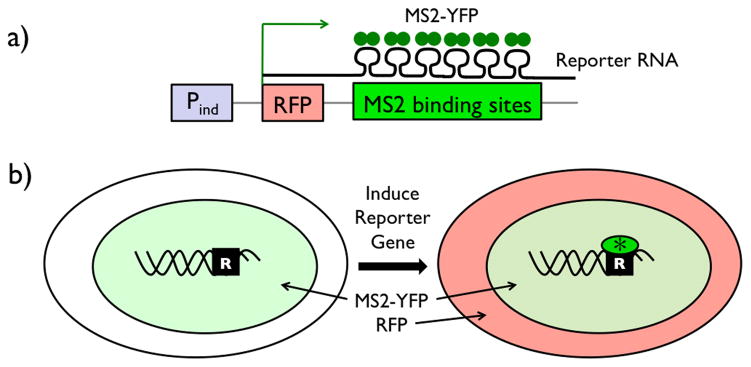
Abstract
The brain is a highly adaptable organ that is capable of converting sensory information into changes in neuronal function. This plasticity allows behavior to be accommodated to the environment, providing an important evolutionary advantage. Neurons convert environmental stimuli into long-lasting changes in their physiology in part through the synaptic activity-regulated transcription of new gene products. Since the neurotransmitter-dependent regulation of Fos transcription was first discovered nearly 25 years ago, a wealth of studies have enriched our understanding of the molecular pathways that mediate activity-regulated changes in gene transcription. These findings show that a broad range of signaling pathways and transcriptional regulators can be engaged by neuronal activity to sculpt complex programs of stimulus-regulated gene transcription. However, the shear scope of the transcriptional pathways engaged by neuronal activity raises the question of how specificity in the nature of the transcriptional response is achieved in order to encode physiologically relevant responses to divergent stimuli. Here we summarize the general paradigms by which neuronal activity regulates transcription while focusing on the molecular mechanisms that confer differential stimulus-, cell-type-, and developmental-specificity upon activity-regulated programs of neuronal gene transcription. In addition, we preview some of the new technologies that will advance our future understanding of the mechanisms and consequences of activity-regulated gene transcription in the brain.
Keywords: neuron, activity-regulated transcription, calcium signaling
1. Introduction
Stimulus-regulated transcription factors play an essential role in cell biology by coupling extracellular stimuli to coordinated intracellular responses. The basic principles of regulated transcription first emerged from the study of growth-factor dependent induction of Fos transcription in fibroblasts (Cochran et al., 1984; Greenberg and Ziff, 1984; Müller et al., 1984). Addition of growth factors promotes cell division, and some of the first growth factor-induced genes to be identified (i.e. Fos and Myc) were known proto-oncogenes, suggesting a role for this transcriptional program in mitogenesis (Stiles, 1985). Thus, it came as something of a surprise when Fos transcription was shown to be induced in neuroendocrine cells by stimuli including Nerve Growth Factor (NGF), which promotes differentiation, not division (Curran and Morgan, 1985; Greenberg and Ziff, 1984). The regulation of Fos transcription by neurotransmitters was quickly generalized to post-mitotic neurons of the central nervous system (CNS), where Fos transcription is robustly induced following seizure (Morgan et al., 1987). These exciting results suggested the possibility that changes in neuronal activity levels might be capable of exerting long-lasting effects on neuronal gene expression, which could in turn influence various aspects of a given neuron’s life and metabolism.
An important breakthrough in the understanding of neuronal activity-regulated transcription came from a series of studies that used differential cloning strategies to screen for activity-induced mRNAs (Hevroni et al., 1998; Lanahan and Worley, 1998; Nedivi et al., 1993; Qian et al., 1993). These findings revealed that many of the genes induced by activity were selectively expressed in neurons and furthermore that some had known functions in neuronal or synaptic physiology (see review by Leslie and Nedivi, this issue). More recently, increasingly comprehensive screens have described complex patterns of gene transcription induced and/or repressed following different kinds of stimuli that act in concert to effect adaptations upon neuronal and synaptic physiology (Flavell et al., 2008; Guan et al., 2005; Kim et al., 2010; Xiang et al., 2007; Zhang et al., 2007). A key theme to emerge from these studies is that of specificity, meaning that different kinds of stimuli up- and down-regulate distinct sets of genes (Bading et al., 1993; Bartel et al., 1989; Hardingham et al., 2002). A major focus of the research effort directed at activity-regulated gene transcription has been to identify the molecular mechanisms that underlie this specificity. These findings have enhanced understanding of how neurons use information from the environment to properly and constantly adapt their development and plasticity to the world outside.
The goal of this review is to summarize major molecular advances that have been made in the understanding of neuronal activity-regulated gene transcription to date, and to describe where the field is heading in the future. We will 1) review the classes of genes and transcription factors that are regulated by neuronal activity, 2) describe how calcium signaling pathways differentially modulate transcription factors to confer specificity upon cellular responses to stimuli, 3) discuss the importance of activity-regulated transcriptional pathways in synaptic homeostasis, and finally 4) describe some of the technological developments that will advance future studies in this field. Because we focus on the idea of signaling specificity at many stages along the synapse to nucleus pathway, we do not cover every step in equal detail. Alternative reviews that offer additional detail about key steps in these pathways will be cited in the course of the discussion as they arise.
2. Transcriptional control of neuronal activity-regulated genes
Many studies have characterized the programs of gene transcription that are induced in response to altered activity states (Guan et al., 2005; Hevroni et al., 1998; Lanahan and Worley, 1998; Nedivi et al., 1993; Qian et al., 1993; Zhang et al., 2007). Although these gene expression programs vary by brain region and stimulus type, there are some important commonalities. Specifically, these studies have shown that there are two particularly important classes of genes that are regulated by neuronal activity: 1) classic immediate-early gene transcription factors, which are general response factors in a broad range of cell types and 2) neuronally-enriched gene products with specific functions at synapses. The evidence that neural activity can change the expression of synaptic gene products suggests that study of this second class in particular may reveal neural-selective mechanisms of transcription that are of particular importance for synaptic plasticity. Here we review the transcriptional mechanisms that regulate key genes in these two classes to exemplify the molecular pathways that differentially control stimulus-regulated gene transcription in neurons.
2.1. Fos: the archetype
In fibroblasts, Fos is only one of a set of genes whose transcription is rapidly induced by growth factors. Since induction of this transcriptional program proceeds without the need for prior protein synthesis, these genes were termed “immediate early genes” (IEGs) in analogy to the protein synthesis-independent gene expression program that underlies viral oncogenesis (Lau and Nathans, 1987). IEGs include members of the Fos, Jun, early growth response (Egr), and nuclear receptor (Nr) families of transcription factors. In addition to Fos, many of the classic growth factor regulated IEGs are part of the neural activity-dependent transcriptional program including Fosb, Fosl1, Fosl2, Jun, Junb, Egr1 (also termed zif/268; Ngfi-a; Krox-24), Egr3, and Nr4a1 (also termed nur77, Ngfi-b) (Herdegen and Leah, 1998). These genes encode transcription factors that are induced in many cells types in response to a broad range of stimuli. Thus, rather than having a specific cellular function, the IEG program appears to play a more general role in coupling extracellular stimuli with intracellular adaptations. The different functional consequences of IEG induction arise through cell type- and stimulus-specific recruitment of complexes of these transcription factors to distinct sets of late response genes promoters (Hill and Treisman, 1999). In the CNS, IEG transcription factors have been shown to contribute to diverse processes, including neurite outgrowth, neurotransmitter fate, and synapse plasticity (Dragunow et al., 2000; Li et al., 2007; Marek et al., 2010; Maze et al., 2010).
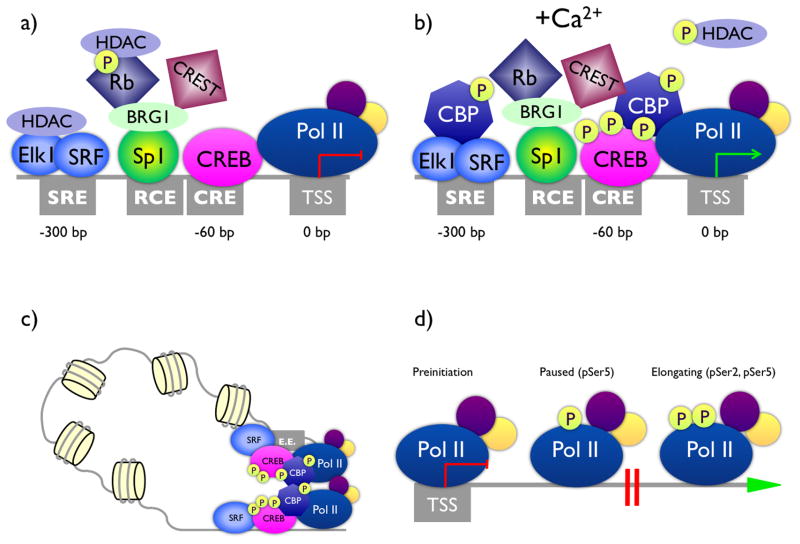
Because Fos was the first of the activity-regulated genes to be identified, and because it is so widely and strongly induced, considerable effort has been devoted to understanding the molecular mechanisms that regulate its transcription. The details of Fos transcription exemplify many of the general principles of activity-dependent transcription, and thus will be described in detail here. Specifically, Fos induction has been shown to be dependent upon 1) the association of sequence-specific DNA binding transcription factors with stimulus-response elements in the proximal promoter, 2) the stimulus-dependent recruitment to these transcription factors of transcriptional co-activators and co-repressors that post-translationally modify promoter-associated histones, 3) the modulation of promoter function by distant enhancer elements, and 4) the regulation of transcriptional elongation (FIGURE 1).
FIGURE 1. Mechanisms of neuronal activity-induced transcription.
The diagrams represent four steps in the process of Fos transcription that are regulated by neural activity. a) Prior to neuronal activity, the Fos promoter is primed for response by the association of sequence-specific DNA binding transcription factors with stimulus-response elements (gray boxes) in the proximal promoter. RNA polymerase II (Pol II) is also pre-associated with the Fos promoter prior to activation. Numbers show base pair distances of each element from the transcription start site (TSS). The promoter is kept off in part by the local recruitment of HDACs. Key sites of calcium-regulated phosphorylation are represented by the circled letter P. b) Calcium induces a switch in cofactors present at the Fos promoter, with recruitment of the co-activator and histone acetyltransferase CBP and loss of the repressive HDACs. c) Distal enhancer elements (E.E.) contribute to activity-dependent transcription. These elements are bound by transcription factors including SRF and CREB, and show calcium-dependent recruitment of CBP and Pol II binding. Chromatin looping may bring the enhancer into physical proximity of the proximal promoter and Fos TSS. d) Transcription elongation is regulated by stimulus-dependent phosphorylation of two sites in the C-terminal domain of the large subunit of RNA polymerase II. The pre-initiation form of RNA Pol II is bound to the Fos promoter but is not competent to drive RNA synthesis. Phosphorylation of serine 5 (pSer5) is sufficient to promote engagement but results in polymerase stalling within the transcribed region of many genes. Productive elongation requires additional phosphorylation of RNA Pol II at serine 2 (pSer2).
Mutagenesis of the Fos promoter revealed two elements that are differentially required for promoter activity in response to a variety of stimuli (Sheng et al., 1990). One element about 300bp upstream of the Fos transcription start site (TSS) is required for serum- and growth factor-dependent induction of Fos and was therefore named the Serum Response Element (SRE) (Sheng et al., 1988; Treisman, 1986). A second element, about 60bp upstream of the TSS is required for calcium- and cAMP-dependent regulation of Fos and was named the Calcium/cAMP-Response Element (CRE) (Hyman et al., 1988; Sheng et al., 1988). Finally, a distinct element just 5′ to the CRE is required for regulation of Fos transcription by the retinoblastoma tumor suppressor (Rb) and was named the Retinoblastoma Control Element (RCE) (Udvadia et al., 1992). DNA elements control transcription because they are binding sites for transcription factors, thus once identified the elements are powerful tools for purifying or cloning the binding proteins. In the case of the SRE, the binding protein was called the Serum Response Factor (SRF) (Norman et al., 1988). Subsequent studies showed that SRF binds the SRE in cooperation with additional proteins called the Ternary Complex Factors (TCF), which are a subfamily of E-twenty six (Ets) domain-containing transcription factors exemplified by Ets like gene 1 (Elk-1) (Buchwalter et al., 2004; Dalton and Treisman, 1992). CRE binding proteins were named the Calcium-Response Element Binding protein (CREB) family (Montminy and Bilezikjian, 1987). The RCE is bound by the zinc-finger transcription factor Sp1 (Udvadia et al., 1993).
Even prior to neural activity, all of these transcription factors as well as the preinitiation form of the RNA polymerase II complex are bound to the Fos promoter (Kim et al., 2010; Sheng et al., 1988). The primed state of this promoter is likely to be important for the very rapid induction of Fos transcription by stimuli. However, this state also implies that active repression of the promoter may be required in the absence of a stimulus to prevent Fos from being constitutively transcribed.
Genomic DNA is wound into a complex secondary and tertiary structure called chromatin via its interactions with structural proteins. The basic repeating unit of chromatin in the nucleosome, which consists of 147bp of DNA wound around an octamer of histone proteins: 2 each of histones H2A, H2B, H3, and H4 (Horn and Peterson, 2002). These histones are subject to extensive post-translational modifications (e.g. acetylation, methylation, phosphorylation, ubiquitination) at specific amino acid residues in their flexible N-terminal tails. The observed correlation between specific modifications and transcriptional activity has given rise to the hypothesis of a “histone code” that determines the transcriptional state of a gene (Strahl and Allis, 2000). Although the details of this relationship between histone modifications and transcription remains an active area of investigation (Lee et al., 2010), substantial evidence suggests that stimulus-dependent regulation of histone H3 and H4 acetylation is a major mechanism of promoter activation and repression (Bernstein et al., 2007; Clayton et al., 2006; Roh et al., 2005). In the case of Fos, one mechanism of promoter repression is via the RCE, which recruits a protein complex consisting of Sp1, the transcriptional co-repressor Rb, and the scaffolding protein Calcium-Responsive Transactivator (CREST). In the absence of synaptic activity, Rb is bound to histone deacetylases (HDACs), which enzymatically remove acetyl groups from the histones surrounding the Fos TSS, repressing transcription (Qiu and Ghosh, 2008). HDACs can also be recruited to the SRF-binding protein Elk-1, although the functional requirement for this interaction at the Fos gene is not known (Yang et al., 2002). Following neuronal activity, calcium-dependent signaling events drive the dissociation of the HDACs from both Rb and Elk, and also induce the active export of class II HDACs (HDAC4 and HDAC5) from the nucleus (Chawla et al., 2003; Qiu and Ghosh, 2008; Yang and Sharrocks, 2006). Simultaneously, signaling cascades lead to the recruitment of the histone acetyltransferase (HAT) CREB-binding protein (CBP) to both CREB and CREST, acetylating promoter histones and promoting transcription (Chrivia et al., 1993; Impey et al., 2002; Qiu and Ghosh, 2008).
In addition to these local events at the Fos promoter, membrane depolarization of neurons is associated with widespread, activity-induced recruitment of both CBP and RNA polymerase II to enhancer elements that neighbor Fos and other activity-regulated genes (Kim et al., 2010). Biochemically, enhancers have been defined across the genome as regions that show a characteristic chromatin signature that is enriched for histone H3 lysine 4 monomethylation and that shows hypersensitivity to cleavage with enzyme DNAseI (Heintzman et al., 2007). Functionally, enhancers are defined as any DNA element that promotes transcription of a given gene regardless of its location in the genome or its orientation with respect to the regulated gene. Enhancers can be found at great distances from the TSS, as is well documented for regulation of the β-globin locus and the Hoxd gene cluster (Hérault et al., 1999; Tuan et al., 1985). How distant enhancers regulate promoter activity remains an active area of investigation, however these mechanisms may involve looping of chromatin to bring the enhancers in close proximity to the gene promoter (Li et al., 2006a), and/or large scale chromosome reorganization that brings multiple actively transcribed regions of chromatin to pre-formed transcriptional “factories” near the nuclear periphery (Sutherland and Bickmore, 2009). Similar to promoter regulatory elements, many enhancer elements are pre-bound by activity-regulated transcription factors including SRF, CREB, and Myocyte Enhancer Factor 2 (MEF2), which may act to recruit CBP (Flavell et al., 2008; Kim et al., 2010). Interestingly, activity-dependent RNA polymerase II recruitment to enhancers is associated with the induced expression of short, non-coding enhancer RNA transcripts (eRNAs) that initiate at these non-coding elements (Kim et al., 2010). Although the functions of eRNAs are not known, they may play a role in recruiting chromatin modifying enzymes to maintain the chromatin landscape in a transcriptionally permissive state.
Finally, in addition to the regulation of transcriptional initiation, evidence suggests that Fos transcription may also be regulated at the level of transcriptional elongation. Productive transcriptional elongation is an active process that requires dynamic CDK9/pTEFb-dependent phosphorylation of amino acids in the C-terminal domain of the large subunit of RNA polymerase II (Buratowski, 2009). For at least a subset of genes, regulation of RNA polymerase II phosphorylation causes the polymerase complex to undergo promoter proximal stalling following initiation until signal-dependent elongation is cued (Nechaev et al., 2010). Consistent with the possibility that Fos transcription may be subject to regulation of elongation, nuclear run-on assays of Fos transcription in quiescent fibroblasts show that these cells have constitutive transcription of short 5′ transcripts that fail to extend past a Fos intragenic regulatory element (FIRE) near the end of exon I prior to serum stimulation (Lamb et al., 1990). Genome-level chromatin immunoprecipitation (ChIP) studies have begun to address the localization and phosphorylation state of the polymerase subunits at large sets of activity-regulated genes (Kim et al., 2010), and further studies of this kind are likely to enhance understanding of the relative importance of the regulation of transcriptional elongation for neuronal activity-dependent changes in mRNA expression.
2.2. Brain-Derived Neurotrophic Factor (Bdnf): promoter complexity
BDNF is a secreted protein of the neurotrophin family that has numerous functions in nervous system development and plasticity (Lewin, 1996; Poo, 2001). Exposure to a wide range of environmental stimuli leads to induced expression of Bdnf mRNA in corresponding activated brain regions, suggesting that the induction of Bdnf transcription may be a common mechanism for environmental modulation of neural function (Lu, 2003). Neural activity also regulates both the local secretion of BDNF protein and the trafficking of the BDNF receptor TrkB to the plasma membrane, allowing BDNF-TrkB signaling to be tightly linked to activity (Balkowiec and Katz, 2000; Meyer- Franke et al., 1998). Mutations that partially reduce either overall levels of BDNF or BDNF secretion have substantial effects on brain development and plasticity (Chen et al., 2006; Egan et al., 2003; Genoud et al., 2004). Taken together, these data suggest that precise temporal and spatial control of BDNF expression is essential to its function.
The mammalian Bdnf gene is comprised of nine exons with at least eight alternative promoters that are differentially used during development, across brain regions, and in different cell types (Aid et al., 2007; Liu et al., 2006). All of the Bdnf promoters are active to at least some degree in the CNS, and transcription from each of these promoters can be induced by neuronal activity (Aid et al., 2007). Promoter IV (formerly referred to as “promoter III” in the five-exon nomenclature of the Bdnf gene, see (Timmusk et al., 1993)) is strongly activity-responsive in cultured embryonic neurons and has been the most extensively studied at the molecular level (Shieh et al., 1998; Tao et al., 1998; West et al., 2001; Pruunsild et al., 2011).
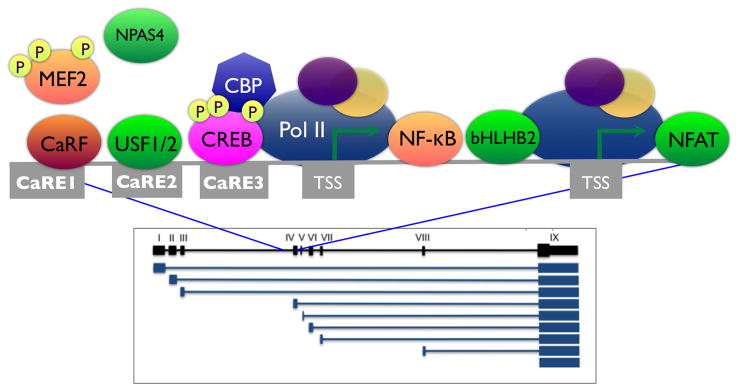
One of the key insights to be derived from studies of Bdnf promoter IV is that the tight temporal, spatial, and stimulus-specific regulation of this single promoter is achieved by a complex interplay between multiple activity-regulated transcriptional pathways (FIGURE 2). Three calcium-response elements (CaREs) within the proximal 170bp of the major embryonic promoter IV TSS cooperatively regulate calcium-induced transcription of Bdnf exon IV (Chen et al., 2003b; Shieh et al., 1998; Tao et al., 1998). Starting at the element most distal to the TSS, these CaREs are selectively bound and regulated by the unique transcription factor Calcium-Response Factor (CaRF) (Tao et al., 2002), the upstream stimulatory factors USF1/2 (Chen et al., 2003b), and members of the CREB family (Shieh et al., 1998; Tao et al., 1998). In addition, the activity-inducible transcription factor Neuronal Per-Arnt-Sim (PAS) Domain Protein 4 (Npas4) has been shown to interact with another activity-response element 5′ to CaRE1 in human Bdnf promoter IV (Pruunsild et al., 2011). An alternative TSS about 100bp downstream of the 5′-most end of Bdnf exon IV is regulated by the association of the Nuclear Factor κB (NF-κB) (Lipsky et al., 2001) and the basic helix-loop-helix (bHLH) protein bHLHB2 (Jiang et al., 2008) with elements that fall between the two TSSs. The Nuclear Factor of Activated T Cells (NFAT) is reported to regulate Bdnf exon IV expression in response to N-methyl-D-aspartate (NMDA) receptor (NMDAR) activation via its association with an intragenic element located 140–156bp 3′ to the alternative TSS (Vashishta et al., 2009). Finally, ChIP studies have shown that Bdnf promoter IV is bound by MEF2 (Hong et al., 2008), however the specific binding site for this factor within Bdnf promoter IV remains to be identified.
FIGURE 2. Multiple activity-regulated transcription factors contribute to inducibility of Bdnf promoter IV.
The box shows spliced mRNAs (blue) encoding Bdnf transcripts driven by the eight alternative Bdnf promoters mapped onto chromosome 2 of Mus musculus (black). Boxes represent the nine exons that comprise the Bdnf gene and the thicker region of exon IX indicates the coding sequence. The gray line shows an expansion of the region just upstream of exon IV. Three calcium-response elements (CaREs) and two transcription start sites (TSSs) are indicated by the gray boxes. Transcription factors demonstrated to regulate Bdnf promoter IV are shown at their binding sites. Npas4 binding has been localized to a PAS response element just 5′ to CaRE1 in human BDNF promoter IV (Pruunsild et al., 2011) and to a region near the CaRE2 element by ChIP-Seq in mouse neurons (Kim et al., 2010). Although MEF2 has been localized to Bdnf promoter IV by chromatin immunoprecipitation, its binding elements have not yet been reported.
Distinct functions of these transcription factors in the regulation of Bdnf promoter IV have been revealed through the generation of transgenic mice that either lack expression of one of these transcription factors or that block the ability of specific factors to regulate Bdnf. For example, studies in mice lacking the CaRE1-binding protein CaRF show that this factor appears to play a brain region-specific role in regulation of Bdnf transcription (McDowell et al., 2010). Carf knockout mice show reduced levels of Bdnf exon IV-containing mRNA transcripts and reduced BDNF protein in the frontal cortex compared with their wildtype littermates, however Bdnf expression is unchanged in the hippocampus and striatum of the knockout mice (McDowell et al., 2010). Furthermore, although CaRE1 is required for activity-dependent transcription of Bdnf exon IV, CaRF is selectively required for the activity-independent regulation of Bdnf promoter IV activity (McDowell et al., 2010). The failure of the Carf knockout to fully phenocopy the loss of the CaRE1 site implies that there likely exist additional CaRE1 binding proteins that are yet to be identified. By contrast with CaRF, the binding of CREB to CaRE3 is selectively required for the activity-dependent regulation of Bdnf exon IV transcription. The functional importance of this interaction was elegantly demonstrated by generation of a mouse strain bearing a mutation knocked into Bdnf promoter IV that selectively mutates the CRE/CaRE3 site (Hong et al., 2008). Neurons from CaRE3 mutant mice have normal basal levels of BDNF but lack activity-inducible transcription from promoter IV, validating the requirement for this CaRE in activity-dependent Bdnf gene regulation in vivo. Interestingly, disruption of CaRE3 is associated with impaired Bdnf promoter IV recruitment of other transcriptional regulators including MEF2, which binds to a DNA sequence distinct from CaRE3. These data provide experimental support for the role of a multifactor transcriptional complex at Bdnf promoter IV, and suggest a function for CREB in nucleating the assembly of this complex.
2.3. Activity-Regulated Cytoskeletal Protein (Arc): signal pathway integration
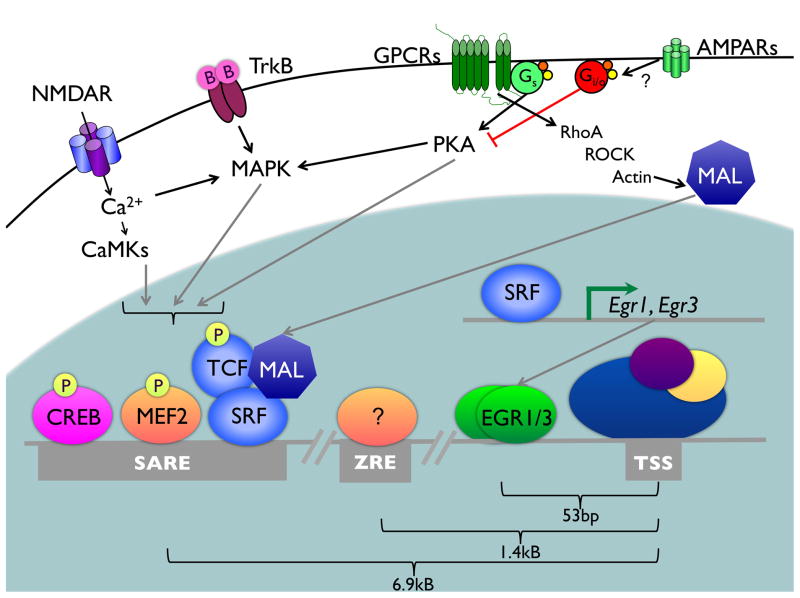
Arc (also known as Arg3.1) was first identified in 1995 as a novel IEG that is rapidly induced in neurons following increased activity (Link et al., 1995; Lyford et al., 1995). Stimulus-inducible transcription of Arc depends on elements within the proximal promoter as well as distal enhancer elements (FIGURE 3). The proximal promoter is not sufficient to confer acute cAMP- or calcium-inducible regulation on a reporter plasmid (Waltereit et al., 2001). However, this region is bound in vivo by the activity-induced transcription factors Egr1 and Egr3 and overexpression of Egr1 or Egr3 is sufficient to drive endogenous Arc expression (Li et al., 2005). Analysis of Arc mRNA expression in Egr1 and Egr3 knockout mice suggests that these transcription factors may function to prolong Arc transcription following its acute induction. These studies showed that although acute stimulus-induced expression of Arc mRNA does not require new protein synthesis, the persistent expression of Arc mRNA 4 hours after kainate-induced seizure is blocked by administration of protein synthesis inhibitors, and this late phase of Arc expression is significantly diminished in Egr3 knockout mice (Li et al., 2005). Intriguingly, the acute induction of Arc transcription by synaptic activity requires distal enhancer elements located several thousand base pairs upstream of the Arc TSS (Kawashima et al., 2009; Pintchovski et al., 2009). The Synaptic Activity Response Element (SARE) located 6.9kB upstream of the TSS is highly conserved across mammalian species, and is comprised of a cluster of binding sites for the activity-regulated transcription factors CREB, MEF2, and SRF (Kawashima et al., 2009). The importance of this regulatory region for the activity-dependent regulation of Arc was elegantly demonstrated in cerebellar Purkinje cells, in an experiment in which the expression of Arc in Arc knockout neurons was rescued by transgenic expression from a BAC construct containing the intact upstream enhancer region, but not by expression from a BAC construct in which the SRE in the SARE had been mutated to a sequence that does not support SRF binding (Smith-Hicks et al., 2010).
FIGURE 3. Convergent signaling cascades regulate Arc transcription.
Signaling cascades that promote (gray arrows) and inhibit (red bars) the transcription of Arc are shown. The light blue region indicates the cell nucleus. B, BDNF; TrkB, the BDNF receptor TrkB; GPCRs, G-protein coupled receptors; Gi/o, heterotrimeric G proteins negatively coupled to adenylate cyclase; Gs, heterotrimeric G proteins positively coupled to adenylate cyclase; ROCK, Rho kinase. Three genetic regulatory regions have been identified in the proximal and distal Arc promoter regions. In addition to the distal SARE element (bound by CREB, SRF, and MEF2) and the proximal promoter (bound by EGR1/3), a region of open chromatin 1.4kB upstream of the TSS has been identified that has homology to a Zeste-like response element (ZRE) and that contributes to activity-dependent regulation of an Arc reporter gene (Pintchovski et al., 2009). The transcription factor(s) that regulate this element in mammalian neurons remain to be identified.
Transcription of Arc is regulated through the integration of multiple synaptic signaling cascades. As a result of the conversion of these signaling pathways, both the levels and distribution of Arc are particularly sensitive to changes in synaptic activity. An important early finding was the observation that although forskolin-induced elevation of intracellular cAMP can drive induction of Arc in the neuroendocrine PC12 cell line in a manner that depends on the protein kinase A (PKA) and mitogen-activated protein kinase (MAPK) signaling pathways, this stimulus is not sufficient to induce Arc in the neuroblastoma line Neuro2a or in NIH3T3 fibroblasts (Waltereit et al., 2001). These data raised the possibility that cAMP signaling might interact with additional neural-selective signaling pathways to regulate Arc transcription. Subsequent studies have suggested that calcium influx through NMDARs can provide the second signal. For example, treatment of neuron cultures with the growth factor BDNF drives robust Arc induction, however this induction is dependent upon synaptic activity and inhibited by antagonists of NMDARs (Rao et al., 2006). Interestingly, BDNF-dependent Arc induction is antagonized by α-amino-3-hydroxyl-5-methyl-4-isoxazole-proprionate (AMPA) receptor (AMPAR) activation, through a mechanism that involves pertussis toxin–sensitive Gi/o proteins (Rao et al., 2006). Further support for coactivation of NMDAR and cAMP pathways in the control of Arc transcription comes from the observation that the cAMP-dependent activation of Arc induced by agonists of Gs-coupled dopamine and beta-adrenergic receptors is blocked in the presence of NMDAR antagonists (Bloomer et al., 2008). However, the specific point at which NMDARs and G-protein coupled signaling pathways converge to regulate Arc transcription remains to be characterized.
2.4. Homer1a: alternative polyadenylation
The Homer family of synaptic scaffolding proteins is comprised of three genes (Homer1-3), each of which has multiple splice isoforms (Bottai et al., 2002). Only the Homer1 gene shows neuronal activity-induced transcription (Brakeman et al., 1997). This gene is comprised of 10 exons spanning over 100kB in the mouse with alternative exon usage giving rise to two full length splice variants called Homer1b and Homer1c (Bottai et al., 2002). Neuronal activity sharply increases the rate of transcriptional initiation at the Homer1 promoter, however the activity-induced transcripts from the Homer1 gene (Homer1a and Ania-3) contain only exons 1–5 and part of intron 5. The short forms of Homer1 lack a C-terminal oligomerization domain and are thought to act in a dominant negative fashion to disrupt the formation or function of synaptic signaling complexes (Kammermeier and Worley, 2007; Sala et al., 2003). These short splice variants are generated through the use of alternative polyadenylation sites within intron 5 of the Homer1 gene (Bottai et al., 2002; Niibori et al., 2007). Interestingly, an mRNA profiling study that analyzed changes in exon usage following neuronal membrane depolarization identified a large number of additional genes that show similar activity-dependent alternative polyadenylation site usage (Flavell et al., 2008). In many of these cases, recognition of the intragenic polyadenylation site leads to the expression of a truncated gene product that lacks functional C-terminal domains. These data raise the possibility that alternative polyadenylation site usage may be a common mechanism to modulate the function of proteins in a neural activity-dependent manner.
3. Transcription factors regulated by neuronal activity
Activity-regulated signaling pathways modulate gene transcription by altering the function, localization, or expression of transcriptional factors in the nucleus (West et al., 2002). A growing number of transcriptional regulators have been shown to be targets of activity-dependent signaling cascades in neurons (FIGURE 4). Here we describe the most well-studied activity-regulated transcription factors in neurons, and review the mechanisms of their regulation.
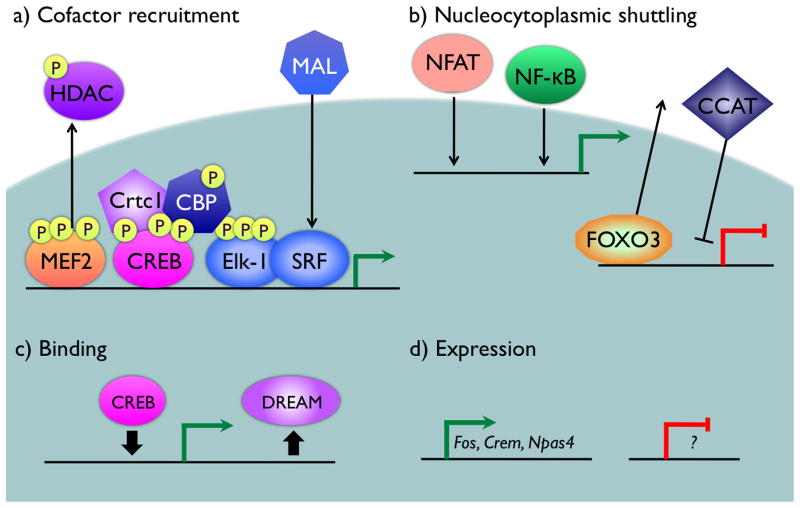
FIGURE 4. Mechanisms of transcription factor regulation by neuronal activity.
Transcription factor function can be regulated by neuronal activity through at least four different mechanisms. Green arrows represent genes that show activity-induced transcriptional activation, red bars represent genes that undergo activity-induced transcriptional repression. a) Recruitment of transcriptional coactivators and corepressors is an important mechanism to regulate the function of pre-bound transcription factors such as CREB, MEF2, and SRF. b) Nuclear translocation of the transcription factors NFAT and NF-κB is induced by a wide variety of stimuli including neuronal activity, allowing these factors to bind to their target gene promoters. By contrast, neuronal activity drives the nuclear export of the forkhead box transcription factor FOXO3 and prevents nuclear translocation of CCAT. c) Regulated binding of transcription factors is controlled through different mechanisms. In the case of CREB, signal-dependent regulation of histone modifying enzymes may lead to a change chromatin structure that alters the accessibility of CREB binding sites. For DREAM, direct binding of calcium to the EF-hands in this repressor protein changes its affinity for DNA, releasing it from its binding site. d) In addition to the classic IEGs transcription factors, such as Fos, Jun, and Egr family members, additional transcription factors are subject to activity-dependent regulation of expression. These transcription factors include ICER, a repressor form of the CREB family member CREM, and the bHLH-PAS domain transcription factor NPAS4. Expression of other transcription factors is likely to be repressed by neuronal activity, however these remain to be identified.
3.1. Activation of prebound transcription factors
One of the most striking features of neuronal activity-regulated transcription is its very rapid initiation. Newly synthesized nuclear RNA transcripts can be detected within one minute of stimuli that induce calcium influx suggesting that some gene promoters are primed for rapid response (Greenberg et al., 1986). At these promoters, such as Fos, transcriptional activators are prebound before stimulation arrives (Kim et al., 2010). Activity-dependent signaling pathways are required to post-translationally modify these factors and/or their associated proteins to change their activity state.
