Abstract
Rectal cancer response to chemoradiation (CRT) varies from no response to a complete pathologic response (pCR). Identifying predictive biomarkers of response would therefore be useful. We assessed whether chromosomal copy-number alterations (CNAs) can assist in predicting pCR. Pre-treatment tumor biopsies and paired normal surgical tissues from the proximal resection margin were collected from 95 rectal cancer patients treated with pre-operative CRT and total mesorectal excision in a prospective Phase II study. Tumor and control DNA was micro-dissected and oligonucleotide array-based comparative genomic hybridization (aCGH) was used to identify CNAs which were correlated with pCR. Ingenuity pathway analysis (IPA) was then used to identify functionally relevant genes in aberrant regions. Finally, a predictive model for pCR was built using Support Vector Machine (SVM) and leave-one-out cross-validation assessed the accuracy of aCGH. Chromosomal regions most commonly affected by gains were 20q11.21–q13.33, 13q11.32–23, 7p22.3-p22.2 and 8q23.3–q24.3, and losses, 18q11.32–q23, 17p13.3-q11.1, 10q23.1, and 4q32.1–q32.3. The 25 (26%) patients who achieved a pCR had significantly fewer high copy gains overall than non-pCR patients (p=0.01). Loss of chromosomal region 15q11.2 was significantly associated with non-pCR (p<0.00002, Q-Bound<0.0391), while loss of 12p13.31 was significantly associated with pCR (p<0.0003; Q-Bound<0.097). IPA identified 8 genes in the imbalanced chromosomal regions that associated with tumor response. SVM identified 58 probes that predict pCR with 76% sensitivity, 97% specificity, and positive and negative predictive values of 91% and 92%. Our data indicate that chromosomal CNAs can help identify rectal cancer patients more likely to develop a pCR to CRT.
INTRODUCTION
Pre-operative chemoradiation (CRT) followed by total mesorectal excision (TME) is the current standard of care for patients with locally advanced rectal cancer. However, not every patient benefits equally from CRT. While some patients have a minimal response, between 15% and 25% of patients treated with pre-operative CRT achieve a pathologic complete response (pCR) and patients with a pCR have better long-term survival compared to patients with resistant tumors (Garcia-Aguilar et al., 2003; Ciccocioppo et al., 2009). However, CRT causes toxicity and increases health care costs, so identifying those patients who are more likely to benefit from CRT is desirable and of considerable clinical interest.
Although tumor response to CRT varies with the dose and fractionation of the radiation, the type of chemotherapy, the CRT-to-surgery interval, and the size and stage of the tumor, substantial evidence suggests that the differences in response observed between patients are mostly related to the biology of the tumor (Smith et al., 2006; Kuremsky et al., 2009). However, there are currently no validated biomarkers that predict pCR. Identifying such predictive biomarkers could help define and direct treatment for this subset of patients.
Most colorectal cancers display a form of genetic instability characterized by the amplification and deletion of entire chromosomes or chromosomal segments (Lengauer et al., 1998; Teixeira and Heim, 2005). Gains and loses of chromosomal segments lead to changes in oncogenes and tumor suppressor genes that are important for the development and progression of colorectal cancer. The hybridization of tumor DNA to normal metaphase chromosomes, a technique known as comparative genomic hybridization (CGH), allows for the simultaneous screening and mapping of gains and losses of specific chromosomal segments (Grade et al., 2009; Postma et al., 2009). New high-throughput approaches, such as array-based comparative genomic hybridization (aCGH) using oligonucleotides allow genome-wide detection of chromosomal changes, also known as copy-number alterations (CNAs), at a higher resolution compared to metaphase-based CGH. Oligonucleotide aCGH is therefore one of the preferred methods to characterize the degree of aneuploidy or chromosomal complexity in cancer.
In this study we used high density whole genome oligonucleotide aCGH to identify CNAs in patients with advanced rectal cancer and determine whether a specific CNA profile associates with tumor response and can predict pCR. Here, we present the CNA profiles for 95 rectal cancer patients treated with CRT and TME and describe the value of this molecular approach for predicting pCR.
MATERIALS AND METHODS
Patients
This study included patients with clinical stage II (T3–4, N0) or stage III (any T, N1–2) invasive adenocarcinoma of the rectum with a distal tumor border within 12cm of the anal verge, as measured on rigid proctoscopic exam, who were enrolled in the Timing of Rectal Cancer Response to Chemoradiation study, a multi-institutional clinical trial investigating the effect of increasing the CRT-to-surgery interval, and adding chemotherapy, modified FOLFOX-6 (mFOLFOX-6) during the waiting period (ClinicalTrials.org Identifier: NCT00335816). This trial was designed in a series of sequential Phase II trials or study groups (SGs), each with a progressively longer CRT-to-surgery interval and increasing cycles of pre-operative mFOLFOX-6. This study was approved by an Institutional Review Board (IRB) at each participating institution as well as a central IRB, and informed written consent was obtained from each patient prior to enrollment in the trial. Patients included in the present study were pooled from SG1 (n=43) and SG2 (n=52). Further details of patient eligibility for this trial are presented elsewhere (Garcia-Aguilar et al., 2011).
Treatment protocol
Patients in both groups were treated with CRT; 5-Fluorouracil (FU) 225mg/m2/day for 7 days in continuous infusion and a total of 50.4Gy of radiation. Patients in SG1 underwent TME an average of 6 weeks after completing CRT (standard of care). Following CRT, patients in SG2 with no evidence of stable disease received 2 cycles of additional chemotherapy (mFOLFOX-6); leucovorin 200mg/m2 or 400mg/m2 plus oxaliplatin 85mg/m2 by 2h infusion, followed by bolus of 5-FU 400mg/m2 and a 46h infusion of 5-FU 2,400mg/m2. Patients in SG2 underwent TME an average of 11 weeks after completing CRT. The clinical outcomes for these patients are presented elsewhere (Garcia-Aguilar et al., 2011).
Tumor response to CRT
Pathologic complete response was defined as the complete absence of tumor cells from the rectal wall and regional lymph nodes. Tumor pathology was assessed by two independent pathologists and graded according to the recommendations of the American Joint Committee of Cancer (AJCC) (Edge et al., 2010). For the purposes of the study, response was classified as either pCR or non-pCR based on the above criteria.
