Abstract
Over the past decades, considerable advances have been made in understanding the crucial role and the regulation of sucrose metabolism in plants. Among the various sucrose-catabolizing enzymes, alkaline/neutral invertases (A/N-Invs) have long remained poorly studied. However, recent findings have demonstrated the presence of A/N-Invs in various organelles in addition to the cytosol, and their importance for plant development and stress tolerance. A cytosolic (At-A/N-InvG, At1g35580) and a mitochondrial (At-A/N-InvA, At1g56560) member of the A/N-Invs have been analysed in more detail in Arabidopsis and it was found that At-A/N-InvA knockout plants show an even more severe growth phenotype than At-A/N-InvG knockout plants. The absence of either A/N-Inv was associated with higher oxidative stress defence gene expression, while transient overexpression of At-A/N-InvA and At-A/N-InvG in leaf mesophyll protoplasts down-regulated the oxidative stress-responsive ascorbate peroxidase 2 (APX2) promoter. Moreover, up-regulation of the APX2 promoter by hydrogen peroxide or abscisic acid could be blocked by adding metabolizable sugars or ascorbate. A hypothetical model is proposed in which both mitochondrial and cytosolic A/N-Invs can generate glucose as a substrate for mitochondria-associated hexokinase, contributing to mitochondrial reactive oxygen species homeostasis.
Keywords: Arabidopsis thaliana, ascorbate peroxidase, hexokinase, mitochondria, neutral invertase, oxidative stress
Introduction
Sucrose (Suc) and its cleavage products glucose (Glc) and fructose (Fru) are central molecules for cellular biosynthesis and signal transduction throughout the plant life cycle (Smeekens et al., 2010). In general, high Glc levels favour cell division and expansion, while high Suc levels are associated with differentiation and maturation (Weber et al., 1995). Suc needs to be cleaved before it can be used as a carbon and energy source, and this is catalysed by two types of enzymes: sucrose synthases (Susys) reversibly hydrolyse Suc to UDPGlc and Fru, whereas invertases (Invs) catalyse an irreversible hydrolysis to Glc and Fru.
Invertases can be further classified into two major groups according to their pH optimum: the acid invertases (Ac-Invs) and neutral/alkaline invertases (A/N-Invs). Ac-Invs are glycosylated proteins occurring in the vacuole (Vac-Invs) or in the apoplast (Cw-Invs) and belong to glycoside hydrolase family 32 (GH32). A/N-Invs, on the other hand, are non-glycosylated and are classified in family GH100 (Lammens et al., 2009). A/N-Invs have been poorly studied at the native protein level, mainly because of apparent protein instability and their low expression levels in plant tissues. Only a few groups have succeeded in a complete purification and characterization of this type of enzyme (Chen and Black, 1992; Van den Ende and Van Laere, 1995; Lee and Sturm, 1996; Ross et al., 1996; Walker et al., 1997). A peculiar characteristic of A/N-Invs is the inhibition of their activity by TRIS, a commonly used buffer (Van den Ende and Van Laere, 1995). Possibly, TRIS mimicks a so far unidentified inhibitor acting in planta. A/N-Inv genes have been studied in Arabidopsis thaliana (Ji et al., 2005), Lolium temulentum (Gallagher and Pollock, 1998), Daucus carota (Sturm et al., 1999), Beta vulgaris (Gonzalez and Cejudo, 2007), Oryza sativa (Murayama and Handa, 2007), and Anabaena cyanobacterium (Vargas et al., 2003).
For decades, A/N-Invs were believed to occur exclusively in the cytosol, although proper localization data to corroborate this assumption were not available. Phylogenetic and in silico analyses, however, suggested the existence of two A/N-Inv subfamilies with different subcellular localization: an α-group with a predicted mitochondrial or plastidic localization and a β-group with a predicted cytosolic localization (Ji et al., 2005). Murayama and Handa (2007) were the first to demonstrate the presence of A/N-Invs experimentally in (or attached to) mitochondria and plastids of rice (OsNIN1 and OsNIN3), while Vargas et al. (2007) provided additional evidence for a chloroplast-targeted A/N-Inv in Arabidopsis (At-A/N-InvE; At5g22510). Similarly, some Susy forms have been localized in mitochondria (Subbaiah et al., 2006). AtSUS2 seems to be localized in (or attached to) plastids of the embryo (Núñez et al., 2008). The unexpected presence of A/N-Invs and Susy forms in organelles and the observed interaction of an A/N-Inv (At-A/N-InvG) with a phosphatidyl monophosphate 5 kinase (PIP5K9; Lou et al., 2007) suggest that some of these proteins are involved in signalling functions, stimulating further research in this area (Vargas and Salerno, 2010). Recently, it was convincingly demonstrated in Arabidopsis that cytosolic A/N-Invs (cA/N-Invs) are indispensable for normal plant growth and development (Barratt et al., 2009; Welham et al., 2009) as postulated in previous studies (Qi et al., 2007; Jia et al., 2008; Yao et al., 2009). Clearly, most recent reports have focused on the cA/N-Invs of the β-group in Arabidopsis and rice, and on a chloroplastic A/N-Inv (of the α-group in Arabidopsis: Vargas et al., 2007), while the mitochondrial A/N-Invs (mtA/N-Invs) have received little attention.
When plants are exposed to abiotic and biotic stresses, reactive oxygen species (ROS) homeostasis is disturbed, resulting in oxidative stress (Asada, 1999; Mittler, 2002; Neill et al., 2002). Chloroplasts, peroxisomes, and mitochondria are the major sources of ROS production in plant cells (Møller, 2001; Mittler et al., 2004; Blokhina and Fagerstedt, 2010). Superoxide dismutase (SOD) is the first line of defence against oxidative stress by catalysing the dismutation of superoxide (O2·–) to molecular oxygen and hydrogen peroxide (H2O2) (Okamoto et al., 2001). Three types of SOD isoenzymes have been reported in various plant species: Mn-SOD (e.g. MSD1), Cu/Zn-SOD (e.g. CSD1), and Fe-SOD (e.g. FSD1). Mn-SOD is located in mitochondria while Cu/Zn-SOD and Fe-SOD are chloroplastic (Alscher et al., 2002). In addition, both catalases (CATs) and ascorbate peroxidases (APXs)—the latter using ascorbate (AsA) as a substrate—play crucial roles in H2O2 scavenging processes, in concert with the different enzymes of the so-called Halliwell–Asada pathway which represents one of the most important antioxidant systems of the cytosol (Noctor and Foyer, 1998), chloroplasts (Arora et al., 2002), and mitochondria (Mittova et al., 2004). Recently, a specific vacuolar antioxidant mechanism has been proposed to work in concert with the well-known cytosolic, chloroplastic, and mitochondrial antioxidant mechanisms in plants (Van den Ende and Valluru, 2009; Bolouri-Moghaddam et al., 2010). Moreover, a scenario is emerging in which the catalytic activity of mitochondria-associated hexokinase (HXK) regulates ROS levels and perhaps also the signalling pathways leading to antioxidant defence responses (Camacho-Pereira et al., 2009; Bolouri-Moghaddam et al., 2010).
A mutant affected in the At-A/N-InvG gene (CINV1, At1g35580) is severely affected in root growth under osmotic stress (Qi et al., 2007). Vargas et al. (2008) also reported extensive increases in A/N-Inv enzymatic activities and gene expression under osmotic and cold stress in wheat leaves. Since all biotic and abiotic stresses are believed to result in oxidative stress responses in planta (Mittler et al., 2004), this led to the hypothesis that both organellar and cytoplasmic A/N-Invs could be directly or indirectly linked to oxidative stress defence responses in plants.
Gene expression analyses (derived from the Arabidopsis eFP browser http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi and Fig. S7 in Barratt et al., 2009) indicated that At-A/N-InvA (At1g56560) and At-A/N-InvG (At1g35580) are the most abundantly expressed A/N-Inv genes of the α (mitochondrial) and β (cytosolic) subgroups, respectively. Here, the subcellular localization of the At-A/N-InvA and At-A/N-InvG gene products in Arabidopsis leaf protoplasts is reported. Analysis of T-DNA knockout plants and transient overexpression in protoplasts further suggested for the first time a direct connection between A/N-Inv activities and the expression levels of genes involved in oxidative stress defence. More particularly, the results suggest that both cytosolic and mitochondrial A/N-Invs might contribute to delivering Glc as a substrate for HXK, in this way contributing to ROS homeostasis.
Materials and methods
Amplification of target clones
Total RNA was extracted with Trizol Reagent from leaves of 30-day-old Arabidopsis plants. RT-PCR (reverse transcriptase PCR) was carried out according to the supplier's instructions (Access RT-PCR System, Promega, USA).