3.1.1. cAMP-Response element binding protein (CREB)
The CREB family of transcription factors is comprised of Creb, cAMP Response Element Modulatory Protein (Crem), and activating transcription factor 1 (Atf1). In the CNS, CREB family members have been shown to be essential for neuronal survival and are thought to modulate both synaptic and intrinsic plasticity in response to neuronal activity (Benito and Barco, 2010; Lonze and Ginty, 2002). Loss-of-function genetic studies show that CREB and CREM have overlapping functions in the developing and mature CNS, whereas ATF1 plays particularly important roles early in development (Bleckmann et al., 2002; Mantamadiotis et al., 2002). All stimuli that activate neuronal CREB-dependent transcription (e.g., receptor tyrosine kinases, calcium signaling pathways, cAMP) do so by inducing phosphorylation of CREB at serine 133 (Ser133) (Shaywitz and Greenberg, 1999). Distinct Ser133 kinases including PKA, Ca2+/calmodulin (CaM)-dependent protein kinases (CaMKs), MAPK, and Akt (also known as protein kinase B, PKB) mediate phosphorylation of CREB in response to different stimuli in different cell types (Gonzalez and Montminy, 1989; Lonze and Ginty, 2002; Mayr and Montminy, 2001; Sheng et al., 1991). In neurons, CaMKs mediate rapid but transient phosphorylation of CREB following membrane depolarization, whereas MAPKs make a major contribution to later, sustained CREB phosphorylation (Dolmetsch et al., 2001; Wu et al., 2001a). Phosphorylation of CREB at Ser133 activates transcription by inducing the association of CREB with the transcriptional coactivators CBP and p300, which promote transcription by acetylating promoter histones (Chrivia et al., 1993; Goodman and Smolik, 2000). CREB can also bind directly to elements of the RNA polymerase complex in a phosphorylation-independent manner, which may allow it to recruit the pre-initiation form of RNA polymerase II onto the promoters of primed IEGs such as Fos (Felinski et al., 2001; Hong et al., 2008).
In addition to phosphorylation of Ser133, activation of neuronal calcium signaling pathways induces phosphorylation of CREB at Ser142 and Ser143 (Kornhauser et al., 2002). These phosphorylation events occur concurrently with activation of CREB-dependent transcription, and mutation of these serines to non-phosphorylatable alanine residues selectively inhibits calcium-regulated CREB activity in a reporter gene assay, while leaving cAMP-dependent transcription unaffected (Kornhauser et al., 2002). Consistent with a role for these phosphorylation sites in regulation of CREB-dependent transcription in vivo, phosphorylation of CREB Ser142 is induced in neurons of the suprachiasmatic nucleus by light exposure, and mice bearing a mutation that changes Ser142 to alanine knocked into the Creb1 gene show altered circadian rhythms (Gau et al., 2002). Interestingly, phosphorylation of CREB at Ser142/143 inhibits the ability of Ser133 phosphorylated CREB to bind to the KID-interacting domain (KIX) domain of CBP (Parker et al., 1998). This raises the possibility that activation of calcium signaling pathways may selectively promote the association of CREB with other transcriptional coactivators than just CBP/p300. Alternate coactivators include the CREB-regulated transcription coactivators (Crtc1-3, previously known as TORCs). The Crtcs are CREB coactivators that bind the C-terminal leucine zipper domain of CREB, a domain distinct from the Ser133-containing kinase-inducible domain (KID) domain bound by CBP/p300 (Conkright et al., 2003). Under basal conditions, Crtc1 and Crtc2 are sequestered in the cytoplasm through a phosphorylation-dependent interaction with the chaperone protein 14-3-3 (Screaton et al., 2004). Calcium-dependent activation of the phosphatase calcineurin combined with cAMP-dependent inhibition of the AMP-activated protein kinase (AMPK) family of Ser/Thr kinases salt-inducible kinase 2 (SIK2), leads to dephosphorylation and nuclear import of Crtc1 and 2, where they co-activate CREB-dependent transcription (Kovács et al., 2007; Sasaki et al., 2011; Screaton et al., 2004). Consistent with a role for these CREB co-activators in neuronal biology, absence of Crtc1 in neurons impairs the late phase of long-term potentiation (LTP), BDNF-dependent dendrite outgrowth, and neuronal survival after ischemic injury (Finsterwald et al., 2010; Kovács et al., 2007; Sasaki et al., 2011). Future studies that dissect the relative roles of CBP/p300 and Crtc-dependent regulation of CREB in different contexts may facilitate new understanding into the distinct roles of these regulatory mechanisms.
3.1.2. Serum-Response Factor (SRF)
SRF is a versatile transcription factor expressed in many cell types that plays an important role in coupling actin signaling to changes in gene expression that control cell motility (Arsenian et al., 1998; Knöll et al., 2006; Olson and Nordheim, 2010). In the CNS, disruption of SRF function is associated with impaired axon pathfinding during development and impaired synaptic plasticity in the adult hippocampus (Etkin et al., 2006; Knöll et al., 2006; Ramanan et al., 2005). Furthermore the inducibility of many of the immediate-early gene transcription factors is abolished in the absence of SRF expression, implying an essential role for this factor in activating the general cellular response to changes in extracellular stimuli (Parkitna et al., 2010; Ramanan et al., 2005).
Unlike the CREB family of transcription factors, SRF acts as a homodimer and is encoded by a single gene (Norman et al., 1988). Although activity-regulated signaling pathways can induce phosphorylation of SRF at Ser103, the functional significance of this phosphorylation event for SRF-dependent transcription remains largely unknown (Rivera et al., 1993). However, SRF interacts with a diverse set of transcriptional co-regulators, many of which are highly modulated by activity-regulated signaling cascades (Knöll and Nordheim, 2009). Evidence suggests that it is the differential regulation and use of these cofactors that confers functional versatility upon SRF-dependent programs of gene transcription.
SRF-dependent transcription is activated by two sets of cofactors: 1) the TCFs (Buchwalter et al., 2004), and 2) the myocardin family of transcriptional co-factors, which includes myocardin, myocardin-related transcription factor A (MKL1, also known as MAL or MRTF-A) and MKL2 (also known as MRTF-B) (Pipes et al., 2006). The TCFs Elk-1, Net, and SRF accessory protein 1 (Sap-1) comprise a subfamily of the Ets domain transcription factors, and were first shown to complex with SRF on the SRE of the Fos gene (Shaw et al., 1989). In addition to being recruited to SRF target genes via their interactions with SRF, the TCFs also contain a well-conserved DNA binding domain that interacts with a DNA sequence element neighboring a subset of SRF binding sites (Treisman, 1992). The N-terminal domain of the TCF factors recruits co-repressors to keep target genes off in the absence of stimuli (Yang et al., 2001). Following neuronal activity- and calcium-dependent activation of MAPK signaling pathways, the TCFs are extensively phosphorylated (Marais et al., 1993; Miranti et al., 1995). Phosphorylation of TCFs regulates their activity by inducing a conformational change that both enhances DNA binding and also activates the TCF-bound HAT CBP (Gille et al., 1992).
In contrast to the dominant role of the MAPKs in regulation of the TCFs, the myocardin family of SRF coactivators is regulated by actin signaling (Miralles et al., 2003; Sotiropoulos et al., 1999). In resting cells, myocardin family members such as MKL1 are bound to monomeric G-actin, which keeps them sequestered in the cytoplasm. Extracellular signals that activate intracellular Rho-GTPases stimulate the assembly of filamentous F-actin, freeing MKL1 to translocate into the nucleus where it can bind to SRF and activate SRF-dependent transcription (Vartiainen et al., 2007). Although the relative importance of these two sets of cofactors in neurons for SRF-dependent transcription is not fully, known, evidence suggests that the myocardins may be particularly important during neuronal development. For example, whereas Elk1 knockout mice show grossly normal brain development (Cesari et al., 2004), mice lacking both Mkl1 and Mkl2 in the brain have abnormal neuronal migration, neurite outgrowth, and SRF-dependent gene expression (Mokalled et al., 2010), phenocopying many of the early neurodevelopmental defects observed in Srf knockout mice (Knöll et al., 2006).
3.1.3. Myocyte Enhancer Factor 2 (MEF2)
The MEF2 family was first identified for its role in muscle differentiation (Molkentin et al., 1995), however the four members of the MEF2 family (MEF2A-D) are also found in distinct but overlapping expression patterns in neurons throughout the developing and adult CNS (Leifer et al., 1993; Lyons et al., 1995). Various members of the MEF2 family have been implicated in the regulation of neuronal survival during development (Mao et al., 1999), in the activity-dependent elimination of excitatory synapses (Flavell et al., 2006), and in behavioral adaptations to drugs of abuse (Pulipparacharuvil et al., 2008).
The transcriptional activity of MEF2 is highly sensitive to regulation by a complex array of stimulus-dependent post-translational modifications that modulate MEF2’s interactions with multiple transcriptional cofactors (McKinsey et al., 2002; Shalizi et al., 2006). The intricate details of these regulatory mechanisms suggest that post-translational modification of MEF2 by various signaling enzymes may provide a way for distinct stimuli to differentially regulate MEF2-dependent transcriptional programs. MEF2 can be either an activator or a repressor of transcription, and stimulus-induced signaling pathways switch it between these states. In its repressor state, class IIa HDACs (HDAC 4,5, 7 and 9) bind to the N-terminal domain of the MEF2s (Lemercier et al., 2000). This interaction contributes to repression of MEF2-dependent transcription both by reducing the acetylation of histones at MEF target gene promoters and by deacetylating a key residue in MEF2, Lys403 (note, all amino acid numbers presented in this section refer to sequence positions in human MEF2A) (Shalizi et al., 2006). Deacetylation of Lys403 is correlated with modification of this residue by the small ubiquitin-like modifier (SUMO) moiety, which appears to stabilize MEF2 in the repressor state (Shalizi et al., 2006; Zhao et al., 2005).
Neuronal activity-induced calcium signaling pathways activate MEF2-dependent transcription by at least three mechanisms. First, CaMK-dependent phosphorylation of the HDACs leads to their release from MEF2 and their export from the nucleus (Chawla et al., 2003; Lu et al., 2000). Since the HDACs compete for binding to the same region on MEF2 as the HATs p300 and CBP, HDAC unbinding promotes the ability of MEF2 to recruit co-activators (McKinsey et al., 2001). Second, calcium-dependent activation of the p38-MAPK leads to phosphorylation of MEF2s at multiple sites (Han et al., 1997; Yang et al., 1999; Zhao et al., 1999). At least three sites of p38-induced phosphorylation contribute to the transcriptional activity of the MEF2s (Thr312, Thr319, and Ser453). These sites are conserved in MEF2A, C, and D, and these isoforms of MEF2 are targets of p38-MAPK regulation (Yang et al., 1999). In addition, the extracellular signal-regulated kinase 5 (Erk5) MAPK is capable of phosphorylating MEF2A, C, and D and has been implicated in neuronal MEF2 regulation downstream of BDNF signaling (Kato et al., 1997; Shalizi et al., 2003). Third, calcium-dependent activation of the phosphatase calcineurin leads to dephosphorylation of Ser and Thr residues on MEF2 (Mao and Wiedmann, 1999; Wu et al., 2001b). Dephosphorylation at Ser408 is particularly important for the ability to switch MEF2 from repressor to activator and is required for activity to induce the switch at Lys403 from SUMOylation to acetylation (Grégoire et al., 2006; Shalizi et al., 2006). Interestingly, these sites of regulation are conserved in MEF2A and D, however they are contained within an alternatively spliced region of MEF2C (the γ domain) that is only present in a fraction of the MEF2C protein expressed in brain (Zhu et al., 2005). Finally, whereas calcium and cAMP cooperate to activate the transcription factor CREB, cAMP can either prevent or inhibit MEF2 activation by blocking HDAC export from the nucleus and by inhibiting import of the MEF2 co-activator NFATc3/c4 (Belfield et al., 2006). Taken together, these data raise the possibility that differential activation of signaling cascades, combined with differential expression of MEF2 isoforms, is a mechanism of transcriptional specificity for MEF2-dependent gene expression.
3.2. Transcription factors that undergo regulated nucleocytoplasmic shuttling
An alternative mechanism by which activity can induce transcription factor activity is via the regulation of nuclear import. Cytoplasmic sequestration of transcription factors provides a robust means to keep transcription off in the absence of stimuli, and local tethering of transcription factors near channels may also allow them to be directly regulated by signaling events that occur near the cell membrane. In the nervous system it has been suggested that transcription factors may be capable of being locally activated at dendritic synapses or in the distal growing axon and then translocating to the nucleus (Cox et al., 2008; Meffert et al., 2003). In addition, for transcription factors that are retained in the nucleus, key transcriptional coactivator and corepressor proteins can be shuttled in and out of the nucleus to regulate transcriptional activity (Parkitna et al., 2010; Soriano et al., 2011). Compared with transcription induced by prebound transcription factors, the transcriptional response to shuttling factors is slower and constitutes a late wave of activity-regulated transcription. Termination of transcription through these pathways is primarily mediated by signaling pathways that induce nuclear export.
3.2.1. Activity-dependent nuclear translocation
The two most widely studied transcription factors that translocate into the nucleus following stimulation are NF-κB and NFAT. The calcium-dependent regulation of these factors was first studied in immune cells, where these pathways play crucial roles is gene expression activated by T and B cell receptors. Although the sources of calcium and the biological context of the signaling pathways are different in neurons, these factors appear to play an important role in regulating intracellular neuronal responses to stimuli that include synaptic activity.
NF-κB refers to the activity of a set of five mammalian Rel-domain DNA binding subunits: NF-κB1 (p50/p105), NF-κB2 (p52/p100), RelA (p65), RelB, and c-Rel (Liou and Baltimore, 1993). In some cell types, most notably activated immune cells, NF-κB activity is constitutive. However, in many cells, NF-κB is held in the cytosol in an inactive form through its association with one of the Inhibitor of NF-κB (IκB) proteins (Meffert and Baltimore, 2005). The ankyrin repeat domains of IκB mask the nuclear localization signal (NLS) of NF-κB, and thus keep this transcription factor in an inactive form in the cytoplasm. Nuclear translocation of NF-κB is induced by stimuli that lead to phosphorylation and subsequent degradation of IκB (Mercurio et al., 1997). Phosphorylation of IκB is mediated primarily by the IκB kinase (IKK), which is composed of the IKKα and IKKβ subunits along with the regulatory protein NF-κB essential modulator (NEMO).
NF-κB is best known for its role in inflammation and immune responses. However, this transcription factor is also expressed in neurons, where the most prominent species are the canonical p50-p65 heterodimer, which is a regulated transcriptional activator, and the inactive p50-p50 homodimer (Meffert and Baltimore, 2005). Mice lacking expression of p65 show defects in a spatial learning task, suggesting that NF-κB-dependent transcription could contribute to activity-induced synaptic plasticity (Meffert and Baltimore, 2005). Some of the same stimuli that activate NF-κB in the immune system also regulate this transcription factor in the CNS. These stimuli include cytokines such as TNFα, viral infections, and oxidative stress. However, in the CNS NF-κB activity can also be induced by glutamate activation of synaptic NMDARs or the opening of L-VGCCs (Guerrini et al., 1995; Meffert et al., 2003). Both Ca2+/CaM-dependent protein kinase II (CaMKII) and MAPKs have been implicated in CNS regulation of NF-κB activity (Lilienbaum and Israël, 2003; Meffert et al., 2003; Takeuchi and Fukunaga, 2004). However, some controversy exists over whether glutamate induces NF-κB activity in neurons as opposed to other CNS cell types such as glia or microglia (Massa et al., 2006). The need to identify the cell types in which transcription is activated is not unique to NF-κB, but rather is a concern for any of the transcription factors that are expressed in both neurons and glia (this concern applies to CREB, for example). However in the case of NF-κB, resolving whether this transcription factor can be inducible in neurons is particularly important because apparently constitutive NF-κB transcriptional activity has been detected throughout the CNS during development in a transgenic mouse strain that expresses β-galactosidase under the control of an NF-κB-responsive promoter (Bhakar et al., 2002). Future development of cell type-specific genetic tools for manipulating NF-κB activity in the CNS will be important for addressing the different biological functions of this transcription factor in neurons, glia, and microglia.
Although evolutionarily related to the Rel/NF-κB family, the NFAT transcription factors (NFAT1-4) are distinguished by their sensitivity to intracellular Ca2+ and their regulation by the Ca2+-dependent serine/threonine phosphatase calcineurin (Hogan et al., 2003). A fifth NFAT family member (NFAT5/TonEBP) shares homology to the NFAT DNA binding domain but lacks calcium sensitivity and instead plays important roles in cellular responses to changes in extracellular tonicity (Miyakawa et al., 1999). NFAT family members have been most highly studied for their role in regulation of cytokine gene expression in T cells, however these proteins are widely expressed in numerous tissues including the nervous system (Vihma et al., 2008). Genetic studies have revealed that NFAT is required for neurotrophin- and netrin-dependent axon outgrowth during embryonic development (Graef et al., 2003) and for serum- and activity-dependent survival of cerebellar granule neurons (Benedito et al., 2005). However, the substantial redundancy in function among the four calcium-regulated members of this family and the requirement for these factors in development of other organ systems have limited genetic analysis of adult brain functions in constitutive knockout mice.
In response to stimuli that elevate intracellular calcium and/or induce the release of calcium from intracellular stores, the phosphatase calcineurin dephosphorylates multiple Ser and Thr residues on NFAT, which facilitates nuclear translocation (Okamura et al., 2000). Although NFAT can bind DNA as a dimer at regulatory elements that resemble NF-kB binding sites, NFAT also binds and cooperates with a number of other nuclear transcription factors including Fos/Jun family members and MEF2 (collectively known as “NFATn” for the NFAT “nuclear” component) to synergistically promote gene transcription (Hogan et al., 2003). NFAT-dependent transcription is terminated by nuclear export following rephosphorylation of the protein by a number of constitutively active kinases including dual specificity tyrosine phosphorylation-regulated kinase 1A (DYRK1A) and glycogen synthase kinase 3 (GSK3) (Graef et al., 1999; Gwack et al., 2006).
Experimental observations combined with mathematical modeling of this molecular regulatory loop have led to intriguing ideas about the ways in which temporal information encoded in neuronal firing patterns may modulate the activation of NFAT-dependent transcription (Arron et al., 2006; Cai et al., 2008; Crabtree and Graef, 2008). Activation of NFAT nuclear translocation and transcriptional activity in both lymphocytes and developing muscles is known to be sensitive to the frequency of calcium oscillations (Dolmetsch et al., 1997; Dolmetsch et al., 1998; McCullagh et al., 2004). Although the specific mechanisms that confer frequency-specific activation of NFAT target genes are not fully understood, they may involve changes in the intensity of the calcium signal such that a threshold for activation of target genes is passed (Fiering et al., 1990). Another possibility is that frequency modulation of the nuclear transport of NFAT could induce “waves” of NFAT translocation to the nucleus, such that bursts of nuclear protein are able to cooperatively activate sets of target genes (Cai et al., 2008).
3.2.2. Synaptic activity-dependent nuclear export
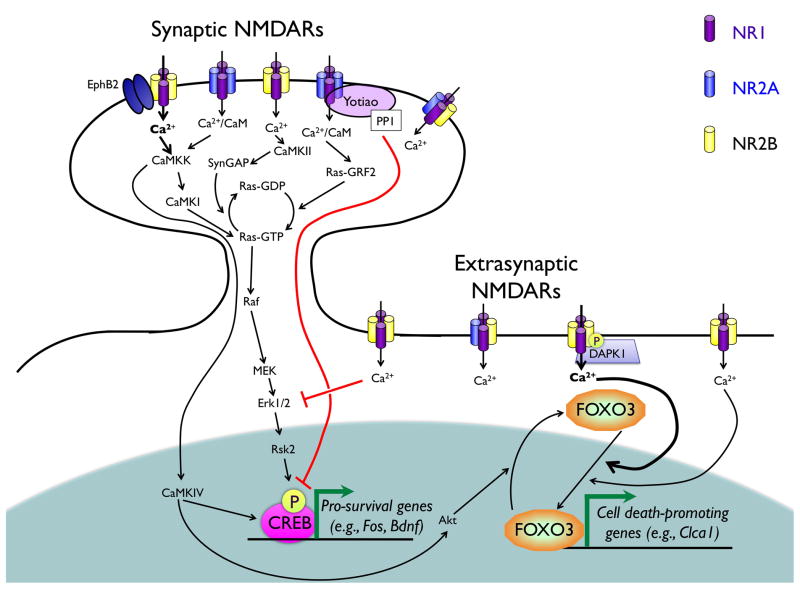
In addition to driving transcription factors into the nucleus, increased activity can also lead to nuclear export. The transcription factor forkhead box O3 (FOXO3) is best known for its regulation of cell death and stress responses (Brunet et al., 1999; Brunet et al., 2004; Tran et al., 2002). FOXO3 is phosphorylated at three sites by the growth factor-regulated Akt signaling pathway, promoting 14-3-3 association and nuclear export (Brunet et al., 1999). By contrast, growth factor withdrawal and oxidative stress are associated with FOXO3 dephosphorylation and nuclear entry, and the activation of FOXO3 target genes that promote cell death. In neurons the NMDAR-dependent activation of calcium signaling pathways can also lead to regulated shuttling of both FOXO3 and FOXO1 (Al-Mubarak et al., 2009; Dick and Bading, 2010; Papadia et al., 2008; Soriano et al., 2006). For example, stimulation of extrasynaptic NMDARs, which can induce cell death, drives nuclear translocation of FOXO3 (Dick and Bading, 2010). By contrast, prolonged synaptic activation prior to growth factor withdrawal reduces nuclear translocation of FOXO3 and protects against cell death through a signaling pathway that requires the activation of synaptic NMDARs, the elevation of nuclear calcium, and Ca2+/CaM-dependent protein kinase IV (CaMKIV). This bidirectional regulation of the FOXO3 and FOXO1 pathways demonstrates one of the many downstream pathways through which synaptic and extrasynaptic NMDARs can have opposing effects on neuronal biology. Importantly, this example highlights the fact that calcium signaling pathways can drive the nuclear export as well as the nuclear import of transcription factors.
A relative newcomer to the field of stimulus-regulated neuronal transcription factors is the calcium channel associated transcription regulator (CCAT). Experiments in both neurons and cardiac myocytes had suggested that the C-terminal domain of the L- VGCC α subunit Cav1.2 could be cleaved from the membrane-associated channel (De Jongh et al., 1994; Gerhardstein et al., 2000). When Gomez-Ospina and colleagues raised an antibody against this C-terminal domain of the protein, they noticed that it strongly localized to the nucleus, especially in glutamic acid decarboxylase 65 (GAD65)-expressing γ-aminobutyric acid (GABA)-ergic interneurons (Gomez-Ospina et al., 2006). In the nucleus, CCAT binds to the transcriptional regulator p54(nrb)/NonO, and acts as a transcriptional activator of a number of genes including that encoding the connexin Cx31.1 (Gjb5). Most interestingly, CCAT localization is regulated by intracellular calcium, and it accumulates in the nucleus under conditions of low intracellular calcium. These data raise the intriguing possibility that CCAT could contribute to transcriptional regulation induced under periods of low rather than high synaptic activity, providing an important counterbalance to the programs of transcription driven by activity-induced factors such as CREB, SRF, and MEF2.
3.2.3. Translocation from dendrites and axons
Transcription factors have been demonstrated to translocate between the nucleus and the cytoplasm in many cell types. Although small molecules can diffuse freely into the nucleus through the nuclear pore complex, larger molecules including most transcription factors are actively transported through the pore by associating with a family of nuclear transport proteins called the importins (Otis et al., 2006). Classically, importin α subunits bind directly to nuclear localization sequences on their target proteins whereas importin β subunits mediate the transport of the complex through the nuclear pore. Given the central importance of this mechanism in cellular physiology, it is not surprising the function of the core components of the importin pathway are conserved in neurons. However, neurons are unique with respect to the shape and the size of their cytoplasmic compartment. Unlike most other kinds of cells, in which no part of the plasma membrane is more than 100 μm from the nucleus, neurons have large and complex shapes. Furthermore, the most important functional parts of the cell, the pre- and post-synaptic regions of the axons and dendrites, can be found at very great distances from the nucleus. This unusual morphology has raised the intriguing possibility that transcription factors might sample environmental information at the neuronal periphery and then, via the traditional transport pathways described above, translocate to the nucleus to effect changes in gene expression.
Consistent with this possibility, several studies have shown that key elements of the nuclear translocation machinery are localized to distal parts of the neuron. For example, in dorsal root ganglion neurons not only are the importins found in the axon at significant distances from the cell body, but importin β protein levels are also rapidly increased in the axon after the nerve is lesioned via a mechanism that involves local translation of axonal importin β mRNA (Hanz et al., 2003). In dendrites, α-importin has been shown to bind to the cytoplasmic tail of the NMDAR subunit NR1-1a at postsynaptic sites in synapses (Jeffrey et al., 2009). This α-importin-NR1 interaction is disrupted by activation of NMDARs in a protein kinase C (PKC)-dependent manner in cultured neurons, allowing α-importin to translocate to the nucleus. Transcription factors that depend on importin translocation are also found at pre- and post-synaptic sites. Activating transcription factor 4 (ATF4 or CREB2) is localized to distal dendrites under resting conditions and translocates to the nucleus in an α-importin-dependent manner following stimuli that induce long-term depression but not long-term facilitation of Aplysia sensory neurons (Lai et al., 2008). mRNA encoding CREB1 has been found in axons of developing neurons, where CREB is both inducibly translated and retrogradely trafficked in response to NGF application (Cox et al., 2008). Finally, several studies have suggested that NF-κB may translocate from neurites to the nucleus in an activity-dependent manner (Meffert et al., 2003; Wellmann et al., 2001).
These data demonstrate that signal-dependent translocation of transcription factors can occur in neurites, but substantial questions about the mechanisms and consequences of this process remain to be solved. Is it possible for a single transcription factor molecule activated at a synapse or axon tip to translocate all the way to the nucleus? Is there something special about the way in which transcription factors become activated in the distal periphery of neurons that alters their transcriptional potential, or would somatic activation of the same transcription factor provide an equivalent signal? These questions as well as other ideas about synapse to nucleus signaling have been discussed in detail elsewhere (Jordan and Kreutz, 2009) and represent intriguing potential areas for the discovery of new mechanisms that could contribute specificity to the control of activity-regulated transcription.
3.3. Transcription factors that show regulated binding
The specificity of transcription factor function is determined in large part by the factor’s sites of DNA binding, which in turn determine which genes that transcription factor can regulate. For this reason, inducible binding of a transcription factor to different sites in the genome would have an obvious impact on its function. Perhaps the most clear example of calcium-dependent regulation of transcription factor binding has been that described for the Downstream Repressor Element Antagonist Modulator (DREAM) (Carrión et al., 1999). DREAM (also known as KChIP3 and calsenillin (An et al., 2000; Buxbaum et al., 1998)) belongs to the Neuronal Calcium Sensor (NCS) family of EF-hand proteins (Burgoyne, 2007). This protein family is related to the ubiquitous calcium effector protein CaM, but NCS family members also have domains distinct from those found in CaM. The 14 members of the NCS family have functions ranging from vesicle trafficking to ion channel regulation to transcriptional control (Burgoyne, 2007). DREAM was first characterized as a transcription factor when it was shown to bind to a repressor element in the first intron of the gene encoding prodynorphin (Pdyn) (Carrión et al., 1999). In the absence of calcium signaling, DREAM acts as a repressor of Pdyn by sterically hindering progression of the polymerase. Upon a rise in nuclear calcium, DREAM binds calcium through its EF-hands and its affinity for DNA is subsequently reduced (Lusin et al., 2008; Osawa et al., 2005; Osawa et al., 2001). Mice lacking DREAM (Kcnip3−/− mice) have elevated Pdyn mRNA in the spinal cord and reduced behavioral responses to painful stimuli, indicating a physiological requirement for DREAM in this pathway (Cheng et al., 2002).
Although a few other factors including some bHLH proteins (Corneliussen et al., 1994) and members of the MEF2 family (Mao and Wiedmann, 1999) have been shown to exhibit regulated DNA binding in vitro, direct regulation of DNA binding capacity has not emerged as a major mechanism of transcriptional regulation for most activity-regulated factors. Instead, it seems likely that inducible binding may be more likely to occur as a consequence of multiple coordinated cellular signaling events that either shift the subcellular localization of transcription factors and cofactors or that change the structure of genomic DNA. A good example of how these kinds of processes may result in regulated DNA binding has recently been described for CREB. Following the initial identification of CREB as the “cAMP response element binding protein,” substantial effort was devoted to understanding the mechanisms by which phosphorylation of CREB at Ser133 led to transcriptional activation. Most in vitro studies concluded that CREB was constitutively bound to the CRE, and that Ser133 phosphorylation did not change CREB’s DNA binding affinity (Montminy and Bilezikjian, 1987; Yamamoto et al., 1988). However, a set of in vivo studies showed evidence for inducible protein binding to at least some CREs (Boshart et al., 1991; Weih et al., 1990; Wölfl et al., 1999). Consistent with these early studies, a recent genome-wide study mapped sites of CREB binding before and after membrane depolarization of neurons and provided support for both of these scenarios (Kim et al., 2010). Although CREB was found prebound at most sites identified across the genome, a subset of binding sites showed enhanced CREB association after membrane depolarization. A potential mechanistic explanation for this observation is that changes in chromatin conformation may underlie the apparent induction of CREB binding by altering the availability of the CREs at some CREB target genes (Riccio et al., 2006). Upon exposure of neurons to BDNF, CREB becomes rapidly bound to some sites of DNA coincident with phosphorylation at its transcriptional regulatory site, Ser133. However, this inducible CREB-DNA binding is independent of CREB Ser133 phosphorylation and is not affected by inhibition of the Erk or phosphatidylinositol 3-kinase (PI3K) signaling pathways. Instead, BDNF appears to regulate CREB binding by initiating a nitric oxide-dependent signaling pathway that leads to S-nitrosylation of nuclear proteins including the histone deactylase HDAC2 (Nott et al., 2008). S-nitrosylation of HDAC2 induces its release from chromatin and is associated with increased histone acetylation and transcriptional induction of neurotrophin-regulated and CREB-dependent target genes. Traditionally most transcription factor binding assays have been conducted in cell-free systems, where this sort of chromatin regulatory mechanism of inducible binding would be missed. However, with the growing ease of performing genome-wide transcription factor binding assays in vivo, it will be of great interest in the future to determine how commonly these kinds of regulated binding site mechanisms are used to modulate transcription factor function following changes in neuronal activity.
3.4. Neuronal activity-regulated transcription factor expression
As initially described for Fos and the other IEGs, an important class of transcription factors are regulated by neuronal activity at the level of their expression. The promoters of these genes are targets of the transcription factors in the categories above. Robust induction of the IEG transcription factors is a cardinal feature of the transcriptional response to a wide variety of stimuli in many cell types (Herdegen and Leah, 1998; Morgan and Curran, 1991). Notably these transcription factors are also subject to additional forms of activity-dependent regulation. For example, after synthesis, Fos and Jun family transcription factors are subject to phosphorylation, which can change their nuclear localization or protein-protein interactions (Chen et al., 1993). Furthermore, the specific complexes of Fos/Jun proteins bound to activator protein 1 (AP-1) elements are subject to change under different conditions (Herdegen and Leah, 1998). For example, through an alternative splicing event that does not appear to be sensitive to neuronal activity, the Fosb locus gives rise to two transcripts that encode either full length FosB or a C-terminally truncated protein termed ΔFosB (Mumberg et al., 1991; Nakabeppu and Nathans, 1991). The short ΔFosB protein lacks a protein domain that promotes FosB degradation and thus has a much longer half-life than full length FosB. Thus, in response to repeated activation of Fosb transcription, over time ΔFosB protein accumulates and becomes the dominant FosB protein isoform in cells (Hope et al., 1994). Although the functional differences between ΔFosB and full length FosB are incompletely understood, these two isoforms show differences in their ability to repress target genes suggesting they may have distinct roles in regulation of genes that contain AP-1 regulatory elements (Mumberg et al., 1991; Nakabeppu and Nathans, 1991).
A more recent addition to the set of activity-induced transcription factors is the bHLH-PAS domain family member Npas4 (Lin et al., 2008). Like other IEGs, Npas4 is very rapidly and robustly induced by membrane depolarization of cultured neurons (Lin et al., 2008). However, unlike the Fos and Jun families of transcription factors, expression of Npas4 is largely restricted to neurons and its transcription is selectively induced by stimuli that activate intracellular calcium signaling pathways (Coba et al., 2008; Lin et al., 2008; Ooe et al., 2004; Zhang et al., 2009). Interestingly, acute RNAi-mediated knockdown of Npas4 during synaptogenesis in cultured hippocampal neurons reduces GABAergic synapse numbers while overexpression of Npas4 selectively increases GABAergic synapses (Lin et al., 2008). By contrast these manipulations have no effect on the number of glutamatergic synapses, indicating a selective role for Npas4 in regulating GABAergic synapse development. Npas4 forms functional heterodimers with other members of the bHLH-PAS domain protein family including Arnt1 and Arnt 2 (Ooe et al., 2009). Genome-wide ChIP-sequencing (ChIP-Seq) for Npas4 shows that this protein is widely found not only at promoters but also at activity-regulated distant enhancers (Kim et al., 2010). The function of Npas4 recruitment to enhancers remains to be determined, however one speculative idea is that late recruitment of Npas4 may be required to prolong the transcription of activity-regulated genes such that they reach critical expression levels for regulation of downstream processes (Greer and Greenberg, 2008). RNA interference (RNAi)-mediated knockdown of Npas4 affects the expression of a large number of gene products, many of which are activity-regulated, consistent with the possibility that this transcription factor contributes to stimulus-dependent regulation of gene expression (Lin et al., 2008).
Finally, the CREB family provides a particularly intriguing example of how the interaction between different regulatory mechanisms can confer specificity upon the control of gene transcription. The inducible cAMP early repressor (ICER) is a stimulus-inducible transcriptional repressor of the CREB family (Foulkes et al., 1996). ICER is inducibly transcribed from an intragenic promoter of the Crem gene (Borlikova and Endo, 2009; Stehle et al., 1993). This promoter contains four CRE elements that are bound by phosphorylated CREB. Once synthesized, the ICER protein can dimerize with other members of the CREB family including both CREB and CREM, changing the nature of the CRE-binding complex. ICER contains a DNA binding domain but it lacks a transcriptional activation domain and thus acts in a dominant negative manner to suppress CREB family-dependent transcription (Stehle et al., 1993). In this way ICER may function as a feedback mechanism to help turn off activity-stimulated transcription of CREB target genes. After its expression is induced by activity, ICER competes with CREB for binding to the CREs in its own promoter, shutting off its own expression and thereby defining the temporal window during which this mechanism is active. Furthermore, after expression, the stability of ICER is subject to regulation by the ubiquitin-proteasome system (Folco and Koren, 1997). Taken together, the mechanisms that regulate ICER provide a means for fine-tuning the temporal regulation of CRE-dependent transcription during physiologically relevant stimuli.
3.5. Regulation of chromatin
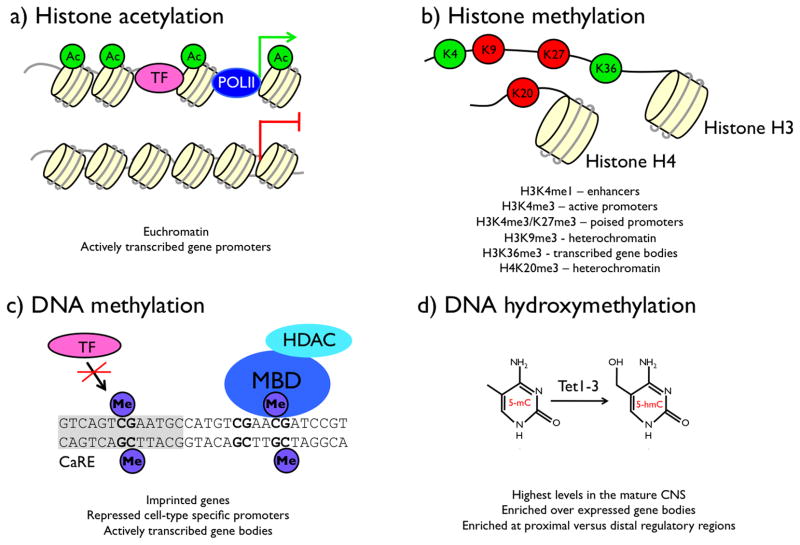
In order to fit into the nucleus, long strands of genomic DNA are twisted into complex secondary and tertiary structures. Although all cells contain the same genomic DNA, the architecture of chromatin structure differs between cell types (Xi et al., 2007). These changes in chromatin structure are established and maintained by epigenetic mechanisms that biochemically alter chromatin, and are thought to underlie cell type specific differences in gene expression by altering the availability of transcription factor binding sites. The major epigenetic mechanisms of transcriptional regulation involve both post-translational modifications of the histone proteins that comprise nucleosomes, which are the basic repeating unit of chromatin structure, along with covalent modifications of genomic DNA itself (Jaenisch and Bird, 2003) (FIGURE 5). Intriguingly, an accumulating body of evidence has shown that these epigenetic modifications are not static components of genomic DNA; instead both histone and DNA modifications are subject to regulation by stimuli that converge to regulate the function of histone- and DNA-modifying enzymes (Dulac, 2010; Riccio, 2010). Furthermore, genetic and pharmacological manipulation of the enzymes that mediate epigenetic regulation of chromatin have been associated with changes in learning and memory as well as alterations in affective- and addictive-like behaviors (Dulac, 2010; Roth and Sweatt, 2009; Tsankova et al., 2007). Our understanding of the relationship between the dynamic regulation of the epigenome and transcriptional potential is still in its infancy. However, here we review evidence for the major activity-regulated processes that have an impact on epigenetic mechanisms of chromatin regulation.
