Abstract
Ntann12, encoding a polypeptide homologous to annexins, was found previously to be induced upon infection of tobacco with the bacterium Rhodococcus fascians. In this study, Ntann12 is shown to bind negatively charged phospholipids in a Ca2+-dependent manner. In plants growing in light conditions, Ntann12 is principally expressed in roots and the corresponding protein was mainly immunolocalized in the nucleus. Ntann12 expression was inhibited following plant transfer to darkness and in plants lacking the aerial part. However, an auxin (indole-3-acetic acid) treatment restored the expression of Ntann12 in the root system in dark conditions. Conversely, polar auxin transport inhibitors such as 1-naphthylphthalamic acid (NPA) or 2,3,5-triiodobenzoic acid (TIBA) inhibited Ntann12 expression in light condition. These results indicate that the expression of Ntann12 in the root is linked to the perception of a signal in the aerial part of the plant that is transmitted to the root via polar auxin transport.
Keywords: Annexin, auxin, Ca2+-binding protein, light, nucleus, polar auxin transport, root
Introduction
Annexins are proteins found in most eukaryotes but are absent from yeasts and prokaryotes (Moss and Morgan, 2004). These proteins contain generally four repeats of a 70 amino acid domain (Gerke and Moss, 1997). Several differences exist between plant and animal annexins. The type II Ca2+-binding site provided by the endonexin fold referred to as the consensus sequence KGXGT-(38 residues)-D/E (Geisow et al., 1986) is highly conserved in 3–4 repeats of animal annexins, whereas the consensus sequence is strictly conserved only in the first repeat in plants (Hofmann, 2004). Furthermore, animal annexins have a long and variable N-terminal domain which is the site for post-translational modifications and interaction with membranes and other proteins (Gerke et al., 2005), whereas plant annexins have a short N-terminal sequence that is involved in the function of the protein (Hofmann et al., 2000; Dabitz et al., 2005). Annexins are characterized by their ability to bind negatively charged phospholipids in a Ca2+-dependent manner (Boustead et al., 1989). Both animal and plant annexins have also been shown to bind phospholipids in a Ca2+-independent manner, suggesting that they can be associated with or inserted into membranes (Lim et al., 1998; Gerke and Moss, 2002; Dabitz et al., 2005; Gorecka et al., 2007).
Animal annexins have been demonstrated to be involved in Ca2+ signalling and transport, membrane dynamics and trafficking, cell proliferation, exocytosis, and endocytosis (reviewed by Gerke et al., 2005). In plants, annexins are described as multifunctional components of growth and adaptation, being associated with diverse cellular processes, and expressed throughout the plant life cycle (reviewed by Mortimer et al., 2008). These proteins have diverse functional domains, including a Ca2+-binding site, a potential F-acting-binding motif, a potential haem-binding domain, and an ATP/GTP-binding domain (reviewed by Mortimer et al., 2008). However, their precise role/function in plants is not clearly understood (Hofmann, 2004; Konopka-Postupolska, 2007). In vitro studies revealed biochemical properties for plant annexins, including nucleotide phosphodiesterase activity (Calvert et al., 1996; Shin and Brown, 1999), peroxidase activity (Gorecka et al., 2005; Laohavisit et al., 2009), callose synthase regulation activity (Andrawis et al., 1993), and F-actin binding activity (Calvert et al., 1996; Hu et al., 2000; Hoshino et al., 2004). In addition, plant annexins have been shown to mediate channel-like Ca2+ transport (Hofmann et al., 2000; Laohavisit et al., 2009) possibly involved in reacive oxygen species signalling (Laohavisit et al., 2010).
Various studies have shown that plant annexins are expressed in tissues/cells associated with secretion and polarized growth, such as pollen tube tips (Blackbourn et al., 1992) and root caps (Clark et al., 1992; Carroll et al., 1998), or differential growth during gravitropism (Clark et al., 2005a). In addition, plant annexin expression is differentially regulated by exposure to salt, gravity, abscisic acid (ABA), drought, high or low temperatures, hydrogen peroxide, phosphate deprivation, and heavy metals (Kovács et al., 1998; Breton et al., 2000; Clark et al., 2001; Cantero et al., 2006; Jami et al., 2009; Konopka-Postupolska et al., 2009; Huh et al., 2010).
The overexpression of AnnAt1 in Arabidopsis has been shown to protect cells against drought stress (Konopka-Postupolska et al., 2009) and oxidative stress (Gorecka et al., 2005), and to play a role in osmotic stress and ABA signalling (Lee et al., 2004). In accordance with this, transgenic cotton and tobacco plants expressing mustard annexin AnnBj1, a homologue to AnnAt1, were shown to be more tolerant towards different abiotic stress treatments (Jami et al., 2008; Divya et al., 2010). In addition, AnnAt1 has been found to rescue the Escherichia coli ΔoxyR mutant from H2O2 stress (Gidrol et al., 1996) and mammalian cells from oxidative stress (Kush and Sabapathy, 2001). Moreover, annAt1 and annAt4, but not annAt2 mutants, were shown to be hypersensitive to ABA and osmotic stress during germination and early seedling growth (Lee et al., 2004). Seedlings of annAt1 and annAt2 mutants grown in the dark showed inhibited root and hypocotyl growth, respectively (Clark et al., 2005b). Recently, Huh et al. (2010) demonstrated that, under long day conditions, the sensitivity to abiotic stress of double annat1 annat4 mutants was lower compared with single mutants and this effect was reversed in transgenic 35S:AnnAt4, suggesting that AnnAt1 and AnnAt4 act as regulators of abiotic stress such as drought and salt.
The isolation and the preliminary characterization of the tobacco Ntann12 gene, encoding a putative annexin whose expression was found to be induced in tobacco BY-2 cells following infection by the phytopathogenic bacterium Rhodoccocus fascians, were previously reported (Vandeputte et al., 2007). The closest homologues to Ntann12 are the annexin-like protein RJ4 (65% identity and 81% similarity) that has been identified to be expressed predominantly in developing and ripening fruit (Wilkinson et al., 1995), and the Medicago truncatula MtAnn1 (58% identity and 77% similarity) that has been shown to be induced during symbotic interactions and suggested to be involved in the Ca2+ response signal elicited by symbiotic signals from rhizobia and mycorrhizal fungi (de Carvalho Niebel et al., 1998; de Carvalho-Niebel et al., 2002). The closest Arabidopsis homologue to Ntann12 is AnnAt8 (57% identity and 78% similarity), which was found to be induced mainly by dehydration and NaCl treatment (Cantero et al., 2006).
In this study, biochemical investigations indicate that Ntann12 binds to negatively charged phospholipids in a Ca2+-dependent manner. Moreover, Ca2+ concentration affects in vitro Ntann12 translocation from cytosolic to membrane-enriched fractions. Ntann12 is highly expressed in root cells and the protein was mainly immunolocalized in the nucleus. Ntann12 expression in the root system was found to be regulated by a light-induced signal coming from plant aerial parts, and polar auxin transport seems to be required for Ntann12 expression in root cells. Taken together, the data presented in this study show the role of light and polar auxin transport in the regulation of the expression of the Ntann12 annexin in tobacco roots.
Materials and methods
Plant materials and growth conditions
Non-transgenic and transgenic tobacco plants (Nicotiana tabacum cv. Havana) were grown aseptically on Murashige and Skoog (MS) medium (Micro and 1/2 concentration Macro elements including vitamins; Duchefa) supplemented with 200 mg l−1 kanamycin (Duchefa) when needed and were grown at 23 °C under a 16 h light photoperiod (70 μmol m−2 s−1, cool white fluorescent lamp, Osram). Sown seeds, or acclimatized plants, were cultivated on soil in a growth chamber at 25 °C under a 16 h light photoperiod.