Plant material and growth conditions
Arabidopsis thaliana ecotype Columbia was used in all these experiments. At-A/N-InvA (Salk_109830, Salk_015233) and At-A/N-InvG (Salk_095807) knockout seeds were acquired from the Nottingham Arabidopsis Stock Center. Arabidopsis seeds were surface-sterilized and sown in soil or on vertically placed plates containing half-strength Murashige and Skoog (MS) medium. Plants were grown under 12 h–12 h conditions (75 μmol s−1 m−2), 21 °C, and 50–60% humidity. MS medium was supplemented with 0, 1, 5, and 10% (w/v) Suc or mannitol (Mtl), respectively.
Transient expression of GFP fusion proteins
Full-length (At-A/N-InvA::GFP) and N-terminal (amino acids 1–31; At-A/N-InvAn::GFP) versions of At-A/N-InvA were amplified from cDNA with the primer pairs At-A/N-InvAFw and At-A/N-InvARev and with At-A/N-InvAnFw and At-A/N-InvAnRev (Supplementary Table S1 available at JXB online), respectively. Both fragments were digested with the BglII and StuI restriction endonucleases, and subsequently ligated in-frame with the green fluorescent protein (GFP) tag in the expression vector HBT35S::GFP::NOSter (Sheen, 1993). Similarly, full-length (At-A/N-InvG::GFP) and N-terminal (amino acids 1–31; At-A/N-InvGn::GFP) versions of At-A/N-InvG were cloned in the same GFP expression vector with BamHI and StuI restriction sites. GFP itself, containing no targeting signal, was used as control.
The constructs were subsequently transformed into Arabidopsis protoplasts as described elsewhere (Yoo et al., 2007). After 12 h of incubation under low light, the Arabidopsis protoplasts were treated with 0.5 μM MitoTracker Orange (Molecular Probes Division, Invitrogen) for 10 min. GFP images of transformed protoplasts were captured by confocal microscopy (Olympus, Germany).
Quantitative RT-PCR
Quantitative RT-PCR (qRT-PCR; FastSYBR Green Master Mix Protocol; Applied Biosystems) was carried out to evaluate the expression level of genes involved in antioxidative defence in leaves of 30-day-old wild-type (wt) and knockout plants with/without oxidative (H2O2) stress. Total RNA was extracted as described above. The first cDNA strands were synthesized from 1 μg of total RNA using a Reverse Transcription System (Promega, USA), and MSD1 (At3g10920), FSD1 (At4g25100), CSD1 (At1g08830), CAT2 (At4g35090), APX2 (At3g09640), HXK1 (At4g29130), At-A/N-InvA (At1g56560), and At-A/N-InvG (At1g35580) expression was followed. The PCR program comprised an initial denaturation for 2 min at 95 °C and amplification by 45 cycles of 3 s at 95 °C and 30 s at 58 °C. The housekeeping genes Actin2 (At3g18780) and UBQ10 (At4g05320) were used as references for normalization. The primer sequences are provided in Supplementary Table S1 at JXB online. All qRT–PCR experiments were performed in biological triplicate reactions and the graph values are means with standard error (SE).
APX2 luciferase reporter assay
The primers APX2proFw and APX2proRev (Supplementary Table S1) were used to PCR amplify the promoter region of the APX2 gene (–2000 bp relative to the translation start site) using genomic DNA of A. thaliana var. Columbia (isolated by a DNeasy Plant Mini Kit, Qiagen). The PCR product was digested with BamHI and NcoI, and cloned into the luciferase reporter vector (pUC-Luc). Arabidopsis protoplasts were isolated and transfected by a modified polyethylene glycol method as described (Yoo et al., 2007). Typically, 0.1 ml of protoplast suspension (106 protoplasts ml−1) was co-transfected with 20 μg of DNA of three plasmids containing At-A/N-InvA/At-A/N-InvG or a control plasmid, APX2::LUC as reporter, and UBQ10::GUS as internal control. The transfected protoplasts were incubated for 4 h before collection under different conditions [30 mM Glc, 30 mM Suc, 30 mM Mtl, 30 mM AsA adjusted to pH 5.7 with KOH, 200 μM H2O2, 10 μM 3-(3,4-dichlorophenyl)-1,1-dimethylurea (DCMU), 200 μM H2O2 plus 30 mM Suc, and 10 μM abscisic acid (ABA)].
Heterologous expression of At-A/N-InvA and At-A/N-InvG in Escherichia coli
At-A/N-InvA and At-A/N-InvG were cloned into the pBAD TOPO TA vector. Transformation, heterologous expression, and protein extraction were carried out according to the manufacturer's instructions (pBAD TOPO TA Expression Kit, Invitrogen). The A/N-Inv activities for the pH optima (pH 5.0–11.0) and the TRIS inhibition experiments (pH 8.2) were assayed in 100 μl reaction mixtures containing 100 mM Suc and 50 mM potassium phosphate buffer, with protein concentrations adjusted to 10 μg ml−1 (Sedmak and Grossberg, 1977). The reaction mixtures were incubated at 30 °C for 15, 30, 45, and 60 min, and the reactions were stopped by keeping an aliquot for 10 min in a water bath at 90 °C. Similar reaction conditions were used to determine the Kms of the recombinant At-A/N-InvA and At-A/N-InvG enzymes, at varying Suc concentrations (1–50 mM range). After centrifugation and appropriate dilution with 0.02% (w/v) Na-azide (to prevent microbial growth), 25 μl was injected onto a HPAEC-PAD column as described (Van den Ende and Van Laere, 1996). Only data from the linear range were used, ensuring that <10% of the original substrate was consumed. All experiments were repeated in duplicate or triplicate. The amount of Fru in the reaction mixtures was quantified by the external standard method and used for calculating the Inv activities and Kms (Hanes plots and Michaelis–Menten kinetics) as described before (Ji et al., 2007).
Total A/N-Inv activity measurements on Arabidopsis leaves
Plant material was blended in an equal volume of extraction buffer [50 mM TEA pH 8.5, 0.1% (w/v) Polyclar, 10 mM NaHSO3, 1 mM phenylmethylsulphonyl fluoride (PMSF), 1 mM mercaptoethanol, and 0.02% (w/v) Na-azide]. After centrifugation (5 min, 13 000 rpm), (NH4)2SO4 was added to 80% saturation. After 30 min incubation on ice, the sample was centrifuged (5 min, 13 000 rpm). The pellet was washed three times by adding 800 μl of 80% (NH4)2SO4-saturated TEA buffer (50 mM, pH 8.5), followed by centrifugation. The pellet was dissolved in 50 μl of TEA buffer. Total A/N-Inv activity was determined in 100 μl reaction mixtures containing 100 mM Suc in 50 mM TEA buffer pH 8.5 also containing 0.02% (w/v) Na-azide. Reaction mixtures were incubated at 30 °C for 10, 20, and 30 min, and the reactions were stopped by keeping an aliquot for 10 min in a water bath at 90 °C. Further analysis was as described above.
Results
Phylogenetic relationships within the A/N-Inv gene family
Figure 1 shows an unrooted phylogenetic tree with nine putative A/N-Invs (At-A/N-InvA–I) from Arabidopsis together with other functionally characterized plant A/N-Invs. The α and β clusters are clearly separated. Within the α cluster, two further subgroups can be discerned (LtINV, OsNIN3, BvINV, and At-A/N-InvE on one hand; OsNIN1, DcINV, At-A/N-InvC, At-A/N-InvH, and At-A/N-InvA on the other hand). Members of the latter subgroup localize in mitochondria as demonstrated experimentally (At-A/N-InvA, this study; OsNIN1, Murayama and Handa, 2007) or as predicted by Target P (DcInv, At-A/N-InvH, and At-A/N-InvC). The other subgroup harbours OsNIN3 and At-A/N-InvE (At5g22510) which are localized in plastids (Murayama and Handa, 2007; Vargas et al., 2008). BvINV is also predicted to be localized in plastids. However, LtINV is predicted to localize in mitochondria.
Fig. 1.
Phylogenetic tree of A/N-Invs. An unrooted phylogenetic tree containing nine Arabidopsis A/N-Invs (At-A/N-Inv A–I) and other functionally studied A/N-Invs, drawn by Clustal W2. The α and β type of A/N-Invs can be discriminated. At-A/N-InvA and At-A/N-InvG, studied in this manuscript, are encircled. At-A/N-InvA, At1g56560; At-A/N-InvG, At1g35580 (Qi et al., 2007; Lou et al., 2007); At-A/N-InvF, At1g72000; At-A/N-InvH, At3g05820; At-A/N-InvC, At4g06500; At-A/N-InvI, At4g09510 (Barratt et al., 2009); At-A/N-InvE, At5g22510 (Vargas et al., 2008); At-A/N-InvD, At1g22650; At-A/N-InvB, At4g34860; AsinvA, Anabaena sp. PCC 7120 (Vargas et al., 2003); BvINV, Beta vulgaris (González and Cejudo, 2007); DcINV, Daucus carota (Sturm et al., 1999); LjInv1, Lotus japonicus (Flemetakis et al., 2006); LtINV, Lolium temulentum (Gallagher and Pollock, 1998); OsNIN1 and OsNIN3, Oryza sativa (Murayama and Handa, 2007; Jia et al., 2008); Ta-A/N-INV, Triticum aestivum (Vargas et al., 2007). Referring to the (putative) subcellular localization of the enzymes, the confidence levels generated by target P are indicated. mTP, mitochondrial; cTP, chloroplastic; OP, other (presumably cytosolic); *, proven by localization studies. For a more extensive phylogenetic tree of (putative) At-A/N Invs, refer to the supplementary data of Vargas et al. (2008).
