Abstract
Background: Nearly 30% of people in the United States are colonized with Staphylococcus aureus and 1% to 2% with methicillin-resistant Staphylococcus aureus (MRSA) in the anterior nares. However, it is not known if lytic bacteriophages against S. aureus are present in the anterior nares, and if they are, what the prevalence rate is, or if they interfere with S. aureus colonization. The aim of this study was to determine the prevalence of nasal carriage of S. aureus and MRSA and to screen for S. aureus lytic bacteriophages in healthcare environment workers and ambulatory patients.
Methods: We enrolled 202 individuals into this study. The anterior nares were swabbed to isolate S. aureus, MRSA, and any lytic S. aureus bacteriophages that may be present. Putative S. aureus colonies on blood agar plates were identified using gram stain, catalase and coagulase tests, and confirmed by S. aureus-specific PCR. Presence of staphylococcal lytic bacteriophages were screened by a plaque assay technique using a methicillin-sensitive reference S. aureus strain ATCC 29213.
Results: Of the 49 (24%) individuals who screened positive for S. aureus, two (1%) were positive for MRSA. None of the samples were positive for lytic bacteriophages against S. aureus. Subgroup analysis found no significant difference in the prevalence of S. aureus in the house staff compared to other healthcare environment workers or ambulatory patients of the clinic. Similarly, no significant difference in colonization was noted across the population with respect to age, sex, body mass index, or presence of diabetes mellitus.
Conclusion: The prevalence of nasal carriage of S. aureus and MRSA in the studied population was 24% and 1%, respectively, similar to the rate of prevalence in the United States. The study also showed that the anterior nares do not seem to harbor lytic bacteriophages against S. aureus.
Keywords: Ambulatory patients, Anterior nares, Healthcare environment workers, Lytic bacteriophages, Staphylococcus aureus
Staphylococcus aureus, frequently part of the normal flora of the anterior nares, is an important opportunistic pathogen capable of causing a number of diseases ranging from skin and soft tissue infections to bacteremia, pneumonia, endocarditis, and sepsis.1 The prevalence rates of S. aureus and methicillin-resistant S. aureus (MRSA) in the anterior nares in the United States population are 28.6% and 1.5%, respectively.2 People colonized with S. aureus have a higher rate of S. aureus infections than people who are not colonized.1,3 Elimination of S. aureus carriage in the anterior nares by mupirocin has been shown to reduce the incidence of post-operative S. aureus infections.4 Unfortunately, resistance to mupirocin has been reported in several studies.5,6 Therefore, bacteriophages with lytic activity against S. aureus have been proposed as an additional tool to treat nasal carriage.7,8
The ecological factors leading to colonization of the anterior nares by S. aureus and MRSA are little-understood. Lina et al9 showed that the S. aureus colonization rate in subjects colonized by Corynebacterium spp. and/or non-aureus staphylococci, especially S. epidermidis, was significantly lower than in subjects not colonized by these species. Sivaraman et al10 and Peacock et al11 reviewed several factors both from the host and bacterium that were involved in the variability of nasal colonization by S. aureus. Some of the host factors included certain HLA types, and polymorphisms in glucocorticoid receptors and the Fc fragment of IgG. Bacterial factors included the roles played by proteins made by sdrE and clfB genes among others.10 A study by Nouwen et al12 showed that the majority of noncarriers and nearly all persistent carriers returned to their original carrier state after artificial inoculation, with a majority of persistent carriers testing positive again for their original resident strain. Epidemiologic studies,13,14 have also reported a negative association between S. pneumoniae nasopharyngeal colonization and S. aureus nasal colonization. In a matched case-control study, Regev-Yochay et al15 reported that the odds of co-colonization with S. aureus were significantly lower for individuals carrying a piliated versus nonpiliated S. pneumoniae strain. Wos-Oxley et al16 and Costello et al17 both suggest that the anterior nares represent a complex ecological niche in which the species interaction may well determine the colonization and prevalence of S. aureus and MRSA.