Sample preparation, whole genome amplification and oligonucleotide aCGH
Tumor DNA from each patient was obtained from pre-treatment biopsy tissue and control DNA was obtained following treatment from paired normal surgical tissue from the proximal resection margin. To extract DNA, 10–20 slides per patient sample from formalin-fixed paraffin-embedded (FFPE) tumor biopsy and normal tissue were de-paraffinized, hydrated, and stained with 0.2% methylene blue. A 27.5 gauge needle was then used to manually micro-dissect cells under inverted microscopy. Genomic DNA was then extracted using the QIAamp DNA FFPE Tissue kit (Qiagen Inc., Valencia, CA) according to manufacturer’s instructions with the following modifications; an extension of digestion time at 56°C from 1 hour to 48 hours, and the addition of three 20µl aliquots of Proteinase-K at 4, 20, and 28 hours during digestion. DNA was quantified by measuring absorbance, and 100–200ng of DNA was amplified using the GenomePlex Complete Whole Genome Amplification (WGA)-2 kit (Sigma Cor., Cream Ridge, NJ). WGA-DNA was purified with the GenElute PCR Clean-Up kit (Sigma Cor., Cream Ridge, NJ) and quantified. Amplicon size was determined by running 2µl of WGA-DNA on a 2% agarose gel. Amplified WGA-DNA with an average size of 200–300 base-pairs was used for aCGH.
The Agilent microarray platform was used for oligonucleotide aCGH (Human Genomic CGH 244A Microarrays), with 8.9kb overall median probe-spacing covering more than 236,000 coding and non-coding human DNA sequences. aCGH assays were conducted according to manufacturer’s instructions (Agilent Cor., Santa Clara, CA). Briefly, for each sample, 2µg of WGA-DNA was labeled with the non-enzymatic Universal Linkage System (ULS). Equal amounts of tumor biopsy and paired normal surgical specimen DNA were labeled with ULS-Cy5 and ULS-Cy3, respectively. The labeled samples were purified using Agilent-KREApure columns, and then combined with the hybridization mixture in a SureHyb chamber. Hybridization of arrays was carried out at 65°C for 40 hours. Arrays were then washed in Wash Buffer-1 and Wash Buffer-2. Scanning and image analysis were performed on an Agilent scanner. Agilent Feature Extraction Software (v.9.5) was used for data extraction from raw microarray image files.
Statistical analysis
Patient characteristics
To determine differences in clinical pathological features between pCR and non-PCR patients, Student’s t test was used to compare means of continuous variables, and the two-sided Fisher’s exact test or the Fisher-Freeman-Halton exact test using Monte Carlo was chosen for categorical variables.
Characterizing chromosomal CNAs
Nexus copy-number software (v.4.0) (BioDiscovery Inc., El Segundo, CA), was used to identify chromosomal CNAs using the rank segmentation algorithm, a modified version of the circular binary segmentation (CBS) algorithm. Briefly, a numerical value based on the median log-ratio of all probes in the region (where the minimum number of probes per segment was 5, p=10−6) was assigned to chromosomal alterations as follows: chromosomal single copy gain (+0.2), chromosomal amplification or high copy gain of ≥2 (+0.6), chromosomal single copy loss, (−0.2), and homozygous chromosomal loss (−1). Copy-number changes were determined for each sample, and the fraction of genome alteration (FGA) was calculated to reflect the degree of genomic instability. The FGA was determined by dividing the overall altered segment size by the genome size using the NCBI hg18 database (Build 36.1) comprising 3,080,436,051 base-pairs.
Identifying CNA signatures in patients with and without a pCR
Unsupervised hierarchical cluster analysis was used to analyze the distribution of whole-genome CNA profiles. The CNAs that differed between pCR and non-pCR patients were identified using Nexus copy-number software (p<0.05, differential threshold >25%). To assess CNA differences between pCR and non-pCR patients, the two-sided Fisher’s exact test was used and Q-bound was utilized to correct for multiple testing by performing false discovery rate (FDR) analysis, defined as the proportion of false positives among all positives (Benjamini and Hochberg, 1995; Storey and Tibshirani, 2003). A Q-bound score of <0.25 was considered statistically significant to minimize the false negative rate and prevent missing valuable biological findings (Storey and Tibshirani, 2003).
Identification of functionally relevant genes
Identification of the functionally relevant genes contained in the CNA regions that were associated with tumor response was determined using Ingenuity Pathway Analysis (IPA) (v.8.7) (Ingenuity Systems, Redwood City, CA).
Predictive model for pCR
Predictive biomarkers for pCR were derived using a combination of methods including feature (candidate biomarker) selection, classification model-fitting and cross-validation, as described previously (Shi et al., 2006; Deng and Campagne, 2010). Differential features were selected from all probes on the aCGH array based on two criteria: FDR - the adjusted p values using the Benjamini and Hochberg method (Benjamini and Hochberg, 1995) based on the p values of the Student’s t test between patients with and without a pCR, and probe signal fold-change between patients with and without a pCR. The features were fed into the linear-kernel Support Vector Machine (SVM) classifier to train the classification model based on the selected features (Bang and Davidian, 2010). Leave-one-out cross-validation was performed to evaluate the classification performance on test samples. ROCR package (Sing et al., 2005) was used to calculate performance measures such as sensitivity, specificity, positive and negative predictive values, and to plot the performance measures using the receiver operating characteristic (ROC) curve. The area under the ROC curve (AUC) was calculated to quantitatively summarize the performance of the model. The final predictive model contained 58 features using the following feature selection cutoffs: FDR threshold ≤0.25; log2 fold-change ≥0.4. Gene annotations for the flanking regions (8kb) of the selected features were carried out using the R biomaRt (interface to the BioMart database) package in bio-conductor (Gentleman et al., 2004).
RESULTS
Patient characteristics and tumor response
A total of 95 patients, 43 in SG1 and 52 in SG2, were included in the aCGH analysis. Patient demographics and tumor characteristics stratified by tumor response (pCR versus non-pCR) and SG are shown in Table 1. Twenty-five out of 95 (26%) patients achieved a pCR; 8 (19%) patients in SG1 and 17 (33%) patients in SG2. There was no significant difference in tumor response rate between SG1 and SG2 (p=0.161). Patients with a pCR were slightly younger and had a higher proportion of stage III tumors compared to patients with non-pCR but the difference did not reach statistical significance. Other demographics such as age, and tumor characteristics such as tumor histology were similar between the two groups.
Table 1.