The cytosolic At-A/N-InvG is in the β cluster. All the members of this cluster are believed to be cytosolic proteins based on computational prediction. Besides At-A/N-InvF, At-A/N-InvD, At-A/N-InvB, Oscyt-Inv (Jia et al., 2008), LjInv1 (Flemetakis et al., 2006), Ta-A/N-Inv (Vargas et al., 2007), and At-A/N-InvI (Barratt et al., 2009) are well-characterized A/N-Invs.
At-A/N-InvA is located in mitochondria
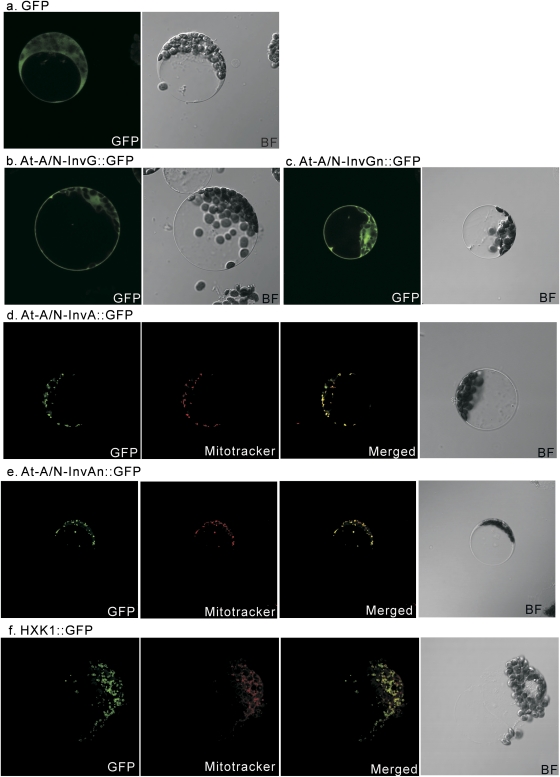
To test the organellar localization of At-A/N-InvA and At-A/N-InvG in the α and β subgroups as predicted by Ji et al. (2005) and Murayama and Handa (2007), full-length and N-terminal versions of At-A/N-InvA and At-A/N-InvG in-frame with GFP were designed. Confocal microscopy was used to localize the resulting GFP fusion proteins (and a GFP only control) in Arabidopsis protoplasts, incubated for 12 h under dim light. In each case, 50 cells were investigated and >95% showed the subcellular localization as described. As expected based on the phylogenetic tree, the protoplasts expressing At-A/N-InvGn::GFP (Fig. 2c) and At-A/N-InvG::GFP (Fig. 2b), as well as the GFP control (Fig. 2a), showed fluorescence in the cytosol. In contrast, At-A/N-InvA::GFP (Fig. 2d) and At-A/N-InvAn::GFP (Fig. 2e) were mainly detected in small vesicle-like structures surrounding the chloroplasts. A clear overlap with the MitoTracker Orange marker demonstrated the mitochondrial location of At-A/N-InvA and suggests that the targeting signal for mitochondrial localization is in the N-terminal part of the protein. HXK1 (At4g29130) associated with the mitochondrial outer membrane (OM; Kim et al., 2006) was used as an additional control for mitochondrial localization (Fig. 2f). The results show that At-A/N-InvA and HXK1 occur remarkably close to each other.
Fig. 2.
Transient expression of fusion proteins in Arabidopsis protoplasts. (a) GFP fluorescence and bright field (BF) of GFP control protein; (b) GFP fluorescence and BF of At-A/N-InvG::GFP fusion protein; (c) GFP fluorescence and BF of At-A/N-InvGn::GFP fusion protein; (d) GFP, mitoTracker fluorescence, merge of GFP and Mitotracker, and BF of At-A/N-InvA::GFP fusion protein; (e) GFP, mitoTracker fluorescence, merge of GFP and mitoTracker, and BF of At-A/N-InvAn::GFP fusion protein; (f) GFP, mitoTracker fluorescence, merge of GFP and mitoTracker, and BF of HXK1::GFP fusion protein. (This figure is available in colour at JXB online.)
Enzymatic activities of recombinant At-A/N-InvA and At-A/N-InvG, and inhibition by TRIS
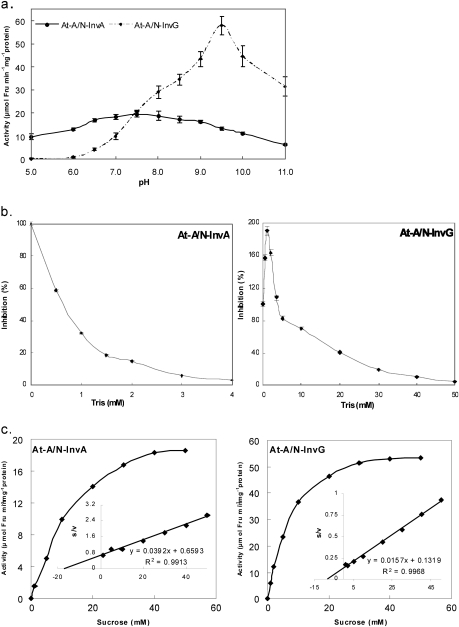
To confirm their catalytic activities, At-A/N-InvA and At-A/N-InvG were heterologously expressed in E. coli. Fully functional recombinant enzymes were obtained. The pH optima, the Km, and the inhibitory effect of TRIS at pH 8.2 were determined. At-A/N-InvA shows a neutral pH optimum at about pH 7.5, and its activity is rather stable over a wide pH range (from pH 5.0 to 10.0: Fig. 3a). In contrast, the pH optimum of At-A/N-InvG is ∼9.5 (Fig. 3a), indicating that it can be classified as an alkaline invertase. A typical inhibitory effect of TRIS on At-A/N-InvA activity was found (Fig. 3b), as observed for other A/N-Invs (Van den Ende and Van Laere, 1995, and references therein). In contrast, the At-A/N-InvG activity increased at the lower TRIS concentrations (0.5–4 mM range; Fig. 3b), 25–30 mM TRIS is needed for 50% inhibition, and the activity is strongly inhibited by 50 mM TRIS. The estimated Km for At-A/N-InvG was 8.4 mM, compared with 17 mM for At-A/N-InvA (Fig. 3c). This is in the same range as the Kms of other A/N-Invs (Vargas et al., 2003).
Fig. 3.
Properties and kinetics of the recombinant At-A/N-InvA and At-A/N-InvG. (a) Invertase activity of At-A/N-InvA and At-A/N-InvG as a function of the pH (5.0–11.0). Reaction conditions: 100 mM Suc, 30 °C, 30 min. Vertical bars represent the SE for n=3. (b) Invertase activity of At-A/N-InvA and At-A/N-InvG (pH 8.2) as a function of TRIS concentration. Vertical bars represent the SE for n=3. (c) Substrate–velocity plots for Fru production by At-A/N-InvA (pH 7.5) and At-A/N-InvG (pH 9.5) at varying Suc concentrations (1–50 mM). Reaction time: 30 min. Reaction temperature: 30 °C. The corresponding linear Hanes plots are shown as inserts.
Growth phenotype and total A/N-Inv activities of atinva and atinvg knockout plants
T-DNA insertion lines for At-A/N-InvA (Salk_109830 and Salk_015233, termed atinva) and At-A/N-InvG (Salk_095807, termed atinvg) (Fig. 4a) were grown on half-strength MS medium supplemented with 0, 1, and 5% (w/v) Suc or Mtl, and growth was compared with that of wt plants up to 15 d post-germination (Fig. 4b). The Mtl treatments resulted in more severe phenotypes compared with the Suc treatments. While Suc and Mtl both cause osmotic stress, Mtl is probably imported much more slowly and cannot be further metabolized once taken up. Consistent with the results of Barratt et al. (2009), atinvg showed a severe root growth defect and a weaker leaf growth compared with the wt, at 0, 1, and 5% Suc and Mtl. Surprisingly, atinva knockout plants showed an even more severe growth phenotype compared with atinvg, affected in both leaf growth and root development (Fig. 4b). However, the main root length of atinva knockouts was relatively less affected at 1% Suc, suggesting that mitochondrial At-A/N-InvA activity is more important under conditions of both sugar starvation (0% Suc) and excess sugar or osmotic stress (5% Suc). Root lengths of wt, atinva, and atinvg plants (MS agar plate, 1.5% Suc, 15 d post-germination) were recorded. Fifty plants of each type were selected. The mean root length of atinva knockouts is remarkably shorter (44% of the wt), while atinvg knockout roots are less affected (Fig. 4c). Soil-grown atinva plants also showed a significantly reduced leaf and shoot development compared with atinvg and wt plants (Fig. 4d).
