Abstract
Alterations in histone lysine methylation and other epigenetic regulators of gene expression contribute to changes in brain transcriptomes in mood and psychosis spectrum disorders, including depression and schizophrenia. Genetic association studies and animal models implicate multiple lysine methyltransferases (KMTs) and demethylases (KDMs) in the neurobiology of emotion and cognition. Here, we review the role of histone lysine methylation and transcriptional regulation in normal and diseased neurodevelopment and discuss various KMTs and KDMs as potential therapeutic targets in the treatment of neuropsychiatric disease.
Epigenetics and Mental Illness
Schizophrenia and depression are major psychiatric disorders that lack consensus neuropathology and, in a large majority of cases, a straightforward genetic risk architecture. Furthermore, many patients on the mood and psychosis spectrum show an incomplete response to conventional pharmacological treatments which are mainly aimed at monoamine signaling pathways in the brain (Box 1).
Box 1. Schizophrenia and Depression.
Schizophrenia affects 1% of the general population and typically begins during young-adult years, although cognitive disturbances could be evident much earlier. The disease is, in terms of genetics and etiology, highly heterogeneous, and increasingly defined as different and partially independent symptom complexes: (i) psychosis with delusions, hallucinations and disorganized thought; (ii) cognitive dysfunction including deficits in attention, memory and executive function; and (iii) depressed mood and negative symptoms including inability to experience pleasure (anhedonia), social withdrawal and poor thought and speech output [42]. Currently prescribed antipsychotics, which are mainly aimed at dopaminergic and/or serotonergic receptor systems, exert therapeutic effects on psychosis in approximately 75% of patients. However, it is the cognitive impairment which is often the more disabling and persistent feature of schizophrenia [42]. Currently there are no established pharmacological treatments for this symptom complex. However, given that cognitive dysfunction is an important predictor for long-term outcome, this area is considered a high priority in schizophrenia research, as reflected by initiatives combining efforts from government agencies, academia and industry, including MATRICS (the Measurement and Treatment Research to Improve Cognition in Schizophrenia) [42].
Affective disorders as a group show, in terms of genetic risk architecture, some overlap with schizophrenia. For example, rare structural variants, including the balanced translocation at the Disrupted-in-Schizophrenia 1 (DISC-1) locus (1q42) or the 22q11 deletion are, in different individuals, associated with either mood disorder or schizophrenia [81, 82].
Depression, including its more severe manifestation, major depressive disorder which has a lifetime risk of 10–15% for the U.S. general population, is associated with excessive sadness, anhedonia, negative thoughts, and neurovegetative symptoms including changes in sleep pattern and appetite [1]. The disorder, which in more severe cases is accompanied by delusions, hallucinations and other symptoms of psychosis, often takes a chronic and recurrent course. Conventional antidepressant therapies primarily target monoamine metabolism and reuptake mechanisms at the terminals of serotonergic, noradrenergic and dopaminergic neurons. Unfortunately, up to 40% of cases show an insufficient response to these pharmacological treatments [1]. In addition, many antipsychotic and antidepressant drugs have significant side effect burden, including weight gain, diabetes and metabolic defects, extrapyramidal symptoms and sexual dysfunction [83, 84].
However, there is evidence that dysregulated gene transcription, indicative of compromised neural circuitry, contributes to disordered brain function in psychosis and mood spectrum disorder [1, 2]. While no single gene transcript is consistently affected, alterations in RNA levels contribute to defects in GABAergic inhibitory neurotransmission and more generally, synapse organization and function, metabolism and mitochondrial functions, and oligodendrocyte pathology [3–5]. While a number of transcriptional and post-transcriptional mechanisms may contribute to these changes, chromatin-associated proteins and epigenetic regulators invoked in sustained alterations of gene expression and function (Box 2) could play a critical role in the pathophysiology, or treatment of mental illness [6, 7]. Indeed, there is evidence that changes in acetylation of histone lysine residues, which are broadly associated with active gene expression [8] and considered a potential therapeutic target for cancer and other medical conditions [9], also impact gene expression patterns in the brain and thereby influence emotional and cognitive functions. For example, mice or rats exposed to systemic treatment, or localized intracranial injections of class I/II histone deacetylase inhibitors (HDACi) exhibit behavioral changes reminiscent of those elicited by conventional antidepressant drugs [10–13]. The short chain fatty acid derivative valproic acid, widely prescribed for its mood-stabilizing and anticonvulsant effects, induces brain histone hyperacetylation at a select set of gene promoters when administered to animals at comparatively high doses [14]. Conversely, overexpression of selected HDACs in neuronal structures implicated in the neurobiology of depression, including the hippocampus, elicit a pro-depressant behavioral phenotype [12]. Similarly, animals treated with class I/II HDACi often show improved performance in learning and memory paradigms and furthermore, drug-induced inhibition or activation of class III HDAC (also known as sirtuins) elicits changes in motivational and reward-related behaviors [15]. Therefore, the orderly balance between histone acetyl-transferase and deacetylase activities is critical for cognitive performance and synaptic and behavioral plasticity [16]. Likewise, However, HDACs interfere with acetylation of many non-histone proteins in the nucleus and cytoplasm [16], and moreover, some of these drugs carry a significant side effect burden [9]. Therefore, in light of the emerging role of epigenetic mechanisms in the neurobiology of these and other psychiatric conditions [6], the therapeutic potential of chromatin modifying drugs, other than the HDACi, warrants further investigations. This review will focus on histone lysine methylation, one of the most highly regulated chromatin markings in brain and other tissues. Multiple methyltransferases (KMTs) and demethylases (KDMs) were recently implicated in emotional and cognitive disorders (Fig. 1), and these types of chromatin modifying enzymes could emerge as novel targets in the treatment of mood and psychosis spectrum disorders.
