Abstract
We describe a transcriptional mechanism regulating the expression of Dnmt1 by nuclear receptors. We show that ERRγ functions as a transcriptional activator of mouse and human Dnmt1 expression by direct binding to its response elements (ERE1/ERE2) in the dnmt1/DNMT1 promoters. The induction of Dnmt1 by ERRγ is repressed by SHP through SHP inhibition of ERRγ transactivity, diminishing ERRγ recruitment to the Dnmt1 promoter, and altering the conformation of local chromatin from an active mode by ERRγ to an inactive mode. Our study provides the first evidence for nuclear receptor mediated regulation of Dnmt1 expression through ERRγ and SHP crosstalk.
Keywords: nuclear receptor, gene regulation, DNA methyltransferase
1. Introduction
DNA cytosine-5-methyltransferases (Dnmts) catalyze the methyl transfer from S-adenosyl-L-methionine to the cytosine in CpG dinucleotides [1]. Gene silencing due to DNA hypermethylation of promoter regions is mainly controlled by three Dnmts (Dnmt1, Dnmt3α and Dnmt3b) [2]. Increased Dnmt expression has been reported in several human cancers, including human hepatocellular carcinoma (HCC) [3]. Despite evidence for their critical roles in carcinogenesis, limited information is available regarding how the expression of Dnmts are transcriptionally regulated [4–7]. Understanding the molecular mechanisms controlling Dnmt expression is essential for learning how changes in DNA methylation result in or predispose to the development of liver cancer.
Nuclear receptor small heterodimer partner (SHP, NROB2) is a transcriptional regulator of a number of genes that play critical roles in hepatic lipid metabolism [8–12]. Our recent studies show that SHP functions as a tumor suppressor in HCC. Deletion of the SHP gene promotes spontaneous hepatoma formation in mice due to hepatocyte hyperproliferation [13] and decreased apoptosis [14], and SHP expression is diminished in HCC in humans due to promoter hypermethylation [15]. These studies provide strong evidence for a critical role of SHP in hepatocarcinogenesis.
In this report, we characterized a novel nuclear receptor-mediated mechanism regulating the transcription of Dnmt1. We show that SHP is a transcriptional repressor of Dnmt1 expression via crosstalk with estrogen related receptor gamma (ERRγ). This is the first report demonstrating that the expression of Dnmt1 is controlled by nuclear receptor signaling.
2. Materials and Methods
2.1. Cell lines and liver biospecimens
Human cervix adenocarcinoma cells (Hela; ATCC CCL-2), human hepatoma cells (Huh7; Health Science Research Resources Bank JCRB0403), and mouse liver cells (Nmuli; ATCC CRL-1638) were maintained as described previously [14,16]. Liver fragments from ERRγ+/+ and liver specific ERRγ−/− mice were obtained from Dr. Ronald Evans’s laboratory.
2.2. Antibodies and siRNAs
Antibodies used in immunoprecipitation, ChIP and immunoblot analyses include M-280 sheep anti-rabbit or mouse IgG Dynabeads (Invitrogen Dynal As), rabbit normal IgG (Sigma, R-2004) and antibodies against Flag (Sigma, F-7425), GFP (Sigma, G-1544), histone H3 acetyl antibody (H3Ac) (Millipore, #06-599), histone H4 acetyl antibody (H4Ac) (Millipore, #06-866), histone H3 dimethyl Lys4 antibody (H3K4Me2) (Millipore, #07-030), histone H3 dimethyl Lys9 antibody (H3K9Me2) (Millipore, #17-648), ERRγ (Sigma, AV-31655), and DNMT1 (Cell signaling, #5032). ERRγ RNAi (PDSIRNA SASI-Hs01-00042292, SASI-Hs01-00042292-AS) and non-specific RNAi (SIC001) were purchased from Sigma. Co-immunoprecipitation (Co-IP) and Western blots (WB) were performed as described previously [14,16].