Bacteriophages are ubiquitous and the most abundant living entities on earth (oceans, soil, deep vents, etc.), with estimates ranging from 1030 to 1032 in total, and they play key roles in regulating the microbial balance in every ecosystem where this has been explored.18 Lytic bacteriophages against different bacterial species have been isolated and even suggested to be part of the complex microbiome of the human gut,19,20 oral cavity,21,22 and vagina.23 The lytic phages are likely to modulate the rate of prevalence for one or more bacteria in different microbiological niches in the body. While lytic bacteriophages against S. aureus and MRSA have been isolated from different sources such as milk,24 sewage influent,25 cow slurry,26 and septic wounds,27 it is not known if they are prevalent in the anterior nares.
The goals of this study were to determine (1) the nasal presence of S. aureus and MRSA in ambulatory patients from the general population and healthcare environment (HCE) workers, including the medical house staff, of a Midwestern rural multispecialty clinic, and (2) to determine the presence of lytic bacteriophages against S. aureus in their anterior nares.
Methods
Study Design
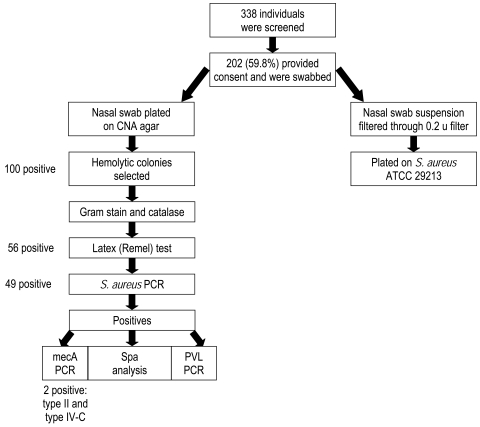
A study of nasal colonization of human subjects by S. aureus was carried out following review and approval by the Institutional Review Board. A flow chart of the overall study is depicted in figure 1▶. From October through November 2008, we screened for potential study participants from the ambulatory patients and HCE workers, including medical house staff, of a Midwestern rural multispecialty clinic. Participants were screened by trained research coordinators. The ambulatory patients group consisted of people that came to seek clinical care at the departments of Internal Medicine, Pediatrics, Internal Medicine-Pediatrics (Med-Peds), and Family Practice. The HCE group consisted of workers at the multispecialty clinic and was not restricted to those having patient contact only. The medical house staff of the clinic included 62 MD or PhD graduate learners in six residency programs (general surgery, internal medicine, pediatrics, med-peds combined residency, dermatology, and transitional year), three fellowship programs (general internal medicine, non-operative spine, and palliative care), one post-doctoral psychology fellowship program, and one pharmacy residency program.
Figure 1.
Study flow chart.
A total of 338 subjects were approached, of which 202 (60%) gave written informed consent to participate in the study. No participant received any remuneration for study participation. The HCE group enrolled 111 participants (including 50 medical house staff), and the ambulatory group recruited and enrolled 91 participants using the following inclusion and exclusion criteria: adult males or females, ages 18 to 100 years old, were included in the study; exclusion criteria included being currently on mupirocin or other nasal medications, having a 2-week history of nose bleed, signs of rhinitis, and/ or using antibiotics at the time of enrollment (table 1▶).
Table 1.
Screened population and exclusion criteria distribution.
| Criteria | n |
|---|---|
| Excluded | 136 (40.2%) |
| Activated POA/<18 or non-English speaking | 10 (3.0%) |
| Refused | 17 (5.0%) |
| Nosebleeds | 4 (1.2%) |
| Nasal spray | 31 (9.2%) |
| Missed/no shows | 25 (7.4%) |
| Signs of rhinitis | 15 (4.4%) |
| Antibiotics | 34 (10.1%) |
| Included (Consented) | 202 (59.8%) |
POA, Power of Attorney
Statistical Basis for Sample Size
Sample size calculations were based on the following: the sample size (n=202) was chosen to have a reasonable likelihood of detecting lytic S. aureus bacteriophages. The reported incidence rate of S. aureus in the anterior nares is 28% to 32%,2,28 and reported prevalence of lytic bacteriophages in certain niches on the human body ranges from 3% to 33%.19–21 With a 30% S. aureus carriage rate and a phage prevalence range of 3% to 33%, we estimated that the likelihood of detecting at least one phage in 202 samples would be 1 – (1 – .3 × .03)202 = 84% (lower estimate), and 1 – (1 – .3 × .3)202 = 99% (upper estimate).