Box 2. Epigenetic regulators and chromatin structure and function.
Epigenetics, in the broader sense, applies both to dividing and postmitotic cells, and refers to a type of cellular memory that involves sustained changes in chromatin structure and function, including gene expression, in the absence of DNA sequence alterations (For in depth discussion, see [85]). Chromatin is essentially a repeating chain of nucleosomes comprised of genomic DNA wrapped around an octamer of core histones H2A/H2B/H3/H4. The histone proteins are intensely decorated with epigenetic information, with more than 70 (amino acid) residue-specific sites subject to various types of post-translational modifications (PTM). These include lysine (K) acetylation, methylation and poly ADP-ribosylation, arginine (R) methylation, and serine (S), threonine (T), tyrosine (Y) and histidine (H) phosphorylation [86]. In addition, a subset of the histone H2A, H2B and H4 lysines are covalently linked to the small protein modifiers ubiquitin and SUMO [87, 88]. Finally, epigenetic markings in genomic DNA include 5-methyl-cytosine and the related form, 5-hydroxy-methyl-cytosine [85]. These DNA and nucleosomal histone markings define the functional architecture of chromatin (see main text).
Proteins associated with methylation and other histone PTM are typically defined either as ‘writers’, ‘erasers’ or ‘readers’, essentially differentiating between the process of establishing or removing a mark as opposed to providing a docking site for chromatin remodeling complexes that regulate transcription, or induce and maintain chromatin condensation [18, 86, 89]. As it pertains to the brain, especially in the context of neuropsychiatric disease, a substantial body of knowledge has been generated for a select set of site-specific (K) methyltransferases and demethylases (Fig. 1A). In contrast, many PTMs are recognized by large numbers of reader proteins [90], but to date only very few of these readers have been explored in the brain. To mention just two examples, there are approximately 75 reader proteins specifically associated with histone H3-trimethyl-lysine 4 (H3K4me3), including several components of the SAGA complex ascribed with a key role for transcriptional initiation at RNA polymerase II target genes [90]. In contrast, H3K9me3, generally considered a repressive mark, provides a central hub for heterochromatin (associated) proteins including several members of the HP1 family and zinc finger domain containing molecules [90]. There is additional complexity because pluripotent stem cells and additional cell types decorate many of their promoters with ‘bivalent domains’ which include both open chromatin-associated (methylated H3K4 and H3/H4 acetylation) and repressive (methylated H3K27) marks [91, 92].
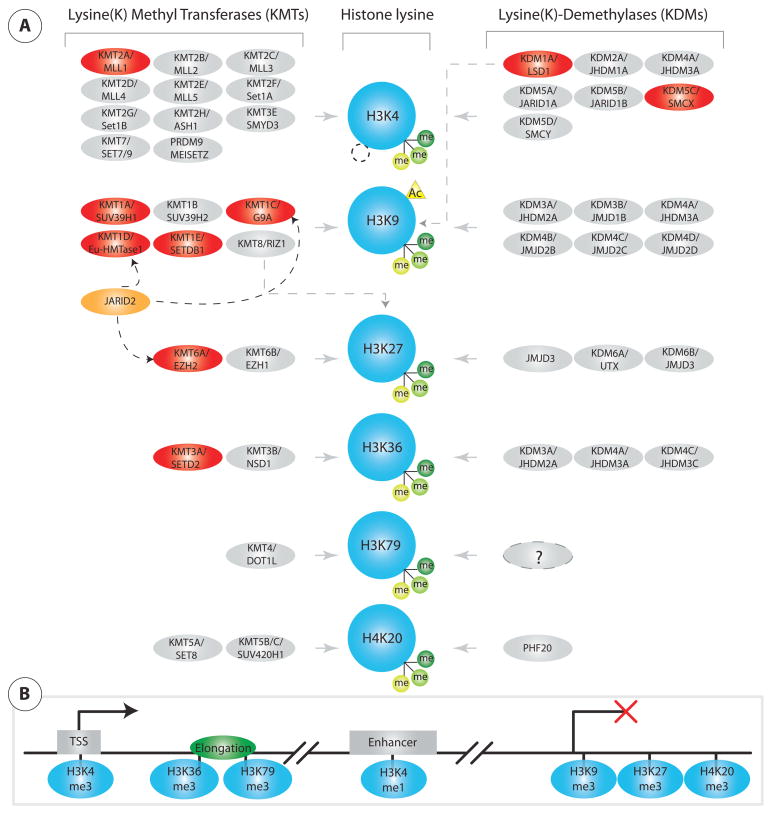
Figure 1.
