Abstract
Nanoparticle-based cancer diagnostics and therapeutics can be significantly enhanced by selective tissue localization, but the strategy can be complicated by the requirement of a targeting ligand conjugated on nanoparticles, that is specific to only one or a limited few types of neoplastic cells, necessitating the development of multiple nanoparticle systems for different diseases. Here, we present a new nanoparticle system that capitalizes on a targeting pretreatment strategy, where a circulating fusion protein (FP) selectively prelabels the targeted cellular epitope, and a biotinylated iron oxide nanoparticle serves as a secondary label that binds to the FP on the target cell. This approach enables a single nanoparticle formulation to be used with any one of existing fusion proteins to bind a variety of target cells. We demonstrated this approach with two fusion proteins against two model cancer cell lines: lymphoma (Ramos) and leukemia (Jurkat), which showed 72.2% and 91.1% positive labeling, respectively. Notably, TEM analysis showed that a large nanoparticle population was endocytosed via attachment to the non-internalizing CD20 epitope.
Keywords: Pretargeting, Nanoparticle, Lymphoma, PEG, Nanomedicine, Cancer
Introduction
Nanoparticle-based targeting agents such as liposomes, quantum dots, and superparamagnetic particles, have been developed that exhibit excellent biomedical imaging and therapeutic capacities.1-3 These nanoparticles capitalize on the specificity of targeting ligands attached at their surfaces, such as antibodies,4,5 peptides,6,7 and small molecules,8 to bind targeted cells,1,9,10 thus providing highly selective cell labeling. Nanoparticles modified with these targeting ligands are able to identify cancerous lesions,6,11 angiogenesis,7 and age-related macular degeneration.12
With a robust pipeline of biomolecular ligands being developed against different diseases, more and more new molecular-targeted nanoparticles are anticipated. However, each ligand is commonly specific to only one or a limited few target receptors, necessitating development of multiple nanoparticle formulations with different targeting ligands for different diseases. The development of each nanoparticle formulation requires extensive, costly design and experimentation to resolve a series of common issues such as possible biomolecule deactivation, low ligand:nanoparticle attachment, and colloidal stability. In addition, the use of large targeting molecules, such as antibodies, can potentially increase nanoparticle immunogenicity, shorten its blood half-life, and limit its vascular extravasation in vivo.13 Thus, new engineering approaches are desirable that can accommodate different targeting ligands without alternating pharmacokinetic profile of the nanoparticle and thus lower the development time and cost.
In this study, we developed a nanoparticle system based on a pretreatment strategy and can bind diverse cell targets. This method is a two-step technique by which targeting functionality and therapeutic/imaging modalities are separated. In this approach, a cell-targeting recombinant fusion protein (FP) composed of a single-chain antibody (scFv) and streptavidin (SA) is utilized to specifically prelabel the targeted cell, followed by application of a biotinylated nanoparticle that binds to the SA of the FP on the target cell (Fig. 1a). This strategy avoids the complications associated with direct conjugation of targeting molecules to nanoparticles and can be used with any one of many available FP targeting constructs without the need of a new nanoparticle system for each disease. Fusion proteins, such as 1F5 (anti-CD20), Lym-1 (anti-HLA-DR) and CC49 (anti-TAG-72), have demonstrated success in binding a range of different cancer types, including B-cell lymphoma,14 gastrointestinal cancers,15 and leukemia.16 These fusion proteins are used in combination with biotinylated radioisotopes (111In, 90Y) in radioimmunotherapy to target cancer cells while limiting radiation exposure to healthy tissue. This targeting strategy has been particularly successful in treating lymphomas.17-21 In this study, we demonstrate this approach with our nanoparticle system in two model cancer cell lines, Ramos and Jurkat, and with two FPs, anti-CD20 and anti-TAG-72 CC49, which specifically target Ramos and Jurkat cancer cells, respectively.
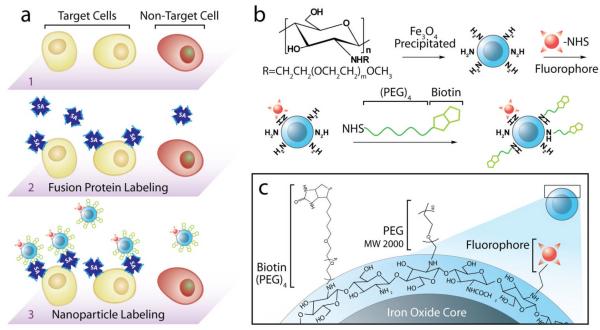
Fig. 1.
