Abstract
ABA-INSENSITIVE (ABI)4 is a transcription factor implicated in response to ABA in maturing seeds, and seedling responses to ABA, salt, and sugar. Previous studies have shown that ABI4 transcripts are high in seeds and in seedlings exposed to high concentrations of glucose and, to a lesser extent, osmotic agents and ABA, but that transcript levels are very low through most of vegetative growth. This study examined ABI4 protein accumulation indirectly, using transgenic lines expressing fusions to GFP and GUS. The GFP fusions were active, but undetectable visually or immunologically. Comparison of transcript and activity levels for GUS expression showed that inclusion of the ABI4 coding sequence reduced the ratio of activity to transcript ∼40-fold when driven by the CaMV 35S promoter, and nearly 150-fold when controlled by the ABI4 promoter. At least part of this discrepancy is due to proteasomal degradation of ABI4, resulting in a half-life of 5–6 h for the ABI4–GUS fusion. Comparison of the spatial localization of transcripts and fusion proteins indicated that the protein preferentially accumulated in roots such that transcript and protein distribution had little similarity. The components mediating targeting to the proteasome or other mechanisms of spatial restriction have not yet been identified, but several domains of ABI4 appear to contribute to its instability.
Keywords: Abscisic acid, ABI4, Arabidopsis, post-transcriptional regulation, proteasome, protein stability
Introduction
Production of healthy viable seedlings depends on a successful transition from seed maturation through developmental arrest to germination and seedling growth. These events are controlled by numerous regulators integrating response to internal signals such as abscisic acid (ABA) and gibberellins, and environmental factors including cold, light, and water availability. Early genetic studies identified the transcription factors ABA-INSENSITIVE(ABI)3, ABI4, and ABI5 as central mediators of this signalling (reviewed in Finkelstein et al., 2002). All three of the ABI transcription factor genes are expressed throughout seed development, reaching their highest transcript levels at seed maturity, but decreasing during germination unless exposed to stresses that inhibit germination such as ABA or dehydrating conditions. Subsequent studies have placed them in a much larger transcriptional hierarchy with extensive cross-regulation among the LEAFY COTYLEDON (LEC) loci, the ABI loci, additional B3-domain loci such as ABI3/VP1-like genes and FUSCA3, and genes encoding the ABI5-related bZIP factors such as the ABF/AREBs controlling the transition from embryogenesis to seed maturity and eventual seedling growth (Finkelstein et al., 2005; To et al., 2006; Suzuki et al., 2007). Some of these factors are also regulated post-transcriptionally: activity of ABI5 and related factors depends on phosphorylation (reviewed in Cutler et al., 2010), FUSCA3 is proteasomally degraded during embryo maturation and germination (Lu et al., 2010), and both ABI3 and ABI5 are degraded via the proteasome in germinating seedlings (Lopez-Molina et al., 2001; Zhang et al., 2005).
Although the ABI4 locus was initially identified on the basis of ABA-resistant germination of mutants (Finkelstein, 1994), additional abi4 alleles have been isolated in screens for defects in salt or sugar signalling in seedlings (Arenas-Huertero et al., 2000; Huijser et al., 2000; Laby et al., 2000; Quesada et al., 2000), and retrograde regulation of plastids (Koussevitzky et al., 2007). Consistent with roles in glucose signalling and expression of plastid proteins, ABI4 expression has been shown to increase dramatically in response to growth-inhibiting concentrations of glucose (Arroyo et al., 2003). Furthermore, the ABI4 protein binds to cis-acting elements mediating both sugar- and ABA-inducible gene expression (Bossi et al., 2009; Reeves et al., 2011) and sugar and ABA repression of photosynthetically active nuclear genes (Acevedo-Hernández et al., 2005).
To analyse ABI4 function and determine whether ABI4 protein accumulation parallels its transcript accumulation, transgenic lines were constructed that overexpressed ABI4 with a variety of fusion tags. These studies revealed that ABI4 is also post-transcriptionally regulated.
Materials and methods
Transgene constructs and plant transformation
35S–GFP–ABI4 fusions were constructed by ligating an EcoRI cDNA fragment encoding all but the first two and last amino acids of ABI4 into the pEGAD vector (accession no. AF218816), as described in Reeves et al. (2011). 35S–ABI4–GR fusions were constructed in pBI-ΔGR, a derivative of pBI121 in which the β-glucuronidase (GUS) gene is replaced with a fragment encoding amino acids (aa) 508–795 of the rat glucocorticoid receptor (Lloyd et al., 1994). 35S–ABI4–GUS and 35S–ABI4domain–GUS fusions were constructed in pBI121 (accession no. AF485783) (Jefferson et al., 1987). The ‘full-length’ ABI4 fusion contains 30 bp of 5'UTR and all but the last two codons of ABI4 (aa 1–326). The N-terminal fusion includes aa 1–224, the C-terminal fusion encodes aa 178–327. The various domains are delimited as follows:
ΔPEST aa 51–326
Δ(PEST-AP2) aa 101–326
PEST aa 1–54
PEST-AP2 aa 1–103
AP2-ST aa 51–187
ST aa 101–187
Q aa 178–213
Plasmids carrying the transgenes were introduced into Agrobacterium tumefaciens line GV3101 by direct transformation, followed by selection for growth on kanamycin. Transgenic lines were constructed by floral dip transformation (Clough and Bent, 1998), followed by selection of transformed seeds on the basis of kanamycin or BASTA resistance.
The ABI4pro–GUS construct was described in Söderman et al. (2000); additional lines with this transgene in the rdr6 background were constructed for comparison with the ABI4pro–ABI4–GUS lines.