Regulation of histone (K) methylation. (A) Listings of residue-specific KMTs and KDMs for H3K4/9/27/36/79 and H4K20. The majority of KMT and KDM are highly specific for a single histone residue, while a few enzymes target multiple residues, as indicated. Red marked KMT/KDM are implicated in neurodevelopment or psychiatric disease as discussed in main text. The non-catalytic JARID2 regulates activity and function of related KMTs. (B) Simplified scheme for selected mono- and trimethylated histone lysine markings implicated in transcriptional regulation, silencing and enhancer functions.
Histone methylation – an overview
The methylation of lysine and arginine residues, like other histone PTM, define chromatin states and function [8, 17]. To date, more than 20 methyl-marks on K and R residues have been described [18, 19]. As it pertains to the lysines, the majority of studies focused on the regulation and methylation-related functions of six specific sites: H3K4, H3K9, H3K27, H3K36, H3K79 and H4K20 [18]. For H3K4 and H3K9/K27, there is additional complexity because specific information is also conveyed (i) for H3K4, the unmethylated lysine effectively serving as a DNA methylation signal [20, 21], and (ii) for H3K9/K27 acetylation as an alternative PTM [22, 23] (Fig. 1A). For the aforementioned H3/H4 residues, specific biological functions and their interrelations with functional chromatin states, including transcriptional initiation and elongation, heterochromatic silencing and other mechanisms, have been described for the trimethyl-, and for some of the mono- and dimethyl-modifications (Representative examples are provided in Fig. 1B. See ref [17] for a detailed description of the histone methylation code and its relation to other types of histone PTM).
The following examples further illustrate the complex regulation of histone lysine methylation. Monomethylation of histone H3-lysine 4 (H3K4me1) plays an important role in neuronal activity-induced transcription at enhancer sequences [24], but the related forms, H3K4me3/2 are primarily found at the 5′ end of genes, with H3K4me3 mostly arranged as distinct and sharp peaks within 1–2Kb of transcription start sites. The H3K4me3 mark provides a docking site at the 5′ end of genes for chromatin remodeling complexes that either facilitate or repress transcription [25]. Furthermore, mono-methyl-H4-K20 shows strong positive correlation with gene expression at promoters enriched with CpGs, which contrasts to the trimethylated form of the same residue which generally is associated with repressed chromatin [23]. Taken together, these examples illustrate that even closely related histone lysine methylation markings are potentially associated with very different chromatin states.
To date, H3K4, H4K9, H3K27 and H4K20 methylation signals were measured at specific loci and genome-wide in human brain, essentially confirming that each of these epigenetic markings defines the same type of chromatin as in the peripheral tissues or animal brain [26–30]. Interestingly, a subset of psychotherapeutic drugs including the mood-stabilizer valproate, the atypical antipsychotic clozapine and some monoamine oxidase inhibitors and stimulant drugs interfere with brain histone methylation (Table 1).
Table 1.
Drug-induced histone methylation changes
| Drug(s) | Type | Histone K residue(s) | Genes epigenetically altered in brain | Comments |
|---|---|---|---|---|
| clozapine | Atypical antipsychotic with broad affinity for dopamine D2, serotonin, muscarinic and other receptors | Trimethyl-H3K4 | GAD1 GABA synthesis enzyme [28] | increased GAD1-H3K4 methylation after clozapine exposure both in mouse and human prefrontal cortex [28] |
| valproate, butyrate | Inhibitors of class I/II histone deacetylase (HDACs); valproate is a mood-stabilizer and anticonvulsant | Trimethyl-H3K4 | Gad1 and other GABAergic gene promoters [28] | Fear conditioning and butyrate treatment in mice induces global increase in hippocampal H3K4 methylation [36] |
| phenelzine, tranylcypromine | Antidepressants inhibiting monoamine oxidases MAO-A/B | Di-, Mono-methylH3K4/9 | unknown | Direct inhibitors of LSD1 histone demethylase (amine oxidase superfamily)[79] |
| cocaine | Stimulant drug-of-abuse and monoamine reuptake inhibitor | DimethylH3K9 Trimethyl-H3K4, Trimethyl-H3K27 | Cdk5, NFkB, Bdnf, Arc, FosB and others [48] | Cocaine induces downregulation of G9a/GLP H3K9 methyltransferase in ventral striatum [48] and global H3K4 and H3K27 demethylation in prefrontal cortex [80] |
Molecular mechanisms of histone (lysine) methylation
A complex system of site-specific methyltransferases, which transfer the methyl-group of S-Adenosyl-Methionine (SAM) to lysine residues, has evolved in the vertebrate cell. There are an estimated 70 human genes harboring the Su(var)3–9, Enhancer of Zeste, Trithorax (SET) domain, which spans approximately 130 amino acids essential for KMT enzymatic activity [31]. The only known exception is the H3K79-specific methyltransferase, KMT4/DOT1L [31, 32], which lacks a SET. Each of the histone K residues discussed above is the preferential target of a distinct set of methyltransferase proteins (Fig. 1A)[19]. Of note, these histone-modifying enzymes are thought not to access histone substrates directly unless recruited by DNA-bound activators and repressors, a mechanism which could target each methyltransferase to a highly specific set of genomic loci [19].