Nanoparticle labeling of target cells pretreated with a fusion protein (FP). a) Illustration of targeting pretreatment strategy: the target cells (1) are pre-labeled with target-specific FP (2), followed by attachment of the biotinylated nanoparticles to the FP (3). b) Synthesis and surface modification of biotinylated nanoparticles conjugated with Alexa Fluor 647 (NPB-AF647). c) Chemical profile of NPB-AF647 surface.
Results and discussion
Nanoparticle synthesis and characterization
The base nanoparticles were produced by iron chloride co-precipitation in the presence of a biocompatible chitosan-poly(ethylene glycol) (PEG) copolymer, yielding chitosan-PEG-coated iron oxide nanoparticles (NP) with a mean core diameter of ~7 nm, as described previously.6 Here, the chitosan-PEG co-polymer provides a protective shell around the iron oxide nanoparticle core for enhanced nanoparticle biostability and amines for further conjugation of biomolecules. Addition of the PEG in the co-polymer also limits the nanoparticle’s overall cationic charge, reducing nonspecific interactions of the material with the cells. The free amino groups on the nanoparticle coat were modified with the Alexa Fluor 647 (AF647) fluorophore and biotin targeting agent using NHS chemistry, yielding the targeting, biotinylated nanoparticle formulation (NPB-AF647, Fig. 1b), while the NP modified with the AF647 only (i.e., without biotin modification) served as controls (NP-AF647). Because the biotin was linked to the chitosan, aside the PEG co-polymer, a (PEO)4 linker group was used between the biotin and chitosan to reduce the possibility of steric hindrance during biotin-streptavidin binding (Fig. 1c).
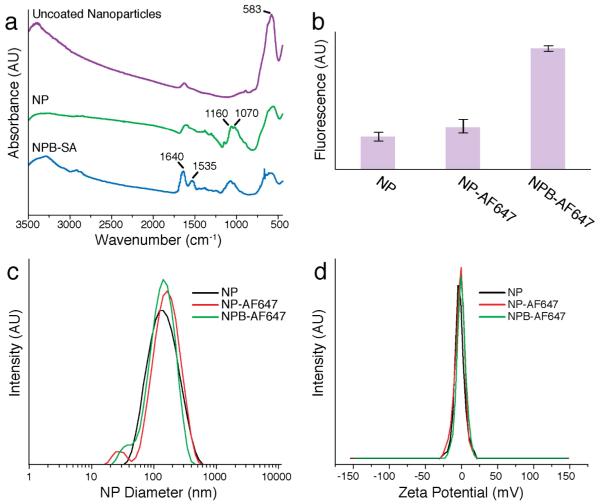
Successful nanoparticle coating and functionalization with the SA-binding molecule, biotin, was confirmed by FTIR spectroscopy (Fig. 2a). The absorption bands of the chitosan-PEG were observed on the spectrum of chitosan-PEG coated nanoparticles (NP) at 1160 (asymmetric stretching of C-O-C) and 1075–1033 cm−1 (C-O stretching), and all nanoparticle samples showed the Fe-O and O-H absorption bands at 583 and 3400 cm−1. To verify biotin-binding activity of NPBs, the NPB was mixed with the biotin-binding protein, SA, rinsed, and then the sample (NPB-SA) was analyzed by FTIR. Amide I (1640 cm−1) and amide II (1535 cm−1) peaks of the SA protein were observed indicating positive SA binding (Fig. 2a). The functional activity of the biotin was further confirmed by the FluoReporter Biotin assay (Fig. 2b), an assay that identifies active biotin labels by the displacement of a quencher dye from biotin binding sites on a fluorescent avidin probe. In this assay, the particles with no biotin, i.e., NP and NP-AF647, served as controls. NP and NP-AF647 samples did not significantly interact with the quenched probe Biotective reagent, while NPB-AF647 showed significant biotin interaction, yielding an increase in fluorescence, indicating specific binding to avidin (Fig. 2b).
Fig. 2.
Nanoparticle physiochemical properties. a) Fourier Transform Infrared (FTIR) of uncoated nanoparticles, chitosan-PEG-coated nanoparticles (NP), and biotinylated-NP binding SA (NPB-SA). b) Fluorescence analysis showing that biotinylated nanoparticles (NPB-AF647) enhanced binding to the avidin of the biotective agent, compared with control nanoparticles (i.e. NP and NP-AF647). c) Hydrodynamic sizes of nanoparticles with and without fluorophores or biotin. d) Zeta potentials of nanoparticles with and without biotin.