Plant growth conditions
Germination and seedling growth assays testing functionality of transgenes were performed as described in Söderman et al. (2000). For testing stability of fusion proteins, seedlings were grown initially on germination medium (GM: 0.5×MS salts and vitamins, 1% sucrose) solidified with 0.7% agar, then transferred to liquid GM in multiwell plates supplemented with cycloheximide, MG132 (Peptides International), or the appropriate solvent controls (EtOH and DMSO, respectively) at the concentrations indicated.
Measurement of GUS activity
GUS activity in intact plants was detected histochemically by vacuum infiltration with 5-bromo-4-chloro-3-indolyl-β-D-glucuronic acid (X-gluc), as described in Jefferson et al. (1987). Plant material was incubated in GUS staining solution containing 50 mM sodium phosphate pH 7.0, 0.1% Triton X-100, 0.5 mM K3/K4 FeCN, and 1 mM X-Gluc at 37 °C for 2–72 h depending on staining intensity. Tissues were cleared of chlorophyll in ethanol. Photographs of whole-mounted tissues were taken using a stereomicroscope.
Soluble extracts of seedlings were assayed fluorometrically for GUS activity, using 4-methylumbelliferyl glucuronide (Rose Scientific Ltd, Canada) as substrate, as described in Jefferson et al. (1987), and normalized relative to total protein content measured by Bradford assays (Bio-Rad).
RNA extraction and hybridization
RNA was extracted from seedling tissues by a modification of the procedure described in Verwoerd et al. (1989), and concentrations were estimated based on absorbance at 260 and 280 nm.
Total RNA was size fractionated on MOPS–formaldehyde gels, then transferred to Magna Nylon membranes (Osmonics, Westborough, MA, USA) using 20×SSPE as blotting buffer, and was bound to the filters by UV-crosslinking (120 mJ cm−2 at 254 nm) as previously described (Söderman et al., 2000). Uniformity of loading and transfer was assayed qualitatively by methylene blue staining of the filters and eventually hybridization to an rDNA probe. Transgene transcripts were detected by hybridization to ABI4 or GUS clones, labelled by random-priming to a specific activity of 108 cpm μg−1. Hybridization conditions and washes were as described in Söderman et al. (2000). Hybridization was quantified by phosphoimager analysis; abundance of individual transcripts was normalized relative to rRNA present in each lane.
Results
Post-transcriptional regulation of ABI4
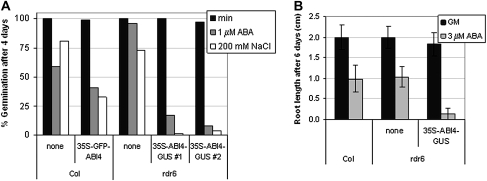
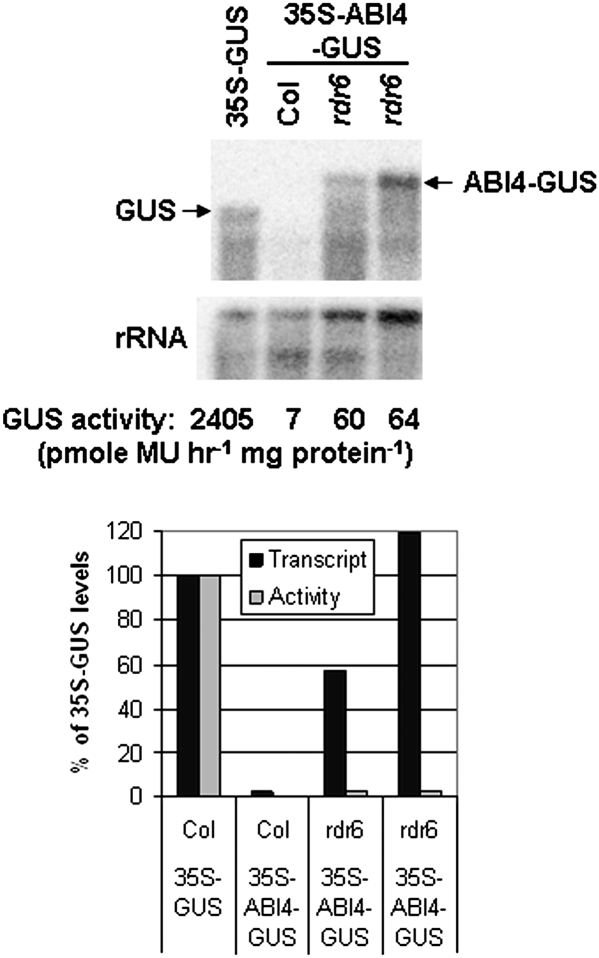
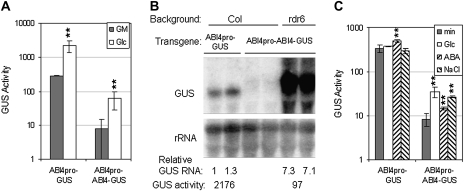
Initial studies of ABI4 overexpression lines demonstrated that this transcription factor was sufficient to confer hypersensitivity to ABA and glucose resulting in reduced root growth, ABA-inducible vegetative expression of genes normally expressed only in seeds, and enhanced glucose-induced accumulation of anthocyanins (Söderman et al., 2000; Finkelstein et al., 2002). However, because the ABI4 protein was undetectable by immunoblotting with antibodies that had been raised against several different epitopes and the initial transgenic lines all inactivated their transgenes over a few generations (data not shown), new lines with fusion proteins that could be readily assayed by activity as well as immunologically were constructed. Function of these transgenes was assayed by their ability to confer hypersensitivity to ABA, salt, and glucose in a wild-type (Col) background and/or complement the ABA resistance of an abi4 mutant. By these criteria, both 35S–GFP–ABI4 and 35S–ABI4–GUS transgenes produced functional ABI4 proteins (Fig. 1 and Reeves et al., 2011), although the overexpression phenotypes were less extreme than those of the original 35S–ABI4 lines (Söderman et al., 2000). In addition, a 35S–ABI4–GR fusion produced steroid-inducible ABI4 activity (Supplementary Fig. S1 available at JXB online), confirming that nuclear localization was required for function. To decrease the likelihood of transgene inactivation, these transgenes were also introduced into the siRNA-reduced rdr6 background (Butaye et al., 2004). Although all of these ABI4 fusion transgenes were similarly highly expressed in a wild-type background, ABI4–GUS transcripts in the rdr6 background were much higher (Supplementary Fig. S1 at JXB online). Consistent with this, the ABI4–GUS fusion in the rdr6 background was detected both histochemically and fluorometrically, albeit at very low levels (Fig. 2 and Supplementary Fig. S2 at JXB online), but the ABI4–GFP fusion was undetectable by either fluorescence or immunoblotting with an anti-GFP antibody (data not shown).