Swabbing the Anterior Nares
Samples were collected from each subject with two dry, un-moistened swabs (Copan Diagnostics, Corona, CA). The swabbing was done by research coordinators trained in the proper technique. The swabs were transported to the research laboratory and immediately stored at 4°C. Most samples were processed within 2 to 16 hours of collection.
Identification of S. aureus and MRSA
One of the nasal swabs was plated on CNA agar (Columbia CNA w/5% Sheep Blood w/Colistin, Nalidixic Acid) and incubated at 37°C for 24 hours. Beta-hemolytic colonies from the CNA plates were subcultured on blood agar plates (BAP) and incubated at 37°C. After 24 hours, any beta-hemolytic colonies on the BAP were screened for S. aureus using gram stain, catalase and coagulase tests (using the Staphaurex Plus latex agglutination test [Remel]). All tentative S. aureus thus identified were confirmed by S. aureus-specific 16S rRNA gene PCR.29 Confirmation of methicillin resistance was by mecA PCR, and the presence of Panton-Valentine leukocidin (PVL) toxin genes by lukSF-PV PCR.30,31 All S. aureus isolates were then genotyped by spa typing32 and multi-locus sequence typing.33
Lytic Bacteriophage Assay
The assay method was modified from Sambrook and Russell.34 Briefly, a nasal swab was suspended into a 5 ml tube containing 3 ml of trypticase soy broth (TSB), vortexed, and incubated at room temperature for 20 minutes. The supernatant was filtered through a 0.22 μm filter into microcentrifuge tubes to remove the bacterial cells and then stored at 4°C. Simultaneously, TSA (tryptic soy agar) base plates were prepared by pouring 15 mls of molten TSA (Trypticase Soy broth with 1.5% bacto-agar and calcium chloride to a final concentration of 400 μg/ml) into sterile petri plates. Two colonies of host cell (methicillin-sensitive S. aureus, ATCC 29213) grown on BAP were inoculated into 2 ml of TSB and incubated at 37°C for 3.5 hours. A 100 μl aliquot of this log phase culture (OD 0.5 at A600) was added to 2.5 ml molten top agar (TSB+0.6% agar+CaCl2 [400 μg/ml]), vortexed briefly, and poured onto the TSA base plates. The plates were allowed to cool. The base of the plate was marked to divide it into 16 squares. A 30 μl aliquot of filtrate was spotted onto each square. The plates were left to stand in a laminar air hood until the spotted filtrates were absorbed into the agar. They were then incubated at 37°C and checked for plaques at 12, 24, 36, and 48 hours. Suspected plaques on plates were photographed.
ATCC 29213 is a reference strain isolated from a human wound that is used in antimicrobial sensitivity testing and quality control in most clinical microbiology laboratories.35 To test the suitability of this reference, we used it to screen for lytic bacteriophages in clinical (abscesses, pustules, and wounds) and sewage materials in preliminary experiments. Three phages (C13–14, C21, and U-11) were thus isolated and used as positive controls in the lytic bacteriophage assay (data not shown). Consequently, it was chosen as a suitable host cell to screen for lytic bacteriophages.
Statistical Analysis
Subgroup analyses for the association between age, gender, and S. aureus prevalence were performed by the Fisher’s exact test using the SAS package (Cary, NC). A P value of <0.05 was considered statistically significant.
Results
Table 1▶ shows that our recruitment rate was 59.8% and resulted in 202 eligible and consenting participants out of the 338 individuals approached. The demographic characteristics of the study population are shown in table 2▶. There were 86 males and 116 females with an average age of 47.15 years. Seventy-seven percent (77%) of the subjects were between 20 to 59 years of age and 22% were ≥ 60 years. Eleven percent (11%) were diabetic and 70% were either overweight or obese. Overweight was defined as a body mass index (BMI) of 25.0 to 29.9; obesity was defined as a BMI of 30.0 or higher.36 The HCE workers population consisted of a younger (average age 40.5 years versus 54.7 years), predominantly female (63% versus 51%) group with fewer known cases of diabetes (4% versus 21%) compared to the ambulatory patient population. The enrolled house staff, which accounted for 50 of the 111 individuals in the HCE workers group, were between the age of 20 to 59 years, with the average age of 33 years. Fifty-two percent (52%) were male, only 2% were diabetic, and 30% were either overweight or obese.