Patient Demographics and Disease Characteristics Stratified by pCRa Status and Study Group
| Demographic or disease characteristic | pCR (n=25) |
Non-pCR (n=70) |
p value |
|---|---|---|---|
| Age, yearsb | 54.4 (10.57) | 57.24 (12.53) | 0.278 |
| Gender | 0.815 | ||
| Male | 13 (25%) | 40 (75%) | |
| Female | 12 (29%) | 30 (71%) | |
| Histological grade | 0.662 | ||
| Well differentiated | 3 (18%) | 14 (82%) | |
| Moderately differentiated | 22 (33%) | 54 (66%) | |
| Poorly differentiated | 0 (0%) | 2 (100%) | |
| Clinical stage | 0.101 | ||
| II | 3 (13%) | 21 (87%) | |
| III | 22 (31%) | 49 (69%) | |
| Study group (SG)c | 0.161 | ||
| SG1 (n=43) | 8 (19%) | 35 (81%) | |
| SG2 (n=52) | 17 (33%) | 35 (67%) | |
Pathologic complete response.
Mean is presented with the standard deviation shown in parentheses.
SG1 had surgery 6 weeks after the completion of CRT (standard of care); SG2 had surgery 11 weeks after the completion of CRT, and had 2 cycles of modified FOLFOX-6 during the waiting period.
Chromosomal copy number alterations
The mean number of gains and losses for all 95 patients and for patients with and without a pCR are presented in Table 2. Patients with a pCR had a lower number of total gains and losses, a lower number of single copy gains and losses, and a lower FGA compared to non-pCR patients but these differences did not reach statistical significance. However, the number of high copy gains was significantly lower in patients who achieved a pCR (p=0.01).
Table 2.
| CNA event | Entire cohortc (n=95) |
pCR tumorsc (n=25) |
Non-pCR tumorsc (n=70) |
p valued |
|---|---|---|---|---|
| FGAe | 31.04% (15.85) | 28.48% (15.90) | 31.95% (15.84) | 0.35 |
| Total gains and losses | 117.59 (99.15) | 98.52 (73.72) | 124.4 (106.41) | 0.19 |
| One copy gain | 49.86 (40.11) | 41.2 (28.12) | 52.96 (43.35) | 0.13 |
| High copy gain | 8.56 (13.29) | 4.48 (6.14) | 10.01 (14.81) | 0.01 * |
| One copy loss | 58.14 (51.44) | 51.68 (50.68) | 60.44 (51.88) | 0.46 |
| Homozygous loss | 1.03 (1.83) | 1.16 (2.61) | 0.99 (1.48) | 0.75 |
Copy number alteration.
Pathologic complete response.
Standard deviation shown in parentheses.
Statistically significant.
Fraction of genomic alteration.
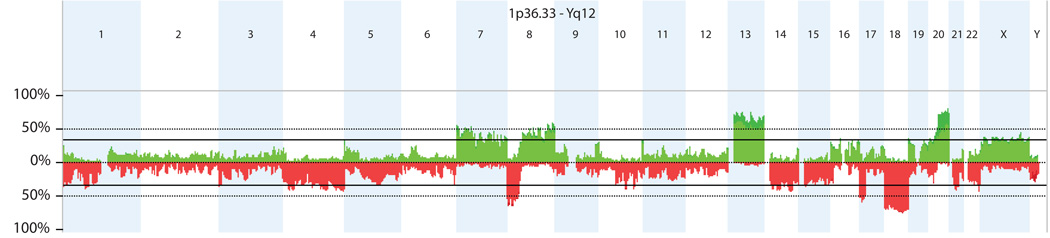
Figure 1 illustrates the specific chromosomal regions where gains and losses were most frequent in the 95 patients assayed. Copy-number gains most frequently affected chromosomal regions 20q11.21–q13.33 (68%), 13q11.32–23 (57%), 7p22.3-p22.2 (36%) and 8q23.3–q24.3 (36%), while losses were most frequently observed in chromosomal regions 18q11.32–q23 (60%), 17p13.3-q11.1 (39%), 10q23.1 (38%) and 4q32.1–q32.3 (37%).
Figure 1.
Overall frequency of copy number alterations (CNAs) detected by oligonucleotide array-based comparative genomic hybridization (aCGH) in 95 rectal cancer patients. The most common alterations were gains in chromosomes 20, 13, 8 and 7 and losses in chromosomes 18, 8, 17 and 4.
Chromosomal regions associated with pCR
Chromosomal regions associated with pCR were identified by comparing the CNA profiles of patients with and without a pCR. A total of 304 regions were found to be different between pCR and non-pCR patients with a minimal differential threshold of >25% and a p value of <0.05 (Supplementary Table 1). When p values were corrected for multiple testing and a Q-bound of <0.25 was considered as the threshold for statistical significance, 65 regions remained associated with pCR (Supplementary Table 2). The majority of these regions were close to one another and given the extensive sequence coverage provided by high density arrays, CNA region boundaries could be accurately defined. Thus, CNAs could be combined to form a number of large consecutive sequences. After combining the consecutive CNAs, loss of chromosomal regions 15q11.2–q26.3, 11q24.3–q25 and 8p12 occurred less frequently in patients with a pCR compared to non-pCR patients, while loss of chromosomal region 12p13.31 occurred more frequently in patients with a pCR compared to non-pCR patients (Table 3).
Table 3.
| Chromosomal loss | Frequency of occurrence | Difference | p value | Q-Bound | |
|---|---|---|---|---|---|
| pCR | Non-pCR | ||||
| 15q11.1–q26.3 | 4% | 37% | 33 | <0.00002 | <0.0391 |
| 11q24.3–q25 | 0% | 29% | 29 | <0.00002 | <0.0391 |
| 8p12 | 20% | 49% | 29 | <0.00002 | <0.0391 |
| 12p13.31 | 32% | 4% | 28 | <0.00003 | <0.097 |
Copy number alteration.
Pathologic complete response.
Ingenuity pathway analysis
The chromosomal regions that differed between pCR and non-pCR patients contain a total of 473 genes, but only 285 of them were significantly different after the p values were corrected for multiple testing (Q-bound <0.25). IPA biomarker filter analysis identified 8 out of the 285 genes as potential candidates of response, diagnosis, prognosis and therapeutic efficacy associated with tumor response (Table 4). Loss of all of these genes except SCG5 was more frequent in pCR patients compared to non-pCR patients.
Table 4.
