Abstract
House fly (Musca domestica) CYP6D1 is a cytochrome P450 involved in metabolism of xenobiotics. CYP6D1 is located on chromosome 1 and its expression is inducible in response to the prototypical P450 inducer phenobarbital (PB) in insecticide susceptible strains. Increased transcription of CYP6D1 confers resistance to permethrin in the LPR strain, and this trait maps to chromosomes 1 and 2. However, the constitutive overexpression of CYP6D1 in LPR is not further increased by PB and the non-responsiveness to PB maps to chromosome 2. It has been suggested that a single factor on chromosome 2 could be responsible for both the constitutive overexpression and lack of PB induction of CYP6D1 in LPR. We examined the PB inducibility of CYP6D1v1 promoter from LPR using dual luciferase reporter assays in Drosophila S2 cells and found the CYP6D1v1 promoter was able to mediate PB induction, similar to the CYP6D1v2 promoter from the insecticide susceptible CS strain. Therefore, variation in promoter sequences of CYP6D1v1 and v2 does not appear responsible for the lack of PB induction of CYP6D1v1 in LPR; this suggests an unidentified trans acting factor is responsible. HR96 has been implicated in having a role in PB induction in Drosophila melanogaster and M. domestica. Therefore, house fly HR96 cDNA was cloned and sequenced to examine if this trans acting factor is responsible for constitutive overexpression of CYP6D1v1 in LPR. Multiple HR96 alleles (v1–v10) were identified, but none were associated with resistance. Expression levels of HR96 were not different between LPR and CS. Thus, HR96 is not the trans acting factor responsible for the constitutive overexpression of CYP6D1 in LPR. The identity of this trans acting factor remains elusive.
Keywords: Pyrethroid resistance, CYP6D1 transcription, Phenobarbital induction, HR96, Musca domestica, S2 cells
1. Introduction
House fly (Musca domestica) cytochrome P450 CYP6D1 can metabolize numerous xenobiotics, including beno(a)pyrene [1], chlorpyrifos [2], methoxyresorufin [1], phenanthrene [3], and phenoxybenzyl pyrethroids such as cypermethrin, deltamethrin, and permethrin [4; 5]. CYP6D1 was sequenced [6] and mapped to chromosome 1 [7]. Phenobarbital (PB) treatment of insecticide susceptible strains results in a 6- to 8-fold increase in CYP6D1 transcript and protein [8; 9]. CYP6D1 is expressed in numerous tissues [10; 11] and expression of CYP6D1 is developmentally regulated, being found only in adults [9].
Increased transcription of CYP6D1 confers metabolism-mediated resistance to phenoxybenzyl pyrethroids (e.g. permethrin) in the Learn Pyrethroid Resistant (LPR) strain of house fly [12]. CYP6D1 transcript and protein levels are overexpressed (~9-fold increase) in LPR relative to insecticide susceptible strains [8; 9; 13]. Overexpression of CYP6D1 in LPR is not due to gene duplication [14] or increased transcript stability [12]. Increased transcription of CYP6D1, measured with nuclear run-on assays, was linked to factors on chromosome 1 and 2 [8; 13]. Multiple CYP6D1 alleles have been identified in pyrethroid susceptible strains, but only one (v1) is found in resistant strains [15; 16]. The promoters of CYP6D1 alleles have been sequenced from five house fly strains. The comparison of promoter sequences showed the LPR specific allele (CYP6D1v1) had a 15 nucleotide insertion located at -29 to -15 relative to the transcription start site [15]. This insertion in the CYP6D1v1 promoter of the LPR disrupts the binding site for a known transcriptional repressor Gfi-1. House fly Gfi-1 is on chromosome 1 [17]. Thus, this deletion in the CYP6D1v1 promoter appears to be the factor on chromosome 1 responsible for the elevated transcription in the LPR strain. However, the factor on chromosome 2 remains unidentified. The constitutive overexpression of CYP6D1 in LPR is not further increased by PB and the non-responsiveness to PB maps to chromosome 2. Whether variation in CYP6D1 promoter sequences is involved in the lack of PB induction in LPR is not known.
The CYP6D1v2 promoter sequence from −330 to −280 (numbers are relative to transcription start site, +1) of the insecticide susceptible CS strain is critical for PB induction [18]. Drosophila HR96 (hormone receptor-like in 96) and BR-C (broad-complex) were identified as a PB dependent transcriptional activator and repressor, respectively, of CYP6D1v2 in Drosophila S2 cells [18]. Based on a chromosome homology map [19; 20], HR96 and BR-C are expected to be on chromosome 2 and 3, respectively, of house fly. Thus, properties of HR96 (activator on chromosome 2) were consistent with the hypothesis that this might be the factor responsible for the overexpression of CYP6D1 in LPR.