Table 2.
Demographics of the study population.
| Total n (%) | HCE Workers n (%) | Ambulatory Patients n (%) | |
|---|---|---|---|
| All Participants | 202 (100%) | 111 (100%) | 91 (100%) |
| Average Age (y) | 47.15 | 40.5 | 54.7 |
| Sex | |||
| Male | 86 (43%) | 41 (37%) | 45 (49%) |
| Female | 116 (57%) | 70 (63%) | 46 (51%) |
| Age | |||
| 20–59 y | 156 (77%) | 101 (91%) | 56 (62%) |
| ≥60 y | 44 (22%) | 9 (8%) | 35 (38%) |
| Diabetes | |||
| Yes | 23 (11%) | 4 (4%) | 19 (21%) |
| No | 179 (89%) | 107 (96%) | 72 (79%) |
| Body Mass Index | |||
| Normal | 35 (17%) | 27 (24%) | 9 (10%) |
| Overweight | 52 (26%) | 26 (23%) | 26 (29%) |
| Obese | 89 (44%) | 34 (31%) | 55 (60%) |
HCE, Healthcare environment
Out of 202 CNA plates (one per individual in the study), 100 showed putative S. aureus as determined by a zone of hemolysis surrounding the colonies. On average, 1.8 putative S. aureus colonies per plate were screened by the Staphaurex Plus latex agglutination test. Of the 56 individuals who had colonies positive by this latex test, 49 (24%) were positively identified as S. aureus by PCR, and 7 of the 56 turned out to be S. lugdunensis by 16S rDNA PCR.
The overall prevalence of S. aureus in the population studied was 24.3% (95% CI, 18.5–30.8) with a MRSA prevalence of 0.99% (95% CI, 0.12–3.53). No significant difference in S. aureus prevalence was found in sub-group analyses (sex, age, diabetes, or BMI) (table 3▶). For example, 27.9% (95% CI, 18.8–38.6) of the males were positive for S. aureus compared to 21.6% (95% CI, 14.5–30.2) of the females, with no statistically significant difference (P=0.3223) by Fischer’s exact test. We also did not find any significant difference between the prevalence of S. aureus in HCE workers versus ambulatory patients, or between the house staff and other HCE workers (table 4▶). Twenty-seven (25.5%; 95% CI, 17.6–34.17) of the HCE workers compared to 22 (23.2%; 95% CI, 15.1–32.9) of the ambulatory patients were positive for S. aureus. This difference was not significant (P=0.87) by Fischer’s exact test. Neither of the two isolated MRSA came from HCE workers nor house staff.
Table 3.
Prevalence of colonization in the study population.
|
Prevalence of colonization |
||||||
|---|---|---|---|---|---|---|
|
S. aureus |
MRSA |
|||||
| Characteristic | n | n | % | Pvalue | n | % |
| All participants | 202 | 49 | 24.3 | 2 | 0.99 | |
| Sex | ||||||
| Male | 86 | 24 | 27.9 | 0.3223 | 1 | 1.16 |
| Female | 116 | 25 | 21.6 | 1 | 0.86 | |
| Age | ||||||
| 20–59 y | 155 | 40 | 25.8 | 0.2279 | ||
| ≥60 y | 44 | 7 | 15.9 | |||
| Diabetes | ||||||
| Yes | 23 | 3 | 13 | 0.2996 | ||
| No | 179 | 46 | 25.7 | |||
| Body Mass Index | ||||||
| Normal | 35 | 8 | 22.9 | 0.7889 | ||
| Overweight | 52 | 10 | 19.2 | 0.8165 | ||
| Obese | 77 | 20 | 26 | |||
MRSA, methicillin-resistant Staphylococcus aureus
Table 4.
Prevalence of nasal carriage of S. aureus in healthcare vs. non-healthcare workers.
| Enrolled (n) | S. aureus positive(n) | % | Pvalue* | |
|---|---|---|---|---|
| Participants | 202 | 49 | 24.3 | |
| Healthcare | 110 | 27 | 25.5 | 0.8704 |
| Non-healthcare | 92 | 22 | 23.2 | |
| Healthcare | 110 | 27 | 25.5 | |
| Residents | 49 | 12 | 24.5 | 1 |
| Non-residents | 61 | 16 | 26.2 |
*Level of significance calculated from Fischer’s exact test.