2.3. Real-time qPCR analysis
The method are provided in our recent publications [14,16,17]. HPRT1 was used as internal control for Dnmt1 mRNA expression. The primer sequences are: forward 5′-CCCCTGAGCCCTACCGAAT-3′ and reverse 5′-CTCGCTGGAGTGGACTTGTG-3′ for DNMT1; forward 5′-CCGGTCTCTTTCGTTTGAGG-3′ and reverse 5′-CATCCTGAAGCTTCTGAACGG-3′ for hERRγ; forward 5′-TGACACTGGCAAAACAATGCA-3′ and reverse 5′-GGTCCTTTTCACCAGCAAGCT-3′ for hHRPT1; forward 5′-CCCAGGGCTTTCCAGATAGC-3′ and reverse 5′-AACTGCAGCTGATGCGCTC-3′ for dnmt1; forward 5′-CACCTGCATCTCACAGCCACT-3′ and reverse 5′-GCCAACCCAAGCAGGAAGA-3′ for mSHP; and forward 5′ - CGTCGTGATTAGCGATGATGA-3′ and reverse 5′-CACACAGAGGGCCACAATGT-3′ for mHPRT1.
2.4. Transient transfection and promoter activity assays
Detailed methods can be found in our recent publications [14,16,18–21]. The mouse dnmt1 (Gene ID: 13433) promoter and its deletion mutation luciferase constructs (mdnmt1 Luc) were engineered in our laboratory. All plasmids were described previously [16].
2.5. ChIP and ChIP-ReChIP assays
Chromatin immunoprecipitation (ChIP) assays were done as described previously [14,16]. To analyze the reciprocal association between ERRγ and SHP immunoprecipitates, first ChIP was performed with antibody against Flag, washed, and the bound immune-DNA complexes were released in 50 μl of elution buffer (1X TE containing 2% SDS, 15 mM DTT and protease inhibitors) for 30 min at 37°C and resuspended in 30 times volume of IP dilution buffer (Tris pH 8.0 50 mM, EDTA 5 mM, NaCl 200 mM, NP40 0.5%) for ReChIP with antibody to GFP. Real-time qPCR was performed using ChIP primer sets specific for the ERRγ binding sites in the dnmt1 and DNMT1 promoters: mouse p1 forward 5′-CTGGCTTTCTCTGTGTGGCTC-3′ and reverse 5′-CATGCCTGCAGTCCCTTACAC-3′, mouse p2 forward 5′-CAAGACCAGAGAGAGGCAGGA-3′ and reverse 5′-CCACATCCAGCCACTCGAA-3′; human p1 forward 5′-TGAGCCGTAGTGACCCACCTA-3′ and reverse 5′-GGGATAAAGCAGCGAGAAGCC-3′, human p2 forward 5′-GTTTGGACTATGGGCACATGC-3′ and reverse 5′-CGCTTGAGCCCAGGAGTTT-3′. A control region from −1489 to −1310 in dnmt1 promoter (forward 5′-CTGAGCCATCTCTCCATCC-3′ and reverse 5′-CCGAGGACCAGAAAGACAGC-3′) or −1364 to −1260 in DNMT1 promoter (forward 5′-TTCTGGCTGGTTACGGTGG-3′ and reverse 5′-CATGTTGGCCAGGCTAGTCTG-3′) serves as negative control.
2.6. Statistical analysis
Data are expressed as mean ± SEM. Statistical analyses were carried out using Student’s unpaired t test; p < 0.01 was considered statistically significant.
3. Results
SHP inhibits ERRγ transactivation of the dnmt1 promoter
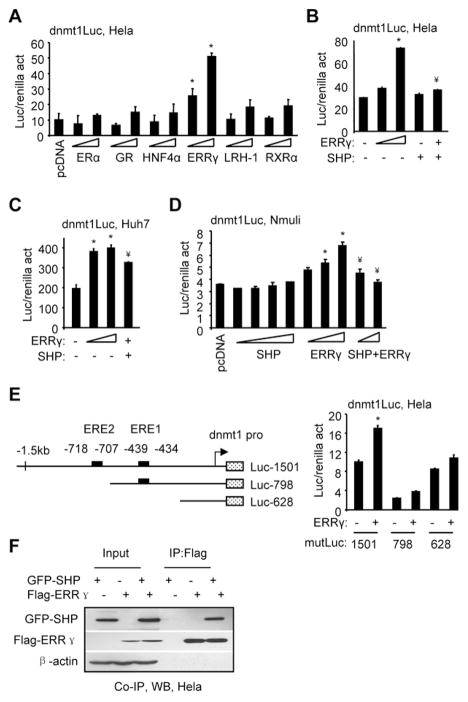
Putative binding elements for nuclear receptors estrogen receptor alpha (ERα), glucocorticoid receptor (GR), hepatocyte nuclear factor 4α (HNF4α), ERRγ, liver receptor homologue 1 (LRH-1), and retinoid X receptor alpha (RXRα) were predicted in the mouse dnmt1 promoter. We cloned a 1.5 kb proximal promoter of dnmt1 into a luciferase (luc) reporter. ERRγ, but not ERα, GR, HNF4α, LRH-1, and RXRα, markedly activated the dnmt1 promoter dose-dependently (Fig. 1A). SHP was previously shown to interact with and inhibit ERRγ transactivation [10,22]. As expected, co-expression of SHP sufficiently abrogated the activity of ERRγ (Fig. 1B). Similar but less striking results were observed in human Huh7 (Fig. 1C) and in mouse Nmuli (Fig. 1D) cells. Two putative ERE sites were located in the dnmt1 promoter (Fig. 1E, left). We generated two mutant (mut) dnmt1 luc constructs (mutLuc 798 and 628). Although the basal luciferase activity of each mutant promoter was different, single deletion of ERE2 (798) or double deletion of ERE2/ERE1 (628) markedly attenuated dnmt1 promoter activation by ERRγ (Fig. 1E, right). The data suggest that both ERE sites may be functional for ERRγ binding. We also confirmed the physical association of SHP with the ERRγ protein in Hela cells using Co-IP and WB (Fig. 1F).
