Abstract
Solanum commersonii and Solanum tuberosum are closely related plant species that differ in their abilities to cold acclimate; whereas S. commersonii increases in freezing tolerance in response to low temperature, S. tuberosum does not. In Arabidopsis thaliana, cold-regulated genes have been shown to contribute to freezing tolerance, including those that comprise the CBF regulon, genes that are controlled by the CBF transcription factors. The low temperature transcriptomes and CBF regulons of S. commersonii and S. tuberosum were therefore compared to determine whether there might be differences that contribute to their differences in ability to cold acclimate. The results indicated that both plants alter gene expression in response to low temperature to similar degrees with similar kinetics and that both plants have CBF regulons composed of hundreds of genes. However, there were considerable differences in the sets of genes that comprised the low temperature transcriptomes and CBF regulons of the two species. Thus differences in cold regulatory programmes may contribute to the differences in freezing tolerance of these two species. However, 53 groups of putative orthologous genes that are cold-regulated in S. commersonii, S. tuberosum, and A. thaliana were identified. Given that the evolutionary distance between the two Solanum species and A. thaliana is 112–156 million years, it seems likely that these conserved cold-regulated genes—many of which encode transcription factors and proteins of unknown function—have fundamental roles in plant growth and development at low temperature.
Keywords: Arabidopsis, CBF regulon, freezing tolerance, low temperature transcriptome, Solanum species
Introduction
Plants differ greatly in their abilities to cope with freezing temperatures (Levitt, 1980). Those plants that have adapted to temperate environments generally have considerable freezing tolerance and can cold acclimate; that is, they increase in freezing tolerance in response to low non-freezing temperatures. In contrast, plants that have adapted to tropical and subtropical climates generally have little, if any, freezing tolerance and do not cold acclimate. Understanding the molecular basis for why some plants can cold acclimate and survive freezing temperatures whereas others cannot is a fundamental goal of cold acclimation research.
In recent years, considerable effort has been directed at determining the role of cold-regulated genes in freezing tolerance. In Arabidopsis thaliana, it has been established that a cascade of changes in gene expression, involving alterations in the transcript levels for hundreds of genes, is initiated within minutes of exposing plants to low temperature (Fowler and Thomashow, 2002; Maruyama et al., 2004; Vogel et al., 2005). Moreover, it has been established that some of these cold-regulated genes contribute to freezing tolerance. The best understood freezing tolerance pathway is the CBF cold response pathway (Van Buskirk and Thomashow, 2006; Chinnusamy et al., 2007). Among the first wave of cold-induced genes are three—CBF1, CBF2, and CBF3 (Stockinger et al., 1997; Gilmour et al., 1998; Medina et al., 1999), also known as DREB1b, DREB1c, and DREB1a, respectively (Shinwari et al., 1998)—that encode members of the AP2/ERF family of DNA-binding proteins (Riechmann and Meyerowitz, 1998). The CBF transcription factors bind to the CRT/DRE DNA regulatory element present in the promoters of cold-regulated genes and induce their expression. Constitutive overexpression of either CBF1, CBF2, or CBF3 in A. thaliana results in expression of the CBF regulon and brings about an increase in freezing tolerance without a cold stimulus, indicating that the CBF regulon has a fundamental role in cold acclimation (Jaglo-Ottosen et al., 1998; Liu et al., 1998; Gilmour et al., 2004; Kasuga et al., 2004).
Changes in gene expression have been demonstrated to occur in response to low temperature in a wide range of plant species, including both freezing-tolerant and freezing-sensitive plants. Moreover, CBF cold response pathways have been demonstrated to exist in plants that cold acclimate, such as Brassica napus (Jaglo et al., 2001), poplar (Benedict et al., 2006), and barley (Choi et al., 2002; Xue, 2002), as well as in plants that do not cold acclimate, such as tomato (Hsieh et al., 2002; Zhang et al., 2004) and rice (Dubouzet et al., 2003). In each of these particular plant species there is at least one CBF gene that is induced in response to low temperature, and constitutive overexpression of CBF genes brings about changes in gene expression without cold treatment. However, whereas CBF overexpression increases the freezing tolerance of A. thaliana (Jaglo-Ottosen et al., 1998; Liu et al., 1998), B. napus (Jaglo et al., 2001), poplar (Benedict et al., 2006), and potato (Pino et al., 2007), it does not result in increased freezing tolerance in tomato (Hsieh et al., 2002; Zhang et al., 2004) and rice (Dubouzet et al., 2003). The reason for this difference is not clear, but it may involve differences in the composition of the CBF regulons. For instance, in A. thaliana and poplar, the CBF regulons include ∼85 (Vogel et al., 2005) and 63 (Benedict et al., 2006) cold-induced genes, respectively, whereas the CBF regulons of tomato (Zhang et al., 2004) and rice (Dubouzet et al., 2003) comprise only ∼10 cold-induced genes.
Given the demonstrated role of cold-regulated genes in plant cold acclimation, it is reasonable to think that differences in gene expression are likely to contribute to differences in the ability of plants to cold acclimate. Such thinking motivates comparative transcriptome studies using plants that differ in freezing tolerance to identify genes with critical roles in freezing tolerance. However, drawing conclusions from such studies requires consideration of the evolutionary distances between the plants being examined and their respective responses to low temperature. For instance, the divergence time between species with cold transcriptome data ranges from ∼20 million years ago (MYA; A. thaliana–Brassica) to ∼200 MYA (monocot–dicot) (Yang et al., 1999). In addition, tomato and rice are not only freezing-sensitive, but are also chilling-sensitive. This adds another factor that could affect gene expression and complicate interpretations of comparative studies.
With these thoughts in mind, a comparative transcriptome study was conducted using the common cultivated potato, Solanum tuberosum, and the wild potato Solanum commersonii. These plants are closely related evolutionarily. Assuming the neutral substitution rate to be ∼7×10−9 (Moniz de Sa and Drouin, 1996), the divergence time between these two species is likely to be only ∼3 million years as the synonymous substitution rate between these two species is ∼0.04 (see Materials and methods). Both plant species are chilling tolerant, but whereas S. commersonii can cold acclimate, S. tuberosum cannot. When S. commersonii and S. tuberosum plants are grown at warm temperature, their freeze-killing temperatures are about –4.5 °C and –3 °C, respectively (Chen and Li, 1980), but after 2 weeks of low temperature treatment (2 °C), the killing temperature of S. commersonii decreases to about –11.5 °C while that of S. tuberosum remains at about –3 °C (Chen and Li, 1980). The results presented here indicate that this difference in freezing tolerance is not due to ‘macro-scale’ differences in gene regulation in response to low temperature or the size of their CBF regulons, but reveal rapid evolution of the CBF pathways in the two plant species that may contribute to their differences in freezing tolerance. The analyses also led to the identification of 40 groups of putative orthologous genes (pOGs) that are cold-induced and 13 pOGs that are cold-repressed in A. thaliana, S. commersonii, and S. tuberosum. Given the relatively large evolutionary distance between these three plant species—they diverged 112–156 MYA (Yang et al., 1999; Bell et al., 2005)—it seems likely that these genes, many of which encode proteins of unknown function, have fundamental roles in plant low temperature biology.
Materials and methods
Plant material, growth conditions, and RNA extraction
Transgenic lines of S. commersonii and S. tuberosum cv. Umatilla that constitutively expressed the A. thaliana CBF3 gene under control of the cauliflower mosaic virus (CaMV) 35S promoter were generated as described by Pino et al. (2007). Wild-type and transgenic plants were grown in an Enconair growth chamber (‘Bigfoot’ GC-20, Enconair Ecological Chambers Inc., Winnipeg, Manitoba, Canada) maintained under a 16 h photoperiod, 350 μmol m−2 s−1 light intensity at 25 °C. Three biological replicates for each wild-type species were grown for 3 weeks under these conditions (each biological replicate consisted of three plants). Eight hours after dawn, wild-type plants were transferred to an environmentally controlled cold room maintained at 2 °C under a 16 h photoperiod with 50 μmol m−2 s−1 light intensity, and leaf tissue was harvested after 2, 8, 24, and 168 h. Warm controls were maintained at 25 °C under a normal growth photoperiod and tissue was harvested at 2, 8, and 24 h in the light for use as reference control samples. In the case of the 168 h cold samples, the 24 h warm control was used as the reference because the 168 h warm plants were already flowering. The leaf tissue of transgenic lines was collected 8 h after dawn.