IPAa Identified Eight Candidate Biomarkers Differentially Expressed Between pCRb and non-pCR Patients
| Gene | Chromosome | Association | Difference | p value | Q-Bound |
|---|---|---|---|---|---|
| ENO2 | 12 | Prognosis, efficacy | 29 | <0.0004 | 0.0964927 |
| TPI1 | 12 | Unspecified application | 29 | <0.0004 | 0.0964927 |
| GAPDH | 12 | Disease progression | 28 | <0.0004 | 0.1793397 |
| CD4 | 12 | Efficacy | 28 | <0.0004 | 0.1793397 |
| ING4 | 12 | Diagnosis | 28 | <0.0004 | 0.1793397 |
| CD27 | 12 | Unspecified application | 28 | <0.0004 | 0.1793397 |
| TNFRSF1A | 12 | Prognosis | 28 | <0.0004 | 0.1793397 |
| SCG5 | 15 | Diagnosis | 33 | <0.003 | 0.2018752 |
Ingenuity pathway analysis.
Pathologic complete response.
We also utilized IPA core analysis to determine the signaling and metabolic pathways, molecular networks, and biological processes that were most significantly changed in our dataset. Through this approach we identified 3 candidate canonical pathways containing new potential candidate genes that were significantly enriched in the regions associated with pCR; the GABA receptor signaling pathway (GABRG3, GABRA5, GABRB3; p=0.001), the glycolysis/gluconeogenesis pathway (GAPDH, ENO2, TPI1; p=0.011), and the glutamate receptor signaling pathway (GNB3, HOMER2; p=0.03).
Performance of pCR prediction
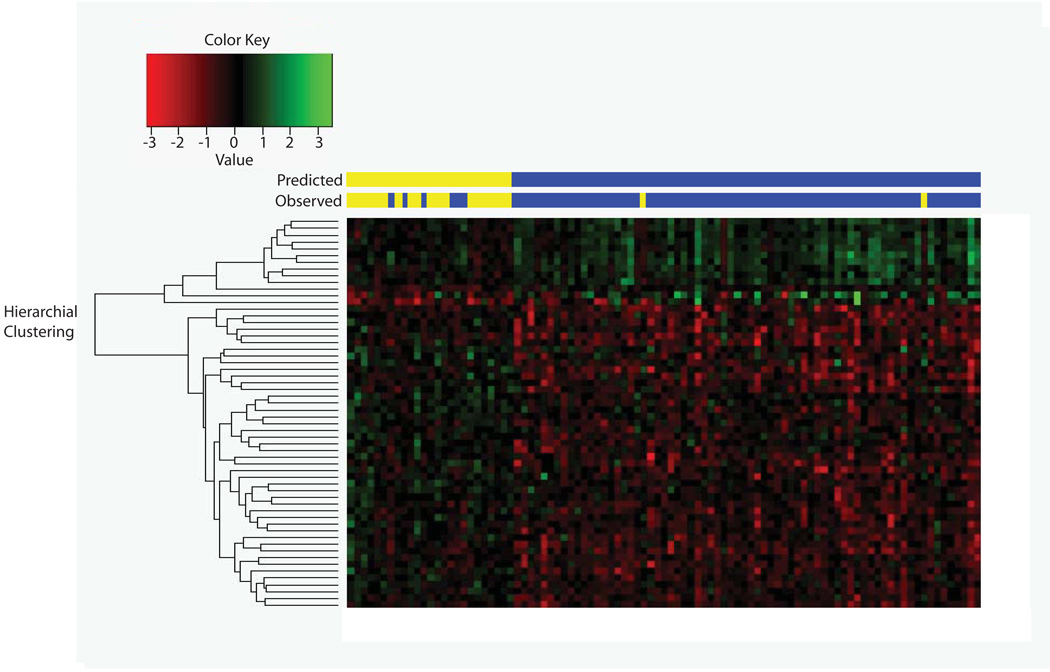
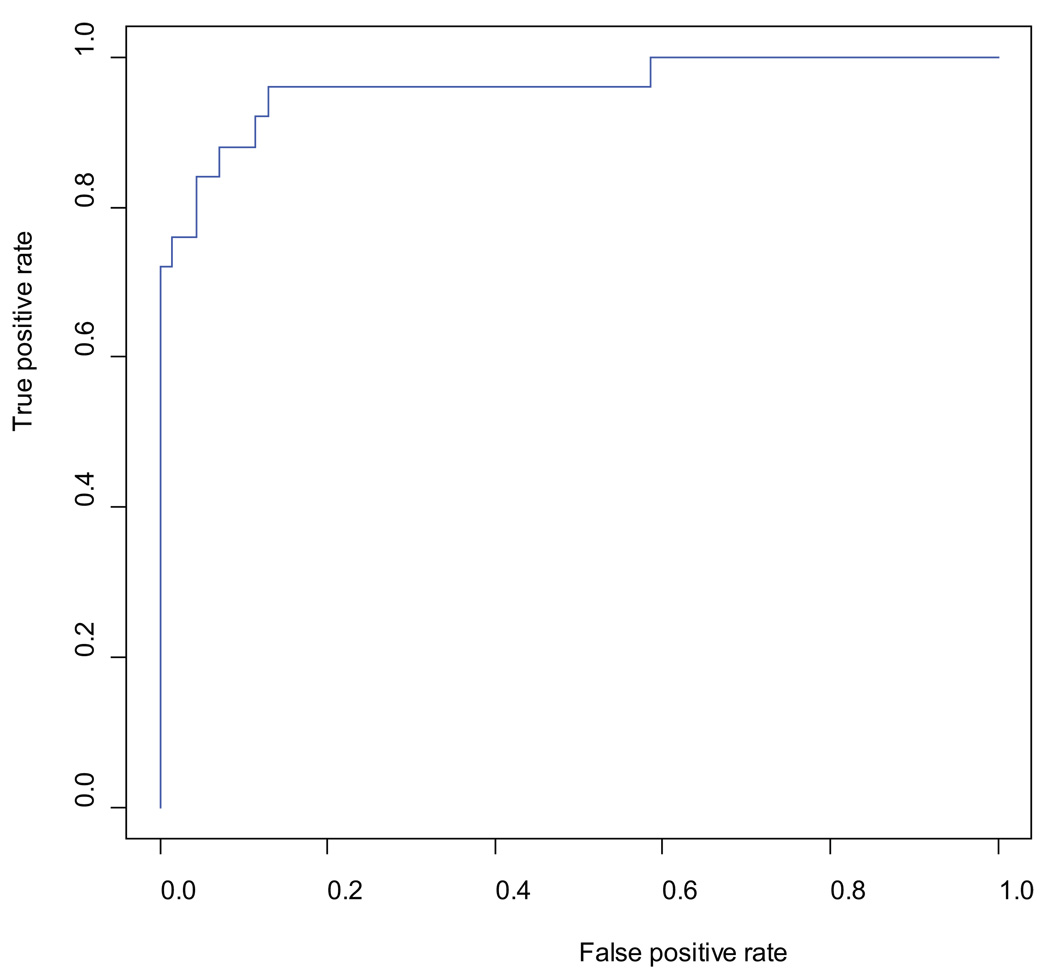
To determine how accurately oligonucleotide aCGH can predict pCR, a biomarker model was built using SVM. Fifty eight probes were selected using FDR and fold-change cutoffs (FDR; threshold ≤0.25; log2 fold-change ≥0.4). Using leave-one-out cross-validation aCGH predicted pCR with a sensitivity of 76%, a specificity of 97%, a positive predictive value of 91%, and a negative predictive value of 92%. Tumor response was predicted accurately by aCGH in 87 out of 95 (92%) patients (Figure 2a) and the performance of the model was plotted as an ROC curve with the AUC value of 0.96 (Figure 2b).