An equally complex system exists for the site-specific lysine demethylases (Fig. 1A). There are at least two different mechanisms for active histone demethylation. The first enzyme type, represented by lysine-specific demethylase 1 (LSD1/KDM1A), contains an amine oxidase domain and requires flavin adenine dinucleotide (FAD) as a cofactor to demethylate di- and mono-methylated lysines. LSD1 and its homologue, LSD2, are primarily H3K4 demethylases, albeit depending on species and context, and activity against H3K9 also has been described [18]. Interestingly, monoamine oxidase inhibitors (MAOi) such as tranylcypromine or phenelzine — powerful antidepressants that exert their therapeutic effects mainly by elevating brain monoamine levels through inhibition of MAO-A/B — also block LSD1 type histone demethylases [18]. While LSD1 is thought to regulate histone methylation at promoters, LSD2 is bound to transcriptional elongation complexes and removes H3K4 methyl markings in gene bodies, thereby facilitating gene expression by reducing spurious transcriptional initiation outside of promoters [33]. The second type of demethylase, which in contrast to LSD1/LSD2 is capable of demethylating trimethyl markings, involves Fe2+-dependent dioxygenation by Jumonji-C (JmJC) domain-mediated catalysis [18]. Given that each of the KMTs and KDMs described has a different combinatorial set of functional domains and (protein) binding partners [18, 34], it is likely that the various site-specific methyltransferases and demethylases are largely non-redundant in function.
KMTs and KDMs with a role in cognition and neuropsychiatric disease
An increasing number of KMTs and KDMs are implicated in neurodevelopment and major psychiatric diseases (marked in red in Fig. 1A).
H3K4
The first histone lysine methyltransferase explored in the nervous system was KMT2A/MLL1, a member of the mixed-lineage leukemia (MLL) family of molecules. Mice heterozygous for an insertional (lacZ) loss-of-function Mll1 mutation show distinct abnormalities in hippocampal plasticity and signaling [35], in conjunction with defects of learning and memory [36]. Of note, the hippocampus, and other portions of the forebrain including prefrontal cortex and ventral striatum, are frequently implicated in the neural circuitry of mood and psychosis spectrum disorders [1]. Furthermore, conditional deletion of Mll1 resulted in defective neurogenesis during the early postnatal period [37]. While the full spectrum of MLL1 target genes in neurons and glia awaits further investigation, dysregulated expression of certain transcription factors such as DLX2, a key regulator for the differentiation of forebrain GABAergic neurons (which are essential for inhibitory neurotransmission and orderly synchronization of neural networks) [38], may contribute to the cognitive phenotype of the Mll1 mutant mice. These observations may be relevant for the pathophysiology of schizophrenia, because some patients show in the prefrontal cortex a deficit in H3K4-trimethylation and gene expression at a subset of GABAergic promoters, including GAD1 encoding a GABA synthesis enzyme [28]. While the timing and age-of-onset for this ‘molecular lesion’ remains unknown, it is of interest that in the normal PFC, H3K4 methylation at the site of GABAergic genes progressively increases during the transition from fetal period to childhood to adulthood [28]. The epigenetic vulnerability of the Gad1 promoter during such prolonged developmental periods is further emphasized by recent animal studies demonstrating that Gad1-DNA methylation and histone acetylation are heavily influenced by the level of maternal care in the neonatal period/pre-weanling period [39].
There is additional evidence that epigenetic fine-tuning of the brain’s H3K4 methyl-markings is critical for orderly neurodevelopment. Of note, loss-of-function mutations in KDM5C/JARID1C/SMCX, an X-linked gene encoding a H3K4 demethylase, have been linked to mental retardation [40] and autism spectrum disorders [41]. The KDM5C gene product operates in a chromatin remodeling complex together with HDAC1/2 histone deacetylases and the transcriptional repressor REST, thereby poising neuron-restrictive silencer elements for H3K4 demethylation and decreased expression of target genes including synaptic proteins and sodium channels [40]. However, because this study was conducted with the HeLa cell line, it remains to be determined whether similar mechanisms operate in the nervous system.