FIGURE 5. Chromatin modifications that regulate transcription.
Post-translational modifications of histones and covalent modifications of genomic DNA regulate transcription. a) Acetylation (Ac) can be added to multiple lysine residues in the N-terminal tail domain of histones H3 and H4. This modification is preferentially associated with transcriptionally active genes. b) Methylation (me) of histones H3 and H4 occurs at specific lysine (K) and arginine residues. The functional consequence of each of these modifications depends on the specific amino acid modified (K4, K9, K27, and K36 in histone H3, K20 in histone H4) (Kouzarides, 2002). Sites of methylation associated with transcriptional activation are shown in green, sites associated with transcriptional repression are shown in red. Each lysine can be mono- (me1), di- (me2), or trimethylated (me3), and the global distribution of these methyl marks varies across different kinds of genetic regulatory elements (Hon et al., 2009; Mikkelsen et al., 2007). c) In differentiated mammalian cells, DNA methylation occurs on cytosines at a subset of CpG dinucleotides. Methylation can regulate transcription by sterically blocking the association of a transcription factor (TF) with its binding site (the gray box represents a CaRE), or by recruiting the association of a protein with a methyl-DNA binding domain (MBD), which can act as a scaffold for additional chromatin regulatory enzymes. d) 5-hydroxymethylation of cytosine (5-hmC) is catalyzed by the enzymes Tet1, Tet2, and Tet3, which add a hydroxyl group to methylated cytosine bases (5-mC) (Ito et al., 2010; Ko et al., 2010; Tahiliani et al., 2009). Little is known about the functional relevance of this modification of DNA, but new chemical methods are beginning to allow its genome-wide distribution to be described (Song et al., 2011).
3.5.1. Regulation of enzymes that modify histones
Histones can be post-translationally modified at specific amino acid residues in their N-terminal tails by several different moieties, including phosphorylation, acetylation and methylation (Strahl and Allis, 2000). Multiple studies have shown that both phosphorylation and acetylation of histones are subject to rapid activity-dependent regulation in neurons (Crosio et al., 2000). It is less clear whether histone methylation can be acutely regulated by changes in neuronal activity (Chen et al., 2003a; Kim et al., 2010).
The ability of activity-regulated signaling pathways to modulate histone phosphorylation and acetylation has been well described. Histone H3 can be phosphorylated at a number of sites including Ser3, Ser10, Thr11, and Ser28. Histone H3 Ser10 phosphorylation was first linked with chromosome condensation during mitosis – a condition under which every Ser10 of every histone H3 is phosphorylated (Gurley et al., 1978; Wei et al., 1999). However, growth factors and neuronal activity can also lead to the rapid, transient, and gene-specific phosphorylation of Ser10 in non-dividing cells, where this histone modification has been associated with transcriptional activation of IEGs including Fos and Jun (Bilang-Bleuel et al., 2005; Crosio et al., 2000). Thus, the context in which phosphorylation of histone H3 Ser10 is induced may determine the functional consequence of this modification for cell physiology. Study of histone H3 Ser 10 phosphorylation by epidermal growth factor (EGF) and the PKC agonist TPA implicated activation of the Erk/MAP kinase pathway (Cheung et al., 2000; Sassone-Corsi et al., 1999). The relevant Ser10 kinase activated by these stimuli was suggested to be the ribosomal S6 kinase 2 (Rsk2), which is mutated in Coffin-Lowry Syndrome, a rare syndromic form of X-linked mental retardation (Sassone-Corsi et al., 1999). MAPK signaling has also been implicated in neuronal activity-induced phosphorylation of histone H3 Ser10 since inhibitors of the MAPK pathway block the inducible Ser10 phosphorylation found in the hippocampus following contextual fear conditioning (Chwang et al., 2006). However, many kinases have been shown to be capable of inducing histone H3 Ser10 phosphorylation in different cell types following distinct stimuli, including a number that are activated by neural activity such as PKA, PKC, the p38 MAPK, Akt, and the mitogen- and stress-induced kinases 1 (MSK1) and 2 (MSK2), among others (Bode and Dong, 2005). The functional consequences of differential kinase regulation of histone H3 Ser10 phosphorylation remain largely unknown.
Histones H3 and H4 are subject to rapid and transient stimulus-dependent changes in acetylation at a number of residues, all of which have been associated with transcriptional activation (Renthal et al., 2009; Riccio, 2010; Wang et al., 2008). The acetylation of histones is regulated by the balance between HATs and HDACs, which are either locally recruited or activated at genetic regulatory elements. As described in the section on transcriptional regulation of Fos above, both HATs and HDACs are targets of activity-regulated signaling cascades. The HAT CBP is recruited to gene promoters and enhancers following membrane depolarization in a manner that depends on its association with transcription factors such as CREB that undergo calcium-regulated phosphorylation (Impey et al., 2002; Kim et al., 2010). In addition, CBP itself is subject to phosphorylation by stimulus-dependent signaling cascades including the calcium-activated kinase CaMKIV, though the functional consequences of these modifications for recruitment or the HAT activity of CBP remain unclear (Chawla et al., 1998; Hardingham et al., 1999; Hu et al., 1999; Impey et al., 2002; Riccio, 2010).
HDACs are a large family of proteins with different regulation and function. The eleven “classical” HDAC proteins remove acetyl groups via hydrolysis and are characterized into three families (class I, class IIa/b, and class IV) based on their structure, enzymatic function, and pattern of expression (Butler and Bates, 2006). HDACs 1, 2, 3, and 8 comprise the class I family and are ubiquitously expressed and predominantly localized to the nucleus. As described in the section on inducible transcription factor binding above, HDAC2 is a target of BDNF signaling which reduces HDAC2 association with its target genes (Nott et al., 2008). HDACs 4, 5, 7, and 9 belong to the class IIa family, while HDACs 6 and 10 form class IIb. Class II HDACs can associate with transcription factors, but unlike class I, they shuttle between the nucleus and the cytoplasm in a calcium-dependent fashion, regulating the function of their transcription factor partners (Chawla et al., 2003). Finally, HDAC11 is the only member of the class IV family and is localized to the nucleus though little is known about its function. Despite their nomenclature, members of the HAT and HDAC family can also regulate acetylation of other proteins in addition to histones, and the non-nuclear HDACs have several biological functions that are unrelated to transcriptional regulation (Pandey et al., 2007). Interestingly, the different HDACs may control distinct groups of genes. By ChIP, HDAC2 but not HDAC1 is found to be bound to the promoters of a number of genes (including Bdnf, Egr1, and Fos) involved in activity-regulated brain plasticity (Guan et al., 2009). Consistent with a functional role for HDAC2 in regulation of these genes, neuron-specific overexpression of HDAC2 but not HDAC1 decreases synapse number, synaptic plasticity, and memory formation. These data raise the possibility that differential regulation of gene subsets by members of the large family of HDACs may provide a mechanism for specificity in the activity-regulated transcription of gene expression programs.
3.5.2. Regulation of DNA methylation
DNA methylation is known to be a key chromatin regulatory mechanism underlying many very long-lasting changes in gene expression (Reik, 2007). For example, DNA methylation is involved in the imprinting of genes, which permits only the maternal or paternal allele of a specific gene to be expressed (Reik and Walter, 1998). DNA methylation is also required for X-chromosome inactivation and it controls the activity of many cell-type specific gene promoters. For these reasons, for many years DNA methylation was largely perceived as a static phenomenon unlikely to be important for transcriptional plasticity. However, a growing body of data has provided increasingly compelling evidence that DNA methylation is subject to stimulus-dependent regulation even in post-mitotic cells. The first evidence for rapid DNA demethylation was shown in sperm cells at the moment of fertilization. Erasure of DNA methylation in the sperm and egg is essential for resetting gene expression potential in the pluripotent cells of the early embryo (Reik et al., 2001). Maternal methylation is lost slowly during early cell divisions of the fertilized egg by failing to transfer methyl marks to the newly synthesized DNA at each cell division. By contrast, paternal methylation is lost rapidly over a period of hours beginning at the moment the sperm invades the egg (Oswald et al., 2000). Rapid demethylation of DNA has also been reported in a stimulus-dependent manner in other cell types. For example, following T-cell receptor activation, multiple sites in the Il2 gene promoter are demethylated within 20 minutes – far too fast to be mediated by passive loss during cell division (Bruniquel and Schwartz, 2003). In the hippocampus, seizure has been shown to drive a significant decrease in DNA methylation within regulatory regions of the fibroblast growth factor (FGF) gene Fgf1 as well as the Bdnf gene, coincident with their activity-dependent transcriptional induction (Ma et al., 2009). Interestingly, genetic and pharmacological manipulations of the enzymes that regulate DNA methylation affect brain plasticity (Feng et al., 2010; Miller and Sweatt, 2007). Thus, these data raise the possibility that stimulus-dependent changes in DNA methylation may contribute to activity-regulated neuronal transcription.
Given the evidence that DNA methylation may be regulated in the nervous system, these data raise the question of the mechanisms that modulate this chromatin modification. DNA is methylated at CpG dinucleotides across the genome by a small family of DNA methyltransferases. These enzymes include the maintenance DNA methyltransferase Dnmt1, which mediates transfer of DNA methylation patterns to the newly synthesized strand of DNA during cell division, and the de novo DNA methyltransferases Dnmt3a and Dnmt3b (Jaenisch and Bird, 2003). There is some evidence suggesting that the expression of Dnmt3a in particular may be regulated by the activation of neuronal signaling cascades (LaPlant et al., 2010; Miller and Sweatt, 2007), however whether these changes in expression are causative for to local activity-dependent differences in DNA methylation has not been determined. Loss of DNA methylation is also presumed to have an enzymatic mediator. The nature of the enzymes that demethylate DNA remains a subject of some controversy but emerging data point toward a role for base excision repair mechanisms in this process (Fritz and Papavasiliou, 2010; Gehring et al., 2009; Wu and Zhang, 2010). Notably, it has recently been discovered that genomic DNA can also be modified by hydroxymethylation (Ito et al., 2010; Kriaucionis and Heintz, 2009; Tahiliani et al., 2009). This modification is induced by the enzymes Tet1-3, which hydryoxylate 5-methyl-cytosine in genomic DNA. Intriguingly, Tet1-dependent cytosine hydroxymethylation promotes DNA demethylation via a mechanism that requires the AID (activation-induced deaminase)/APOBEC (apolipoprotein B mRNA-editing enzyme complex) family of cytidine deaminases (Guo et al., 2011). Viral short hairpin RNA (shRNA)-mediated knockdown of Tet1 or Apobec1 in the dentate gyrus of the hippocampus inhibits electroconvulsive stimulation-induced demethylation of the Fgf1 and Bdnf genes suggesting that this mechanism could be engaged by activity-dependent signaling pathways to demethylate neuronal DNA (Guo et al., 2011). Future study of the mechanisms by which neural activity regulates these enzymes will be an essential step toward understanding the importance of this regulatory process for brain plasticity.
DNA methylation can have an impact on transcription through a number of different mechanisms, most commonly either by sterically hindering the association of transcription factors with their binding sites, or by recruiting the binding of proteins with methyl-DNA binding domains (Klose and Bird, 2006). Biochemical purification of methyl-DNA binding proteins has led to the identification of a small family of methyl-DNA binding domain (MBD) proteins comprised of MBD1, MBD2, MBD4, and the founding member methyl CpG binding protein 2 (MeCP2). A fifth methyl-DNA binding protein named Kaiso lacks an MBD, and instead recognizes methylated DNA through a zinc- nger domain. These proteins act as effectors of the DNA methyl mark by recruiting complexes of chromatin regulatory proteins to methylated regions of the genome.
MeCP2 is of particular interest in the nervous system because loss-of-function mutations in human MECP2 cause the neurodevelopmental disorder Rett Syndrome (Chahrour and Zoghbi, 2007). Rett Syndrome is characterized by motor dysfunction, lack of language development, and severe mental retardation. Many children with Rett Syndrome show grossly normal neurological function at birth but begin to lose developmental milestones at 6–18 months of age. This is the peak period of synapse formation in the brain, which suggested the hypothesis that Rett Syndrome might be a disorder of synapse development (Zoghbi, 2003). Consistent with this possibility, mice bearing loss-of-function mutations in Mecp2 show multiple abnormalities of synapse development, including decreased strength of excitatory glutamatergic synapses and alterations in the number of GABAergic synapses (Dani and Nelson, 2009; Deng et al., 2010; Moretti et al., 2006; Wood et al., 2009; Zhang et al., 2010b). Furthermore, loss of MeCP2 appears to trap cortical synapses in an immature state of development (Tropea et al., 2009). Specifically, it has been shown in mice lacking MeCP2, that monocular deprivation induces an ocular dominance shift in the visual cortex long after the critical period for cortical plasticity has closed in their wildtype littermates.
MeCP2 is subject to phosphorylation by calcium-activated signaling cascades, raising a potential mechanism to link function of this protein with activity-dependent plasticity. Mass spectrometry has identified calcium-regulated sites of MeCP2 phosphorylation at Ser80 and Ser421 (Tao et al., 2009; Zhou et al., 2006). MeCP2 has also been shown to be modified by O-GlcNAc glycosylation, and the fraction of glycosylated MeCP2 is more likely to be phosphorylated at Ser80, suggesting a potential interaction between these two modifications (Rexach et al., 2010). MeCP2 Ser80 is rapidly dephosphorylated following membrane depolarization of cultured neurons (Tao et al., 2009), whereas MeCP2 Ser421 is inducibly phosphorylated by CaMKs following L-VGCC or NMDAR activation (Zhou et al., 2006). Overexpression of a nonphosphorylatable Ser421Ala mutant MeCP2 blocks activity-dependent Bdnf transcription, suggesting that phosphorylation of this site is required for functional regulation of a least a subset of MeCP2-sensitive genes. However, the mechanisms by which Ser421 phosphorylation may alter MeCP2 function remain largely unknown. Phosphorylation at this site is correlated with the dissociation of MeCP2 from Bdnf promoter IV (Chen et al., 2003a), raising the possibility that phosphorylation could be associated with a change in the genomic distribution of MeCP2. However, MeCP2 is very highly expressed in neurons and it is bound widely across the genome essentially wherever DNA is methylated (Skene et al., 2010). Whether Ser421 phosphorylation of MeCP2 occurs at specific genomic loci and/or whether this modification has a selective impact on the transcription of a subset of gene products remains unknown. Future studies that address the functional relevance of activity-dependent regulation of MeCP2 will be expected to increase understanding of this neurologically important epigenetic transcriptional regulator.
4. Calcium channel-dependent regulation of transcription
Transcription occurs in the cell nucleus, and up to this point, we have been focused on the nuclear proteins that regulate transcription in response to neuronal activity. However, the signals that set these nuclear events in motion begin at the synaptic plasma membrane, where neurotransmission occurs. In this section we turn our attention toward the synaptic and cytoplasmic mechanisms by which calcium signaling pathways encode specificity toward the transcriptional pathways they activate.
Presynaptic action potentials release glutamate into the synaptic cleft, activating glutamate receptors of NMDA-, AMPA-, and kainate-types. This postsynaptic glutamate receptor activation then sets into motion a series of intracellular signaling events that culminate in the regulation of the transcription factors and stimulus-regulated genes described above. One of the most salient second messengers in cell biology is the calcium ion (Clapham, 2007). The importance of calcium signaling pathways for activity-regulated gene transcription was first demonstrated by experiments that showed removing calcium from the extracellular solution ablates neurotransmitter-induced gene induction in cultured cells (Greenberg et al., 1986; Morgan and Curran, 1986). Over the course of the following decade, it became apparent that this single signal – the calcium ion – is capable of exerting diverse effects on neuronal physiology and metabolism depending upon the mode of calcium entry and the cellular context in which the calcium signal acts (Ghosh and Greenberg, 1995). The two most physiologically relevant modes of calcium entry into neurons in terms of regulating changes in gene transcription are entry through L-VGCCs and NMDARs (Bading et al., 1993; Ghosh et al., 1994b; Ghosh and Greenberg, 1995; Lerea et al., 1992). Additional sources of calcium include other voltage- and ligand-gated plasma membrane calcium channels as well as the release of calcium from intracellular stores.
Calcium can activate a number of different signaling cascades, and the differential activation of these pathways by distinct pools of intracellular calcium contributes to the specificity of transcriptional responses. Calcium acts on cell physiology by virtue of its ability to bind to proteins, which then changes their conformations and functions. One of the most important calcium adaptors in the cell is the protein CaM, which can coordinate up to four calcium ions with its four EF hand domains (Clapham, 2007). The Ca2+/CaM complex is then capable of activating several different types of effector molecules, including the family of CaMKs and the Ca2+/CaM-dependent protein phosphatase calcineurin (Wayman et al., 2008b). Other Ca2+/CaM binding proteins including Ras-specific guanine nucleotide-releasing factor (Ras-GRF), along with CaMKIV, lead to activation of MAPK signaling pathways. Additionally, Ca2+/CaM-dependent adenylate cyclases elevate cellular cAMP levels to activate PKA. Activation of these kinase cascades drives the changes in transcription factor activation discussed above that regulate gene transcription. Factors such as calcium channel location, channel subunit composition, calcium microdomain dynamics, and protein-protein interactions can all play a role in determining which signaling cascades – and therefore which genes – are differentially regulated following neuronal stimulation (Bading et al., 1993; Ginty, 1997). In this section, we will discuss the different sources of calcium that, upon activation, can lead to alterations in gene transcription in neurons, and we will discuss the mechanisms by which these various routes of calcium influx convey specificity to the nucleus. We note that much has been written about the potential mechanisms that underlie long-distance synapse to nucleus signaling in neurons, and we refer the reader to other, more comprehensive reviews for the details of this discussion (Adams and Dudek, 2005; Carrasco and Hidalgo, 2006).
4.1. NMDA Receptors (NMDARs)
NMDARs are glutamate-gated non-selective cation channels that play a key role in the activation of intracellular calcium signaling cascades following synaptic activity (Dingledine et al., 1999; McBain and Mayer, 1994). Because NMDARs require both presynaptic depolarization (for neurotransmitter release into the synaptic cleft) and postsynaptic depolarization (for the removal of the Mg2+ ion that usually blocks the channel pore) for activation, they play an essential role as coincidence detectors in LTP, a Hebbian form of synaptic plasticity that is a cellular mechanism for learning and memory (Collingridge and Singer, 1990; McBain and Mayer, 1994). The ability of NMDARs to allow calcium to enter the cell has been shown to be important for many neurobiological processes including neuronal survival, migration, neurite outgrowth, synapse formation, and other forms of plasticity such as homeostatic plasticity and metaplasticity (Choi, 1988a, b). Importantly, it has been demonstrated by a myriad of studies that one critical way in which NMDARs are capable of exerting their effects on a neuron’s life and metabolism is via regulation of gene transcription (Hardingham and Bading, 2003; Wayman et al., 2006; West et al., 2002).
4.1.1. NMDARs regulate transcription in vitro and in vivo
There is a substantial body of evidence in support of the idea that calcium influx through NMDARs contributes to induction of gene transcription. The first evidence for NMDARs being critical for the induction of IEGs following electrical stimulation came from a study published by Cole and colleagues (Cole et al., 1989). Briefly, the authors showed that high frequency stimulation of the perforant path-granule cell synapse in vivo leads to robust induction of Egr1 mRNA in an NMDAR-dependent manner. A later study showed that in cultured hippocampal neurons, glutamate-induced increases in mRNA transcript levels of the IEGs Fos, FosB, Jun, JunB, Egr1 and Nr4a1 were almost entirely due to NMDAR activation (Bading et al., 1995). Interestingly, while high frequency stimulation – which is known to induce LTP – is capable of triggering Egr1 mRNA induction, low frequency stimulation – which does not induce LTP – is not sufficient to induce Egr1 transcription (Cole et al., 1989). These correlational findings were also among the first to suggest that IEG transcription might play a role in synaptic plasticity.
NMDAR-regulated gene transcription has been implicated in numerous cellular processes. NMDARs are the critical source of calcium influx for the initiation of LTP (Collingridge et al., 1983a, b; Kauer et al., 1988; Malenka et al., 1988; Muller et al., 1988), and subsequent work has suggested that the late phase of LTP relies on NMDAR-induced, transcription-dependent cellular events (Frey and Morris, 1997; Nguyen et al., 1994). NMDARs also contribute to transcription that promotes neural development. For example, NMDAR activation regulates Wnt-2 transcription in a manner dependent upon CaMK kinase (CaMKK), the Ca2+/CaM-dependent protein kinase I (CaMKI) α and γ isoforms, MAPK, and CREB activation (Wayman et al., 2006). This increase in Wnt2 transcription following synaptic NMDAR stimulation results in increased Wnt-2 protein secretion and a subsequent increase in dendrite outgrowth and branching in hippocampal neurons. Finally, NMDAR activation has been shown to be essential for the regulation of neuronal survival during development (Hardingham and Bading, 2002, 2003; Hardingham et al., 2002; Sala et al., 2000). Interestingly, cell survival can be bidirectionally regulated by NMDARs, and this process appears to rely largely on which specific NMDARs are activated on a given neuron. We will discuss this phenomenon in greater detail below.
4.1.2. Molecular biology and patterns of expression of the NMDARs
NMDAR channels are heterotetramers that can be made up of different subunit compositions. There are seven known NMDAR subunits: NR1, which can be alternatively spliced to yield eight different variants; NR2A-D, which are encoded by four different genes; and NR3A-B, which are also encoded by distinct genes (Ciabarra et al., 1995; Dingledine et al., 1999; Matsuda et al., 2002; McBain and Mayer, 1994; Nishi et al., 2001; Sucher et al., 1995). Functional glutamate-sensing NMDARs require at least one NR1 and one NR2 subunit (Cull-Candy et al., 2001; Köhr, 2006; Rosenmund et al., 1998), however the exact subunit composition of individual receptors varies. The NR1 and NR3A/B subunits contain glycine-binding sites, while the four NR2 subunits all contain glutamate-binding sites. In order to achieve full opening of the ion channel pore, co-activation by both glutamate and glycine is required (Clements and Westbrook, 1991; McGurk et al., 1990). Most CNS NMDARs are thought to contain two NR1 subunits and two NR2 subunits (Dingledine et al., 1999). NMDARs that contain NR3 subunits probably contain NR1, NR2, and NR3 (Sasaki et al., 2002).
NR1 is expressed throughout the nervous system and is an obligate NMDAR subunit; without it, functional NMDARs cannot form (Ishii et al., 1993; Kutsuwada et al., 1992; Meguro et al., 1992; Monyer et al., 1992). Several important, functional differences have been identified that distinguish the four NR2 subunits (NR2A-D) from each other. NR2A and NR2B are most highly expressed in brain structures in the forebrain, while NR2C and NR2D are more highly expressed in the hindbrain and the cerebellum (Monyer et al., 1994; Monyer et al., 1992). The subunits NR2B and NR2D are both expressed in embryonic CNS structures, while there is no notable expression of either NR2A or NR2C until some time after birth (Monyer et al., 1994; Monyer et al., 1992). After birth, expression of NR2A and NR2C slowly increases until their levels plateau shortly before adulthood, while NR2B and NR2D levels either remain unchanged or decrease. The developmental “switch” that occurs between NR2B and NR2A over the course of development is thought to be critical for a number of neurological developmental milestones (Shi et al., 1997). Furthermore, there is evidence that this switch is not only regulated by developmental progression, but also by neuronal activity, as visual deprivation retards this switch in the visual cortex (Nase et al., 1999; Quinlan et al., 1999; Roberts and Ramoa, 1999). Functionally, this switch is important because NR2B-containing NMDARs exhibit longer-lasting currents and carry more calcium per unit of current (Monyer et al., 1994; Sobczyk et al., 2005). This means that for each episode of presynaptic glutamate release, NR2B-containing NMDARs allow more calcium to enter the cell than do NR2A-containing NMDARs. Finally, like NR2A and NR2B, NR3A is most highly expressed in regions of the forebrain, while NR3B is more highly expressing in the midbrain, hindbrain, and in peripheral motor neurons (Al-Hallaq et al., 2002; Matsuda et al., 2002; Nishi et al., 2001; Sasaki et al., 2002; Wong et al., 2002). Both NR3 subunits are highly developmentally regulated, with expression increasing in the early postnatal period and peaking ~P8–10 in the rodent brain before decreasing down to low levels that persist through adulthood.
4.1.3. Different pools of NMDARs differentially regulate transcription
The subunit composition of NMDARs has a strong effect on their function (Hardingham and Bading, 2010). While all NMDARs must contain an NR1 subunit, one key distinction between different receptors is which NR2 subunit they contain, as well as whether they contain an NR3 subunit. The combinations of NMDAR subunit assemblages allows for NMDARs with distinct pharmacological, physiological, and signaling properties (Monyer et al., 1994; Monyer et al., 1992). Subunit composition can significantly affect subcellular localization as well as which signaling cascades are activated downstream of NMDAR stimulation, and subsequently which transcriptional programs are induced in response to that stimulation (Cheng et al., 1999; Hardingham and Bading, 2002, 2003; Hardingham et al., 2002; Martel et al., 2009; Yashiro and Philpot, 2008).
One transcription factor that is particularly important for NMDAR-regulated transcription is CREB (Hardingham and Bading, 2002, 2003; Hardingham et al., 2002; Sala et al., 2000). Interestingly, CREB is not only activated by NMDAR signaling, its transcriptional activity can also be turned off by NMDARs (Hardingham et al., 2002). Specifically, synaptic NMDAR stimulation induces CREB Ser133 phosphorylation, which is required for transcriptional activation. By contrast, extrasynaptic NMDAR stimulation results in CREB Ser133 dephosphorylation, which leads to transcriptional deactivation (Hardingham and Bading, 2002; Hardingham et al., 2002). One possible mechanism by which extrasynaptic NMDARs might induce CREB dephosphorylation is via the nuclear translocation of the recently-identified protein Jacob (Dieterich et al., 2008). Jacob has been shown to be alternately bound by either the NCS Caldendrin – during periods then dendritic calcium levels are very high, such as would occur during synaptic NMDAR stimulation – or by α-importin, which following extrasynaptic NMDAR stimulation localizes Jacob to the nucleus (Dieterich et al., 2008; Thompson et al., 2004). Nuclear localization of Jacob has been shown to be both necessary and sufficient for extrasynaptic NMDAR-dependent CREB dephosphorylation and subsequent cell death pathway initiation (Dieterich et al., 2008).
Importantly, even though they inactivate CREB under some conditions, extrasynaptic NMDARs are capable of inducing other programs of gene transcription (Hardingham and Bading, 2002; Hardingham et al., 2002; Wahl et al., 2009). However, unlike synaptic NMDAR activity, which promotes cell survival, transcription regulated by extrasynaptic glutamate receptors is able to trigger activation of cell death pathways in neurons. For example, as described in section 3.2.2, stimulation of extrasynaptic NMDARs has been shown to drive nuclear translocation of the FOXO family of transcription factors, which functions as a trigger for apoptosis through upregulation of cell-death promoting genes (Brunet et al., 1999; Dick and Bading, 2010), whereas synaptic NMDAR activation induces FOXO export (Papadia et al., 2008; Soriano et al., 2006). Among the extrasynaptically-regulated genes that might damage neurons are Clca1, which encodes a putative calcium-activated chloride channel that has been implicated in other forms of cellular injury (Wahl et al., 2009), and Txnip, a thioredoxin inhibitor that leaves cells more vulnerable to oxidative stress (Papadia et al., 2008).
An important point of uncertainty lies in the identity of the subunits that comprise synaptic versus extrasynaptic NMDARs, and furthermore to what degree the subunit composition of these different pools of receptors matters for downstream transcriptional regulation. In the studies done on synaptic versus extrasynaptic regulation of CREB phosphorylation and CREB-dependent gene transcription, it has been demonstrated that blockade of NR2B-containing NMDARs by ifenprodil greatly impairs the ability of extrasynaptic NMDARs to effect CREB Ser133 dephosphorylation (Hardingham et al., 2002). This raises the intriguing possibility that NMDAR NR2 subunit composition might dictate the specificity of signaling cascades activated downstream of NMDAR stimulation (Cheng et al., 1999; Hardingham et al., 2002; Martel et al., 2009), and specifically suggests that NR2B is critical in mediating extrasynaptic NMDAR-dependent gene transcription programs. Some studies have suggested that synaptic NMDARs might be predominantly NR2A-containing, while extrasynaptic NMDARs are predominantly NR2B-containing (Tovar and Westbrook, 1999). This is interesting given that at birth, NMDARs are almost exclusively NR1/NR2B heteromers, and NR2A subunit expression in the rodent CNS doesn’t begin until 6–10 days after birth, a particularly dynamic period in synapse development (Flint et al., 1997; Kew et al., 1998; Kirson and Yaari, 1996; Li et al., 1998; Monyer et al., 1994; Sheng et al., 1994; Tovar and Westbrook, 1999). However, more recent evidence suggests that this model may be overly simplistic. In addition, at least one study has suggested that there are roughly equal proportions of NR2A and NR2B localized both synaptically and extrasynaptically (Thomas et al., 2006). In dissociated hippocampal neuron cultures NMDARs have been found to be mobile and to traffic in and out of synapses via lateral diffusion (Groc et al., 2006; Tovar and Westbrook, 2002), although a similar study in acutely dissected hippocampal slices reported contrary results (Harris and Pettit, 2007). One possible explanation for this apparent discrepancy could be that the extracellular matrix plays a role in NMDAR stabilization at the plasma membrane, which would only be left intact in whole tissue preps. More work is required to determine where NR2A and NR2B are localized, how their localization is regulated developmentally, and to what degree subunit composition versus localization is the deciding factor in what kind of signal is transmitted from NMDARs to the nucleus.
4.1.4. Protein interactions that underlie NMDAR signaling specificity
The large intracellular C-termini of the NMDAR subunits are variable and have been shown to confer differential protein-protein interactions with NMDARs of distinct subunit composition (FIGURE 6). These interactions have important functional implications for NMDAR localization as well as interaction with signaling molecules and may be responsible for the specificity upon receptor activation of downstream events including transcription. Given the differential effects of NR2A- and NR2B-containing receptors on many biological processes, including the regulation of CREB Ser133 phosphorylation, considerable effort has been devoted to determining whether these two NR2 subunits may differentially interact with signaling proteins.
FIGURE 6. Different pools of NMDARs are capable of effecting distinct changes in gene transcription.
Synaptic NMDARs are capable of signaling through the CaMKs and the Ras/Raf/MEK/Erk/Rsk2 pathway to phosphorylate CREB and initiate transcription of a variety of pro-survival genes. Additionally, synaptic NMDAR activation of CaMKK and CaMKIV leads to the activation of Akt and subsequently the phosphorylation and nuclear export of the transcription factor FOXO3, thereby inhibiting transcription of various cell death-inducing genes. In contrast, activation of extrasynaptic NMDARs opposes the effects of synaptic NMDAR activation. Activation of extrasynaptic NMDARs inhibits the activation of Erk1/2, thereby reducing CREB phosphorylation. Additionally, extrasynaptic NMDARs induce the nuclear import of FOXO3 and the subsequent transcription of pro-death genes; this is potentiated by DAPK1. Subunit composition can also lend specificity to synapse-to-nucleus signaling by NMDARs. NR2A-containing NMDARs are capable of interacting with and selectively activating Ras-GRF2; activation of NR2B-containing NMDARs leads to the activation of CaMKII and the subsequent phosphorylation of SynGAP. While NR2B-containing NMDARs have also been shown to interact with Ras-GRF1 (not shown), how that signal is integrated with SynGAP activation remains to be determined. The EphB2 receptor has been shown to interact with the NR1 subunit and potentiate NMDAR-mediated calcium influx and downstream signaling cascades by phosphorylating the NR2B subunit. Lastly, the scaffolding protein Yotiao can bind to NR1 splice variants containing the C1 cassette and tether PKA and PP1 to the channel complex. While it is not known if this tethering is important for PKA-mediated NMDAR signaling (not shown), this interaction could mediate PP1-dependent signaling to the nucleus and dephosphorylation of CREB.
Some signaling proteins bind to both NR2A and NR2B, however they may show differential regulation at these subunits. NR2A and NR2B both have PDZ-binding domains at the C-terminal tails, allowing them to interact with the membrane-associated guanylate kinase (MAGUK) family of synaptic scaffolding proteins (Kennedy, 2000). These scaffolds in turn associate with important synaptic signaling molecules and tether NMDARs to intracellular signaling pathways. The mammalian MAGUK family is comprised of four members: postsynaptic density-95 (PSD-95), synapse-associated protein-97 (SAP-97), postsynaptic density-93 (PSD-93), and synapse-associated protein-102 (SAP-102) (Sheng and Sala, 2001). Although some studies suggested that NR2A-containing NMDARs might be more likely to bind PSD-95, while NR2B-containing NMDARs could preferentially bind SAP102 (Sans et al., 2000; Townsend et al., 2003), others found that PSD-95 and SAP102 interact with diheteromeric NR1/NR2A and NR1/NR2B receptors in the adult brain in vivo at roughly comparable levels (Al-Hallaq et al., 2007). However, NR2A-containing receptors co-immunoprecipitate with a greater fraction of the synaptic proteins neuronal nitric oxide synthase, Homer, and β-catenin (Al-Hallaq et al., 2007). None of these proteins bind directly to NMDARs; instead they are thought to interact indirectly via their association with the MAGUKs or other scaffolding molecules such as the Shank family (Sheng, 2001). These data are consistent with the possibility that the MAGUKs may be key for differential formation of signaling complexes at distinct NMDARs, although the mechanism of specificity remains to be determined. Additionally, whether these differential interactions with scaffolding molecules influences specificity of signals transmitted to the nucleus remains an unanswered question.
Another signaling molecule that binds to both NR2A and NR2B but exhibits distinct properties at these subunits is CaMKII. NR2A and NR2B have both been shown to bind CaMKIIα and both subunits can be substrates for CaMKII phosphorylation (Bayer et al., 2001; Gardoni et al., 1999; Strack and Colbran, 1998; Strack et al., 2000). However, the affinity of CaMKIIα binding to NR2B is greatly enhanced upon CaMKIIα autophosphorylation, suggesting a mechanism that may explain the stimulus-dependent trafficking of CaMKII to synapses (Bayer and Schulman, 2001; Shen and Meyer, 1999). Furthermore, the sites of CaMKIIα binding to the subunits differ – in NR2A the CaMKII binding site is near the C-terminus and competes with the PDZ domain interactions of the C-terminal tail (Gardoni et al., 2001), whereas in NR2B the binding site is farther away from the C-terminus (Bayer et al., 2001), potentially allowing NR2B to simultaneously interact with both CaMKII and PSD-95. Thus, while NR2A and NR2B can both bind to CaMKII, their specific modes of interaction with this important signaling molecule are fundamentally different, and this may differentially affect CaMKII-dependent gene transcription.