Production of the recombinant Ntann12 protein in Escherichia coli
The pBAD-DEST49 expression system (Invitrogen, Merelbeke, Belgium) was used to produce a recombinant Ntann12 protein with His-Patch (HP) thioredoxin as an N-terminal fusion partner of the cloned gene product and a hexahistidine tag as a C-terminal fusion partner. According to the manufacturer, a fusion partner with HP thioredoxin may improve the translation and the solubility of the fusion protein. The Ntann12 cDNA (Vandeputte et al., 2007) was flanked by attB1 and attB2 recombination sites by two successive PCRs, the first one using the primers F 5′-AAAAAGCAGGCTATGGCTACAATCAATTA-3′ and R 5′-AGAAAGCTGGGTTTAGTTATCATTTCCC-3′ and the second with the primers containing attB1 and attB2 sites for Gateway cloning by recombination (Invitrogen). After the generation of the entry clone (BPNtann12) in plasmid pDONR-221 (Invitrogen), a second recombination reaction was performed with pBAD-DEST49 according to the manufacturer's instructions and cloned into E. coli TOP10 (Invitrogen).
Production of recombinant proteins in TOP10 cells was induced by the addition of 0.2% L-(+)-arabinose to cultures at an optical density at 600 nm of ∼0.8, and cultivation was continued for an additional 6–7 h at 37 °C. Cells were harvested by centrifugation and cell pellets were frozen at –80 °C. Subsequently, cells were extracted using a Qproteome™ Bacterial Protein Prepkit (Qiagen, Hilden, Germany), containing lysozyme and benzonase (Qiagen) supplemented with protease inhibitor cocktail (Sigma). Lysates were centrifuged at 16 000 g for 30 min at 4 °C and the supernatant (soluble fraction of the bacterial proteins) was collected and used immediately.
Protein analysis
Tobacco seedlings were grown for 4 weeks in solid MS medium. Roots and leaves were harvested separately, immediately frozen and ground to a fine powder in liquid nitrogen using a mortar and pestle, and stored at –80 °C until required. The powder was homogenized and incubated in extraction buffer [50 mM TRIS, pH 7.5, 5 mM EDTA, 2 mM dithiothreitol (DTT), 2% benzonase, and protease inhibitor cocktail for native conditions or 100 mM TRIS-HCl, pH 7.5, 10 mM EDTA, 100 mM LiCl2, 1% SDS, and protease inhibitor cocktail for non-native [conditions](1 g powder ml−1 extraction buffer), and centrifuged at 3220 g (Eppendorf 5810R, rotor A-4-81) for 30 min at 4 °C. To assess the Ca2+ response of Ntann12 proteins in plant cells, the total protein extract was treated with either CaCl2 or EDTA before separation of membrane and soluble protein fractions by ultracentrifugation at 125 000 g (Beckman Optima™ LE-80K, rotor SW60) for 1 h at 4 °C. After centrifugation, the supernatant (cytosolic fraction) was recovered, and the pellet (membrane-enriched fraction) was resuspended in an appropriate volume of extraction buffer (Lee et al., 2004). The crude protein samples, and the cytosolic and the membrane-enriched fractions were divided into aliquots and preferably used immediately, or frozen at –80 °C. Protein concentrations were measured using Dye reagent (Bio-Rad, Hercules, CA, USA) according to the manufacturer's instructions.
Preparation of anti-Ntann12 antibodies
A polyclonal antibody was raised in rabbit against a mix of two Ntann12-specific peptides (ATINYPENPSPVADAC and DPQKYYEKVIRYAI) (Eurogentec Inc., Belgium) and was named anti-Ntann12. Non-purified antibodies were used for western blotting. Affinity-purified antibodies using the synthesized peptides (TOYOPEARL AF-Amino-650M, Eurogentec Inc.) were utilized for immunolocalization. An enzyme-linked immunosorbent assay (ELISA) was performed to demonstrate the specificity of the anti-Ntann12 antibodies for the synthesized peptides as well as the absence of cross-reactivity with the pre-immune serum (Eurogentec). The protein concentration of the purified antibody is 0.25 μg μl−1.
Electrophoresis and immunoblotting
Analysis by SDS–PAGE was performed on 10% acrylamide gels (Invitrogen) with molecular mass standards from Invitrogen (SeeBlue Plus 2), for 50 min at 200 V in 5% (v/v) running buffer (Invitrogen), 0.5% (v/v) antioxidant (Invitrogen). Proteins separated by SDS–PAGE were stained with Coomassie blue (Invitrogen), or were electrophoretically transferred to a polyvinylidene difluoride (PVDF) membrane (GE Healthcare) for 1 h at 30 V in 10% (v/v) methanol, 5% (v/v) transfer buffer (Invitrogen), and 0.5% (v/v) antioxidant (Invitrogen). The PVDF membrane was processed for immunoblotting by incubating for 1 h at 20 °C in phosphate-buffered saline–Tween (PBS-T) (10 mM Na2HPO4, 1.8 mM KH2PO4, 137 mM NaCl, 2.7 mM KCl, 0.05% Tween, pH 7.4) containing 5% (w/v) low fat milk. The blot was incubated overnight at 4 °C in the same buffer containing antiserum raised against Ntann12 at a dilution of 1:5000, washed three times in PBS-T, and then incubated with protein A–peroxidase (Sigma) at a dilution of 1:50 000 for 4 h at 4 °C. Following three washes in PBS-T, peroxidase activity was made visible by incubating the blot with Lumigen™ solutions (GE Healthcare) for 5 min and then scanning (Typhoon 9200). Ntann12 proteins from plants and from bacteria were detected using the above protocol. Western blot using pre-immune serum was performed at a similar dilution, and no cross-reactive band was observed (data not shown).
Phospholipid binding assay
Liposomes were prepared using a 2:1 mixture (mol/mol) of L-α-phosphatidylcholine (PC) and L-α-phosphatidyl-L-serine (PS) (Sigma). Large unilamellar vesicles (100–200 nm) were prepared as follows. Lipids (30 mg) were dissolved in chloroform. A dried film was obtained by evaporation of chloroform under a flow of nitrogen, followed by overnight drying under vacuum. The lipid film was vortexed in 7.5 ml of HEPES buffer (100 mM KCl, 2 mM MgCl2, 25 mM HEPES pH 7.5) at 37 °C, frozen in liquid nitrogen, and thawed (five cycles). Finally, the suspension was extruded 10 times through two stacked 100 nm (pore diameter) polycarbonate filters (Nuclepore, Costar, Cambridge, MA, USA) in a thermobarrel extruder (Lipex Biomembranes, Vancouver, Canada) at 37 °C. Proteins (50 μg) were incubated for 30 min at 20 °C with liposomes (400 μg of phospholipids) in 1 ml of HEPES buffer containing 0 or 2 mM Ca2+. After centrifugation at 86 000 g (Beckman Optima™ LE-80K, rotor SW60) for 1 h at 4 °C, the supernatant containing liposomes was fractionated over a discontinuous sucrose gradient (35–30–25–20%–Sample). Sucrose was diluted in HEPES buffer containing 0 or 2 mM Ca2+. Following centrifugation overnight at 86 000 g (Beckman, rotor SW60) and 4 °C, liposomes were harvested in the upper layer and in the 20% sucrose layer. Liposomes were washed twice with HEPES buffer containing 0 or 2 mM Ca2+ by centrifuging for 1 h at 86 000 g and 4 °C, and suspended in an appropriate volume of HEPES buffer containing 0 or 2 mM Ca2+. The liposomal fractions were analysed by 10% SDS–PAGE and gel blotting with the anti-Ntann12 antibody.