In addition to its role in neurodevelopment, MLL-mediated H3K4 methylation could play a potential role for the treatment of psychosis. The atypical antipsychotic clozapine, which has a somewhat higher therapeutic efficacy when compared to conventional antipsychotics that function primarily as dopamine D2 receptor antagonists [42], upregulates H3K4 tri-methylation at the Gad1/GAD1 GABA synthesis enzyme gene promoter. These effects were not mimicked in dopamine receptors D2/D3 (Drd2/3) compound null mutant mice, suggesting that blockade of dopamine D2-like receptors is not sufficient for clozapine-induced H3K4 methylation [28]. In the human PFC, GAD1-associated H3K4 methylation was increased in subjects exposed to clozapine, as compared to subjects treated with conventional antipsychotics. Conversely, mice heterozygous for the H3K4-specific KMT, mixed-lineage leukemia 1 (MLL1), exhibited decreased H3K4 methylation at brain Gad1 [28]. Therefore, it is possible that MLL1, which is highly expressed in GABAergic and other neurons of the adult cerebral cortex [28], will in the future emerge as a novel target for the treatment of psychosis. Questions that remain to be resolved include (i) the molecular pathways linking clozapine — a drug that impacts dopaminergic, serotonergic, muscarinic and other signaling pathways — to MLL1-mediated histone methylation, and (ii) whether or not the clozapine-induced changes in H3K4 methylation are restricted to GABAergic gene promoters or, alternatively, the reflection of more widespread epigenetic changes throughout the genome. Of note, clozapine’s effects on H3K4 methylation require intact brain circuitry and cannot be mimicked in cultured neurons differentiated from forebrain progenitor cells [43]. This finding is in good agreement with the recent observation that some of clozapine’s therapeutic effects require an intact serotonergic system, particularly its presynaptic components [44].
H3K9
The 9q34 subtelomeric deletion syndrome, which includes mental retardation and other developmental defects, is caused by deleterious mutations and haploinsufficiency of euchromatin histone methyltransferase 1 (EHMT1, also known as GLP and KMT1D) [45]. This gene encodes a H3K9-specific methyltransferase that operates in a multimeric complex that includes its closest homologue, G9a/KMT1C, and additional H3K9-specific HMTs [46]. Studies in mutant mice suggest that the GLP/G9a complex is important for suppression of non-neuronal and progenitor genes in mature neurons, and loss of this complex has deleterious effects on cognition and other higher brain functions [47]. Furthermore, G9a-mediated H3K9 methylation events within the reward circuitry, including the ventral striatum, are critical intermediates for the long-term effects of cocaine on reward behavior and neuronal morphology [48]. This would suggest that GLP/G9a, and proper regulation of H3K9 levels, is important for orderly brain function both in developing and mature brain.
Furthermore, changes in motivational and affective behaviors could be elicited by overexpression of the H3K9-HMT, SET domain bifurcated 1 (KMT1C/SETDB1/ESET), in adult forebrain neurons [49]. Interestingly, SETDB1 occupancy in neuronal chromatin is highly restricted, and may be confined to less than 0.75% of annotated genes [49]. However, among these are several NMDA and other ionotropic glutamate receptor subunit genes, including Grin2a/b (Nr2a/b)[49]. Mild to moderate inhibition of NMDA receptor-mediated (including Grin2b) neurotransmission elicits a robust improvement of depressive symptoms in some mood disorder patients [50], and, indeed, SETDB1-mediated H3K9 methylation and repressive chromatin remodeling at the Grin2b locus was associated with antidepressant-like behavioral phenotypes in the Setdb1 transgenic mice [49]. Of note, NMDA receptor antagonists, including GRIN2B-specific drugs, elicit significant therapeutic benefits even in subjects who failed multiple trials of selective serotonin reuptake inhibitors (SSRI) and other conventional antidepressants [50]. However, drugs directly acting at the NMDA receptor site have an unfavorable side effect profile, and therapeutic strategies aimed at SETDB1 expression and activity may therefore provide an alternative strategy.
Interestingly, mice with a genetic ablation of Kap1, encoding the SETDB1 binding partner KRAB-associated protein 1, also known as TRIM28/TIF1b/KRIP1)[51], show increased anxiety and deficits in cognition and memory [52], which are phenotypes that are broadly opposite from those observed in mice with increased Setdb1 expression in brain [49]. These findings further speak to the therapeutic potential of the Kap1-Setdb1 repressor complex in the context of neuropsychiatric disease.
Finally, the H3K9-specific demethylase, KDM3A/Jmjd1A, showed increased H3K9-methylation at its own promoter in the ventral striatum of mice exposed to social defeat (a type of stressor associated with a depression-like syndrome in these animals), while mice that were treated with a conventional antidepressant or that were resilient to this type of stress did not show changes in KDM3A promoter methylation [53]. While it is unclear whether KDM3A or some other demethylase acitivity is altered in the depressed animals, the same study [53] reported widespread repressive histone methylation changes, including increased dimethyl-H3K9 and methylated H3K27 at hundreds of gene promoters in stress susceptible animals, which further emphasizes the importance of these PTMs for the epigenetics of mood disorder.