The goals of this study were: 1) determine if the CYP6D1v1 promoter retained its responsiveness to PB, 2) clone and sequence HR96 from house fly, and 3) determine if HR96 played a role in resistance. We conducted PB responsive promoter assays using dual luciferase reporter assays in Drosophila S2 cells to examine the CYP6D1v1 promoter of LPR. Our results showed the CYP6D1v1 promoter of LPR was able to mediate PB induction, just like the CYP6D1v2 promoter from CS [18]; although there are four single nucleotide polymorphisms (SNPs) in the PB responsive promoter region in LPR compared to CS [15]. These results indicated variations in promoter sequences did not significantly affect of PB inducibility of the CYP6D1v1 promoter in S2 cells. Therefore, the lack of PB induction of CYP6D1 in LPR appears due to an unidentified trans acting factor on chromosome 2. House fly HR96 was cloned and sequenced in order to examine if it is this trans acting factor. Multiple HR96 alleles (v1–v10) were identified, but neither a specific allele nor levels of expression were associated with resistance.
2. Materials and methods
2.1. House flies
Four house fly strains were used. The CS and aabys are insecticide susceptible strains [21]. The LPR is a permethrin resistant strain originally collected from a dairy near Horseheads, New York in 1982 [22]. The OCR is a cyclodiene resistant (Rdl) [23] and pyrethroid susceptible strain [24]. House fly larvae were reared on mixed media made of 500 g of calf manna (Agway, Ithaca, NY), 120 g of wood chips (Agway), 60 g of Baker’s yeast (MP Biomedicals, Solon, OH), 1210 g of wheat bran (Agway) and 2000 ml of water. Adults were fed on powdered milk:granulated sugar (1:1) and water ad libum. Larvae and adults were reared at 28°C, 60% relative humidity, with a 12: 12 hr light/dark photoperiod.
2.2. Drosophila S2 cells
Drosophila S2 cells were maintained and grown in serum free cell culture medium of HyQ SFX-Insect (HyClone, Logan, UT) in 75 cm2 of tissue culture flask (BD Falcon, Bedford, MA). Cells were subcultured every 2–3 days as they reached confluency.
2.3. CYP6D1v1 promoter constructs and PB responsive promoter assays
Progressive 5' deletions of the CYP6D1v1 promoter from the LPR strain [15] were generated by PCR amplification. Promoter regions −925/+85, −365/+85, −267/+85, and −57/+85 (numbers are relative to transcription start site, defined as +1) were constructed into restriction enzyme sites Sac I and Bgl II of pGL3-Basic vector (Promega, Madison, WI) as previously described [18]. PB responsive promoter assays were performed using a dual luciferase reporter assay system (Promega) in Drosophila S2 cells as previously described [18]. Three independent transfections (PB or control) of each promoter construct were conducted in each replicate. The entire experiment was replicated four times. Statistical analysis of pairwise comparisons of difference of [(PB induced promoter activity) – (basal promoter activity) relative to the next shorter CYP6D1 promoter construct] was conducted using Student’s t-test to indicate promoter regions critical for PB induction.
2.4. Isolation of gDNA or mRNA, gel extraction, TA cloning, plasmid DNA purification, and DNA sequencing
DNA was isolated from individual adult house flies as previously described [25]. Purification of mRNA was done using an Illustra QuickPrep™ micro mRNA purification kit (GE healthcare, Little Chalfont, UK). Gel extraction of PCR products was done using a QIAEX II kit (Qiagen, Valencia, CA). Cloning of PCR products was performed using a TOPO TA kit with pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA) and TOP 10 competent cells (Invitrogen). Plasmid DNA was isolated using a QIAprep Miniprep system (Qiagen). Plasmid DNAs were sequenced at the Cornell University Life Sciences Core Laboratories Center.
2.5. Cloning of house fly HR96
Degenerate primers were selected using CODEHOP (http://blocks.fhcrc.org/codehop.html) [26] with an alignment of the HR96 peptide sequences of Drosophila melanogaster (NP_524493.1), Drosophila yakuba (XP_002099134.1), Drosophila virilus (XP_002054249.1), Culex quinquefasciatus (XP_001866050.1), and Anopheles gambiae (XP_313130.4). PCR reactions included 0.5 µl of genomic DNA (aabys adults), 1 µl of 10 µM forward primer, 1 µl of 10 µM reverse primer, 10 µl of ddH2O, and 12.5 µl of 2X GoTaq® Green Master Mix (Promega) and was carried out in a iCycler thermal cycler (Bio-Rad, Hercules, CA) with following temperature program: 95°C for 3 min; 35 cycles of 95°C for 30 sec, 60°C for 45 sec (decrease temperature by 0.3°C every 1 cycle), and 72°C for 1 min 20 sec; and 72°C for 5 min. Three distinct degenerate primer pairs produced partial sequences of HR96: (i) forward: 5'-GTG GTG ATA AAG CCT TGG GTT AYA AYT TYA A-3' and reverse: 5'-GGC ATT AAA GGG GGA ATT CAT DAW YTT-3', (ii) forward: 5'-AAA ATT ACC GCC TTT AGA AAT ATG TGY CAR GA-3' and reverse: 5'-GGT AAT GGC ACA CAT AAT CAA AAT AAT RTT YTC RTC-3', and (iii) forward: 5'-CTT GTT GAA AGG TGG TTG TAC AGA RAT GAT GAT -3' and reverse: 5'-GGT AAT GGC ACA CAT AAT CAA