Fig. 4.
Phenotypes of wt, atinva, and atinvg knockout plants. (a) Gene and PCR data (primer pairs: LBb1.3, LP, and RP, according to http://signal.salk.edu/tdnaprimers.2.html) of wt, atinva (Salk_109830, Salk_015233), and atinvg (Salk_095807). (b) Seedling phenotypes (15 d after germination) of wt, atinva, and atinvg at 0, 1, and 5% Suc and Mtl, respectively. (c) Root length of wt, atinva, and atinvg plants (15 d after germination). Vertical bars represent the SE for n=3. (d) Soil-grown phenotypes of wt, atinva, and atinvg plants (5 and 14 weeks, respectively). (e) Relative total A/N-Inv activities of atinva and atinvg compared with the wt leaves (5 weeks). Vertical bars represent the SE for n=3. (This figure is available in colour at JXB online.)
A/N-Invs were extracted from freshly harvested leaf material of atinva, atinvg, and wt plants. Total A/N-Inv activities were measured at pH 8.5 (to minimize side activities of acid invertases). By comparison with wt plants, the total A/N-Inv activities of atinva and atinvg were ∼40% and 35% lower (Fig. 4e). Knocking out only one out of nine A/N-Invs considerably affects both the growth phenotype and the total A/N-Inv activity, suggesting that both At-A/N-InvA and At-A/N-InvG make up a considerable part of the total activity. Alternatively, the At-A/N-InvA and At-A/N-InvG proteins themselves (independently of their catalytic activities) might play a role in controlling the total A/N-Inv activity and overall plant development, as suggested before by Lou et al. (2007) for At-A/N-InvG.
Expression of genes involved in oxidative stress defence
A clear connection between mitochondria-associated HXK (mtHXK) activity and ROS homeostasis was recently demonstrated (Camacho-Pereira et al., 2009; Bolouri-Moghaddam et al., 2010). It was hypothesized that A/N-Invs might assist in delivering Glc to mtHXK. Therefore, it was decided to investigate the expression levels of At-A/N-InvA, At-A/N-InvG, HXK1, and the oxidative stress-responsive MSD1, FSD1, CSD1, CAT2, and APX2 genes in response to H2O2 by qRT-PCR. For this purpose, detached wt, atinva, and atinvg mature leaves were incubated in water or 20 mM H2O2 for 10 h.
Except for FSD1, all transcript levels significantly increased by the application of exogenous H2O2 in wt leaves (Table 1). Addition of H2O2 to the mutants generally resulted in even higher transcript levels, but untreated mutant leaves already showed enhanced basal levels of oxidative stress-associated gene expression. In line with previous observations (Fryer et al., 2003; Costa et al., 2010), APX2 seems to be the most sensitive to H2O2 treatment. Therefore, this gene was investigated further, and an APX2 promoter–luciferase (LUC) construct was created as an oxidative stress reporter for cellular assays (see Discussion).
Table 1.
qRT-PCR data of antioxidative gene expression for wt and knockout plants, with (+H2O2) or without H2O2 treatment
| wt | atinva | atinvg | wt+H2O2 | atinva+ H2O2 | atinvg+H2O2 | |
| APX2 | 1.00±0.03 | 3.78± 0.43 | 4.67±0.29 | 16.77±2.48 | 13.95±1.18 | 9.25±1.20 |
| CAT2 | 1.00±0.02 | 1.55±0.12 | 1.50±0.09 | 4.73±1.05 | 2.78±0.25 | 2.25±0.18 |
| CSD1 | 1.00± 0.03 | 1.80±0.12 | 1.18±0.14 | 2.86±0.42 | 2.26±0.30 | 2.45±0.42 |
| FSD1 | 1.00±0.01 | 1.62±0.08 | 2.66±0.39 | 1.42±0.58 | 2.06±0.07 | 1.12±0.15 |
| MSD1 | 1.00±0.04 | 1.31±0.15 | 1.63±0.10 | 2.30±0.24 | 1.71±0.21 | 1.65±0.21 |
| HXK1 | 1.00±0.02 | 1.29±0.12 | 1.42±0.18 | 2.46±0.33 | 2.12±0.16 | 2.45±0.17 |
| At-A/N-InvA | 1.00±0.03 | 0.15±0.03 | 1.04±0.19 | 2.05 ±0.53 | 0.18±0.02 | 1.14±0.02 |
| At-A/N-InvG | 1.00±0.03 | 1.13±0.09 | 0.10±0.01 | 2.91±0.32 | 1.41±0.10 | 0.14±0.01 |
Actin2 (At3g18780) and UBQ10 (At4g05320) were used as reference genes. The values in the table are fold changes in transcript levels normalized to the two reference genes with respect to the wt control. Means ±SE (n=3).
The effect of transient At-A/N-InvA and At-A/N-InvG overexpression was analysed in Arabidopsis mesophyll protoplasts incubated for 4 h in the light or in darkness (Table 2) with addition of Glc (30 mM), Suc (30 mM), Mtl (30 mM), H2O2 (200 μM), ABA (10 μM), AsA (30 mM), DCMU (10 μM), and a combination of H2O2 and Suc (HS). Both H2O2 and ABA activated the APX2 promoter in transfected wt protoplasts (Table 2). In contrast, the addition of the metabolizable sugars Glc and Suc as well as AsA down-regulated APX2 promoter activity, while Mtl had no effect. Interestingly, the addition of Suc together with H2O2 resulted in a less pronounced activation of the APX2 promoter compared with H2O2 alone. Furthermore, overexpressing At-A/N-InvA and At-A/N-InvG down-regulated the APX2 promoter, in both the control and treated conditions. DCMU as an inhibitor of photosynthesis worked as an inhibitor of APX2 promoter activity in the light, and as an activator in the dark (Table 2).
Table 2.
APX2 promoter luciferase assay of protoplasts derived from wt plants with or without overexpression of At-A/N-InvA or At-A/N-InvG under different treatments (30 mM Glc, 30 mM Suc, 30 mM AsA (adjusted to pH 5.7 with KOH), 30 mM Mtl, 200 μM H2O2, 10 μM DCMU, 200 μM H2O2+30 mM Suc (+HS), and 10 μM ABA)
| Control | +Glc | +H2O2 | +Suc | +AsA | +ABA | +Mtl | +DCMU | +HS | |
| wt L | 1.00±0.04 | 0.71±0.11 | 3.28±1.27 | 0.64±0.09 | 0.74±0.05 | 1.70±0.13 | 1.13±0.27 | 0.42±0.04 | 1.52±0.04 |
| At-A/N-InvA L | 0.41±0.06 | 0.34±0.06 | 0.96±0.39 | 0.38±0.08 | 0.31±0.04 | 1.04±0.19 | 0.36±0.07 | 0.29±0.04 | 0.54±0.07 |
| At-A/N-InvG L | 0.35±0.04 | 0.21±0.03 | 0.78±0.21 | 0.26±0.03 | 0.27±0.01 | 0.97±0.11 | 0.28±0.04 | 0.23±0.03 | 0.30±0.06 |
| wt D | 1.00±0.04 | 0.60±0.17 | 2.45±0.47 | 0.82±0.08 | 0.77±0.02 | 3.48±0.91 | 1.17±0.14 | 1.88±0.54 | 2.12±0.46 |
| At-A/N-InvA D | 0.76±0.18 | 0.53±0.12 | 1.52±0.30 | 0.62±0.05 | 0.65±0.03 | 2.76±0.65 | 0.74±0.10 | 1.23±0.63 | 1.18±0.12 |
| At-A/N-InvG D | 0.54±0.10 | 0.33±0.14 | 1.01±0.08 | 0.36±0.07 | 0.46±0.12 | 1.50±0.45 | 0.50±0.11 | 0.85±0.37 | 0.56±0.18 |
The experiment was executed with (L) or without (D) light. The values are fold changes normalized to the wt control. Means ±SE (n=3).
