR396a, and ptc-miR396c were up-regulated following the degree of drought (Groups A, B, and C) while suddenly down-regulated in Group D. (iii) ptc-miR169z, ptc-miR166n, and peu-miR102* have their specific expression pattern that cannot be clustered with others. Among them, ptc-miR169z was gradually down-regulated following drought stress intensification, suggesting that its regulating target genes were consistently promoted.
Fig. 6.
MiRNA sequence tag expression patterns in all treatment samples. All 23 sequence tags that had consistent expression between the high-throughput sequencing and microarray at the P<0.01 level were clustered using miRNA expressing microarray data generated from four drought treatment groups.
Discussion
Although the miRNA total counts in these high-throughput sequencing results were reduced under drought stress (Fig. 2B), it was found that actually most conserved and newly identified P. euphratica miRNAs were up-regulated (Supplementary Table S7 at JXB online). This was because a few miRNAs dominated most of the miRNA reads and the changes in these miRNA reads resulted in a reduction in the total miRNA reads in drought-stressed plants. For most of the other miRNA genes, expression was up-regulated, although their sequence reads were small in both drought-stressed and control leaf samples. This observation could be explained by a previous hypothesis that highly expressed miRNAs were mainly responsible for control of the basic cellular and developmental pathways common to most eukaryotes, whereas the little-expressed miRNAs were involved in regulation of lineage-specific pathways and functions (Glazov et al., 2008). Accordingly, it could be supposed that, under drought stress, basic cellular and development pathways were repressed by a large amount of up-regulated miRNA, while P. euphratica lineage-specific drought resistance pathways were promoted by few down-regulated miRNAs.
The present studies also demonstrated a putative mirtron found in P. euphratica. Previous research has reported introns that can cross-pair each terminus (5' and 3') are easily cleaved by RNA polymerase. HnRNP, a nucleic acid binding protein, is involved in this process, and functions by pulling each side of the intron close (Martinez-Contreras et al., 2006). It was found an hnRNP binding site UGGGGU (Buratti et al., 2004) near the 5' end of the newly discovered putative mirtron. Since this UGGGGU was located right in the loop of the putative mirtron secondary structure near its 5' site, this mirtron might be easily cleaved with the help of hnRNP, meanwhile miRNA* in the 5' end might also be constrained by hnRNP, prevented from loading to RICE (Supplementary Fig. S4 at JXB online). Considering the large number of introns in plant genomes, it is dpropose that other yet unknown miRNAs may be generated through this pathway.
In the previously published Arabidopsis degradome sequencing research by German et al. (2009), ∼800,000 acquired unique sequences confirmed 57 of 103 previously validated targets, 14 previously predicted but not validated, and 6 previously neither predicted nor validated targets. Although these degradome sequencing results showed more unique sequences in the numbers of 1,055,227, only 12 of the 129 predicted new miRNA and target pairs were verified, the confirmation rate was 9.3%. The absence of other miRNA and targets in the degradome sequencing result may be due to the lower abundance of these newly identified miRNAs or genomic differences between P. euphratica and P. trichocarpa. Meanwhile, two (peu-miR115a and peu-miR123a, Table 2) candidate miRNAs and their target pairs were certified by degradome sequencing, indicating that the candidate miRNAs from these results have a high possibility of being new P. euphratica miRNAs. Previously published research verified 26 experimental miRNA and target pairs by 5'RACE (Lu et al., 2005, 2008); the degradome sequencing result reported herein confirmed only one of them (miR1444a targeted gw1.182.27.1). However, among the 21 verified conserved miRNA and target pairs, 19 were previously predicted computationally by PopGenIE (http://www.popgenie.org/). The results of ptc-miR482-1 targeted eugene3.02710006 and grail3.0250000201 were not previously predicted or validated. The target mRNA cleavage fragments were short lived in the cell; compared with the identification of miRNA, verification of miRNA targets was more difficult. These degradome sequencing results contributed to the number of verified miRNA targets.