Figure 2.
a: 58 probes were used to build a predictive model using Support Vector Machine (SVM) to predict pathologic complete response (pCR). Sample identification - Red: Loss; Green: Gain; Yellow: pCR; Blue: Non-pCR.
b: Receiver-Operator Characteristics (ROC) curve using the true positive rate and false positive rate to evaluate the prediction ability of pathologic complete response (pCR) using 58 probes selected from significant discriminated genomic aberrant regions between pCR and non-pCR patients. The area under the curve (AUC) value for the performance of the prediction is 0.96.
To identify additional novel genes associated with pCR, we used gene annotation software to identify the genes in the flanking regions (8kb) of the 58 selected probes contained in our predictive model. Through this analysis we identified 32 genes as potential candidate genes related to tumor response (Supplementary Table 3).
DISCUSSION
Our results indicate that CNAs detected by aCGH may help identify rectal cancer patients with and without a pCR in response to CRT. We found that patients with a pCR had significantly fewer high copy gains compared to non-pCR patients and that loss of chromosomal region 15q11.2 was found to be significantly associated with non-pCR, while loss of 12p13.31 was significantly associated with pCR. Finally, we built a pCR prediction model that contained 58 probes and predicted pCR with a high degree of accuracy.
Our oligonucleotide aCGH analysis of 95 patients prior to CRT treatment revealed that chromosomal gains occurred most commonly at 20q11.21–q13.33, 3q11.32–23, 7p22.3-p22.2 and 8q23.3–q24.3, and losses were present at 18q11.32–q23, 17p13.3-q11.1, 10q23.1 and 4q32.1–q32.3. The number and frequency of copy-number gains in our patients were almost identical to the gains that have been reported previously in other series of colorectal cancer patients (Ried et al., 1996; Meijer et al., 1998; De Angelis et al., 1999; Nakao et al., 2004; Camps et al., 2008). With respect to copy-number losses, 18q and 4q have been reported in most series (Ried et al., 1996; Meijer et al., 1998; De Angelis et al., 1999; Nakao et al., 2004; Camps et al., 2008), but losses of 17p and 10q were more frequent in our patients compared to other series (Ried et al., 1996; Meijer et al., 1998; De Angelis et al., 1999); these differences may be explained by the type of specimen used, differences in the method used to assess CNAs, or the location of the tumors.
Formalin fixation and the process of preservation can introduce modifications into nucleic acids such as cross-linking, fragmentation, and base changes that may limit the use of CGH (Srinivasan et al., 2002). These alterations in DNA integrity secondary to fixation and preservation have minimal impact on the low resolution offered by metaphase CGH (5–10Mb), which was the CGH methodology used in the initial studies reporting on CNAs in colorectal cancer (Ried et al., 1996). However, DNA integrity is more important when using high density aCGH that utilizes hundreds of thousands of long oligonucleotides and provides resolution in the range of 6.5kb–30kb. Consequently, most aCGH CNA studies in colorectal cancer have utilized fresh frozen tissue (Ried et al., 1996; Meijer et al., 1998; De Angelis et al., 1999; Nakao et al., 2004; Camps et al., 2008). The problem of tissue preservation in rectal cancer patients treated with CRT is further magnified by the small size of most diagnostic biopsies, and the need to enrich the cancer-cell population by micro-dissection. Bearing all of this in mind, recent direct comparison has in fact shown that oligonucleotide aCGH using FFPE tissues captures the majority of the CNAs detected in frozen tissue (Johnson et al., 2006; van Beers et al., 2006; Tuefferd et al., 2008; Hostetter et al., 2010). In our study we performed oligonucleotide aCGH using FFPE samples and WGA products and demonstrated that the limited DNA extracted from small FFPE biopsy specimens followed by whole genome amplification can produce sufficient DNA for aCGH testing without obvious bias, and that this approach detects CNAs that are consistent with previous reports (Ried et al., 1996; Meijer et al., 1998; De Angelis et al., 1999; Nakao et al., 2004; Camps et al., 2008). Our study therefore further verifies that small biopsy FFPE specimens can serve as a valuable resource for genome-wide studies such as aCGH and importantly that the differences we observed in the frequency of some CNA losses between our study and other reports is not likely due to our use of FFPE tissue.
It is well documented that the tumor location can influence the mutation profile of the tumor. In general, tumors in the colon have a higher level of microsatellite instability (MSI), (up to 15%), compared to tumors in the rectum. In contrast, tumors in the rectum have a higher proportion of TP53 gene mutations and less MSI (Kim et al., 1994; Iacopetta et al., 2006; Gafà et al., 2000; Soong et al., 2000; Samowitz et al., 2001; Young et al., 2001). This may explain some of the copy-number differences we observed in our series, compared to others, as all of our patients had rectal cancer, while most other series included patients with colon and rectal cancers.
A comparison of pCR and non-pCR patient CNA profiles revealed that patients with a pCR have fewer CNA events compared to patients without a pCR, but the differences between pCR and non-pCR tumors were only statistically significant for high copy gains. These data suggest that an increase in the CNAs, a feature previously associated with tumor progression in colorectal cancer (Korn et al., 1999; Ghadimi et al., 2003; Diep et al., 2006), may also be associated with rectal cancer resistance to CRT.