AAT AAT RTT YTC RTC-3'. A FirstChoice® RLM-RACE kit (Ambion, Austin, TX) was used for 5' and 3' RACE of HR96 according to the manufacture’s instructions using mRNA derived from 10 abdomens of 3-day-old male adult aabys flies. PCR for 5' RACE was performed using the 5' RACE outer primer: 5'-GCT GAT GGC GAT GAA TGA ACA CTG-3' and a gene-specific reverse primer: 5'-TCT CGC TCT TCA TGC CGA TGT CT-3' with the following thermal cycler program: 95°C for 3 min; 40 cycles of 95°C for 30 sec, 55°C for 30 sec, and 72°C for 1 min; and 72°C for 5 min. PCR for 3' RACE was conducted using the 3' RACE outer primer: 3'-GCG AGC ACA GAA TTA ATA CGA CT-3' plus a gene-specific forward outer primer: 5'-GCC AAG AGG ATC AGG TTG CCT T-3'. The nested PCR for 3' RACE was conducted using the 3' RACE inner primer: 5'-CGC GGA TCC GAA TTA ATA CGA CTC ACT ATA GG-3' plus a gene-specific forward inner primer: 5'-GCC AAG GGC AAT GTC TAT GAA GAA C-3'. The thermal cycler program for 3' RACE PCRs were identical to the 5' RACE PCR described above. The complete coding sequence of HR96 was obtained from four strains (aabys, CS, OCR, and LPR) by PCR using forward primer 5'-CAA AGA TGT CAC CAA TTA ATA AAG TCT GTG C-3', reverse primer 5'-ATG ATG TAG GAA TTA AGG ACA TTT GAG GTA AC-3' and the following thermal cycler program: 95°C for 3 min; 40 cycles of 95°C for 30 sec, 62°C for 45 sec, and 72°C for 2 min 30 sec; and 72°C for 5 min. The cDNA product used for the above PCR was synthesized from mRNA of 10 abdomens of three-day-old male adults using the SuperScript™ III first-strand synthesis system for RT-PCR (Invitrogen). The PCR products were analyzed on a 1.6% agarose gel and then subjected to gel extraction, TA cloning, plasmid DNA purification, and DNA sequencing as described above.
2.6. Permethrin selection of the LPR strain
Technical grade of permethrin, 60% cis and 39% trans, (ChemService, West Chester, PA) was dissolved in acetone (AR grade) (Mallinckrodt Chemicals, Phillipsburg, NJ). Serial doses (3.35, 0.838, 0.209, and 0.052 µg/fly) were tested to determine the proper dose which could result in 90–95% mortality in LPR strain. Doses of 3.35 and 0.052 µg/fly were also applied to insecticide susceptible aabys strain as a control and resulted in 100% mortality. For permethrin selection of the LPR strain, 300 male adults were treated with 6.25 µg/fly permethrin. Survivors (n = 11) were subjected to DNA isolation and PCR to amplify polymorphic region containing E82V and G110D substitutions as described in Section 2.5.
2.7. HR96 genotyping
The polymorphic region of HR96 (from position -5 to 602, numbers are relative to translation start site ATG as +1) was amplified and sequenced using genomic DNA of individual flies to determine if HR96 was at the permethrin resistance locus on chromosome 2. The PCR was performed in 50 µl of reaction volume including 1 µl of gDNA, 2 µl of 10 µM forward primer (5'-CAA AGA TGT CAA TTA ATA AAG TCT GTG C-3'), 2 µl of 10 µM reverse primer (5'-GCT TGT GAG GCA CGG TCC-3'), 20 µl of ddH2O, and 25 µl of 2X GoTaq® Green Master Mix (Promega). Reactions were carried out in an iCycler thermal cycler (Bio-Rad) with following temperature program: 95°C for 5 min; 30 cycles of 95°C for 30 sec, 58°C for 30 sec, and 72°C for 50 sec; and 72°C for 4 min. PCR products were purified using Wizard SV gel and PCR clean-up system (Promega) and sequenced at the Cornell University Life Sciences Core Laboratories Center using the forward primer described in above.
2.8. Quantitative real-time RT-PCR of HR96
Relative quantitation of HR96 transcript level was measured by normalizing to Actin (GenBank: ES652303.1) using real-time RT-PCR with the comparative CT method. Purified mRNA (500 ng), derived from 10 abdomens of three-day-old male adults, was treated with DNase to remove gDNA (DNA free™ kit, Applied Biosystems, Foster City, CA), and cDNA was synthesized using the SuperScript™ III first-strand synthesis system (Invitrogen). Each real-time PCR reaction included 0.5 µl of above cDNA product, 1 µl of 10 µM forward primer, 1 µl of 10 µM reverse primer, 7.5 µl of ddH2O, and 10 µl of Power SYBR Green PCR Master Mix (2X) (Applied Biosystems). Primers used were HR96-forward: 5'-CGG ACC GTG CCT CAC AAG C-3', HR96-reverse: 5'-TCT TCA AAG CAT CGC CTG GAT AGT-3', ACTIN-forward: 5'-TCT GGC ATC ACG CTT TCT ACA ATG-3', and ACTIN-reverse: 5'-GGA GAG AAC AGC TTG GAA GGC A-3'. Reactions were carried out using Applied Biosystems 7900HT Real-Time PCR system at the Cornell University Life Sciences Core Laboratories Center with following temperature program: 50°C for 2 min; 95°C for 10 min; 40 cycles of 95°C for 15 sec and 60°C for 1 min. Data was processed and analyzed using SDS software (version: 2.1). Three independent real-time PCR reactions of three independent biological samples (i.e. cDNA of 10 three-day-old male abdomens prepared independently) of CS and LPR strains (n=9) were acquired. Statistic analysis was conducted using Student’s t-test. PCR products were further analyzed in a 2% agarose gel to confirm the size of product and were DNA sequenced.