Comparing previously discovered genes comprising drought stress response in different poplar species (Brosche et al., 2005; Plomion et al., 2006; Street et al., 2006; Bogeat-Triboulot et al., 2007; Kreuzwieser et al., 2009; Wilkins et al., 2009; Cohen et al., 2010), it is found that six degradome sequencing-verified miRNA targets were identified as drought responsive previously (Table 4). None of these six miRNA targets was reported to be regulated by miRNA. The homologue of target gene gw1.VII.2722.1 in Arabidopsis is AT1G56010, a gene reported to be an NAC transcription factor involved in auxin and stress response (Riechmann et al., 2000). Moreover, many of the verified miRNA target genes reported herein were found to participate in drought or dehydration stress in other genera. Three new miRNAs (peu-miR101a, peu-miR131, and peu-miR58) and four conserved miRNAs (ptc-miR156a, ptc-miR156i, ptc-miR156k, and ptc-miR159f) targeted four transcription factors belonging to the Myb superfamily or containing an SBP-box. In Arabidopsis, Myb transcription factors were found to recognize the Responses to Dehydration 22 (rd22) promoter region, and function as cis-acting elements in the drought- and ABA-induced gene expression of rd22 (Abe et al., 2003). Many SBP transcription factors were also previously discovered to be stress responsive but less miRNA associated in Arabidopsis (Wang et al., 2009). Five new miRNAs (peu-miR30a, peu-miR30ba, peu-miR71*, peu-miR131, and peu-miR58) were identified to target four signal receptor-like genes, all of them had 2Fe–2S ferredoxin, a von willebrand factor domain, and an epidermal growth factor-like region. The 2Fe–2S ferredoxin iron–sulphur binding site mediates electron transfer in a range of metabolic reactions (Pauff et al., 2008; Tomasiak et al., 2008). vWF domain can be involved in membrane transport and the EGF-like region is found in the extracellular domain of membrane-bound proteins (O'Leary et al., 2004). The conserved miRNAs of ptc-miR164f and ptc-miR164d were verified to target the No Apical Meristem (NAM) protein. This kind of protein was previously found to be involved in plant hormonal control and defence (Xie et al., 2000; Duval et al., 2002). MiRNA targets with other biological functions were also qualified by P. euphratica degradome sequencing, such as a NADH-ubiquinone oxidoreductase and a cytochrome c oxidase that were targeted by peu-miR93aa and Peu-miR106*, respectively.
Table 4.
Degradome sequencing-verified miRNA targets that were also identified in other Populus drought studies
| MiRNA | Target | Arabidopsis homologue | Annotation | Reference |
| peu-miR84* | fgenesh4_pm.C_LG_XIII000061 | AT3G10020.1 | Electron carrier activity | Cohen et al., 2010 |
| peu-miR93a | grail3.0010018301 | AT3G47510.1 | Function unknown | Kreuzwieser et al., 2009 |
| peu-miR93b | grail3.0010018301 | AT3G47510.1 | Function unknown | Kreuzwieser et al., 2009 |
| peu-miR123a | estExt_fgenesh4_pg.C_LG_III1182 | AT5G42050.1 | Development/cell death domain | Cohen et al., 2010 |
| ptc-miR164f | gw1.V.3536.1 | AT1G56010.2 | NAM protein | Cohen et al., 2010 |
| ptc-miR164f | gw1.VII.2722.1 | AT1G56010.2 | NAM protein | Wilkins et al., 2009 |
| ptc-miR164d | gw1.V.3536.1 | AT1G56010.2 | NAM protein | Cohen et al., 2010 |
| ptc-miR164d | gw1.VII.2722.1 | AT1G56010.2 | NAM protein | Wilkins et al., 2009 |
| ptc-miR1444a | gw1.182.27.1 | AT1G08170.1 | Oxidoreductase activity | Cohen et al., 2010 |
High-throughput sRNA sequencing has great potential to identify new members of known sRNA classes, especially in tissues or under environmental conditions that have not been investigated yet. This technology can also be used to compare miRNA profiles, thus gaining further insights into miRNA biogenesis and function. The research reported here has benefited greatly from the high-throughput sequencing and microarray technologies, and has given a deep cross-comparison analysis of the data generated by these two technologies. The accuracy of the sequencing and the three replicates microarray-probe characters made reliable miRNA discoveries and also miRNA expression profile results. However, there were many limitations in the use of these two technologies. On one aspect, compared with previous work, although it could generate >1,000,000 sequences, the sRNA covering rate of high-throughput sequencing seemed still insufficient to cover all miRNAs in a species (Glazov et al., 2008), partly because a PCR process existed in the library preparation steps. On another aspect, microarrays had the advantage of covering all the designed miRNA sequence tag probes, but the accuracy was insufficient and false-positive signals were high (Lee et al., 2008). Meanwhile, the microarray technology was limited to detect only known mature miRNA sequence signal intensity, whereas high-throughput sequencing could evaluate miRNA hairpin sequence counts with the assistance of mature miRNA adjacent sequence reads (±2 nt in this research). The combination and comparison of data from both these technologies could generate reliable miRNA discovery and expression results. Interestingly, both the P. euphratica high-throughput sequencing and microarray profiling found 62 conserved miRNAs in other non-poplar plant species (Supplementary Table S7 at JXB online), these 62 conserved miRNAs could not locate their hairpin sequence in the P. trichocarpa genome. This result indicated the genomic diversity between P. euphratica and P. trichocarpa and some species of conserved miRNA remain undiscovered in poplar.