The ability of NMDARs to regulate CREB is known to require activation of the MAPK signaling pathway; thus, several studies have examined the differential recruitment to NR2 subunits of the upstream regulators of MAPK activation. The classic calcium-regulated MAPK cascade in neurons begins with signaling events that change the ratio of GTP:GDP-bound Ras (Rosen et al., 1994). Ras then activates the MAP kinase kinase Raf, which in turn activates the MAP kinase MEK, which then activates the MAP kinases Erk1 and Erk2. Erk1/2 activate various kinases including Rsk2, which then translocate to the nucleus to phosphorylate CREB at Ser133. The ratio of GTP:GDP-bound Ras is determined by the balance in the activation of the GTPase activating proteins (GAPs) that enhance the ability of Ras to hydrolyze bound GTP, and the GTP exchange factors (GEFs) that catalyze the release of GDP from Ras in exchange for binding GTP.
Two of the most important calcium-regulated Ras modulators in neurons are the synaptic Ras GTPase activating protein (SynGAP) and the GEF Ras-GRF1/2 (Cullen and Lockyer, 2002). SynGAP is highly concentrated in the PSD of excitatory glutamatergic synapses in the CNS and the GAP activity of this protein is enhanced upon phosphorylation by CaMKII (Chen et al., 1998; Oh et al., 2004). Ras-GRF1 contains a CaM-binding IQ domain that is required for the regulation of its ability to active MAPK signaling, suggesting that calcium-dependent regulation is conferred on Ras-GRF1 indirectly through calcium binding to CaM (Farnsworth et al., 1995). Ras-GRF2 also contains an IQ domain, but has a somewhat more complex mechanism of activation by calcium that involves additional protein-protein and protein-lipid interactions (Cullen and Lockyer, 2002). SynGAP is selectively found in synaptic complexes with NR2B-containing NMDARs where it may contribute to the ability of these receptors to suppress activation of MAPK signaling (Kim et al., 2005). Intriguingly, Ras-GRF1 is also enriched at NR2B-containing receptors (Krapivinsky et al., 2003), whereas Ras-GRF2 has been shown to selectively mediate signaling to MAPKs downstream of NR2A-containing receptors (Li et al., 2006b). How the effects of Ras-GRF1 and SynGAP signaling toward MAPK pathways are integrated at NR2B-containing receptors is not entirely clear, but combinatorial signaling through these Ras regulators appears to largely have a net effect of inhibiting Erk1/2 activation (Kim et al., 2005). By contrast, the Ras-GRF2 is required for NMDAR-dependent LTP (Li et al., 2006b), a cellular phenomenon largely ascribed as a downstream action of NR2A-containing NMDARs (Liu et al., 2004).
Finally, there are a few classes of proteins that do appear to be specifically bound to one or the other of the two NR2 subunits NR2A and NR2B. One protein that may be involved in the pro-death-associated signaling capabilities of extrasynaptic NMDARs (as apposed to the pro-survival signaling of synaptic NMDARs) is death-associated protein kinase 1 (DAPK1). DAPK1 has been shown to directly bind to the NMDAR subunit NR2B and to phosphorylate NR2B at Ser1303, thereby enhancing NR2B-containing NMDAR channel conductance (Tu et al., 2010). Importantly, genetic deletion of DAPK1 appears to uncouple NR2B-containing, extrasynaptic NMDARs from cell death-associated signaling pathways, while leaving NMDAR-mediated synaptic transmission unaffected. In mice deficient for DAPK1, activation of extrasynaptic NMDARs does not activate cell death, thus protecting them from ischemic insults (Tu et al., 2010).
In addition to proteins that associate with the different NR2 subunits, it is also possible to generate diversity amongst NMDAR-associated signaling complexes via proteins that differentially associate with alternatively spliced regions of the C-terminus of the NR1 subunit. The Grin1 gene encoding NR1 contains three alternatively spliced exons: exon 5 in the N terminus (also called the N1 cassette) and exons 21 and 22 in the C terminus (also known as the C1 and C2 cassettes) (Dingledine et al., 1999). There is evidence that the different C-terminal NR1 splice variants may couple differentially to downstream signaling cascades and subsequent changes in gene transcription. In conventional nomenclature, NR1-1 is the full-length NR1 variant containing both C-terminal exons, NR1-2 lacks exon 21 (the C1 cassette), NR1-3 lacks exon 22 (the C2 cassette), and NR1-4 lacks both (Dingledine et al., 1999; Hollmann et al., 1993). Exclusion of exon 22 by splicing removes the stop codon present in exon 22 and introduces a new coding region that encodes an unrelated domain termed C2′ (Zukin and Bennett, 1995). Bradley and colleagues have found that NMDARs lacking the C1 cassette (aka exon 21; NR1-2 and NR1-4 splice variants) are much less efficacious in their ability to activate gene transcription than are NMDARs that contain the C1 cassette (NR1-1 and NR1-3) (Bradley et al., 2006). Additionally, it has been shown that the splicing of the C2 cassette is activity-dependent, with increased activity leading to more C2-containing NR1 subunits and activity blockade yielding more C2′-containing NR1 subunits (Mu et al., 2003). This activity-dependent splicing could provide a mechanism of homeostatic control, as the C2′ domain is much more efficiently trafficked out of the ER and to the cell surface than the C2 domain.
A yeast two-hybrid screen for proteins that bind to NR1 identified Yotiao as a protein that interacts with the alternatively spliced C-terminal C1 exon cassette of the ion channel (Lin et al., 1998). Yotiao binds to both the cAMP-dependent protein kinase PKA and the protein phosphatase 1 (PP1) and is thought to be important for anchoring these proteins to NR1-1 and 1-3 containing NMDARs (Westphal et al., 1999). It has been suggested that activation of PP1, which can dephosphorylate CREB, is important for limiting the duration of NMDAR-mediated transcription (Bito et al., 1996). However, what effect the interaction of Yotiao with alternatively spliced NR1 subunits might have on NMDAR dependent transcription remains to be determined. Finally, two other NR1-interacting proteins that may contribute to the regulation of transcriptional pathways are the EF-hand-containing transcriptional repressor DREAM and the Eph family receptor tyrosine kinase EphB2. DREAM has been shown to bind to the membrane proximal region of the NR1 C-terminal domain (the “C0” domain) (Zhang et al., 2010a). The interaction of DREAM with NR1 significantly impairs NMDAR surface expression in neurons and is neuroprotective against NMDA-induced excitotoxic neuronal injury, however the functional consequence of this interaction for NMDAR-mediated transcription, and the regulation of DREAM-mediated transcription, is not known. Unlike the other interactions discussed so far, which are associated with the intracellular domains of the NMDA-R, EphB2 binds to the extracellular domain of NR1 (Dalva et al., 2000). EphrinB2-mediated activation of EphB2 receptors in primary cortical neurons potentiates NMDAR-dependent influx of calcium and subsequent CREB-dependent gene transcription (Takasu et al., 2002). The effects of EphB2 on NMDAR function are mediated by phosphorylation of the NR2B subunit at Tyr1252, Tyr1336, and Tyr1472 through the activity Src tyrosine kinase family members. The cooperative regulation of transcription by Eph receptors and NMDARs may be a mechanism to couple activity-independent and activity-dependent signaling pathways in early neuronal development (Takasu et al., 2002).
Finally, NR3A has been shown to engage in numerous protein-protein interactions that could have important implications for the function of NMDARs that contain this subunit. The C-terminus of NR3A has been shown to interact with several intracellular proteins, including PACSIN1/syndapin-1 (Pérez-Otaño et al., 2006), protein phosphatase 2A (PP2A) (Chan and Sucher, 2001), and the small GTPase Ras homologue enriched in brain (Rheb) (Sucher et al., 2010). The interaction with PACSIN1/syndapin-1 has been the most extensively studied, and has been shown to mediate activity-dependent endocytosis of NR3A-containing NMDARs, which may be important for the downregulation of NR3A surface expression during development (Pérez-Otaño et al., 2006). Association of PP2A with the C-terminal tail of NR3A increases the phosphatase activity of PP2A and leads to dephosphorylation of the NR1 subunit at the PKA-regulated site at Ser897 (Chan and Sucher, 2001). The association of NR3A and PP2A is inhibited by NMDAR stimulation, suggesting a mechanism by which control of this signaling event could be regulated by synaptic activity. The C-terminal tail of NR3B differs substantially from that of NR3A, suggesting that the protein-protein interactions of these two related subunits may be distinct. The role of either of the NR3 subunits in influencing NMDAR-dependent gene transcription remains unknown.
4.2. L-type Voltage-Gated Calcium Channels (L-VGCCs)
AMPARs comprise the other major class of glutamate receptors at synapses, and activation of these receptors by glutamate results in a large current influx that drives postsynaptic membrane depolarization. This depolarization then induces the opening of voltage-gated ion channels, including voltage-gated calcium channels (VGCCs). The VGCCs are commonly referred to by names that arose from aspects of the physiological properties of their currents – for example there are L-type (long-lasting activation), N-type (neurotransmitter release), T-type (transient activation), P/Q-type (found in Purkinje cells), and R-type (drug-resistant) channels. Molecular cloning of VGCC subunits has shown that a set of distinct α1 subunits confers the specific physiological properties upon each of these calcium channel types. Among the classes of VGCCs expressed in neurons, L-VGCCs have been shown to be a particularly salient mode of calcium entry in terms of their ability to bring about alterations in gene transcription (Bading et al., 1993).
4.2.1. Molecular biology of the L-VGCCs
All VGCCs are made up of a single, central α1 subunit, which both forms the ion- conducting pore and defines the channel type, along with a variable number of auxiliary α2-δ, β, and γ subunits (Dai et al., 2009) L-VGCCs are categorized as being high-voltage-activated channels (calcium channels that require strong membrane depolarization for activation) and are pharmacologically defined by their sensitivity to dihydropyridine agonists or antagonists (Dai et al., 2009; Dolphin, 2009; Lipscombe et al., 2004). The best-characterized L-VGCC α1 subunit in terms of ability to regulate gene transcription in neurons is CaV1.2 (Dolmetsch et al., 2001; Weick et al., 2003). Additionally, several biochemical and functional studies indicate that CaV1.2-containing channels account for at least 75–80% of all L-VGCCs in the brain (Hell et al., 1993a; Hell et al., 1993b; Koschak et al., 2007). The Cav1.3 α1 subunit is also highly expressed in the brain where it has been shown to contribute to activity-dependent induction of nuclear CREB Ser133 phosphorylation (Zhang et al., 2006). The α2-δ, β, and γ auxiliary subunits function to modulate channel surface expression, localization, biophysical properties, and intracellular protein-protein interactions (Chen et al., 2004; Dai et al., 2009; Lipscombe et al., 2004; Opatowsky et al., 2004; Van Petegem et al., 2004).
4.2.2. Physiological relevance of L-VGCCs for transcription
L-VGCCs are present along the dendrites and the somata of neurons. They have been shown to couple membrane depolarization in neurons to numerous cellular processes, and are thought to exert many of these actions through their effects on activity- and calcium-regulated gene transcription. Significant data implicate L-VGCCs in the activity-dependent regulation of neuronal survival. For example, Ghosh and colleagues demonstrated that activation of L-VGCCs promotes cell survival in cultured cortical neurons in a manner that requires BDNF, which can be transcriptionally induced by L-VGCC signaling (Ghosh et al., 1994a). L-VGCCs have also been shown to regulate dendritic growth in cultured cortical neurons in a calcium-, CREB-, and transcription-dependent manner (Redmond and Ghosh, 2005; Redmond et al., 2002). Interestingly, evidence suggests that the unique biophysical properties of L-VGCCs–namely their ability to activate at relatively negative potentials, and their slow activation kinetics as compared with other VGCCs–may allow them to act as a kinetic filter to support spike–EPSP discrimination in the activation of CREB-dependent gene transcription (Mermelstein et al., 2000).
Genetic evidence for a critical role of L-VGCCs in neuronal and other cellular physiology came when the CaV1.2 gene was shown to be mutated in Timothy Syndrome. This syndrome is characterized by multiorgan dysfunction including lethal arrhythmias, webbing of fingers and toes, congenital heart disease, immune deficiency, intermittent hypoglycemia, cognitive abnormalities, and autism (Splawski et al., 2005; Splawski et al., 2004). Patients with Timothy syndrome have a critical mutation in an alternatively spliced exon of the Cacna1c gene encoding CaV1.2, which results in the coding sequence mutation G406R. The G406R mutation produces maintained inward Ca2+ currents by causing nearly complete loss of voltage-dependent channel inactivation, a key property of normal L-VGCCs (Splawski et al., 2004). This could hypothetically lead to calcium overload in many excitable cell types, including neurons. Intriguingly, while cardiac arrhythmia due to delayed cardiomyocyte repolarization is the primary cause of death in Timothy syndrome (Splawski et al., 2005; Splawski et al., 2004), it remains to be determined what effect the prolonged inward Ca2+ currents might have on calcium-dependent gene transcription – particularly in the nervous system – and what role that could then play in the manifestation of plasticity-related neurological deficits such as autism.
4.2.3. Signaling pathways and protein interactions that regulate L-VGCCs
Like NMDARs, L-VGCCs have long been known to play an important role in activity-regulated gene transcription (Greenberg et al., 1986). However, several early studies demonstrated important differences in the ability of calcium influx through L-VGCCs versus NMDARs to activate specific transcriptional pathways (Bading et al., 1993; Ghosh et al., 1994b). For example, whereas the activation of transcription through the SRF-binding SRE element can be driven by calcium influx through NMDARs, the calcium rises that activate transcription through the CREB-binding CRE-element are more selectively coupled to L-VGCCs (Bading et al., 1993; Hardingham et al., 1997). Like other plasma membrane receptors, the specificity of L-VGCC signaling is thought to arise in part through the nature of the specific protein-protein interactions between these channels and components of intracellular signaling pathways. Strong evidence for this idea of local calcium signaling at the L-type channel came from an elegant study that examined the ability of calcium chelators with different calcium binding affinities and KON rates to block membrane depolarization induced Ser133 phosphorylation of CREB (Deisseroth et al., 1996). These studies showed that only the very rapid chelation of calcium, which inhibits the elevation of calcium with the microdomain at the mouth of the channel, is sufficient to block CREB Ser133 phosphorylation following L-VGCC activation. These data suggested that a local submembranous calcium sensor bound at or near the channel was required for transmission of the calcium signal to CREB.
One way in which L-VGCCs may be capable of coupling to downstream transcriptional signaling pathways is via their direct interaction with the calcium sensor CaM. An isoleucine-glutamine (“IQ”) motif in the C-terminus of the L-VGCC α1 subunit CaV1.2 allows the channel to bind CaM and this interaction is critical for conveying the Ca2+ signal through the channel to the nucleus (Dolmetsch et al., 2001). Specifically, cultured neurons transfected with a construct encoding a CaV1.2 in which the IQ domain has been mutated are deficient for activation of the Ras/MAPK pathway, which is normally activated following L-VGCC activation. Additionally, these neurons fail to exhibit CREB- or MEF2-dependent gene transcription following a stimulus that normally leads to L-VGCC activation (Dolmetsch et al., 2001). How the local activation of CaM might selectively couple L-VGCCs to CREB-dependent transcription is not entirely clear. CaM is involved in gating the calcium-dependent inactivation of L-VGCCs (Peterson et al., 1999), which could influence the amplitude or kinetics of downstream signaling pathway activation. In addition, some studies have suggested that CaM locally activated at calcium channels may translocate to the nucleus (Deisseroth et al., 1998). Nuclear CaM levels are elevated within 15 seconds of calcium channel activation, and this elevation precedes the phosphorylation of CREB (Mermelstein et al., 2001). However, in isolated preparations of hippocampal nuclei, elevation of calcium levels is sufficient to induce CREB phosphorylation in a CaMK-dependent manner indicating that CaM translocation from the synapse is not required to activate this signaling pathway (Hardingham et al., 2001). Furthermore, nuclear expression of a CaM-binding peptide that inhibits nuclear CaM blocks 43% of all genes induced by action potential firing in cultured hippocampal neurons, indicating that elevation of nuclear calcium is essential for controlling the expression of a large subset of activity-regulated genes (Zhang et al., 2009).
Protein-protein interactions that affect L-VGCC channel function may serve to regulate gene transcription programs by regulating the location or function of L-VGCCs themselves. For example, the tumor suppressor eukaryotic initiation factor 3 subunit E (eIF3E, also known as Int6) binds to the II-III loop of CaV1.2 in a calcium-dependent manner following electrical stimulation. This interaction facilitates the activity-dependent internalization of L-VGCCs into endosomes following neuronal activity (Green et al., 2007), and thus has the potential to function in the homeostatic regulation of L-VGCCs and their downstream signaling effects on the neuron. Interestingly, the lipid kinase phosphatidylinositol 3-phosphate 5-kinase (PIKfyve) has also been shown to bind to CaV1.2 in an activity-dependent manner (Tsuruta et al., 2009). Following NMDAR- induced binding of PIKfyve to CaV1.2, CaV1.2 is endocytosed and targeted to lysosomes for degradation. Tsuruta and colleagues suggest that this phenomenon could be a novel mechanism by which neurons guard against excitotoxicity, as knockdown of PIKfyve prevents CaV1.2 degradation and increases neuronal susceptibility to excitotoxicity (Tsuruta et al., 2009). It is important to note that the stimulus used in this study to activate NMDARs was the bath application of high concentrations of glutamate to cortical neurons in culture. It has been shown previously that this stimulus not only activates synaptic NMDARs (which induce pro-survival signaling pathways and gene transcription paradigms) but also activates extrasynaptic NMDARs, which have been shown to induce cell death in a manner that is dominant over the synaptic NMDAR-mediated signals (Hardingham and Bading, 2002, 2003; Hardingham et al., 2002). It would be interesting to determine whether the binding of PIKfyve to CaV1.2 and the subsequent internalization and degradation of CaV1.2 is specifically induced by activation of extrasynaptic NMDARs (which would indicate a possible compensatory role for this interaction) or by synaptic NMDARs (which would indicate that it functions as part of a larger pro-survival signaling paradigm induced by synaptic activation).
Finally, the activity of L-VGCCs is also subject to modulation downstream of other signaling cascades in addition to changes in cellular calcium. Thus, modulation of L-VGCC function may be a pathway through which other signaling cascades can alter gene transcription. For example, it has long been known that activity of L-VGCCs in neurons was subject to modulation by cAMP signaling (Gray and Johnston, 1987). Phosphorylation of the L-VGCC α1 subunit CaV1.2 by PKA enhances L-type Ca2+ currents by increasing the open probability (PO) of the individual channels in cardiac myocytes (Bean et al., 1984; Yue et al., 1990), and PKA modulates L-VGCCs in the dendrites of hippocampal neurons as well (Hoogland and Saggau, 2004; Kavalali et al., 1997). A substantial body of evidence indicates that the PKA-mediated enhancement of L-VGCC current specifically relies on the phosphorylation of Ser1928 in the C-terminal tail of the pore-forming CaV1.2 subunit (Catterall, 2000). The A kinase-anchoring protein 15 (AKAP15) is capable of directly interacting with the distal C terminus of the L-VGCC α1 subunit CaV1.2 via a leucine zipper-like motif, and this interaction is important for the β-adrenergic regulation of CaV1.2 channels via the PKA pathway (Hulme et al., 2003). However, while AKAP15 can direct PKA to its protein substrates, other AKAPs can function as signaling integrators by binding additional regulators other than PKA to target proteins. Recently, Oliveria and colleagues have shown that AKAP79/150 is capable of anchoring both PKA and calcineurin to CaV1.2 via a direct interaction between modified leucine zipper motifs in both proteins, and that this association is capable of modulating neuronal L-VGCC-mediated calcium currents (Oliveria et al., 2007). Interestingly, these authors also showed that AKAP79/150 targeting of calcineurin is necessary to couple Ca2+ influx via L-VGCCs to activation of the transcription factor NFATc4, thus providing a novel mechanism by which L-VGCCs might be capable of signaling to the nucleus.
4.3. Other sources of calcium
Although NMDARs and L-VGCCs appear to play the major roles in the regulation of calcium-dependent transcription in neurons, other sources of calcium may be important under select conditions. For example, although N-type voltage-gated calcium channels (N-VGCCs), are generally inactivated during persistent neuronal depolarization, transient activation of these channels has been shown to contribute to the induction of tyrosine hydroxylase expression that occurs following 5 Hz electrical stimulation of primary sensory neurons (Brosenitsch and Katz, 2001).
In addition, subsets of AMPARs can provide a source of calcium entry at the synapse. AMPARs are tetramers comprised of various combinations of the GluA1-4 subunits (formerly called GluR1-4 (Collingridge et al., 2009)). In the mature brain, the majority of AMPARs contain the GluA2 subunit and are impermeable to calcium due to the presence of a positively-charged arginine (R) residue in a key position in the GluA2 pore domain that blocks calcium influx. Notably, however, some GluA2-containing AMPARs are calcium permeable. This is because the calcium permeability of GluA2 is regulated post-transcriptionally by RNA editing. In the genome, the DNA sequence of the Gria2 gene encodes a negatively charged glutamine (Q) residue at the key selectivity residue in the GluA2 pore, rather than the R found in edited Gria2 mRNA (Lomeli et al., 1994). The Q/R switch occurs by editing of the Gria2 mRNA that is mediated by the enzyme RNA-dependent adenosine deaminase 2 (ADAR2) (Melcher et al., 1996). In addition to AMPARs with unedited GluA2, some AMPARs pass calcium because they lack GluA2. For example, early in development a significant fraction of AMPARs lack GluA2 (Kumar et al., 2002), and GluA2-lacking, calcium-permeable AMPARs are also expressed in interneurons of the adult cortex (Geiger et al., 1995). Furthermore, following the induction of LTP, the selective insertion of GluA1-only AMPA receptors has been reported, leading to the transient appearance of GluA2-lacking, calcium permeable AMPARs at potentiated synapses (Liu and Cull-Candy, 2000; Plant et al., 2006). This calcium could contribute to transcriptional regulation. It has been shown that pharmacological activation of AMPARs in striatal and cortical neurons leads to a MAPK- and calcium-dependent phosphorylation of the transcription factor CREB, raising the possibility that AMPARs could function in some cases to activate changes in gene transcription (Perkinton et al., 1999; Tian and Feig, 2006). However, whether calcium influx through AMPARs contributes to the regulation of specific transcriptional programs has not been well established.
Finally, neuronal transcription can also be regulated by calcium release from intracellular calcium stores (Hardingham et al., 2001). In order to utilize the highly abundant ion calcium as a second messenger, all cells tightly regulate intracellular calcium levels by binding or sequestering the vast majority of free calcium (Clapham, 2007). The endoplasmic reticulum (ER) serves as a particularly large and important reservoir of stored calcium. In response to extracellular signals that activate receptors coupled to Gq-type G proteins or phospholipase C (PLC), calcium is released from the ER into the cytoplasm through IP3- and ryanodine receptors (Berridge, 2002). In non-excitable cells such as lymphocytes, this depletion of intracellular calcium stores is coupled to the opening of calcium release-activated calcium (CRAC) channels in the plasma membrane. Lymphocytes lack the VGCCs that are so important for activity-dependent transcription in neurons, and in non-excitable cells it is the CRAC channels that serve as the major source of calcium for stimulus-dependent transcriptional regulation (Hogan et al., 2010). Store-operated calcium entry depends on the proteins known as stromal interaction molecules 1 and 2 (Stim1 and Stim2), which sense the depletion of calcium in the ER. The Stim proteins then form a complex with the plasma membrane protein Orai, which forms the pore subunit of the CRAC channel.
Stim1 is expressed in the brain, where it has been shown to interact directly with the C-terminal tail of the L-VGCC subunit Cav1.2 (Park et al., 2010). Interestingly, the association of Stim1 with Cav1.2 suppresses depolarization-induced opening of the L- VGCC channel and causes long-term depletion of L-type channels from the plasma membrane by promoting their internalization. These data suggest that the actions of Stim1 and L-VGCCs are reciprocally regulated in neurons, raising the possibility that they could signal the activation of distinct downstream events. One stimulus that may utilize this interplay between these calcium sources to differentially regulate transcription is the activation of nicotinic acetylcholine receptors (nAChRs). Activated nAChRs are capable of inducing CREB Ser133 phosphorylation and activating Fos expression (Chang and Berg, 2001; Hu et al., 2002). This transcriptional response requires calcium influx across the plasma membrane as well as subsequent release of calcium from internal stores, and is thought to signal to the nucleus through CaMK and MAPK pathways. Interestingly, however, sustained CREB phosphorylation is only driven by nAChRs if L-VGCCs are silent (Chang and Berg, 2001). As described above, Stim1 is one potential mechanism that could couple intracellular calcium release and silencing of L-VGCCs (Park et al., 2010). Future studies that address the requirements for Stim1/2 in stimulus-regulated transcription in the nervous system may reveal selective functions for intracellular calcium stores in response to different kinds of extracellular stimuli.
5. Roles for activity-regulated transcription in homeostatic synaptic plasticity
We have concentrated on describing the signaling pathways that confer specificity upon transcriptional regulatory processes induced by neuronal activity. Now we turn our attention to considering how this biochemical selectivity is translated into differential biological outcomes. As an example, we consider the transcription-dependent processes that regulate homeostatic synaptic plasticity in response to elevations versus reductions in synaptic activity.
Neuronal activity-regulated transcription has been shown to contribute to many biological processes in the brain including neurite outgrowth (Spitzer, 2006; Wayman et al., 2006), cell fate determination (Borodinsky et al., 2004), synapse development (Greer and Greenberg, 2008), critical period plasticity (Sugiyama et al., 2008), long-lasting changes in synaptic strength (Alberini, 2009), and neural adaptations to drugs of abuse (Nestler, 2001). In some cases such as cortical development, the activity-dependent processes appear to be inductive. For example, in the visual cortex, the onset of patterned visual input from the two eyes and the subsequent activity-regulated, transcription-dependent maturation of inhibition induces the closure of the critical period for experience-dependent rewiring of inputs from the two eyes (Fagiolini et al., 2004). However, more often the actions of activity-regulated transcription appear to be largely homeostatic. The term homeostasis refers to phenomena by which a single cell, an organ system, or an entire organism maintains a stable internal environment in the face of changing external stimuli (Cannon, 1932).
While some homeostatic mechanisms in the nervous system occur at the level of the neural network or circuit, there also exist cellular homeostatic plasticity mechanisms that are able to mediate the scaling of excitatory synaptic strength following changes in synaptic activity within a single cell (Burrone and Murthy, 2003; Turrigiano, 2007). It has been proposed that for a molecular mechanism to function as a homeostatic regulator of synaptic scaling that the integrator needs to be able to sample the overall activity of the cell’s synapses and feedback to modulate synaptic function globally (Turrigiano, 2007; Turrigiano and Nelson, 2004). Activity-regulated transcription factors are well-suited to play this role, as they serve as substrates for converging signaling pathways, they are centrally located in the cell nucleus, where they respond to signals integrated from all over the cell, and they function on a slow time course relative to synaptic signaling, allowing them to respond to accumulated changes in activity over time. Furthermore, many activity-regulated transcription factors control the expression of gene expression programs that function at synapses (West et al., 2001).
However, one additional tenet of the integrator hypothesis is that a homeostatic mechanism is expected to be bidirectionally responsive – such that when activity is enhanced the integrator mechanism can scale the strength of synapses down, whereas when activity is reduced the mechanism can scale synapses up. Early gene expression studies primarily focused on the ability of neuronal transcription to be induced by increases in activity and even seemed to suggest that activity-dependent decreases in transcription were less common than increases in gene expression (Nedivi et al., 1993). However, with improved sensitivity in the techniques for the detection of mRNA levels and an increasing range in the kinds of stimuli that can be used to alter neural activity, it has become clear that transcription can be bidirectionally regulated by changes in neural activity, expanding the range of biological stimuli for which this mechanism could be relevant (Kim et al., 2010; Zhang et al., 2007). Intriguingly, two recent complementary studies have demonstrated the importance of transcriptional mechanisms for both up- and downregulation of synaptic strength following changes in cell firing (Goold and Nicoll, 2010; Ibata et al., 2008) (FIGURE 7). These studies highlight the idea that specificity in the regulation of transcriptional pathways can have highly specific, and in this case opposing, effects on cellular physiology.
FIGURE 7. Transcriptional regulation of homeostatic synaptic scaling.
Changes in synaptic activity drive homeostatic compensatory changes in the number of surface AMPARs at synapses. Downregulation of synaptic AMPARs in response to increased activity requires the influx of calcium (Ca2+) through L-VGCCs and activation of a CaMKIV-dependent transcriptional pathway (green arrow). Upregulation of synaptic AMPARs in response to decreased synaptic activity is driven by reduced calcium influx through L-VGCCs, and a decrease in activity of the CaMKK/CaMKIV pathway (red bar). Because transcriptional activity is required for scaling induced by either increases or decreases in synaptic activity, these data that suggest that distinct classes of calcium-regulated transcription factors (one group activated by CaMKIV and one repressed) and distinct sets of target genes (Gene A and Gene B) mediate the two sides of the pathway.
In 2008, Ibata and colleagues published a landmark study demonstrating that transcription is required for the ability of cortical neurons to homeostatically adjust synaptic strengths in response to changes in their own firing rates (Ibata et al., 2008). To monitor synaptic strength, cells were transfected with GluR2 tagged at the extracellular N-terminus with enhanced yellow fluorescent protein (eYFP). Since the fluorescence of eYFP is highly pH-sensitive, the signal is quenched when GluA2 is held in intracellular endosomes. Thus, plasma membrane insertion of GluA2 can be detected as an acute increase in eYFP fluorescence at synapses. Although synaptic scaling has previously been studied on longer time scales, using this live imaging method the authors discovered that bath application of the sodium channel blocker tetrodotoxin (TTX) for as little as 4 hours resulted in the accumulation of AMPARs at synapses. Local application of TTX to the soma of a single neuron in the culture was sufficient and necessary to induce this synaptic scaling effect, demonstrating that it is somatic action potential firing that underlies the change in synaptic AMPAR number. Inhibition of somatic calcium signaling by local application of either the broad spectrum calcium channel blocker NiCl2 or the L-VGCC antagonist nifedipine was also sufficient to induce the increase in synaptic AMPARs, indicating that a decrease in somatic calcium signaling could underlie this effect. Phosphorylation of the calcium-regulated nuclear kinase CaMKIV was reduced when TTX was applied to the cells, and overexpression of a dominant-negative CaMKIV occluded the effects of TTX on AMPAR accumulation at synapses. Most strikingly, co-application of the transcriptional inhibitor actinomycin D (ActD) blocked the ability of either TTX or the CaMKK2-CaMKIV inhibitor Sto-609 to increase synaptic AMPARs. Taken together, these results demonstrate that these effects of reducing activity and calcium signaling on synaptic scaling depend on the activation of transcription.
In an aesthetically pleasing complement to this study, in 2010, Goold and Nicoll examined the cellular homeostatic mechanisms that are involved in synaptic scaling during periods of increased neuronal activity (Goold and Nicoll, 2010). To change the firing of single neurons within an organotypic hippocampal slice, the authors used optogenetic techniques, transfecting single cells with the light-gated ion channel channel rhodopsin 2 (ChR2). ChR2 is a non-selective cation channel that is opened upon exposure to blue light. Neurons expressing ChR2 can be made to fire following ChR2 activation at rates up to 50Hz (Boyden et al., 2005). Using electrophysiological measures of synaptic function, the authors found that cell autonomous activation of neuronal firing resulted in a downregulation of both synaptic AMPARs and synaptic NMDARs. Pharmacologically, the scaling of these synaptic proteins relied on a CaMKK2- and CaMKIV-dependent signaling cascade. However, while the synaptic depression of AMPARs was blocked by inhibitors of transcription and translation, the synaptic depression of NMDARs was not, indicating a divergence in the cellular mechanisms responsible for the downregulation of these two different types of glutamate receptors following increased neuronal activity (Goold and Nicoll, 2010).
Taken together, these two studies point toward CaMKIV and GluA2 as key nodes in the signaling pathway that controls bi-directional transcription regulation of homeostatic synaptic scaling in single neurons. However, the transcription factors and gene targets that mediate this process remain unknown. CREB is a known target of regulation by CaMKIV, but most experimental manipulations of CREB appear to be positively coupled to synapse formation and thus seem unlikely to be homeostatic (Impey et al., 2002; Wayman et al., 2008a). MEF2 and SRF have been highly linked to homeostatic regulation of excitatory synapse number or strength (Flavell et al., 2006; Smith-Hicks et al., 2010), however the regulation of these factors by CaMKIV has not been reported. Furthermore, very little is known about transcriptional mechanisms that activate transcription during periods of decreased synaptic activity. As described in the transcription factor section above, nuclear translocation of the transcription factor CCAT has been observed following treatment of cells with TTX (Gomez-Ospina et al., 2006). CCAT is derived from the C-terminal tail of the L-VGCC subunit Cav1.2 and it accumulates in the nucleus under conditions of low intracellular calcium (Gomez-Ospina et al., 2006). It is highly likely that there are additional factors that respond to decreases in neuronal activity. Future studies that identify these factors and study their roles in synaptic scaling will expand our understanding of how transcriptional specificity is achieved in this paradigm.
6. Technological advances for studying neuronal activity-regulated transcription
Despite substantial progress in identifying the molecular components of neuronal activity-regulated transcription in neurons, important questions regarding the mechanisms and consequences of this biological process remain to be addressed. As indicated by the theme of this review, one of the most significant directions of recent research has been to elucidate mechanisms that can confer specificity upon transcriptional regulation. However, even though the activation of a transcription factor might be subject to differential phosphorylation downstream of calcium influx through L-VGCCs versus NMDARs, for many factors it remains to be resolved what the precise biological consequence of this specificity might be for a neuron in vivo. In addition, since the regulation and function of single transcriptional pathways are often studied in experimental isolation, much remains to be learned about how single transcription factors integrate into the highly interconnected signaling networks that comprise the native cellular context. Finally, because many of the methods used to study transcriptional regulatory pathways are biochemical techniques that average the effect of experimental manipulations over a population of cells, significant questions arise about whether there exist transcriptional variations between individual neurons and how these variations could influence brain function. These questions of biological significance, and mechanistic or cellular specificity, are likely to dominate the field of neuronal activity-regulated transcription in coming years. Here we review some of the technological innovations that are helping to advance each of these areas of inquiry.
6.1. Genetic techniques to study the function of transcription factors
Transcription factors are like any other gene product in that understanding the biology of these proteins has been greatly advanced by the developments in experimental molecular genetics that allow the generation of knockout mouse strains. However, activity-regulated transcription factors present three specific problems that limit the usefulness of conventional mouse knockouts. First, most transcription factors belong to large protein families with related DNA binding domains and overlapping functions. Thus, compensation by related family members can limit the detection of phenotypes in single transcription factor knockout strains, whereas embryonic lethality is a frequent complication of double or triple knockout combinations. Second, knockout technology has poor temporal resolution and thus is not well suited to isolating the activity-dependent component of transcription factor function independent of functions in brain development. Finally, since transcription factors regulate large sets of target genes, knockout of any given factor is likely to have effects on a large number of these genes, challenging the ability of this loss-of-function genetic approach to identify the biological significance of any given transcription factor-target gene interaction.
6.1.1. RNA interference (RNAi) – spatial, temporal, and cell-type specific gene disruption
The genetic analysis of protein function has been dramatically enhanced with the advent and widespread use of RNAi technology (Novina and Sharp, 2004; Zeringue and Constantine-Paton, 2004). A significant advantage of RNAi is that it is easier using this technology to disrupt several RNA species at the same time, compared with crossing multiple strains of knockout mice (Paradis et al., 2007), and thus could be useful for the functional study of transcription factor families.
One key application of RNAi technology for neuronal activity-dependent transcription studies is virally-mediated RNAi delivery into the brain of adult animals to study the function of specific transcription factors in stimulus-dependent behavioral plasticity. The regional knockdown of neuronal proteins using stereotactically injected viruses allows the functional contributions of these pathways to be isolated to specific parts of defined neuronal circuits. This spatial specificity has revealed that altering activity-dependent transcription in different brain regions can have substantially different effects on behavior. For example, genetic manipulations that reduce BDNF expression in subcortical regions (nucleus accumbens, caudate/putamen, ventral tegmental area) are associated with impaired susceptibility to addictive-like behaviors in rodents treated with psychostimulant drugs of abuse (Graham et al., 2007; Im et al., 2010; Pu et al., 2006), whereas genetically reducing BDNF expression in the prefrontal cortex enhances addictive-like behaviors in these same paradigms (Sadri-Vakili et al., 2010).