Fig. 1.
35S–GFP–ABI4 and 35S–ABI4–GUS confer hypersensitivity to ABA and salt stress. (A) Hypersensitivity to ABA and NaCl inhibition of germination due to 35S–GFP–ABI4 and 35S–ABI4–GUS transgenes in Col and rdr6 backgrounds, respectively. Germination was scored as radicle emergence after 4 d of incubation on minimal nutrient salt medium (min), or min supplemented with 1 μM ABA or 200 mM NaCl. (B) 35S–ABI4–GUS confers hypersensitivity to ABA for inhibition of root growth in rdr6 background. Root lengths were measured 6 d after transfer from GM to fresh GM with or without 3 μM ABA. Genotypes are indicated by genetic background (Col or rdr6) and transgene present (none, 35S–GFP–ABI4, or 35S–ABI4–GUS).
Fig. 2.
Post-transcriptional control of GUS activity in transgenic lines. (Top) Comparison of GUS transcript levels and GUS activities of 35S–GUS and 35S–ABI4–GUS lines in wild-type and rdr6 backgrounds. The rdr6 lines are derived from independent transformants. (Bottom) Transcript and activity levels are displayed normalized to the levels in the 35S–GUS line.
The reduced activity of the ABI4 fusion proteins could reflect impaired expression at many levels, including transcription, mRNA stability, or translation of the transgene. To distinguish between these possibilities, relative levels of transcripts and GUS activity for 35S–GUS and 35S–ABI4–GUS lines in a wild-type background were compared, as were 35S–ABI4–GUS expression in wild-type and rdr6 backgrounds (Fig. 2). These studies showed at least 50-fold differences in transcript levels, but >300-fold differences in activity levels between 35S–GUS and 35S–ABI4–GUS transgenes in the wild-type background, indicating that transcript levels were not sufficient to explain the differences in activity. Although 35S–ABI4–GUS transcripts in the rdr6 background accumulated to levels similar to those of the 35S–GUS transcripts, GUS activity was still ∼40-fold lower in the 35S–ABI4–GUS fusion line, again supporting regulation at a post-transcript stage. Although all lines showed multiple GUS-homologous degradation products, the differences in ABI4–GUS transcript levels between wild-type and rdr6 lines suggested that the transgene was being aggressively silenced in the wild-type background. Interestingly, the lines with the most active 35S–ABI4–GUS transgenes grew very slowly and either failed to bolt and set seed, or inactivated their transgenes while doing so (data not shown). Lines with slightly lower transgene activity remained active, but homozygous progeny that could complete development and set seed could not be obtained. Consequently, even the lines with ‘active’ transgenes tend to have variable expression as they are comprised of mixtures of plants with different numbers of transgenes, some of which are inactivating.
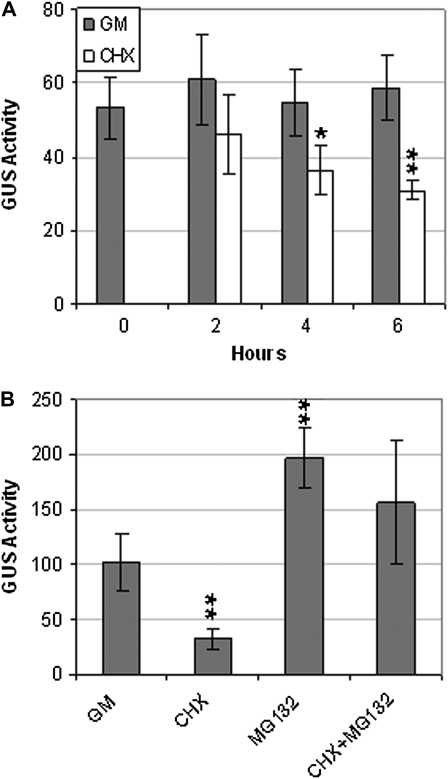
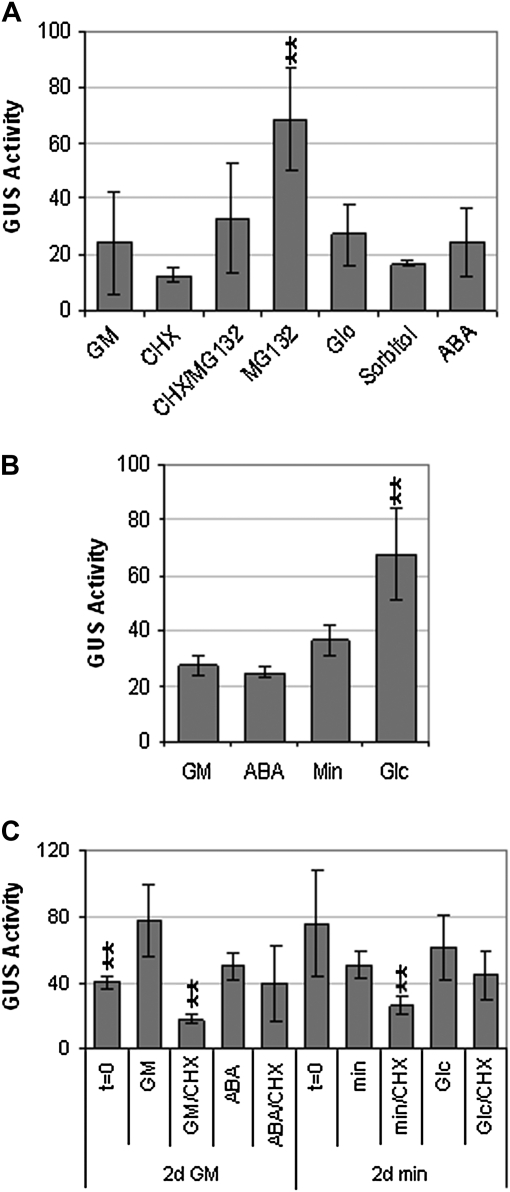
Another possible cause of reduced GUS activity in the transgenic lines was reduced stability of the ABI4–GUS protein. This was tested by assaying GUS activity in the presence or absence of the protein synthesis inhibitor cycloheximide (Fig. 3). Our results showed that the ABI4–GUS fusion protein had a half-life of between 4 and 6 h (Fig. 3), ∼10-fold less than the 50 h reported for GUS itself (Jefferson et al., 1987). However, inclusion of the proteasome inhibitor MG132 largely reversed the effects of cycloheximide, indicating that ABI4 turnover is mediated at least partially by proteasomal degradation.