Immunolabelling
Roots were fixed with 4% paraformaldehyde and 0.3% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.2) for 3 h at room temperature and incubated overnight at 4 °C. Samples were then serially dehydrated in increasing concentrations of methanol, embedded in Lowicryl K4M (Electron Microscopy Sciences) at –20 °C, and left to polymerize for 2 d at –20 °C under low wavelength UV light. Ultrathin sections (50–70 nm thick) were collected (i) on glass slides and processed for immunofluorescence (see below) and (ii) on formvar carbon-coated nickel grids that were subsequently floated on drops of 1% bovine serum albumin (BSA) in PBS buffer for 30 min at room temperature. After overnight incubation at 4 °C with anti-Ntann12 rabbit antibodies (diluted 100 times in PBS/1% BSA), grids were washed with PBS and treated for 2 h at room temperature with goat anti-rabbit antibodies conjugated to 5 nm colloidal gold particles (diluted 100 times) (British BioCell). Finally, grids were washed in PBS and then stained with uranyl acetate and lead citrate. Observations were made on a Tecnai 10 electron microscope and images were captured with a MegaView II camera and processed with AnalySIS and Adobe Photoshop softwares. For immunofluorescence, the same protocol was used, except that goat anti-rabbit antibodies conjugated to AlexaFluor 488 (Invitrogen) (diluted 500 times) were used. Samples were observed in a Zeiss fluorescence microscope (Axioscope) coupled to an Axiocam CCD camera. Captured images were processed by Axio and Photoshop software. Appropriate controls using either pre-immune serum or no first antibody were performed and no signal was observed (data not shown).
Quantitative RT-PCR (qRT-PCR) analysis
Total RNA from leaves of 6-week-old plants was prepared using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) then treated with DNase I (DNA-free™ from Ambion, Austin, TX, USA). RNA quality and quantity were assessed with a Bioanalyzer 2100 (Agilent). Single-stranded cDNA was synthesized from 1 μg of mRNA using the Reverse Transcription System (Promega, Madison, WI, USA).
qRT-PCR analysis was performed in an ABI 7900 system (Applied Biosystems) using 1 μl of each cDNA. Transcriptional changes were calculated based on the comparative ΔΔCT method as described by Livak and Schmittgen (2001) and are reported as ratios between expression in roots and in leaves of wild-type plants. The CT value of each gene was normalized to the CT value of the reference gene EF1α. The expression of each gene was investigated in three biological replicates and six technical repeats. Expression levels of Ntann12 and EF1α were assessed using the primers F 5′-CTTCTCTGCCCTTGTAACTAT-3′ and R 5′-CAACCGCTACAAGGGTGATTA-3′ for Ntann12 and F 5′-TGCTACCACCCCCAAGTACTC-3′ and R 5′-TAAAGCTGGCAGCACCCTTAG-3′ for EF1α. Conditions for qRT-PCR were as follows: 2 min at 50 °C, 10 min at 95 °C, 40 cycles of 30 s at 95 °C, 1 min at 60 °C, and 1 min at 72 °C. As a final step, a dissociation curve consisting of a 15 s denaturation at 95 °C, 15 s at 60 °C, and 15 s at 95 °C was performed to detect unwanted primer dimers or PCR products that could interfere with the fluorescence data.
pNtann12-GUS expression
The amplification of the Ntann12 promoter region, the construction of pNtann12-GUS construct, and the plant transformation were described previously (Vandeputte et al., 2007). Plants were cultured in solid or liquid MS medium with 50 μM indole-3-acetic acid (IAA), 50 μM 1-naphthylphthalamic acid (NPA), or 50 μM 2,3,5-triiodobenzoic acid (TIBA) (Sigma). β-Glucuronidase (GUS) staining was perfomed as described by Hemerly et al. (1993).
Results
Recombinant tobacco Ntann12 is a Ca2+-dependent phospholipid-binding protein
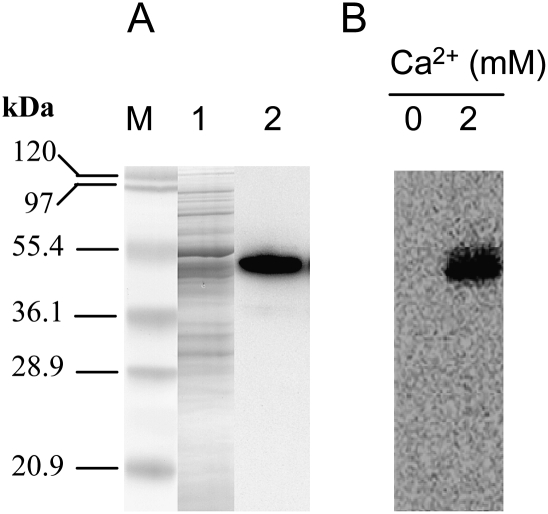
The phospolipid binding property of Ntann12 was investigated using the recombinant Ntann12 protein produced in E. coli. Effective expression of the recombinant Ntann12 protein was monitored by SDS–PAGE and western blot analysis. As shown in Fig. 1A, anti-Ntann12 polyclonal antibodies clearly cross-react with a polypeptide of ∼54 kDa, corresponding to the expected molecular weight of the fusion protein obtained in the pBAD-Dest49 destination vector based on 36 kDa for the native Ntann12 plus 14 kDa for the N-terminal His-Patch thioredoxin plus 4 kDa for the C-terminal tag. Subsequently, recombinant Ntann12 proteins were incubated with liposomes either in the presence or in the absence of Ca2+. Following sucrose gradient centrifugation, liposome fractions were analysed by immunoblot with the anti-Ntann12 antibody. As shown in Fig. 1B, in the absence of Ca2+, no association of the proteins with liposomes was detected, whereas in the presence of 2 mM Ca2+, Ntann12 proteins were detected in the liposome fraction, indicating that recombinant Ntann12 is a Ca2+-dependent phospholipid-binding protein.
Fig. 1.
Production of recombinant Ntann12 and its Ca2+-dependent phospholipid binding property. (A) Western blot analysis of recombinant proteins produced in E. coli using anti-Ntann12 antibodies. Lane 1, gel migration of 0.1 mg of total bacterial extract stained with Coomassie; lane 2, gel migration of 5 ng of bacterial extract revealed by anti-Ntann12 antibodies. (B) Phospholipid binding property of recombinant Ntann12. Proteins were incubated with liposomes (2:1 PC/PS) in buffer containing 0 or 2 mM Ca2+. Proteins (40 μg) were analysed by western blot using the anti-Ntann12 antibodies. M, molecular marker.
Ntann12 is highly expressed in the root system
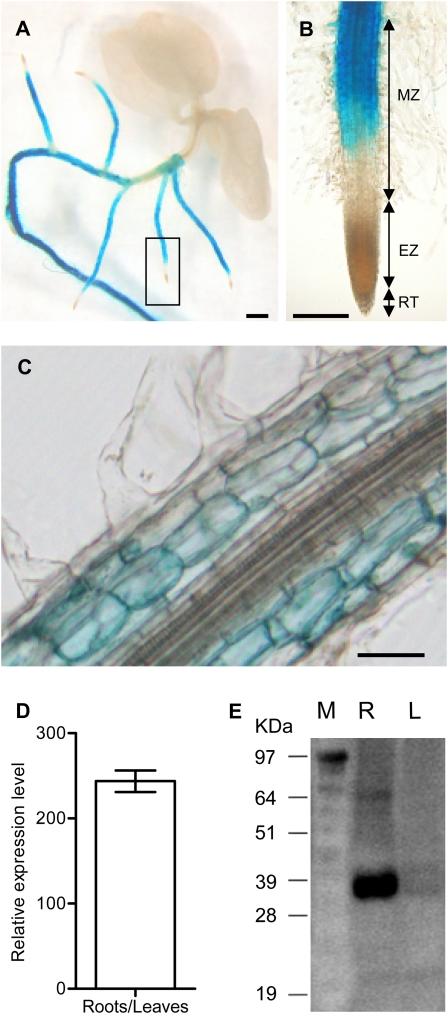
Ntann12 expression was investigated by histochemical GUS staining in pNtann12-GUS transgenic plants and by qRT-PCR in wild-type plants. Protein levels were compared in both aerial parts and in roots by western blot using anti-Ntann12 antibodies. As shown in Fig. 2A, Ntann12 is highly expressed in the root system of 1-month-old pNtann12-GUS transgenic plants but not in the aerial part. A close-up picture of the root extremity shows that the GUS activity is highly visible within the root maturation zone but not in the root cap, or in the root apical meristem and the elongation zone (Fig. 2B). In longitudinal cross-sections, the GUS activity is detected mainly in the cortex cells of the roots (Fig. 2C). As determined by qRT-PCR analysis, the Ntann12 expression level was ∼250 times more abundant in the roots than in the aerial parts of 1-month old plants (Fig. 2D). To investigate whether the Ntann12 expression level was correlated with the abundance of the protein in the different plant parts, a western blot analysis was performed. The anti-Ntann12 antibodies cross-react with a protein of ∼36 kDa in roots, but not in leaves, confirming that Ntann12 is more abundant in roots than in leaves (Fig. 2E).