H3K27
The H3K27-selective methyltransferase, KMT6A, also known as Enhancer of zeste homolog2 (EZH2), is associated with the polycomb repressive chromatin remodeling complex 2 (PRC2) [54], and essential for cortical progenitor cell and neuron production. Consequently, loss of EZH2 function is associated with severe thinning of the cerebral cortex and a disproportionate loss of neurons residing in upper cortical layers I–IV [55]. Likewise, the H3K27-specific demethylase, JMJD3, is important for neurogenesis and neuronal lineage commitment [56]. Furthermore, H3K27 methylation is dynamically regulated in mature brain and involved in the neurobiology of major psychiatric disease. For example, changes in expression of brain-derived neurotrophic factor (Bdnf) in hippocampus of mice exposed to environmental enrichment or chronic stress are associated with opposite changes in the H3K27me3 mark at a subset of Bdnf gene promoters [12, 57]. In addition, acute stress leads to an overall decrease in hippocampal H3K27me3 and H3K9me3 [58]. Furthermore, in the orbitofrontal cortex of suicide completers, alterations in H3K27 methylation were described at the TRKB gene, encoding the high affinity receptor for the nerve growth factor molecule, BDNF [27]. Changes in the balance between histone H3K4 and H3K27 methylation, or DNA cytosine and H3K27 methylation may also contribute to GABAergic gene expression deficits in schizophrenia [28, 43]. To date it remains unclear which of the various H3K27-specific KMTs and KDMs (Fig. 1) are involved in these disease-related alterations in postmortem brain tissue. Of note, the Jumonji and Arid containing protein 2 (JARID2), which by itself lacks catalytic activity but is crucial for subsequent H3K27 or H3K9 methylation by recruiting the polycomb PRC2 complex to its target genes [59, 60], is located within the schizophrenia susceptibility locus on chromosome 6p22 and confers genetic risk in multiple populations of different ethnic origin [61, 62]. While the biological functions of JARID2 have been studied primarily in the context of transcriptional regulation in stem cells [63, 64], this gene shows widespread expression in the mature nervous system [65], implying JARID2-mediated control over polycomb repressive chromatin remodeling in the adult brain.
H3K36 and H4K20
Epigenetic dysregulation of nuclear receptor-binding SET domain containing protein 1/KMT3B could play a role in some neuro- and glioblastomas [66], but like for other H3K36 and H4K20 regulating enzymes (Fig. 1), to date little is known about their role in neurodevelopment, cognition and psychiatric disease. Strikingly, however, KMT3A/HYPB/SETD2, a member of the SET2 family of KMTs mediating H3K36 methylation [67], is also known as huntingtin-interacting protein 1 (HIP-1) or huntingtin(yeast)-interacting protein B (HYPB) [68]. Huntington’s is a triplet repeat disorder and chronic neurodegenerative condition with motor symptoms and cognitive defects, and significant changes in mood and affect [69]. Whether or not there is altered H3K36 methylation in the neuronal populations that are at risk for degeneration is unclear. Furthermore, the huntingtin/KMT3A interaction has been documented for yeast [68] but not brain. Of note, wildtype huntingtin is a facilitator of polycomb complex PCR2-mediated H3K27 methylation [70], and furthermore, H3K4 and H3K9 methylation changes have been reported in preclinical model systems and postmortem brains with Huntington’s disease [71, 72]. Therefore, it is possible that transcriptional dysregulation in this condition is associated with aberrant methylation patterns of multiple lysine residues.
KMTs and KDMs as Novel Drug Targets
Given the emerging role of histone methylation in the neurobiology of psychiatric disease, the next obvious question is whether this type of PTM could provide a target for a new generation of psychotropic therapeutics. In principle, KMTs and KDMs should provide fertile ground for the development of novel drugs, because these enzymes are considered more specific than, for example, HDACs, because each HDAC enzyme is likely to affect a much larger number of histone residues as compared to KMTs/KDMs [73]. However, like for other histone modifying enzymes, the specificity of KMTs and KDMs is not limited to histones but includes the (de)methylation of lysines of non-histone proteins, including the p53 tumor suppressor protein and the VEGF growth factor [74]. Druggable domains within the KMTs and KDMs could involve not only their catalytic sites, such as the SET domain for the KMTs or the amino oxidase and JmjC domains for the LSD1 and JMJD subtypes of KDMs, respectively, but also some of the many other functional domains that are specific to subsets of these proteins [75]. One potential candidate would be the bromodomain of the MLLs and other H3K4-specific methyltransferases [75]. Bromodomains, which are present in many different types of nuclear proteins, bind to acetylated histones and small molecules interfering with some of these interactions recently emerged as powerful modulators of systemic inflammation [76].