Physical analysis of the NP and NPB formulations showed that little size change occurred during the functionalization of the nanoparticles with the fluorochrome and biotin small molecules (Fig. 2c). The average hydrodynamic size of NP-AF647 and NPB-AF647 increased by < 20 nm over NP. Importantly, the addition of the biotin small molecule to the surface of the NP caused little to no change in the zeta potential of the nanoparticle (Fig. 2d).
Selective uptake of biotinylated nanoparticles by target cells
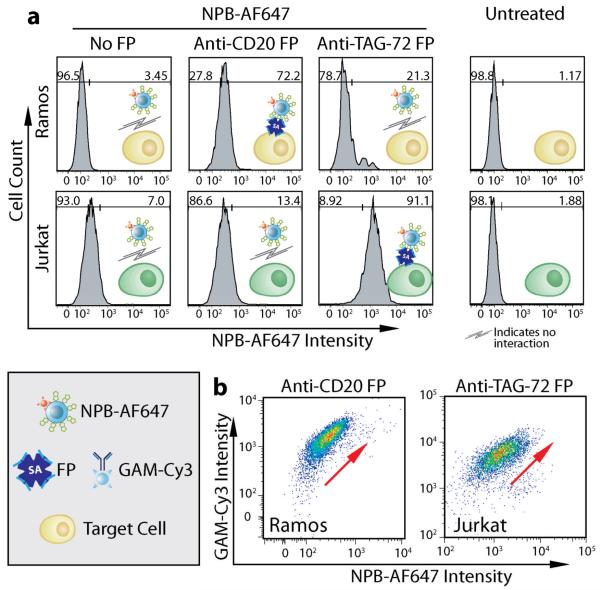
Binding specificity of the prepared NPB-AF647 for cells pretreated with FPs was tested using CD20+/TAG72− Ramos and CD20−/TAG72+ Jurkat cell lines (American Type Culture Collection, Manassas, VA). Each cell line was treated with either anti-CD20 or anti-TAG-72 CC49 FPs which are routinely used as the targeting agents for labeling non-Hodgekin’s lymphoma22,23 and adenocarcinoma,21 respectively. Cells treated with the FPs or receiving no FP pretreatment were then exposed to NPB-AF647 and evaluated by flow cytometry (Fig. 3a). Cells that received neither FP nor NPB-AF647 treatment were also analyzed as references (Fig. 3a, fourth column “Untreated”).
Fig. 3.
Cell targeting with NPB-AF647. a) Flow cytometry analysis of Ramos and Jurkat cells treated with NPB-AF647 after pretreatment with different FPs or receiving no FP pretreatment. Ramos and Jurkat cells receiving neither FP nor NP treatment are given as references (right column). b) Dependence of NPB-AF647 cell-binding on FP pre-labeling, demonstrated by flow cytometry, where the secondary antibody, goat-antimouse-Cy3 (GAM-Cy3), indicates FP labeling.
Here, Ramos cells (CD20+/TAG72−) showed positive NPB-AF647 binding (72.2%) when pretreated with the anti-CD20 FP, but limited NPB-AF647 labeling (21.3%) when pretreated with the anti-TAG-72 FP, and minimal NPB-AF647 labeling (3.45%) when receiving no FP pretreatment. The difference in labeling between these two controls might be due to the non-specific binding of the anti-TAG-72 FP to the Ramos cells. Alternatively, Jurkat cells (CD20−/TAG72+) showed positive labeling (91.1%) when pretreated with anti-TAG-72 CC49 FP, but limited NPB-AF647 binding (13.4%) when pretreated with the anti-CD20 FP, and minimal NPB-AF647 labeling (7%) when receiving no FP pretreatment. Control nanoparticles (NP-AF647), with no biotin functionalization, showed no specific cell binding regardless of cells’ FP pretreatment (Supplementary Fig. S1). Since anti-CD20 FP binds to Ramos, but not Jurkat cells, while anti-TAG-72 FP binds to Jurkat, but not Ramos cells, the above results suggests that NPB-AF647 preferentially binds to cells that bear their complementary FPs. This was further supported by the fact that NPB-AF647 bound minimally to both cell lines that received no FP pretreatment (Fig. 3a, first column “No FP”). These results also confirmed the binding activity of the biotin conjugated on the nanoparticle in addition to the confirmation by the FTIR and fluorescence analyses (Figs. 2a and 2b, respectively).