Fig. 2.
Ntann12 expression in 4-week-old plants. (A) pNtann12-GUS expression in whole tobacco plant; bar=1 mm. (B) Close-up of (A) showing pNtann12-GUS expression in the root tip; bar=0.5 mm. (C) GUS staining in a longitudinal section of the root; bar=100 μm. (D) Relative expression of Ntann12 in roots and in aerial parts of plants, as determined by qRT-PCR analysis. (E) Western blot analysis of total protein extracts (10 μg) from roots and leaves using anti-Ntann12 antibodies. EZ, elongation zone; L, leaf; M, molecular marker; MZ, maturation zone; RT, root tip; R, root.
Subcellular distribution of native Ntann12 is modulated by Ca2+ concentration
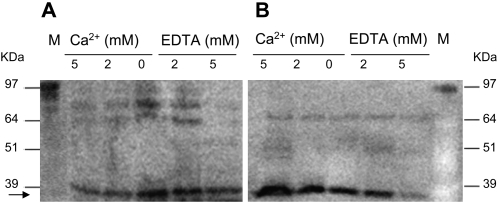
In order to determine the intracellular distribution of Ntann12 in roots and to evaluate whether its distribution is regulated by Ca2+ concentration, immunoblots were made using both cytosolic and membrane-enriched fractions from tobacco root extracts. Total native protein extracts were incubated with either Ca2+ or EDTA before separation of the cytosolic fraction from the microsomal fraction. As shown in Fig. 3, a band corresponding to the molecular weight of Ntann12 was detected in both the cytosolic (Fig. 3A) and membrane-enriched fractions (Fig. 3B) at all tested Ca2+ or EDTA concentrations.
Fig. 3.
Subcellular distribution and Ca2+ response of native Ntann12 proteins. The total protein extract from roots of 4-week-old tobacco plants was treated with either CaCl2 or EDTA before separation of the membrane-enriched and cytosolic fractions and western blot analysis using the anti-Ntann12 antibodies. (A) Cytosolic fraction. (B) Membrane-enriched fraction. For the analysis, 40 μg of membrane-enriched fraction proteins and 60 μg of cytosolic fraction proteins were loaded into gels. M, molecular marker. The arrow denotes the position of the expected size for Ntann12.
Ntann12 was detectable in the membrane-enriched fraction even when no Ca2+ was added, indicating that Ntann12 is associated to some extent with the membrane in a Ca2+-independent manner (Fig. 3B). The amount of Ntann12 bound to or associated with the membrane-enriched fraction increased with increasing Ca2+ concentration, whereas the Ntann12 level decreased when EDTA was added, becoming almost undetectable in 5 mM EDTA buffer, supporting the fact that Ntann12 phospholipid binding is Ca2+ dependent. An opposite effect was noticed in the cytosolic fraction where the Ntann12 level decreased with increasing Ca2+ concentration (Fig. 3A). This result is consistent with a Ca2+-mediated translocation of Ntann12 from the cytosolic to the membrane-enriched fraction.
Ntann12 is localized in the nucleus of root cells
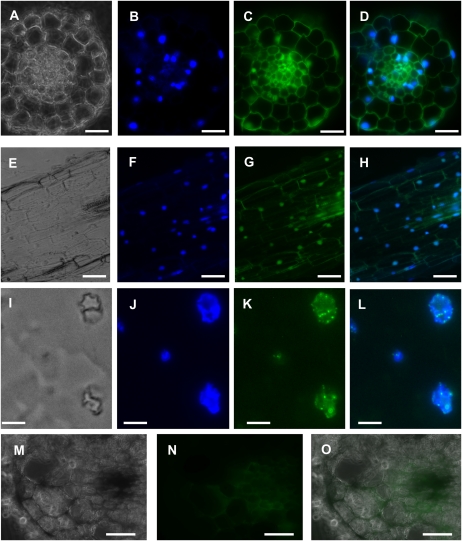
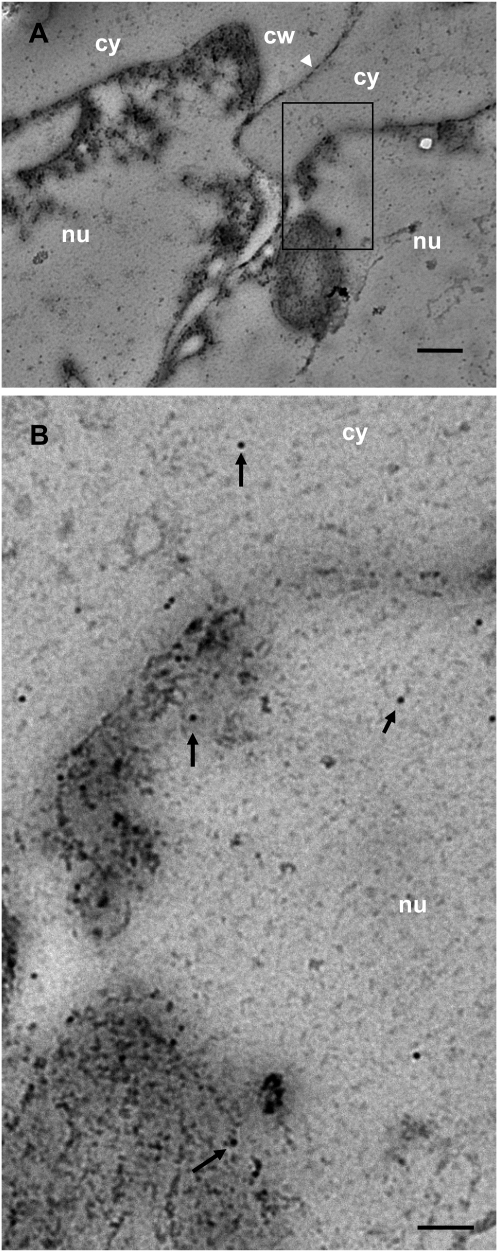
To investigate the subcellular localization of Ntann12 in roots, immunohistochemical localization experiments were performed using both fluorescence and electron microscopy. Fig. 4 shows the results of an immunolabelling experiment on 100 μm cross- (Fig. 4A–D) or longitudinal (Fig. 4E–H) sections as well as on 50–70 nm (Fig. 4I–L) sections through 4-week-old tobacco roots. Both cell walls and the vascular system display a background signal due to autofluorescence (Fig. 4M–O). The co-localization of both 4',6-diamidino-2-phenylindole (DAPI) and fluorescent signals indicate that Ntann12 was present in the nucleus of almost all cell types within the root (Fig. 4A–H). However, due to the intensive background fluorescence (Fig. 4O), it cannot be excluded that Ntann12 might be present in the cell periphery (cytoplasm and plasma membrane). To localize the Ntann12 signal more precisely within the nucleus, 50 nm cross-sections were labelled in the same conditions. The labelling was found to be localized as discrete spots in the nuclei (Fig. 4I–L). Subsequently, immunolabelling was investigated by immunotransmission electron microscopy. As shown in Fig. 5, and Supplementary Fig. S2 available at JXB online, within root cells, gold particles were detected in both the nucleus and the cytoplasm. The labelling was present in the whole nucleus and might be associated with the nucleoli, euchromatin, and heterochromatin.
Fig. 4.