3. Results and discussion
3.1 CYP6D1v1 (LPR) promoter is able to mediate PB induction in S2 cells
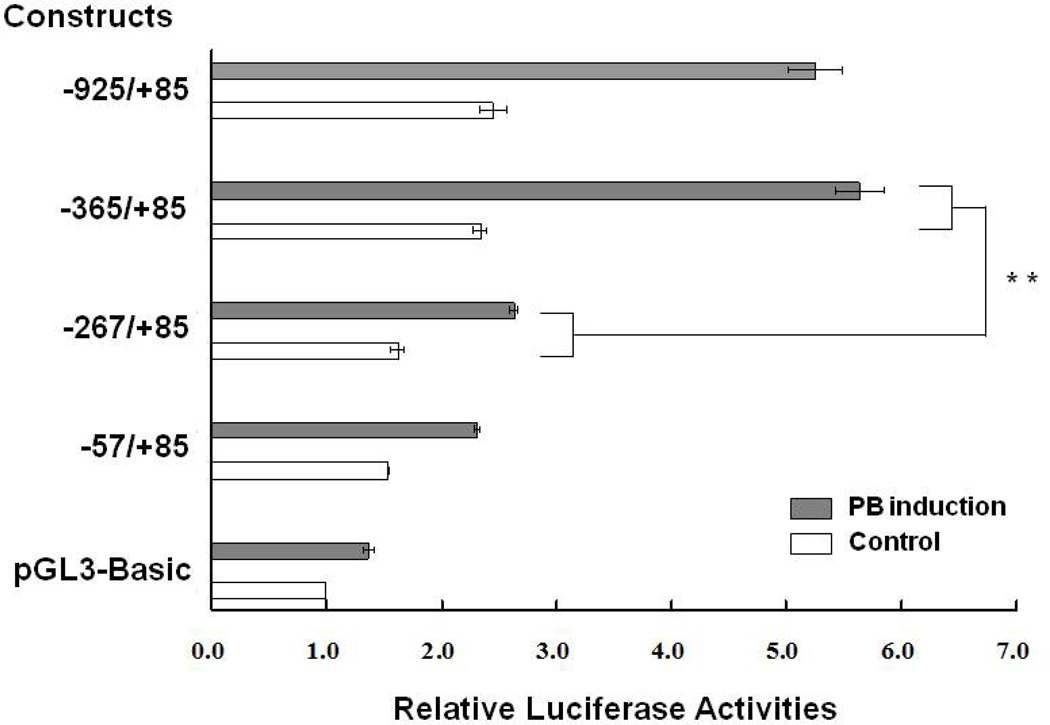
The CYP6D1v2 (CS) promoter sequence from −330 to −280 has been identified to be critical for mediating PB induction in Drosophila S2 cells [18]. The equivalent CYP6D1v1 promoter sequence (from −351 to −301) from LPR contains four SNPs compared to CYP6D1v2 promoter sequence from CS [15]. Promoter constructs of progressive 5' deletions of the CYP6D1v1 promoter of LPR (−925/+85, −365/+85, −267/+85, and −57/+85, numbers are relative to transcription start site, defined as +1) were used to determine if a region of CYP6D1v1 controlled PB responsiveness. Four independent replicates indicated the promoter construct −365/+85 had increased PB induced luciferase activity compared to promoter construct −267/+85 (Fig. 1), indicating the presence of critical cis-regulatory sequence for PB induction within CYP6D1v1 promoter region (−365 to −267) of LPR. The fold change in PB induction of CYP6D1v1 (2.03 ± 0.27, n = 12) did not show significant difference (p< 0.3774, Student’s t-test) compared to the fold change in PB induction of CYP6D1v2 (1.87 ± 0.07, n = 9) [18]. This indicates that despite polymorphisms between the CYP6D1v1 (from LPR) and CYP6D1v2 (from CS) promoters, both are able to mediate induction in response to PB. Thus, the lack of PB induction of CYP6D1 in the LPR strain appears to be attributed to an unknown trans acting factor, rather than sequence variations in the CYP6D1v1 promoter rendering it unresponsive to PB.
Fig. 1.