The temporally defined gene knockdown afforded by this methodology allows the function of proteins in the adult brain to be isolated from roles they may have during development. For example, both the activity-regulated transcriptional activator MEF2 and the methyl-DNA binding protein MeCP2 are known to play important roles in synapse development (Barbosa et al., 2008; Dani and Nelson, 2009; Deng et al., 2010; Wood et al., 2009; Zhang et al., 2010b). Yet these proteins are also expressed in the adult brain where both of these transcription factors were identified as targets of psychostimulant-induced signaling cascades, raising the possibility that they might contribute to the transcriptional programs that underlie psychostimulant-induced behavioral modifications (Deng et al., 2010; Pulipparacharuvil et al., 2008). Using virally-mediated RNAi in adult mice, knockdown of MEF2 in the nucleus accumbens was shown to impair behavioral sensitization to repeated cocaine treatment, while knockdown of MeCP2 enhanced the rewarding properties of amphetamine as measured by the expression of conditioned place preference (Deng et al., 2010; Pulipparacharuvil et al., 2008).
Other variants of viral vectors allow for cell-type specificity of the knockdown by combining overexpression or RNAi with cell-type specific Cre recombinase expression. In this method, the cDNA or RNAi target sequence is cloned into a viral vector either after a floxed stop codon or in reverse orientation between two sets of loxP sites (Atasoy et al., 2008; Stern et al., 2008). When injected into the brain, although the virus infects many cells, only in those cells expressing the Cre recombinase will the overexpression or RNA interference cassette be correctly positioned with respect to the promoter to drive expression. This is an important new addition to the toolkit for the study of specificity in activity-regulated transcription.
6.1.2. Strategies for isolating the activity-dependent functions of transcription factors
Perhaps the most significant genetic challenge for this field has been discovering ways to selectively disrupt the activity-dependent functions of transcription factors without altering their basal activities. There have been essentially two approaches to this problem. One is to alter the transcription factor or its cofactors in ways that selectively disrupt the activity-dependent component of their function. The other is to dissociate the actions of the transcription factor from its ability to confer activity-dependent regulation on a target gene.
Capitalizing on the fact that the stimulus-inducible activity of many transcription factors depends on the local recruitment of the HAT CBP, Korzus and colleagues devised a genetic strategy to inducibly block CBP function in the adult brain. This mouse strain overexpresses a tetracycline-inducible allele of full-length CBP that bears two amino acid substitutions that abolish the HAT activity of CBP (CBP[HAT−/−]). Animals in which CBP[HAT−/−] transgene expression is activated during adulthood exhibit long term, but not short term, memory deficits that can be fully rescued by subsequent suppression of transgene expression or administration of a HAT inhibitor (Korzus et al., 2004). These data provide important support for the possibility that CBP-dependent stimulus-induced gene transcription is important for learning and memory.
Another strategy for disrupting stimulus-dependent functions of transcription factors that are targets of calcium signaling cascades is to generate knockin mice that bear germline mutations of activity-regulated transcription factor phosphorylation sites. For example, activation of calcium signaling cascades leads to phosphorylation of CREB on several serine residues (Shaywitz and Greenberg, 1999). Whereas phosphorylation of Ser133 is required for most CREB activity and is broadly induced by all stimuli that activate CREB-dependent transcription, induced phosphorylation of CREB at Ser142 and Ser143 is selective for calcium signaling pathways and may provide a modulatory influence on CREB function (Kornhauser et al., 2002). In vivo, as described in section 3.1.1, CREB Ser142 phosphorylation is induced in neurons of the suprachiasmatic nucleus (SCN) by light exposure. Thus to address the potential functions of this modification in light-induced plasticity of SCN function, Gau and colleagues generated a transgenic mouse strain in which a mutation was knocked into the endogenous Creb1 locus that changes Ser142 to an alanine residue, rendering CREB non-phosphorylatable at this site (Gau et al., 2002). Light-induced phase shifts of locomotion and expression of c-Fos and Per1 in the SCN are significantly attenuated in the knockin mutants, demonstrating that Ser142 phosphorylation of CREB is involved in the entrainment of the mammalian clock.
The phosphorylation site knockin and RNAi concepts for studying activity-regulated transcription factors can be combined into viral replacement strategies. For example, Zhou and colleagues developed a bicistronic lentiviral vector that simultaneously expresses an shRNA to knockdown endogenous MeCP2 while also expressing an RNAi-resistant version of either wildtype or Ser421Ala mutant MeCP2 (Zhou et al., 2006). Using this vector, the authors found that overexpression of wildtype MeCP2 in hippocampal neurons in organotypic slice culture leads to a decrease in dendritic spine density. By contrast, replacement of endogenous MeCP2 with overexpressed Ser421Ala mutant MeCP2 has no effect on spines. These data suggest that phosphorylation of MeCP2 is important for its ability to regulate dendritic spine density, supporting a role for the activity-dependent modulation of this protein in synapse development and plasticity.
Rather than targeting the transcription factor, knockin strategies can also be used to disconnect the activation of a transcriptional signaling pathway from its activity-regulated target gene. Transcription factors act on their target genes by binding to specific sequence elements in gene promoters or other genomic regulatory regions. Mutation of a genetic regulatory element to a sequence that does not permit transcription factor binding disrupts this r egulatory interaction without affecting other functions of the transcription factor and leaving intact alternative genomic regulatory elements through which the target gene can be regulated by other factors. For example, in the case of the Bdnf gene, to demonstrate the requirement for the CREB-regulated CaRE3 site in activity-dependent regulation of Bdnf exon IV transcription, Hong and colleagues generated a mouse strain bearing a mutation knocked into Bdnf promoter IV that changes the CaRE3 sequence (TCACGTCA) to a sequence that does not support CREB family protein binding (CAGCTGCA) (Hong et al., 2008). Neurons from CaRE3 mutant mice have normal basal levels of BDNF but selectively lack activity-inducible transcription from promoter IV. Interestingly, mice bearing this mutation have reduced numbers of GABAergic synapses. Although BDNF levels had been linked with GABAergic synapse development prior to this study (Genoud et al., 2004; Huang et al., 1999), the experiments in this mouse strain were the first to demonstrate that it is the activity-dependent component of BDNF expression that is selectively required for this synapse development process.
Finally, whereas transgenic mouse generation is a laborious process, the role of activity-regulated transcriptional elements can also be more rapidly assayed using the expression of proteins from recombineered bacterial artificial chromosomes (BACs). BACs are large DNA constructs that can be used to clone, propagate, and transfer genomic DNA regions that range from 150–300kbp. Thus, for any given gene, the BAC can contain the complete coding region as well as the large majority of the genetic regulatory elements required for proper regulated expression of the gene. Transfection of a BAC into a cell can be used to drive expression of a gene of interest within the genomic regions carried in the BAC, and mutation of genetic regulatory elements within the BAC can be used to test the function of these elements. For example, Smith-Hicks and colleagues recently used this strategy to study the role of SRF-dependent Arc transcription in cerebellar LTD. In this study, the authors showed that the impaired LTD found in cultured cerebellar neurons from Arc knockout mice can be rescued by re-expression of Arc from a transfected BAC (Smith-Hicks et al., 2010). However, if the SRF binding site in the synaptic activity-regulated enhancer of the Arc gene was mutated to a sequence which does not support wildtype SRF binding, the BAC was no longer able to drive stimulus-dependent Arc expression or to rescue LTD. Most elegantly, when an SRF engineered to bind the mutated SRF binding site was co-transfected with the mutated BAC, Arc expression as well as LTD were once again restored. Additional future studies of this kind will undoubtedly expand our understanding of the functional consequences of activity-regulated gene transcription.
6.2. A systems biology approach to neuronal activity-regulated gene transcription
Advances in high-throughput sequencing technologies are revolutionizing the quantitative study of transcription. Large-scale sequencing efforts started evolving rapidly at the time of the publicly-funded International Human Genome Project (Watson, 1990). Although this project, and the parallel effort led by Celera Genomics, led to the development of many of the sequence alignment, sequence data visualization, and data storage tools still used today, the actual sequencing of the human genome was accomplished with incremental modifications of the Sanger sequencing methods first developed in the 1970s (Sanger et al., 1977). However, in 2004, a new and far more rapid method of sequencing was introduced. Instead of sequencing a single strand of DNA at a time, the next generation sequencers (which include the Roche 454, the Solexa/Illumina Genome Analyzer, the SOLiD platform from Applied Biosystems, and the new single molecule platform from Helicos called the HeliScope) use massively parallel sequencing platforms that allow them to simultaneously sequence up to several hundred million DNA fragments in parallel, generating a very large number of short (50–500bp) sequence reads (McPherson, 2009).
There are three main advantages to next-generation sequencing. First, the large volume of sequence data from these techniques means that the content and dynamic range of the information obtained can outstrip that obtained with other technologies such as gene expression microarrays, for example. Second, large volumes of sequence information can be obtained very rapidly permitting analysis of large sets of experimental samples. Finally, the cost of next-generation sequencing has fallen to the point where it is within the reach of individual investigators and thus can be applied to a broad range of experimental questions. For these reasons, high-throughput sequencing represents one of the most rapidly expanding current experimental technologies. We will discuss three areas where sequencing has influenced the understanding of neuronal activity-regulated transcription – comparative genome sequencing, whole transcriptome sequencing (RNA-Seq), and genome-wide chromatin immunoprecipitation (ChIP-Seq).
6.2.1. Comparative genomics for identification of genetic regulatory elements
One of the first applications of genomic technology for the study of transcription was the use of evolutionary comparisons between multiple species to identify conserved blocks of non-coding DNA sequences that contain gene regulatory elements (e.g., promoters, enhancers, insulators, silencers) (Pennacchio and Rubin, 2001). Like coding sequences, gene regulatory elements constitute a small fraction of the total genome. However, unlike coding sequences, which can be identified by aligning sequenced cDNA with genomic DNA, traditional methods for identifying regulatory elements have relied on labor-intensive functional evaluation in biochemical or cell biological laboratories. Once the genomes of multiple species became available (the first comparisons were of human and mouse, then human, mouse, and pufferfish), cross-species comparisons of sequence showed that there were significant regions of conservation not only within coding but also in non-coding regions of the genome. Analysis of these conserved regions has helped to localize certain kinds of gene regulatory elements, such as distal enhancers of transcription that control cell-type specific gene transcription (De Val et al., 2008; Visel et al., 2008). In addition, analysis of single-nucleotide polymorphisms within these conserved regions has increased understanding of the mechanisms by which variations within non-coding regions of the genome may confer individual risk for disease susceptibility (Visel et al., 2009). An important limitation of this analysis is that sequence conservation does not necessarily imply conserved function, and many studies have shown that evolutionarily conserved sequences are utilized in a tissue-specific and/or species-specific manner (Heintzman et al., 2009; Schmidt et al., 2010).
6.2.2. Transcriptional complexity revealed by whole transcriptome sequencing
Quantitative analysis of RNA expression, whether performed by RNAse protection, quantitative polymerase chain reaction (PCR), or microarray hybridization, has always been an essential part of transcriptional studies. RNA-Seq is the latest addition to this arsenal (Wang et al., 2009; Wilhelm and Landry, 2009). Like other RNA analysis techniques, this process begins with the synthesis of cDNA. However, in most of the methods described above, only specific target segments within the RNA are quantified. For RNAse protection, the 5′ end of the RNA is the target sequence; for quantitative PCR, whatever fragment lies between the gene-specific primers is measured; for microarray hybridization, it is usually a small region of homology to a short oligonucleotide that is quantified, though there are often several oligonucleotides on a microarray that recognize the RNA transcribed from any single gene. By contrast, in RNA-Seq the entire cDNA for each transcript is fragmented into small pieces and subjected to quantitative sequencing.
A major advantage of this technique is that it facilitates the identification and quantification of RNA variants, such as alternative transcriptional start sites, alternative exon usage, and different sites of polyadenylation (Cloonan et al., 2008; Mortazavi et al., 2008). Furthermore, because the sensitivity of the technique can be enhanced by increasing the number of sequence reads, it can potentially be sensitive enough for use with small amounts of starting material (Tang et al., 2009). These are very early days for RNA-Seq, and to date only one landmark study has utilized the technique for the study of activity-regulated gene expression in neurons (Kim et al., 2010). However, significant technological advances are likely to come quickly that will enhance the use of this technology. Specifically the large amount of information about RNA complexity that can be obtained from a single RNA-Seq experiment means that there remain significant bioinformatic hurdles to be crossed, such as the development of software tools for processing this information, before the technique can be widely applied. As software programs become available and the cost of sequencing continues to fall, it is likely that RNA-Seq will begin to replace microarray hybridization as the technique of choice for RNA profiling experiments, opening the potential to significantly expand our understanding of the complexity of transcriptional regulation.
6.2.3. ChIP-Seq describes the chromatin landscape
Gene regulation by transcription factors depends on the binding of these proteins to specific sequences in genomic DNA. Where any given transcription factor binds largely determines its set of potential target genes, thus a comprehensive knowledge of the position of transcription factor binding sites across the genome is essential for understanding transcription factor biology. The major biochemical method used to study transcription factor-genomic DNA interactions in vivo is that of chromatin immunoprecipitation (ChIP) (Aparicio et al., 2004) (FIGURE 8).
FIGURE 8. ChIP-Seq defines transcription factor binding sites genome-wide.
The steps in a ChIP-Seq protocol are diagrammed. Blue and red circles represent two different DNA binding proteins. An antibody specific for one of the transcription factors selectively co-immunoprecipitates the subset of DNA fragments to which that factor is crosslinked. For the final step, the black line diagrams represent genes visualized across a region of a chromosome. Vertical lines represent exons and arrows show the direction of transcription. The blue and red diagrams represent the distribution of sequences obtained after co-immunoprecipitation with either the red or blue transcription factors. In this example, the red protein is preferentially bound immediately upstream of the TSS for each of the three genes shown, whereas the blue protein shows additional binding sites outside of proximal promoters (gray boxes).
Traditionally, quantitative PCR has been used to measure the co-immunoprecipitation of specific target sequences with a transcription factor of interest. However, when combined with high-throughput sequencing techniques (ChIP-Seq) it is now possible to obtain an unbiased view of transcription factor binding across the genome (Schmidt et al., 2009). In this variation of the technique, all of the genomic DNA fragments co-precipitated with the transcription factor are purified, size selected, and cloned with adaptors for sequencing on the SOLiD or Solexa/Illumina platforms to a depth of 20–40 million reads. Mapping these fragments to the reference genome gives a profile of the DNA regions that are enriched in the pulldown. An important challenge is to determine which of the enriched regions (“peaks”) are statistically significant at a given threshold and which correspond to the genomic background.
Because ChIP-Seq makes no assumptions about where transcription factors are likely to be bound, the results of these studies have revealed new fundamental aspects of transcription factor biology (Farnham, 2009). For example, because traditional biochemical assays of transcription factor function rely on short regions of genomic regulatory sequences to drive the expression of reporter genes, the vast majority of studies in the literature have focused on the ability of transcription factors to bind sequences within a few hundred base pairs of the transcription start site for a candidate target gene. Genome-wide studies have shown that some transcription factors, such as the cell-cycle regulating factor E2F, CREB, and USF1/2 do in fact appear to preferentially bind to proximal gene promoters (Bieda et al., 2006; Kim et al., 2010; Rada-Iglesias et al., 2008). However, other factors including SRF, Npas4, and CaRF have far more diverse binding patterns in which they associate with both extragenic and intragenic (exonic and intronic) regions as well as proximal gene promoters (Kim et al., 2010; Pfenning et al., 2010). A second theme that has emerged is that transcription factors do not always bind to strict consensus sites. The binding site for the transcription factor can be inferred by examining the sequences within the co-immunoprecipitated peak. Some transcription factors such as the neuronal repressor RE1-silencing transcription factor (REST) show high enrichment for a specific motif (Johnson et al., 2007). Some other transcription factors such as SRF have multiple subsets of binding sites (Valouev et al., 2008). Others such as CREB and MEF2 bind both consensus and non-consensus sites (Flavell et al., 2008; Impey et al., 2004), whereas some such as E2F show no sequence specificity whatsoever (Rabinovich et al., 2004). Although the mechanisms of these interactions are not known, this knowledge increases the diversity of potential regulatory mechanisms.
Studying the sites of transcription factor binding can yield new insights into transcription factor function. For CREB, the observation that CREB was binding near annotated positions of microRNAs led to the understanding of the role of CREB in regulation of non-coding as well as coding RNA sequences in the genome (Cheng et al., 2007; Impey et al., 2004; Vo et al., 2005). For the unique transcription factor CaRF, for which prior to ChIP-Seq only a single target gene (Bdnf) was known, the distribution of CaRF binding sites suggested a role for this factor in coordinating the expression of a number of important neuronal signaling pathways including a number of proteins that regulate calcium signaling (Pfenning et al., 2010).
However, ChIP-Seq still has several important limitations that remain areas for development in the future. One concern is to develop techniques to resolve the cell-type specificity of transcription factor binding profiles. Current methods for ChIP-Seq require large amounts of chromatin that are usually derived from pooled sources of cells and from heterogeneous brain tissue. Interestingly, the BacTRAP technology pioneered by Nathaniel Heintz and colleagues may offer a solution to this problem. The BacTRAP system was developed to isolate the full complement of actively translating mRNAs from specific sets of cells (Heiman et al., 2008). In these transgenic mice, BACs are used to drive cell type-specific expression of a ribosomal protein fused with enhanced green fluorescent protein (eGFP). Because ribosomal proteins are assembled in the nucleolus of all cells, the eGFP fluorescence can also be used for fluorescence-activated cell sorting of specific subsets of nuclei. Although this technique has not yet been applied to ChIP-Seq, it has been used for the isolation of Purkinje cell nuclei that led to the identification of the novel chromatin modification 5-hydroxymethylcytosine (Kriaucionis and Heintz, 2009). Another significant issue that need to be resolved is that of target gene assignment for ChIP peaks. Most often it is assumed that a site of transcription factor binding will allow that factor to modulate transcription of the nearest gene. However since genomic DNA is wound into a complex secondary and tertiary structure, transcription factor-gene interactions could be brought together across large expanses of primary sequence. Future insight into these long-distance interactions is likely to come from detailed studies of the spatial organization of chromosomes using techniques such as chromosome conformation capture (3C) and its genome-wide variant Hi-C (Dekker et al., 2002; Lieberman-Aiden et al., 2009). Advances in this area will be aided by close collaboration between neurobiologists, biochemists, genome biologists, and computational biologists.
6.3. Imaging approaches to neuronal activity-regulated transcription
Transcription studies are most often conducted using large-scale biochemical techniques like the RNA analyses and ChIP studies described in the section above. These are very effective techniques for the identification of molecular mechanisms that transduce synaptic activity into new gene transcription. However, as biochemical methodology yields static snapshots of transcriptional activity averaged across a cell population, these studies provide little insight into the temporal response properties of activity-regulated genes and no information about the variation in the transcriptional response between individual cells. Understanding the kinetic parameters of induced transcription is of particular importance in neurons, where tightly regulated patterns of synaptic activity are the key source of information that drives plasticity. Furthermore, studies in single living cells have revealed that the expression of individual genes is highly variable, even within a clonal population of cells (Chubb et al., 2006; Golding et al., 2005; Janicki et al., 2004; Raj et al., 2006; Raser and O’Shea, 2004). Since many of the genes induced by neuronal activity feed back to alter synaptic function, if in the nervous system there is similar randomness and variation in transcriptional activation from cell to cell this would have direct phenotypic consequences for synaptic plasticity within interconnected neuronal networks.
Developments in live imaging technologies have been particularly useful for studying the processes by which synaptic signals reach the nucleus. The refinement of these techniques through their application to new imaging platforms along with the development of genetic tools for imaging continues to open new avenues of understanding. For example, Ryohei Yasuda and colleagues are using a two-photon technique called fluorescence lifetime imaging microscopy (FLIM) to study the temporal and spatial dynamics of synaptic signaling cascade activation following stimulation of single dendritic spines (Yasuda, 2006). Their results suggest that single spine activation of CaMKII drives transient activation of this pathway that is restricted to a single spine (Lee et al., 2009), whereas Ras activated by the same stimulus spreads into the neighboring dendritic shaft (Yasuda et al., 2006). The Ras pathway drives transcription by activating the Erk1/2 MAPKs, which have long been known to translocate to the nucleus in response to many stimuli that lead to their activation including neural activity (Impey et al., 1998). Tagging Erk1/2 with the photoactivatable compound Dronpa, Weigert and colleagues examined the translocation of Erk1/2 in different parts of cultured hippocampal neurons (dendrites versus soma) in response to BDNF stimulation or action potential bursting (Wiegert et al., 2007). Their results indicate that propagation of Erk1/2 in dendrites is passive and not signal-inducible. By contrast, translocation of somatic Erk1/2 to the nucleus is induced by stimulation and occurs by facilitated diffusion. Imaging techniques can also be applied to study intranuclear signaling. Hilmar Bading and colleagues capitalized on the development of genetically encoded calcium indicators (Inverse Pericam and GCaMP2.0) to study the specificity of stimuli that induce nuclear calcium transients by targeting these reporters to the nucleus (Bengtson et al., 2010). In acute hippocampal slices, they found that although a burst of synaptic activity elicits a nuclear calcium rise, repetition of the action potential burst (using classical late-LTP-inducing stimuli such as high-frequency stimulation and theta-burst stimulation) greatly amplified the magnitude of the nuclear calcium increase. These data may help to explain why repeated stimulation is required to induce the transcription-dependent phase of late-LTP. Taken together, these data help to define distinct cellular compartments where different stages in the synapse-nucleus signaling pathway may occur to complete information transfer.
Imaging has also been used to identify the specific cells where transcription has occurred, facilitating study of the cellular consequences of activity-regulated transcription (Barth, 2007). Barth and colleagues were the first to develop mice for this kind of experiment, by expressing a Fos-GFP fusion protein from a transgene under the control of the activity-regulated proximal Fos promoter (Barth et al., 2004). GFP expression is robustly induced in neurons that have had a history of elevated activity, permitting the exploration of the physiological properties of these highly active neurons (Barth et al., 2004; Yassin et al., 2010). Wang and colleagues extended this methodology by generating knockin mice in which GFP replaces the coding sequence of the Arc gene (Wang et al., 2006). In mice heterozygous for the knockin mutation, Arc is expressed at normal levels and GFP can again be used to mark populations of neurons in which the neural activity levels and/or patterns have been sufficient to induce Arc transcription. However, homozygous knockin mice lack all Arc protein expression, thus in these mice GFP can be used to focus studies of cellular physiology on the time points and in the specific cells in which Arc expression should have been induced but was not.
It has been more challenging to develop live imaging reagents that directly get at the process of transcription itself. Perhaps the closest reagents have been those that allow the fluorescent detection of the stimulus-induced interaction between CREB and its co-activator CBP. The direct physical interaction of these two proteins can be monitored by fluorescence-resonance energy transfer (FRET) between compatible fluorophores fused to the KID domain of CREB and the KIX domain of CBP. The challenge for FRET is optimizing the development of the interacting partners to give a maximal signal to noise ratio. One recent study used such a CREB-CBP FRET system as a readout to study the role of the PKA anchoring protein AKAP79 in the L-VGCC-dependent activation of CREB (Friedrich et al., 2010). Because this FRET pair allows dynamic and reversible imaging of CREB-related transcription in live cells, it may have great usefulness for studying the molecular components and cell-type specificity of activity-regulated gene expression. Alternatively, transcription itself can be monitored in single cells in fixed tissue using the cellular compartment analysis of temporal activity by fluorescent in situ hybridization (catFISH) technique (Guzowski et al., 1999, 2001). This method relies on the fact that newly synthesized RNA first appears in the nucleus before being transported to the cytoplasm. By using oligonucleotide-based FISH, which has subcellular resolution for localizing RNA, it is possible to derive information about the temporal profile of transcription induction after a stimulus and prior to tissue fixation.
A fascinating challenge that remains for the field of neuronal activity-regulated transcription is to develop ways to visualize transcription itself as it occurs in single cells in real time. Imaging techniques based on fluorescent proteins have been successfully applied for this purpose in other cell types, yielding substantial information about the kinetics of transcription and the consequences of stochasticity for transcriptional regulatory processes (Darzacq et al., 2009). One of the most useful technologies for imaging transcription relies on the tagging of the target RNA with multiple copies of a stem-loop binding site for the phage RNA-binding protein MS2 (Chubb et al., 2006; Janicki et al., 2004) (FIGURE 9). When nuclear-targeted MS2 is fused to a fluorophore such as YFP and expressed in cells in the absence of a tagged target gene, it is found diffusely throughout the nucleus. However, when transcription of the tagged gene is induced, MS2-YFP binds to the stem-loop sequence in the newly synthesized RNA and it clusters in the nucleus at the site of active transcription, causing the appearance of a bright spot of nuclear fluorescence. Important properties of transcription can then be inferred by studying the timing of spot appearance after cell simulation, the frequency of spot formation and disappearance, the intensity of spot fluorescence (which correlates with RNA copy number), and the likelihood that spots appear in any single cell within a cell population. Furthermore, since MS2-YFP stays bound to the RNA as it is processed and exported from the nucleus, this technique has been widely used to study cytoplasmic RNA trafficking, and would be well applied to the study of dendritic RNA targeting in neurons (Grünwald and Singer, 2010; Querido and Chartrand, 2008). Application of the MS2-based technology to the study of one or more of the activity-regulated genes in neurons would represent an important new path for inquiry into the mechanisms of neuronal activity-regulated transcription.
FIGURE 9. Imaging RNA synthesis with the RNA binding protein MS2.
a) A representative reporter gene for MS2-based visualization of new RNA synthesis. Pind, stimulus-inducible promoter; RFP, red fluorescent protein. The black line represents the RNA transcribed from the reporter locus. A dimer of the phage protein MS2 fused to the fluorescent protein YFP binds each stem loop made by one of the series of MS2 binding sites encoded at the reporter locus. b) Representation of MS2-based transcriptional imaging. When reporter gene expression is induced, MS2-YFP binds to the cluster of stem-loop sequences in the new RNA synthesized at the site of reporter gene integration (R). Relocalization of MS2-YFP in the nucleus to this binding site is detected as a bright spot of nuclear fluorescence (asterisk).
7. Concluding remarks
Substantial progress has been made in the identification of molecular mechanisms that mediate neuronal activity-dependent changes in gene transcription. As described here, these data have shown that neurons use a complex array of signaling pathways and transcriptional mechanisms to orchestrate the expression of diverse gene expression programs in response to a wide range of extracellular stimuli. Among the key principles of activity-dependent transcriptional regulation are the following: 1) transcription factors can be regulated at the level of post-translational modifications that either activate or repress transcription, nuclear versus non-nuclear localization, protein-DNA binding, or expression; 2) regulation of chromatin structure adds an additional layer of context that can modulate the outcome of transcription factor activation; and 3) different neurotransmitter receptors and ion channels in the plasma membrane – both at the synapse and outside the synapse – activate distinct signaling molecules that differentially regulate the transcriptional machinery in the nucleus. The net outcome of these pathways is the tight and appropriate control of cellular responses to extracellular signals, as exemplified by the ability of distinct transcriptional regulatory pathways to drive opposite effects of synaptic scaling under conditions of high versus low synaptic activity.
Under optimal conditions, this dynamic and flexible network permits the brain to rapidly initiate physiologically relevant cellular and behavioral responses to a constantly changing environment. By contrast, disruption of this transcriptional network by genetic mutations that impair the function of activity-regulated transcription factors is all too often associated with mental retardation and neuropsychiatric illness (Hong et al., 2005; Tsankova et al., 2007). The challenge for the future is to harness new experimental techniques in genetic, genomics, and imaging that will allow for an enhanced understanding of the functional importance and biological consequences of fine-tuning transcriptional regulation in the CNS.
Research Highlights.
In this review we cover the following:
The classes of genes and transcription factors that are regulated by neuronal activity.
How calcium-signaling pathways confer specificity upon cellular responses to stimuli.
Importance of activity-regulated transcriptional pathways for synaptic homeostasis.
Technological developments that are advancing studies in this field.