Discussion
Recently, cytosolic A/N-Invs have been recognized as important regulators of plant growth and development, possibly involved in metabolic signalling processes, especially under stress conditions (Barratt et al., 2009; Vargas and Salerno, 2010). However, to date no research has been dedicated to studying the links between A/N-Invs and oxidative stress defence. In the model plant A. thaliana, At-A/N-InvA and At-A/N-InvG are expressed proteins belonging to α and β subgroups (Fig. 1) and with a different localization (Fig. 2). The optimal pH and TRIS inhibition differed strongly between the two recombinant enzymes, but the physiological implications of these differences, if any, remain unclear. Depending on conditions, both At-A/N-InvA and At-A/N-InvG knockouts showed smaller roots (Fig. 4b, c), which are typical phenotypes for plants suffering from oxidative stress, limited nitrate availability or signalling (Foreman et al., 2003; Pnueli et al., 2003; Rizhsky et al., 2003; Qi et al., 2007). In atinva seedlings grown on 1% Suc, typically resulting in optimal growth, the phenotype was less prominent compared with 0% and 5% Suc, respectively (Fig. 4b). These findings are in line with previous observations suggesting that soluble sugars can help protect plants prior to or under oxidative stress (Sulmon et al., 2006; Nishizawa et al., 2008; Ramel et al., 2009; Bolouri-Moghaddam et al., 2010). When plants are stressed, the steady-state level of ROS usually increases, but ROS (specifically H2O2) can also act as a signal for turning on stress-related genes (Mittler et al., 2004). Exogenous application of H2O2 is widely used to induce oxidative stress-related gene expression. Table 1 shows the induction of several of these marker genes. Intriguingly, the H2O2 treatment also induced At-A/N-InvA and At-A/N-InvG gene expression (Table 1). In particular, the APX2 promoter responds significantly and can be used as an oxidative stress reporter. Indeed, it is widely accepted that the APX2 promoter acts as an integrator of various stress-related stimuli including high light, the redox status of the plastoquinone pool associated with photosynthesis, cellular H2O2 levels, as well as responses to ABA and osmotic stress (Karpinski et al., 1999; Shigeoka et al., 2002; Fryer et al., 2003; Ball et al., 2004; Chang et al., 2004; Rossel et al., 2004, 2006; Bechtold et al., 2008; Barba-Espin et al., 2010). Besides the fact that control of APX2 expression and leaf water status is mediated by ABA, a certain concentration of APX2 seems necessary for ABA sensing and response (Fryer et al., 2003).
Consistent with the results under light (Table 2), DCMU, an inhibitor of photosystem II, abolishes the production of chloroplastic H2O2 and subsequently the induction of APX2 in Arabidopsis leaves (Karpinski et al., 1999; Chang et al., 2004). These observations confirm the idea that APX2 promoter activity, at least to some extent, can be used as a rough indicator of endogenous H2O2 levels (Fryer et al., 2003). In the dark, chloroplastic ROS production is hampered and cytosolic H2O2 levels are thus more likely to be determined by the level of mitochondrial ROS production. Nonetheless, an up-regulation of APX2 expression upon addition of H2O2 and ABA, and down-regulation by metabolizable sugars is still observed in the dark albeit to a lesser extent (Table 2), suggesting that the decrease in APX2 promoter activity reflects the decreasing mitochondrial ROS production in the At-A/N-Inv-overexpressing cells.
The importance of A/N-Inv activity may reside in regulating the Suc concentration in the cytosol and/or in organelles, or in the production of Glc which could be sensed by mitochondria-associated HXKs (Rolland et al,. 2006; Li et al., 2007; Bolouri-Moghaddam et al., 2010) or serve as a substrate for HXK to control mitochondrial ROS production (Camacho-Pereira et al., 2009). The increased expression of antioxidant defence-related genes in atinva and atinvg knockout plants, the reported increased expression of A/N-Invs and HXK under stress conditions (Vargas et al., 2008; Table 1), and the decrease in APX2 promoter activity in protoplasts overexpressing At-A/N-InvA and At-A/N-InvG strongly suggest that A/N-Invs are part of the antioxidant system involved in cellular ROS homeostasis. Lou et al. (2007) demonstrated that At-A/N-InvG (AtCYTINV1 in their terminology) interacts with an Arabidopsis phosphatidylinositol monophosphate kinase (PIP5K9), and that it can localize in the nucleus, in contrast to the exclusive cytosolic localization that was observed here in Arabidopsis protoplasts. Similarly, it was suggested that AtHXK1 may be involved in a nuclear protein complex that directly modulates specific target genes in a Glc-dependent manner (Cho et al., 2009). Therefore, one hypothesis could be that the presence of At-A/N-InvG in the nucleus may be associated with the production of Suc-derived Glc for the AtHXK1-mediated control of gene expression (Vargas and Salerno, 2010). However, the fact that Glc probably can diffuse freely through nuclear pores argues against the need for a Suc-hydrolysing enzyme activity in the nucleus.
Arabidopsis mesophyll protoplasts are widely used as a versatile cellular system for transient gene expression analysis (Yoo et al., 2007). However, protoplasts immediately initiate new cellulose biosynthesis requiring massive amounts of UDPGlc derived from the activity of sucrose synthase (Fujii et al., 2010). Since this requires an enormous investment of ATP and carbon skeletons, mesophyll protoplasts have a tendency to enter into sugar starvation conditions quite rapidly, and as such are sensitive to additional (oxidative) stresses such as exogenous H2O2 and ABA (Table 2). Exogenously supplied metabolizable sugars and AsA can counteract the oxidative stress as judged from the decreased APX2 gene expression (Table 2). The increase in APX2 expression upon addition of 30 mM Mtl, however, suggests that this non-metabolizable sugar might generate additional stress, although Qi et al. (2007) reported that Mtl could also enhance endogenous At-A/N-InvG activity levels.
Sugar supplementation or enhanced Glc production by overexpressing A/N-Invs might help to increase the number of mitochondria, the respiration rate, and ATP generation (Giegé et al., 2005), both in the light and in the dark (Table 2). However, it cannot be excluded that sugar-mediated feedback regulation of photosynthesis occurs in the light, thus partly decreasing the chloroplast-generated H2O2. Recently, a chloroplastic A/N-Inv has been suggested as a putative player in this process (Tamoi et al., 2010). However, this reasoning seems inconsistent with the fact that somewhat weaker but otherwise similar results were recorded in the dark (Table 2), suggesting that mitochondrial ROS-producing processes are important both in the light and in the dark, as recently discussed in other manuscripts (Dinakar et al., 2010; Rossouw et al., 2010; Nunes-Nesi et al., 2011).
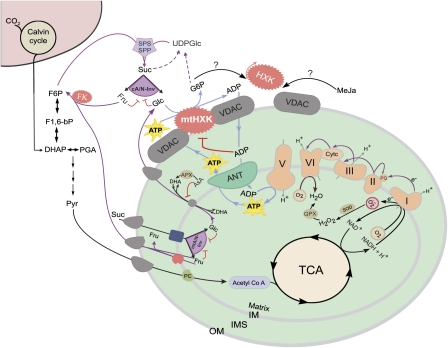
It has been demonstrated that cytosolic HXK isoforms are typically associated with the mitochondrial OM (Graham et al., 2007; Balasubramanian et al., 2007, Fig. 2f). It is suggested here that a continuous and efficient supply of Glc to mtHXK is necessary to maintain its activity at a rather constant level, which itself is needed to control the flux through the mitochondrial electron transport chain, influencing mitochondrial ROS production (Fig. 5; Camacho-Pereira et al., 2009). In particular, it has been demonstrated that mtHXK can contribute to the steady-state recycling of ADP (ADP production by mtHXK, bound to the mitochondrial OM; ADP consumption through oxidative phosphorylation) which regulates H2O2 formation in the electron transport chain on the mitochondrial inner membrane (IM; Fig. 5). Importantly, this mitochondrial ADP recycling mechanism leads to a decrease in the mitochondrial membrane potential while an inhibition of the associated mtHXK causes an increase in H2O2 production. Thus, a tightly bound OM mtHXK produces ADP that can be transported through both mitochondrial membranes to reach the F0F1ATP synthase complex (Fig. 5). In conclusion, a tightly bound OM mtHXK could provide a basal level of ADP for ATP synthesis, preventing overpolarization of the mitochondrial IM and consequent accumulation of ROS, including H2O2. This mechanism would be particularly useful when other sources of ADP become limiting for oxidative phosphorylation. Interestingly, both methyl jasmonate (MeJa) and Glc-6-P (G6P) are known to induce the detachment of mammalian HXK from the mitochondrial OM (da-Silva et al., 2004; Goldin et al., 2008), but additional research is needed to verify whether MeJa and G6P fulfil similar roles in the vicinity of plant mitochondria (Fig. 5).
Fig. 5.