The catalytic activity of the SET domain containing KMTs requires the universal methyl donor, S-adenosyl-methionine (also known as AdoMET). Crystallographic and functional studies revealed that the SAM binding pocket of KMTs is different from the SAM pockets of other proteins, which may increase the chance to develop compounds which specifically target histone methyltransferases but not other enzymes and proteins [31]. Currently, however, no KMT or KDM related drug is in clinical trials. However, several of these compounds show therapeutic promise in preclinical studies. For example, the S-adenosylhomocysteine hydrolase inhibitor, 3-deazaneplanocin A (DZNep) induces apoptosis in breast cancer cells [77]. This drug alters H3K27 and H4K20 trimethylation via interference with polycomb PRC2 repressive chromatin remodeling [73]. Antioncogenic effects were also observed with BIX-01294, a drug that downregulates H3K9 methylation levels by binding to the SET domain of the G9a/GLP(EHMT1) methyltransferases [73]. The same drug was shown to alter addictive behaviors and H3K9 methylation when infused locally into the brain of cocaine-exposed mice [48]. As discussed above, while tranylcypromine and other monoamine oxidase inhibitors used for the treatment of depression are weak inhibitors of the LSD1 type of KDM, recently several compounds emerged with much stronger activity against LSD1/LSD2 [18]. It will be extremely interesting to explore these drugs in preclinical models for mood and psychosis spectrum disorders. Finally, microRNA-based therapeutic strategies, aimed at decreasing levels and expression of chromatin remodeling complexes, including some of the histone modifying enzymes discussed here, are gaining increasing prominence in the field of cancer therapy [73] and may in the future emerge as a novel therapeutic option in the context of neuropsychiatric disease.
Synopsis and Outlook
Alterations in histone methylation, both on the level of global (‘bulk’) chromatin and more specifically at select genes and loci, were reported in postmortem brain of subjects diagnosed with schizophrenia or depression [27, 28, 43, 78]. Histone methylation serves as a complex epigenetic regulator in the brain and other tissues, with multiple sets of residue-specific methyltransferases and demethylases, further diversified in terms of the composition and structures of their catalytic and other functional domains. Genetic studies in mouse and human implicate multiple lysine methyltransferases and demethylases, including MLL1, G9a/GLP(EHMT1), SETDB1, SMCX/JARID1c and a related gene JARID2, in the regulation of cognition and motivational and affective behaviors and could in the near future become a therapeutic target for a variety of psychiatric conditions. With an estimated 100 lysine and arginine residue-specific histone methyltransferases and demethylases encoded in the genome, these epigenetic enzymes are expected to provide plenty of targets for drug discovery [31].
Acknowledgments
Work conducted in the author’s laboratory is supported by grants from the National Institutes of Health, the International Mental Health Research Organization and Autism Speaks.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Krishnan V, Nestler EJ. Linking molecules to mood: new insight into the biology of depression. Am J Psychiatry. 2010;167(11):1305–20. doi: 10.1176/appi.ajp.2009.10030434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balu DT, Coyle JT. Neuroplasticity signaling pathways linked to the pathophysiology of schizophrenia. Neurosci Biobehav Rev. 2011;35(3):848–70. doi: 10.1016/j.neubiorev.2010.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lewis DA, Gonzalez-Burgos G. Pathophysiologically based treatment interventions in schizophrenia. Nat Med. 2006;12(9):1016–22. doi: 10.1038/nm1478. [DOI] [PubMed] [Google Scholar]
- 4.Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2010 doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim S, Webster MJ. Correlation analysis between genome-wide expression profiles and cytoarchitectural abnormalities in the prefrontal cortex of psychiatric disorders. Mol Psychiatry. 2010;15(3):326–36. doi: 10.1038/mp.2008.99. [DOI] [PubMed] [Google Scholar]
- 6.Tsankova N, et al. Epigenetic regulation in psychiatric disorders. Nat Rev Neurosci. 2007;8(5):355–67. doi: 10.1038/nrn2132. [DOI] [PubMed] [Google Scholar]
- 7.Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry. 2009;65(3):198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447(7143):407–12. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- 9.Marks PA. The clinical development of histone deacetylase inhibitors as targeted anticancer drugs. Expert Opin Investig Drugs. 2010;19(9):1049–66. doi: 10.1517/13543784.2010.510514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covington HE, 3rd, et al. Antidepressant actions of histone deacetylase inhibitors. J Neurosci. 2009;29(37):11451–60. doi: 10.1523/JNEUROSCI.1758-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schroeder FA, et al. Antidepressant-like effects of the histone deacetylase inhibitor, sodium butyrate, in the mouse. Biol Psychiatry. 2007;62(1):55–64. doi: 10.1016/j.biopsych.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 12.Tsankova NM, et al. Sustained hippocampal chromatin regulation in a mouse model of depression and antidepressant action. Nat Neurosci. 2006;9(4):519–25. doi: 10.1038/nn1659. [DOI] [PubMed] [Google Scholar]
- 13.Zhu H, et al. Antidepressant-like effects of sodium butyrate in combination with estrogen in rat forced swimming test: involvement of 5-HT(1A) receptors. Behav Brain Res. 2009;196(2):200–6. doi: 10.1016/j.bbr.2008.08.039. [DOI] [PubMed] [Google Scholar]