Fig. 3.
35S–ABI4–GUS activity in rdr6 background. (A) Comparison of GUS activities during 6 h incubation in GM, with or without cycloheximide (CHX). (B) Comparison of GUS activities after 5 h exposure to the indicated treatments. Seedlings were incubated in GM, supplemented with CHX and/or MG132, or the appropriate solvent controls. GUS activity units are pmol MU h−1 mg protein−1. ** and * indicate statistically different from activity in GM (P<0.01 and P<0.02, respectively, based on two-tailed Student's t-test).
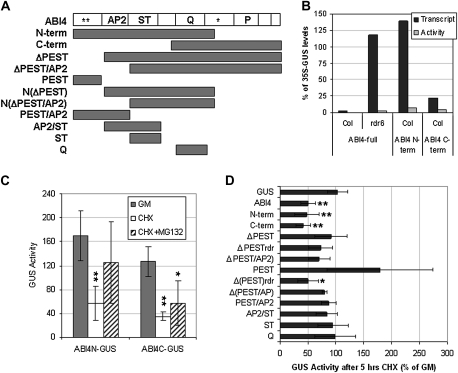
Domains involved in instability
Analysis of the predicted amino acid sequence of ABI4 revealed no clear degradation-associated motifs other than a possible PEST domain near the amino terminus (aa 22–40, PESTfind score: +13.48) and two poor PEST sequences in the carboxy half (aa 218–236, PESTfind score –2.65, and aa 274–311, PESTfind score –1.51) (Rechsteiner and Rogers, 1996). To test the relative stability of different domains of the protein, a series of 35S-(ABI4domain)–GUS fusion lines was constructed (Fig. 4A). Comparison of GUS activity levels in these lines showed that fusions containing either the amino or carboxy halves of the protein were more active than those with the full-length protein, but still much less active than GUS alone (Fig. 4B). Differences in GUS fusion transcript levels were not sufficient to account for the different activities (Fig. 4B), indicating that fusion accumulation was still regulated at a post-transcript stage. Cycloheximide treatment for 5 h reduced all three of these fusions to ∼50% of their levels in control treatments. However, MG132 suppressed this effect only for fusions containing the N-terminal half of ABI4, suggesting that proteasomal degradation depended on motif(s) in this half of the protein (Fig. 4C).
Fig. 4.
Mapping ABI4 domains contributing to instability. (A) Domain structure and subclones; * potential PEST domains; AP2, APETALA2 domain; ST, serine/threonine-rich domain; Q, glutamine-rich domain; P, proline-rich domain. (B) Comparison of GUS transcript and activity in full-length, N-terminal, and C-terminal domain fusions relative to 35S–GUS expression. (C) Comparison of GUS activity (pmol MU h−1 mg protein−1) of N-terminal and C-terminal domain fusions following 5 h exposure to the indicated treatments. Seedlings were incubated in GM, supplemented with cycloheximide (CHX) with or without MG132, or the appropriate solvent controls. ** and * indicate statistically different from activity in GM (P<0.01 and P<0.03, respectively, based on ANOVA). (D) Effect of CHX treatment on GUS activity of all deletion transgenes. ** and * indicate fusions with statistically different stability in CHX compared with 35S–GUS (P<0.01 and P<0.02, respectively, based on ANOVA).
The GUS activities of the fusion lines varied over several orders of magnitude, even for a single construct, as is common for independent transformants. Part of this variability was due to differences in transcript level, but several of the fusions containing smaller regions of ABI4 also had higher ratios of activity to transcript (Supplementary Fig. S3 at JXB online). The fusions with the highest activity were those containing just the potentially destabilizing PEST domain or the Q-rich domain, and these remained at high levels in the presence of cycloheximide (Fig. 4D). Fusion proteins lacking the PEST and AP2 domains were slightly more stable than the full-length or N-terminal fusions, retaining ∼80% of their activity after 5 h exposure to cycloheximide. Although this suggested that the AP2 domain contributed to instability, fusions containing both the AP2 and ST-rich domains were not significantly less stable than GUS.