CYP6D1v1 promoter sequence from −365 to −267 (from LPR) is able to mediate PB induction. Promoter constructs are numbered relative to the transcription start site (TSS) at +1. Relative luciferase activity was estimated by normalizing each signal of each promoter construct to the mean of signals of pGL3-Basic vector in the same replicate. Bars represent the mean relative luciferase activity ± S.D. of three independent transfections. Gray bars represent the signal in the presence of PB and white bars represent the control. Double asterisks indicate a greater PB induction relative to the next shorter promoter construct (p<0.01, Student’s t-test).
Comparison of the current results with our previous study shows basal promoter luciferase activities among parallel promoter constructs between CS and LPR did not significantly differ, except the −900/+85 construct from CS had significantly greater luciferase activities compared to the −925/+85 construct from LPR. This contrasts with the 9-fold greater transcription of CYP6D1 found in LPR house flies, relative to CS. This suggests that Drosophila S2 cells lack the factor(s) found in LPR house flies that cause the enhanced transcription or that another region of DNA is responsible for the increased transcription of CYP6D1 in LPR (e.g. other 5' flanking sequences, intron sequences, or 3' flanking sequences).
Previously, Drosophila HR96 and BR-C were identified as a key transcription factors regulating PB induction of CYP6D1v2 in Drosophila S2 cells [18]. Based on a chromosome homology map [19; 20], HR96 is expected to be on chromosome 2 of house fly, while BR-C is expected to be on chromosome 3. Therefore, house fly HR96 could be the trans acting factor responsible for constitutive overexpression of CYP6D1 in LPR, and it was studied further.
3.2. Cloning of house fly HR96 complete coding sequence
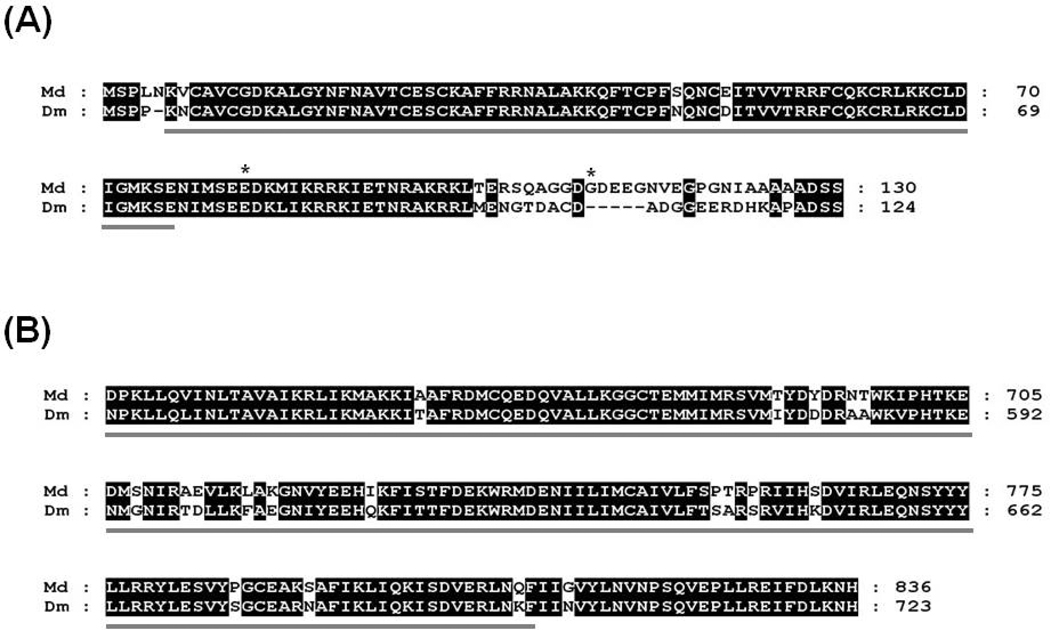
The full length cDNA of house fly HR96 gene contains the ORF of 2508 nucleotides encoding 836 amino acids, a 5' UTR of 6 nucleotides, and a 3' UTR of 440 nucleotides (GenBank accession number: HM150722). The DNA binding domain (aa: 6–76, zinc finger, C4 type, Fig. 2A) and ligand binding domain (aa: 638–810, ligand-binding domain of nuclear receptor family 1, Fig. 2B) were 94% and 84 % identical to domains of HR96 of D. melanogaster, respectively.
Fig. 2.
Alignment of D. melanogaster (Dm) and M. domestica (Md) HR96 deduced protein sequences. (A) Protein sequence alignment of the DNA binding domain (DBD). Sequences representing the DBD were underlined. Locations of two amino acid substitutions (E82V and G110D) identified in alleles v8–v10 from the LPR strain were denoted with asterisks above the alignment. (B) Protein sequence alignment of the ligand binding domain (LBD). Sequences representing the LBD were underlined.