For the mechanical agitation treatment, 3-week-old plants of both species grown under continuous light in magenta vessels were secured together in a cardboard box and dropped ∼15 cm every 2 s for 15 min. Samples were collected at 0, 15, and 30 min after mechanical treatment. Two replicates of two plants from each species were randomly selected from each vessel and immediately frozen in liquid nitrogen. A control without agitation was included.
Total RNA was isolated from leaf tissue using RNeasy Plant Mini Kits (Qiagen, Valencia, CA, USA). For quantitative real-time PCR (qRT-PCR) experiments, samples were treated with RNase-free DNase (Qiagen) using the on-column DNase digestion method provided by the manufacturer.
qRT-PCR
An amount of RNA that fell within the linear range for all genes tested (generally 100–250 ng) was reverse transcribed using a reverse transcription system (Promega, Madison, WI, USA) according to the manufacturer's instructions. The 20 μl final reaction was diluted to 200 μl. A 1 μl aliquot of each cDNA was used in a qRT-PCR, with the addition of 0.4 μM of each primer and Fast SYBR Green master mix (Applied Biosystems, Foster City, CA, USA) to make a final reaction volume of 10 μl. The qRT-PCRs were performed using a FAST 7500 Real Time PCR System (Applied Biosystems). The primers used are listed in Supplementary Table S1 available at JXB online. Relative expression was calculated using the potato 60S gene [expressed sequence tag (EST) clone STMCK67] as reference. This gene was confirmed not to change in levels under the test conditions used. Standard curves for each gene were included in each qRT-PCR run as a measure of efficiencies. Data were analysed by analysis of variance (ANOVA) using SAS program 9.1 (SAS Institute Inc., Cary, NC, USA) with mixed procedures; when appropriate, least significant difference was used for multiple comparisons.
RNA labelling and hybridization of potato microarrays
cDNA microarray experiments were conducted using the 10K potato cDNA microarray (TIGR, http://www.jcvi.org/potato/sol_ma_microarrays.shtml). RNA (20 μg) was labelled by the indirect labelling aminoallyl method. The slides were hybridized using the indirectly labelled aminoallyl probes hybridization method (http://www.jcvi.org/potato/sol_ma_protocols.shtml). To avoid bias due to dye-related differences, labelling dyes for each sample pair (cold/warm or transgenic line/wild type) were swapped in one of the three independent hybridizations (three biological replicates for cold treatments and two or three transgenic lines for the CBF regulon experiment).
Data processing and analysis
The TIFF images were quantified using Genepix 3.0 (Axon Instruments, Union City, CA, USA). The software automatically flags spots that cannot be found in one of the channels. Spots with aberrant shapes were checked manually and flagged as bad. Spots with lower signal intensity than the background (spots with ≥55% of the pixels with lower signal intensities than background) were also flagged as bad. All these ‘bad’ flagged clones were excluded from further analysis.
The data were normalized using the print tip loess method in the LIMMA package (Smyth, 2004). cDNA clones were regarded as differentially expressed if their modulated P-values from LIMMA were <0.05. The modulated P-values were derived from false discovery rate (FDR)-corrected raw P-values (Storey and Tibshirani, 2003). An average fold-change value was calculated for the two duplicates of each clone on the array. In cases where one duplicate of a clone did not pass the threshold P-value and/or was flagged as low quality, the value of the other duplicate was used if it had a qualifying P-value and was not flagged as low quality.
Hierarchical clustering was generated with Cluster (Eisen et al., 1998) using normalized log ratios. Gene Ontology (GO) designations for S. tuberosum EST clones were obtained from http://jcvi.org/potato/sol_ma_microarrays.shtml.
Arabidopsis thaliana cold-regulated and CBF regulon genes
A list of cold-regulated A. thaliana genes was generated from a large number of previously published microarray experiments (Vogel et al., 2005; Kilian et al., 2007) and new experiments (unpublished arrays have been submitted to Array Express) that used the Affymetrix ATH1 gene chip to monitor transcript levels (Supplementary Table S2 at JXB online). The cel files from previous experiments (submission numbers ME00320 and ME00325) were downloaded from TAIR (http://www.arabidopsis.org), imported into R, and analysed using Bioconductor Affy and SimpleAffy packages (Gautier et al., 2004; Wilson and Miller, 2005). Eighty Affymetrix cel files that passed quality control analysis were used to identify cold-regulated transcripts in the Columbia ecotype (Gentleman et al., 2005). Arrays were RMA (robust multichip average) normalized and the expression values for 15 220 probes that were present in at least half of the arrays by MAS5.0 present calls were retained for further analysis (Irizarry et al., 2003). Probes were analysed for differential expression between low temperature treatment and controls using LIMMA (Smyth, 2004). Contrasts between cold-treated Columbia samples and controls were performed for early cold response (0.5, 1, 2, and 3 h), 24 h cold response, and 168 h cold response. Differentially expressed probes were identified using LIMMA's decide tests function to select probe sets with a ≥2-fold difference between treatment and control with a significance after FDR adjustment of <0.05. Probe sets were mapped to A. thaliana transcripts using the ath1121501.db Bioconductor package (http://www.bioconductor.org/packages/2.5/data/annotation/html/ath1121501.db.html). Transcripts were identified as differentially expressed in each category: early cold response, 24 h cold response, and 7 d cold response. This cold-regulated gene list of 1151 cold-induced and 1095 cold-repressed transcripts was used for subsequent analysis (Supplementary Table S3 at JXB online).
To identify CBF-regulated transcripts, contrasts between arrays of CBF2-overexpressing seedlings and wild-type seedlings that were grown at warm temperature were made (Vogel et al., 2005). LIMMA's decide test was performed to identify differentially expressed transcripts with a fold-change >2, and a BH (Benjamini and Hochberg)-adjusted P-value of <0.05. Probe sets were mapped to A. thaliana transcripts using the ath1121501.db Bioconductor package (http://www.bioconductor.org/packages/2.5/data/annotation/html/ath1121501.db.html). Transcripts appearing in both the cold-regulated gene list and the CBF-regulated gene list were designated members of the A. thaliana CBF regulon.
Inference of putative orthologous groups (pOGs)
A list of pOGs between the putative unique transcripts (PUTs) sequences assembled from ESTs of S. tuberosum (PlantGDB, http://www.plantgdb.org/, version 157a) and A. thaliana protein sequences was obtained from TAIR.
pOGs have been previously established using protein sequences from four plant species with complete genomes (A. thaliana, TAIR6; Populus trichocarpa, v1.1; Oryza sativa japonica, version 2; and Physcomitrella patens, version 1.1) (Hanada et al., 2008). From all these four genomes, a best matching protein for PUTs was identified with BLAST (Altschul et al., 1997) using only matches with E-values <10−5. A PUT was assumed to be in the pOG of its best matching protein from these four species only if the evolutionary distance between PUTs and the rice member(s) and PUTs and the A. thaliana member(s) in the pOG is less than the distance between the rice and the A. thaliana members. The evolutionary distances of all sequences were calculated using the protdist program in the Phylogeny Inference Package (PHYLIP) with Gamma correction (Felsenstein, 2005).