Developmental and environmental regulation
ABI4 transcripts have been shown to be highly expressed in seeds and in seedlings exposed to stresses such as high glucose, and to a lesser extent ABA and osmoticum (Arroyo et al., 2003). However, if the ABI4 protein is unstable, these major fluctuations in transcript levels may not result in substantial changes in ABI4 activity. To determine whether any of the environmental signals inducing ABI4 transcript accumulation could also enhance protein stability, the effects of ABA, glucose, and sorbitol on GUS activity were tested (Fig. 5A). None of these signals stabilized the 35S–ABI4–GUS fusion product in 8-d seedlings to the same extent as seen with MG132. ABA and glucose effects on the stability of the 35S–ABI4–GUS product at up to 2 d post-stratification were also tested because previous studies had shown that seedlings are most sensitive to ABA and stress-induced growth arrest during the first 48 h post-imbibition (Gibson et al., 2001; Lopez-Molina et al., 2001). Although both ABA and glucose reduced germination and growth of these seeds, only glucose-treated seedlings had slightly higher ABI4–GUS activity (Fig. 5B). However, transfer to media for a 6-h exposure to ABA or glucose did not significantly stabilize the ABI4–GUS fusion, which was still substantially degraded in the presence of cycloheximide (Fig. 5C). This suggests that the large increase reported for ABI4 transcript levels in 3-d seedlings exposed to 7% glucose (Arroyo et al., 2003) might not actually result in a comparable increase in ABI4 protein.
Fig. 5.
Developmental or stress regulation of 35S–ABI4–GUS activity. (A) GUS activity in 8-d seedlings exposed to the indicated treatments for 5 h (100 μM CHX, 100 μM MG132, 6% glucose (Glc), 6% sorbitol, 100 μM ABA); (B) GUS activity in seedlings stratified and incubated for an additional 2 d on indicated medium (GM, GM + 3 μM ABA, min, min + 6% glucose), (C) GUS activity in 2 d seedlings germinated on either GM or min medium, then transferred for an additional 6 h to the indicated medium (as in B, with or without 100 μM CHX). GUS activity units, media, and treatment abbreviations as described in Figs 1, 3. ** indicates statistically different from activity on GM or minimal medium (P<0.01, based on ANOVA).
It is possible that plants can tolerate only a limited amount of ABI4, such that all 35S-driven expression exceeds this level and they are unable to stabilize fusions to such high levels except by pharmacological inhibition of the degradation machinery. To test this possibility, transgenic lines with ABI4–GUS under control of the ABI4 promoter were constructed. ABI4pro–ABI4–GUS transgenes partially complemented an abi4 mutant (Supplementary Fig. S4 at JXB online), but the GUS activity was undetectable. The levels were higher in an rdr6 background, permitting comparison of glucose-induced transcript and activity levels in ABI4pro–GUS and ABI4pro–ABI4–GUS lines. ABI4pro–GUS lines in the rdr6 background had similar activities to those in the Col background (data not shown). Although seedlings with either transgene had ∼8-fold higher GUS activity after 6 d on 5% glucose than when grown on 1% glucose, they differed in that these levels were ∼20-fold higher in the ABI4pro–GUS lines (Fig. 6A). This might reflect the stronger expression of ABI4pro–GUS in the shoots, or a higher total protein concentration in the ABI4pro–ABI4–GUS seedlings due to their minimal growth on glucose (Supplementary Fig. S5 at JXB online). Two possible explanations for the limited activity of the ABI4–GUS fusion protein are limited transcript accumulation or limited protein accumulation. To distinguish between these, GUS transcript levels were measured and, surprisingly, showed that the ABI4pro–ABI4–GUS transcripts were actually ∼7-fold higher than the ABI4pro–GUS transcripts (Fig. 6B). Consequently, the GUS activity per transcript was nearly 150-fold higher for ABI4pro–GUS than for the ABI4pro–ABI4–GUS transgene.
Fig. 6.
Glucose regulation of transcriptional and translational ABI4–GUS fusions. (A) GUS activity of ABI4pro–GUS and ABI4pro–ABI4–GUS seedlings after 6 d incubation on GM with or without 5% glucose (Glc). (B) GUS transcript levels in seedlings grown and harvested in parallel with those used for GUS assays in (A). (C) GUS activity of ABI4pro–GUS and ABI4pro–ABI4–GUS seedlings after 2 d incubation on minimal medium with or without 6% glucose, 2 μM ABA, or 200 mM NaCl. GUS activity units, displayed on a log scale, are pmol MU h−1 mg protein−1. ** indicates statistically different from activity on minimal medium (P<0.01, based on two-tailed Student's t-test).
Comparison of ABI4pro–GUS and ABI4pro–ABI4–GUS function after only 2 d showed similar GUS activities for the transcriptional fusion with or without high glucose or NaCl, but both stresses induced a 3- to 4-fold increase in fusion protein activity (Fig. 6C), suggesting that they primarily affect protein accumulation. In contrast, exposure to 2 μM ABA induced mild (∼1.5-fold) increases in GUS activity of both fusion lines, indicating that ABA primarily affected transcript accumulation. However, the fusion protein activities were ∼15-fold lower than those of the transcriptional fusion under all conditions. As at 6 d, the promoter was active in both shoots and roots, even without glucose, but the ABI4–GUS fusion protein did not accumulate in unstressed shoots (Supplementary Fig. S5 at JXB online).