To further confirm that NPB-AF647 was bound to cells labeled with the murine-derived FPs, Ramos and Jurkat cells were pretreated with anti-CD20 and anti-TAG-72 CC49 FPs, respectively, and then exposed to NPB-AF647. The cells were then co-stained with a fluorescent goat anti-mouse (GAM-Cy3) secondary antibody, which binds to the SA of the FPs, and analyzed by flow cytometry (Fig. 3b). As observed in both cell lines, higher GAM-Cy3 staining correlated with increased nanoparticle binding (denoted by the red arrows), confirming specific targeting of NPB-AF647 to cells with positive FP expression.
Endocytosis of Nanoparticles by Target Cells
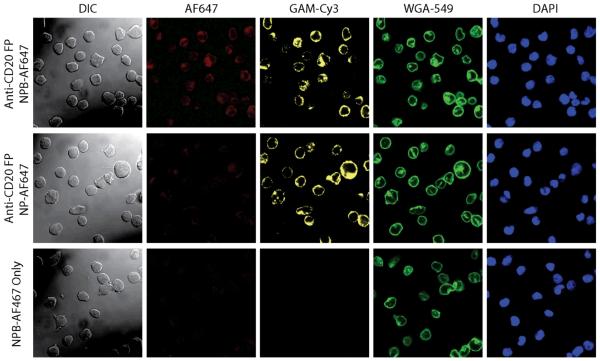
Selective binding of the NPB-AF647 to Ramos cells pretreated with anti-CD20 FP was further visualized by confocal and differential interference (DIC) microscopy; here, confocal imaging showed nanoparticle-cell co-localization, while DIC imaging corroborated cell location (Fig. 4). Ramos cells pretreated with anti-CD20 FP (target cells) or receiving no pretreatment (control cells) were exposed to either NPB-AF647 (targeting particles) or NP-AF647 (non-targeting particle controls). Fluorescence imaging of the nanoparticles (AF647, red), FPs (GAM-Cy3, yellow), cell membranes (WGA-594, green), and nuclei (DAPI, blue) was performed. The yellow fluorescence observed on Ramos cells pretreated with anti-CD20 FP (rows 1 and 2) and the co-localization of yellow and green fluorescence indicate the positive FP labeling on the cell surfaces. Strong red fluorescence was observed on the Ramos cells pretreated with anti-CD20 FP (row 1) but not on the cells receiving no anti-CD20 FP pretreatment (row 3), after both were exposed to NPB-AF647, indicating that NPB-AF647 bound specifically to the target cells. On the other hand, little to no red fluorescence was observed on Ramos cells exposed to NP-AF647 controls (row 2), as expected, even though the cells were pretreated with anti-CD20 FP. Notably, nanoparticle fluorescence (AF647) was co-localized with GAM-Cy3, indicating the targeted cell binding by NPB-AF647 via the SA of the FPs bound to the cell (row 1).
Fig. 4.
Nanoparticle interaction with cells. Nanoparticle-Ramos cell interactions analyzed by Differential Interference Contrast (DIC, column 1) and confocal fluorescence microscopy (columns 2–5). Row 1: Ramos cells pretreated with anti-CD20 FP and exposed to NPB-AF647; row 2: Ramos cells pretreated with anti-CD20 FP and exposed to NP-AF674 (control particles); row 3: Ramos cells with no pretreatment (control cells) exposed to NPB-AF674. Here, the nanoparticles were labeled red (AF674), anti-CD20 on cells labeled yellow (GAM-Cy3), cell membranes labeled green (WGA-594), and cell nuclei labeled blue (DAPI). NPB showed no cell binding to Ramos cells receiving no FP pretreatment (row 3).