Results
Identification of cold-regulated genes in S. commersonii and S. tuberosum
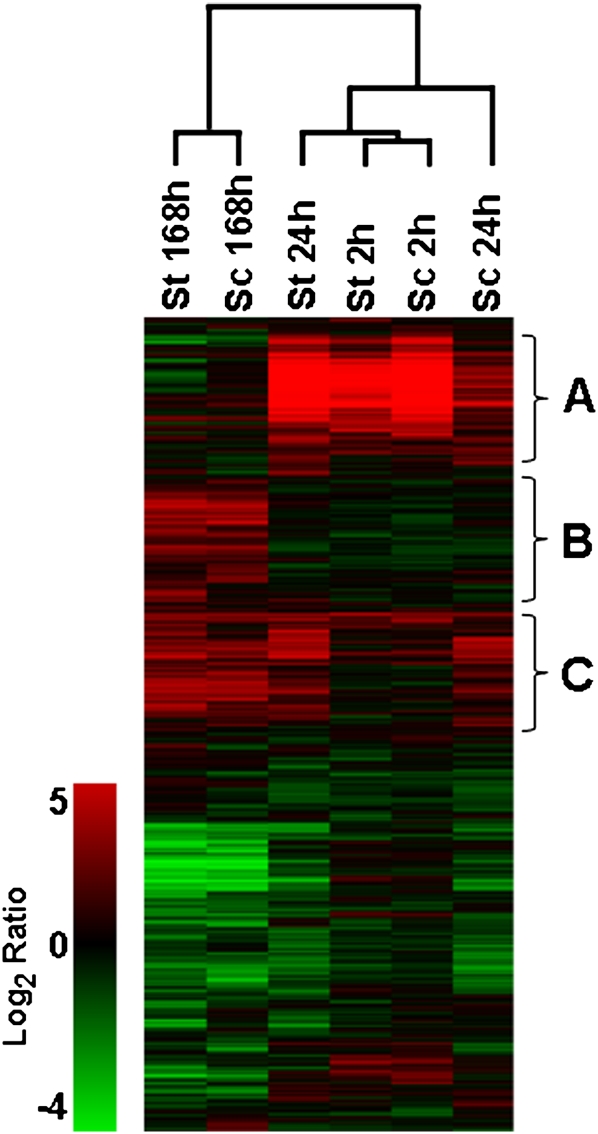
The low temperature transcriptomes of S. commersonii and S. tuberosum cv. Umatilla were compared using the TIGR potato cDNA array (10K, version 4) which has ESTs for ∼10,000 genes. Plants were grown for 3 weeks at 25 °C, transferred to 2 °C for 2, 24, and 168 h, and the RNA was isolated and analysed (there were three biological replicates for each time point). As a first assessment of the similarities and differences between the low temperature transcriptomes of S. commersonii and S. tuberosum, a heat map presenting the transcript levels for all probes at 2, 24, and 168 h without regard for fold-change or statistical significance was prepared (Fig. 1). The results indicated that the transcript levels for the vast majority of the genes represented on the array either increased or decreased at one or more time points during the experiment in both S. commersonii and S. tuberosum. Moreover, the expression patterns for both species were similar. For instance, the group of genes designated as cluster A were induced early in both species—at 2 h and 24 h—and returned to non-induced levels by 168 h; cluster B genes were unaffected by low temperature during the first 24 h of treatment, but were highly induced at 168 h; and cluster C genes were little affected after 2 h of cold treatment, but were induced at 24 h and 168 h. Nevertheless, the patterns were far from identical, with both qualitative and quantitative differences clearly evident.
Fig. 1.
Hierarchical clustering of S. commersonii (Sc) and S. tuberosum (St) transcripts at 2, 24, and 168 h of cold treatment at 2 °C. Data are shown as the average log ratio from three biological replicates. The figure shows all spots on the array (including bad flagged spots) prior to statistical selection.
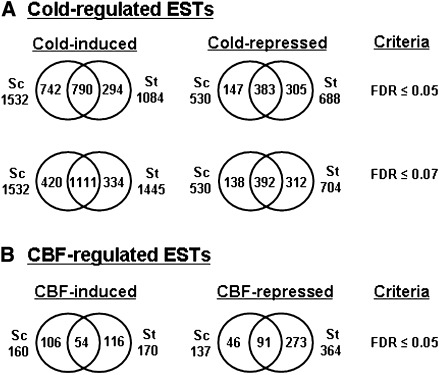
Next two filtering criteria were applied to the data: a 2-fold change in transcript level and an FDR of ≤0.05. Using these criteria, ∼15–20% of the ESTs were considered cold-regulated in each species. In S. commersonii and S. tuberosum, respectively, ∼13% and ∼10% of the ESTs were cold-induced and 5% and 6% were cold repressed at one or more of the time points tested. Transcripts for 1532 and 1084 ESTs increased in S. commersonii and S. tuberosum, respectively, of which 790 increased in both plant species (Fig. 2A); transcripts for 530 and 688 ESTs decreased in S. commersonii and S. tuberosum, respectively, of which 383 decreased in both plant species (Fig. 2A). However, the apparent differences in gene expression for the cold-induced genes were very sensitive to the filtering criteria used. For instance, if the fold-change criterion was kept at 2, but the statistical significance was slightly relaxed to FDR ≤0.07, then 1531 and 1445 ESTs increased in levels in S. commersonii and S. tuberosum, respectively, with an overlap of 1111 transcripts (Fig. 2A). Thus, by making a small change in the statistical significance level, the overlap in cold-induced genes increased from 52% to 73%.
Fig. 2.
Cold- and CBF-regulated ESTs in both S. commersonii (Sc). and S. tuberosum (St). (A) The number of total ESTs that were either cold-induced or cold-repressed in Sc or St using 2-fold change and either FDR ≤0.05 or FDR ≤0.07. The results from 2, 24, and 168 h of cold treatment at 2 °C were combined. A given EST was counted only once regardless of the number of time points at which it was determined to be cold-regulated. (B) CBF regulons in Sc and St. CBF-regulated genes were selected as being differentially expressed in 35S::AtCBF3 transgenic lines compared with non-transformed plants with a 2-fold change cut-off and FDR ≤0.05.
In sum, the expression profiling experiments indicated that in S. commersonii and S. tuberosum plants, gene expression was altered in response to low temperature to similar degrees with similar kinetics and that there was considerable overlap in the genes that were cold-regulated in both plant species. The responses, however, were clearly not identical, indicating divergence in the cold regulatory programmes of the two closely related species.
Identification of CBF regulon genes in S. commersonii and S. tuberosum
Genes that comprise the CBF regulons of S. commersonii and S. tuberosum were defined as those that were determined to be cold-regulated in the experiments described above and correspondingly induced or repressed in response to constitutive overexpression of the A. thaliana CBF3 gene (AtCBF3) in transgenic plants grown at warm temperature. CBF-regulated genes were identified by transforming plants with the A. thaliana CBF3 gene placed under control of the constitutive CaMV 35S promoter (Fig. 3) and using the potato microarrays to identify the genes that were differentially expressed in the transformed plants using the lines shown in Fig. 3. Using a 2-fold change and FDR ≤0.05 as cut-off criteria, it was determined that of the 1532 cold-induced ESTs of S. commersonii and 1048 cold-induced ESTs of S. tuberosum, 160 (10%) and 170 (16%), respectively, were members of the CBF regulon (Fig. 2B). Of these, 54 ESTs were members of the CBF regulon in both S. commersonii and S. tuberosum. With regard to the 530 cold-repressed ESTs of S. commersonii and 688 cold-repressed ESTs of S. tuberosum, 137 (26%) and 364 (51%), respectively, were CBF regulon members (Fig. 2B). Of these, 91 were members of the CBF regulons of both S. commersonii and S. tuberosum (Fig. 2B). These results indicate that many of the CBF-regulated genes in S. commersonii are also members of the CBF regulon of S. tuberosum, but, additionally, that there is considerable divergence in the CBF regulatory programmes between the two species.
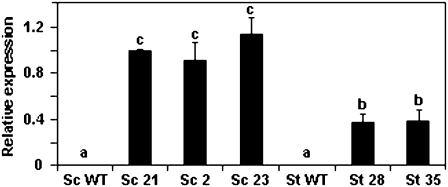
Fig. 3.
AtCBF3 transcript levels in S. tuberosum (St) and S. commersonii (Sc) transformed with 35S::AtCBF3. AtCBF3 transcript levels were determined by qRT-PCR in three Sc and two St 35S::AtCBF3 transgenic lines grown at 25 °C. qRT-PCR was performed using 100 ng of total RNA per sample. Relative expression was calculated using the potato 60S gene (clone STMCK67) as an internal reference. Relative expression of Sc 21 was set to 1 and the other lines were adjusted accordingly. The letters a, b, and c indicate statistically significant differences (ANOVA, P <0.05, n=2). Error bars indicate the standard deviation. WT, non-transformed plants.