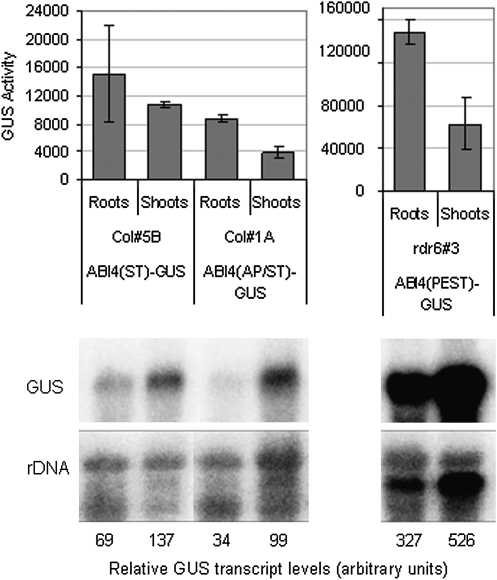
Histochemical staining of 35S–(ABI4domain)–GUS transgenic lines also revealed non-uniform expression, with GUS activities higher in roots than shoots for many lines (Supplementary Fig. S6 at JXB online), in contrast to the constitutively high expression of 35S–GUS fusions throughout the plant. Comparison of transcript levels shows that the ABI4–GUS fusion transcripts are often 2- to 3-fold more abundant in shoots (Fig. 7), indicating that the GUS activity disparities are due to tissue-specific differences in translation or protein stability.
Fig. 7.
Organ-specific differences in ABI4–GUS activity. (Top) GUS activity levels (pmol MU h−1 mg protein−1) in roots and shoots of the indicated transgenic lines. AP/ST- and PEST-domain fusions have statistically different activity in roots and shoots (P=0.00024 and P= 0.0028, respectively, based on two-tailed Student's t-test) (Bottom) RNA gel blots showing GUS-fusion transcript levels in parallel samples aligned with their activity levels.
Discussion
Numerous transgenic lines with ABI4 fusions under control of either the CaMV 35S promoter or the ABI4 promoter have transgene expression levels sufficient for complementation of the abi4 mutation, yet are often undetectable by GFP or GUS activity. Lines that achieve higher levels of transgene expression display very low ratios of activity relative to the transcripts encoding these fusions. In fact, even constitutive expression via the CaMV 35S promoter was not sufficient to raise ABI4–GUS activity levels above those produced by glucose-inducible expression via the ABI4 promoter. Although these experiments do not exclude the possibility of poor translation or improper folding, the fact that similar physiological phenotypes have been produced by 35S-driven ABI4–GUS, GFP–ABI4, and GR–ABI4 fusions, as well as 35S–ABI4, yet most are undetectable immunologically and these transgenes tend to inactivate rapidly, suggests that these proteins are simply accumulated to low levels. Our studies show that in the case of the GUS fusions the low activity reflects protein instability, at least partly via the proteasome. The instability of ABI4 is reminiscent of similar regulation of ABI3 and ABI5, but differs in that ABA can stabilize those transcription factors (Lopez-Molina et al., 2001; Zhang et al., 2005), but not ABI4. However, high glucose is a more effective inducer of ABI4 expression than ABA (Arroyo et al., 2003) and also promotes ABI4 accumulation within 2 d after stratification, as do growth-inhibiting levels of NaCl and ABA.
Proteasomal regulation of transcriptional regulators has been well-characterized for the AUX/IAA repressors of auxin response, the JAZ repressors of jasmonate response, the DELLA protein repressors of GA response, the EIN3 regulator of ethylene response factors, and two ABA response factors: ABI3 and ABI5 (reviewed in Vierstra, 2009). For most of these, the half-lives have been documented to be as little as an hour or less, which is substantially shorter than that observed for ABI4. F-box subunits of the E3 ligases required for ubiquitination leading to degradation are known for the auxin, jasmonic acid, gibberellin and ethylene regulators, and specific conserved domains have been identified as essential for instability in the DELLA and AUX/IAA proteins. Two RING E3 ligases involved in ABI factor degradation have also been identified: KEEP ON GOING (KEG), which ubiquitinates ABI5, and an ABI3-interacting protein (AIP2) (Zhang et al., 2005; Stone et al., 2006). AIP2 is highly expressed in freshly stratified seeds, where it can induce destruction of ABI3 as part of dormancy release. In addition, AIP2 levels increase in vegetative tissues exposed to ABA, leading to ABI3 degradation and decreased ABA signalling at later stages. Surprisingly, ABA has the opposite effect on KEG; by promoting self-ubiquitination and degradation of KEG, it inhibits destruction of ABI5 (Liu and Stone, 2010). ABI5 action is also regulated by sumoylation, which both represses its activity and increases its stability (Miura et al., 2009). An additional class of ABI5-interacting proteins, the AFPs, have been implicated in altering stability of ABI5, but the mechanism is not clear (Garcia et al., 2008; Lopez-Molina et al., 2003) and recent studies suggest that they may actually function as transcriptional co-repressors (Pauwels et al., 2010). ABA sensitivity of germination, seedling sugar sensitivity, and lipid breakdown are also regulated by the N-end rule pathway of protein degradation, but the specific substrates involved have not yet been identified (Holman et al., 2009). A recurring theme is the existence of multiple regulators responsible for controlling stability of a given protein or protein family in a variety of tissues or conditions.
Superficially one might expect reciprocal abundance of destabilizing factors and their targets, but many (e.g. AIP2, EBF1 and EBF2, AFP1 and AFP2), are components of negative feedback loops such that their accumulation is induced by the signals whose action they will inhibit. Furthermore, many of the destabilizing factors are post-transcriptionally regulated themselves. For example, the auxin receptor F box genes are broadly transcribed, but protein accumulation is under miRNA control (Parry et al., 2009). Consequently, it is not possible to predict candidate regulators based on expression patterns.
Our current study implicates several regions contributing to the instability of ABI4, none of which resemble previously characterized destabilizing domains. Although the susceptibility to proteasomal degradation is likely to involve ubiquitination, some proteins are targeted by ubiquitin-independent mechanisms. The targeting mechanism for ABI4 has not yet been identified.