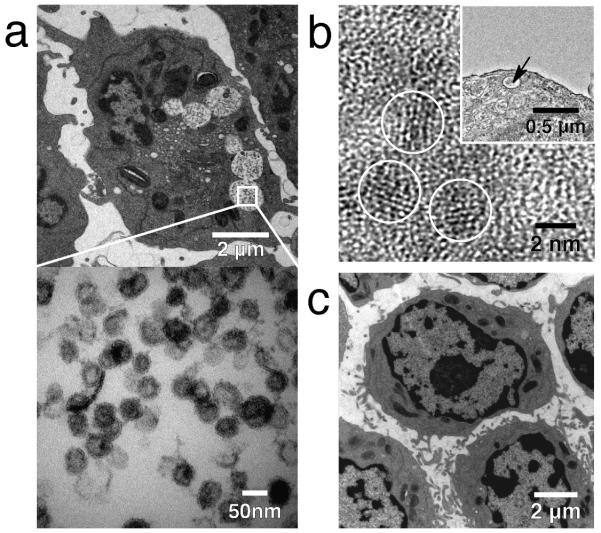
TEM was used to examine the localization of the nanoparticles bound to target cells. The TEM micrograph showed that a large population of nanoparticles were internalized by the Ramos cells and localized in the endosomes having diameters on the order of 1 μm (Fig. 5a, top image). TEM imaging at higher magnification further revealed that the nanoparticles were well dispersed in the endosomes and exhibited a uniform size distribution (Fig. 5a, bottom image). At even higher magnification (Fig. 5b), TEM analysis showed that the nanoparticles in the endosome (inset of Fig. 5b) had characteristic lattice fringe patterns of the iron oxide crystals (circled in Fig. 5b). Further confirmation of nanoparticle internalization by cells was made by energy-dispersive X-ray spectroscopy (EDX), where a cluster of nanoparticles inside a cellular endosome (Supplementary Fig. S2, inset) was probed, showing characteristic iron peaks (Supplementary Fig. S2). Similar probing elsewhere in the cell showed the presence of minimal or no iron (data not shown). Electron micrographs of Ramos cells treated with NPB-AF647, but with no FP pretreatment, showed no evidence of nanoparticle internalization in cells (Fig. 5c). This visually validated the flow cytometry results shown above.
Fig. 5.
Nanoparticle internalization by target cells. a) Transmission electron microscopy (TEM) analysis of Ramos cells treated with the anti-CD20 FP and exposed to NPB-AF647 showing a large population of nanoparticles (black dots) inside cellular endosomes (circular white areas with black dots). The white square frame (top image) marks the region of interest in an endosome shown in the higher magnification micrograph (bottom image) showing the dispersed nanoparticles in the endosome. b) TEM image at high magnification showing the characteristic lattice planes of the nanoparticle crystal (circled areas) taken from a cellular endosome (marked by the arrow in the inset) containing nanoparticles. The scale bar is 2 nm in the image and 0.5 μm in the inset. c) TEM micrograph of Ramos cells receiving no anti-CD20 FP pretreatment, showing no particle internalization after exposure to NPB-AF647.
While the underlying mechanism of the particle internalization by Ramos cells that bear CD20, a non-internalizing epitope target as shown in previous studies in radioimmunotherapy,24 is not the subject of this study, we believe that it was likely attributed the physicochemical profile of the nanoparticle. In this study, we coated the nanoparticle with a chitosan-PEG graft co-polymer for nanoparticle stabilization, functionalization, and cell membrane interactivity. Chitosan, a naturally derived cationic polysaccharide, has demonstrated success in many biomedical applications as a biocompatible, biodegradable material, and has been shown to induce endocytosis mediated by the anionic proteoglycans at the cell surface.6,25-29 Nanoparticle internalization by targeted cells is often desirable because many chemotherapeutics require cellular internalization before they can effectively treat the targeted cells.
Conclusions
The endurance and functionality of a nanoparticle system in vivo depends on its pharmacokinetic profile for which the targeting ligand plays a key role. Therefore, the targeting ligand on a nanoparticle system specific to one disease cannot be readily replaced with another for treatment of a different disease without significantly altering the nanoparticle’s pharmacokinetic profile. This necessitates significant research in design, development and testing of a new nanoparticle system for each disease or biomolecular marker that is to be targeted. In this study, we have developed a nanoparticle system based on a pretreatment regimen that can be matched with a range of SA-enabled FPs. We demonstrated this approach with two model cancer cell lines that display different cell epitopes. We showed that this nanoparticle system binds preferentially to target cells pretreated with their complementary FPs. This approach enables the nanoparticle system to retain a common physiochemical profile and used against multiple disease types.
Experimental
Nanoparticle preparation
The iron oxide nanoparticles were synthesized and coated with the chitosan-PEG polymer by coprecipitating Fe(II) and Fe(III) with the addition of sodium hydroxide in the presence of the polymer, as previously reported.6 The chitosan-PEG coated nanoparticles (NPs) were then transferred into a 0.1M NaBicarbonate pH 8.5 buffer by GPC using Sephacryl S200 medium (GE Healthcare, Piscataway, NJ). The NPs were then fluorescently labeled with the Alexa Fluor 647 (AF647) fluorochrome by dissolving 1 mg AF647 succinimidyl ester (Invitrogen, Carlsbad, CA) into 0.15 mL dimethylsulfoxide (DMSO) and mixing them with the NPs (3.5 mg Fe; ~2 mg Fe/mL) at room temperature for 2 hrs. Free AF647 was removed by GPC purification into PBS pH 7.4 (Sephacryl S200) to form control nanoparticles (NP-AF647).