Identification of cold-regulated genes conserved in both Solanum species and A. thaliana
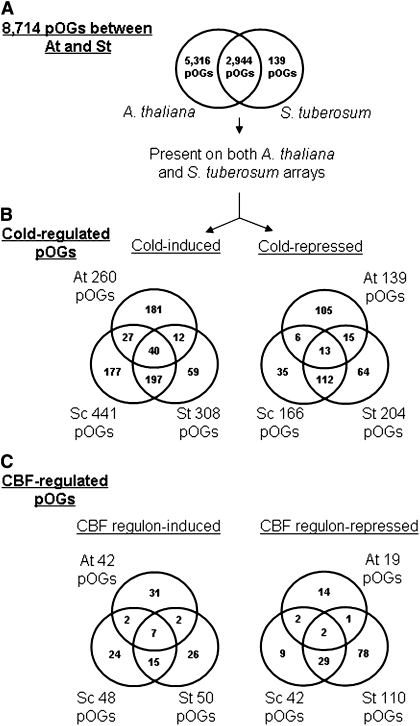
The microarray results identified 790 ESTs that were cold-induced in both S. commersonii and S. tuberosum, and another 383 ESTs that were cold-repressed in both potato species (Fig. 2A). A question raised was whether A. thaliana had orthologues of these potato ESTs and, if so, whether they too were cold-regulated. Given the relatively large evolutionary distance between A. thaliana and Solanum species [they diverged 112–156 MYA (Yang et al., 1999; Bell et al., 2005)], orthologous genes that are commonly responsive to cold conditions are likely to be conserved components of a fundamental cold response programme in dicots. To address this issue, pOGs between S. tuberosum ESTs and A. thaliana genes were first defined (see Materials and methods). A total of 8714 pOGs (Fig. 4A; Supplementary Table S4 at JXB online) were identified including 18 420 S. tuberosum ESTs (26% of the total) and 14 647 A. thaliana protein-coding genes (55% of the total). The low number of S. tuberosum ESTs assigned to pOGs is probably due to the fact that the pOGs were constructed using protein-encoding genes while the S. tuberosum ESTs not only include protein-encoding regions of mRNAs, but also probably included untranslated regions of mRNAs and transcripts from RNA genes. In addition, some of the ESTs may represent lineage-specific genes (Yang et al., 2009; Lin et al., 2010).
Fig. 4.
Comparison of cold- and CBF-regulated pOGs of S. tuberosum (St), S. commersonii (Sc), and A. thaliana (At). (A) A total of 8714 putative orthologous groups (pOGs) were identified between At and St. The Venn diagram shows pOGs with at least one At gene on the ATH1 chip, at least one potato clone on the potato cDNA array, or at least one gene from each species in both arrays (overlap). (B) Overlaps of cold-induced and cold-repressed pOGs in Sc, St, and At are shown from the pOGs present in both arrays in A. (C) Comparison of CBF regulon pOGs in Sc, St, and At. Overlaps of CBF-induced and CBF-repressed pOGs are shown based on the pOGs present in both arrays.
Of the 8714 pOGs between S. tuberosum and A. thaliana, 2944 (34%) (Fig. 4A) had at least one S. tuberosum EST present on the potato array and at least one A. thaliana gene represented on the ATH1 Affymetrix gene chip. These 2944 pOGs could thus be tested for conserved cold-regulated genes, though the estimate of conservation would be a minimal value as not all members of a given pOG were represented on both arrays. Analysis of the potato gene expression data indicated that 441 and 308 pOGs included at least one cold-induced gene from S. commersonii and S. tuberosum, respectively, and that, of these, 237 pOGs had at least one cold-induced gene from both species (Fig. 4B). Similarly, 166 and 204 pOGs included at least one cold-repressed gene in S. commersonii and S. tuberosum, respectively, with 125 pOGs having at least one cold-repressed gene in both species (Fig. 4B).
To determine whether A. thaliana had cold-regulated genes that belonged to these same pOGs, it was first necessary to develop a list of A. thaliana cold-regulated genes. This was accomplished by analysing the microarray data from previously published and new experiments comparing warm- and cold-treated plants (see Materials and methods). This resulted in the identification of 1151 cold-induced genes and 1095 cold-repressed genes (Supplementary Table S3 at JXB online). Of the 2944 pOGs that had at least one gene from A. thaliana represented on the ATH1 array, 260 had at least one cold-induced gene and 139 had at least one cold-repressed gene (Fig. 4B).
The results presented in Fig. 4B show that 40 pOGs included one or more cold-induced gene from each plant species and that 13 pOGs included one or more cold-repressed gene from all three species. The 40 conserved cold-induced pOGs included genes with a wide range of functions (Table 1). A number of the encoded proteins had previously been associated with ‘stress’, including two LEA (late embryogenesis abundant) proteins [LEA14 (Choi et al., 2002) and ERD10 (early responsive to desiccation) (Kiyosue et al., 1994)], ELIP1 (early light-inducible protein 1) (Hutin et al., 2003), HSP60 (Rikhvanov et al., 2007), sucrose synthase 1 (Dejardin et al., 1999), and protein phosphatase 2C (Umezawa et al., 2009). In addition, four transcription factors—Agamous-like 20 (AGL20)/Suppressor of Overexpression of CO (SOC1) (Lee et al., 2000), ADOF1 (Lijavetzky et al., 2003), HSFA8 (Chawade et al., 2007), and NAC019 (Jensen et al., 2010)—and nine proteins of unknown function were found to be conserved. As for the 13 conserved cold-repressed pOGs, six were associated with photosynthesis; they were either proteins located in the chloroplast or enzymes involved in photosynthesis (Table 2). Low temperature has been shown to down-regulate the expression of photosynthesis-related genes in a number of plant species (Wisniewski et al., 2008; Zhang et al., 2009).
Table 1.
Forty cold-induced pOGs from S. tuberosum (St), S.commersonii (Sc), and A. thaliana (At)
| St up | Sc up | At up | At description |
| STMDJ69 | STMGE83, STMDJ69 | AT2G18900 | Transducin family protein/WD-40 repeat family protein |
| STMIW06 | STMIW06 | AT4G25990 | Chloroplast import apparatus CIA2-like |
| STMIU11 | STMIU11 | AT5G60680, AT2G28400 | Unknown protein, unknown protein |
| STMCG52 | STMGV17, STMCG52 | AT2G45660 | AGL20 (Agamous-like 20) |
| STMCN22 | STMCN22 | AT5G65280 | GCL1 (GCR2-like 1); catalytic |
| STMEK16 | STMIQ63, STMEK16 | AT3G12670 | EMB2742 (embryo defective 2742) |
| STMIV71, STMIY51 | STMIV71, STMIY51 | AT1G27760 | Interferon-related developmental regulator family protein/FRD protein family |
| STMJG77, STMIU74 | STMIU74 | AT5G01880 | Zinc finger (C3HC4-type RING finger) family protein |
| STMEP26 | STMEP26 | AT5G26920 | Calmodulin binding |
| STMEO27 | STMEO27 | AT2G33210, AT3G23990 | Chaperonin, putative HSP60 (heat shock protein 60) |
| STMIU32, STMID24 | STMIU32, STMID24 | AT3G53230 | Cell division cycle protein 48 putative (CDC48) |
| STMIQ26, STMJI56, STMGA34 | STMIQ26, STMJI56, STMGA34 | AT1G01470 | LEA14 (late embryogenesis abundant 14) |
| STMDH66, STMJL22 | STMJL22 | AT4G35940 | Unknown protein |
| STMEI36, STMEQ55 | STMEI36 | AT1G31660 | Unknown protein |
| STMET41 | STMET41 | AT1G25400 | Unknown protein |
| STMGF95 | STMGF95 | AT1G51700 | ADOF1 (Arabidopsis dof zinc finger protein 1) |
| STMIY82 | STMIY82 | AT1G52890, AT4G27410 | ANAC019 (Arabidopsis NAC domain-containing protein 19), RD26 (responsive to dessication 26) |
| STMDO86 | STMDS75, STMDO86 | AT3G16810 | APUM24 (Arabidopsis pumilio 24) |
| STMEW81, STMCB90 | STMEW81, STMCB90 | AT5G62360, AT5G62350 | Invertase/pectin methylesterase inhibitor family protein invertase/pectin methylesterase inhibitor family protein (DC 1.2 homologue) |
| STMHE19 | STMHE19 | AT5G20830 | SUS1 (sucrose synthase 1) |
| STMDP77 | STMDP77 | AT4G29780 | Unknown protein |
| STMGH65, STMJJ17 | STMHG34, STMJJ17 | AT4G30290, AT5G48070 | ATXTH19, ATXTH20 (xyloglucan endotransglucosilase hydrolase 19 and 20) |
| STMHA92 | STMHA92 | AT1G67970 | AT-HSFA8 (Arabidopsis thaliana heat shock transcription factor A8) |
| STMHS29 | STMHS29 | AT1G42440 | Unknown protein |
| STMGG79 | STMGG79 | AT3G55510 | Unknown protein |
| STMCX87 | STMCX87 | AT5G16010 | 3-Oxo-5-alpha-steroid 4-dehydrogenase family protein/steroid 5-alpha-reductase family protein |
| STMDU38 | STMHN39, STMDU38 | AT1G80270 | DNA-binding protein putative |
| STMHO64 | STMHO64 | AT2G17270 | Mitochondrial substrate carrier family protein |
| STMIH78, STMGL16 | STMIH78, STMGL16 | AT4G27940 | Mitochondrial substrate carrier family protein |
| STMED50 | STMED50 | AT4G33905, AT2G14860 | Peroxisomal membrane protein 22 kDa putative |
| STMGR56 | STMGR56 | AT4G28450 | Transducin family protein/WD-40 repeat family protein |
| STMIP59 | STMIP59 | AT1G32860 | Glycosyl hydrolase family 17 protein |
| STMHT66 | STMHT66 | AT4G31140 | Glycosyl hydrolase family 17 protein |
| STMIO48 | STMIO48 | AT4G00640 | Unknown protein |
| STMGU17 | STMGU17 | AT4G12000 | Unknown protein |
| STMHS17 | STMHS17 | AT3G11410, AT1G07430 | ATPP2CA (Arabidopsis protein phosphatase 2CA), protein phosphatase 2C putative |
| STMJO29 | STMJO29 | AT1G20450, AT1G20440, AT1G76180 | ERD10/LTI45 (early responsive to dehydration 10), COR47 (cold regulated 47), ERD14 (early responsive to dehydration 14) |
| STMJO47, STMIX48 | STMJO47, STMIX48 | AT3G22840, AT4G14690 | ELIP1 (early light-inducible protein) chlorophyll binding, ELIP2 (early light-inducible protein 2) chlorophyll binding |
| STMHO88, STMHT73 | STMHO88, STMHT73 | AT5G07990 | TT7 (transparent testa 7) |
| STMGJ81 | STMGJ81 | AT1G53645 | Hydroxyproline-rich glycoprotein family protein |
Table 2.