Using the Fisher’s exact test we identified a large number of chromosomal regions of significant difference between pCR and non-pCR patients, but when the data was sorted by the Q-bound value (the FDR of the p value), only four chromosomal regions remained different. Loss of 15q11.1–q26.3, 11q24.3–q25, and 8p12 were more common in non-pCR patients compared to pCR patients, while loss of 12p13.31 was more common in pCR patients. The Q-bound value suggests that it is unlikely that these aberrations were identified by chance. The study by Grade et al., also analyzed the association of CNAs with tumor response in rectal cancer patients treated with CRT (Grade et al., 2009). They found that gains of chromosomal regions 7q32–q36 and 7q11–q31, and amplification of 20q11–q13 were associated with tumor response, but that the probability of detecting these CNAs by chance was high (p=0.21). However, their study only included 51 patients, less than half than in our series. Furthermore, in their study, response was defined as tumor down-staging which is less precise and less biologically relevant than pCR. Finally, they only analyzed chromosomal imbalances in 260 chromosome bands, rather than the ~244,000 probes used in our study.
The unsupervised hierarchical cluster analysis initially failed to discriminate pCR from non-pCR profiles using all probes across the genome, likely due to the high variability of DNA copy-number profiling in rectal cancer. In addition, the total number of probes (~244,000) was much greater than the number of samples (n=95) in this study. Therefore, to build a model that predicted pCR versus non-pCR, a feature selection procedure was performed to narrow down the number of features (probes) by using adjusted p values coupled with mean fold-change. In building our model we adopted measures to ensure that the model was not over-fit or self-fulfilling. Two of the most common causes of over-fitting a high-dimension model such as the model used in our study, are sample size limitation and the leaking of training samples to test samples during model validation. To combat sample size limitation, we collected and analyzed a large cohort of rectal cancer patients (n=95); to our knowledge this is the largest rectal cancer patient series studied by oligonucleotide aCGH. Additionally, we used leave-one-out cross validation, an approach which maximizes preservation of the training sample size. Finally, we applied rigorous feature selection and cross-validation procedures to minimize the risk of leaking information, to ensure that the training samples were completely hidden from the test samples during all iterations of feature selection and cross-validation. Using this approach we identified 58 probes that might predict pCR. Our predictive model built on those 58 probes from significant aberrant regions could be used to identify pCR with high accuracy (92%) and distinguish between patient samples with and without pCR with high specificity (97%). These are promising results suggesting that aCGH could be used to predict pCR to CRT. However, these studies need to be validated in an independent cohort of patients.
Several studies have demonstrated that genomic alterations can lead to corresponding gene expression changes in colorectal cancer using integrated gene expression profiling and CNA profiling (Lips et al., 2008; Camps et al., 2009). The CNAs covering large regions may affect the expression of one or more genes by altering gene levels, unmasking recessive alleles, disrupting the gene-coding sequence or promoting alternatively-spliced or fusion genes, and perturbing long-range gene regulation (Kleinjan and van Heyningen, 2005). While correlation of altered gene expression with CNA profile was not experimentally verified in our study, we used IPA to identify functionally relevant genes and pathways contained in the CNA regions that were associated with pCR. Among the eight candidate genes, we identified at least three, GAPDH, ENO2 and TPI1, that have been previously observed to be up-regulated in colorectal cancer (Yeh et al., 2008), and down-regulation of TNFRSF1B has been shown to predict chemosensitivity to 5-FU in colorectal cancer (Lin et al., 2007). However, to validate our results, we need to study the expression of the genes of interest that we identified to determine if in fact their expression is altered and has an effect on response to CRT.
There are a number of limitations to our study that deserve mention. While our aCGH analysis was based on the largest rectal cancer cohort reported thus far, the patient group was not completely homogeneous; there were two treatment arms in our study and while all patients received pre-operative CRT, some patients also received additional chemotherapy which may have influenced their clinical outcome. Additionally, as mentioned above, it will be important to validate the expression of the genes contained within the CNA regions in independent studies with fresh tissue to determine if indeed their expression profile is altered in patients, based on their response to CRT. Finally, we compared DNA from pre-treatment tumor to post-treatment DNA from normal surgical margins. This raises the issue of whether the post-treatment control tissue may have been affected by the CRT and additional neoadjuvant chemotherapy. However, the control tissue was obtained from the proximal resection margin, which is usually outside the radiation field and there is no data indicating that CNAs arise following chemotherapy treatment of normal tissue. Furthermore, we have recently shown that mutations in KRAS and TP53, two genes which play a role in the pathogenesis of rectal cancer, remain largely unchanged after CRT in these patients (Chen et al., 2011).
In conclusion, locally advanced rectal cancer has a specific pattern of CNAs that can now be analyzed using high-throughput techniques in tumor samples obtained during diagnostic biopsies. Some CNAs are specifically more common in tumors that respond to CRT compared to non-responsive tumors. These CNAs contain genes that belong to pathways involved in colorectal cancer response to therapy. This technology may therefore help to predict rectal cancer response to CRT with a high degree of accuracy, identifying both a subset of rectal cancer patients likely to achieve a pCR to CRT who may not need radical surgery, and similarly predicting which patients are likely to respond CRT, therefore potentially avoiding CRT-related toxicity in patients not expected to respond.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Nicola Solomon, PhD, for assistance in writing and editing the manuscript, and Karin Avila for specimen collection and study management.
Supported by: This study was supported by the National Institutes of Health (NIH), National Cancer Institute (NCI), R01 Grant CA090559 (JGA). ClinicalTrials.org Identifier: NCT00335816.