Ntann12 immunolocalization in tobacco root sections visualized by fluorescence microscopy. (A–D) A 100μm cross-section. (E–H) A 100 μm longitudinal section. (I–L) A 50–70 nm cross-section. (M–O) A 100μm cross-section. (A), (E), (I), (M) Phase contrast. (B), (F), (J) Staining with 4',6-diamidine 2-phenyl indole (DAPI). (C), (G), (K) Immunostaining with affinity-purified antiserum against Ntann12. (D), (H), (L) Superposition of DAPI staining and immunostaining with affinity-purified antiserum against Ntann12. The green fluorescence is indicative of the presence of Ntann12, whereas DNA is represented by blue colour resulting from DAPI staining. (N), (O) The green fluorescence shows the background autofluorescence without immunostaining. Bar=50 μm (A–H and M–O), 5 μm (I–L).
Fig. 5.
Transmission electron micrographs of Ntann12 immunogold labelling in a section of root cells. (A) A section in two root cells. (B) Detail of (A) showing labelling in the cytoplasm and in the nucleus (see arrows). Cw, cell wall; cy, cytoplasm; nu, nucleus. Bars=400 nm (A), 50 nm (B).
In conclusion, the data presented in Figs 3–5 clearly indicate that Ntann12 is present in the nucleus (Fig. 4) but also in the cytoplasm (Fig. 5 and Supplementary Fig. S2), and can be associated with the membrane system according to the physiological conditions (Fig. 3).
Light and polar auxin transport regulate Ntann12 expression in the tobacco root system
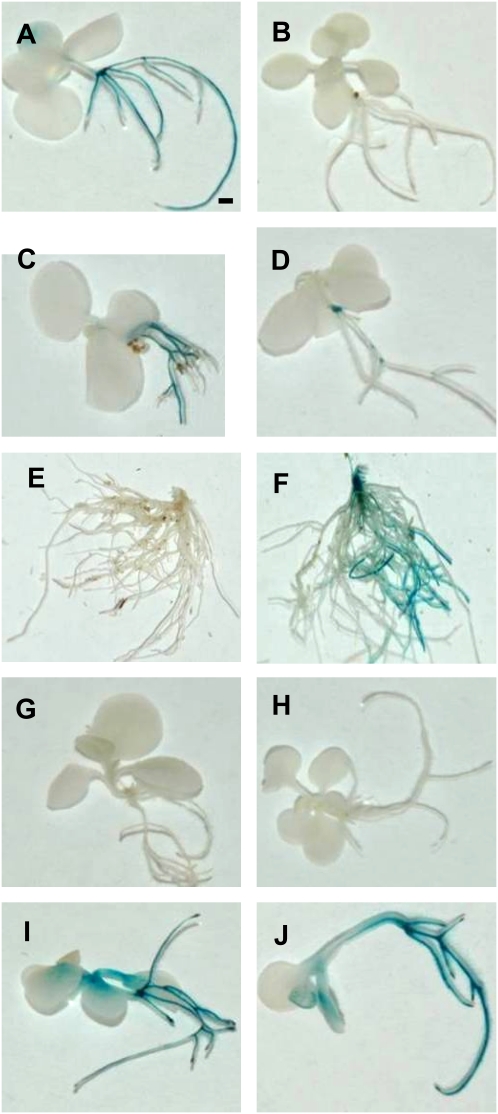
In silico analysis of pNtann12 using PlantPAN (Chang et al., 2008) indicates the occurrence of nine cis-element sequences possibly implicated in light responses: three GT1 consensus sequences and six GATABOX sequences (data not shown). Therefore, 1-month-old pNtann12-GUS transgenic plants were transferred for 48 h to darkness. As compared with the control kept in light conditions (16 h/8 h photoperiod) (Fig. 6A), GUS activity was no longer detected in the roots of the plants transferred to darkness (Fig. 6B). In accordance with this, Ntann12 proteins were not detected in roots of plants exposed for 48 h to darkness as compared with roots exposed to light conditions (Fig. 7). In addition, Ntann12 expression was no longer detected in roots of pNtann12-GUS transgenic plants 48 h after removal of the aerial part either in light or in dark conditions (data not shown). To check whether light was the signal activating the expression of Ntann12 in the roots, two treatments were carried out on plants grown in a greenhouse. The first treatment consisted of covering the soil with aluminium foil to ensure complete darkness for the root system and the second was to place aluminium foil on the whole aerial part of the plant to ensure complete darkness. As shown in Fig. 6C, when the leaves were submitted to light and the roots to darkness, the expression pattern was similar to that observed in in vitro conditions (Fig. 6A). In contrast, when the leaves were placed in darkness for 48 h (Fig. 6D), the GUS activity in the root system dramatically decreased, indicating that the expression of Ntann12 is regulated by a light-activated messenger in the aerial part of the plant.
Fig. 6.
pNtann12 responses to light, darkness, IAA, and NPA. (A) A whole plant exposed to light. (B) A whole plant maintained for 48 h in darkness. (C) Aerial part exposed to light and roots to darkness. (D) A whole plant exposed to darkness. (E) A water-containing agar block fixed on the stem section of an aerial part-free plant. (F) An IAA-containing agar block fixed on the stem section of an aerial part-free plant. (G) A plant incubated for 48 h with 50 μM NPA and exposed to light. (H) A plant incubated for 48 h with 50 μM NPA in the darkness. (I) Whole plants treated with 50 μM IAA and exposed to light. (J) Whole plants treated with 50 μM IAA in darkness. (A–D) and (G–J) Two-week-old plants. (E-F) Roots of 6-week-old plants. Bar=1 mm.
Fig. 7.
Western blot analysis of Ntann12 expression in roots of 4-week-old-plants grown in solid MS and exposed to light (L) or to darkness (D) for 48 h. Total protein extracts (10 μg) of roots were separated by SDS–PAGE and western blot analysis with the anti-Ntann12 antibodies.
As auxin has been associated with several light-regulated processes (reviewed by Swarup et al., 2002), the possibility that auxin could be linked to the signal coming from aerial parts exposed to light was investigated. The aerial parts of pNtann12-GUS plants grown in soil were cut off, small agar blocks containing either IAA or water were fixed to the cut stems section, and the soil was covered with aluminium foil. As shown in Fig. 6E, after 48 h treatment, no GUS activity was detected in the roots when the cut stem section was covered with the agar block containing water. In contrast, when IAA was applied to the cut stem section, a strong GUS signal was detected in the whole root system (Fig. 6F). Consequently, 4-week-old pNtann12-GUS transgenic plants were subcultured in liquid MS medium containing the polar auxin transport inhibitor NPA. No GUS activity was detected in plants treated with NPA either in light (Fig. 6G) or in dark (Fig. 6H) conditions. These results indicate that polar auxin transport is a limiting factor for Ntann12 expression in the root system under light conditions. Similar results were obtained with TIBA (data not shown). Moreover, plants treated with IAA in the light (Fig. 6I) or in darkness (Fig. 6J) showed GUS staining in all parts, clearly indicating that Ntann12 is induced by auxin.
Discussion
In this report, it is shown that the recombinant Ntann12 is a Ca2+-dependent phospholipid-binding protein, confirming that it has the biochemical property of annexins (Fig. 1). Furthermore, the distribution of native Ntann12 in the cytosol or in the cell membrane fraction is demonstrated to be modulated by the Ca2+ concentration (Fig. 3). Similarly to Ntann12, Arabidopsis AnnAt1 and AnnAt2 have been found to be localized in both the cytosol and microsome compartments (Clark et al., 2005b). In addition, the translocation of plant annexins from the cytosol to membranes has been shown to occur in response to specific stimuli (for a review, see Mortimer et al., 2008), such as touch (Thonat et al., 1997), cold (Breton et al., 2000), and salinity (Lee et al., 2004), suggesting a role for annexins as signalling molecules associated with multifunctional components of plant growth and adaptation to environmental stimuli.