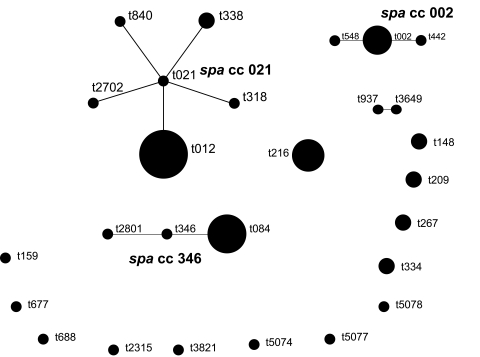
Multiple genotypes of S. aureus were identified in our study, but none of the S. aureus strains were positive for PVL genes. Of the 26 different spa types identified, t012 (16%) and t084 (10%) were the most common. All isolates showing spa type t012 belonged to sequence type ST30. Isolates with spa type t084 were represented by two STs: ST15 and ST18. Three new spa types, t5074, t5077, and t5087 were identified. Based upon Repeat Patterns (BURP) analysis of 49 S. aureus strains, the strains were grouped into four spa Clonal Complexes (spaCC) and 13 singletons (figure 2▶). The two MRSA isolates belonged to spa type t002 (SCCmec type II) and t012 (SCCmec type IVc). Interestingly, MRSA isolates were recovered from two ambulatory patients who were pre-diabetic with a BMI of 31.1 and 37.5, and aged 58 and 51 years, respectively.
Figure 2.
Based upon Repeat Patterns (BURP) analysis of the 49 S. aureus strains isolated.
None of the 202 samples were positive for lytic bacteriophages against S. aureus. However, 11 samples (5.4%) were positive for lytic bacteriophages against S. epidermidis (unpublished data).
Discussion
The primary goal of this study was to answer the question of whether lytic bacteriophages against S. aureus are present in the anterior nares of humans. A sample size was chosen to adequately power the study to answer this question. Out of 338 individuals who were approached and screened, 202 (59.8%) were eligible and consented to participate. Of the approached subjects, those excluded from participation in the study were excluded mostly because of being on nasal medications or antibiotics.
The demographics of our entire study population (table 2▶) are similar to that of the U.S. population at large for age distribution,28 diabetes,37 and obesity.38 This study’s overall prevalence of S. aureus (24.3%) and MRSA (0.99%) is also similar to those noted for the U.S. population and published by Kuehnert et al,28 who reported S. aureus and MRSA colonization prevalence estimates as 32.4% (95% CI, 30.7–34.1) and 0.8% (95% CI, 0.4–1.4), respectively.
However, our study showed a lower prevalence rate (25.5% for S. aureus and 0% for MRSA) among HCE workers than previous studies, such as Elie-Turenne et al39 who reported a 43.8% prevalence for S. aureus and a 15.2% prevalence for MRSA in healthcare professionals, and Barbosa et al40 who reported a 5% MRSA prevalence in house staff in their study, all from surgical rather than medical house officers. In addition, Halablab et al41 reported contact with healthcare workers as a risk factor for nasal colonization. Typically, the rate of MRSA colonization is reported to be higher in coastal cities and teaching hospitals due to a higher patient population. The lower rate of S. aureus and MRSA nasal colonization in the HCE workers in our study could have been due to increased compliance with infectious control policies and procedures that are monitored in this clinic.
We identified 26 different spa types in this population, suggesting a diversity of S. aureus genotypes present in the anterior nares. The t012 spa type had a frequency of 16% in our study population, compared to a frequency of 1.38% in the Ridon SpaServer (http://spaserver2.ridom.de) database. Similarly, the t084 spa type had a frequency of 10% in our study population compared to a frequency of 1.37% in the Ridon SpaServer. Both of these spa types belonged to two of the major clonal complexes of S. aureus (CC15 and CC30). The two MRSA isolates belonged to spa type t002 (SCCmec type II) and t012 (SCCmec type IVc) representing common healthcare-associated and community-associated genotypes, respectively.