Biotinylated NP-AF647 (NPB-AF647) was prepared by dissolving 2 mg EZ-link NHS-PEG4-Biotin (Thermo Fisher Scientific, Rockford, IL) in 100 μL 0.1M NaBicarbonate pH 8.5 and adding it to 1.75 mg Fe of the unpurified NP/AF647 mixture. This mixture was allowed to sit at room temperature for 2 hrs, then purified into PBS pH 7.4 by GPC (Sephacryl S200), yielding the NPB-AF647.
Nanoparticle characterization
For FTIR analysis, nanoparticle samples were milled with KBr, and pressed into pellets. FTIR spectra were acquired with a Nicolet 5-DXB spectrometer with a resolution of 4 cm−1. For the NPB-AF647 sample, the nanoparticles were mixed with SA (0.1 mg/mL; Invitrogen) for 30 min, washed of unbound SA on a rare earth magnet 3×, and analyzed by FTIR. For FluoReporter Biotin Assay analysis, nanoparticles were dissolved in 1M HCl for 2 hrs at 35°C before being neutralized. Dissolved nanoparticle samples had a final Fe concentration of 50 μg/mL. All samples were mixed with the Biotective Green reagent for 10 min, fluorescence was measured at 495/519 nm (ex/em; SpectraMax microplate reader). Nanoparticle sizes and zeta-potentials were determined by light scattering using a Zetasizer ZS (Malvern Instruments, Southborough, MA). All samples were analyzed in PBS pH 7.4, and sizing was completed using z-average data acquisition.
Fusion protein production
The heavy-chain (VH) and light-chain (VL) genes were cloned from the 1F5 (anti-CD20) and CC49 (anti-TAG-72) hybridomas. The single-chain antibody-SA genes from the current study were engineered by fusing the scFv gene to the full-length genomic SA of Streptomyces avidinii as described.30 The FPs were expressed from an IPTG-inducible lac promoter. An E coli XL1 Blue (Stratagene, La Jolla, CA) transformant of the antibody-SA construct was grown in shaker flasks for qualitative expression of the FPs and subsequently in a 4 L fermentor (BioFlo 3000; New Brunswick Scientific, Edison, NJ) using methods similar to those described.31 Cultures were induced with 0.1 mM IPTG and cells harvested after 44 hrs. The cell paste was washed 3× with PBS and the lysate was purified by iminobiotin column chromatography.31 The eluted FPs were treated with 20% DMSO for 2 hrs to reduce aggregates, extensively dialyzed in PBS, filter-sterilized, formulated in 5% sorbitol, and stored at −80°C.
Cell prelabeling
Ramos and Jurkat cells were cultured in RPMI media modified with 10% FBS and 1% penicillin and streptomycin (Invitrogen) at 37°C under 5% CO2. To test nanoparticle affinity and specificity, 106 cells/sample were washed into 100 μL PBS and incubated with 10 μg of either anti-CD20 [1F5-(sFv)4-streptavidin] or anti-TAG-72 [CC49-(scFv)4-streptavidin] FP for 30 min at 4°C and washed 3× with PBS by centrifugation. Cell samples were then incubated with either NP-AF647 or NPB-AF647 at 0.1 mg Fe/mL in PBS for 1 hr at 4°C. Cells were washed 3× with PBS, and incubated with 10 μg goat-antimouse-Cy3 (GAM-Cy3) for 30 min at 4°C. Samples were then analyzed by flow cytometry (FACS Canto, BD Biosciences, San Jose, CA). 20,000 cells were analyzed from each sample. For each condition, n = 3 samples were analyzed in one experiment, and each experiment was at least repeated twice to confirm the result.
Confocal microscopy
Three million Ramos cells, incubated with either NPB-AF647 (with and without anti-CD20 FP pretreatment) or NP-AF647 (with anti-CD20 FP pretreatment), were washed with PBS 3×, stained with GAM-Cy3, washed again 3×, and fixed in a 4% formaldehyde/PBS solution (methanol free, Polysciences Inc., Warrington, PA) for 30 min. The fixative was then removed and cells washed with PBS 3×. The cell membranes were then stained with wheat germ agglutinin-AlexaFluor 594 (WGA-AF594; Invitrogen) according the manufacturer’s instructions, and incubated with DAPI-containing Prolong Gold antifade solution (Invitrogen) for cellular nuclei staining and fluorescence preservation. Cell samples were then dried on cover slips and imaged using a Zeiss LSM 510 Meta confocal fluorescence microscope (Peabody, MA).