3.3. HR96 alleles
The complete coding sequence of HR96 was cloned and sequenced from four house fly strains: aabys, CS, OCR, and LPR. Forty-five full length cDNA clones were sequenced and 10 HR96 alleles (v1–v10) were identified (Table 1). The deduced amino acid sequences of alleles v1–v7 were all identical (alleles were identified by synonymous SNPs and/or intron polymorphisms). However, alleles v8, v9, and v10 all contained two non-synonymous SNPs (E82V and G110D; Table 1). The E82 residue is highly conserved among insect HR96 orthologs (even the local region is highly conserved) and is located right after the zinc finger DNA binding domain (aa: 6–76) (Fig. 2A). The G110 residue is located in a region which is less conserved among insect HR96 orthologs. Alleles v8, v9, and v10 were found only in the LPR strain, although LPR also contained the v4–7 alleles which were found in susceptible strains (Table 1).
Table 1.
HR96 alleles, based on full length cDNA, from four house fly strains.
The presence of multiple HR96 alleles in LPR (including those found in susceptible strains) suggested that either this locus was not linked to resistance or that the LPR strain had been inadvertently contaminated. To test these competing hypotheses, 300 adult male LPR flies were topically treated with permethrin (6.25µg/fly), which resulted in 96.3% mortality (11 survivors). The HR96 alleles present in these 11 survivors were compared to the alleles found in 10 non-treated and randomly sampled LPR flies. There was no difference in the frequency of the alleles between the permethrin treated survivors and the non-treated flies. The permethrin survivors had 14 wild-type alleles (v4–7) and 8 alleles containing the E82V and G110D mutations (v8–10), compared to 14 v4–7 and 6 v8–10 alleles in the non-treated flies. These results indicate there was no selective sweep at the HR96 locus associated with permethrin resistance in LPR.
3.4. Expression of HR96 in LPR and CS
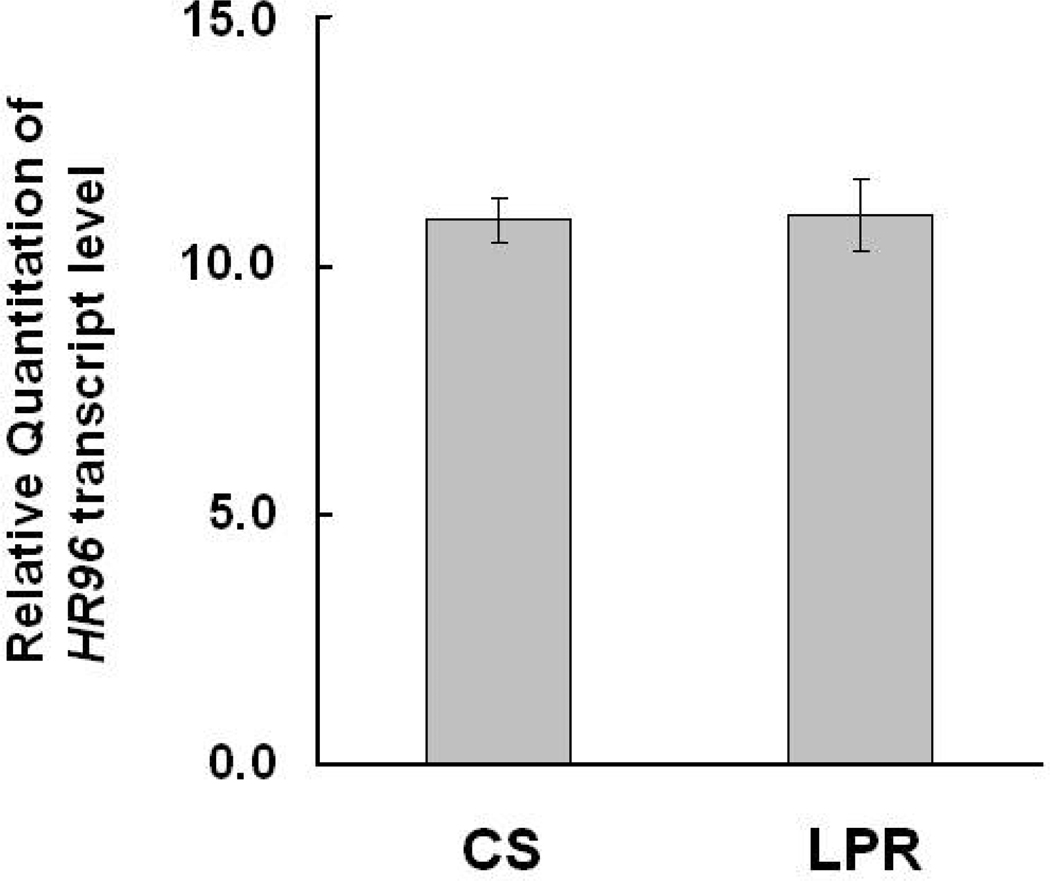
Quantitative real-time RT-PCR was used to measure HR96 transcript level in the resistant LPR and susceptible CS strains. No significant difference in HR96 expression was found between these strains (Fig. 3). Therefore, differences in HR96 expression are not associated with constitutive overexpression of CYP6D1 (nor resistance) in the LPR strain.
Fig. 3.
Relative quantitation of HR96 transcript level of the CS and LPR strains. Abdomens of ten three-day-old male adults of CS and LPR were used to measure HR96 transcript level relative to Actin transcript level using real-time RT-PCR with comparative CT method. Bars represent means of relative quantitation of HR96 (normalized by Actin transcript level) ± S.D. of three PCR reactions of three biological sample pools (n = 9). The HR96 transcript levels were not significantly different between CS and LPR (by Student’s t-test).