Abbreviations
- NGF
Nerve Growth Factor
- CNS
central nervous system
- IEG
immediate-early gene
- Egr
early growth response factor
- Nr
nuclear receptor
- TSS
transcription start site
- SRE
Serum Response Element
- CRE
Calcium/cAMP-Response Element
- Rb
retinoblastoma tumor suppressor
- RCE
Retinoblastoma Control Element
- SRF
Serum Response Factor
- TCF
Ternary Complex Factors
- Ets
E-twenty six domain
- Elk-1
Ets like gene 1
- CREB
Calcium-Response Element Binding protein
- CREST
Calcium-Responsive Transactivator
- HDAC
histone deacetylase
- HAT
histone acetyltransferase
- CBP
CREB-binding protein
- MEF2
Myocyte Enhancer Factor 2
- eRNA
enhancer RNA
- FIRE
Fos intragenic regulatory element
- ChIP
chromatin immunoprecipitation
- BDNF
Brain-Derived Neurotrophic Factor
- CaRE
calcium-response element
- CaRF
Calcium-Response Factor
- USF
upstream stimulatory factor
- PAS
Per-Arnt-Sim
- Npas4
Neuronal PAS Domain Protein 4
- NF-κB
Nuclear Factor κB
- bHLH
basic helix-loop-helix
- NFAT
Nuclear Factor of Activated T Cells
- NMDA
N-methyl-D-aspartate
- NMDAR
NMDA receptor
- Arc
activity-regulated cytoskeletal-associated protein
- SARE
Synaptic Activity Response Element
- LTD
long-term depression
- PKA
protein kinase A
- MAPK
mitogen-activated protein kinase
- AMPA
α-amino-3-hydroxyl-5-methyl-4-isoxazole-proprionate
- AMPAR
AMPA receptor
- PSD
postsynaptic density
- CREM
cAMP Response Element Modulatory Protein
- ATF1
activating transcription factor 1
- Ser133
serine 133
- CaM
calmodulin
- CaMK
Ca2+/CaM-dependent protein kinase
- Akt or PKB
protein kinase B
- Crtc
CREB-regulated transcription coactivator
- KID
kinase-inducible domain
- KIX
KID-interacting domain
- AMPK
AMP-activated protein kinase
- SIK2
salt-inducible kinase 2
- LTP
long-term potentiation
- MKL1
myocardin-related transcription factor A
- MKL2
myocardin-related transcription factor B
- Sap-1
SRF accessory protein 1
- SUMO
small ubiquitin-like modifier
- Erk5
extracellular signal-regulated kinase 5
- IκB
Inhibitor of NF-κB
- IKK
IκB kinase
- NEMO
NF-κB essential modulator
- L-VGCC
L-type voltage-gated calcium channel
- CaMKII
Ca2+/CaM-dependent protein kinase II
- NFATn
NFAT nuclear component
- DYRK1A
dual specificity tyrosine phosphorylation-regulated kinase 1A
- GSK3
glycogen synthase kinase 3
- FOXO3
forkhead box O3
- CaMKIV
Ca2+/CaM-dependent protein kinase IV
- CCAT
calcium channel associated transcription regulator
- GAD65
glutamic acid decarboxylase 65
- GABA
γ-aminobutyric acid
- PKC
protein kinase C
- ATF4 or CREB2
activating transcription factor 4
- DREAM
Downstream Repressor Element Antagonist Modulator
- NCS
Neuronal Calcium Sensor
- Pdyn
prodynorphin
- PI3K
phosphatidylinositol 3-kinase
- AP-1
activator protein 1
- ChIP-Seq
whole genome ChIP sequencing
- RNAi
RNA interference
- ICER
inducible cAMP early repressor
- EGF
epidermal growth factor
- Rsk2
ribosomal S6 kinase 2
- MSK1
mitogen- and stress-induced kinase 1
- MSK2
mitogen- and stress-induced kinase 2
- FGF
fibroblast growth factor
- Dnmt1
DNA methyltransferase 1
- Dnmt3a
DNA methyltransferase 3a
- Dnmt3b
DNA methyltransferase 3b
- AID
activation-induced deaminase
- APOBEC
apolipoprotein B mRNA-editing enzyme complex
- shRNA
short hairpin RNA
- MBD
methyl-DNA binding domain
- MBD1, MBD2, and MBD4
MBD proteins 1, 2, and 4
- MeCP2
methyl CpG binding protein 2
- Ras-GRF
Ras-specific guanine nucleotide-releasing factor
- CaMKK
CaMK kinase
- CaMKI
Ca2+/CaM-dependent protein kinase I
- TM
transmembrane domain
- Clca1
calcium-activated chloride channel regulator 1
- MAGUK
membrane-associated guanylate kinase
- PSD-95
postsynaptic density-95
- SAP-97
synapse-associated protein-97
- PSD-93
postsynaptic density-93
- SAP-102
synapse-associated protein-102
- MEK
Erk/MAPK kinase
- GAP
GTPase activating protein
- GEF
GTP exchange factor
- SynGAP
synaptic Ras GTPase activating protein
- DAPK1
death-associated protein kinase 1
- PP1
protein phosphatase 1
- EphB2
Ephrin type-B receptor 2
- PP2A
protein phosphatase 2A
- Rheb
small GTPase Ras homologue enriched in brain
- VGCCs
voltage-gated calcium channels
- eIF3E
eukaryotic initiation factor 3 subunit E
- PIKfyve
phosphatidylinositol 3-phosphate 5-kinase
- PO
open probability
- AKAP15
A kinase-anchoring protein 15
- N-VGCCs
N-type voltage-gated calcium channels
- ADAR2
RNA-dependent adenosine deaminase 2
- ER
endoplasmic reticulum
- PLC
phospholipase C
- IP3
inositol trisphosphate
- CRAC channels
calcium release-activated calcium channels
- Stim1
stromal interaction molecule 1
- Stim2
stromal interaction molecule 2
- nAChRs
nicotinic acetylcholine receptors
- YFP
yellow fluorescent protein
- eYFP
enhanced YFP
- TTX
tetrodotoxin
- ActD
actinomycin D
- ChR2
channel rhodopsin 2
- SCN
suprachiasmatic nucleus
- BAC
bacterial artificial chromosome
- RNA-Seq
whole transcriptome RNA sequencing
- PCR
polymerase chain reaction
- REST
RE1-silencing transcription factor
- GFP
green fluorescent protein
- eGFP
enhanced GFP
- 3C
chromosome conformation capture
- FLIM
fluorescence lifetime imaging microscopy
- FRET
fluorescence-resonance energy transfer
- catFISH
cellular compartment analysis of temporal activity by fluorescent in situ hybridization
- ZRE
Zeste-like response element
- RFP
red fluorescent protein
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JP, Dudek SM. Late-phase long-term potentiation: getting to the nucleus. Nat Rev Neurosci. 2005;6:737–743. doi: 10.1038/nrn1749. [DOI] [PubMed] [Google Scholar]
- Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hallaq RA, Conrads TP, Veenstra TD, Wenthold RJ. NMDA di-heteromeric receptor populations and associated proteins in rat hippocampus. J Neurosci. 2007;27:8334–8343. doi: 10.1523/JNEUROSCI.2155-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Hallaq RA, Jarabek BR, Fu Z, Vicini S, Wolfe BB, Yasuda RP. Association of NR3A with the N-methyl-D-aspartate receptor NR1 and NR2 subunits. Mol Pharmacol. 2002;62:1119–1127. doi: 10.1124/mol.62.5.1119. [DOI] [PubMed] [Google Scholar]
- Al-Mubarak B, Soriano FX, Hardingham GE. Synaptic NMDAR activity suppresses FOXO1 expression via a cis-acting FOXO binding site: FOXO1 is a FOXO target gene. Channels (Austin) 2009;3:233–238. doi: 10.4161/chan.3.4.9381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An WF, Bowlby MR, Betty M, Cao J, Ling HP, Mendoza G, Hinson JW, Mattsson KI, Strassle BW, Trimmer JS, Rhodes KJ. Modulation of A-type potassium channels by a family of calcium sensors. Nature. 2000;403:553–556. doi: 10.1038/35000592. [DOI] [PubMed] [Google Scholar]
- Aparicio O, Geisberg JV, Struhl K. Curr Protoc Cell Biol. Unit 17. Chapter 17. 2004. Chromatin immunoprecipitation for determining the association of proteins with specific genomic sequences in vivo; p. 17. [DOI] [PubMed] [Google Scholar]
- Arron JR, Winslow MM, Polleri A, Chang CP, Wu H, Gao X, Neilson JR, Chen L, Heit JJ, Kim SK, Yamasaki N, Miyakawa T, Francke U, Graef IA, Crabtree GR. NFAT dysregulation by increased dosage of DSCR1 and DYRK1A on chromosome 21. Nature. 2006;441:595–600. doi: 10.1038/nature04678. [DOI] [PubMed] [Google Scholar]
- Arsenian S, Weinhold B, Oelgeschläger M, Rüther U, Nordheim A. Serum response factor is essential for mesoderm formation during mouse embryogenesis. The EMBO Journal. 1998;17:6289–6299. doi: 10.1093/emboj/17.21.6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atasoy D, Aponte Y, Su HH, Sternson SM. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci. 2008;28:7025–7030. doi: 10.1523/JNEUROSCI.1954-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. doi: 10.1126/science.8097060. [DOI] [PubMed] [Google Scholar]
- Bading H, Segal MM, Sucher NJ, Dudek H, Lipton SA, Greenberg ME. N-methyl-D-aspartate receptors are critical for mediating the effects of glutamate on intracellular calcium concentration and immediate early gene expression in cultured hippocampal neurons. Neuroscience. 1995;64:653–664. doi: 10.1016/0306-4522(94)00462-e. [DOI] [PubMed] [Google Scholar]
- Balkowiec A, Katz DM. Activity-dependent release of endogenous brain-derived neurotrophic factor from primary sensory neurons detected by ELISA in situ. J Neurosci. 2000;20:7417–7423. doi: 10.1523/JNEUROSCI.20-19-07417.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, McAnally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci USA. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP, Sheng M, Lau LF, Greenberg ME. Growth factors and membrane depolarization activate distinct programs of early response gene expression: dissociation of fos and jun induction. Genes Dev. 1989;3:304–313. doi: 10.1101/gad.3.3.304. [DOI] [PubMed] [Google Scholar]
- Barth AL. Visualizing circuits and systems using transgenic reporters of neural activity. Curr Opin Neurobiol. 2007;17:567–571. doi: 10.1016/j.conb.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer KU, De Koninck P, Leonard AS, Hell JW, Schulman H. Interaction with the NMDA receptor locks CaMKII in an active conformation. Nature. 2001;411:801–805. doi: 10.1038/35081080. [DOI] [PubMed] [Google Scholar]
- Bayer KU, Schulman H. Regulation of signal transduction by protein targeting: the case for CaMKII. Biochem Biophys Res Commun. 2001;289:917–923. doi: 10.1006/bbrc.2001.6063. [DOI] [PubMed] [Google Scholar]
- Bean BP, Nowycky MC, Tsien RW. Beta-adrenergic modulation of calcium channels in frog ventricular heart cells. Nature. 1984;307:371–375. doi: 10.1038/307371a0. [DOI] [PubMed] [Google Scholar]
- Belfield JL, Whittaker C, Cader MZ, Chawla S. Differential effects of Ca2+ and cAMP on transcription mediated by MEF2D and cAMP-response element-binding protein in hippocampal neurons. J Biol Chem. 2006;281:27724–27732. doi: 10.1074/jbc.M601485200. [DOI] [PubMed] [Google Scholar]
- Benedito AB, Lehtinen M, Massol R, Lopes UG, Kirchhausen T, Rao A, Bonni A. The transcription factor NFAT3 mediates neuronal survival. J Biol Chem. 2005;280:2818–2825. doi: 10.1074/jbc.M408741200. [DOI] [PubMed] [Google Scholar]
- Bengtson CP, Freitag HE, Weislogel JM, Bading H. Nuclear calcium sensors reveal that repetition of trains of synaptic stimuli boosts nuclear calcium signaling in CA1 pyramidal neurons. Biophys J. 2010;99:4066–4077. doi: 10.1016/j.bpj.2010.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benito E, Barco A. CREB’s control of intrinsic and synaptic plasticity: implications for CREB-dependent memory models. Trends Neurosci. 2010;33:230–240. doi: 10.1016/j.tins.2010.02.001. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- Berridge MJ. The endoplasmic reticulum: a multifunctional signaling organelle. Cell Calcium. 2002;32:235–249. doi: 10.1016/s0143416002001823. [DOI] [PubMed] [Google Scholar]
- Bhakar AL, Tannis LL, Zeindler C, Russo MP, Jobin C, Park DS, MacPherson S, Barker PA. Constitutive nuclear factor-kappa B activity is required for central neuron survival. J Neurosci. 2002;22:8466–8475. doi: 10.1523/JNEUROSCI.22-19-08466.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieda M, Xu X, Singer MA, Green R, Farnham PJ. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 2006;16:595–605. doi: 10.1101/gr.4887606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilang-Bleuel A, Ulbricht S, Chandramohan Y, De Carli S, Droste SK, Reul JMHM. Psychological stress increases histone H3 phosphorylation in adult dentate gyrus granule neurons: involvement in a glucocorticoid receptor-dependent behavioural response. Eur J Neurosci. 2005;22:1691–1700. doi: 10.1111/j.1460-9568.2005.04358.x. [DOI] [PubMed] [Google Scholar]
- Bito H, Deisseroth K, Tsien RW. CREB phosphorylation and dephosphorylation: a Ca(2+)- and stimulus duration-dependent switch for hippocampal gene expression. Cell. 1996;87:1203–1214. doi: 10.1016/s0092-8674(00)81816-4. [DOI] [PubMed] [Google Scholar]
- Bleckmann SC, Blendy JA, Rudolph D, Monaghan AP, Schmid W, Schütz G. Activating transcription factor 1 and CREB are important for cell survival during early mouse development. Mol Cell Biol. 2002;22:1919–1925. doi: 10.1128/MCB.22.6.1919-1925.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomer WAC, VanDongen HMA, VanDongen AMJ. Arc/Arg3.1 translation is controlled by convergent N-methyl-D-aspartate and Gs-coupled receptor signaling pathways. J Biol Chem. 2008;283:582–592. doi: 10.1074/jbc.M702451200. [DOI] [PubMed] [Google Scholar]
- Bode AM, Dong Z. Inducible covalent posttranslational modification of histone H3. Sci STKE. 2005:re4. doi: 10.1126/stke.2812005re4. [DOI] [PubMed] [Google Scholar]
- Borlikova G, Endo S. Inducible cAMP early repressor (ICER) and brain functions. Mol Neurobiol. 2009;40:73–86. doi: 10.1007/s12035-009-8072-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodinsky LN, Root CM, Cronin JA, Sann SB, Gu X, Spitzer NC. Activity-dependent homeostatic specification of transmitter expression in embryonic neurons. Nature. 2004;429:523–530. doi: 10.1038/nature02518. [DOI] [PubMed] [Google Scholar]
- Boshart M, Weih F, Nichols M, Schütz G. The tissue-specific extinguisher locus TSE1 encodes a regulatory subunit of cAMP-dependent protein kinase. Cell. 1991;66:849–859. doi: 10.1016/0092-8674(91)90432-x. [DOI] [PubMed] [Google Scholar]
- Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Bradley J, Carter SR, Rao VR, Wang J, Finkbeiner S. Splice variants of the NR1 subunit differentially induce NMDA receptor-dependent gene expression. J Neurosci. 2006;26:1065–1076. doi: 10.1523/JNEUROSCI.3347-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- Brosenitsch TA, Katz DM. Physiological patterns of electrical stimulation can induce neuronal gene expression by activating N-type calcium channels. J Neurosci. 2001;21:2571–2579. doi: 10.1523/JNEUROSCI.21-08-02571.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–240. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Buchwalter G, Gross C, Wasylyk B. Ets ternary complex transcription factors. Gene. 2004;324:1–14. doi: 10.1016/j.gene.2003.09.028. [DOI] [PubMed] [Google Scholar]
- Buratowski S. Progression through the RNA polymerase II CTD cycle. Molecular Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne RD. Neuronal calcium sensor proteins: generating diversity in neuronal Ca2+ signalling. Nat Rev Neurosci. 2007;8:182–193. doi: 10.1038/nrn2093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrone J, Murthy VN. Synaptic gain control and homeostasis. Curr Opin Neurobiol. 2003;13:560–567. doi: 10.1016/j.conb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- Butler R, Bates GP. Histone deacetylase inhibitors as therapeutics for polyglutamine disorders. Nat Rev Neurosci. 2006;7:784–796. doi: 10.1038/nrn1989. [DOI] [PubMed] [Google Scholar]
- Buxbaum JD, Choi EK, Luo Y, Lilliehook C, Crowley AC, Merriam DE, Wasco W. Calsenilin: a calcium-binding protein that interacts with the presenilins and regulates the levels of a presenilin fragment. Nat Med. 1998;4:1177–1181. doi: 10.1038/2673. [DOI] [PubMed] [Google Scholar]
- Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485–490. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon W. The wisdom of the body. The American Journal of the Medical Sciences 1932
- Carrasco MA, Hidalgo C. Calcium microdomains and gene expression in neurons and skeletal muscle cells. Cell Calcium. 2006;40:575–583. doi: 10.1016/j.ceca.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Carrión AM, Link WA, Ledo F, Mellström B, Naranjo JR. DREAM is a Ca2+-regulated transcriptional repressor. Nature. 1999;398:80–84. doi: 10.1038/18044. [DOI] [PubMed] [Google Scholar]
- Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol. 2000;16:521–555. doi: 10.1146/annurev.cellbio.16.1.521. [DOI] [PubMed] [Google Scholar]
- Cesari F, Brecht S, Vintersten K, Vuong LG, Hofmann M, Klingel K, Schnorr JJ, Arsenian S, Schild H, Herdegen T, Wiebel FF, Nordheim A. Mice deficient for the ets transcription factor elk-1 show normal immune responses and mildly impaired neuronal gene activation. Mol Cell Biol. 2004;24:294–305. doi: 10.1128/MCB.24.1.294-305.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56:422–437. doi: 10.1016/j.neuron.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Chan SF, Sucher NJ. An NMDA receptor signaling complex with protein phosphatase 2A. J Neurosci. 2001;21:7985–7992. doi: 10.1523/JNEUROSCI.21-20-07985.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang KT, Berg DK. Voltage-gated channels block nicotinic regulation of CREB phosphorylation and gene expression in neurons. Neuron. 2001;32:855–865. doi: 10.1016/s0896-6273(01)00516-5. [DOI] [PubMed] [Google Scholar]
- Chawla S, Hardingham GE, Quinn DR, Bading H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- Chawla S, Vanhoutte P, Arnold FJL, Huang CLH, Bading H. Neuronal activity-dependent nucleocytoplasmic shuttling of HDAC4 and HDAC5. J Neurochem. 2003;85:151–159. doi: 10.1046/j.1471-4159.2003.01648.x. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- Chen RH, Abate C, Blenis J. Phosphorylation of the c-Fos transrepression domain by mitogen-activated protein kinase and 90-kDa ribosomal S6 kinase. Proc Natl Acad Sci USA. 1993;90:10952–10956. doi: 10.1073/pnas.90.23.10952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WG, Chang Q, Lin Y, Meissner A, West AE, Griffith EC, Jaenisch R, Greenberg ME. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003a;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons. J Neurosci. 2003b;23:2572–2581. doi: 10.1523/JNEUROSCI.23-07-02572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Li MH, Zhang Y, He LL, Yamada Y, Fitzmaurice A, Shen Y, Zhang H, Tong L, Yang J. Structural basis of the alpha1-beta subunit interaction of voltage-gated Ca2+ channels. Nature. 2004;429:675–680. doi: 10.1038/nature02641. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, Hempstead BL, Lee FS. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C, Fass DM, Reynolds IJ. Emergence of excitotoxicity in cultured forebrain neurons coincides with larger glutamate-stimulated [Ca(2+)](i) increases and NMDA receptor mRNA levels. Brain Res. 1999;849:97–108. doi: 10.1016/s0006-8993(99)01995-2. [DOI] [PubMed] [Google Scholar]
- Cheng HYM, Papp JW, Varlamova O, Dziema H, Russell B, Curfman JP, Nakazawa T, Shimizu K, Okamura H, Impey S, Obrietan K. microRNA modulation of circadian-clock period and entrainment. Neuron. 2007;54:813–829. doi: 10.1016/j.neuron.2007.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HYM, Pitcher GM, Laviolette SR, Whishaw IQ, Tong KI, Kockeritz LK, Wada T, Joza NA, Crackower M, Goncalves J, Sarosi I, Woodgett JR, Oliveira-dos-Santos AJ, Ikura M, van der Kooy D, Salter MW, Penninger JM. DREAM is a critical transcriptional repressor for pain modulation. Cell. 2002;108:31–43. doi: 10.1016/s0092-8674(01)00629-8. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–915. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Choi DW. Calcium-mediated neurotoxicity: relationship to specific channel types and role in ischemic damage. Trends Neurosci. 1988a;11:465–469. doi: 10.1016/0166-2236(88)90200-7. [DOI] [PubMed] [Google Scholar]
- Choi DW. Glutamate neurotoxicity and diseases of the nervous system. Neuron. 1988b;1:623–634. doi: 10.1016/0896-6273(88)90162-6. [DOI] [PubMed] [Google Scholar]
- Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- Chubb JR, Trcek T, Shenoy SM, Singer RH. Transcriptional pulsing of a developmental gene. Curr Biol. 2006;16:1018–1025. doi: 10.1016/j.cub.2006.03.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chwang WB, O’Riordan KJ, Levenson JM, Sweatt JD. ERK/MAPK regulates hippocampal histone phosphorylation following contextual fear conditioning. Learn Mem. 2006;13:322–328. doi: 10.1101/lm.152906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciabarra AM, Sullivan JM, Gahn LG, Pecht G, Heinemann S, Sevarino KA. Cloning and characterization of chi-1: a developmentally regulated member of a novel class of the ionotropic glutamate receptor family. J Neurosci. 1995;15:6498–6508. doi: 10.1523/JNEUROSCI.15-10-06498.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 2007;131:1047–1058. doi: 10.1016/j.cell.2007.11.028. [DOI] [PubMed] [Google Scholar]
- Clayton AL, Hazzalin CA, Mahadevan LC. Enhanced histone acetylation and transcription: a dynamic perspective. Molecular Cell. 2006;23:289–296. doi: 10.1016/j.molcel.2006.06.017. [DOI] [PubMed] [Google Scholar]
- Clements JD, Westbrook GL. Activation kinetics reveal the number of glutamate and glycine binding sites on the N-methyl-D-aspartate receptor. Neuron. 1991;7:605–613. doi: 10.1016/0896-6273(91)90373-8. [DOI] [PubMed] [Google Scholar]
- Cloonan N, Forrest ARR, Kolle G, Gardiner BBA, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, Robertson AJ, Perkins AC, Bruce SJ, Lee CC, Ranade SS, Peckham HE, Manning JM, McKernan KJ, Grimmond SM. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods. 2008;5:613–619. doi: 10.1038/nmeth.1223. [DOI] [PubMed] [Google Scholar]
- Coba MP, Valor LM, Kopanitsa MV, Afinowi NO, Grant SGN. Kinase networks integrate profiles of N-methyl-D-aspartate receptor-mediated gene expression in hippocampus. J Biol Chem. 2008;283:34101–34107. doi: 10.1074/jbc.M804951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran BH, Zullo J, Verma IM, Stiles CD. Expression of the c-fos gene and of an fos-related gene is stimulated by platelet-derived growth factor. Science. 1984;226:1080–1082. doi: 10.1126/science.6093261. [DOI] [PubMed] [Google Scholar]
- Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- Collingridge GL, Kehl SJ, McLennan H. Excitatory amino acids in synaptic transmission in the Schaffer collateral-commissural pathway of the rat hippocampus. The Journal of Physiology. 1983a;334:33–46. doi: 10.1113/jphysiol.1983.sp014478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Kehl SJ, McLennan H. The antagonism of amino acid-induced excitations of rat hippocampal CA1 neurones in vitro. The Journal of Physiology. 1983b;334:19–31. doi: 10.1113/jphysiol.1983.sp014477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Olsen RW, Peters J, Spedding M. A nomenclature for ligand-gated ion channels. Neuropharmacology. 2009;56:2–5. doi: 10.1016/j.neuropharm.2008.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collingridge GL, Singer W. Excitatory amino acid receptors and synaptic plasticity. Trends Pharmacol Sci. 1990;11:290–296. doi: 10.1016/0165-6147(90)90011-v. [DOI] [PubMed] [Google Scholar]
- Conkright MD, Canettieri G, Screaton R, Guzman E, Miraglia L, Hogenesch JB, Montminy M. TORCs: transducers of regulated CREB activity. Molecular Cell. 2003;12:413–423. doi: 10.1016/j.molcel.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Corneliussen B, Holm M, Waltersson Y, Onions J, Hallberg B, Thornell A, Grundström T. Calcium/calmodulin inhibition of basic-helix-loop-helix transcription factor domains. Nature. 1994;368:760–764. doi: 10.1038/368760a0. [DOI] [PubMed] [Google Scholar]
- Cox LJ, Hengst U, Gurskaya NG, Lukyanov KA, Jaffrey SR. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat Cell Biol. 2008;10:149–159. doi: 10.1038/ncb1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crabtree GR, Graef IA. Bursting into the nucleus. Sci Signal. 2008;1:pe54. doi: 10.1126/scisignal.151pe54. [DOI] [PMC free article] [PubMed]
- Crosio C, Cermakian N, Allis CD, Sassone-Corsi P. Light induces chromatin modification in cells of the mammalian circadian clock. Nat Neurosci. 2000;3:1241–1247. doi: 10.1038/81767. [DOI] [PubMed] [Google Scholar]
- Cull-Candy S, Brickley S, Farrant M. NMDA receptor subunits: diversity, development and disease. Curr Opin Neurobiol. 2001;11:327–335. doi: 10.1016/s0959-4388(00)00215-4. [DOI] [PubMed] [Google Scholar]
- Cullen PJ, Lockyer PJ. Integration of calcium and Ras signalling. Nat Rev Mol Cell Biol. 2002;3:339–348. doi: 10.1038/nrm808. [DOI] [PubMed] [Google Scholar]
- Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- Dai S, Hall DD, Hell JW. Supramolecular assemblies and localized regulation of voltage-gated ion channels. Physiol Rev. 2009;89:411–452. doi: 10.1152/physrev.00029.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- Dalva MB, Takasu MA, Lin MZ, Shamah SM, Hu L, Gale NW, Greenberg ME. EphB receptors interact with NMDA receptors and regulate excitatory synapse formation. Cell. 2000;103:945–956. doi: 10.1016/s0092-8674(00)00197-5. [DOI] [PubMed] [Google Scholar]
- Dani VS, Nelson SB. Intact long-term potentiation but reduced connectivity between neocortical layer 5 pyramidal neurons in a mouse model of Rett syndrome. J Neurosci. 2009;29:11263–11270. doi: 10.1523/JNEUROSCI.1019-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzacq X, Yao J, Larson DR, Causse SZ, Bosanac L, de Turris V, Ruda VM, Lionnet T, Zenklusen D, Guglielmi B, Tjian R, Singer RH. Imaging transcription in living cells. Annu Rev Biophys. 2009;38:173–196. doi: 10.1146/annurev.biophys.050708.133728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jongh KS, Colvin AA, Wang KK, Catterall WA. Differential proteolysis of the full-length form of the L-type calcium channel alpha 1 subunit by calpain. J Neurochem. 1994;63:1558–1564. doi: 10.1046/j.1471-4159.1994.63041558.x. [DOI] [PubMed] [Google Scholar]
- De Val S, Chi NC, Meadows SM, Minovitsky S, Anderson JP, Harris IS, Ehlers ML, Agarwal P, Visel A, Xu SM, Pennacchio LA, Dubchak I, Krieg PA, Stainier DYR, Black BL. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell. 2008;135:1053–1064. doi: 10.1016/j.cell.2008.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- Deisseroth K, Heist EK, Tsien RW. Translocation of calmodulin to the nucleus supports CREB phosphorylation in hippocampal neurons. Nature. 1998;392:198–202. doi: 10.1038/32448. [DOI] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- Deng JV, Rodriguiz RM, Hutchinson AN, Kim IH, Wetsel WC, West AE. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick O, Bading H. Synaptic activity and nuclear calcium signaling protect hippocampal neurons from death signal-associated nuclear translocation of FoxO3a induced by extrasynaptic N-methyl-D-aspartate receptors. J Biol Chem. 2010;285:19354–19361. doi: 10.1074/jbc.M110.127654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieterich DC, Karpova A, Mikhaylova M, Zdobnova I, König I, Landwehr M, Kreutz M, Smalla KH, Richter K, Landgraf P, Reissner C, Boeckers TM, Zuschratter W, Spilker C, Seidenbecher CI, Garner CC, Gundelfinger ED, Kreutz MR. Caldendrin-Jacob: a protein liaison that couples NMDA receptor signalling to the nucleus. PLoS Biol. 2008;6:e34. doi: 10.1371/journal.pbio.0060034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingledine R, Borges K, Bowie D, Traynelis SF. The glutamate receptor ion channels. Pharmacol Rev. 1999;51:7–61. [PubMed] [Google Scholar]
- Dolmetsch RE, Lewis RS, Goodnow CC, Healy JI. Differential activation of transcription factors induced by Ca2+ response amplitude and duration. Nature. 1997;386:855–858. doi: 10.1038/386855a0. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science. 2001;294:333–339. doi: 10.1126/science.1063395. [DOI] [PubMed] [Google Scholar]
- Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933–936. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- Dolphin AC. Calcium channel diversity: multiple roles of calcium channel subunits. Curr Opin Neurobiol. 2009;19:237–244. doi: 10.1016/j.conb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- Dragunow M, Xu R, Walton M, Woodgate A, Lawlor P, MacGibbon GA, Young D, Gibbons H, Lipski J, Muravlev A, Pearson A, During M. c-Jun promotes neurite outgrowth and survival in PC12 cells. Brain Res Mol Brain Res. 2000;83:20–33. doi: 10.1016/s0169-328x(00)00191-1. [DOI] [PubMed] [Google Scholar]
- Dulac C. Brain function and chromatin plasticity. Nature. 2010;465:728–735. doi: 10.1038/nature09231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, Lu B, Weinberger DR. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Etkin A, Alarcón JM, Weisberg SP, Touzani K, Huang YY, Nordheim A, Kandel ER. A role in learning for SRF: deletion in the adult forebrain disrupts LTD and the formation of an immediate memory of a novel context. Neuron. 2006;50:127–143. doi: 10.1016/j.neuron.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Fagiolini M, Fritschy JM, Löw K, Möhler H, Rudolph U, Hensch TK. Specific GABAA circuits for visual cortical plasticity. Science. 2004;303:1681–1683. doi: 10.1126/science.1091032. [DOI] [PubMed] [Google Scholar]
- Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10:605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth CL, Freshney NW, Rosen LB, Ghosh A, Greenberg ME, Feig LA. Calcium activation of Ras mediated by neuronal exchange factor Ras-GRF. Nature. 1995;376:524–527. doi: 10.1038/376524a0. [DOI] [PubMed] [Google Scholar]
- Felinski EA, Kim J, Lu J, Quinn PG. Recruitment of an RNA polymerase II complex is mediated by the constitutive activation domain in CREB, independently of CREB phosphorylation. Mol Cell Biol. 2001;21:1001–1010. doi: 10.1128/MCB.21.4.1001-1010.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Zhou Y, Campbell SL, Le T, Li E, Sweatt JD, Silva AJ, Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci. 2010;13:423–430. doi: 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiering S, Northrop JP, Nolan GP, Mattila PS, Crabtree GR, Herzenberg LA. Single cell assay of a transcription factor reveals a threshold in transcription activated by signals emanating from the T-cell antigen receptor. Genes Dev. 1990;4:1823–1834. doi: 10.1101/gad.4.10.1823. [DOI] [PubMed] [Google Scholar]
- Finsterwald C, Fiumelli H, Cardinaux JR, Martin JL. Regulation of dendritic development by BDNF requires activation of CRTC1 by glutamate. J Biol Chem. 2010;285:28587–28595. doi: 10.1074/jbc.M110.125740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint AC, Maisch US, Weishaupt JH, Kriegstein AR, Monyer H. NR2A subunit expression shortens NMDA receptor synaptic currents in developing neocortex. J Neurosci. 1997;17:2469–2476. doi: 10.1523/JNEUROSCI.17-07-02469.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folco EJ, Koren G. Degradation of the inducible cAMP early repressor (ICER) by the ubiquitin-proteasome pathway. Biochem J. 1997;328 ( Pt 1):37–43. doi: 10.1042/bj3280037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes NS, Duval G, Sassone-Corsi P. Adaptive inducibility of CREM as transcriptional memory of circadian rhythms. Nature. 1996;381:83–85. doi: 10.1038/381083a0. [DOI] [PubMed] [Google Scholar]
- Frey U, Morris RG. Synaptic tagging and long-term potentiation. Nature. 1997;385:533–536. doi: 10.1038/385533a0. [DOI] [PubMed] [Google Scholar]
- Friedrich MW, Aramuni G, Mank M, Mackinnon JAG, Griesbeck O. Imaging CREB activation in living cells. J Biol Chem. 2010 doi: 10.1074/jbc.M110.124545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz EL, Papavasiliou FN. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 2010;24:2107–2114. doi: 10.1101/gad.1963010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardoni F, Schrama LH, Kamal A, Gispen WH, Cattabeni F, Di Luca M. Hippocampal synaptic plasticity involves competition between Ca2+/calmodulin-dependent protein kinase II and postsynaptic density 95 for binding to the NR2A subunit of the NMDA receptor. J Neurosci. 2001;21:1501–1509. doi: 10.1523/JNEUROSCI.21-05-01501.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardoni F, Schrama LH, van Dalen JJ, Gispen WH, Cattabeni F, Di Luca M. AlphaCaMKII binding to the C-terminal tail of NMDA receptor subunit NR2A and its modulation by autophosphorylation. FEBS Lett. 1999;456:394–398. doi: 10.1016/s0014-5793(99)00985-0. [DOI] [PubMed] [Google Scholar]
- Gau D, Lemberger T, von Gall C, Kretz O, Le Minh N, Gass P, Schmid W, Schibler U, Korf HW, Schütz G. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 2002;34:245–253. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- Gehring M, Reik W, Henikoff S. DNA demethylation by DNA repair. Trends Genet. 2009;25:82–90. doi: 10.1016/j.tig.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Geiger JR, Melcher T, Koh DS, Sakmann B, Seeburg PH, Jonas P, Monyer H. Relative abundance of subunit mRNAs determines gating and Ca2+ permeability of AMPA receptors in principal neurons and interneurons in rat CNS. Neuron. 1995;15:193–204. doi: 10.1016/0896-6273(95)90076-4. [DOI] [PubMed] [Google Scholar]
- Genoud C, Knott GW, Sakata K, Lu B, Welker E. Altered synapse formation in the adult somatosensory cortex of brain-derived neurotrophic factor heterozygote mice. J Neurosci. 2004;24:2394–2400. doi: 10.1523/JNEUROSCI.4040-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhardstein BL, Gao T, Bünemann M, Puri TS, Adair A, Ma H, Hosey MM. Proteolytic processing of the C terminus of the alpha(1C) subunit of L-type calcium channels and the role of a proline-rich domain in membrane tethering of proteolytic fragments. J Biol Chem. 2000;275:8556–8563. doi: 10.1074/jbc.275.12.8556. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Carnahan J, Greenberg ME. Requirement for BDNF in activity-dependent survival of cortical neurons. Science. 1994a;263:1618–1623. doi: 10.1126/science.7907431. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Ginty DD, Bading H, Greenberg ME. Calcium regulation of gene expression in neuronal cells. J Neurobiol. 1994b;25:294–303. doi: 10.1002/neu.480250309. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Greenberg ME. Calcium signaling in neurons: molecular mechanisms and cellular consequences. Science. 1995;268:239–247. doi: 10.1126/science.7716515. [DOI] [PubMed] [Google Scholar]
- Gille H, Sharrocks AD, Shaw PE. Phosphorylation of transcription factor p62TCF by MAP kinase stimulates ternary complex formation at c-fos promoter. Nature. 1992;358:414–417. doi: 10.1038/358414a0. [DOI] [PubMed] [Google Scholar]
- Ginty DD. Calcium regulation of gene expression: isn’t that spatial? Neuron. 1997;18:183–186. doi: 10.1016/s0896-6273(00)80258-5. [DOI] [PubMed] [Google Scholar]
- Golding I, Paulsson J, Zawilski SM, Cox EC. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- Goodman RH, Smolik S. CBP/p300 in cell growth, transformation, and development. Genes Dev. 2000;14:1553–1577. [PubMed] [Google Scholar]
- Goold CP, Nicoll RA. Single-cell optogenetic excitation drives homeostatic synaptic depression. Neuron. 2010;68:512–528. doi: 10.1016/j.neuron.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graef IA, Mermelstein PG, Stankunas K, Neilson JR, Deisseroth K, Tsien RW, Crabtree GR. L-type calcium channels and GSK-3 regulate the activity of NF-ATc4 in hippocampal neurons. Nature. 1999;401:703–708. doi: 10.1038/44378. [DOI] [PubMed] [Google Scholar]
- Graef IA, Wang F, Charron F, Chen L, Neilson J, Tessier-Lavigne M, Crabtree GR. Neurotrophins and netrins require calcineurin/NFAT signaling to stimulate outgrowth of embryonic axons. Cell. 2003;113:657–670. doi: 10.1016/s0092-8674(03)00390-8. [DOI] [PubMed] [Google Scholar]
- Graham DL, Edwards S, Bachtell RK, DiLeone RJ, Rios M, Self DW. Dynamic BDNF activity in nucleus accumbens with cocaine use increases self-administration and relapse. Nat Neurosci. 2007;10:1029–1037. doi: 10.1038/nn1929. [DOI] [PubMed] [Google Scholar]
- Gray R, Johnston D. Noradrenaline and beta-adrenoceptor agonists increase activity of voltage-dependent calcium channels in hippocampal neurons. Nature. 1987;327:620–622. doi: 10.1038/327620a0. [DOI] [PubMed] [Google Scholar]
- Green EM, Barrett CF, Bultynck G, Shamah SM, Dolmetsch RE. The tumor suppressor eIF3e mediates calcium-dependent internalization of the L-type calcium channel CaV1.2. Neuron. 2007;55:615–632. doi: 10.1016/j.neuron.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos proto-oncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Ziff EB, Greene LA. Stimulation of neuronal acetylcholine receptors induces rapid gene transcription. Science. 1986;234:80–83. doi: 10.1126/science.3749894. [DOI] [PubMed] [Google Scholar]
- Greer PL, Greenberg ME. From synapse to nucleus: calcium-dependent gene transcription in the control of synapse development and function. Neuron. 2008;59:846–860. doi: 10.1016/j.neuron.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Grégoire S, Tremblay AM, Xiao L, Yang Q, Ma K, Nie J, Mao Z, Wu Z, Giguère V, Yang XJ. Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J Biol Chem. 2006;281:4423–4433. doi: 10.1074/jbc.M509471200. [DOI] [PubMed] [Google Scholar]