Transmission electron microscopy
One million Ramos cells, pretreated with the FP and incubated with NPB-AF647, were washed 3× with PBS and immersed in ice cold Karnovsky’s fixative for 24 hrs. Following fixation, the cells were palletized, sectioned and stained with osmium tetroxide, lead citrate, and uranyl acetate for TEM-contrast enhancement. Cell samples were mounted onto carbon-coated copper grids. Micrographs were taken with a Philips CM100 operating at 100 kV (low magnification) and a Tecnai G2 F20 TEM operating at 200 kV (high magnification). EDX spectra were obtained on the Tecnai TEM equipped with an EDAX detector.
Supplementary Material
Acknowledgements
This work is supported in part by NIH grants R01CA119408, R01EB006043, R01CA134213 (M. Zhang) and R01CA76287 (O. Press). J. Gunn and O. Veiseh would like to acknowledge the support of NIH training grants T32GM065098 and T32CA138312, respectively. We would like to thank Dr. Narayan Bhattarai for his assistance in acquiring the TEM image (Fig. 5b), and acknowledge the use of facilities at the Diagnostic Imaging Sciences Center, Center for Nanotechnology, and Keck Microscopy Imaging Facility at the University of Washington.
Footnotes
Electronic supplementary Information (ESI) available: Supplementary figures.
References
- 1.Debbage P, Jaschke W. Histochem Cell Biol. 2008;130:845–875. doi: 10.1007/s00418-008-0511-y. [DOI] [PubMed] [Google Scholar]
- 2.Nie SM, Xing Y, Kim GJ, Simons JW. Annual Review Of Biomedical Engineering. 2007;9:257–288. doi: 10.1146/annurev.bioeng.9.060906.152025. [DOI] [PubMed] [Google Scholar]
- 3.Sun C, Lee JSH, Zhang MQ. Advanced Drug Delivery Reviews. 2008;60:1252–1265. doi: 10.1016/j.addr.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee JH, Huh YM, Jun YW, Seo JW, Jang JT, Song HT, Kim S, Cho EJ, Yoon HG, Suh JS, Cheon J. Nat Med. 2007;13:95–99. doi: 10.1038/nm1467. [DOI] [PubMed] [Google Scholar]
- 5.Peer D, Park EJ, Morishita Y, Carman CV, Shimaoka M. Science. 2008;319:627–630. doi: 10.1126/science.1149859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Veiseh O, Sun C, Fang C, Bhattarai N, Gunn J, Kievit F, Du K, Pullar B, Lee D, Ellenbogen RG, Olson J, Zhang M. Cancer Res. 2009;69:6200–6207. doi: 10.1158/0008-5472.CAN-09-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai W, Chen X. Nat Protoc. 2008;3:89–96. doi: 10.1038/nprot.2007.478. [DOI] [PubMed] [Google Scholar]
- 8.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Nat Biotech. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 9.Weissleder R, Pittet MJ. Nature. 2008;452:580–589. doi: 10.1038/nature06917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davis ME, Chen Z, Shin DM. Nature Reviews Drug Discovery. 2008;7:771–782. doi: 10.1038/nrd2614. [DOI] [PubMed] [Google Scholar]
- 11.Veiseh O, Sun C, Gunn J, Kohler N, Gabikian P, Lee D, Bhattarai N, Ellenbogen R, Sze R, Hallahan A, Olson J, Zhang M. Nano Lett. 2005;5:1003–1008. doi: 10.1021/nl0502569. [DOI] [PubMed] [Google Scholar]