Fig. 1.
SHP inhibition of dnmt1 promoter activity by ERRγ. (A) Transient transfection assays to determine dnmt1 promoter luciferase (luc) activity (act) by expression of ERα (50, 200 ng), GR (50, 200 ng), HNF4α (50, 200 ng), ERRγ (50, 200 ng), LRH-1 (50, 200 ng), or RXRα (50, 200 ng) in Hela cells. (B–D) Transient transfection assays to determine dnmt1 promoter luciferase activity by co-expression of ERRγ and SHP in Hela (B) (ERRγ: 50, 200 ng; SHP, 200 ng; ERRγ+SHP, 200 ng+200 ng), Huh7 (C) (ERRγ: 100, 400 ng; ERRγ+SHP: 400 ng+100 ng), and Nmuli cells (D) (SHP: 100, 200, 400, 800 ng; ERRγ: 200, 400, 800 ng; ERRγ+SHP: 800 ng+200 ng, 800 ng+400 ng). (E) Left: Diagram showing the location of EREs in the dnmt1 promoter and its deletion constructs. Right: Transient transfection assays to determine dnmt1 promoter lucifease activity by ERRγ (100 ng) in Hela cells. A–E: The luciferase activities were normalized by renilla activities. Data is represented as mean ± SEM (*p<0.01 vs. control; ¥p<0.01 vs. ERRγ alone). The experiments were repeated three times (triplicate wells/time) with similar results. One representative result is shown. (F) Immunoprecipitation (IP) and Western blots (WB) to determine the direct association of SHP with the ERRγ protein. Hela cells were transfected with Flag-ERRγ (1 μg, 3 cm plate) and/or GFP-SHP (1 μg, 3 cm plate) expression vectors. Anti-Flag antibodies were used to immunoprecipitate ERRγ, and the protein levels of SHP and ERRγ were detected by WB using anti-GFP or anti-Flag antibodies, respectively.
SHP decreases ERRγ recruitment to the Dnmt1 promoter
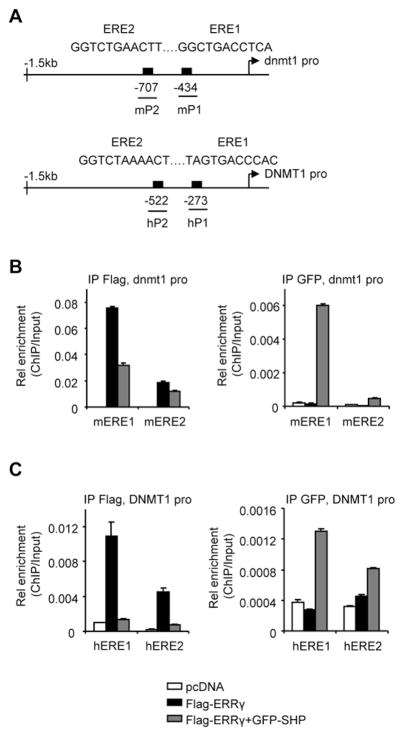
Putative ERE sites were also located in the human DNMT1 promoter (Fig. 2A). ChIP analysis revealed that ERRγ had an approximately 4 fold higher affinity with mERE1 than with mERE2 in the dnmt1 promoter, which was markedly diminished by SHP co-expression (Fig. 2B, left). The association of SHP with mERE1 was stronger as compared to mERE2 (Fig. 2B, right). The physical association of ERRγ and SHP with the endogenous DNMT1 promoter was examined. Similar to what was observed with the dnmt1 promoter, ERRγ bound preferentially to hERE1 than to hERE2, which was abrogated by SHP (Fig. 2C, left). In addition, SHP exhibited favorable binding to hERE1 than to hERE2 (Fig. 2C, right) in the DNMT1 promoter.