- Groc L, Heine M, Cousins SL, Stephenson FA, Lounis B, Cognet L, Choquet D. NMDA receptor surface mobility depends on NR2A-2B subunits. Proc Natl Acad Sci USA. 2006;103:18769–18774. doi: 10.1073/pnas.0605238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grünwald D, Singer RH. In vivo imaging of labelled endogenous β-actin mRNA during nucleocytoplasmic transport. Nature. 2010;467:604–607. doi: 10.1038/nature09438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan JS, Haggarty SJ, Giacometti E, Dannenberg JH, Joseph N, Gao J, Nieland TJF, Zhou Y, Wang X, Mazitschek R, Bradner JE, DePinho RA, Jaenisch R, Tsai LH. HDAC2 negatively regulates memory formation and synaptic plasticity. Nature. 2009;459:55–60. doi: 10.1038/nature07925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- Guerrini L, Blasi F, Denis-Donini S. Synaptic activation of NF-kappa B by glutamate in cerebellar granule neurons in vitro. Proc Natl Acad Sci USA. 1995;92:9077–9081. doi: 10.1073/pnas.92.20.9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JU, Su Y, Zhong C, Ming GL, Song H. Hydroxylation of 5-Methylcytosine by TET1 Promotes Active DNA Demethylation in the Adult Brain. Cell. 2011;145:423–434. doi: 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurley LR, D’Anna JA, Barham SS, Deaven LL, Tobey RA. Histone phosphorylation and chromatin structure during mitosis in Chinese hamster cells. Eur J Biochem. 1978;84:1–15. doi: 10.1111/j.1432-1033.1978.tb12135.x. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Environment-specific expression of the immediate-early gene Arc in hippocampal neuronal ensembles. Nat Neurosci. 1999;2:1120–1124. doi: 10.1038/16046. [DOI] [PubMed] [Google Scholar]
- Guzowski JF, McNaughton BL, Barnes CA, Worley PF. Imaging neural activity with temporal and cellular resolution using FISH. Curr Opin Neurobiol. 2001;11:579–584. doi: 10.1016/s0959-4388(00)00252-x. [DOI] [PubMed] [Google Scholar]
- Gwack Y, Sharma S, Nardone J, Tanasa B, Iuga A, Srikanth S, Okamura H, Bolton D, Feske S, Hogan PG, Rao A. A genome-wide Drosophila RNAi screen identifies DYRK-family kinases as regulators of NFAT. Nature. 2006;441:646–650. doi: 10.1038/nature04631. [DOI] [PubMed] [Google Scholar]
- Han J, Jiang Y, Li Z, Kravchenko VV, Ulevitch RJ. Activation of the transcription factor MEF2C by the MAP kinase p38 in inflammation. Nature. 1997;386:296–299. doi: 10.1038/386296a0. [DOI] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa Ra, Huerta JJ, Koltzenburg M, Kohler M, van-Minnen J, Twiss JL, Fainzilber M. Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron. 2003;40:1095–1104. doi: 10.1016/s0896-6273(03)00770-0. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Arnold FJ, Bading H. Nuclear calcium signaling controls CREB-mediated gene expression triggered by synaptic activity. Nat Neurosci. 2001;4:261–267. doi: 10.1038/85109. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. Coupling of extrasynaptic NMDA receptors to a CREB shut-off pathway is developmentally regulated. Biochim Biophys Acta. 2002;1600:148–153. doi: 10.1016/s1570-9639(02)00455-7. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. The Yin and Yang of NMDA receptor signalling. Trends Neurosci. 2003;26:81–89. doi: 10.1016/S0166-2236(02)00040-1. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Bading H. Synaptic versus extrasynaptic NMDA receptor signalling: implications for neurodegenerative disorders. Nat Rev Neurosci. 2010;11:682–696. doi: 10.1038/nrn2911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardingham GE, Chawla S, Cruzalegui FH, Bading H. Control of recruitment and transcription-activating function of CBP determines gene regulation by NMDA receptors and L-type calcium channels. Neuron. 1999;22:789–798. doi: 10.1016/s0896-6273(00)80737-0. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Chawla S, Johnson CM, Bading H. Distinct functions of nuclear and cytoplasmic calcium in the control of gene expression. Nature. 1997;385:260–265. doi: 10.1038/385260a0. [DOI] [PubMed] [Google Scholar]
- Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5:405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- Harris AZ, Pettit DL. Extrasynaptic and synaptic NMDA receptors form stable and uniform pools in rat hippocampal slices. J Physiol (Lond) 2007;584:509–519. doi: 10.1113/jphysiol.2007.137679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, Greengard P, Heintz N. A translational profiling approach for the molecular characterization of CNS cell types. Cell. 2008;135:738–748. doi: 10.1016/j.cell.2008.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M, Ren B. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- Hell JW, Westenbroek RE, Warner C, Ahlijanian MK, Prystay W, Gilbert MM, Snutch TP, Catterall WA. Identification and differential subcellular localization of the neuronal class C and class D L-type calcium channel alpha 1 subunits. J Cell Biol. 1993a;123:949–962. doi: 10.1083/jcb.123.4.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell JW, Yokoyama CT, Wong ST, Warner C, Snutch TP, Catterall WA. Differential phosphorylation of two size forms of the neuronal class C L-type calcium channel alpha 1 subunit. J Biol Chem. 1993b;268:19451–19457. [PubMed] [Google Scholar]
- Hérault Y, Beckers J, Gérard M, Duboule D. Hox gene expression in limbs: colinearity by opposite regulatory controls. Dev Biol. 1999;208:157–165. doi: 10.1006/dbio.1998.9179. [DOI] [PubMed] [Google Scholar]
- Herdegen T, Leah JD. Inducible and constitutive transcription factors in the mammalian nervous system: control of gene expression by Jun, Fos and Krox, and CREB/ATF proteins. Brain Res Brain Res Rev. 1998;28:370–490. doi: 10.1016/s0165-0173(98)00018-6. [DOI] [PubMed] [Google Scholar]
- Hevroni D, Rattner A, Bundman M, Lederfein D, Gabarah A, Mangelus M, Silverman MA, Kedar H, Naor C, Kornuc M, Hanoch T, Seger R, Theill LE, Nedivi E, Richter-Levin G, Citri Y. Hippocampal plasticity involves extensive gene induction and multiple cellular mechanisms. J Mol Neurosci. 1998;10:75–98. doi: 10.1007/BF02737120. [DOI] [PubMed] [Google Scholar]
- Hill CS, Treisman R. Growth factors and gene expression: fresh insights from arrays. Sci STKE. 1999;1999:PE1. doi: 10.1126/stke.1999.3.pe1. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Chen L, Nardone J, Rao A. Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev. 2003;17:2205–2232. doi: 10.1101/gad.1102703. [DOI] [PubMed] [Google Scholar]
- Hogan PG, Lewis RS, Rao A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Annu Rev Immunol. 2010;28:491–533. doi: 10.1146/annurev.immunol.021908.132550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollmann M, Boulter J, Maron C, Beasley L, Sullivan J, Pecht G, Heinemann S. Zinc potentiates agonist-induced currents at certain splice variants of the NMDA receptor. Neuron. 1993;10:943–954. doi: 10.1016/0896-6273(93)90209-a. [DOI] [PubMed] [Google Scholar]
- Hon G, Wang W, Ren B. Discovery and annotation of functional chromatin signatures in the human genome. PLoS Comput Biol. 2009;5:e1000566. doi: 10.1371/journal.pcbi.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, Mccord AE, Greenberg ME. A biological function for the neuronal activity-dependent component of Bdnf transcription in the development of cortical inhibition. Neuron. 2008;60:610–624. doi: 10.1016/j.neuron.2008.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong EJ, West AE, Greenberg ME. Transcriptional control of cognitive development. Curr Opin Neurobiol. 2005;15:21–28. doi: 10.1016/j.conb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- Hoogland TM, Saggau P. Facilitation of L-type Ca2+ channels in dendritic spines by activation of beta2 adrenergic receptors. Journal of Neuroscience. 2004;24:8416–8427. doi: 10.1523/JNEUROSCI.1677-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hope BT, Kelz MB, Duman RS, Nestler EJ. Chronic electroconvulsive seizure (ECS) treatment results in expression of a long-lasting AP-1 complex in brain with altered composition and characteristics. J Neurosci. 1994;14:4318–4328. doi: 10.1523/JNEUROSCI.14-07-04318.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn PJ, Peterson CL. Molecular biology. Chromatin higher order folding--wrapping up transcription. Science. 2002;297:1824–1827. doi: 10.1126/science.1074200. [DOI] [PubMed] [Google Scholar]
- Hu M, Liu Q-s, Chang KT, Berg DK. Nicotinic regulation of CREB activation in hippocampal neurons by glutamatergic and nonglutamatergic pathways. Mol Cell Neurosci. 2002;21:616–625. doi: 10.1006/mcne.2002.1202. [DOI] [PubMed] [Google Scholar]
- Hu SC, Chrivia J, Ghosh A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- Hulme JT, Lin TWC, Westenbroek RE, Scheuer T, Catterall WA. Beta-adrenergic regulation requires direct anchoring of PKA to cardiac CaV1.2 channels via a leucine zipper interaction with A kinase-anchoring protein 15. Proc Natl Acad Sci USA. 2003;100:13093–13098. doi: 10.1073/pnas.2135335100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman SE, Comb M, Lin YS, Pearlberg J, Green MR, Goodman HM. A common trans-acting factor is involved in transcriptional regulation of neurotransmitter genes by cyclic AMP. Mol Cell Biol. 1988;8:4225–4233. doi: 10.1128/mcb.8.10.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibata K, Sun Q, Turrigiano GG. Rapid synaptic scaling induced by changes in postsynaptic firing. Neuron. 2008;57:819–826. doi: 10.1016/j.neuron.2008.02.031. [DOI] [PubMed] [Google Scholar]
- Im H-I, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nature neuroscience. 2010 doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impey S, Fong AL, Wang Y, Cardinaux JR, Fass DM, Obrietan K, Wayman GA, Storm DR, Soderling TR, Goodman RH. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- Impey S, Obrietan K, Wong ST, Poser S, Yano S, Wayman G, Deloulme JC, Chan G, Storm DR. Cross talk between ERK and PKA is required for Ca2+ stimulation of CREB-dependent transcription and ERK nuclear translocation. Neuron. 1998;21:869–883. doi: 10.1016/s0896-6273(00)80602-9. [DOI] [PubMed] [Google Scholar]
- Ishii T, Moriyoshi K, Sugihara H, Sakurada K, Kadotani H, Yokoi M, Akazawa C, Shigemoto R, Mizuno N, Masu M. Molecular characterization of the family of the N-methyl-D-aspartate receptor subunits. J Biol Chem. 1993;268:2836–2843. [PubMed] [Google Scholar]
- Ito S, D’Alessio AC, Taranova OV, Hong K, Sowers LC, Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003;33(Suppl):245–254. doi: 10.1038/ng1089. [DOI] [PubMed] [Google Scholar]
- Janicki SM, Tsukamoto T, Salghetti SE, Tansey WP, Sachidanandam R, Prasanth KV, Ried T, Shav-Tal Y, Bertrand E, Singer RH, Spector DL. From silencing to gene expression: real-time analysis in single cells. Cell. 2004;116:683–698. doi: 10.1016/s0092-8674(04)00171-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey RA, Ch’ng TH, O’Dell TJ, Martin KC. Activity-dependent anchoring of importin alpha at the synapse involves regulated binding to the cytoplasmic tail of the NR1–1a subunit of the NMDA receptor. J Neurosci. 2009;29:15613–15620. doi: 10.1523/JNEUROSCI.3314-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X, Tian F, Du Y, Copeland NG, Jenkins NA, Tessarollo L, Wu X, Pan H, Hu XZ, Xu K, Kenney H, Egan SE, Turley H, Harris AL, Marini AM, Lipsky RH. BHLHB2 controls Bdnf promoter 4 activity and neuronal excitability. J Neurosci. 2008;28:1118–1130. doi: 10.1523/JNEUROSCI.2262-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- Jordan BA, Kreutz MR. Nucleocytoplasmic protein shuttling: the direct route in synapse-to-nucleus signaling. Trends Neurosci. 2009;32:392–401. doi: 10.1016/j.tins.2009.04.001. [DOI] [PubMed] [Google Scholar]
- Kammermeier PJ, Worley PF. Homer 1a uncouples metabotropic glutamate receptor 5 from postsynaptic effectors. Proc Natl Acad Sci USA. 2007;104:6055–6060. doi: 10.1073/pnas.0608991104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato Y, Kravchenko VV, Tapping RI, Han J, Ulevitch RJ, Lee JD. BMK1/ERK5 regulates serum-induced early gene expression through transcription factor MEF2C. EMBO J. 1997;16:7054–7066. doi: 10.1093/emboj/16.23.7054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer JA, Malenka RC, Nicoll RA. NMDA application potentiates synaptic transmission in the hippocampus. Nature. 1988;334:250–252. doi: 10.1038/334250a0. [DOI] [PubMed] [Google Scholar]
- Kavalali ET, Hwang KS, Plummer MR. cAMP-dependent enhancement of dihydropyridine-sensitive calcium channel availability in hippocampal neurons. J Neurosci. 1997;17:5334–5348. doi: 10.1523/JNEUROSCI.17-14-05334.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci USA. 2009;106:316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MB. Signal-processing machines at the postsynaptic density. Science. 2000;290:750–754. doi: 10.1126/science.290.5492.750. [DOI] [PubMed] [Google Scholar]
- Kew JN, Richards JG, Mutel V, Kemp JA. Developmental changes in NMDA receptor glycine affinity and ifenprodil sensitivity reveal three distinct populations of NMDA receptors in individual rat cortical neurons. J Neurosci. 1998;18:1935–1943. doi: 10.1523/JNEUROSCI.18-06-01935.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MJ, Dunah AW, Wang YT, Sheng M. Differential roles of NR2A- and NR2B-containing NMDA receptors in Ras-ERK signaling and AMPA receptor trafficking. Neuron. 2005;46:745–760. doi: 10.1016/j.neuron.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Kim T-K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010 doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirson ED, Yaari Y. Synaptic NMDA receptors in developing mouse hippocampal neurones: functional properties and sensitivity to ifenprodil. J Physiol (Lond) 1996;497 ( Pt 2):437–455. doi: 10.1113/jphysiol.1996.sp021779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci. 2006;31:89–97. doi: 10.1016/j.tibs.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Knöll B, Kretz O, Fiedler C, Alberti S, Schütz G, Frotscher M, Nordheim A. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat Neurosci. 2006;9:195–204. doi: 10.1038/nn1627. [DOI] [PubMed] [Google Scholar]
- Knöll B, Nordheim A. Functional versatility of transcription factors in the nervous system: the SRF paradigm. Trends Neurosci. 2009;32:432–442. doi: 10.1016/j.tins.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, An J, Lamperti ED, Koh KP, Ganetzky R, Liu XS, Aravind L, Agarwal S, Maciejewski JP, Rao A. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature. 2010;468:839–843. doi: 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhr G. NMDA receptor function: subunit composition versus spatial distribution. Cell Tissue Res. 2006;326:439–446. doi: 10.1007/s00441-006-0273-6. [DOI] [PubMed] [Google Scholar]
- Kornhauser JM, Cowan CW, Shaywitz AJ, Dolmetsch RE, Griffith EC, Hu LS, Haddad C, Xia Z, Greenberg ME. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- Korzus E, Rosenfeld MG, Mayford M. CBP histone acetyltransferase activity is a critical component of memory consolidation. Neuron. 2004;42:961–972. doi: 10.1016/j.neuron.2004.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koschak A, Obermair GJ, Pivotto F, Sinnegger-Brauns MJ, Striessnig J, Pietrobon D. Molecular nature of anomalous L-type calcium channels in mouse cerebellar granule cells. J Neurosci. 2007;27:3855–3863. doi: 10.1523/JNEUROSCI.4028-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- Kovács KA, Steullet P, Steinmann M, Do KQ, Magistretti PJ, Halfon O, Cardinaux JR. TORC1 is a calcium- and cAMP-sensitive coincidence detector involved in hippocampal long-term synaptic plasticity. Proc Natl Acad Sci USA. 2007;104:4700–4705. doi: 10.1073/pnas.0607524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Krapivinsky L, Manasian Y, Ivanov A, Tyzio R, Pellegrino C, Ben-Ari Y, Clapham DE, Medina I. The NMDA receptor is coupled to the ERK pathway by a direct interaction between NR2B and RasGRF1. Neuron. 2003;40:775–784. doi: 10.1016/s0896-6273(03)00645-7. [DOI] [PubMed] [Google Scholar]
- Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar SS, Bacci A, Kharazia V, Huguenard JR. A developmental switch of AMPA receptor subunits in neocortical pyramidal neurons. J Neurosci. 2002;22:3005–3015. doi: 10.1523/JNEUROSCI.22-08-03005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutsuwada T, Kashiwabuchi N, Mori H, Sakimura K, Kushiya E, Araki K, Meguro H, Masaki H, Kumanishi T, Arakawa M. Molecular diversity of the NMDA receptor channel. Nature. 1992;358:36–41. doi: 10.1038/358036a0. [DOI] [PubMed] [Google Scholar]
- Lai KO, Zhao Y, Ch’ng TH, Martin KC. Importin-mediated retrograde transport of CREB2 from distal processes to the nucleus in neurons. Proc Natl Acad Sci USA. 2008;105:17175–17180. doi: 10.1073/pnas.0803906105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb NJ, Fernandez A, Tourkine N, Jeanteur P, Blanchard JM. Demonstration in living cells of an intragenic negative regulatory element within the rodent c-fos gene. Cell. 1990;61:485–496. doi: 10.1016/0092-8674(90)90530-r. [DOI] [PubMed] [Google Scholar]
- Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iñiguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolaños CA, Kabbaj M, Xiao G, Neve RL, Hurd YL, Oosting RS, Fan G, Morrison JH, Nestler EJ. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau LF, Nathans D. Expression of a set of growth-related immediate early genes in BALB/c 3T3 cells: coordinate regulation with c-fos or c-myc. Proc Natl Acad Sci USA. 1987;84:1182–1186. doi: 10.1073/pnas.84.5.1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Smith E, Shilatifard A. The language of histone crosstalk. Cell. 2010;142:682–685. doi: 10.1016/j.cell.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SJR, Escobedo-Lozoya Y, Szatmari EM, Yasuda R. Activation of CaMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299–304. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leifer D, Krainc D, Yu YT, McDermott J, Breitbart RE, Heng J, Neve RL, Kosofsky B, Nadal-Ginard B, Lipton SA. MEF2C, a MADS/MEF2-family transcription factor expressed in a laminar distribution in cerebral cortex. Proc Natl Acad Sci USA. 1993;90:1546–1550. doi: 10.1073/pnas.90.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemercier C, Verdel A, Galloo B, Curtet S, Brocard MP, Khochbin S. mHDA1/HDAC5 histone deacetylase interacts with and represses MEF2A transcriptional activity. J Biol Chem. 2000;275:15594–15599. doi: 10.1074/jbc.M908437199. [DOI] [PubMed] [Google Scholar]
- Lerea LS, Butler LS, McNamara JO. NMDA and non-NMDA receptor-mediated increase of c-fos mRNA in dentate gyrus neurons involves calcium influx via different routes. J Neurosci. 1992;12:2973–2981. doi: 10.1523/JNEUROSCI.12-08-02973.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin GR. Neurotrophins and the specification of neuronal phenotype. Philos Trans R Soc Lond, B, Biol Sci. 1996;351:405–411. doi: 10.1098/rstb.1996.0035. [DOI] [PubMed] [Google Scholar]
- Li JH, Wang YH, Wolfe BB, Krueger KE, Corsi L, Stocca G, Vicini S. Developmental changes in localization of NMDA receptor subunits in primary cultures of cortical neurons. Eur J Neurosci. 1998;10:1704–1715. doi: 10.1046/j.1460-9568.1998.00169.x. [DOI] [PubMed] [Google Scholar]
- Li L, Carter J, Gao X, Whitehead J, Tourtellotte WG. The neuroplasticity-associated arc gene is a direct transcriptional target of early growth response (Egr) transcription factors. Mol Cell Biol. 2005;25:10286–10300. doi: 10.1128/MCB.25.23.10286-10300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, Radulovic J, Tourtellotte WG. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci. 2007;35:76–88. doi: 10.1016/j.mcn.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Barkess G, Qian H. Chromatin looping and the probability of transcription. Trends Genet. 2006a;22:197–202. doi: 10.1016/j.tig.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Li S, Tian X, Hartley DM, Feig LA. Distinct roles for Ras-guanine nucleotide-releasing factor 1 (Ras-GRF1) and Ras-GRF2 in the induction of long-term potentiation and long-term depression. J Neurosci. 2006b;26:1721–1729. doi: 10.1523/JNEUROSCI.3990-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, Sandstrom R, Bernstein B, Bender MA, Groudine M, Gnirke A, Stamatoyannopoulos J, Mirny LA, Lander ES, Dekker J. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilienbaum A, Israël A. From calcium to NF-kappa B signaling pathways in neurons. Mol Cell Biol. 2003;23:2680–2698. doi: 10.1128/MCB.23.8.2680-2698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JW, Wyszynski M, Madhavan R, Sealock R, Kim JU, Sheng M. Yotiao, a novel protein of neuromuscular junction and brain that interacts with specific splice variants of NMDA receptor subunit NR1. J Neurosci. 1998;18:2017–2027. doi: 10.1523/JNEUROSCI.18-06-02017.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou HC, Baltimore D. Regulation of the NF-kappa B/rel transcription factor and I kappa B inhibitor system. Curr Opin Cell Biol. 1993;5:477–487. doi: 10.1016/0955-0674(93)90014-h. [DOI] [PubMed] [Google Scholar]
- Lipscombe D, Helton TD, Xu W. L-type calcium channels: the low down. J Neurophysiol. 2004;92:2633–2641. doi: 10.1152/jn.00486.2004. [DOI] [PubMed] [Google Scholar]
- Lipsky RH, Xu K, Zhu D, Kelly C, Terhakopian A, Novelli A, Marini AM. Nuclear factor kappaB is a critical determinant in N-methyl-D-aspartate receptor-mediated neuroprotection. J Neurochem. 2001;78:254–264. doi: 10.1046/j.1471-4159.2001.00386.x. [DOI] [PubMed] [Google Scholar]
- Liu L, Wong TP, Pozza MF, Lingenhoehl K, Wang Y, Sheng M, Auberson YP, Wang YT. Role of NMDA receptor subtypes in governing the direction of hippocampal synaptic plasticity. Science. 2004;304:1021–1024. doi: 10.1126/science.1096615. [DOI] [PubMed] [Google Scholar]
- Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Liu SQ, Cull-Candy SG. Synaptic activity at calcium-permeable AMPA receptors induces a switch in receptor subtype. Nature. 2000;405:454–458. doi: 10.1038/35013064. [DOI] [PubMed] [Google Scholar]
- Lomeli H, Mosbacher J, Melcher T, Höger T, Geiger JR, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg PH. Control of kinetic properties of AMPA receptor channels by nuclear RNA editing. Science. 1994;266:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- Lonze BE, Ginty DD. Function and regulation of CREB family transcription factors in the nervous system. Neuron. 2002;35:605–623. doi: 10.1016/s0896-6273(02)00828-0. [DOI] [PubMed] [Google Scholar]
- Lu B. BDNF and activity-dependent synaptic modulation. Learn Mem. 2003;10:86–98. doi: 10.1101/lm.54603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, McKinsey TA, Nicol RL, Olson EN. Signal-dependent activation of the MEF2 transcription factor by dissociation from histone deacetylases. Proc Natl Acad Sci USA. 2000;97:4070–4075. doi: 10.1073/pnas.080064097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusin JD, Vanarotti M, Li C, Valiveti A, Ames JB. NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization. Biochemistry. 2008;47:2252–2264. doi: 10.1021/bi7017267. [DOI] [PubMed] [Google Scholar]
- Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- Lyons GE, Micales BK, Schwarz J, Martin JF, Olson EN. Expression of mef2 genes in the mouse central nervous system suggests a role in neuronal maturation. J Neurosci. 1995;15:5727–5738. doi: 10.1523/JNEUROSCI.15-08-05727.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma DK, Jang MH, Guo JU, Kitabatake Y, Chang ML, Pow-Anpongkul N, Flavell RA, Lu B, Ming GL, Song H. Neuronal activity-induced Gadd45b promotes epigenetic DNA demethylation and adult neurogenesis. Science. 2009;323:1074–1077. doi: 10.1126/science.1166859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenka RC, Kauer JA, Zucker RS, Nicoll RA. Postsynaptic calcium is sufficient for potentiation of hippocampal synaptic transmission. Science. 1988;242:81–84. doi: 10.1126/science.2845577. [DOI] [PubMed] [Google Scholar]
- Mantamadiotis T, Lemberger T, Bleckmann SC, Kern H, Kretz O, Martin Villalba A, Tronche F, Kellendonk C, Gau D, Kapfhammer J, Otto C, Schmid W, Schütz G. Disruption of CREB function in brain leads to neurodegeneration. Nat Genet. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- Mao Z, Bonni A, Xia F, Nadal-Vicens M, Greenberg ME. Neuronal activity-dependent cell survival mediated by transcription factor MEF2. Science. 1999;286:785–790. doi: 10.1126/science.286.5440.785. [DOI] [PubMed] [Google Scholar]
- Mao Z, Wiedmann M. Calcineurin enhances MEF2 DNA binding activity in calcium-dependent survival of cerebellar granule neurons. J Biol Chem. 1999;274:31102–31107. doi: 10.1074/jbc.274.43.31102. [DOI] [PubMed] [Google Scholar]
- Marais R, Wynne J, Treisman R. The SRF accessory protein Elk-1 contains a growth factor-regulated transcriptional activation domain. Cell. 1993;73:381–393. doi: 10.1016/0092-8674(93)90237-k. [DOI] [PubMed] [Google Scholar]
- Marek KW, Kurtz LM, Spitzer NC. cJun integrates calcium activity and tlx3 expression to regulate neurotransmitter specification. Nature neuroscience. 2010 doi: 10.1038/nn.2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel MA, Wyllie DJA, Hardingham GE. In developing hippocampal neurons, NR2B-containing N-methyl-D-aspartate receptors (NMDARs) can mediate signaling to neuronal survival and synaptic potentiation, as well as neuronal death. Neuroscience. 2009;158:334–343. doi: 10.1016/j.neuroscience.2008.01.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massa PT, Aleyasin H, Park DS, Mao X, Barger SW. NFkappaB in neurons? The uncertainty principle in neurobiology. J Neurochem. 2006;97:607–618. doi: 10.1111/j.1471-4159.2006.03810.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda K, Kamiya Y, Matsuda S, Yuzaki M. Cloning and characterization of a novel NMDA receptor subunit NR3B: a dominant subunit that reduces calcium permeability. Brain Res Mol Brain Res. 2002;100:43–52. doi: 10.1016/s0169-328x(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBain CJ, Mayer ML. N-methyl-D-aspartic acid receptor structure and function. Physiol Rev. 1994;74:723–760. doi: 10.1152/physrev.1994.74.3.723. [DOI] [PubMed] [Google Scholar]
- McCullagh KJA, Calabria E, Pallafacchina G, Ciciliot S, Serrano AL, Argentini C, Kalhovde JM, Lømo T, Schiaffino S. NFAT is a nerve activity sensor in skeletal muscle and controls activity-dependent myosin switching. Proc Natl Acad Sci USA. 2004;101:10590–10595. doi: 10.1073/pnas.0308035101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell KA, Hutchinson AN, Wong-Goodrich SJE, Presby MM, Su D, Rodriguiz RM, Law KC, Williams CL, Wetsel WC, West AE. Reduced cortical BDNF expression and aberrant memory in carf knockout mice. J Neurosci. 2010;30:7453–7465. doi: 10.1523/JNEUROSCI.3997-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGurk JF, Bennett MV, Zukin RS. Polyamines potentiate responses of N-methyl-D-aspartate receptors expressed in xenopus oocytes. Proc Natl Acad Sci USA. 1990;87:9971–9974. doi: 10.1073/pnas.87.24.9971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev. 2001;11:497–504. doi: 10.1016/s0959-437x(00)00224-0. [DOI] [PubMed] [Google Scholar]
- McKinsey TA, Zhang CL, Olson EN. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem Sci. 2002;27:40–47. doi: 10.1016/s0968-0004(01)02031-x. [DOI] [PubMed] [Google Scholar]
- McPherson JD. Next-generation gap. Nat Methods. 2009;6:S2–5. doi: 10.1038/nmeth.f.268. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Baltimore D. Physiological functions for brain NF-kappaB. Trends Neurosci. 2005;28:37–43. doi: 10.1016/j.tins.2004.11.002. [DOI] [PubMed] [Google Scholar]
- Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-kappa B functions in synaptic signaling and behavior. Nat Neurosci. 2003;6:1072–1078. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- Meguro H, Mori H, Araki K, Kushiya E, Kutsuwada T, Yamazaki M, Kumanishi T, Arakawa M, Sakimura K, Mishina M. Functional characterization of a heteromeric NMDA receptor channel expressed from cloned cDNAs. Nature. 1992;357:70–74. doi: 10.1038/357070a0. [DOI] [PubMed] [Google Scholar]
- Melcher T, Maas S, Herb A, Sprengel R, Seeburg PH, Higuchi M. A mammalian RNA editing enzyme. Nature. 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- Mercurio F, Zhu H, Murray BW, Shevchenko A, Bennett BL, Li J, Young DB, Barbosa M, Mann M, Manning A, Rao A. IKK-1 and IKK-2: cytokine-activated IkappaB kinases essential for NF-kappaB activation. Science. 1997;278:860–866. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- Mermelstein PG, Bito H, Deisseroth K, Tsien RW. Critical dependence of cAMP response element-binding protein phosphorylation on L-type calcium channels supports a selective response to EPSPs in preference to action potentials. Journal of Neuroscience. 2000;20:266–273. doi: 10.1523/JNEUROSCI.20-01-00266.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermelstein PG, Deisseroth K, Dasgupta N, Isaksen AL, Tsien RW. Calmodulin priming: nuclear translocation of a calmodulin complex and the memory of prior neuronal activity. Proc Natl Acad Sci USA. 2001;98:15342–15347. doi: 10.1073/pnas.211563998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer-Franke A, Wilkinson GA, Kruttgen A, Hu M, Munro E, Hanson MG, Reichardt LF, Barres BA. Depolarization and cAMP elevation rapidly recruit TrkB to the plasma membrane of CNS neurons. Neuron. 1998;21:681–693. doi: 10.1016/s0896-6273(00)80586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O’Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller CA, Sweatt JD. Covalent modification of DNA regulates memory formation. Neuron. 2007;53:857–869. doi: 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Miralles F, Posern G, Zaromytidou AI, Treisman R. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell. 2003;113:329–342. doi: 10.1016/s0092-8674(03)00278-2. [DOI] [PubMed] [Google Scholar]
- Miranti CK, Ginty DD, Huang G, Chatila T, Greenberg ME. Calcium activates serum response factor-dependent transcription by a Ras- and Elk-1-independent mechanism that involves a Ca2+/calmodulin-dependent kinase. Mol Cell Biol. 1995;15:3672–3684. doi: 10.1128/mcb.15.7.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokalled MH, Johnson A, Kim Y, Oh J, Olson EN. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development. 2010;137:2365–2374. doi: 10.1242/dev.047605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molkentin JD, Black BL, Martin JF, Olson EN. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 1995;83:1125–1136. doi: 10.1016/0092-8674(95)90139-6. [DOI] [PubMed] [Google Scholar]
- Montminy MR, Bilezikjian LM. Binding of a nuclear protein to the cyclic-AMP response element of the somatostatin gene. Nature. 1987;328:175–178. doi: 10.1038/328175a0. [DOI] [PubMed] [Google Scholar]
- Monyer H, Burnashev N, Laurie DJ, Sakmann B, Seeburg PH. Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 1994;12:529–540. doi: 10.1016/0896-6273(94)90210-0. [DOI] [PubMed] [Google Scholar]
- Monyer H, Sprengel R, Schoepfer R, Herb A, Higuchi M, Lomeli H, Burnashev N, Sakmann B, Seeburg PH. Heteromeric NMDA receptors: molecular and functional distinction of subtypes. Science. 1992;256:1217–1221. doi: 10.1126/science.256.5060.1217. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26:319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Role of ion flux in the control of c-fos expression. Nature. 1986;322:552–555. doi: 10.1038/322552a0. [DOI] [PubMed] [Google Scholar]
- Morgan JI, Curran T. Stimulus-transcription coupling in the nervous system: involvement of the inducible proto-oncogenes fos and jun. Annu Rev Neurosci. 1991;14:421–451. doi: 10.1146/annurev.ne.14.030191.002225. [DOI] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Mu Y, Otsuka T, Horton AC, Scott DB, Ehlers MD. Activity-dependent mRNA splicing controls ER export and synaptic delivery of NMDA receptors. Neuron. 2003;40:581–594. doi: 10.1016/s0896-6273(03)00676-7. [DOI] [PubMed] [Google Scholar]
- Muller D, Joly M, Lynch G. Contributions of quisqualate and NMDA receptors to the induction and expression of LTP. Science. 1988;242:1694–1697. doi: 10.1126/science.2904701. [DOI] [PubMed] [Google Scholar]
- Müller R, Bravo R, Burckhardt J, Curran T. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 1984;312:716–720. doi: 10.1038/312716a0. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Lucibello FC, Schuermann M, Müller R. Alternative splicing of fosB transcripts results in differentially expressed mRNAs encoding functionally antagonistic proteins. Genes Dev. 1991;5:1212–1223. doi: 10.1101/gad.5.7.1212. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y, Nathans D. A naturally occurring truncated form of FosB that inhibits Fos/Jun transcriptional activity. Cell. 1991;64:751–759. doi: 10.1016/0092-8674(91)90504-r. [DOI] [PubMed] [Google Scholar]
- Nase G, Weishaupt J, Stern P, Singer W, Monyer H. Genetic and epigenetic regulation of NMDA receptor expression in the rat visual cortex. Eur J Neurosci. 1999;11:4320–4326. doi: 10.1046/j.1460-9568.1999.00859.x. [DOI] [PubMed] [Google Scholar]
- Nechaev S, Fargo DC, dos Santos G, Liu L, Gao Y, Adelman K. Global analysis of short RNAs reveals widespread promoter-proximal stalling and arrest of Pol II in Drosophila. Science. 2010;327:335–338. doi: 10.1126/science.1181421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- Nestler EJ. Molecular basis of long-term plasticity underlying addiction. Nat Rev Neurosci. 2001;2:119–128. doi: 10.1038/35053570. [DOI] [PubMed] [Google Scholar]
- Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- Niibori Y, Hayashi F, Hirai K, Matsui M, Inokuchi K. Alternative poly(A) site-selection regulates the production of alternatively spliced vesl-1/homer1 isoforms that encode postsynaptic scaffolding proteins. Neurosci Res. 2007;57:399–410. doi: 10.1016/j.neures.2006.11.014. [DOI] [PubMed] [Google Scholar]
- Nishi M, Hinds H, Lu HP, Kawata M, Hayashi Y. Motoneuron-specific expression of NR3B, a novel NMDA-type glutamate receptor subunit that works in a dominant-negative manner. J Neurosci. 2001;21:RC185. doi: 10.1523/JNEUROSCI.21-23-j0003.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman C, Runswick M, Pollock R, Treisman R. Isolation and properties of cDNA clones encoding SRF, a transcription factor that binds to the c-fos serum response element. Cell. 1988;55:989–1003. doi: 10.1016/0092-8674(88)90244-9. [DOI] [PubMed] [Google Scholar]
- Nott A, Watson PM, Robinson JD, Crepaldi L, Riccio A. S- Nitrosylation of histone deacetylase 2 induces chromatin remodelling in neurons. Nature. 2008;455:411–415. doi: 10.1038/nature07238. [DOI] [PubMed] [Google Scholar]
- Novina CD, Sharp PA. The RNAi revolution. Nature. 2004;430:161–164. doi: 10.1038/430161a. [DOI] [PubMed] [Google Scholar]
- Oh JS, Manzerra P, Kennedy MB. Regulation of the neuron-specific Ras GTPase-activating protein, synGAP, by Ca2+/calmodulin-dependent protein kinase II. J Biol Chem. 2004;279:17980–17988. doi: 10.1074/jbc.M314109200. [DOI] [PubMed] [Google Scholar]
- Okamura H, Aramburu J, García-Rodríguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- Oliveria SF, Dell’Acqua ML, Sather WA. AKAP79/150 anchoring of calcineurin controls neuronal L-type Ca2+ channel activity and nuclear signaling. Neuron. 2007;55:261–275. doi: 10.1016/j.neuron.2007.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson EN, Nordheim A. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat Rev Mol Cell Biol. 2010;11:353–365. doi: 10.1038/nrm2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooe N, Saito K, Kaneko H. Characterization of functional heterodimer partners in brain for a bHLH-PAS factor NXF. Biochim Biophys Acta. 2009;1789:192–197. doi: 10.1016/j.bbagrm.2009.01.003. [DOI] [PubMed] [Google Scholar]
- Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol Cell Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opatowsky Y, Chen CC, Campbell KP, Hirsch JA. Structural analysis of the voltage-dependent calcium channel beta subunit functional core and its complex with the alpha 1 interaction domain. Neuron. 2004;42:387–399. doi: 10.1016/s0896-6273(04)00250-8. [DOI] [PubMed] [Google Scholar]
- Osawa M, Dace A, Tong KI, Valiveti A, Ikura M, Ames JB. Mg2+ and Ca2+ differentially regulate DNA binding and dimerization of DREAM. J Biol Chem. 2005;280:18008–18014. doi: 10.1074/jbc.M500338200. [DOI] [PubMed] [Google Scholar]
- Osawa M, Tong KI, Lilliehook C, Wasco W, Buxbaum JD, Cheng HY, Penninger JM, Ikura M, Ames JB. Calcium-regulated DNA binding and oligomerization of the neuronal calcium-sensing protein, calsenilin/DREAM/KChIP3. J Biol Chem. 2001;276:41005–41013. doi: 10.1074/jbc.M105842200. [DOI] [PubMed] [Google Scholar]
- Oswald J, Engemann S, Lane N, Mayer W, Olek A, Fundele R, Dean W, Reik W, Walter J. Active demethylation of the paternal genome in the mouse zygote. Curr Biol. 2000;10:475–478. doi: 10.1016/s0960-9822(00)00448-6. [DOI] [PubMed] [Google Scholar]
- Otis KO, Thompson KR, Martin KC. Importin-mediated nuclear transport in neurons. Curr Opin Neurobiol. 2006;16:329–335. doi: 10.1016/j.conb.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- Papadia S, Soriano FX, Léveillé F, Martel MA, Dakin KA, Hansen HH, Kaindl A, Sifringer M, Fowler J, Stefovska V, McKenzie G, Craigon M, Corriveau R, Ghazal P, Horsburgh K, Yankner BA, Wyllie DJA, Ikonomidou C, Hardingham GE. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat Neurosci. 2008;11:476–487. doi: 10.1038/nn2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradis S, Harrar DB, Lin Y, Koon AC, Hauser JL, Griffith EC, Zhu L, Brass LF, Chen C, Greenberg ME. An RNAi-based approach identifies molecules required for glutamatergic and GABAergic synapse development. Neuron. 2007;53:217–232. doi: 10.1016/j.neuron.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CY, Shcheglovitov A, Dolmetsch R. The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science. 2010;330:101–105. doi: 10.1126/science.1191027. [DOI] [PubMed] [Google Scholar]