Although most plant annexins are found in the cytosol, several have also been shown to be located in various compartments, such as the cell wall (Kwon et al., 2005), the plasma membrane (Santoni et al., 1998), the vacuole (Seals and Randall, 1997), the chloroplast envelope membranes (Seigneurin-Berny et al., 2000), or the endoplasmic reticulum (Huh et al., 2010), or to be extracellular (Laohavisit et al., 2009). Similarly to other plant annexins, by means of immunolocalization, Ntann12 was shown to be mainly present in the nucleus (Figs 4, 5). For instance, P35 annexin from pea has been immunolocalized in the nucleus (Clark et al., 1998). As shown by these authors, a band was detected by immunoblot in fractions corresponding to purified nuclei, nuclear envelope matrix, nucleoli, and chromatin. Futher studies showed that M. sativa ANNMS2 annexin was immunolocalized in the nucleoli (Kovács et al., 1998). As far as is known, no nuclear localization signal has been found for any of the annexin localized in the nucleus, including Ntann12, and the mechanism of their nuclear import has not been elucidated yet. The recruitment of a specific annexin in a particular membrane has been proposed to be dependent on the cell type, the Ca2+ concentration, the pH, by post-translational modification, the lipid composition, and membrane oxidation, but the targeting mechanisms are unknown (reviewed by Gerke and Moss, 2002: Laohavisit and Davies, 2011).
Several mammal annexins have been shown to be localized in the nucleus. For instance, ANXA2 has been functionally involved in DNA replication (Liu and Vishwanatha, 2007). ANX11 has been shown to translocate from the nucleus to the spindle poles in metaphase and to the spindle midzone in anaphase, and to be recruited to the midbody in late telophase (Tomas et al., 2004). In addition, these authors showed that cells silenced for Anx11 fail to establish a functional midbody, suggesting a fundamental role for this annexin in the terminal phase of cytokinesis. Finally, suppression of AnxA3 by RNA interference (RNAi) in primary cultured parenchymal rat hepatocytes has been shown to inhibit DNA synthesis (Niimi et al., 2005). Therefore, as suggested for human annexins (Rick et al., 2005), it may be assumed that plant nuclear annexins, including Ntann12, participate in a nuclear response to cell stimulation or to Ca2+ transient signalling, presumably by regulating DNA replication or transcription.
As shown by qRT-PCR, immunoblot, and promoter–GUS analyses carried out in 1-month-old tobacco plants grown in vitro or in vivo, Ntann12 is highly expressed in roots but almost not detected in the aerial parts (Fig. 2). Within the root system, GUS staining indicated that Ntann12 is mainly expressed within the root maturation zone, but not in the root cap or in the root elongation zone. Several other plant annexins have been shown to be expressed in root tissues, such as AnnAt1 (Lee et al., 2004). By in situ mRNA hybridization studies, AnnAt1 was found to be expressed in all cells of the root, except at the root tip where the expression is restricted to the root cap, and AnnAt2 was expressed in endodermal cells near the root hypocotyl junction (Clark et al., 2001). MtAnn1 was shown to be expressed in cortical and endodermal cell layers of the roots (de Carvalho-Niebel et al., 2002) and the annexin Gh1 was found to be one of the major proteins in the root proteome of Gossypium hirsutum and G. arboreum (Mavlonov et al., 2009). As suggested by Huh et al. (2010), the annexin expression patterns may depend on both culture conditions and developmental stage.
The present study also indicates that the expression of Ntann12 in the root system is regulated by a signal produced in the aerial parts of the plant under light conditions, a signal that is possibly auxin. Actually, GUS staining almost disappeared in the roots of in vitro pNtann12-GUS transgenic plants that were placed in darkness for 48 h (Figs 6B, 7) and when the aerial part was removed from plants grown in soil (Fig. 6E). However, GUS staining in the roots was restored when auxin was applied to a stem section of an aerial part-free root system (Fig. 6F) or to dark-growing or light-growing plants (Fig. 6I, J). The involvement of IAA in the regulation of Ntann12 is supported by the observation that the auxin efflux carrier inhibitor NPA repressed Ntann12 in both light and dark conditions (Fig. 6G, H), indicating that the expression level of Ntann12 in roots is regulated by polar auxin transport. According to the literature, light affects the expression of several Arabidopsis annexins (Cantero et al., 2006; Huh et al., 2010). Moreover, a role in nyctinastic movement has been suggested for the p35 Mimosa pudica annexin, which accumulated in the cytosol at night but was redistributed to the outermost periphery of the motor cells in the pulvinus during the daytime (Hoshino et al., 2004).
Molecular links between light and auxin signalling pathways are well documented since light-regulated transcription factors have been shown to affect auxin responses, and mutations in Aux/IAA genes affect light signalling (reviewed by Tian and Reed, 2001). More basically, several results suggest that light regulates the amount of auxin or polar auxin transport, and this leads to quantitative changes in local auxin concentrations that correlate with local tissue growth rates and/or particular physiological processes. For instance, dim red light increased auxin transport in cucumber hypocotyls relative to dark-grown hypocotyls (Shinkle et al., 1998). Additional facts suggest that auxin has a central role in shoot to root relationships (reviewed by Sachs, 2005). Indeed, lateral root development has been shown to be modulated by shoot-localized light signalling and requires shoot-derived transport of auxin (Reed et al., 1998; Bhalerao et al., 2002). Auxin formed in leaves is transported to the roots, both by phloem and by polar transport (Friml, 2003). In this latter case, IAA is directionally transported by plasma membrane-localized auxin influx and efflux carriers in transporting cells.
To gain knowledge of the function of Ntann12, transgenic tobacco plants overexpressing or down-regulating Ntann12 were produced (Supplementary data S3 at JXB online). Overall, no phenotypic differences between transgenic and wild-type plants were observed during germination and growth, or after biotic (R. fascians infection) and abiotic stress (NaCl, light) or hormonal treatment (auxin) (data not shown). As discussed by Lee et al. (2004), members of the annexin family may function at a particular developmental stage or exhibit functional redundancy, possibly explaining the absence of phenotype. Further investigations on multiple mutants are required to shed light on the biological function of the diversity of plant annexins.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Ntann12 immunolocalization in tobacco root 100 μm cross-sections visualized by fluorescence microscopy.
Figure S2. Transmission electron micrographs of Ntann12 immunogold labelling in a section of several root cells.
Supplementary data S3. Characterization of T2 transgenic tobacco progeny overexpressing and down-regulating Ntann12.
Acknowledgments
MB is Senior Research Associate of the FRS-FNRS (Belgian Fund for Scientific Research), FH is Research Director of the FRS-FNRS, and OMV is a Post-Doctoral Researcher of the FRS-FNRS. JM is a scholar of the Lebanese National Council for Scientific Research (CNRS-L).