Thirteen cold-repressed pOGs from S. tuberosum (St), S. commersonii (Sc), and A. thaliana (At)
| St down | Sc down | At down | At description |
| STMDV46 | STMDV46 | AT1G09750 | Chloroplast nucleoid DNA-binding protein-related |
| STMIV36 | STMIV36 | AT2G39470 | PPL2 (PSBP-like protein 2) |
| STMCX38 | STMCX38 | AT3G16150 | L-Asparaginase putative |
| STMER63 | STMDB57, STMER63, STMDB57 | AT3G23730 | Xyloglucan:xyloglucosyl transferase putative |
| STMGO23 | STMGO23 | AT4G14540 | CCAAT-box-binding transcription factor subunit B (NF-YB) (HAP3) |
| STMCR16, STMCL01, STMCV75, STMIV24, STMCK44 | STMCR16, STMCL01, STMCV75, STMIV24, STMCK44 | AT1G70410, AT3G01500 | Carbonic anhydrase putative, carbonate dehydratase putative, CA1 (carbonic anhydrase 1) |
| STMEP82, STMCS89 | STMCS89 | AT1G48600 | Phosphoethanolamine N-methyltransferase 2 putative (NMT2) |
| STMCD65 | STMCD65 | AT5G56870 | BGAL4 (beta-galactosidase 4) |
| STMGX24 | STMGX24 | AT5G35790 | G6PD1 (glucose-6-phosphate dehydrogenase 1) |
| STMJH69 | STMJH69 | AT3G15840 | PIFI (post-illumination chlorophyll fluorescence increase) |
| STMIM55 | STMIM55 | AT1G32080 | Membrane protein putative |
| STMDB78, STMCQ55 | STMCQ55 | AT1G73330 | ATDR4 (Arabidopsis thaliana drought-repressed 4) |
| STMJD18 | STMJD18 | AT4G25260, AT4G12390 | Invertase/pectin methylesterase inhibitor family protein PME1; pectinesterase inhibitor |
Identification of the CBF regulon genes conserved in both Solanum species and A. thaliana
The A. thaliana CBF regulon was defined as those genes for which transcript levels either increased or decreased at least 2-fold (FDR ≤0.05) in response to constitutive overexpression of CBF2 at warm temperature in A. thaliana transgenic lines (Vogel et al., 2005) and were also included in the list of cold-induced or cold-repressed genes described above. This resulted in the assignment of 169 cold-induced genes and 58 cold-repressed genes to the A. thaliana CBF regulon (Supplementary Tables S5, S6 at JXB online). These genes, as well as the genes that were determined to comprise the CBF regulons of S. commersonii and S. tuberosum (Fig. 2B), were then assigned to pOGs (Fig. 4C). An inspection of the resulting Venn diagrams indicated that of the 22 cold-induced CBF regulon pOGs conserved in both Solanum species, only seven were members of the A. thaliana CBF regulon, and that of the 31 cold-repressed CBF regulon pOGs conserved in both Solanum species, only two were members of the A. thaliana CBF regulon. The seven conserved cold-induced CBF regulon pOGs encoded ELIP1, LEA14, ERD10, COR47, ADOF1, RD26, SUS1, and an invertase/pectin methylesterase inhibitor family protein. The two conserved cold-repressed CBF regulon pOGs encoded a phosphoethanolamine N-methyltransferase and β-galactosidase. These data indicate that there is considerable divergence in the CBF regulons of A. thaliana and the two Solanum species.
Conservation of rapidly induced transcription factors in both Solanum species and A. thaliana
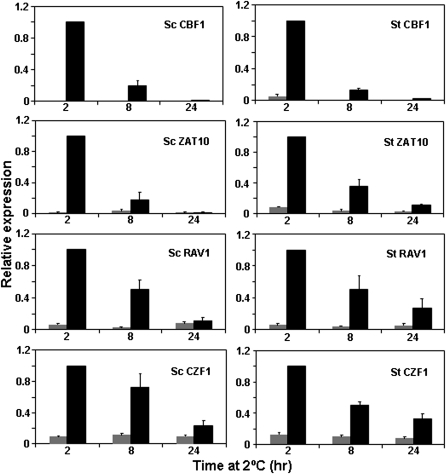
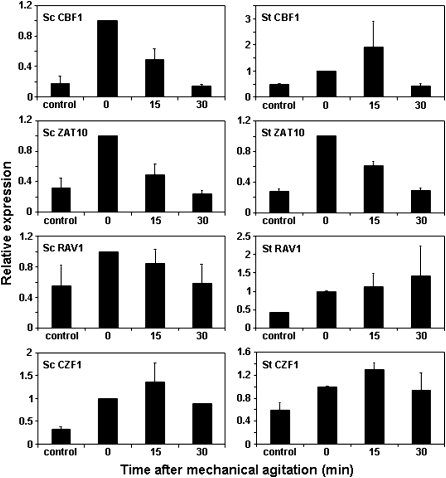
In A. thaliana, there are a number of genes encoding transcription factors that are induced rapidly in response to low temperature with similar kinetics to CBF1, CBF2, and CBF3. These genes include CZF1, ZAT10, and RAV1 (Vogel et al., 2005). Like CBF1, CBF2, and CBF3, these genes are also induced in response to mechanical agitation. To explore the conservation of low temperature regulatory programmes, it was of interest to determine whether S. tuberosum and S. commersonii had potential orthologues of CZF1, ZAT10, and RAV1 and, if so, whether they were induced quickly in response to low temperature and in response to mechanical agitation. Among the potato ESTs, potential orthologues of these genes were identified and their regulation was tested. The results indicated that all three of these genes were quickly induced in response to low temperature (Fig. 5) and mechanical agitation (Fig. 6) in both potato species.
Fig. 5.