None of the 202 study samples were positive for lytic bacteriophages against the methicillin-sensitive S. aureus reference strain ATCC 29213. We noticed that coagulase-negative staphylococci were the predominant flora on the CNA plates (data not shown), while only 24% grew S. aureus. Typically, bacteriophages are best isolated from environments where the host cells are present in abundance.18 Indeed, Furuse et al42 reported a higher percent of stool samples positive for the coliphages from patients with travelers’ diarrhea than in healthy patients. Absence of lytic bacteriophage against S. aureus in the anterior nares could, therefore, be explained in the context of possible dynamic interaction between host and its phages. When the numbers of host cells are high, the number of phages could be low, and vice versa. It is possible, but not likely, that our sampling time escaped the periods of the host and phage cycle when bacteriophage concentration was high.
An additional reason for not isolating S. aureus bacteriophages could have been that we used only one host cell strain, ATTC 29213, a standard reference S. aureus strain used in clinical microbiology for antimicrobial testing.35 A known weakness of isolating bacteriophages from their natural habitat is their detection using a specific host strain.43 Furthermore, the limited amount of material from the anterior nares of each study participant made testing against multiple strains of S. aureus difficult.
Interestingly, we found 11 samples that were positive for lytic bacteriophages against S. epidermidis which is very commonly found in the anterior nares.44 These bacteriophages belonged to the families Podoviridae and Siphoviridae. Indeed, we had more clinical samples positive for S. epidermidis than S. aureus (data not shown).
In conclusion, our study showed a lack of detectable levels of lytic bacteriophages against S. aureus in the anterior nares, probably due to the low prevalence of lytic bacteriophages in this region.
Acknowledgements
The authors would like to thank the MCRF’s research coordinators for their help with this study, Po-Huang Chyou, PhD for statistical analysis, Pravin Kaldhone and Jennifer Kislow for their technical assistance, and the Office of Scientific Writing & Publication for writing and editorial assistance.
Disclosure: This project was funded by the Marshfield Clinic Research Foundation Resident Research Funds, grant no. ASW10108.
References
- 1.Lowy FD. Staphylococcus aureus infections. N Eng J Med 1998;339:520–532. [DOI] [PubMed] [Google Scholar]
- 2.Gorwitz RJ, Kruszon-Moran D, McAllister SK, McQuillan G, McDougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. J Infect Dis 2008;197:1226–1234. [DOI] [PubMed] [Google Scholar]
- 3.Wertheim HF, Melles DC, Vos MC, van Leeuwen W, van Belkum A, Verbrugh HA, Nouwen JL. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis 2005;5:751–762. [DOI] [PubMed] [Google Scholar]
- 4.Bode LG, Kluytmans JA, Wertheim HF, Bogaers D, Vandenbroucke-Grauls CM, Roosendaal R, Troelstra A, Box AT, Voss A, van der Tweel I, van Belkum A, Verbrugh HA, Vos MC. Preventing surgical-site infections in nasal carriers of Staphylococcus aureus. N Engl J Med 2010;362:9–17. [DOI] [PubMed] [Google Scholar]
- 5.Rossney A, O’Connell S. 2008. Emerging high-level mupirocin resistance among MRSA isolates in Ireland. Euro Surveill 2008;13.pii:8084. [PubMed] [Google Scholar]
- 6.Jones JC, Rogers TJ, Brookmeyer P, Dunne WM Jr, Storch GA, Coopersmith CM, Fraser VJ, Warren DK. Mupirocin resistance in patients colonized with methicillin-resistant Staphylococcus aureus in a surgical intensive care unit. Clin Infect Dis 2007;45:541–547. [DOI] [PubMed] [Google Scholar]
- 7.Górski A, Miedzybrodzki R, Borysowski J, Weber-Dabrowska B, Lobocka M, Fortuna W, Letkiewicz S, Zimecki M, Filby G. Bacteriophage therapy for the treatment of infections. Curr Opin Investig Drugs 2009;10:766–774. [PubMed] [Google Scholar]