Hypothetical model showing the putative role of A/N-Invs and mtHXKs in oxidative defence-related processes in plant mitochondria. Cytosolic Suc can serve as a substrate for cA/N-Invs. However, Suc can also enter the matrix via a Suc transporter in the inner membrane (IM). Glc and Fru are subsequently produced by mtA/N-Inv. Glc is transported back into the intermembrane space (IMS) through a Glc transporter that is regulated by AsA and through the outer membrane (OM) to serve as a substrate for HXK bound to the OM (termed mtHXK). mtHXK contributes to a steady-state ADP recycling via voltage-dependent anion channels (VDACs) and adenine nucleotide transporters (ANTs) to regulate H2O2 formation in the electron transport chain (ETC) on the IM. mtHXK activity is inhibited by ADP. It is not known whether methyl jasmonate (MeJa) and glucose 6-phosphate (G6P) can induce the detachement of mtHXK from the OM, as observed in animals. The G6P generated by mtHXK (as well as chloroplastic triose phosphates), can enter glycolysis, and the pyruvate (Pyr) produced crosses the IM via Pyr carboxylase (PC) to enter the Krebs (TCA) cycle. Alternatively, G6P can be used to produce UDPGlc for the resynthesis of carbohydrates (e.g. Suc or cellulose). Fru resulting from mtA/N-Inv or cA/N-Inv activity can be transformed into fructose 6-phosphate (F6P) by fructokinase (FK), which can enter glycolysis or can be used for Suc synthesis by sucrose phosphate synthetase (SPS) and sucrose phosphate phosphatase (SPP). Mitochondrial SOD and glutathione peroxidase (GPX) can assist in ROS-scavenging processes within the matrix. AsA is synthesized within the IMS, and can serve as a substrate for APX, to produce dehydroxyascorbate (DHA), which can be imported into the mitochondrial matrix by a transporter in the IM which also can function as a Glc transporter, regulated by AsA. Acetyl CoA, acetyl coenzyme A; Cytc, cytochrome c; DHAP, dihydroxyacetone phosphate; F1,6-bP, fructose 1,6-bisphosphate; PGA, phosphoglycolic acid; Q, plastoquinone pool; TCA, tricarboxylic acid.
What could be the source of the Glc substrate for HXKs in leaf mesophyll cells? First, Glc can originate from the activity of cA/N-Invs (Fig. 5). Secondly, cytosolic Glc might result from starch breakdown during the night, and, finally, it is proposed here that Glc could originate from the action of mtA/N-Invs (Fig. 5). Indeed, it was demonstrated in Helianthus tuberosus that mtA/N-Inv is present in the mitochondrial matrix (Szarka et al., 2008). In this species, separate, bidirectional Suc, Glc, and Fru transporters are presumably present in the IM (Szarka et al., 2008; Fig. 5). The mitochondrial OM contains numerous porins (e.g. voltage-dependent anion channels; Fig. 5) and is permeable to all molecules of ≤5000 Da (Colombini, 1979). Therefore, cytosolic Suc could enter through these porins and enter the matrix via a Suc transporter in the IM (Fig. 5), leading to the formation of Glc and Fru by the activity of mtA/N-Inv. It could be hypothesized that Glc is transported back into the intermembrane space (IMS; Fig. 5) and through pores of the OM to serve as a substrate for mtHXK (Fig. 5). Intriguingly, the backflow of Glc into the IMS might be tightly regulated by AsA (Fig. 5). Indeed, AsA very specifically inhibits the Glc transporter and not the Suc and Fru transporters in the IM (Fig. 5; Szarka et al., 2008). In case mitochondrial AsA contents become too low (e.g. as a result of oxidative stress), Glc outflow and mtHXK activity are promoted, ADP recycling is stimulated, ATP synthesis-related limitation of respiration is avoided, and subsequent H2O2 release is reduced/avoided (Camacho-Pereira et al., 2009). This mtHXK/mtA/N-Inv-controlled regulatory mechanism might be less critical when sufficient AsA and antioxidant-related enzymes are present to prevent ROS accumulation. Therefore, consistent with the data presented here, it is suggested that the inhibition of Glc outflow by AsA can be considered as an efficient feedback mechanism to control mtHXK activity and mitochondrial ROS production (Fig. 5).
Consistent with the proposed A/N-Inv/mtHXK model, it is well known that A/N-Inv enzymes are inhibited by their own hexose products (Van den Ende and Van Laere, 1995), which provides an elegant system to synchronize the A/N-Inv activities (cytosolic and/or mitochondrial) to mtHXK activities. Similar mechanisms, involving chloroplastic HXKs and A/N-Invs, might also occur in chloroplasts.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Names and sequence of the primers used in this manuscript.
Acknowledgments
The authors thank Rudy Vergauwen, Lies Vandesteene, Veerle Cammaer, and Ingeborg Millet for technical support, and Johan Thevelein and Koen Geuten for the use of their equipment. This work is supported by grants from FWO Vlaanderen.
Glossary
Abbreviations
- A/N-Inv
neutral/alkaline invertase
- APX
ascorbate peroxidase
- AsA
ascorbate
- CAT
catalase
- cA/N-Inv
cytosolic neutral/alkaline invertase
- DCMU
3-(3,4-dichlorophenyl)-1,1-dimethylurea
- Fru
fructose
- Glc
glucose
- HXK
hexokinase
- IM
inner membrane
- IMS
intermembrane space
- Inv
invertase
- MeJa
methyl jasmonate
- mtHXK
mitochondrial hexokinase
- Mtl
mannitol
- mtA/N-Inv
mitochondrial neutral/alkaline invertase
- OM
outer membrane
- ROS
reactive oxygen species
- SE
standard error
- SOD
superoxide dismutase
- Suc
sucrose
- Susy
sucrose synthase
- wt
wild type
References
- Alscher RG, Erturk N, Heath LS. Role of superoxide dismutases (SODs) in controlling oxidative stress in plants. Journal of Experimental Botany. 2002;53:1331–1341. [PubMed] [Google Scholar]
- Arora A, Sairam RK, Srivastava VA. Oxidative stress and antioxidative system in plants. Current Science. 2002;82:1227–1238. [Google Scholar]
- Asada K. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Ball L, Accotto G-P, Bechtold U, et al. Evidence for a direct link between glutathione biosynthesis and stress defense gene expression in Arabidopsis. The Plant Cell. 2004;16:2448–2462. doi: 10.1105/tpc.104.022608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian R, Karve A, Kandasamy M, Meagher RB, Moore BD. A role for F-actin in hexokinase-mediated glucose signaling. Plant Physiology. 2007;145:1423–1434. doi: 10.1104/pp.107.108704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barba-Espin G, Diaz-Vivancos P, Clemente-Moreno M, Albacete A, Faize L, Faize M, Pérez-Alfocea F, Hernández JA. Interaction between hydrogen peroxide and plant hormones during germination and the early growth of pea seedlings. Plant, Cell and Enviroment 33. 2010:981–994. doi: 10.1111/j.1365-3040.2010.02120.x. [DOI] [PubMed] [Google Scholar]
- Barratt DHP, Derbyshire P, Findlay K, Pike M, Wellner N, Lunn J, Feil R, Simpson C, Maule AJ, Smith AM. Normal growth of Arabidopsis requires cytosolic invertase but not sucrose synthase. Proceedings of the National Academy of Sciences, USA. 2009;106:13124–13129. doi: 10.1073/pnas.0900689106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold U, Richard O, Zamboni A, Gapper C, Geisler M, Pogson B, Karpinski S, Mullineaux PM. Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. Journal of Experimental Botany. 2008;59:121–133. doi: 10.1093/jxb/erm289. [DOI] [PubMed] [Google Scholar]
- Blokhina O, Fagerstedt KV. Oxidative metabolism, ROS and NO under oxygen deprivation. Plant Physiology and Biochemistry. 2010;48:359–373. doi: 10.1016/j.plaphy.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Bolouri-Moghaddam M-R, Le Roy K, Xiang L, Rolland F, Van den Ende W. Sugar signalling and antioxidant network connections in plant cells. FEBS Journal. 2010;277:2022–2037. doi: 10.1111/j.1742-4658.2010.07633.x. [DOI] [PubMed] [Google Scholar]
- Camacho-Pereira J, Meyer LE, Machado LB, Oliveira MF, Galina A. Reactive oxygen species production by potato tuber mitochondria is modulated by mitochondrially bound hexokinase activity. Plant Physiology. 2009;149:1099–1110. doi: 10.1104/pp.108.129247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CCC, Ball L, Fryer MJ, Baker NR, Karpinski S, Mullineaux PM. Induction of ASCORBATE PEROXIDASE 2 expression in wounded Arabidopsis leaves does not involve known wound-signaling pathways but is associated with changes in photosynthesis. The Plant Journal. 2004;38:499–511. doi: 10.1111/j.1365-313X.2004.02066.x. [DOI] [PubMed] [Google Scholar]