The research presented here showed a functional way to find differentially expressed miRNAs by combining the methods of high-throughput sequencing and microarray profiling. This combination brought both efficiency in saving experimental work and the challenge of bioinformatics and statistical data analysis. The results did not represent all the existing undiscovered miRNAs and all the drought stress-induced miRNAs in P. euphratica; however, this research significantly increased the number of new miRNAs and annotated a large number of those induced by drought stress. Further studies, such as transgenic phenotype analysis, are required to enlarge understanding of miRNA function and P. euphratica long-term stress resistance mechanisms.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. The nucleotide bias in each position of the newly identified miRNAs in P. euphratica.
Supplementary Fig. S2. Cleavage site distribution of all degradome sequencing-verified new miRNA targets.
Supplementary Fig. S3. Cleavage site distribution of all degradome sequencing-verified conserved miRNA targets.
Supplementary Fig. S4. P. euphratica putative mirtron sequencing reads and expression model.
Supplementary Table S1. Conserved miRNAs between P. euphratica and P. trichocarpa.
Supplementary Table S2. New miRNAs identified in P. euphratica by high-throughput sequencing.
Supplementary Table S3. Summary of P. euphratica degradome sequencing.
Supplementary Table S4. New P. euphratica miRNA predicted targets.
Supplementary Table S5. Targets of conserved P. euphratica miRNA verified by degradome sequencing.
Supplementary Table S6. MiRNA expression in drought stress-treated P. euphratica.
Supplementary Table S7. All detectable miRNA sequence tags.
Acknowledgments
This work was funded by “948” Project of State Forestry Administration of China (2007-4-01); Program for New Century Excellent Talents in University of China(NCET-07-0083); National Natural Science Foundation of China (30730077, 30972339); and Beijing Forestry University Technology Innovation Program (BLYJ200902).
Glossary
Abbreviations
- AGO
Argonaute
- DCL
Dicer-like
- PARE
parallel analysis of RNA ends
- RISC
RNA-induced silencing complex
- RSMC
relative soil moisture content
- WP
water potential
- WUE
water use efficiency
References
- Abe H, Urao T, Ito T, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. The Plant Cell. 2003;15:63–78. doi: 10.1105/tpc.006130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abdelali B, Wall PK, DiLoreto S, dePamphilis CW, Carlson JE. Conservation and divergence of microRNAs in Populus. BMC Genomics. 2007;8:481. doi: 10.1186/1471-2164-8-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C, Miller W, Axtell MJ. CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics. 2009;25:130–131. doi: 10.1093/bioinformatics/btn604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audic S, Claverie JM. The significance of digital gene expression profiles. Genome Research. 1997;7:986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. The Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Baumberger N, Baulcombe DC. Arabidopsis ARGONAUTE1 is an RNA slicer that selectively recruits microRNAs and short interfering RNAs. Proceedings of the National Academy of Sciences, USA. 2005;102:11928–11933. doi: 10.1073/pnas.0505461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Molecular Cell. 2007;28:328–336. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogeat-Triboulot MB, Brosche M, Renaut J, et al. Gradual soil water depletion results in reversible changes of gene expression, protein profiles, ecophysiology, and growth performance in Populus euphratica, a poplar growing in arid regions. Plant Physiology. 2007;143:876–892. doi: 10.1104/pp.106.088708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Brosche M, Vinocur B, Alatalo ER, et al. Gene expression and metabolite profiling of Populus euphratica growing in the Negev desert. Genome Biology. 2005;6:R101. doi: 10.1186/gb-2005-6-12-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buratti E, Baralle M, De Conti L, Baralle D, Romano M, Ayala YM, Baralle FE. hnRNP H binding at the 5' splice site correlates with the pathological effect of two intronic mutations in the NF-1 and TSHβ genes. Nucleic Acids Research. 2004;32:4224–4236. doi: 10.1093/nar/gkh752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Puryear J, Cairney J. A simple and efficient method for isolating RNA from pine trees. Plant Molecular Biology Reporter. 1993;11:113–116. [Google Scholar]