Fig. 2.
SHP inhibition of ERRγ recruitment to the Dnmt1 promoter. (A) Schematic showing the putative ERRγ binding sites (EREs) in the dnmt1 and DNMT1 promoters. (B–C) ChIP assays to determine ERRγ and SHP co-immunoprecipitation (Co-IP) to the ERE region of the dnmt1 (B) or DNMT1 (C) promoter. ERRγ and SHP were exogenously expressed in Hela cells with Flag-ERRγ (5 μg, 10 cm plate) and/or GFP-SHP (5 μg, 10 cm plate) plasmids, along with the dnmt1 promoter. The chromatin was co-immunoprecipitated using anti-Flag or anti-GFP antibodies. Real-time qPCR was used to determine the association of ERRγ and SHP to the exogenous dnmt1 promoter using mP1 and mP2 primers (B), and their association to the endogenous DNMT1 promoter was determined by hP1 and hP2 primers (C). PCR products were amplified from input-positive controls, IgG-negative controls, and antibody immunoprecipitates. Histograms show antibody/input ratios of PCR products quantified using qPCR, expressed as Relative (Rel) enrichment. Error bars represent SEM from three independent measurements.
We performed primary ChIP followed by a secondary immunoprecipitation (ReChIP) to determine whether ERRγ and SHP were enriched on similar ERE region of the dnmt1/DNMT1 promoter or whether the two receptors were recruited independently of each other. Consistent with the results in Fig. 2, ChIP analysis revealed a stronger binding of ERRγ to mERE1/hERE1 than to mERE2/hERE2 by both PCR (Fig. 3A, 3C, 1° ChIP vs. Input) and q-PCR (Fig. 3B, 3D, left). The ReChIP experiments showed binding of SHP on ERRγ immunoprecipitates (Fig. 3A, 3C, 2° ReChIP; Fig. 3B, 3D, right).
Fig. 3.
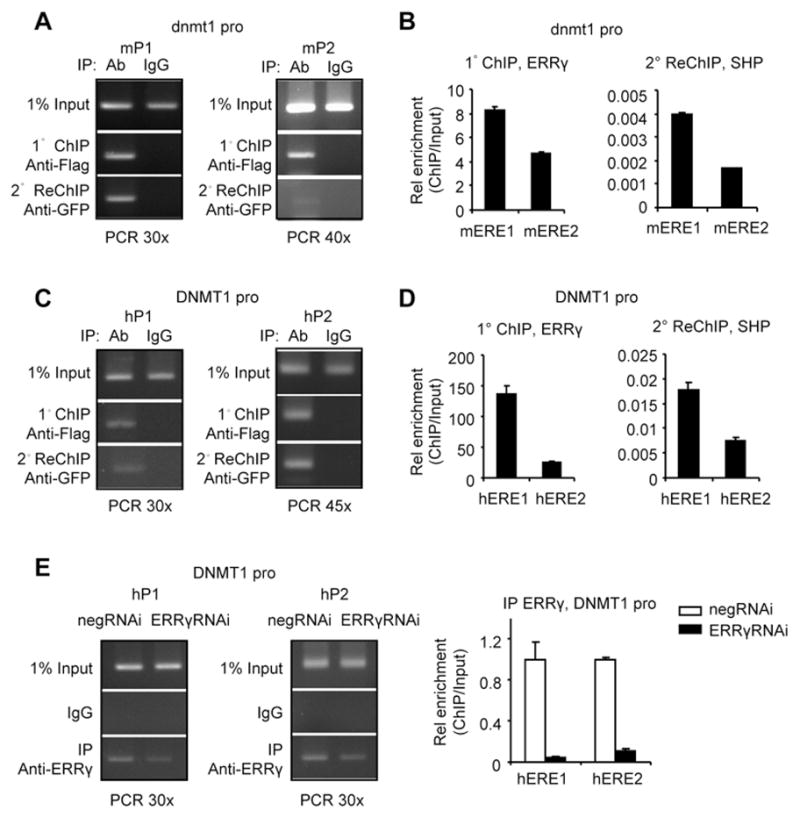
Co-recruitment of ERRγ and SHP to the Dnmt1 promoter. (A–D) ChIP-ReChIP analysis by PCR (A, C) or qPCR (B, D) to illustrate the enrichment of ERRγ and SHP to the dnmt1 (A–B) or DNMT1 (C–D) promoter. Soluble chromatin was prepared and immunoprecipitated (1° ChIP) with anti-Flag antibodies. Immunocomplexes were collected and eluted, and soluble chromatin fraction was reimmunoprecipitated (2° ReChIP) with anti-GFP antibodies. Other procedures were done as described in Fig. 2. (E) ChIP assays by PCR (left and middle) and q-PCR (right) to determine ERRγ recruitment to the endogenous DNMT1 promoter in Hela cells with ERRγ knockdown using siRNA (ERRγRNAi, 100 pmol). ChIP analysis was done as described in Fig. 2.