Transcript accumulation of S. commersonii (Sc) and S. tuberosum (St) orthologues of CBF1, ZAT10, RAV1, and CZF1 in response to low temperature. Sc and St plants were grown for 3 weeks under a 16 h photoperiod. Eight hours after dawn plants were either transferred to 2 °C (black bars) for 2, 8, and 24 h or kept at 25 °C (grey bars) for the same periods of time. qRT-PCR analysis was performed to determine the transcript levels of Sc and St genes. Average values of three different experiments are shown. The relative expression level of each transcript was normalized using the potato 60S gene (clone STMCK67) as an internal reference. Relative expression of the Sc 2 h cold sample was set to 1 and the other values were adjusted accordingly. Error bars indicate the SE.
Fig. 6.
Transcript accumulation of S. commersonii (Sc) and S. tuberosum (St) orthologues of CBF1, ZAT10, RAV1, and CZF1 in response to mechanical agitation. Three-week-old plants grown in magenta boxes were mechanically agitated for 15 min (see Materials and methods). Tissue was collected from two plants at 0, 15, and 30 min after the mechanical agitation treatment. The control sample received no mechanical agitation. qRT-PCR analysis was performed to determine the transcript levels of Sc and St genes. Average values of two different experiments are shown. The relative expression level of each transcript was normalized using the potato 60S gene (clone STMCK67) as an internal reference. The relative expression of the Sc 0 min sample was set to 1 and the other values were adjusted accordingly. Error bars indicate the SE.
Discussion
One of the fundamental goals of cold acclimation research is to understand the molecular basis for the differences in freezing tolerance between plant species that are evolutionarily closely related. Here this issue is addressed through a comparative transcriptome analysis of two closely related potato species—S. commersonii, which cold acclimates, and S. tuberosum (cv. Umatilla), which does not. The present focus on the transcriptome was motivated by the fact that cold acclimation is known to involve changes in gene expression. The goal of the study was to explore whether there might be an obvious ‘macro-scale’ explanation for the differences in these two potato species in their ability to cold acclimate. In short, the expression profiling experiments did not provide such an explanation. Both plants altered gene expression in response to low temperature to similar degrees with similar kinetics. Moreover, there was considerable overlap in the cold-regulated gene sets. Thus, the differences in freezing tolerance between S. tuberosum and S. commersonii must trace to specific differences in their responses to low temperature. Indeed, although there was considerable overlap in the genes that were cold-regulated in both plant species, the results point to both qualitative and quantitative differences in the cold-regulated gene sets of the two species (Figs 1, 2). However, at this point, the specific differences observed must be viewed with caution. Some of these differences are likely to be only apparent, as a small change in statistical criteria used for the analysis had a large effect on the percentage overlap of cold-induced genes (Fig. 2). Moreover, the microarray that was used to monitor gene expression was based on ESTs from S. tuberosum. Given that there are notable differences in the nucleotide sequences of S. tuberosum and S. commersonii—the synonymous substitution rate between these two species is ∼0.04 (see Materials and methods)—it is possible that some of the genes that were observed to be cold-regulated in S. tuberosum, but not in S. commersonii, might be due to nucleotide mismatches between the S. commersonii transcripts and the probe ESTs (those genes that were found to be cold regulated in S. commersonii, but not S. tuberosum, would be less likely to be subject to this possible limitation). Future studies will be directed at detailing the qualitative and quantitative differences between the transcriptomes of these two potato species. These efforts will be greatly facilitated by the recently completed sequencing of the potato genome (http://potatogenomics.plantbiology.msu.edu/) and the rapidly developing technologies for RNA sequencing.
The present findings are similar to those of Oufir et al. (2008) who used the same potato array to compare the low temperature transcriptome of S.tuberosum (cv. Desiree), which they showed does not cold acclimate, with that of S. phureja CHS (diploid potato), which they showed does cold acclimate. They found that 213 genes were cold-induced and 101 cold-repressed ≥2-fold in S. tuberosum cv. Desiree and that 92 genes were induced and 159 repressed in S. phureja. Only 22 genes were found to be cold-induced and 15 to be cold-repressed in both species.
A second objective of this study was to determine whether macro-scale differences in the composition of the CBF regulons of S. tuberosum and S. commersonii might account for their differences in ability to cold acclimate. One specific question was whether the CBF regulon of S. tuberosum was much smaller than that of S. commersonii. As noted earlier, the CBF regulons of freezing-sensitive tomato (Zhang et al., 2004) and rice (Dubouzet et al., 2003) have been reported to be composed of as few as 10 cold-induced genes, whereas the CBF regulon of A. thaliana includes at least 85 cold-induced genes (Vogel et al., 2005). The present results, however, indicate that the sizes of the S. tuberosum and S. commersonii CBF regulons are not appreciably different; the number of cold-induced genes assigned to the regulons was 170 and 160, respectively (Fig. 2B). Moreover, the results indicate that the CBF regulons of both S. tuberosum and S. commersonii are much larger than that of A. thaliana. Indeed, the potato arrays used here included probes for only ∼10 000 genes. Thus, if the probes on this array are reflective of the overall genome, it could be that the CBF regulons of these two potato species each comprise nearly a thousand cold-induced genes.
While the difference in freezing tolerance between S. commersonii and S. tuberosum does not appear to involve a significant difference in size of their CBF regulons, the results suggest that the regulons have diverged considerably (Fig. 2B). The results indicate that of the 160 cold-induced genes that were up-regulated by AtCBF3 overexpression in S. commersonii, only 54 were up-regulated by AtCBF3 overexpression in S. tuberosum (as alluded to above, those genes that were found to be induced in S. commersonii, but not in S. tuberosum, are not likely to be artefacts of using ESTs from S. tuberosum). One might therefore speculate that the genes that are members of the CBF regulon of S. commersonii, but not members of the S. tuberosum CBF regulon, have critical roles in freezing tolerance. However, this must not be the case as constitutive overexpression of AtCBF3 has been shown to increase the freezing tolerance of S. tuberosum without a cold treatment; whereas the LT50 for non-acclimated wild-type S. tuberosum plants was –3.0 °C, non-acclimated transgenic S. tuberosum plants expressing AtCBF3 had an LT50 of –5.0 °C (Pino et al., 2007). Thus, S. tuberosum has genes that can impart freezing tolerance, and at least some of these are members of the AtCBF3 regulon.
The basic question thus raised is: why does S. tuberosum not increase in freezing tolerance in response to low temperature? At this point it is known that the answer is not simply a matter of S. commersonii having CBF genes that are cold-induced and S. tuberosum not having such genes. Both S. tuberosum and S. commersonii have two CBF loci (Pennycooke et al., 2008). At one locus, both S. commersonii and S. tuberosum have four CBF genes, one of which, CBF1, is induced in response to low temperature. At the second locus, S. commersonii has one CBF gene, CBF4, and S. tuberosum has two CBF genes, CBF4 and CBF5, and in both species the CBF4 gene is cold-induced. The implication is that the genes that are targeted by the CBF1 and CBF4 proteins of S. tuberosum do not include all of those targeted by AtCBF3 and that this difference is the fundamental reason why S. tuberosum does not cold acclimate. Transgenic S. tuberosum plants that express each of the S. commersonii and S. tuberosum CBF genes are currently being made to test this possibility.
One note of caution is that the genes that were identified here as members of the S. tuberosum AtCBF3 regulon are not likely to be sufficient for freezing tolerance. The first obvious reason is that the arrays that were used to monitor gene expression only included probes for ∼10 000 genes. Thus, there are certain to be many genes that comprise the AtCBF3 regulon that are not on the current list and might have roles in freezing tolerance. However, even more important is that the definition of the AtCBF3 regulon used here is likely specifically to exclude the genes with roles in freezing tolerance. For genes to be assigned to the regulon, they must be induced or repressed in response to AtCBF3 overexpression in transgenic plants grown at warm non-acclimating temperatures and also be correspondingly induced or repressed in response to low temperature. Therefore, all of the genes in the present CBF regulon list are cold-regulated genes in wild-type S. tuberosum. However, given that AtCBF3 overexpression imparts freezing tolerance and low temperature does not, it would appear that the genes that are regulated by AtCBF3 and impart freezing tolerance are not cold-regulated in S. tuberosum and, therefore, would not be included in the present CBF regulon gene list. One might then think that those genes that are induced or repressed in response to AtCBF3 expression, but are not cold regulated, would be among those that are most critical to freezing tolerance. The problem in identifying these critical genes is that AtCBF3 overexpression in S. tuberosum (Pino et al., 2007), as is the case in A. thaliana (Liu et al., 1998; Gilmour et al., 2000), affects multiple aspects of plant growth and development; AtCBF3-overexpressing plants have a dwarf stature, for instance. Thus, many of the genes that are affected in plants overexpressing AtCBF3 are likely to be secondary effects of CBF expression and not related directly to freezing tolerance.