- Cohen D, Bogeat-Triboulot MB, Tisserant E, Balzergue S, Martin-Magniette ML, Lelandais G, Ningre N, Renou JP, Tamby JP. Comparative transcriptomics of drought responses in Populus: a meta-analysis of genome-wide expression profiling in mature leaves and root apices across two genotypes. BMC Genomics. 2010;11:630. doi: 10.1186/1471-2164-11-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duval M, Hsieh TF, Kim SY, Thomas TL. Molecular characterization of AtNAM: a member of the Arabidopsis NAC domain superfamily. Plant Molecular Biology. 2002;50:237–248. doi: 10.1023/a:1016028530943. [DOI] [PubMed] [Google Scholar]
- Edwards A, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- German MA, Luo S, Schroth G, Meyers BC, Green PJ. Construction of parallel analysis of RNA ends (PARE) libraries for the study of cleaved miRNA targets and the RNA degradome. Nature Protocols. 2009;4:356–362. doi: 10.1038/nprot.2009.8. [DOI] [PubMed] [Google Scholar]
- Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML. A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Research. 2008;18:957–964. doi: 10.1101/gr.074740.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gries DZF, Foetzki A, Arndt SK, Bruelheide H, Thomas FM, Zhang X, Runge M. Growth and water relations of Tamarix ramosissima and Populus euphratica on Taklamakan desert dunes in relation to depth to a permanent water table. Plant, Cell and Environment. 2003;26:725–736. [Google Scholar]
- Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Research. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasio T. Plant responses to water stress. Annunal Review of Plant Physiology and Plant Molecular Biology. 1973;24:519–570. [Google Scholar]
- Jones-Rhoades MW, Bartel DP. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Molecular Cell. 2004;14:787–799. doi: 10.1016/j.molcel.2004.05.027. [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annual Review of Plant Biology. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- Kreuzwieser J, Hauberg J, Howell KA, Carroll A, Rennenberg H, Millar AH, Whelan J. Differential response of gray poplar leaves and roots underpins stress adaptation during hypoxia. Plant Physiology. 2009;149:461–473. doi: 10.1104/pp.108.125989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proceedings of the National Academy of Sciences, USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak PB, Wang QQ, Chen XS, Qiu CX, Yang ZM. Enrichment of a set of microRNAs during the cotton fiber development. BMC Genomics. 2009;10:457. doi: 10.1186/1471-2164-10-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Current Biology. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Ajay SS, Chen H, Maruyama A, Wang N, McInnis MG, Athey BD. Discriminating single-base difference miRNA expressions using microarray Probe Design Guru (ProDeG) Nucleic Acids Research. 2008;36:e27. doi: 10.1093/nar/gkm1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Yin W, Xia X. Identification of microRNAs and their targets from. Populus euphratica. Biochemical and Biophysical Research Communications. 2009;388:272–277. doi: 10.1016/j.bbrc.2009.07.161. [DOI] [PubMed] [Google Scholar]
- Li R, Li Y, Kristiansen K, Wang J. SOAP: short oligonucleotide alignment program. Bioinformatics. 2008;24:713–714. doi: 10.1093/bioinformatics/btn025. [DOI] [PubMed] [Google Scholar]
- Lu S, Sun YH, Chiang VL. Stress-responsive microRNAs in. Populus. The Plant Journal. 2008;55:131–151. doi: 10.1111/j.1365-313X.2008.03497.x. [DOI] [PubMed] [Google Scholar]
- Lu S, Sun YH, Shi R, Clark C, Li L, Chiang VL. Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. The Plant Cell. 2005;17:2186–2203. doi: 10.1105/tpc.105.033456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man MZ, Wang X, Wang Y. POWER_SAGE: comparing statistical tests for SAGE experiments. Bioinformatics. 2000;16:953–959. doi: 10.1093/bioinformatics/16.11.953. [DOI] [PubMed] [Google Scholar]