REFERENCES
- Bang H, Davidian M. Experimental statistics for biological sciences. Methods Mol Biol. 2010;620:3–104. doi: 10.1007/978-1-60761-580-4_1. [DOI] [PubMed] [Google Scholar]
- Benjamini YH, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Statistical Methodology. 1995;57:289–300. [Google Scholar]
- Camps J, Grade M, Nguyen QT, Hörmann P, Becker S, Hummon AB, Rodriguez V, Chandrasekharappa S, Chen Y, Difilippantonio MJ, Becker H, Ghadimi BM, Ried T. Chromosomal breakpoints in primary colon cancer cluster at sites of structural variants in the genome. Cancer Res. 2008;68:1284–1295. doi: 10.1158/0008-5472.CAN-07-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camps J, Nguyen QT, Padilla-Nash HM, Knutsen T, McNeil NE, Wangsa D, Hummon AB, Grade M, Ried T, Difilippantonio MJ. Integrative genomics reveals mechanisms of copy number alterations responsible for transcriptional deregulation in colorectal cancer. Genes Chromosomes Cancer. 2009;48:1002–1017. doi: 10.1002/gcc.20699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Duldulao MP, Li W, Lee W, Kim J, Garcia-Aguilar J. Molecular Diagnosis of Response to Neoadjuvant Chemoradiation Therapy in Patients with Locally Advanced Rectal Cancer. J Am Coll Surg. 2011 Mar 30; doi: 10.1016/j.jamcollsurg.2011.02.024. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccocioppo A, Stephens JH, Hewett PJ, Rieger NA. Complete pathologic response after preoperative rectal cancer chemoradiotherapy. ANZ J Surg. 2009;79:481–484. doi: 10.1111/j.1445-2197.2009.04950.x. [DOI] [PubMed] [Google Scholar]
- De Angelis PM, Clausen OP, Schjølberg A, Stokke T. Chromosomal gains and losses in primary colorectal carcinomas detected by CGH and their associations with tumour DNA ploidy, genotypes and phenotypes. Br J Cancer. 1999;80:526–535. doi: 10.1038/sj.bjc.6690388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Campagne F. Introduction to the development and validation of predictive biomarker models from high-throughput data sets. Methods Mol Biol. 2010;620:435–470. doi: 10.1007/978-1-60761-580-4_15. [DOI] [PubMed] [Google Scholar]
- Diep CB, Kleivi K, Ribeiro FR, Teixeira MR, Lindgjaerde OC, Lothe RA. The order of genetic events associated with colorectal cancer progression inferred from meta-analysis of copy number changes. Genes Chromosomes Cancer. 2006;45:31–41. doi: 10.1002/gcc.20261. [DOI] [PubMed] [Google Scholar]
- Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A. American Joint Committee on Cancer (AJCC) cancer staging manual. 7th ed. Chicago: Springer, Inc.; 2010. [Google Scholar]
- Gafà R, Maestri I, Matteuzzi M, Santini A, Ferretti S, Cavazzini L, Lanza G. Sporadic colorectal adenocarcinomas with high-frequency microsatellite instability. Cancer. 2000;89:2025–2037. [PubMed] [Google Scholar]
- Garcia-Aguilar J, Hernandez de Anda E, Sirivongs P, Lee SH, Madoff RD, Rothenberger DA. A pathologic complete response to preoperative chemoradiation is associated with lower local recurrence and improved survival in rectal cancer patients treated by mesorectal excision. Dis Colon Rectum. 2003;46:298–304. doi: 10.1007/s10350-004-6545-x. [DOI] [PubMed] [Google Scholar]
- Garcia-Aguilar J, Smith DD, Avila K, Bergsland EK, Chu P, Krieg RM Timing of Rectal Cancer Response to Chemoradiation Consortium. Optimal timing of surgery after chemoradiation for advanced rectal cancer: Preliminary results of a prospective trial. Ann Surg. 2011 doi: 10.1097/SLA.0b013e3182196e1f. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bio-conductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadimi BM, Grade M, Liersch T, Langer C, Siemer A, Füzesi L, Becker H. Gain of chromosome 8q23–24 is a predictive marker for lymph node positivity in colorectal cancer. Clin Cancer Res. 2003;9:1808–1814. [PubMed] [Google Scholar]
- Grade M, Gaedcke J, Wangsa D, Varma S, Beckmann J, Liersch T, Hess C, Becker H, Difilippantonio MJ, Ried T, Ghadimi BM. Chromosomal copy number changes of locally advanced rectal cancers treated with preoperative chemoradiotherapy. Cancer Genet Cytogenet. 2009;193:19–28. doi: 10.1016/j.cancergencyto.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hostetter G, Kim SY, Savage S, Gooden GC, Barrett M, Zhang J, Alla L, Watanabe A, Einspahr J, Prasad A, Nickoloff BJ, Carpten J, Trent J, Alberts D, Bittner M. Random DNA fragmentation allows detection of single-copy, single-exon alterations of copy number by oligonucleotide array CGH in clinical FFPE samples. Nucleic Acids Res. 2010;38:e9. doi: 10.1093/nar/gkp881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacopetta B, Russo A, Bazan V, Dardanoni G, Gebbia N, Soussi T, Kerr D, Elsaleh H, Soong R, Kandioler D, Janschek E, Kappel S, Lung M, Leung CS, Ko JM, Yuen S, Ho J, Leung SY, Crapez E, Duffour J, Ychou M, Leahy DT, O'Donoghue DP, Agnese V, Cascio S, Di Fede G, Chieco-Bianchi L, Bertorelle R, Belluco C, Giaretti W, Castagnola P, Ricevuto E, Ficorella C, Bosari S, Arizzi CD, Miyaki M, Onda M, Kampman E, Diergaarde B, Royds J, Lothe RA, Diep CB, Meling GI, Ostrowski J, Trzeciak L, Guzinska-Ustymowicz K, Zalewski B, Capellá GM, Moreno V, Peinado MA, Lönnroth C, Lundholm K, Sun XF, Jansson A, Bouzourene H, Hsieh LL, Tang R, Smith DR, Allen-Mersh TG, Khan ZA, Shorthouse AJ, Silverman ML, Kato S, Ishioka C TP53-CRC Collaborative Group. Functional categories of TP53 mutation in colorectal cancer: results of an International Collaborative Study. Ann Oncol. 2006;17:842–847. doi: 10.1093/annonc/mdl035. [DOI] [PubMed] [Google Scholar]
- Johnson NA, Hamoudi RA, Ichimura K, Liu L, Pearson DM, Collins VP, Du MQ. Application of array CGH on archival formalin-fixed paraffin-embedded tissues including small numbers of microdissected cells. Lab Invest. 2006;86:968–978. doi: 10.1038/labinvest.3700441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Jen J, Vogelstein B, Hamilton SR. Clinical and pathological characteristics of sporadic colorectal carcinomas with DNA replication errors in microsatellite sequences. Am J Pathol. 1994;145:148–156. [PMC free article] [PubMed] [Google Scholar]