- 8.Mann NH. The potential of phages to prevent MRSA infections. Res Microbiol 2008;159:400–405. [DOI] [PubMed] [Google Scholar]
- 9.Lina G, Boutite F, Tristan A, Bes M, Etienne J, Vandenesch F. Bacterial competition for human nasal cavity colonization: role of Staphylococcal agr alleles. Appl Environ Microbiol 2003;69:18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivaraman K, Venkataraman N, Cole AM. Staphylococcus aureus nasal carriage and its contributing factors. Future Microbiol 2009;4:999–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peacock SJ, de Silva I, Lowy FD. What determines nasal carriage of Staphylococcus aureus? Trends Microbiol 2001;9:605–610. [DOI] [PubMed] [Google Scholar]
- 12.Nouwen J, Boelens H, van Belkum A, Verbrugh H. Human factor in Staphylococcus aureus nasal carriage. Infect Immun 2004;72:6685–6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Regev-Yochay G, Dagan R, Raz M, Carmeli Y, Shainberg B, Derazne E, Rahav G, Rubinstein E. Association between carriage of Streptococcus pneumoniae and Staphylococcus aureus in Children. JAMA 2004;292:716–720. [DOI] [PubMed] [Google Scholar]
- 14.Bogaert D, van Belkum A, Sluijter M, Luijendijk A, de Groot R, Rümke HC, Verbrugh HA, Hermans PW. Colonisation by Streptococcus pneumoniae and Staphylococcus aureus in healthy children. Lancet 2004;363:1871–1872. [DOI] [PubMed] [Google Scholar]
- 15.Regev-Yochay G, Lipsitch M, Basset A, Rubinstein E, Dagan R, Raz M, Malley R. The pneumococcal pilus predicts the absence of Staphylococcus aureus co-colonization in pneumococcal carriers. Clin Infect Dis 2009;48:760–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wos-Oxley ML, Plumeier I, von Eiff C, Taudien S, Platzer M, Vilchez-Vargas R, Becker K, Pieper DH. A poke into the diversity and associations within human anterior nare microbial communities. ISME J 2010;4:839–851. [DOI] [PubMed] [Google Scholar]
- 17.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science 2009;326:1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kutter E, Sulakvelidze A. Bacteriophages: Biology and Applications. Boca Raton, FL: CRC Press; 2004.
- 19.Breitbart M, Hewson I, Felts B, Mahaffy JM, Nulton J, Salamon P, Rohwer F. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol 2003;185:6220–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chibani-Chennoufi S, Sidoti J, Bruttin A, Kutter E, Sarker S, Brüssow H. In vitro and in vivo bacteriolytic activities of Escherichia coli phages: implications for phage therapy. Antimicrob Agents Chemother 2004;48:2558–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tylenda CA, Calvert C, Kolenbrander PE, Tylenda A. Isolation of Actinomyces bacteriophage from human dental plaque. Infect Immun 1985;49:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bachrach G, Leizerovici-Zigmond M, Zlotkin A, Naor R, Steinberg D. Bacteriophage isolation from human saliva. Lett Appl Microbiol 2003;36:50–53. [DOI] [PubMed] [Google Scholar]
- 23.Kilic AO, Pavlova SI, Alpay S, Kilic SS, Tao L. Comparative study of vaginal Lactobacillus phages isolated from women in the United States and Turkey: prevalence, morphology, host range, and DNA homology. Clin Diagn Lab Immunol 2001;8:31–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.García P, Martínez B, Obeso JM, Lavigne R, Lurz R, Rodríguez A. Functional genomic analysis of two Staphylococcus aureus phages isolated from the dairy environment. Appl Environ Microbiol 2009;75:7663–7673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Synnott AJ, Kuang Y, Kurimoto M, Yamamichi K, Iwano H, Tanji Y. Isolation from sewage influent and characterization of novel Staphylococcus aureus bacteriophages with wide host ranges and potent lytic capabilities. Appl Environ Microbiol 2009;75:4483–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O’Flaherty S, Ross RP, Flynn J, Meaney WJ, Fitzgerald GF, Coffey A. Isolation and characterization of two anti-staphylococcal bacteriophages specific for pathogenic Staphylococcus aureus associated with bovine infections. Lett Appl Microbiol 2005;41:482–486. [DOI] [PubMed] [Google Scholar]