Previous studies of ABI4 regulation have shown strong induction by glucose in seedlings, with preferential promoter activity in shoots and root tips (Arroyo et al., 2003; Bossi et al., 2009). The current study confirms this result by histochemical staining of ABI4pro–GUS seedlings, but ABI4pro–ABI4–GUS lines show stronger activity in roots than shoots. Similarly, CaMV 35S-driven expression generally resulted in higher ABI4–GUS transcript levels in shoots, based on RNA gel blot analyses, yet GUS activity was usually higher in roots. This difference in the ratio of activity to transcript between roots and shoots implies preferential translation or stability in roots such that the levels of functional ABI4 do not reflect the transcript levels. To date, searches of small RNA databases (available at http://asrp.cgrb.oregonstate.edu/db/) have not shown any likely candidates for regulators of ABI4. However, a variety of RNA-binding proteins have been implicated in stress responses (reviewed in Lorković, 2009), including a zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, with mutant and overexpression phenotypes very similar to those for ABI4 (Kim et al., 2007). Although ABI4 transcript levels are unaffected in these loss- and gain-of-function lines, this does not preclude the possibility of effects on translation.
In summary, these studies show that ABI4 is subject to stringent post-transcriptional regulation that prevents the protein from accumulating to high levels, and restricts its action to a subset of the tissues where the gene is expressed. The specific regulatory components remain unknown, but at least part of the mechanism involves proteasomal degradation.
Supplementary data
Supplementary Fig. S1. 35S–ABI4–GR transgenes confer dexamethasone (Dex)-dependent hypersensitivity to ABA inhibition of germination and root growth, and glucose (Glc) inhibition of germination and seedling growth. Transcript levels for these ABI4 fusion transgenes are similar to those for the GFP– and –GUS fusions in a wild-type background.
Supplementary Fig. S2. 35S–ABI4–GUS activity in Col (left) and rdr6 (right) backgrounds. Fluorometrically assayed GUS activity is ∼10-fold higher in the rdr6 background.
Supplementary Fig. S3. Comparison of GUS transcript and activity levels shows that all ABI4 domain fusion constructs displayed except that containing only the PEST domain accumulate fusion proteins relatively inefficiently.
Supplementary Fig. S4. ABI4pro–ABI4–GUS weakly complements the abi4 mutation, suppressing the glucose resistance of this background.
Supplementary Fig. S5. Histochemical staining of GUS activity in seedlings of the indicated genotypes (ABI4pro–GUS and ABI4pro–ABI4–GUS) grown for 6 d on GM with or without 5% glucose, or 2 d on minimal medium with or without 6% glucose.
Supplementary Fig. S6. Histochemical staining of GUS activity in a variety of 35S-(ABI4domain)–GUS transgenic seedlings. Activity varied substantially between independent transgenic lines for each fusion, and even between individual progeny of each line, but the shoots were much more likely to lose activity than the roots.
Acknowledgments
The authors thank Drs Chris Rock for the rdr6 line, Sean Cutler for the pEGAD vector, Eva Soderman for the pBI-ΔGR vector, and Douglas Bush for helpful discussions. This work was supported by the National Science Foundation (Grant #0446048 to R.R.F.) and the UC Leadership Excellence through Advanced DegreeS (UC LEADS) programme (fellowship to M.P.).
Glossary
Abbreviations
- ABA
abscisic acid
- ABI
ABA insensitive
- GFP
green fluorescent protein
- GM
germination medium
- GR
glucocorticoid receptor
- GUS
β-glucuronidase
References
- Acevedo-Hernández GJ, León P, Herrera-Estrella LR. Sugar and ABA responsiveness of a minimal RBCS light-responsive unit is mediated by direct binding of ABI4. The Plant Journal. 2005;43:506–519. doi: 10.1111/j.1365-313X.2005.02468.x. [DOI] [PubMed] [Google Scholar]
- Arenas-Huertero F, Arroyo A, Zhou L, Sheen J, León P. Analysis of Arabidopsis glucose insensitive mutants, gin5 and gin6, reveals a central role of the plant hormone ABA in the regulation of plant vegetative development by sugar. Genes and Development. 2000;14:2085–2096. [PMC free article] [PubMed] [Google Scholar]
- Arroyo A, Bossi F, Finkelstein R, León P. Three genes that affect sugar sensing: ABA INSENSITIVE4, ABA INSENSITIVE5 and CONSTITUTIVE TRIPLE RESPONSE1, are differentially regulated by glucose in Arabidopsis thaliana. Plant Physiology. 2003;133:231–242. doi: 10.1104/pp.103.021089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi F, Cordoba E, Dupré P, Mendoza MS, Román CS, León P. The Arabidopsis ABA-INSENSITIVE (ABI) 4 factor acts as a central transcription activator of the expression of its own gene, and for the induction of ABI5 and SBE2.2 genes during sugar signaling. The Plant Journal. 2009;59:359–374. doi: 10.1111/j.1365-313X.2009.03877.x. [DOI] [PubMed] [Google Scholar]
- Butaye KMJ, Goderis IJWM, Wouters PFJ, Pues JMTG, Delauré SL, Broekaert WF, Depicker A, Cammue BPA, De Bolle MFC. Stable high-level transgene expression in Arabidopsis thaliana using gene silencing mutants and matrix attachment regions. The Plant Journal. 2004;39:440–449. doi: 10.1111/j.1365-313X.2004.02144.x. [DOI] [PubMed] [Google Scholar]
- Clough S, Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: emergence of a core signaling network. Annual Review of Plant Biology. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Gampala SSL, Lynch TJ, Thomas TL, Rock CD. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Molecular Biology. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- Finkelstein R, Gampala S, Rock C. Abscisic acid signaling in seeds and seedlings. The Plant Cell. 2002;14(Suppl. 1):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. The Plant Journal. 1994;5:765–771. [Google Scholar]