To further validate the finding that ERRγ binding was specific to the DNMT1 promoter, ERRγ-siRNA was used to knockdown the endogenous ERRγ in Hela cells. The enrichment of ERRγ to hERE1 and hERE2 was markedly reduced by ERRγRNAi (Fig. 3E).
SHP reverses active histone modifications of the dnmt1 promoter by ERRγ
Gene expression could be regulated by epigenetic modification and more particularly by posttranslational modification of histones [23]. We examined histone modifications in the dnmt1 promoter by ERRγ and SHP using antibodies specific to transcriptionally active (H3Ac, H4Ac, and H3K4Me2) and inactive (H3K9Me2) histone marks [24]. We observed a marked increase in dnmt1 promoter occupancy by ERRγ, as indicated by the increased levels of H3Ac and H3K4Me2 and decreased levels of H3K9Me2 (Fig. 4). ERRγ did not alter H4Ac occupancy in the dnmt1 promoter. On the contrary, SHP co-expression reversed the effect of ERRγ by decreasing H3K4Me2 and increasing H3K9Me2. SHP also markedly decreased H3Ac and H4Ac occupancy in the dnmt1 promoter. The results suggest that ERRγ results in transcriptionally active configuration of the dnmt1 promoter which is inhibited by SHP.
Fig. 4.
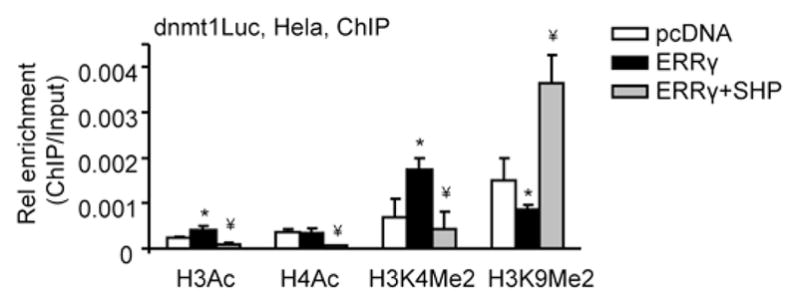
Alteration of histone modifications of the dnmt1 promoter by ERRγ and SHP. The chromatin was co-immunoprecipitated using anti-H3Ac, anti-H4Ac, anti-H3K4Me2 or anti-H3K9Me2 antibodies. ChIP analysis was done as described in Fig. 2. *p<0.01 vs. pcDNA control; ¥p<0.01 vs. ERRγ group.