3.5. Conclusion
The CYP6D1v1 (LPR) promoter is able to mediate PB induction similar to the CYP6D1v2 (CS) promoter. Thus, the constitutive overexpression of CYP6D1 in LPR is attributed to a trans acting factor on chromosome 2, rather than differences in the promoter. House fly HR96 was cloned, sequenced and 10 alleles were identified. However, analysis of the alleles present, and of HR96 levels in LPR compared to CS, did not support the hypothesis that HR96 was the transcription factor responsible for the constitutive overexpression of CYP6D1 in LPR. The identification of this factor remains elusive. It has recently been proposed that the house fly genome should be sequenced [27]. Such an effort would offer tremendous new insights into the biology of the house fly, including offering potential genes that could be investigated for their role in regulating CYP6D1 (and other P450s).
Highlights.
House fly (Musca domestica) CYP6D1 is a cytochrome P450 involved in metabolism of xenobiotics and resistance to pyrethroid insecticides.
Expression of CYP6D1 is inducible in response to the prototypical phenobarbital in insecticide susceptible, but not in insecticide susceptible strains.
Variation in promoter sequences of CYP6D1 between resistant ad susceptible strains does not explain the differential response to phenobarbital.
House fly HR96 cDNA was cloned and sequenced to examine if this trans acting factor is responsible for constitutive overexpression of CYP6D1v1 in LPR.
Multiple HR96 alleles (v1–v10) were identified, but neither a specific allele nor levels of expression were associated with resistance.
Acknowledgements
We thank Dr. John Lis (Molecular Biology and Genetics, Cornell University) for providing the Drosophila S2 cells and valuable advice. We thank Dr. Brian Lazzaro (Entomology, Cornell University) for providing valuable advice and Dr. Toshinori Kozaki for providing some of the constructs used. This work was supported by a Sarkaria Fellowship (to G.G.H.L.), a Griswold Fellowship (to G.G.H.L.), and the National Institutes of Health (GM47835).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wheelock GD, Scott JG. Anti-P450lpr antiserum inhibits specific monooxygenase activities in LPR house fly microsomes. J. Exp. Zool. 1992;264:153–158. doi: 10.1002/jez.1402640206. [DOI] [PubMed] [Google Scholar]
- 2.Hatano R, Scott JG. Anti-P450lpr antiserum inhibits the activation of chlorpyrifos to chlorpyrifos oxon in house fly microsomes. Pestic. Biochem. Physiol. 1993;45:228–233. [Google Scholar]
- 3.Korytko PJ, Quimby FW, Scott JG. Metabolism of phenanthrene by house fly CYP6D1 and dog liver cytochrome P450. J. Biochem. Molec. Toxicol. 2000;14:20–25. doi: 10.1002/(sici)1099-0461(2000)14:1<20::aid-jbt3>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 4.Zhang M, Scott JG. Cytochrome b5 is essential for cytochrome P450 6D1-mediated cypermethrin resistance in LPR house flies. Pestic. Biochem. Physiol. 1996;55:150–156. doi: 10.1006/pest.1996.0044. [DOI] [PubMed] [Google Scholar]
- 5.Wheelock GD, Scott JG. The role of cytochrome P450lpr in deltamethrin metabolism by pyrethroid resistant and susceptible strains of house flies. Pestic. Biochem. Physiol. 1992;43:67–77. [Google Scholar]
- 6.Tomita T, Scott JG. cDNA and deduced protein sequence of CYP6D1: the putative gene for a cytochrome P450 responsible for pyrethroid resistance in house fly. Insect Biochem. Molec. Biol. 1995;25:275–283. doi: 10.1016/0965-1748(94)00066-q. [DOI] [PubMed] [Google Scholar]
- 7.Liu N, Tomita T, Scott JG. Allele-specific PCR reveals that the cytochrome P450lpr gene is on chromosome 1 in the house fly, Musca domestica. Experientia. 1995;51:164–167. doi: 10.1007/BF01929363. [DOI] [PubMed] [Google Scholar]
- 8.Liu N, Scott JG. Phenobarbital induction of CYP6D1 is due to a trans acting factor on autosome 2 in house flies, Musca domestica. Insect Molec. Biol. 1997;6:77–81. doi: 10.1046/j.1365-2583.1997.00160.x. [DOI] [PubMed] [Google Scholar]