- Parker D, Jhala US, Radhakrishnan I, Yaffe MB, Reyes C, Shulman AI, Cantley LC, Wright PE, Montminy M. Analysis of an activator:coactivator complex reveals an essential role for secondary structure in transcriptional activation. Molecular Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- Parkitna JR, Bilbao A, Rieker C, Engblom D, Piechota M, Nordheim A, Spanagel R, Schütz G. Loss of the serum response factor in the dopamine system leads to hyperactivity. FASEB J. 2010;24:2427–2435. doi: 10.1096/fj.09-151423. [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat Rev Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- Pérez-Otaño I, Luján R, Tavalin SJ, Plomann M, Modregger J, Liu XB, Jones EG, Heinemann SF, Lo DC, Ehlers MD. Endocytosis and synaptic removal of NR3A-containing NMDA receptors by PACSIN1/syndapin1. Nat Neurosci. 2006;9:611–621. doi: 10.1038/nn1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkinton MS, Sihra TS, Williams RJ. Ca(2+)-permeable AMPA receptors induce phosphorylation of cAMP response element-binding protein through a phosphatidylinositol 3-kinase-dependent stimulation of the mitogen-activated protein kinase signaling cascade in neurons. J Neurosci. 1999;19:5861–5874. doi: 10.1523/JNEUROSCI.19-14-05861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BZ, DeMaria CD, Adelman JP, Yue DT. Calmodulin is the Ca2+ sensor for Ca2+-dependent inactivation of L-type calcium channels. Neuron. 1999;22:549–558. doi: 10.1016/s0896-6273(00)80709-6. [DOI] [PubMed] [Google Scholar]
- Pfenning AR, Kim TK, Spotts JM, Hemberg M, Su D, West AE. Genome-Wide Identification of Calcium-Response Factor (CaRF) Binding Sites Predicts a Role in Regulation of Neuronal Signaling Pathways. PLoS ONE. 2010;5:e10870. doi: 10.1371/journal.pone.0010870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintchovski SA, Peebles CL, Kim HJ, Verdin E, Finkbeiner S. The serum response factor and a putative novel transcription factor regulate expression of the immediate-early gene Arc/Arg3.1 in neurons. Journal of Neuroscience. 2009;29:1525–1537. doi: 10.1523/JNEUROSCI.5575-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipes GCT, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- Plant K, Pelkey KA, Bortolotto ZA, Morita D, Terashima A, McBain CJ, Collingridge GL, Isaac JTR. Transient incorporation of native GluR2-lacking AMPA receptors during hippocampal long-term potentiation. Nat Neurosci. 2006;9:602–604. doi: 10.1038/nn1678. [DOI] [PubMed] [Google Scholar]
- Poo MM. Neurotrophins as synaptic modulators. Nat Rev Neurosci. 2001;2:24–32. doi: 10.1038/35049004. [DOI] [PubMed] [Google Scholar]
- Pruunsild P, Sepp M, Orav E, Koppel I, Timmusk T. Identification of cis-Elements and Transcription Factors Regulating Neuronal Activity-Dependent Transcription of Human BDNF Gene. J Neurosci. 2011;31:3295–3308. doi: 10.1523/JNEUROSCI.4540-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pu L, Liu Q-s, Poo M-m. BDNF-dependent synaptic sensitization in midbrain dopamine neurons after cocaine withdrawal. Nat Neurosci. 2006;9:605–607. doi: 10.1038/nn1687. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Nairn AC, Nestler EJ, Cowan CW. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–457. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- Qiu Z, Ghosh A. A calcium-dependent switch in a CREST-BRG1 complex regulates activity-dependent gene expression. Neuron. 2008;60:775–787. doi: 10.1016/j.neuron.2008.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Querido E, Chartrand P. Using fluorescent proteins to study mRNA trafficking in living cells. Methods Cell Biol. 2008;85:273–292. doi: 10.1016/S0091-679X(08)85012-1. [DOI] [PubMed] [Google Scholar]
- Quinlan EM, Olstein DH, Bear MF. Bidirectional, experience-dependent regulation of N-methyl-D-aspartate receptor subunit composition in the rat visual cortex during postnatal development. Proc Natl Acad Sci USA. 1999;96:12876–12880. doi: 10.1073/pnas.96.22.12876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich GA, Toscano MA, Ilarregui JM, Rubinstein N. Shedding light on the immunomodulatory properties of galectins: novel regulators of innate and adaptive immune responses. Glycoconj J. 2004;19:565–573. doi: 10.1023/B:GLYC.0000014087.41914.72. [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Ameur A, Kapranov P, Enroth S, Komorowski J, Gingeras TR, Wadelius C. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 2008;18:380–392. doi: 10.1101/gr.6880908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanan N, Shen Y, Sarsfield S, Lemberger T, Schütz G, Linden DJ, Ginty DD. SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat Neurosci. 2005;8:759–767. doi: 10.1038/nn1462. [DOI] [PubMed] [Google Scholar]
- Rao VR, Pintchovski SA, Chin J, Peebles CL, Mitra S, Finkbeiner S. AMPA receptors regulate transcription of the plasticity-related immediate-early gene Arc. Nat Neurosci. 2006;9:887–895. doi: 10.1038/nn1708. [DOI] [PubMed] [Google Scholar]
- Raser JM, O’Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redmond L, Ghosh A. Regulation of dendritic development by calcium signaling. Cell Calcium. 2005;37:411–416. doi: 10.1016/j.ceca.2005.01.009. [DOI] [PubMed] [Google Scholar]
- Redmond L, Kashani AH, Ghosh A. Calcium regulation of dendritic growth via CaM kinase IV and CREB-mediated transcription. Neuron. 2002;34:999–1010. doi: 10.1016/s0896-6273(02)00737-7. [DOI] [PubMed] [Google Scholar]
- Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- Reik W, Dean W, Walter J. Epigenetic reprogramming in mammalian development. Science. 2001;293:1089–1093. doi: 10.1126/science.1063443. [DOI] [PubMed] [Google Scholar]
- Reik W, Walter J. Imprinting mechanisms in mammals. Curr Opin Genet Dev. 1998;8:154–164. doi: 10.1016/s0959-437x(98)80136-6. [DOI] [PubMed] [Google Scholar]
- Renthal W, Kumar A, Xiao G, Wilkinson M, Covington HE, Maze I, Sikder D, Robison AJ, LaPlant Q, Dietz DM, Russo SJ, Vialou V, Chakravarty S, Kodadek TJ, Stack A, Kabbaj M, Nestler EJ. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexach JE, Rogers CJ, Yu SH, Tao J, Sun YE, Hsieh-Wilson LC. Quantification of O-glycosylation stoichiometry and dynamics using resolvable mass tags. Nat Chem Biol. 2010;6:645–651. doi: 10.1038/nchembio.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccio A. Dynamic epigenetic regulation in neurons: enzymes, stimuli and signaling pathways. Nat Neurosci. 2010;13:1330–1337. doi: 10.1038/nn.2671. [DOI] [PubMed] [Google Scholar]
- Riccio A, Alvania RS, Lonze BE, Ramanan N, Kim T, Huang Y, Dawson TM, Snyder SH, Ginty DD. A nitric oxide signaling pathway controls CREB-mediated gene expression in neurons. Mol Cell. 2006;21:283–294. doi: 10.1016/j.molcel.2005.12.006. [DOI] [PubMed] [Google Scholar]
- Rivera VM, Miranti CK, Misra RP, Ginty DD, Chen RH, Blenis J, Greenberg ME. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol Cell Biol. 1993;13:6260–6273. doi: 10.1128/mcb.13.10.6260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts EB, Ramoa AS. Enhanced NR2A subunit expression and decreased NMDA receptor decay time at the onset of ocular dominance plasticity in the ferret. J Neurophysiol. 1999;81:2587–2591. doi: 10.1152/jn.1999.81.5.2587. [DOI] [PubMed] [Google Scholar]
- Roh TY, Cuddapah S, Zhao K. Active chromatin domains are defined by acetylation islands revealed by genome-wide mapping. Genes Dev. 2005;19:542–552. doi: 10.1101/gad.1272505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen LB, Ginty DD, Weber MJ, Greenberg ME. Membrane depolarization and calcium influx stimulate MEK and MAP kinase via activation of Ras. Neuron. 1994;12:1207–1221. doi: 10.1016/0896-6273(94)90438-3. [DOI] [PubMed] [Google Scholar]
- Rosenmund C, Stern-Bach Y, Stevens CF. The tetrameric structure of a glutamate receptor channel. Science. 1998;280:1596–1599. doi: 10.1126/science.280.5369.1596. [DOI] [PubMed] [Google Scholar]
- Roth TL, Sweatt JD. Regulation of chromatin structure in memory formation. Curr Opin Neurobiol. 2009;19:336–342. doi: 10.1016/j.conb.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadri-Vakili G, Kumaresan V, Schmidt HD, Famous KR, Chawla P, Vassoler FM, Overland RP, Xia E, Bass CE, Terwilliger EF, Pierce RC, Cha JHJ. Cocaine-induced chromatin remodeling increases brain-derived neurotrophic factor transcription in the rat medial prefrontal cortex, which alters the reinforcing efficacy of cocaine. J Neurosci. 2010;30:11735–11744. doi: 10.1523/JNEUROSCI.2328-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Futai K, Yamamoto K, Worley PF, Hayashi Y, Sheng M. Inhibition of dendritic spine morphogenesis and synaptic transmission by activity-inducible protein Homer1a. Journal of Neuroscience. 2003;23:6327–6337. doi: 10.1523/JNEUROSCI.23-15-06327.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sala C, Rudolph-Correia S, Sheng M. Developmentally regulated NMDA receptor-dependent dephosphorylation of cAMP response element-binding protein (CREB) in hippocampal neurons. J Neurosci. 2000;20:3529–3536. doi: 10.1523/JNEUROSCI.20-10-03529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sans N, Petralia RS, Wang YX, Blahos J, Hell JW, Wenthold RJ. A developmental change in NMDA receptor-associated proteins at hippocampal synapses. J Neurosci. 2000;20:1260–1271. doi: 10.1523/JNEUROSCI.20-03-01260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Takemori H, Yagita Y, Terasaki Y, Uebi T, Horike N, Takagi H, Susumu T, Teraoka H, Kusano K-i, Hatano O, Oyama N, Sugiyama Y, Sakoda S, Kitagawa K. SIK2 Is a Key Regulator for Neuronal Survival after Ischemia via TORC1-CREB. Neuron. 2011;69:106–119. doi: 10.1016/j.neuron.2010.12.004. [DOI] [PubMed] [Google Scholar]
- Sasaki YF, Rothe T, Premkumar LS, Das S, Cui J, Talantova MV, Wong HK, Gong X, Chan SF, Zhang D, Nakanishi N, Sucher NJ, Lipton SA. Characterization and comparison of the NR3A subunit of the NMDA receptor in recombinant systems and primary cortical neurons. J Neurophysiol. 2002;87:2052–2063. doi: 10.1152/jn.00531.2001. [DOI] [PubMed] [Google Scholar]
- Sassone-Corsi P, Mizzen CA, Cheung P, Crosio C, Monaco L, Jacquot S, Hanauer A, Allis CD. Requirement of Rsk-2 for epidermal growth factor-activated phosphorylation of histone H3. Science. 1999;285:886–891. doi: 10.1126/science.285.5429.886. [DOI] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, Talianidis I, Flicek P, Odom DT. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science. 2010;328:1036–1040. doi: 10.1126/science.1186176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Spyrou C, Brown GD, Hadfield J, Odom DT. ChIP-seq: using high-throughput sequencing to discover protein-DNA interactions. Methods. 2009;48:240–248. doi: 10.1016/j.ymeth.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screaton RA, Conkright MD, Katoh Y, Best JL, Canettieri G, Jeffries S, Guzman E, Niessen S, Yates JR, Takemori H, Okamoto M, Montminy M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Gaudillière B, Yuan Z, Stegmüller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- Shalizi A, Lehtinen M, Gaudilliere B, Donovan N, Han J, Konishi Y, Bonni A. Characterization of a neurotrophin signaling mechanism that mediates neuron survival in a temporally specific pattern. J Neurosci. 2003;23:7326–7336. doi: 10.1523/JNEUROSCI.23-19-07326.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw PE, Schröter H, Nordheim A. The ability of a ternary complex to form over the serum response element correlates with serum inducibility of the human c-fos promoter. Cell. 1989;56:563–572. doi: 10.1016/0092-8674(89)90579-5. [DOI] [PubMed] [Google Scholar]
- Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- Shen K, Meyer T. Dynamic control of CaMKII translocation and localization in hippocampal neurons by NMDA receptor stimulation. Science. 1999;284:162–166. doi: 10.1126/science.284.5411.162. [DOI] [PubMed] [Google Scholar]
- Sheng M. Molecular organization of the postsynaptic specialization. Proc Natl Acad Sci USA. 2001;98:7058–7061. doi: 10.1073/pnas.111146298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Cummings J, Roldan LA, Jan YN, Jan LY. Changing subunit composition of heteromeric NMDA receptors during development of rat cortex. Nature. 1994;368:144–147. doi: 10.1038/368144a0. [DOI] [PubMed] [Google Scholar]
- Sheng M, Dougan ST, McFadden G, Greenberg ME. Calcium and growth factor pathways of c-fos transcriptional activation require distinct upstream regulatory sequences. Mol Cell Biol. 1988;8:2787–2796. doi: 10.1128/mcb.8.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- Sheng M, Sala C. PDZ domains and the organization of supramolecular complexes. Annu Rev Neurosci. 2001;24:1–29. doi: 10.1146/annurev.neuro.24.1.1. [DOI] [PubMed] [Google Scholar]
- Sheng M, Thompson MA, Greenberg ME. CREB: a Ca(2+)-regulated transcription factor phosphorylated by calmodulin-dependent kinases. Science. 1991;252:1427–1430. doi: 10.1126/science.1646483. [DOI] [PubMed] [Google Scholar]
- Shi J, Aamodt SM, Constantine-Paton M. Temporal correlations between functional and molecular changes in NMDA receptors and GABA neurotransmission in the superior colliculus. J Neurosci. 1997;17:6264–6276. doi: 10.1523/JNEUROSCI.17-16-06264.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shieh PB, Hu SC, Bobb K, Timmusk T, Ghosh A. Identification of a signaling pathway involved in calcium regulation of BDNF expression. Neuron. 1998;20:727–740. doi: 10.1016/s0896-6273(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Skene PJ, Illingworth RS, Webb S, Kerr ARW, James KD, Turner DJ, Andrews R, Bird AP. Neuronal MeCP2 Is Expressed at Near Histone-Octamer Levels and Globally Alters the Chromatin State. Molecular Cell. 2010;37:457–468. doi: 10.1016/j.molcel.2010.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Hicks C, Xiao B, Deng R, Ji Y, Zhao X, Shepherd JD, Posern G, Kuhl D, Huganir RL, Ginty DD, Worley PF, Linden DJ. SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nat Neurosci. 2010;13:1082–1089. doi: 10.1038/nn.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczyk A, Scheuss V, Svoboda K. NMDA receptor subunit-dependent [Ca2+] signaling in individual hippocampal dendritic spines. J Neurosci. 2005;25:6037–6046. doi: 10.1523/JNEUROSCI.1221-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CX, Szulwach KE, Fu Y, Dai Q, Yi C, Li X, Li Y, Chen CH, Zhang W, Jian X, Wang J, Zhang L, Looney TJ, Zhang B, Godley LA, Hicks LM, Lahn BT, Jin P, He C. Selective chemical labeling reveals the genome-wide distribution of 5-hydroxymethylcytosine. Nat Biotechnol. 2011;29:68–72. doi: 10.1038/nbt.1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, Léveillé F, Papadia S, Bell KFS, Puddifoot C, Hardingham GE. Neuronal activity controls the antagonistic balance between peroxisome proliferator-activated receptor-γ coactivator-1α and silencing mediator of retinoic acid and thyroid hormone receptors in regulating antioxidant defenses. Antioxid Redox Signal. 2011;14:1425–1436. doi: 10.1089/ars.2010.3568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano FX, Papadia S, Hofmann F, Hardingham NR, Bading H, Hardingham GE. Preconditioning doses of NMDA promote neuroprotection by enhancing neuronal excitability. J Neurosci. 2006;26:4509–4518. doi: 10.1523/JNEUROSCI.0455-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotiropoulos A, Gineitis D, Copeland J, Treisman R. Signal-regulated activation of serum response factor is mediated by changes in actin dynamics. Cell. 1999;98:159–169. doi: 10.1016/s0092-8674(00)81011-9. [DOI] [PubMed] [Google Scholar]
- Spitzer NC. Electrical activity in early neuronal development. Nature. 2006;444:707–712. doi: 10.1038/nature05300. [DOI] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Decher N, Kumar P, Sachse FB, Beggs AH, Sanguinetti MC, Keating MT. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci USA. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. discussion 8086–8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Splawski I, Timothy KW, Sharpe LM, Decher N, Kumar P, Bloise R, Napolitano C, Schwartz PJ, Joseph RM, Condouris K, Tager-Flusberg H, Priori SG, Sanguinetti MC, Keating MT. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Stehle JH, Foulkes NS, Molina CA, Simonneaux V, Pévet P, Sassone-Corsi P. Adrenergic signals direct rhythmic expression of transcriptional repressor CREM in the pineal gland. Nature. 1993;365:314–320. doi: 10.1038/365314a0. [DOI] [PubMed] [Google Scholar]
- Stern P, Astrof S, Erkeland SJ, Schustak J, Sharp PA, Hynes RO. A system for Cre-regulated RNA interference in vivo. Proc Natl Acad Sci USA. 2008;105:13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiles CD. The biological role of oncogenes--insights from platelet-derived growth factor: Rhoads Memorial Award lecture. Cancer Res. 1985;45:5215–5218. [PubMed] [Google Scholar]
- Strack S, Colbran RJ. Autophosphorylation-dependent targeting of calcium/calmodulin-dependent protein kinase II by the NR2B subunit of the N-methyl- D-aspartate receptor. J Biol Chem. 1998;273:20689–20692. doi: 10.1074/jbc.273.33.20689. [DOI] [PubMed] [Google Scholar]
- Strack S, McNeill RB, Colbran RJ. Mechanism and regulation of calcium/calmodulin-dependent protein kinase II targeting to the NR2B subunit of the N-methyl-D-aspartate receptor. J Biol Chem. 2000;275:23798–23806. doi: 10.1074/jbc.M001471200. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Sucher NJ, Akbarian S, Chi CL, Leclerc CL, Awobuluyi M, Deitcher DL, Wu MK, Yuan JP, Jones EG, Lipton SA. Developmental and regional expression pattern of a novel NMDA receptor-like subunit (NMDAR-L) in the rodent brain. J Neurosci. 1995;15:6509–6520. doi: 10.1523/JNEUROSCI.15-10-06509.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucher NJ, Yu E, Chan SF, Miri M, Lee BJ, Xiao B, Worley PF, Jensen FE. Association of the Small GTPase Rheb with the NMDA Receptor Subunit NR3A. Neurosignals. 2010 doi: 10.1159/000322206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiyama S, Di Nardo AA, Aizawa S, Matsuo I, Volovitch M, Prochiantz A, Hensch TK. Experience-dependent transfer of Otx2 homeoprotein into the visual cortex activates postnatal plasticity. Cell. 2008;134:508–520. doi: 10.1016/j.cell.2008.05.054. [DOI] [PubMed] [Google Scholar]
- Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takasu MA, Dalva MB, Zigmond RE, Greenberg ME. Modulation of NMDA receptor-dependent calcium influx and gene expression through EphB receptors. Science. 2002;295:491–495. doi: 10.1126/science.1065983. [DOI] [PubMed] [Google Scholar]
- Takeuchi Y, Fukunaga K. Different effects of five dopamine receptor subtypes on nuclear factor-kappaB activity in NG108–15 cells and mouse brain. J Neurochem. 2004;88:41–50. doi: 10.1046/j.1471-4159.2003.02129.x. [DOI] [PubMed] [Google Scholar]
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, Wang X, Bodeau J, Tuch BB, Siddiqui A, Lao K, Surani MA. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Tao J, Hu K, Chang Q, Wu H, Sherman NE, Martinowich K, Klose RJ, Schanen C, Jaenisch R, Wang W, Sun YE. Phosphorylation of MeCP2 at Serine 80 regulates its chromatin association and neurological function. Proc Natl Acad Sci USA. 2009;106:4882–4887. doi: 10.1073/pnas.0811648106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- Tao X, West AE, Chen WG, Corfas G, Greenberg ME. A calcium-responsive transcription factor, CaRF, that regulates neuronal activity-dependent expression of BDNF. Neuron. 2002;33:383–395. doi: 10.1016/s0896-6273(01)00561-x. [DOI] [PubMed] [Google Scholar]
- Thomas CG, Miller AJ, Westbrook GL. Synaptic and extrasynaptic NMDA receptor NR2 subunits in cultured hippocampal neurons. J Neurophysiol. 2006;95:1727–1734. doi: 10.1152/jn.00771.2005. [DOI] [PubMed] [Google Scholar]
- Thompson KR, Otis KO, Chen DY, Zhao Y, O’Dell TJ, Martin KC. Synapse to nucleus signaling during long-term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron. 2004;44:997–1009. doi: 10.1016/j.neuron.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Tian X, Feig LA. Age-dependent participation of Ras-GRF proteins in coupling calcium-permeable AMPA glutamate receptors to Ras/Erk signaling in cortical neurons. J Biol Chem. 2006;281:7578–7582. doi: 10.1074/jbc.M512060200. [DOI] [PubMed] [Google Scholar]
- Timmusk T, Palm K, Metsis M, Reintam T, Paalme V, Saarma M, Persson H. Multiple promoters direct tissue-specific expression of the rat BDNF gene. Neuron. 1993;10:475–489. doi: 10.1016/0896-6273(93)90335-o. [DOI] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. The incorporation of NMDA receptors with a distinct subunit composition at nascent hippocampal synapses in vitro. Journal of Neuroscience. 1999;19:4180–4188. doi: 10.1523/JNEUROSCI.19-10-04180.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovar KR, Westbrook GL. Mobile NMDA receptors at hippocampal synapses. Neuron. 2002;34:255–264. doi: 10.1016/s0896-6273(02)00658-x. [DOI] [PubMed] [Google Scholar]
- Townsend M, Yoshii A, Mishina M, Constantine-Paton M. Developmental loss of miniature N-methyl-D-aspartate receptor currents in NR2A knockout mice. Proc Natl Acad Sci USA. 2003;100:1340–1345. doi: 10.1073/pnas.0335786100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Brunet A, Grenier JM, Datta SR, Fornace AJ, DiStefano PS, Chiang LW, Greenberg ME. DNA repair pathway stimulated by the forkhead transcription factor FOXO3a through the Gadd45 protein. Science. 2002;296:530–534. doi: 10.1126/science.1068712. [DOI] [PubMed] [Google Scholar]
- Treisman R. Identification of a protein-binding site that mediates transcriptional response of the c-fos gene to serum factors. Cell. 1986;46:567–574. doi: 10.1016/0092-8674(86)90882-2. [DOI] [PubMed] [Google Scholar]
- Treisman R. The serum response element. Trends Biochem Sci. 1992;17:423–426. doi: 10.1016/0968-0004(92)90013-y. [DOI] [PubMed] [Google Scholar]
- Tropea D, Giacometti E, Wilson NR, Beard C, McCurry C, Fu DD, Flannery R, Jaenisch R, Sur M. Partial reversal of Rett Syndrome-like symptoms in MeCP2 mutant mice. Proc Natl Acad Sci USA. 2009;106:2029–2034. doi: 10.1073/pnas.0812394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankova N, Renthal W, Kumar A, Nestler EJ. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8:355–367. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- Tsuruta F, Green EM, Rousset M, Dolmetsch RE. PIKfyve regulates CaV1.2 degradation and prevents excitotoxic cell death. J Cell Biol. 2009;187:279–294. doi: 10.1083/jcb.200903028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu W, Xu X, Peng L, Zhong X, Zhang W, Soundarapandian MM, Balel C, Wang M, Jia N, Zhang W, Lew F, Chan SL, Chen Y, Lu Y. DAPK1 interaction with NMDA receptor NR2B subunits mediates brain damage in stroke. Cell. 2010;140:222–234. doi: 10.1016/j.cell.2009.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuan D, Solomon W, Li Q, London IM. The “beta-like-globin” gene domain in human erythroid cells. Proc Natl Acad Sci USA. 1985;82:6384–6388. doi: 10.1073/pnas.82.19.6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol. 2007;17:318–324. doi: 10.1016/j.conb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- Udvadia AJ, Rogers KT, Higgins PD, Murata Y, Martin KH, Humphrey PA, Horowitz JM. Sp-1 binds promoter elements regulated by the RB protein and Sp-1-mediated transcription is stimulated by RB coexpression. Proc Natl Acad Sci USA. 1993;90:3265–3269. doi: 10.1073/pnas.90.8.3265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udvadia AJ, Rogers KT, Horowitz JM. A common set of nuclear factors bind to promoter elements regulated by the retinoblastoma protein. Cell Growth Differ. 1992;3:597–608. [PubMed] [Google Scholar]
- Valouev A, Johnson DS, Sundquist A, Medina C, Anton E, Batzoglou S, Myers RM, Sidow A. Genome-wide analysis of transcription factor binding sites based on ChIP-Seq data. Nat Methods. 2008;5:829–834. doi: 10.1038/nmeth.1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Petegem F, Clark KA, Chatelain FC, Minor DL. Structure of a complex between a voltage-gated calcium channel beta-subunit and an alpha-subunit domain. Nature. 2004;429:671–675. doi: 10.1038/nature02588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartiainen MK, Guettler S, Larijani B, Treisman R. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science. 2007;316:1749–1752. doi: 10.1126/science.1141084. [DOI] [PubMed] [Google Scholar]
- Vashishta A, Habas A, Pruunsild P, Zheng JJ, Timmusk T, Hetman M. Nuclear factor of activated T-cells isoform c4 (NFATc4/NFAT3) as a mediator of antiapoptotic transcription in NMDA receptor-stimulated cortical neurons. Journal of Neuroscience. 2009;29:15331–15340. doi: 10.1523/JNEUROSCI.4873-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vihma H, Pruunsild P, Timmusk T. Alternative splicing and expression of human and mouse NFAT genes. Genomics. 2008;92:279–291. doi: 10.1016/j.ygeno.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Prabhakar S, Akiyama JA, Shoukry M, Lewis KD, Holt A, Plajzer-Frick I, Afzal V, Rubin EM, Pennacchio LA. Ultraconservation identifies a small subset of extremely constrained developmental enhancers. Nat Genet. 2008;40:158–160. doi: 10.1038/ng.2007.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O, Keller DM, Yamamoto T, Goodman RH, Impey S. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–16431. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahl AS, Buchthal B, Rode F, Bomholt SF, Freitag HE, Hardingham GE, Rønn LCB, Bading H. Hypoxic/ischemic conditions induce expression of the putative pro-death gene Clca1 via activation of extrasynaptic N-methyl-D-aspartate receptors. Neuroscience. 2009;158:344–352. doi: 10.1016/j.neuroscience.2008.06.018. [DOI] [PubMed] [Google Scholar]
- Waltereit R, Dammermann B, Wulff P, Scafidi J, Staubli U, Kauselmann G, Bundman M, Kuhl D. Arg3.1/Arc mRNA induction by Ca2+ and cAMP requires protein kinase A and mitogen-activated protein kinase/extracellular regulated kinase activation. Journal of Neuroscience. 2001;21:5484–5493. doi: 10.1523/JNEUROSCI.21-15-05484.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KH, Majewska A, Schummers J, Farley B, Hu C, Sur M, Tonegawa S. In vivo two-photon imaging reveals a role of arc in enhancing orientation specificity in visual cortex. Cell. 2006;126:389–402. doi: 10.1016/j.cell.2006.06.038. [DOI] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, Zhao K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD. The human genome project: past, present, and future. Science. 1990;248:44–49. doi: 10.1126/science.2181665. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Davare M, Ando H, Fortin D, Varlamova O, Cheng HYM, Marks D, Obrietan K, Soderling TR, Goodman RH, Impey S. An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc Natl Acad Sci USA. 2008a;105:9093–9098. doi: 10.1073/pnas.0803072105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayman GA, Impey S, Marks D, Saneyoshi T, Grant WF, Derkach V, Soderling TR. Activity-dependent dendritic arborization mediated by CaM-kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron. 2006;50:897–909. doi: 10.1016/j.neuron.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Wayman GA, Lee YS, Tokumitsu H, Silva AJ, Silva A, Soderling TR. Calmodulin-kinases: modulators of neuronal development and plasticity. Neuron. 2008b;59:914–931. doi: 10.1016/j.neuron.2008.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Weick JP, Groth RD, Isaksen AL, Mermelstein PG. Interactions with PDZ proteins are required for L-type calcium channels to activate cAMP response element-binding protein-dependent gene expression. J Neurosci. 2003;23:3446–3456. doi: 10.1523/JNEUROSCI.23-08-03446.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weih F, Stewart AF, Boshart M, Nitsch D, Schütz G. In vivo monitoring of a cAMP-stimulated DNA-binding activity. Genes Dev. 1990;4:1437–1449. doi: 10.1101/gad.4.8.1437. [DOI] [PubMed] [Google Scholar]
- Wellmann H, Kaltschmidt B, Kaltschmidt C. Retrograde transport of transcription factor NF-kappa B in living neurons. J Biol Chem. 2001;276:11821–11829. doi: 10.1074/jbc.M009253200. [DOI] [PubMed] [Google Scholar]
- West AE, Chen WG, Dalva MB, Dolmetsch RE, Kornhauser JM, Shaywitz AJ, Takasu MA, Tao X, Greenberg ME. Calcium regulation of neuronal gene expression. Proc Natl Acad Sci USA. 2001;98:11024–11031. doi: 10.1073/pnas.191352298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AE, Griffith EC, Greenberg ME. Regulation of transcription factors by neuronal activity. Nat Rev Neurosci. 2002;3:921–931. doi: 10.1038/nrn987. [DOI] [PubMed] [Google Scholar]
- Westphal RS, Tavalin SJ, Lin JW, Alto NM, Fraser ID, Langeberg LK, Sheng M, Scott JD. Regulation of NMDA receptors by an associated phosphatase-kinase signaling complex. Science. 1999;285:93–96. doi: 10.1126/science.285.5424.93. [DOI] [PubMed] [Google Scholar]
- Wiegert JS, Bengtson CP, Bading H. Diffusion and not active transport underlies and limits ERK1/2 synapse-to-nucleus signaling in hippocampal neurons. J Biol Chem. 2007;282:29621–29633. doi: 10.1074/jbc.M701448200. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Landry JR. RNA-Seq-quantitative measurement of expression through massively parallel RNA-sequencing. Methods. 2009;48:249–257. doi: 10.1016/j.ymeth.2009.03.016. [DOI] [PubMed] [Google Scholar]
- Wölfl S, Martinez C, Majzoub JA. Inducible binding of cyclic adenosine 3′,5′-monophosphate (cAMP)-responsive element binding protein (CREB) to a cAMP-responsive promoter in vivo. Mol Endocrinol. 1999;13:659–669. doi: 10.1210/mend.13.5.0282. [DOI] [PubMed] [Google Scholar]
- Wong HK, Liu XB, Matos MF, Chan SF, Pérez-Otaño I, Boysen M, Cui J, Nakanishi N, Trimmer JS, Jones EG, Lipton SA, Sucher NJ. Temporal and regional expression of NMDA receptor subunit NR3A in the mammalian brain. J Comp Neurol. 2002;450:303–317. doi: 10.1002/cne.10314. [DOI] [PubMed] [Google Scholar]
- Wood L, Gray NW, Zhou Z, Greenberg ME, Shepherd GMG. Synaptic circuit abnormalities of motor-frontal layer 2/3 pyramidal neurons in an RNA interference model of methyl-CpG-binding protein 2 deficiency. J Neurosci. 2009;29:12440–12448. doi: 10.1523/JNEUROSCI.3321-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GY, Deisseroth K, Tsien RW. Activity-dependent CREB phosphorylation: convergence of a fast, sensitive calmodulin kinase pathway and a slow, less sensitive mitogen-activated protein kinase pathway. Proc Natl Acad Sci USA. 2001a;98:2808–2813. doi: 10.1073/pnas.051634198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Rothermel B, Kanatous S, Rosenberg P, Naya FJ, Shelton JM, Hutcheson KA, DiMaio JM, Olson EN, Bassel-Duby R, Williams RS. Activation of MEF2 by muscle activity is mediated through a calcineurin-dependent pathway. EMBO J. 2001b;20:6414–6423. doi: 10.1093/emboj/20.22.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nat Rev Mol Cell Biol. 2010;11:607–620. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi H, Shulha HP, Lin JM, Vales TR, Fu Y, Bodine DM, McKay RDG, Chenoweth JG, Tesar PJ, Furey TS, Ren B, Weng Z, Crawford GE. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang G, Pan L, Xing W, Zhang L, Huang L, Yu J, Zhang R, Wu J, Cheng J, Zhou Y. Identification of activity-dependent gene expression profiles reveals specific subsets of genes induced by different routes of Ca(2+) entry in cultured rat cortical neurons. J Cell Physiol. 2007;212:126–136. doi: 10.1002/jcp.21008. [DOI] [PubMed] [Google Scholar]
- Yamamoto KK, Gonzalez GA, Biggs WH, Montminy MR. Phosphorylation-induced binding and transcriptional efficacy of nuclear factor CREB. Nature. 1988;334:494–498. doi: 10.1038/334494a0. [DOI] [PubMed] [Google Scholar]
- Yang SH, Bumpass DC, Perkins ND, Sharrocks AD. The ETS domain transcription factor Elk-1 contains a novel class of repression domain. Mol Cell Biol. 2002;22:5036–5046. doi: 10.1128/MCB.22.14.5036-5046.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Sharrocks AD. PIASxalpha differentially regulates the amplitudes of transcriptional responses following activation of the ERK and p38 MAPK pathways. Molecular Cell. 2006;22:477–487. doi: 10.1016/j.molcel.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Yang SH, Galanis A, Sharrocks AD. Targeting of p38 mitogen-activated protein kinases to MEF2 transcription factors. Mol Cell Biol. 1999;19:4028–4038. doi: 10.1128/mcb.19.6.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Vickers E, Brehm A, Kouzarides T, Sharrocks AD. Temporal recruitment of the mSin3A-histone deacetylase corepressor complex to the ETS domain transcription factor Elk-1. Mol Cell Biol. 2001;21:2802–2814. doi: 10.1128/MCB.21.8.2802-2814.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yashiro K, Philpot BD. Regulation of NMDA receptor subunit expression and its implications for LTD, LTP, and metaplasticity. Neuropharmacology. 2008;55:1081–1094. doi: 10.1016/j.neuropharm.2008.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassin L, Benedetti BL, Jouhanneau JS, Wen JA, Poulet JFA, Barth AL. An embedded subnetwork of highly active neurons in the neocortex. Neuron. 2010;68:1043–1050. doi: 10.1016/j.neuron.2010.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda R. Imaging spatiotemporal dynamics of neuronal signaling using fluorescence resonance energy transfer and fluorescence lifetime imaging microscopy. Curr Opin Neurobiol. 2006;16:551–561. doi: 10.1016/j.conb.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Yasuda R, Harvey CD, Zhong H, Sobczyk A, van Aelst L, Svoboda K. Supersensitive Ras activation in dendrites and spines revealed by two-photon fluorescence lifetime imaging. Nat Neurosci. 2006;9:283–291. doi: 10.1038/nn1635. [DOI] [PubMed] [Google Scholar]
- Yue DT, Herzig S, Marban E. Beta-adrenergic stimulation of calcium channels occurs by potentiation of high-activity gating modes. Proc Natl Acad Sci USA. 1990;87:753–757. doi: 10.1073/pnas.87.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeringue HC, Constantine-Paton M. Post-transcriptional gene silencing in neurons. Curr Opin Neurobiol. 2004;14:654–659. doi: 10.1016/j.conb.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Zhang H, Fu Y, Altier C, Platzer J, Surmeier DJ, Bezprozvanny I. Ca1.2 and CaV1.3 neuronal L-type calcium channels: differential targeting and signaling to pCREB. Eur J Neurosci. 2006;23:2297–2310. doi: 10.1111/j.1460-9568.2006.04734.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Steijaert MN, Lau D, Schütz G, Delucinge-Vivier C, Descombes P, Bading H. Decoding NMDA receptor signaling: identification of genomic programs specifying neuronal survival and death. Neuron. 2007;53:549–562. doi: 10.1016/j.neuron.2007.01.025. [DOI] [PubMed] [Google Scholar]
- Zhang SJ, Zou M, Lu L, Lau D, Ditzel DAW, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Su P, Liang P, Liu T, Liu X, Liu XY, Zhang B, Han T, Zhu YB, Yin DM, Li J, Zhou Z, Wang KW, Wang Y. The DREAM protein negatively regulates the NMDA receptor through interaction with the NR1 subunit. J Neurosci. 2010a;30:7575–7586. doi: 10.1523/JNEUROSCI.1312-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZW, Zak JD, Liu H. MeCP2 is required for normal development of GABAergic circuits in the thalamus. J Neurophysiol. 2010b;103:2470–2481. doi: 10.1152/jn.00601.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, New L, Kravchenko VV, Kato Y, Gram H, di Padova F, Olson EN, Ulevitch RJ, Han J. Regulation of the MEF2 family of transcription factors by p38. Mol Cell Biol. 1999;19:21–30. doi: 10.1128/mcb.19.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Sternsdorf T, Bolger TA, Evans RM, Yao TP. Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol Cell Biol. 2005;25:8456–8464. doi: 10.1128/MCB.25.19.8456-8464.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HYH, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JAJ, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52:255–269. doi: 10.1016/j.neuron.2006.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Ramachandran B, Gulick T. Alternative pre-mRNA splicing governs expression of a conserved acidic transactivation domain in myocyte enhancer factor 2 factors of striated muscle and brain. J Biol Chem. 2005;280:28749–28760. doi: 10.1074/jbc.M502491200. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302:826–830. doi: 10.1126/science.1089071. [DOI] [PubMed] [Google Scholar]
- Zukin RS, Bennett MV. Alternatively spliced isoforms of the NMDARI receptor subunit. Trends Neurosci. 1995;18:306–313. doi: 10.1016/0166-2236(95)93920-s. [DOI] [PubMed] [Google Scholar]