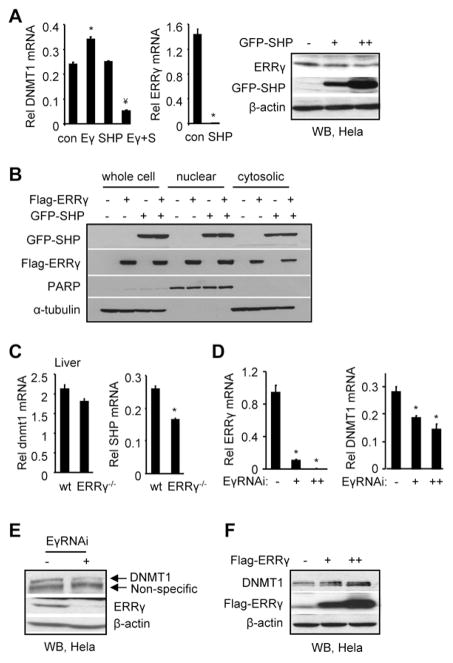
SHP inhibits ERRγ induction of Dnmt1 expression
The endogenous DNMT1 mRNA was induced by ERRγ which was down-regulated by SHP (Fig. 5A, left). Interestingly, SHP also decreased the endogenous ERRγ mRNA (Fig. 5A, middle) to undetectable levels, suggesting that SHP may counteract the effect of ERRγ by repressing ERRγ gene expression, in addition to interacting with ERRγ to repress the transactivation of its target gene [22]. ERRγ protein was not completely repressed by SHP (Fig. 5A, right) compared to its mRNA, suggesting that a post-translational mechanism may be involved in regulating ERRγ protein stability. The levels of exogenously expressed ERRγ protein were not inhibited by SHP co-expression (Fig. 5B), confirming that the SHP inhibition of ERRγ activity in Fig. 1 represents the effect of a direct protein-protein interaction between ERRγ and SHP. The mRNA of dnmt1 was moderately but not significantly decreased in liver of ERRγ−/− mice (Fig. 5C, left). SHP can be constitutively activated by ERRγ [22] and SHP mRNA was markedly decreased in ERRγ−/− mice (Fig. 5C, right). The reduced SHP expression may result in a derepression of Dnmt1 therefore mask the lost activation of ERRγ on dnmt1. ERRγ knockdown using siRNA reduced the levels of DNMT1 mRNA (Fig. 5D) and protein (Fig. 5E), whereas ERRγ overexpression increased DNMT1 protein (Fig. 5F). These results further suggest a crosstalk between ERRγ and SHP in regulating the expression of dnmt1 and DNMT1.
Fig. 5.
SHP inhibition of Dnmt1 expression through ERRγ. (A) qPCR analysis of DNMT1 (left) and ERRγ (middle) mRNA in Hela cells that were overexpressed with ERRγ (3 μg, 6 cm plate) and/or SHP (3 μg, 6 cm plate) expression vectors. Right: WB analysis of ERRγ and GFP-SHP protein in Hela cells that were overexpressed with GFP-SHP plasmids (0, 0.5, 1 μg, 12 well plate). Anti-ERRγ and anti-GFP antibodies were used, respectively. (B) WB analysis of ERRγ and SHP protein in Hela cells that were overexpressed with Flag-ERRγ and GFP-SHP plasmids. The protein levels of ERRγ and SHP in the whole cell lysate, nuclear fraction and cytosolic fraction were detected using anti-Flag and anti-GFP antibodies, respectively. PARP was used as a marker for nuclear proteins and α-tubulin was used as a marker for whole cell and cytosolic proteins. (C) qPCR analysis of hepatic dnmt1 and SHP mRNA in liver specific ERRγ-knockout (ERRγ−/−) mice. (D) qPCR analysis of ERRγ and DNMT1 mRNA in Hela cells with ERRγ knockdown using siRNA (−, non-specific siRNA control; +, 100 pmol ERRγRNAi; ++, 200 pmol ERRγRNAi). A, C, D: Data are represented as mean ± SEM of three independent assays (n=three). *p<0.01 vs. control; ¥p<0.01 vs. ERRγ alone. Rel, relative. (E) WB analysis of DNMT1 and ERRγ protein in Hela cells with ERRγ knockdown using siRNA (−, non-specific siRNA control; +, 100 pmol ERRγRNAi). Anti- DNMT1 and anti-ERRγ antibodies were used, respectively. (F) WB analysis of DNMT1 and Flag-ERRγ protein in Hela cells with Flag-ERRγ overexpression (0, 1, 4 μg, 6 cm plate). Anti-DNMT1 and anti-Flag antibodies were used, respectively. A weak non-specific band was seen in the first lane.
4. Discussion
Previous work has shown that the dnmt1 promoter of mouse is independently activated by Sp1/Sp3 [4] and E2F [5,25] transcription factors, and that human DNMT1 is activated by STAT3 [7]. A recent study demonstrated that p53 is a negative regulator of Dnmt1 promoter activity [26]. However, no information is available whether Dnmt1 gene transcription is regulated by nuclear receptors.
In this study, we identified nuclear receptor SHP as a transcriptional repressor of both mouse dnmt1 and human DNMT1 expression by inhibiting the activity of ERRγ. Such regulation was observed in several cancer cell lines including Hela and Huh7 cells, but was stronger in Hela cells, suggesting that this regulatory mechanism may play an important role in controlling DNMT1 function in cervical cancer. However, it should be noted that the expression of Dnmt1 was only partially decreased by ERRγ knockdown, indicating that in addition to the ERRγ/SHP pathway, other mechanisms may exist to control the expression of Dnmt1.