The third objective of this study was to identify cold-regulated genes that were conserved between the two Solanum species and A. thaliana. Given the relatively large evolutionary distance between these plants, such genes might have fundamental roles in low temperature biology. Due to the fact that the potato array used includes <25% of the potato genome, the present analysis was significantly limited in scope. Nevertheless, 53 pOGs were identified, 40 of which include cold-induced genes (Table 1) and 13 of which include cold-repressed genes (Table 2). Some of the conserved genes had previously been associated with stress, including genes encoding LEA and ELIP proteins. However, the largest group of conserved genes encoded proteins of unknown function. The study of these genes may reveal novel mechanisms fundamental to plants coping with low temperature. In addition, it was found that the low temperature regulatory programmes of these three species have seven transcription factors in common in addition to CBF genes. Moreover, four of these genes—CBF1, ZAT10, RAV1, and CZF1—were found to be induced not only in response to low temperature (Fig. 5), but also in response to mechanical agitation (Fig. 6). Thus, not only are the genes conserved, but regulatory mechanisms controlling expression of these genes have conserved elements beyond low temperature per se.
Comparative studies of genome-wide transcript profiles have revealed conserved components in core biological processes and divergent expression patterns across species that may be important for taxa-specific processes. For example, transcript profiling data from three highly divergent species including Caenorhabditis elegans, Drosohphila melanogaster, and budding yeast have been used to dissect expression conservation and divergence among orthologous genes (McCarroll et al., 2004). Expression patterns remain conserved among these three species for genes involved in fundamental biological processes such as mitochondrial metabolism, DNA repair, and cellular transport. However, most expression profiles are species specific. Even between relatively closely related species, a substantial number of genes show significant differences in expression levels. In a comparison between D. melanogaster, D. simulans, and D. yakuba, species that diverged ∼2.5–5 MYA, ∼20–50% of the orthologous genes showed expression differences (Ranz et al., 2003; Rifkin et al., 2003). Similarly, in a comparison of Saccharomyces cerevisiae and S. paradoxus, species that diverged ∼5 MYA, the expression patterns of orthologous genes under various environmental stresses showed extensive differences (Tirosh et al., 2006). In addition, due to a high rate of gene duplication, losses and gains of stress responses have occurred readily among plant duplicate genes that contribute to expression divergence between species (Zou et al., 2009). These findings are largely consistent with the apparent divergence among cold-responsive genes of S. tuberosum and S. commersonii. Moreover, given the large evolutionary distance between A. thaliana and the two Solanum species (diverged ∼112–156 MYA; Yang et al., 1999; Bell et al., 2005), it was not surprising to find a low degree of conservation of cold-responsive genes between these species. Indeed, the genes that were found to be conserved over such evolutionary time are likely to be members of a core cold response network that have fundamental roles in the low temperature biology of plants.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in qRT-PCR.
Table S2. Affymetrix cel files used to identify A. thaliana cold-regulated genes.
Table S3. Arabidopsis thaliana cold-regulated genes.
Table S4. List of 8714 pOGs between A. thaliana and S. tuberosum.
Table S5. Arabidopsis thaliana up-regulated CBF regulon genes.
Table S6. Arabidopsis thaliana down-regulated CBF regulon genes.
Acknowledgments
We thank Sarah Gilmour for assistance in preparing this manuscript for publication. The research reported was supported by grants from the National Science Foundation Plant Genome Program (DBI 0110124), the Chemical Sciences, Geosciences and Biosciences Division, Office of Basic Energy Sciences, US Department of Energy (DE-FG02-91ER20021), and the Michigan Agricultural Experiment Station.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell CD, Soltis DE, Soltis PS. The age of the angiosperms: a molecular timescale without a clock. Evolution. 2005;59:1245–1258. [PubMed] [Google Scholar]
- Benedict C, Skinner JS, Meng R, Chang Y, Bhalerao R, Huner NP, Finn CE, Chen TH, Hurry V. The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant, Cell and Environment. 2006;29:1259–1272. doi: 10.1111/j.1365-3040.2006.01505.x. [DOI] [PubMed] [Google Scholar]
- Chawade A, Brautigam M, Lindlof A, Olsson O, Olsson B. Putative cold acclimation pathways in Arabidopsis thaliana identified by a combined analysis of mRNA co-expression patterns, promoter motifs and transcription factors. BMC Genomics. 2007;8:304. doi: 10.1186/1471-2164-8-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Li PH. Characteristics of cold acclimation and deacclimation in tuber-bearing Solanum species. Plant Physiology. 1980;65:1146–1148. doi: 10.1104/pp.65.6.1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V, Zhu J, Zhu JK. Cold stress regulation of gene expression in plants. Trends in Plant Science. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Choi DW, Rodriguez EM, Close TJ. Barley Cbf3 gene identification, expression pattern, and map location. Plant Physiology. 2002;129:1781–1787. doi: 10.1104/pp.003046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin A, Sokolov LN, Kleczkowski LA. Sugar/osmoticum levels modulate differential abscisic acid-independent expression of two stress-responsive sucrose synthase genes in Arabidopsis. Biochemical Journal. 1999;344:503–509. [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. The Plant Journal. 2003;33:751–763. doi: 10.1046/j.1365-313x.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Using the quantitative genetic threshold model for inferences between and within species. Philosophical Transactions of the Royal Society B: Biological Sciences. 2005;360:1427–1434. doi: 10.1098/rstb.2005.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler S, Thomashow MF. Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. The Plant Cell. 2002;14:1675–1690. doi: 10.1105/tpc.003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier L, Cope L, Bolstad BM, Irizarry RA. affy—analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- Gentleman R, Carey VJ, Huber WH, Irizarry RA, Dudoit S. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer Science; 2005. pp. 15–46. [Google Scholar]
- Gilmour SJ, Fowler SG, Thomashow MF. Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Molecular Biology. 2004;54:767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4. [DOI] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Zarka DG, Stockinger EJ, Salazar MP, Houghton JM, Thomashow MF. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. The Plant Journal. 1998;16:433–442. doi: 10.1046/j.1365-313x.1998.00310.x. [DOI] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiology. 2008;148:993–1003. doi: 10.1104/pp.108.122457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh TH, Lee JT, Yang PT, Chiu LH, Charng YY, Wang YC, Chan MT. Heterology expression of the Arabidopsis C-repeat/dehydration response element binding factor 1 gene confers elevated tolerance to chilling and oxidative stresses in transgenic tomato. Plant Physiology. 2002;129:1086–1094. doi: 10.1104/pp.003442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutin C, Nussaume L, Moise N, Moya I, Kloppstech K, Havaux M. Early light-induced proteins protect Arabidopsis from photooxidative stress. Proceedings of the National Academy of Sciences, USA. 2003;100:4921–4926. doi: 10.1073/pnas.0736939100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen KR, Gilmour SJ, Zarka DG, Schabenberger O, Thomashow MF. Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science. 1998;280:104–106. doi: 10.1126/science.280.5360.104. [DOI] [PubMed] [Google Scholar]
- Jaglo KR, Kleff S, Amundsen KL, Zhang X, Haake V, Zhang JZ, Deits T, Thomashow MF. Components of the Arabidopsis C-repeat/dehydration-responsive element binding factor cold-response pathway are conserved in Brassica napus and other plant species. Plant Physiology. 2001;127:910–917. [PMC free article] [PubMed] [Google Scholar]