References
- Andrawis A, Solomon M, Delmer DP. Cotton fiber annexins: a potential role in the regulation of callose synthase. The Plant Journal. 1993;3:763–772. doi: 10.1111/j.1365-313x.1993.00763.x. [DOI] [PubMed] [Google Scholar]
- Bhalerao R, Eklöf J, Marchant A, Bennett M, Sandberg G. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. The Plant Journal. 2002;29:325–332. doi: 10.1046/j.0960-7412.2001.01217.x. [DOI] [PubMed] [Google Scholar]
- Blackbourn HD, Barker PJ, Huskisson NS, Battey NH. Properties and partial protein sequence of plant annexins. Plant Physiology. 1992;99:864–871. doi: 10.1104/pp.99.3.864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G, Vazquez-Tello A, Danyluk J, Sarhan F. Two novel intrinsic annexins accumulate in wheat membranes in response to low temperature. Plant and Cell Physiology. 2000;41:177–184. doi: 10.1093/pcp/41.2.177. [DOI] [PubMed] [Google Scholar]
- Boustead CM, Smallwood M, Small H, Bowles DJ, Walker JH. Identification of calcium-dependent phospholipid-binding proteins in higher plant cells. FEBS Letters. 1989;244:456–460. [Google Scholar]
- Calvert CM, Gant SJ, Bowles DJ. Tomato annexins p34 and p35 bind to F-actin and display nucleotide phosphodiesterase activity inhibited by phospholipid binding. The Plant Cell. 1996;8:333–342. doi: 10.1105/tpc.8.2.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantero A, Barthakur S, Bushart TJ, Chou S, Morgan RO, Fernandez MP, Clark GB, Roux SJ. Expression profiling of the Arabidopsis annexin gene family during germination, de-etiolation and abiotic stress. Plant Physiology and Biochemistry. 2006;44:13–24. doi: 10.1016/j.plaphy.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Carroll AD, Moyen C, Van Kesteren P, Tooke F, Battey NH, Brownlee C. Ca2+, annexins, and GTP modulate exocytosis from maize root cap protoplasts. The Plant Cell. 1998;10:1267–1276. doi: 10.1105/tpc.10.8.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang W-C, Lee T-T, Huang H-D, Huang H-Y, Pan R- L. PlantPAN: plant promoter analysis navigator, for identifying combinatorial cis-regulatory elements with distance constraint in plant gene groups. BMC Genomics. 2008;9:561. doi: 10.1186/1471-2164-9-561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark G, Cantero-Garcia A, Butterfield T, Dauwalder M, Roux SJ. Secretion as a key component of gravitropic growth: implications for annexin involvement in differential growth. Gravitational and Space Biology. 2005a;18:113–114. [PubMed] [Google Scholar]
- Clark GB, Dauwalder M, Roux SJ. Purification and immunolocalization of an annexin-like protein in pea seedlings. Planta. 1992;187:1–9. doi: 10.1007/BF00201617. [DOI] [PubMed] [Google Scholar]
- Clark GB, Dauwalder M, Roux SJ. Immunological and biochemical evidence for nuclear localization of annexin in peas. Plant Physiology and Biochemistry. 1998;36:621–627. doi: 10.1016/s0981-9428(98)80010-7. [DOI] [PubMed] [Google Scholar]
- Clark GB, Lee D, Dauwalder M, Roux SJ. Immunolocalization and histochemical evidence for the association of two different Arabidopsis annexins with secretion during early seedling growth and development. Planta. 2005b;220:621–631. doi: 10.1007/s00425-004-1374-7. [DOI] [PubMed] [Google Scholar]
- Clark GB, Sessions A, Eastburn DJ, Roux SJ. Differential expression of members of the annexin multigene family in Arabidopsis. Plant Physiology. 2001;126:1072–1084. doi: 10.1104/pp.126.3.1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dabitz N, Hu N-J, Yusof AM, Tranter N, Winter A, Daley M, Zschörnig O, Brisson A, Hofmann A. Structural determinants for plant annexin–membrane interactions. Biochemistry. 2005;44:16292–16300. doi: 10.1021/bi0516226. [DOI] [PubMed] [Google Scholar]
- de Carvalho Niebel F, Lescure N, Cullimore JV, Gamas P. The Medicago truncatula Mtann1 gene encoding an annexin is induced by Nod factors and during the symbiotic interaction with Rhizobium meliloti. Molecular Plant-Microbe Interactactions. 1998;11:504–513. doi: 10.1094/MPMI.1998.11.6.504. [DOI] [PubMed] [Google Scholar]
- de Carvalho-Niebel F, Timmers ACJ, Chabaud M, Defaux-Petras A, Barker DG. The Nod factor-elicited annexin MtAnn1 is preferentially localised at the nuclear periphery in symbiotically activated root tissues of Medicago truncatula. The Plant Journal. 2002;32:343–352. doi: 10.1046/j.1365-313x.2002.01429.x. [DOI] [PubMed] [Google Scholar]
- Divya K, Jami SK, Kirtti PB. Constitutive expression of mustard annexin, AnnBj1 enhances abiotic stress tolerance and fiber quality in cotton under stress. Plant Molecular Biology. 2010;73:293–308. doi: 10.1007/s11103-010-9615-6. [DOI] [PubMed] [Google Scholar]
- Friml J. Auxin transport—shaping the plant. Current Opinion in Plant Biology. 2003;6:7–12. doi: 10.1016/s1369526602000031. [DOI] [PubMed] [Google Scholar]
- Geisow MJ, Fritsche U, Hexham JM, Dash B, Johnson T. A consensus amino-acid sequence repeat in Torpedo and mammalian Ca2+-dependent membrane-binding proteins. Nature. 1986;320:636–638. doi: 10.1038/320636a0. [DOI] [PubMed] [Google Scholar]
- Gerke V, Creutz CE, Moss SE. Annexins: linking Ca2+ signalling to membrane dynamics. Nature Reviews Molecular Cell Biology. 2005;6:449–461. doi: 10.1038/nrm1661. [DOI] [PubMed] [Google Scholar]
- Gerke V, Moss SE. Annexins and membrane dynamics. Biochimica et Biophysica Acta. 1997;1357:129–154. doi: 10.1016/s0167-4889(97)00038-4. [DOI] [PubMed] [Google Scholar]
- Gerke V, Moss SE. Annexins: from structure to function. Physiological Reviews. 2002;82:331–371. doi: 10.1152/physrev.00030.2001. [DOI] [PubMed] [Google Scholar]
- Gidrol X, Sabelli PA, Fern YS, Kush AK. Annexin-like protein from Arabidopsis thaliana rescues Δ oxyR mutant of Escherichia coli from H2O2 stress. Proceedings of the National Academy of Sciences, USA. 1996;93:11268–11273. doi: 10.1073/pnas.93.20.11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorecka KM, Konopka-Postulpolska D, Hennig J, Buchet R, Pikula S. Peroxidase activity of annexin 1 from Arabidopsis thaliana. Biochemical and Biophysical Research Communications. 2005;336:868–875. doi: 10.1016/j.bbrc.2005.08.181. [DOI] [PubMed] [Google Scholar]
- Gorecka KM, Thouverey C, Buchet R, Pikula S. Potential role of annexin AnnAt1 from Arabidopsis thaliana in pH-mediated cellular response to environmental stimuli. Plant and Cell Physiology. 2007;48:792–803. doi: 10.1093/pcp/pcm046. [DOI] [PubMed] [Google Scholar]
- Hemerly AS, Ferreira P, de Almeida Engler J, Van Montagu M, Engler G, Inzé D. cdc2a expression in Arabidopsis is linked with competence for cell division. The Plant Cell. 1993;5:1711–1723. doi: 10.1105/tpc.5.12.1711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A. Annexins in the plant kingdom: perspectives and potentials. Annexins. 2004;1:51–61. [Google Scholar]
- Hofmann A, Proust J, Dorowski A, Schantz R, Huber R. Annexin 24 from Capsicum annuum. X-ray structure and biochemical characterization. Journal of Biological Chemistry. 2000;275:8072–8082. doi: 10.1074/jbc.275.11.8072. [DOI] [PubMed] [Google Scholar]
- Hoshino D, Hayashi A, Temmei Y, Kanzawa N, Tsuchiya T. Biochemical and immunohistochemical characterization of Mimosa annexin. Planta. 2004;219:867–875. doi: 10.1007/s00425-004-1285-7. [DOI] [PubMed] [Google Scholar]
- Hu S, Brady SR, Kovar DR, Staiger CJ, Clark GB, Roux SJ, Muday GK. Identification of plant actin-binding proteins by F-actin affinity chromatography. The Plant Journal. 2000;24:127–137. doi: 10.1046/j.1365-313x.2000.00852.x. [DOI] [PubMed] [Google Scholar]
- Huh SM, Noh EK, Kim HG, Jeon BW, Bae K, Hu H-C, Kwak JM, Park OK. Arabidopsis annexins AnnAt1 and Annat4 interact with each other and regulate drought and salt stress responses. Plant and Cell Physiology. 2010;51:1499–1514. doi: 10.1093/pcp/pcq111. [DOI] [PubMed] [Google Scholar]