Another observation was that the ERRγ protein was moderately repressed by SHP, which is distinct from the complete repression of its mRNA by SHP. We also observed that SHP overexpression itself did not inhibit DNMT1 mRNA and protein (not shown) in Hela cells. This could be in part attributed to the moderate inhibitory effect of SHP on the ERRγ protein. Nevertheless, our results provide strong evidence for a novel role of SHP in repressing gene transcription of Dnmt1 via ERRγ.
Importantly, the co-recruitment of SHP with ERRγ to the dnmt1/DNMT1 promoters significantly decreased the enrichment of ERRγ. In addition, binding of ERRγ to the dnmt1 promoter induces transcriptionally active configuration of the promoter, which is reversed by co-expression of SHP. This suggests that the major mechanism for SHP inhibition of dnmt1 promoter activity is through alteration of local chromatin structure by ERRγ. SHP-mediated repression is not limited to the action of ERRγ, because SHP also decreases H4Ac occupancy which is not significantly affected by ERRγ. The results suggest that SHP may have a general function to keep the local chromatin configuration of its target genes in a repressive mode, which is consistent with its role as a universal transcriptional repressor [27].
Over sixty chromatin modifying enzymes have been identified [28,29], and many of them are associated with post-translational modification of H3Ac, H4Ac, H3K4Me2, and H3K9Me2. It remains to be determined in future studies whether SHP and ERRγ are co-recruited to a multiprotein complex including proteins governing the post-translational modification of histones, and how this would regulate the gene transcription of Dnmt1.
Overall, our findings are important because they link SHP function to DNA methylation at the molecular levels. Considering the importance of Dnmt1 in DNA methylation associated silencing of tumor suppressors [2], and the critical role of SHP in liver cancer progression [13–15], the identification of the regulatory role of SHP in controlling Dnmt1 expression improves our understanding of the epigenetic mechanisms governing the development of liver cancer.
Acknowledgments
We thank Dr. Curt Hagedorn for reading the manuscript. Y.Z. is supported by NIH T32CA092347 Multidisciplinary Cancer Research Training Program (MCRTP). This work is in part supported by National Institutes of Health grant DK080440 to L.W.
Footnotes
No conflicts of interest exist for all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Turek-Plewa J, Jagodzinski PP. The role of mammalian DNA methyltransferases in the regulation of gene expression. Cell Mol Biol Lett. 2005;10:631–47. [PubMed] [Google Scholar]
- 2.Park HJ, Yu E, Shim YH. DNA methyltransferase expression and DNA hypermethylation in human hepatocellular carcinoma. Cancer Lett. 2006;233:271–8. doi: 10.1016/j.canlet.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 3.Rountree MR, Bachman KE, Herman JG, Baylin SB. DNA methylation, chromatin inheritance, and cancer. Oncogene. 2001;20:3156–65. doi: 10.1038/sj.onc.1204339. [DOI] [PubMed] [Google Scholar]
- 4.Kishikawa S, Murata T, Kimura H, Shiota K, Yokoyama KK. Regulation of transcription of the Dnmt1 gene by Sp1 and Sp3 zinc finger proteins. Eur J Biochem. 2002;269:2961–70. doi: 10.1046/j.1432-1033.2002.02972.x. [DOI] [PubMed] [Google Scholar]
- 5.Kimura H, Nakamura T, Ogawa T, Tanaka S, Shiota K. Transcription of mouse DNA methyltransferase 1 (Dnmt1) is regulated by both E2F-Rb-HDAC-dependent and -independent pathways. Nucleic Acids Res. 2003;31:3101–13. doi: 10.1093/nar/gkg406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jinawath A, Miyake S, Yanagisawa Y, Akiyama Y, Yuasa Y. Transcriptional regulation of the human DNA methyltransferase 3A and 3B genes by Sp3 and Sp1 zinc finger proteins. Biochem J. 2005;385:557–64. doi: 10.1042/BJ20040684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Q, Wang HY, Woetmann A, Raghunath PN, Odum N, Wasik MA. STAT3 induces transcription of the DNA methyltransferase 1 gene (DNMT1) in malignant T lymphocytes. Blood. 2006;108:1058–64. doi: 10.1182/blood-2005-08-007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Han Y, Kim CS, Lee YK, Moore DD. Resistance of SHP-null mice to bile acid-induced liver damage. J Biol Chem. 2003;278:44475–81. doi: 10.1074/jbc.M305258200. [DOI] [PubMed] [Google Scholar]