- Jensen MK, Kjaersgaard T, Nielsen MM, Galberg P, Petersen K, O'Shea C, Skriver K. The Arabidopsis thaliana NAC transcription factor family: structure–function relationships and determinants of ANAC019 stress signalling. Biochemical Journal. 2010;426:183–196. doi: 10.1042/BJ20091234. [DOI] [PubMed] [Google Scholar]
- Kasuga M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. A combination of the Arabidopsis DREB1A gene and stress-inducible rd29A promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant and Cell Physiology. 2004;45:346–350. doi: 10.1093/pcp/pch037. [DOI] [PubMed] [Google Scholar]
- Kilian J, Whitehead D, Horak J, Wanke D, Weinl S, Batistic O, D'Angelo C, Bornberg-Bauer E, Kudla J, Harter K. The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. The Plant Journal. 2007;50:347–363. doi: 10.1111/j.1365-313X.2007.03052.x. [DOI] [PubMed] [Google Scholar]
- Kiyosue T, Yamaguchi-Shinozaki K, Shinozaki K. Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant and Cell Physiology. 1994;35:225–231. [PubMed] [Google Scholar]
- Lee H, Suh SS, Park E, Cho E, Ahn JH, Kim SG, Lee JS, Kwon YM, Lee I. The AGAMOUS-LIKE 20 MADS domain protein integrates floral inductive pathways in Arabidopsis. Genes and Development. 2000;14:2366–2376. doi: 10.1101/gad.813600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt J. Responses of plants to environmental stresses. 2nd edn. New York: Academic Press; 1980. [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J. Genome-wide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionary Biology. 2003;3:17. doi: 10.1186/1471-2148-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Moghe G, Ouyang S, Iezzoni A, Shiu SH, Gu X, Buell CR. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evolutionary Biology. 2010;10:41. doi: 10.1186/1471-2148-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama K, Sakuma Y, Kasuga M, Ito Y, Seki M, Goda H, Shimada Y, Yoshida S, Shinozaki K, Yamaguchi-Shinozaki K. Identification of cold-inducible downstream genes of the Arabidopsis DREB1A/CBF3 transcriptional factor using two microarray systems. The Plant Journal. 2004;38:982–993. doi: 10.1111/j.1365-313X.2004.02100.x. [DOI] [PubMed] [Google Scholar]
- McCarroll SA, Murphy CT, Zou S, Pletcher SD, Chin CS, Jan YN, Kenyon C, Bargmann CI, Li H. Comparing genomic expression patterns across species identifies shared transcriptional profile in aging. Nature Genetics. 2004;36:197–204. doi: 10.1038/ng1291. [DOI] [PubMed] [Google Scholar]
- Medina J, Bargues M, Terol J, Perez-Alonso M, Salinas J. The Arabidopsis CBF gene family is composed of three genes encoding AP2 domain-containing proteins whose expression is regulated by low temperature but not by abscisic acid or dehydration. Plant Physiology. 1999;119:463–469. doi: 10.1104/pp.119.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moniz de Sa M, Drouin G. Phylogeny and substitution rates of angiosperm actin genes. Molecular Biology and Evolution. 1996;13:1198–1212. doi: 10.1093/oxfordjournals.molbev.a025685. [DOI] [PubMed] [Google Scholar]
- Oufir M, Legay S, Nicot N, Van Moer K, Hoffmann L, Renaut J, Hausman J, Evers D. Gene expression in potato during cold exposure: changes in carbohydrate and polyamine metabolisms. Plant Science. 2008;175:839–852. [Google Scholar]
- Pennycooke JC, Cheng H, Roberts SM, Yang Q, Rhee SY, Stockinger EJ. The low temperature-responsive, Solanum CBF1 genes maintain high identity in their upstream regions in a genomic environment undergoing gene duplications, deletions, and rearrangements. Plant Molecular Biology. 2008;67:483–497. doi: 10.1007/s11103-008-9333-5. [DOI] [PubMed] [Google Scholar]
- Pino MT, Skinner JS, Park EJ, Jeknic Z, Hayes PM, Thomashow MF, Chen TH. Use of a stress inducible promoter to drive ectopic AtCBF expression improves potato freezing tolerance while minimizing negative effects on tuber yield. Plant Biotechnology Journal. 2007;5:591–604. doi: 10.1111/j.1467-7652.2007.00269.x. [DOI] [PubMed] [Google Scholar]
- Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–1745. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Meyerowitz EM. The AP2/EREBP family of plant transcription factors. Biological Chemistry. 1998;379:633–646. doi: 10.1515/bchm.1998.379.6.633. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Kim J, White KP. Evolution of gene expression in the Drosophila melanogaster subgroup. Nature Genetics. 2003;33:138–144. doi: 10.1038/ng1086. [DOI] [PubMed] [Google Scholar]
- Rikhvanov EG, Gamburg KZ, Varakina NN, Rusaleva TM, Fedoseeva IV, Tauson EL, Stupnikova IV, Stepanov AV, Borovskii GB, Voinikov VK. Nuclear–mitochondrial cross-talk during heat shock in Arabidopsis cell culture. The Plant Journal. 2007;52:763–778. doi: 10.1111/j.1365-313X.2007.03275.x. [DOI] [PubMed] [Google Scholar]
- Shinwari ZK, Nakashima K, Miura S, Kasuga M, Seki M, Yamaguchi-Shinozaki K, Shinozaki K. An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochemical and Biophysical Research Communications. 1998;250:161–170. doi: 10.1006/bbrc.1998.9267. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Statistical Applications in Genetics and Molecular Biology. 2004;3 doi: 10.2202/1544-6115.1027. article3. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proceedings of the National Academy of Sciences, USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proceedings of the National Academy of Sciences, USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Weinberger A, Carmi M, Barkai N. A genetic signature of interspecies variations in gene expression. Nature Genetics. 2006;38:830–834. doi: 10.1038/ng1819. [DOI] [PubMed] [Google Scholar]
- Umezawa T, Sugiyama N, Mizoguchi M, Hayashi S, Myouga F, Yamaguchi-Shinozaki K, Ishihama Y, Hirayama T, Shinozaki K. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buskirk HA, Thomashow MF. Arabidopsis transcription factors regulating cold acclimation. Physiologia Plantarum. 2006;126:72–80. [Google Scholar]
- Vogel JT, Zarka DG, Van Buskirk HA, Fowler SG, Thomashow MF. Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. The Plant Journal. 2005;41:195–211. doi: 10.1111/j.1365-313X.2004.02288.x. [DOI] [PubMed] [Google Scholar]
- Wilson CL, Miller CJ. Simpleaffy: a BioConductor package for Affymetrix quality control and data analysis. Bioinformatics. 2005;21:3683–3685. doi: 10.1093/bioinformatics/bti605. [DOI] [PubMed] [Google Scholar]
- Wisniewski M, Bassett C, Norelli J, Macarisin D, Artlip T, Gasic K, Korban S. Expressed sequence tag analysis of the response of apple (Malus×domestica ‘Royal Gala’) to low temperature and water deficit. Physiologia Plantarum. 2008;133:298–317. doi: 10.1111/j.1399-3054.2008.01063.x. [DOI] [PubMed] [Google Scholar]
- Xue GP. An AP2 domain transcription factor HvCBF1 activates expression of cold-responsive genes in barley through interaction with a (G/a) (C/t)CGAC motif. Biochimica et Biophysica Acta. 2002;1577:63–72. doi: 10.1016/s0167-4781(02)00410-4. [DOI] [PubMed] [Google Scholar]
- Yang X, Jawdy S, Tschaplinski TJ, Tuskan GA. Genome-wide identification of lineage-specific genes in Arabidopsis, Oryza and Populus. Genomics. 2009;93:473–480. doi: 10.1016/j.ygeno.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Yang YW, Lai KN, Tai PY, Li WH. Rates of nucleotide substitution in angiosperm mitochondrial DNA sequences and dates of divergence between Brassica and other angiosperm lineages. Journal of Molecular Evolution. 1999;48:597–604. doi: 10.1007/pl00006502. [DOI] [PubMed] [Google Scholar]
- Zhang C, Fei SZ, Warnke S, Li L, Hannapel D. Identification of genes associated with cold acclimation in perennial ryegrass. Journal of Plant Physiology. 2009;166:1436–1445. doi: 10.1016/j.jplph.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Zhang X, Fowler SG, Cheng H, Lou Y, Rhee SY, Stockinger EJ, Thomashow MF. Freezing-sensitive tomato has a functional CBF cold response pathway, but a CBF regulon that differs from that of freezing-tolerant Arabidopsis. The Plant Journal. 2004;39:905–919. doi: 10.1111/j.1365-313X.2004.02176.x. [DOI] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH. Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genetics. 2009;5:e1000581. doi: 10.1371/journal.pgen.1000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.