- 12.Takeda A, Baffi JZ, Kleinman ME, Cho WG, Nozaki M, Yamada K, Kaneko H, Albuquerque RJ, Dridi S, Saito K, Raisler BJ, Budd SJ, Geisen P, Munitz A, Ambati BK, Green MG, Ishibashi T, Wright JD, Humbles AA, Gerard CJ, Ogura Y, Pan Y, Smith JR, Grisanti S, Hartnett ME, Rothenberg ME, Ambati J. Nature. 2009;460:225–230. doi: 10.1038/nature08151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veiseh O, Gunn JW, Zhang M. Adv Drug Deliv Rev. 2009 doi: 10.1016/j.addr.2009.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pagel JM, Orgun N, Hamlin DK, Wilbur DS, Gooley TA, Gopal AK, Park SI, Green DJ, Lin Y, Press OW. Blood. 2009;113:4903–4913. doi: 10.1182/blood-2008-11-187401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen S, Forero A, LoBuglio AF, Breitz H, Khazaeli MB, Fisher DR, Wang W, Meredith RF. J Nucl Med. 2005;46:642–651. [PubMed] [Google Scholar]
- 16.Pagel JM, Hedin N, Drouet L, Wood BL, Pantelias A, Lin Y, Hamlin DK, Wilbur DS, Gopal AK, Green D, Appelbaum FR, Press OW. Blood. 2008;111:2261–2268. doi: 10.1182/blood-2007-06-097451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharkey RM, Karacay H, Litwin S, Rossi EA, McBride WJ, Chang CH, Goldenberg DM. Cancer Res. 2008;68:5282–5290. doi: 10.1158/0008-5472.CAN-08-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagel JM, Matthews DC, Kenoyer A, Hamlin DK, Wilbur DS, Fisher DR, Gopal AK, Lin Y, Saganic L, Appelbaum FR, Press OW. Cancer Res. 2009;69:185–192. doi: 10.1158/0008-5472.CAN-08-2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Green DJ, Pagel JM, Pantelias A, Hedin N, Lin Y, Wilbur DS, Gopal A, Hamlin DK, Press OW. Clin Cancer Res. 2007;13:5598s–5603s. doi: 10.1158/1078-0432.CCR-07-1223. [DOI] [PubMed] [Google Scholar]
- 20.Weiden PL, Breitz HB. Crit Rev Oncol Hematol. 2001;40:37–51. doi: 10.1016/s1040-8428(01)00133-0. [DOI] [PubMed] [Google Scholar]
- 21.Forster GJ, Santos EB, Smith-Jones PM, Zanzonico P, Larson SM. J Nucl Med. 2006;47:140–149. [PubMed] [Google Scholar]
- 22.Pantelias A, Pagel JM, Hedin N, Saganic L, Wilbur S, Hamlin DK, Wilbur DS, Lin Y, Stone D, Axworthy D, Gopal AK, Press OW. Blood. 2007;109:4980–4987. doi: 10.1182/blood-2006-11-056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goodwin DA, Meares CF. Biotechnol Adv. 2001;19:435–450. doi: 10.1016/s0734-9750(01)00065-9. [DOI] [PubMed] [Google Scholar]
- 24.Press OW, Corcoran M, Subbiah K, Hamlin DK, Wilbur DS, Johnson T, Theodore L, Yau E, Mallett R, Meyer DL, Axworthy D. Blood. 2001;98:2535–2543. doi: 10.1182/blood.v98.8.2535. [DOI] [PubMed] [Google Scholar]
- 25.Kumar M, Muzzarelli RAA, Muzzarelli C, Sashiwa H, Domb AJ. Chem. Rev. 2004;104:6017–6084. doi: 10.1021/cr030441b. [DOI] [PubMed] [Google Scholar]
- 26.Berger J, Reist M, Mayer JM, Felt O, Gurny R. Eur. J. Pharm. Biopharm. 2004;57:35–52. doi: 10.1016/s0939-6411(03)00160-7. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Ramay HR, Hauch KD, Xiao D, Zhang M. biomaterials. 2005;26:3919–3928. doi: 10.1016/j.biomaterials.2004.09.062. [DOI] [PubMed] [Google Scholar]
- 28.Zhang Y, Zhang MQ. J. Mater. Sci.-Mater. Med. 2004;15:255–260. doi: 10.1023/b:jmsm.0000015485.94665.25. [DOI] [PubMed] [Google Scholar]
- 29.Li Z, Leung M, Hopper R, Ellenbogen R, Zhang M. biomaterials. 2010;31:404–412. doi: 10.1016/j.biomaterials.2009.09.070. [DOI] [PubMed] [Google Scholar]
- 30.Shan D, Press OW, Tsu TT, Hayden MS, Ledbetter JA. J Immunol. 1999;162:6589–6595. [PubMed] [Google Scholar]
- 31.Schultz J, Lin Y, Sanderson J, Zuo Y, Stone D, Mallett R, Wilbert S, Axworthy D. Cancer Res. 2000;60:6663–6669. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.