- 9.Wang L, et al. Redundant pathways for negative feedback regulation of bile acid production. Dev Cell. 2002;2:721–31. doi: 10.1016/s1534-5807(02)00187-9. [DOI] [PubMed] [Google Scholar]
- 10.Wang L, Liu J, Saha P, Huang J, Chan L, Spiegelman B, Moore DD. The orphan nuclear receptor SHP regulates PGC-1alpha expression and energy production in brown adipocytes. Cell Metab. 2005;2:227–38. doi: 10.1016/j.cmet.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 11.Huang J, Iqbal J, Saha PK, Liu J, Chan L, Hussain MM, Moore DD, Wang L. Molecular characterization of the role of orphan receptor small heterodimer partner in development of fatty liver. Hepatology. 2007;46:147–57. doi: 10.1002/hep.21632. [DOI] [PubMed] [Google Scholar]
- 12.Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am J Physiol Gastrointest Liver Physiol. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y, Xu P, Park K, Choi Y, Moore DD, Wang L. Orphan receptor small heterodimer partner suppresses tumorigenesis by modulating cyclin D1 expression and cellular proliferation. Hepatology. 2008;48:289–98. doi: 10.1002/hep.22342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y, Soto J, Park K, Viswanath G, Kuwada S, Abel ED, Wang L. Nuclear receptor SHP, a death receptor that targets mitochondria, induces apoptosis and inhibits tumor growth. Mol Cell Biol. 30:1341–56. doi: 10.1128/MCB.01076-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He N, Park K, Zhang Y, Huang J, Lu S, Wang L. Epigenetic inhibition of nuclear receptor small heterodimer partner is associated with and regulates hepatocellular carcinoma growth. Gastroenterology. 2008;134:793–802. doi: 10.1053/j.gastro.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Zhou T, et al. Novel polymorphisms of nuclear receptor SHP associated with functional and structural changes. J Biol Chem. 285:24871–81. doi: 10.1074/jbc.M110.133280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weaver BP, et al. Zip4 (Slc39a4) Expression is Activated in Hepatocellular Carcinomas and Functions to Repress Apoptosis, Enhance Cell Cycle and Increase Migration. PLoS One. 5 doi: 10.1371/journal.pone.0013158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song G, Wang L. Nuclear receptor SHP activates miR-206 expression via a cascade dual inhibitory mechanism. PLoS One. 2009;4:e6880. doi: 10.1371/journal.pone.0006880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Song G, Wang L. Transcriptional mechanism for the paired miR-433 and miR-127 genes by nuclear receptors SHP and ERRgamma. Nucleic Acids Res. 2008;36:5727–35. doi: 10.1093/nar/gkn567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Song G, Wang L. A conserved gene structure and expression regulation of miR-433 and miR-127 in mammals. PLoS One. 2009;4:e7829. doi: 10.1371/journal.pone.0007829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song G, Zhang Y, Wang L. MicroRNA-206 targets notch3, activates apoptosis, and inhibits tumor cell migration and focus formation. J Biol Chem. 2009;284:31921–7. doi: 10.1074/jbc.M109.046862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanyal S, Kim JY, Kim HJ, Takeda J, Lee YK, Moore DD, Choi HS. Differential regulation of the orphan nuclear receptor small heterodimer partner (SHP) gene promoter by orphan nuclear receptor ERR isoforms. J Biol Chem. 2002;277:1739–48. doi: 10.1074/jbc.M106140200. [DOI] [PubMed] [Google Scholar]
- 23.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 24.Brunmeir R, et al. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 6:e1000927. doi: 10.1371/journal.pgen.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McCabe MT, Low JA, Imperiale MJ, Day ML. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene. 2006;25:2727–35. doi: 10.1038/sj.onc.1209266. [DOI] [PubMed] [Google Scholar]
- 26.Lin RK, et al. Dysregulation of p53/Sp1 control leads to DNA methyltransferase-1 overexpression in lung cancer. Cancer Res. 70:5807–17. doi: 10.1158/0008-5472.CAN-09-4161. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Hagedorn CH, Wang L. Role of nuclear receptor SHP in metabolism and cancer. Biochim Biophys Acta. doi: 10.1016/j.bbadis.2010.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouzarides T. SnapShot: Histone-modifying enzymes. Cell. 2007;131:822. doi: 10.1016/j.cell.2007.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Allis CD, et al. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]