- Garcia M, Lynch T, Peeters J, Snowden C, Finkelstein R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Molecular Biology. 2008;67:643–658. doi: 10.1007/s11103-008-9344-2. [DOI] [PubMed] [Google Scholar]
- Gibson SI, Laby RJ, Kim D. The sugar-insensitive1 (sis1) mutant of Arabidopsis is allelic to ctr1. Biochemical and Biophysical Research Communications. 2001;280:196–203. doi: 10.1006/bbrc.2000.4062. [DOI] [PubMed] [Google Scholar]
- Holman TJ, Jones PD, Russell L, et al. The N-end rule pathway promotes seed germination and establishment through removal of ABA sensitivity in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2009;106:4549–4554. doi: 10.1073/pnas.0810280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijser C, Kortstee A, Pego J, Weisbeek P, Wisman E, Smeekens S. The Arabidopsis SUCROSE UNCOUPLED-6 gene is identical to ABSCISIC ACID INSENSITIVE-4: involvement of abscisic acid in sugar responses. The Plant Journal. 2000;23:577–585. doi: 10.1046/j.1365-313x.2000.00822.x. [DOI] [PubMed] [Google Scholar]
- Jefferson R, Kavanagh T, Bevan M. GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO Journal. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-O, Pan S, Jung C-H, Kang H. A zinc finger-containing glycine-rich RNA-binding protein, atRZ-1a, has a negative impact on seed germination and seedling growth of Arabidopsis thaliana under salt or drought stress conditions. Plant and Cell Physiology. 2007;48:1170–1181. doi: 10.1093/pcp/pcm087. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. [PubMed] [Google Scholar]
- Laby R, Kincaid M, Kim D, Gibson S. The Arabidopsis sugar-insensitive mutants sis4 and sis5 are defective in abscisic acid synthesis and response. The Plant Journal. 2000;23:587–596. doi: 10.1046/j.1365-313x.2000.00833.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Stone SL. Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. The Plant Cell. 2010;22:2630–2641. doi: 10.1105/tpc.110.076075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd A, Schena M, Walbot V, Davis R. Epidermal cell fate determination in Arabidopsis: patterns defined by a steroid-inducible regulator. Science. 1994;266:436–439. doi: 10.1126/science.7939683. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua N- H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Kinoshita N, Chua N- H. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes and Development. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković ZJ. Role of plant RNA-binding proteins in development, stress response and genome organization. Trends in Plant Science. 2009;14:229–236. doi: 10.1016/j.tplants.2009.01.007. [DOI] [PubMed] [Google Scholar]
- Lu QS, Dela Paz J, Pathmanathan A, Chiu RS, Tsai AYL, Gazzarrini S. The C-terminal domain of FUSCA3 negatively regulates mRNA and protein levels, and mediates sensitivity to the hormones abscisic acid and gibberellic acid in Arabidopsis. The Plant Journal. 2010;64:100–113. doi: 10.1111/j.1365-313X.2010.04307.x. [DOI] [PubMed] [Google Scholar]
- Miura K, Lee J, Jin JB, Yoo CY, Miura T, Hasegawa PM. Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proceedings of the National Academy of Sciences, USA. 2009;106:5418–5423. doi: 10.1073/pnas.0811088106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry G, Calderon-Villalobos LI, Prigge M, Peret B, Dharmasiri S, Itoh H, Lechner E, Gray WM, Bennett M, Estelle M. Complex regulation of the TIR1/AFB family of auxin receptors. Proceedings of the National Academy of Sciences, USA. 2009;106:22540–22545. doi: 10.1073/pnas.0911967106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Barbero GF, Geerinck J, et al. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature. 2010;464:788–791. doi: 10.1038/nature08854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce M, Micol J. Genetic analysis of salt-tolerant mutants in Arabidopsis thaliana. Genetics. 2000;154:421–436. doi: 10.1093/genetics/154.1.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechsteiner M, Rogers SW. PEST sequences and regulation by proteolysis. Trends in Biochemical Sciences. 1996;21:267–271. [PubMed] [Google Scholar]
- Reeves WM, Lynch TJ, Mobin R, Finkelstein RR. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Molecular Biology. 2011;75:347–363. doi: 10.1007/s11103-011-9733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman E, Brocard I, Lynch T, Finkelstein R. Regulation and function of the Arabidopsis ABA-insensitive4 (ABI4) gene in seed and ABA response signaling networks. Plant Physiology. 2000;124:1752–1765. doi: 10.1104/pp.124.4.1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Williams LA, Farmer LM, Vierstra RD, Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. The Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Wang HHY, McCarty DR. Repression of the LEAFY COTYLEDON 1/B3 regulatory network in plant embryo development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 genes. Plant Physiology. 2007;143:902–911. doi: 10.1104/pp.106.092320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To A, Valon C, Savino G, Guilleminot J, Devic M, Giraudat J, Parcy F. A network of local and redundant gene regulation governs Arabidopsis seed maturation. The Plant Cell. 2006;18:1642–1651. doi: 10.1105/tpc.105.039925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verwoerd TC, Dekker BMM, Hoekema A. A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Research. 1989;17:2362. doi: 10.1093/nar/17.6.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierstra RD. The ubiquitin–26S proteasome system at the nexus of plant biology. Nature Reviews in Molecular and Cellular Biology. 2009;10:385–397. doi: 10.1038/nrm2688. [DOI] [PubMed] [Google Scholar]
- Zhang X, Garreton V, Chua N- H. The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes and Development. 2005;19:1532–1543. doi: 10.1101/gad.1318705. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.