- Jami SK Clark GB, Turlapati SA, Handley C, Roux SJ, Kirti PB. Ectopic expression of an annexin from Brassica juncea confers tolerance to abiotic and biotic stress treatments in transgenic tobacco. Plant Physiology and Biochemistry. 2008;46:1019–1030. doi: 10.1016/j.plaphy.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Jami SK, Dalal A, Divya K, Kirti PB. Molecular cloning and characterization of five annexin genes from Indian mustard (Brassica juncea L. Czern and Coss. Plant Physiology and Biochemistry. 2009;47:977–990. doi: 10.1016/j.plaphy.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Konopka-Postupolska D. Annexins: putative linkers in dynamic membrane–cytoskeleton interactions in plant cells. Protoplasma. 2007;230:203–215. doi: 10.1007/s00709-006-0234-7. [DOI] [PubMed] [Google Scholar]
- Konopka-Postupolska D, Clark G, Goch G, Debski J, Floras K, Cantero A, Fijolek B, Roux S, Hennig J. The role of annexin 1 in drought stress in Arabidopsis. Plant Physiology. 2009;150:1394–1410. doi: 10.1104/pp.109.135228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovács I, Ayaydin F, Oberschall A, Ipacs I, Bottka S, Pongor S, Dudits D, Tóth ÉC. Immunolocalization of a novel annexin-like protein encoded by a stress and abscisic acid responsive gene in alfalfa. The Plant Journal. 1998;15:185–197. doi: 10.1046/j.1365-313x.1998.00194.x. [DOI] [PubMed] [Google Scholar]
- Kush A, Sabapathy K. Oxy5, a novel protein from Arabidopsis thaliana, protects mammalian cells from oxidative stress. International Journal of Biochemistry and Cell Biology. 2001;33:591–602. doi: 10.1016/s1357-2725(01)00040-1. [DOI] [PubMed] [Google Scholar]
- Kwon H-K, Yokoyama R, Nishitani K. A proteomic approach to apoplastic proteins involved in cell wall regeneration in protoplasts of Arabidopsis suspension-cultured cells. Plant and Cell Physiology. 2005;46:843–857. doi: 10.1093/pcp/pci089. [DOI] [PubMed] [Google Scholar]
- Laohavisit A, Brown AT, Cicuta P, Davies JM. Annexins—components of the calcium and reactive oxygen signaling network. Plant Physiology. 2010;152:1824–1829. doi: 10.1104/pp.109.145458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laohavisit A, Davies JM. Annexins. New Phytologist. 2011;189:40–53. doi: 10.1111/j.1469-8137.2010.03533.x. [DOI] [PubMed] [Google Scholar]
- Laohavisit A, Mortimer JC, Demidchik V, et al. Zea mays annexins modulate cytosolic free Ca2+ and generate a Ca2+-permeable conductance. The Plant Cell. 2009;21:479–493. doi: 10.1105/tpc.108.059550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee EJ, Yang EJ, Lee JE, Park AR, Song WH, Park OK. Proteomic identification of annexins, calcium-dependent membrane binding proteins that mediate osmotic stress and abscisic acid signal transduction in Arabidopsis. The Plant Cell. 2004;16:1378–1391. doi: 10.1105/tpc.021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim E-K, Roberts MR, Bowles DJ. Biochemical characterization of tomato annexin p35. Journal of Biological Chemistry. 1998;273:34920–34925. doi: 10.1074/jbc.273.52.34920. [DOI] [PubMed] [Google Scholar]
- Liu J, Vishwanatha JK. Regulation of nucleo-cytoplasmic shuttling of human annexin A2—a proposed mechanism. Molecular and Cellular Biochemistry. 2007;303:211–220. doi: 10.1007/s11010-007-9477-7. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the –2ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Mavlonov GT, Abdurakhmonov IY, Abdukarimov A, Kantety R, Sharma GC. The characterization of major proteins expressed in roots of four Gossypium species. Journal of Cotton Science. 2009;13:256–264. [Google Scholar]
- Mortimer JC, Laohavisit A, Macpherson N, Webb A, Brownlee C, Battey NH, Davies JM. Annexins: multifunctional components of growth and adaptation. Journal of Experimental Botany. 2008;59:533–544. doi: 10.1093/jxb/erm344. [DOI] [PubMed] [Google Scholar]
- Moss SE, Morgan RO. The annexins. Genome Biology. 2004;5(219):1–8. doi: 10.1186/gb-2004-5-4-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niimi S, Harashima M, Gamou M, Hyuga M, Seki T, Ariga T, Kawanishi T, Hayakawa T. Expression of annexin A3 in primary cultured parenchymal rat hepatocytes and inhibition of DNA synthesis by suppression of annexin A3 expression using RNA intereference. Biological and Pharmaceutical Bulletin. 2005;28:424–428. doi: 10.1248/bpb.28.424. [DOI] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiology. 1998;118:1369–1378. doi: 10.1104/pp.118.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick M, Ramos Garrido SI, Herr C, Thal DR, Noegel AA, Clemen CS. Nuclear localization of annexin A7 during murine brain development. BMC Neuroscience. 2005;6:25. doi: 10.1186/1471-2202-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs T. Auxin's role as an example of the mechanisms of shoot/root relations. Plant and Soil. 2005;268:13–19. [Google Scholar]
- Santoni V, Rouquié D, Doumas P, et al. Use of a proteome strategy for tagging proteins present at the plasma membrane. The Plant Journal. 1998;16:633–641. doi: 10.1046/j.1365-313x.1998.00335.x. [DOI] [PubMed] [Google Scholar]
- Seals DF, Randall SK. A vacuole-associated annexin protein, VCaB42, correlates with the expansion of tobacco cells. Plant Physiology. 1997;115:753–761. doi: 10.1104/pp.115.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneurin-Berny D, Rolland N, Dorne A-J, Joyard J. Sulfolipid is a potential candidate for annexin binding to the outer surface of chloroplast. Biochemical and Biophysical Research Communications. 2000;272:519–524. doi: 10.1006/bbrc.2000.2805. [DOI] [PubMed] [Google Scholar]
- Shin H, Brown RM., Jr. GTPase activity and biochemical characterization of a recombinant cotton fiber annexin. Plant Physiology. 1999;119:925–934. doi: 10.1104/pp.119.3.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkle JR, Kadakia R, Jones AM. Dim-red-light-induced increase in polar auxin transport in cucumber seedlings. I. Development of altered capacity, velocity, and response to inhibitors. Plant Physiology. 1998;116:1505–1513. doi: 10.1104/pp.116.4.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup R, Parry G, Graham N, Allen T, Bennet M. Auxin cross-talk: integration of signalling pathways to control plant development. Plant Molecular Biology. 2002;49:411–426. doi: 10.1007/978-94-010-0377-3_12. [DOI] [PubMed] [Google Scholar]
- Thonat C, Mathieu C, Crevecoeur M, Penel C, Gaspar T, Boyer N. Effects of a mechanical stimulation on localization of annexin-like proteins in Bryonia dioica internodes. Plant Physiology. 1997;114:981–988. doi: 10.1104/pp.114.3.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Q, Reed JW. Molecular links between light and auxin signaling pathways. Journal of Plant Growth Regulation. 2001;20:274–280. [Google Scholar]
- Tomas A, Futter C, Moss SE. Annexin 11 is required for midbody formation and completion of the terminal phase of cytokinesis. Journal of Cell Biology. 2004;165:813–822. doi: 10.1083/jcb.200311054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte O, Oukouomi Lowe Y, Burssens S, van Raemdonck D, Hutin D, Boniver D, Geelen D, El Jaziri M, Baucher M. The tobacco Ntann12 gene, encoding an annexin, is induced upon Rhodoccocus fascians infection and during leafy gall development. Molecular Plant Pathology. 2007;8:185–194. doi: 10.1111/j.1364-3703.2007.00385.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson JQ, Lanahan MB, Conner TW, Klee HJ. Identification of mRNAs with enhanced expression in ripening strawberry fruit using polymerase chain reaction differential display. Plant Molecular Biology. 1995;27:1097–1108. doi: 10.1007/BF00020883. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.