- Kleinjan DA, van Heyningen V. Long-range control of gene expression: emerging mechanisms and disruption in disease. Am J Hum Genet. 2005;76:8–32. doi: 10.1086/426833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn WM, Yasutake T, Kuo WL, Warren RS, Collins C, Tomita M, Gray J, Waldman FM. Chromosome arm 20q gains and other genomic alterations in colorectal cancer metastatic to liver, as analyzed by comparative genomic hybridization and fluorescence in situ hybridization. Genes Chromosomes Cancer. 1999;25:82–90. doi: 10.1002/(sici)1098-2264(199906)25:2<82::aid-gcc2>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- Kuremsky JG, Tepper JE, McLeod HL. Biomarkers for response to neoadjuvant chemoradiation for rectal cancer. Int J Radiat Oncol Biol Phys. 2009;74:673–688. doi: 10.1016/j.ijrobp.2009.03.003. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lin YH, Friederichs J, Black MA, Mages J, Rosenberg R, Guilford PJ, Phillips V, Thompson-Fawcett M, Kasabov N, Toro T, Merrie AE, van Rij A, Yoon HS, McCall JL, Siewert JR, Holzmann B, Reeve AE. Multiple gene expression classifiers from different array platforms predict poor prognosis of colorectal cancer. Clin Cancer Res. 2007;13:498–507. doi: 10.1158/1078-0432.CCR-05-2734. [DOI] [PubMed] [Google Scholar]
- Lips EH, van Eijk R, de Graaf EJ, Oosting J, de Miranda NF, Karsten T, van de Velde CJ, Eilers PH, Tollenaar RA, van Wezel T, Morreau H. Integrating chromosomal aberrations and gene expression profiles to dissect rectal tumorigenesis. BMC Cancer. 2008;8:314. doi: 10.1186/1471-2407-8-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer GA, Hermsen MA, Baak JP, van Diest PJ, Meuwissen SG, Beliën JA, Hoovers JM, Joenje H, Snijders PJ, Walboomers JM. Progression from colorectal adenoma to carcinoma is associated with non-random chromosomal gains as detected by comparative genomic hybridization. J Clin Pathol. 1998;51:901–909. doi: 10.1136/jcp.51.12.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao K, Mehta KR, Fridlyand J, Moore DH, Jain AN, Lafuente A, Wiencke JW, Terdiman JP, Waldman FM. High-resolution analysis of DNA copy number alterations in colorectal cancer by array-based comparative genomic hybridization. Carcinogenesis. 2004;25:1345–1357. doi: 10.1093/carcin/bgh134. [DOI] [PubMed] [Google Scholar]
- Postma C, Koopman M, Buffart TE, Eijk PP, Carvalho B, Peters GJ, Ylstra B, van Krieken JH, Punt CJ, Meijer GA. DNA copy number profiles of primary tumors as predictors of response to chemotherapy in advanced colorectal cancer. Ann Oncol. 2009;20:1048–1056. doi: 10.1093/annonc/mdn738. [DOI] [PubMed] [Google Scholar]
- Ried T, Knutzen R, Steinbeck R, Blegen H, Schröck E, Heselmeyer K, du Manoir S, Auer G. Comparative genomic hybridization reveals a specific pattern of chromosomal gains and losses during the genesis of colorectal tumors. Genes Chromosomes Cancer. 1996;15:234–245. doi: 10.1002/(SICI)1098-2264(199604)15:4<234::AID-GCC5>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Samowitz WS, Holden JA, Curtin K, Edwards SL, Walker AR, Lin HA, Robertson MA, Nichols MF, Gruenthal KM, Lynch BJ, Leppert MF, Slattery ML. Inverse relationship between microsatellite instability and K-ras and p53 gene alterations in colon cancer. Am J Pathol. 2001;158:1517–1524. doi: 10.1016/S0002-9440(10)64102-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sing T, Sander O, Beerenwinkel N, Lengauer T. ROCR: visualizing classifier performance in R. Bioinformatics. 2005;21:3940–3941. doi: 10.1093/bioinformatics/bti623. [DOI] [PubMed] [Google Scholar]
- Smith FM, Reynolds JV, Miller N, Stephens RB, Kennedy MJ. Pathological and molecular predictors of the response of rectal cancer to neoadjuvant radiochemotherapy. Eur J Surg Oncol. 2006;32:55–64. doi: 10.1016/j.ejso.2005.09.010. [DOI] [PubMed] [Google Scholar]
- Soong R, Powell B, Elsaleh H, Gnanasampanthan G, Smith DR, Goh HS, Joseph D, Iacopetta B. Prognostic significance of TP53 gene mutation in 995 cases of colorectal carcinoma. Influence of tumour site, stage, adjuvant chemotherapy and type of mutation. Eur J Cancer. 2000;36:2053–2060. doi: 10.1016/s0959-8049(00)00285-9. [DOI] [PubMed] [Google Scholar]
- Srinivasan M, Sedmak D, Jewell S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am J Pathol. 2002;161:1961–1971. doi: 10.1016/S0002-9440(10)64472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixeira MR, Heim S. Multiple numerical chromosome aberrations in cancer: what are their causes and what are their consequences? Semin Cancer Biol. 2005;15:3–12. doi: 10.1016/j.semcancer.2004.09.006. [DOI] [PubMed] [Google Scholar]
- Tuefferd M, De Bondt A, Van Den Wyngaert I, Talloen W, Verbeke T, Carvalho B, Clevert DA, Alifano M, Raghavan N, Amaratunga D, Göhlmann H, Broët P, Camilleri-Broët S. Genome-wide copy number alterations detection in fresh frozen and matched FFPE samples using SNP 6.0 arrays. Genes Chromosomes Cancer. 2008;47:957–964. doi: 10.1002/gcc.20599. [DOI] [PubMed] [Google Scholar]
- van Beers EH, Joosse SA, Ligtenberg MJ, Fles R, Hogervorst FB, Verhoef S, Nederlof PM. A multiplex PCR predictor for aCGH success of FFPE samples. Br J Cancer. 2006;94:333–337. doi: 10.1038/sj.bjc.6602889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh CS, Wang JY, Chung FY, Lee SC, Huang MY, Kuo CW, Yang MJ, Lin SR. Significance of the glycolytic pathway and glycolysis related-genes in tumorigenesis of human colorectal cancers. Oncol Rep. 2008;19:81–91. [PubMed] [Google Scholar]
- Young J, Simms LA, Biden KG, Wynter C, Whitehall V, Karamatic R, George J, Goldblatt J, Walpole I, Robin SA, Borten MM, Stitz R, Searle J, McKeone D, Fraser L, Purdie DR, Podger K, Price R, Buttenshaw R, Walsh MD, Barker M, Leggett BA, Jass JR. Features of colorectal cancers with high-level microsatellite instability occurring in familial and sporadic settings: parallel pathways of tumorigenesis. Am J Pathol. 2001;159:2107–2116. doi: 10.1016/S0002-9440(10)63062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.