- Chen JQ, Black CC. Biochemical and immunological properties of alkaline invertase isolated from sprouting soybean hypocotyls. Archives of Biochemistry and Biophysics. 1992;295:61–69. doi: 10.1016/0003-9861(92)90488-i. [DOI] [PubMed] [Google Scholar]
- Cho Jl Ryoo N, Hahn T-R, Jeon J- S. Evidence for a role of hexokinases as conserved glucose sensors in both monocot and dicot plant species. Plant Signaling and Behaviour. 2009;4:908–910. doi: 10.4161/psb.4.9.9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombini M. A candidate for the permeability pathway of the outer mitochondrial membrane. Nature. 1979;279:643–645. doi: 10.1038/279643a0. [DOI] [PubMed] [Google Scholar]
- Costa A, Drago I, Behera S, Zottini M, Pizzo P, Schroeder JI, Pozzan T, Schiavo FL. H2O2 in plant peroxisomes: an in vivo analysis uncovers a Ca2+-dependent scavenging system. The Plant Journal. 2010;62:760–772. doi: 10.1111/j.1365-313X.2010.04190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da-Silva WS, Gómez-Puyou A, de Gómez-Puyou MT, Moreno-Sanchez R, De Felice FG, de Meis L, Oliveira MF, Galina A. Mitochondrial bound hexokinase activity as a preventive antioxidant defense steady-state ADP formation as a regulatory mechanism of membrane potential and reactive oxygen species generation in mitochondria. Journal of Biological Chemistry. 2004;279:39846–39855. doi: 10.1074/jbc.M403835200. [DOI] [PubMed] [Google Scholar]
- Dinakar C, Abhaypratap V, Yearla SR, Raghavendra AS, Padmasree K. Importance of ROS and antioxidant system during the beneficial interactions of mitochondrial metabolism with photosynthetic carbon assimilation. Planta. 2010;231:461–474. doi: 10.1007/s00425-009-1067-3. [DOI] [PubMed] [Google Scholar]
- Flemetakis E, Efrose RC, Ott T, Stedel C, Aivalakis G, Udvardi MK, Katinakis P. Spatial and temporal organization of sucrose metabolism in Lotus japonicus nitrogen-fixing nodules suggests a role for the elusive alkaline/neutral invertase. Plant Molecular Biology. 2006;62:53–69. doi: 10.1007/s11103-006-9003-4. [DOI] [PubMed] [Google Scholar]
- Foreman J, Demidchik V, Bothwell JHF, Mylona P, Miedema H, Torres MA, Linstead P, Costa S, Brownlee C, Jones JDG. Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature. 2003;422:442–446. doi: 10.1038/nature01485. [DOI] [PubMed] [Google Scholar]
- Fryer MJ, Ball L, Oxborough K, Karpinski S, Mullineaux PM, Baker NR. Control of ascorbate peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. The Plant Journal. 2003;33:691–705. doi: 10.1046/j.1365-313x.2003.01656.x. [DOI] [PubMed] [Google Scholar]
- Fujii S, Hayashi T, Mizuno K. Sucrose synthase is an integral component of the cellulose synthesis machinery. Plant and Cell Physiology. 2010;51:294–301. doi: 10.1093/pcp/pcp190. [DOI] [PubMed] [Google Scholar]
- Gallagher J, Pollock C. Isolation and characterization of a cDNA clone from Lolium temulentum L. encoding for a sucrose hydrolytic enzyme which shows alkaline/neutral invertase activity. Journal of Experimental Botany. 1998;49:789–795. [Google Scholar]
- Giegé P, Sweetlove LJ, Cognat V, Leaver CJ. Coordination of nuclear and mitochondrial genome expression during mitochondrial biogenesis in Arabidopsis. The Plant Cell. 2005;17:1497–1512. doi: 10.1105/tpc.104.030254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldin N, Arzoine L, Heyfets A, Israelson A, Zaslavsky Z, Bravman T, Bronner V, Notcovich A, Shoshan-Barmatz V, Flescher E. Methyl jasmonate binds to and detaches mitochondria-bound hexokinase. Oncogene. 2008;27:4636–4643. doi: 10.1038/onc.2008.108. [DOI] [PubMed] [Google Scholar]
- Gonzalez MC, Cejudo FJ. Gibberellin-regulated expression of neutral and vacuolar invertase genes in petioles of sugar beet plants. Plant Science. 2007;172:839–846. [Google Scholar]
- Graham JWA, Williams TCR, Morgan M, Fernie AR, Ratcliffe RG, Sweetlove LJ. Glycolytic enzymes associate dynamically with mitochondria in response to respiratory demand and support substrate channeling. The Plant Cell. 2007;19:3723–3738. doi: 10.1105/tpc.107.053371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Van den Ende W, Schroeven L, Clerens S, Geuten K, Cheng SH, Bennett J. The rice genome encodes two vacuolar invertases with fructan exohydrolase activity but lacks the related fructan biosynthesis genes of the Pooideae. New Phytologist. 2007;173:50–62. doi: 10.1111/j.1469-8137.2006.01896.x. [DOI] [PubMed] [Google Scholar]
- Ji X, Van den Ende W, Van Laere A, Cheng S, Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. Journal of Molecular Evolution. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Jia L, Zhang B, Mao C, Li J, Wu Y, Wu P, Wu Z. OsCYT-INV1 for alkaline/neutral invertase is involved in root cell development and reproductivity in rice (Oryza sativa L.) Planta. 2008;288:51–59. doi: 10.1007/s00425-008-0718-0. [DOI] [PubMed] [Google Scholar]
- Karpinski S, Reynold H, Karpinska B, Wingsle G, Mullineaux PM. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science. 1999;284:654–657. doi: 10.1126/science.284.5414.654. [DOI] [PubMed] [Google Scholar]
- Kim M, Lim JH, Chang SA, Park K, Kim GT, Kim WT, Paic HS. Mitochondria-associated hexokinases play a role in the control of programmed cell death in. Nicotiana benthamiana. The Plant Cell. 2006;18:2341–2355. doi: 10.1105/tpc.106.041509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammens W, Le Roy K, Schroeven L, Van Laere A, Rabijns A, Van den Ende W. Structural insights into glycoside hydrolase family 32 and 68 enzymes: functional implications. Journal of Experimental Botany. 2009;60:727–740. doi: 10.1093/jxb/ern333. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Sturm A. Purification and characterization of neutral and alkaline invertase from carrot. Plant Physiology. 1996;112:1513–1522. doi: 10.1104/pp.112.4.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Smith C, Corke F, Zheng L, Merali Z, Ryden P, Derbyshire P, Waldron K, Bevan MW. Signaling from an altered cell wall to the nucleus mediates sugar-responsive growth and development in. Arabidopsis thaliana. The Plant Cell. 2007;19:2500–2515. doi: 10.1105/tpc.106.049965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou Y, Gou JY, Xue HW. PIP5K9, an Arabidopsis phosphatidylinositol monophosphate kinase, interacts with a cytosolic invertase to negatively regulate sugar mediated root growth. The Plant Cell. 2007;19:163–181. doi: 10.1105/tpc.106.045658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends in Plant Science. 2002;7:405–410. doi: 10.1016/s1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- Mittler R, Vanderauwera S, Gollery M, Van Breusegem F. The reactive oxygen gene network of plants. Trends in Plant Science. 2004;9:490–498. doi: 10.1016/j.tplants.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Mittova V, Guy M, Tal M, Volokita M. Salinity upregulates the antioxidative system in root mitochondria and peroxisomes of the wild salt-tolerant tomato species Lycopersicon pennellii. Journal of Experimental Botany. 2004;55:1105–1113. doi: 10.1093/jxb/erh113. [DOI] [PubMed] [Google Scholar]
- Møller IM. Plant mitochondria and oxidative stress: electron transport, NADPH turnover and metabolism of reactive oxygen species. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:561–591. doi: 10.1146/annurev.arplant.52.1.561. [DOI] [PubMed] [Google Scholar]
- Murayama S, Handa H. Genes for alkaline/neutral invertase in rice: alkaline/neutral invertases are located in plant mitochondria and also in plastids. Planta. 2007;225:1193–1203. doi: 10.1007/s00425-006-0430-x. [DOI] [PubMed] [Google Scholar]
- Neill SJ, Desikan D, Clarke A, Hancock JT. Nitric oxide is a novel component of abscisic acid signaling in stomatal guard cells. Plant Physiology. 2002;128:13–16. [PMC free article] [PubMed] [Google Scholar]
- Nishizawa A, Yukinori Y, Shigeoka S. Galactinol and raffinose as a novel function to protect plants from oxidative damage. Plant Physiology. 2008;147:1251–1263. doi: 10.1104/pp.108.122465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor G, Foyer CH. A re-evaluation of the ATP: NADPH budget during C3 photosynthesis. A contribution from nitrate assimilation and its associated respiratory activity? Journal of Experimental Botany. 1998;49:1895–1908. [Google Scholar]