- 9.Scott JG, Sridhar P, Liu N. Adult specific expression and induction of cytochrome P450lpr in house flies. Arch. Insect Biochem. Physiol. 1996;31:313–323. doi: 10.1002/(SICI)1520-6327(1996)31:3<313::AID-ARCH6>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 10.Korytko PJ, Scott JG. CYP6D1 protects thoracic ganglia of house flies from the neurotoxic insecticide cypermethrin. Arch. Insect Biochem. Physiol. 1998;37:57–63. doi: 10.1002/(SICI)1520-6327(1998)37:1<57::AID-ARCH7>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 11.Scott JG, Lee SST. Tissue distribution of microsomal cytochrome P-450 monooxygenases and their inducibility by phenobarbital in the insecticide resistant LPR strain of house fly, Musca domestica L. Insect Biochem. Molec. Biol. 1993;23:729–738. doi: 10.1016/0965-1748(93)90047-v. [DOI] [PubMed] [Google Scholar]
- 12.Liu N, Scott JG. Increased transcription of CYP6D1 causes cytochrome P450-mediated insecticide resistance in house fly. Insect Biochem. Molec. Biol. 1998;28:531–535. doi: 10.1016/s0965-1748(98)00039-3. [DOI] [PubMed] [Google Scholar]
- 13.Liu N, Scott JG. Inheritance of CYP6D1-mediated pyrethroid resistance in house fly (Diptera: Muscidae) J. Econ. Entomol. 1997;90:1478–1481. doi: 10.1093/jee/90.6.1478. [DOI] [PubMed] [Google Scholar]
- 14.Tomita T, Liu N, Smith FF, Sridhar P, Scott JG. Molecular mechanisms involved in increased expression of a cytochrome P450 responsible for pyrethroid resistance in the housefly, Musca domestica. Insect Molec. Biol. 1995;4:135–140. doi: 10.1111/j.1365-2583.1995.tb00018.x. [DOI] [PubMed] [Google Scholar]
- 15.Scott JG, Liu N, Wen Z, Smith FF, Kasai S, Horak CE. House fly cytochrome P450 CYP6D1: 5 prime flanking sequences and comparison of alleles. Gene. 1999 226;:347–353. doi: 10.1016/s0378-1119(98)00545-9. [DOI] [PubMed] [Google Scholar]
- 16.Seifert J, Scott JG. The CYP6D1v1 allele is associated with pyrethroid resistance in the house fly, Musca domestica. Pestic. Biochem. Physiol. 2002;72:40–44. [Google Scholar]
- 17.Gao J, Scott JG. Use of quantitative real-time PCR to estimate the size of the house fly (Musca domestica) genome. Insect Molec. Biol. 2006;15:835–837. doi: 10.1111/j.1365-2583.2006.00690.x. [DOI] [PubMed] [Google Scholar]
- 18.Lin GG-H, Kozaki T, Scott JG. Hormone receptor-like in 96 and Broad-Complex modulate phenobarbital induced transcription of cytochrome P450 CYP6D1 in Drosophila S2 cells. Insect Mol. Biol. 2011;20:87–95. doi: 10.1111/j.1365-2583.2010.01047.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster GG, Whitten MJ, Konovalov C, Arnold JTA, Maffi G. Autosomal genetic maps of the Australian sheep blowfly, Lucilia cuprina dorsalis R.-D. (Diptera: Calliphoridae), and possible correlations with the linkage maps of Musca domestica L. and Drosophila melanogaster (Mg.) Genet. Res. Camb. 1981;37:55–69. [Google Scholar]
- 20.Weller GL, Foster GC. Genetic maps of the sheep blowfly Lucilia cuprina: linkage-group correlations with other dipteran genera. Genome. 1993;36:495–506. doi: 10.1139/g93-068. [DOI] [PubMed] [Google Scholar]
- 21.Hamm R, Shono T, Scott JG. A cline in frequency of autosomal males is not associated with insecticide resistance in house fly (Diptera: Muscidae) J. Econ. Entomol. 2005;98:171–176. doi: 10.1093/jee/98.1.171. [DOI] [PubMed] [Google Scholar]
- 22.Scott JG, Shono T, Georghiou GP. Genetic analysis of permethrin resistance in the house fly, Musca domestica L. Experientia. 1984;40:1416–1418. [Google Scholar]
- 23.Shono T, Scott JG. Spinosad resistance in the house fly, Musca domestica, is due to a recessive factor on autosome 1. Pestic. Biochem. Physiol. 2003;75:1–7. [Google Scholar]
- 24.Gao J-R, Kozaki T, Leichter CA, Rinkevich FD, Shono T, Scott JG. The A302S mutation in Rdl that confers resistance to cyclodienes and limited cross-resistance to fipronil is undectable in field populations of house flies from the USA. Pestic. Biochem. Physiol. 2007;88:66–70. [Google Scholar]
- 25.Hamm RL, Scott JG. A high frequency of male determining factors in male Musca domestica L. (Diptera: Muscidae) from Ipswich, Australia. J. Med. Entomol. 2009;46:169–172. doi: 10.1603/033.046.0121. [DOI] [PubMed] [Google Scholar]
- 26.Rose TM, Henikoff JG, Henikoff S. CODEHOP (COnsensus-DEgenerate Hybrid Oligonucleotide Primer) PCR primer design. Nucleic Acids Res. 2003;31:3763–3766. doi: 10.1093/nar/gkg524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scott JG, Liu N, Kristensen M, Clark AG. A case for sequencing the genome of the house fly, Musca domestica (Diptera: Muscidae) J. Med. Entomol. 2009;46:175–182. doi: 10.1603/033.046.0202. [DOI] [PubMed] [Google Scholar]