- Martinez-Contreras R, Fisette JF, Nasim FU, Madden R, Cordeau M, Chabot B. Intronic binding sites for hnRNP A/B and hnRNP F/H proteins stimulate pre-mRNA splicing. PLoS Biology. 2006;4:e21. doi: 10.1371/journal.pbio.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers BC, Axtell MJ, Bartel B, et al. Criteria for annotation of plant microRNAs. The Plant Cell. 2008;20:3186–3190. doi: 10.1105/tpc.108.064311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, et al. Sorting of small RNAs into Arabidopsis Argonaute complexes is directed by the 5' terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, et al. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Research. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Hagen JW, Duan H, Tyler DM, Lai EC. The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell. 2007;130:89–100. doi: 10.1016/j.cell.2007.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary JM, Hamilton JM, Deane CM, Valeyev NV, Sandell LJ, Downing AK. Solution structure and dynamics of a prototypical chordin-like cysteine-rich repeat (von Willebrand Factor type C module) from collagen IIA. Journal of Biological Chemistry. 2004;279:53857–53866. doi: 10.1074/jbc.M409225200. [DOI] [PubMed] [Google Scholar]
- Ottow EA, Polle A, Brosche M, Kangasjarvi J, Dibrov P, Zorb C, Teichmann T. Molecular characterization of PeNhaD1: the first member of the NhaD Na+/H+ antiporter family of plant origin. Plant Molecular Biology. 2005;58:75–88. doi: 10.1007/s11103-005-4525-8. [DOI] [PubMed] [Google Scholar]
- Pan W. A comparative review of statistical methods for discovering differentially expressed genes in replicated microarray experiments. Bioinformatics. 2002;18:546–554. doi: 10.1093/bioinformatics/18.4.546. [DOI] [PubMed] [Google Scholar]
- Pauff JM, Zhang J, Bell CE, Hille R. Substrate orientation in xanthine oxidase: crystal structure of enzyme in reaction with 2-hydroxy-6-methylpurine. Journal of Biological Chemistry. 2008;283:4818–4824. doi: 10.1074/jbc.M707918200. [DOI] [PubMed] [Google Scholar]
- Plomion C, Lalanne C, Claverol S, et al. Mapping the proteome of poplar and application to the discovery of drought-stress responsive proteins. Proteomics. 2006;6:6509–6527. doi: 10.1002/pmic.200600362. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature. 2007;448:83–86. doi: 10.1038/nature05983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schattner P, Brooks AN, Lowe TM. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Research. 2005;33:W686–W689. doi: 10.1093/nar/gki366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- Street NR, Skogstrom O, Sjodin A, Tucker J, Rodriguez-Acosta M, Nilsson P, Jansson S, Taylor G. The genetics and genomics of the drought response in Populus. The Plant Journal. 2006;48:321–341. doi: 10.1111/j.1365-313X.2006.02864.x. [DOI] [PubMed] [Google Scholar]
- Tomasiak TM, Maklashina E, Cecchini G, Iverson TM. A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II. Journal of Biological Chemistry. 2008;283:15460–15468. doi: 10.1074/jbc.M801372200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Hu Z, Yang Y, Chen X, Chen G. Function annotation of an SBP-box gene in Arabidopsis based on analysis of co-expression networks and promoters. International Journal of Molecular Sciences. 2009;10:116–132. doi: 10.3390/ijms10010116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Waldron L, Nahal H, Provart NJ, Campbell MM. Genotype and time of day shape the Populus drought response. The Plant Journal. 2009;60:703–715. doi: 10.1111/j.1365-313X.2009.03993.x. [DOI] [PubMed] [Google Scholar]
- Xie Q, Frugis G, Colgan D, Chua NH. Arabidopsis NAC1 transduces auxin signal downstream of TIR1 to promote lateral root development. Genes and Development. 2000;14:3024–3036. doi: 10.1101/gad.852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Spriggs A, Matthew L, Fan L, Kennedy G, Gubler F, Helliwell C. A diverse set of microRNAs and microRNA-like small RNAs in developing rice grains. Genome Research. 2008;18:1456–1465. doi: 10.1101/gr.075572.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.