- Nunes-Nesi A, Araújo WL, Fernie AR. UPDATE: targeting mitochondrial metabolism and machinery as a means to enhance photosynthesis. Plant Physiology. 2011;155:101–107. doi: 10.1104/pp.110.163816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez JGA, Kronenberger J, Wuillème S, Lepiniec L, Rochat C. Study of AtSUS2 localization in seeds reveals a strong association with plastids. Plant and Cell Physiology. 2008;49:1621–1626. doi: 10.1093/pcp/pcn117. [DOI] [PubMed] [Google Scholar]
- Okamoto OK, Pinto E, Latorre LR, Bechara EJH, Colepicolo P. Antioxidant modulation in response to metal induced oxidative stress in algal chloroplast. Archives of Environmental Contamination and Toxicology. 2001;40:18–24. doi: 10.1007/s002440010144. [DOI] [PubMed] [Google Scholar]
- Pnueli L, Liang H, Rozenberg M, Mittler R. Growth suppression, altered stomatal responses, and augmented induction of heat shock proteins in cytosolic ascorbate peroxidase (Apx1)-deficient Arabidopsis plants. The Plant Journal. 2003;34:187–203. doi: 10.1046/j.1365-313x.2003.01715.x. [DOI] [PubMed] [Google Scholar]
- Qi X, Wu Z, Li J, Mo X, Wu S, Chu J, Wu P. AtCYT-INV1, a neutral invertase, is involved in osmotic stress-induced inhibition on lateral root growth in Arabidopsis. Plant Molecular Biology. 2007;64:575–587. doi: 10.1007/s11103-007-9177-4. [DOI] [PubMed] [Google Scholar]
- Ramel F, Sulmon C, Bogard M, Couée I, Gouesbet G. Differential patterns of reactive oxygen species and antioxidative mechanisms during atrazine injury and sucrose-induced tolerance in Arabidopsis thaliana plantlets. BMC Plant Biology. 2009;9:28–45. doi: 10.1186/1471-2229-9-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Mittler R. The water–water cycle is essential for chloroplast protection in the absence of stress. Journal of Biological Chemistry. 2003;278:38921–38925. doi: 10.1074/jbc.M304987200. [DOI] [PubMed] [Google Scholar]
- Rolland F, Baena-Gonzalez E, Sheen J. Sugar sensing and signaling in plants: conserved and novel mechanisms. Annual Review of Plant Biology. 2006;57:675–709. doi: 10.1146/annurev.arplant.57.032905.105441. [DOI] [PubMed] [Google Scholar]
- Ross HA, McRae D, Davies HV. Sucrolytic enzyme activities in cotyledons of the faba bean. Plant Physiology. 1996;111:329–338. doi: 10.1104/pp.111.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossel JB, Cuttriss A, Pogson BJ. Identifying photoprotection mutants in Arabidopsis thaliana. Methods in Molecular Biology. 2004;274:287–299. doi: 10.1385/1-59259-799-8:287. [DOI] [PubMed] [Google Scholar]
- Rossel JB, Walter PB, Hendrickson L, Chow WS, Poole A, Mullineaux PM, Pogson BJ. A mutation affecting ASCORBATE PEROXIDASE 2 gene expression reveals a link between responses to high light and drought tolerance. Plant, Cell and Environment. 2006;29:269–281. doi: 10.1111/j.1365-3040.2005.01419.x. [DOI] [PubMed] [Google Scholar]
- Rossouw D, Kossmann J, Botha FC, Groenewald JH. Reduced neutral invertase activity in the culm tissues of transgenic sugarcane plants results in a decrease in respiration and sucrose cycling and an increase in the sucrose to hexose ratio. Functional Plant Biology. 2010;37:22–31. [Google Scholar]
- Sedmak J, Grossberg E. A rapid, sensitive and versatile assay for protein using Coomassie Brilliant Blue G250. Analytical Biochemistry. 1977;79:544–546. doi: 10.1016/0003-2697(77)90428-6. [DOI] [PubMed] [Google Scholar]
- Sheen J. Protein phosphatase activity is required for light-inducible gene expression in maize. EMBO Journal. 1993;12:3497–3505. doi: 10.1002/j.1460-2075.1993.tb06024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shigeoka S, Ishikawa T, Tamoi M, Miyagawa Y, Takeda T, Yabuta Y, Yoshimura K. Regulation and function of ascorbate peroxidase isoenzymes. Journal of Experimental Botany. 2002;53:1305–1319. [PubMed] [Google Scholar]
- Smeekens S, Ma J, Hanson J, Rolland F. Sugar signals and molecular networks controlling plant growth. Current Opinion in Plant Biology. 2010;3:273–278. doi: 10.1016/j.pbi.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Sturm A, Lienhard S, Schatt S, Hardegger M. Tissue-specific expression of two genes for sucrose synthase in carrot (Daucus carota L.) Plant Molecular Biology. 1999;39:349–360. doi: 10.1023/a:1006199003756. [DOI] [PubMed] [Google Scholar]
- Subbaiah CC, Palaniappan A, Duncan K, Rhoads MD, Huber CD, Sachs MM. Mitochondrial localization and putative signaling function of sucrose synthase in maize. Journal of Biological Chemistry. 2006;281:15625–15635. doi: 10.1074/jbc.M600355200. [DOI] [PubMed] [Google Scholar]
- Sulmon C, Gouesbet G, El Amrani A, Couée I. Sugar-induced tolerance to the herbicide atrazine in Arabidopsis seedlings involves activation of oxidative and xenobiotic stress responses. Plant Cell Reports. 2006;25:489–498. doi: 10.1007/s00299-005-0062-9. [DOI] [PubMed] [Google Scholar]
- Szarka A, Horemans N, Passarella S, Tarcsay A, Orsi F, Salgó A, Bánhegyi G. Demonstration of an intramitochondrial invertase activity and the corresponding sugar transporters of the inner mitochondrial membrane in Jerusalem artichoke (Helianthus tuberosus L.) tubers. Planta. 2008;228:765–775. doi: 10.1007/s00425-008-0778-1. [DOI] [PubMed] [Google Scholar]
- Tamoi M, Tabuchi T, Demuratani M, Otori K, Tanabe N, Maruta T, Shigeoka S. Point mutation of a plastidic invertase inhibits development of the photosynthetic apparatus and enhances nitrate assimilation in sugar-treated Arabidopsis seedlings. Journal of Biological Chemistry. 2010;285:15399–15407. doi: 10.1074/jbc.M109.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Ende W, Van Laere A. Purification and properties of a neutral invertase from the roots of Cichorium intybus L. Physiologia Plantarum. 1995;93:241–248. [Google Scholar]
- Van den Ende W, Van Laere A. De-novo synthesis of fructans from sucrose in vitro by a combination of two purified enzymes (sucrose:sucrose 1-fructosyltransferase and fructan:fructan 1-fructosyltransferase) from chicory roots (Cichorium intybus L.) Planta. 1996;200:335–342. [Google Scholar]
- Van den Ende W, Valluru R. Sucrose, sucrosyl oligosaccharides, and oxidative stress: scavenging and salvaging? J Exp Bot. 2009;60:9–18. doi: 10.1093/jxb/ern297. [DOI] [PubMed] [Google Scholar]
- Vargas WA, Cumino A, Salerno GL. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta. 2003;216:951–960. doi: 10.1007/s00425-002-0943-x. [DOI] [PubMed] [Google Scholar]
- Vargas WA, Pontis HG, Salerno GL. Differential expression of alkaline and neutral invertases in response to environmental stresses: characterization of an alkaline isoform as a stress-response enzyme in wheat leaves. Planta. 2007;226:1535–1545. doi: 10.1007/s00425-007-0590-3. [DOI] [PubMed] [Google Scholar]
- Vargas WA, Pontis HG, Salerno GL. New insights on sucrose metabolism: evidence for an active A/N Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta. 2008;227:795–807. doi: 10.1007/s00425-007-0657-1. [DOI] [PubMed] [Google Scholar]
- Vargas WA, Salerno GL. The Cinderella story of sucrose hydrolysis: alkaline/neutral invertases, from cyanobacteria to unforeseen roles in plant cytosol and organelles. Plant Science. 2010;178:1–8. [Google Scholar]
- Walker RP, Winters AL, Pollock CJ. Purification and characterization of invertases from leaves of Lolium temulentum. New Phytologist. 1997;135:259–266. [Google Scholar]
- Weber H, Borisjuk L, Heim U, Buchner P, Wobus U. Seed coat-associated invertases of fava bean control both unloading and storage functions: cloning of cDNAs and cell type-specific expression. The Plant Cell. 1995;7:1835–1846. doi: 10.1105/tpc.7.11.1835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welham T, Pike J, Horst I, et al. A cytosolic invertase is required for normal growth and cell development in the model legume. Lotus japonicus. Journal of Experimental Botany. 2009;60:3353–3365. doi: 10.1093/jxb/erp169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S-G, Kodama R, Wang H, Ichii M, Taketa S, Yoshida H. Analysis of the rice SHORT-ROOT5 gene revealed functional diversification of plant neutral/alkaline invertase family. Plant Science. 2009;176:627–634. [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.