- 27.Twort FW. An investigation on the nature of ultra-microscopic viruses. Lancet 1915;186:1241–1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuehnert MJ, Kruszon-Moran D, Hill HA, McQuillan G, McAllister SK, Fosheim G, McDougal LK, Chaitram J, Jensen B, Fridkin SK, Killgore G, Tenover FC. Prevalence of Staphylococcus aureus nasal colonization in the United States, 2001–2002. J Infect Dis 2006;193:172–179. [DOI] [PubMed] [Google Scholar]
- 29.Shukla SK, Karow ME, Brady JM, Stemper ME, Kislow J, Moore N, Wroblewski K, Chyou PH, Warshauer DM, Reed KD, Lynfield R, Schwan WR. Virulence genes and genotypic associations in nasal carriage, community-associated methicillin-susceptible and methicillin-resistant USA400 Staphylococcus aureus isolates. J Clin Microbiol 2010;48:3582–3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lina G, Jarraud S, Ji G, Greenland T, Pedraza A, Etienne J, Novick RP, Vandenesch F. Transmembrane topology and histidine protein kinase activity of AgrC, the agr signal receptor in Staphylococcus aureus. Mol Microbiol 1998;28:655–662. [DOI] [PubMed] [Google Scholar]
- 31.Shukla SK, Ramaswamy SV, Conradt J, Stemper ME, Reich R, Reed KD, Graviss EA. Novel polymorphisms in mec genes and a new mec complex type in methicillin-resistant Staphylococcus aureus isolates obtained in rural Wisconsin. Antimicrob Agents Chemother 2004;48:3080–3085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koreen L, Ramaswamy SV, Graviss EA, Naidich S, Musser JM, Kreiswirth BN. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J Clin Microbiol 2004;42:792–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 2000;38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. 3rd ed. Woodbury, NY: Cold Spring Harbor Laboratory Press; 2001.
- 35.Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically; Approved Standard–Eighth Edition. Clinical and Laboratory Standards Institute. CLSI M07-A8:29; 2009.
- 36.Clinical Guidelines on the Identification, Evaluation, and Treatment of Overweight and Obesity in Adults: The Evidence Report. National Institutes of Health. Obes Res 1998;6:51S–209S. [PubMed] [Google Scholar]
- 37.Cowie CC, Rust KF, Byrd-Holt DD, Eberhardt MS, Flegal KM, Engelgau MM, Saydah SH, Williams DE, Geiss LS, Gregg EW. Prevalence of diabetes and impaired fasting glucose in adults in the U.S. population: National Health and Nutrition Examination Survey 1999–2002. Diabetes Care 2006;29:1263–1268. [DOI] [PubMed] [Google Scholar]
- 38.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–1555. [DOI] [PubMed] [Google Scholar]
- 39.Elie-Turenne M, Fernandes H, Mediavilla JR, Rosenthal M, Mathema B, Singh A, Cohen TR, Pawar KA, Shahidi H, Kreiswirth BN, Deitch EA. Prevalence and characteristics of Staphylococcus aureus colonization among healthcare professionals in an urban teaching hospital. Infect Control Hosp Epidemiol 2010;31:574–580. [DOI] [PubMed] [Google Scholar]
- 40.Barbosa A, Chapin K, Mermel LA. Methicillin-resistant Staphylococcus aureus colonization of house officers. Infect Control Hosp Epidemiol 2009;30:912–914. [DOI] [PubMed] [Google Scholar]
- 41.Halablab MA, Hijazi SM, Fawzi MA, Araj GF. Staphylococcus aureus nasal carriage rate and associated risk factors in individuals in the community. Epidemiol Infect 2010;138:702–706. [DOI] [PubMed] [Google Scholar]
- 42.Furuse K. Distribution of coliphages in the general environment: general considerations. In: Goyal SM, Gerba CP, Bitton G, eds. Phage Ecology. New York: Wiley; 1987. 87–124.
- 43.Kutter E. Phage host range and efficiency of plating. In: Clockie MRJ, Kropinski AM, eds. Bacteriophages: Methods and Protocols, Volume 1: Isolation, Characterization and Interactions, Vol. 501. New York: Springer/Humana Press; 2009. 141–149. [DOI] [PubMed]
- 44.Kloos WE. Natural populations of the genus Staphylococcus. Annu Rev Microbiol 1980;34:559–592. [DOI] [PubMed] [Google Scholar]