- 14.Dong E, et al. Reelin and glutamic acid decarboxylase67 promoter remodeling in an epigenetic methionine-induced mouse model of schizophrenia. Proc Natl Acad Sci U S A. 2005;102(35):12578–83. doi: 10.1073/pnas.0505394102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62(3):335–48. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer A, et al. Targeting the correct HDAC(s) to treat cognitive disorders. Trends Pharmacol Sci. 2010;31(12):605–17. doi: 10.1016/j.tips.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 17.Zhou VW, Goren A, Bernstein BE. Charting histone modifications and the functional organization of mammalian genomes. Nat Rev Genet. 2011;12(1):7–18. doi: 10.1038/nrg2905. [DOI] [PubMed] [Google Scholar]
- 18.Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–79. doi: 10.1146/annurev.biochem.78.070907.103946. [DOI] [PubMed] [Google Scholar]
- 19.Kouzarides T. Histone methylation in transcriptional control. Curr Opin Genet Dev. 2002;12(2):198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 20.Hu JL, et al. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc Natl Acad Sci U S A. 2009;106(52):22187–92. doi: 10.1073/pnas.0905767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ooi SK, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448(7154):714–7. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondo Y, et al. Chromatin immunoprecipitation microarrays for identification of genes silenced by histone H3 lysine 9 methylation. Proc Natl Acad Sci U S A. 2004;101(19):7398–403. doi: 10.1073/pnas.0306641101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karlic R, et al. Histone modification levels are predictive for gene expression. Proc Natl Acad Sci U S A. 2010;107(7):2926–31. doi: 10.1073/pnas.0909344107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465(7295):182–7. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shilatifard A. Molecular implementation and physiological roles for histone H3 lysine 4 (H3K4) methylation. Curr Opin Cell Biol. 2008;20(3):341–348. doi: 10.1016/j.ceb.2008.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stadler F, et al. Histone methylation at gene promoters is associated with developmental regulation and region-specific expression of ionotropic and metabotropic glutamate receptors in human brain. J Neurochem. 2005;94(2):324–36. doi: 10.1111/j.1471-4159.2005.03190.x. [DOI] [PubMed] [Google Scholar]
- 27.Ernst C, Chen ES, Turecki G. Histone methylation and decreased expression of TrkB.T1 in orbital frontal cortex of suicide completers. Mol Psychiatry. 2009;14(9):830–2. doi: 10.1038/mp.2009.35. [DOI] [PubMed] [Google Scholar]
- 28.Huang HS, et al. Prefrontal dysfunction in schizophrenia involves mixed-lineage leukemia 1-regulated histone methylation at GABAergic gene promoters. J Neurosci. 2007;27(42):11254–62. doi: 10.1523/JNEUROSCI.3272-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y, et al. Isolation of neuronal chromatin from brain tissue. BMC Neurosci. 2008;9:42. doi: 10.1186/1471-2202-9-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheung I, et al. Developmental regulation and individual differences of neuronal H3K4me3 epigenomes in the prefrontal cortex. Proc Natl Acad Sci U S A. 2010;107(19):8824–9. doi: 10.1073/pnas.1001702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8(9):724–32. doi: 10.1038/nrd2974. [DOI] [PubMed] [Google Scholar]
- 32.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 33.Fang R, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol Cell. 2010;39(2):222–33. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selvi BR, et al. Small molecule modulators of histone acetylation and methylation: A disease perspective. Biochim Biophys Acta. 2010 doi: 10.1016/j.bbagrm.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Kim SY, et al. Developmental regulation of Eed complex composition governs a switch in global histone modification in brain. J Biol Chem. 2007;282(13):9962–72. doi: 10.1074/jbc.M608722200. [DOI] [PubMed] [Google Scholar]
- 36.Gupta S, et al. Histone methylation regulates memory formation. J Neurosci. 2010;30(10):3589–99. doi: 10.1523/JNEUROSCI.3732-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lim DA, et al. Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells. Nature. 2009;458(7237):529–33. doi: 10.1038/nature07726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anderson S, et al. Differential origins of neocortical projection and local circuit neurons: role of Dlx genes in neocortical interneuronogenesis. Cereb Cortex. 1999;9(6):646–54. doi: 10.1093/cercor/9.6.646. [DOI] [PubMed] [Google Scholar]
- 39.Zhang TY, et al. Maternal care and DNA methylation of a glutamic acid decarboxylase 1 promoter in rat hippocampus. J Neurosci. 2010;30(39):13130–7. doi: 10.1523/JNEUROSCI.1039-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tahiliani M, et al. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447(7144):601–5. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- 41.Adegbola A, et al. A novel mutation in JARID1C/SMCX in a patient with autism spectrum disorder (ASD) Am J Med Genet A. 2008;146A(4):505–11. doi: 10.1002/ajmg.a.32142. [DOI] [PubMed] [Google Scholar]
- 42.Ibrahim HM, Tamminga CA. Schizophrenia: Treatment Targets Beyond Monoamine Systems. Annu Rev Pharmacol Toxicol. 2010 doi: 10.1146/annurev.pharmtox.010909.105851. [DOI] [PubMed] [Google Scholar]
- 43.Huang HS, Akbarian S. GAD1 mRNA expression and DNA methylation in prefrontal cortex of subjects with schizophrenia. PLoS One. 2007;2(8):e809. doi: 10.1371/journal.pone.0000809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yadav PN, et al. The Presynaptic Component of the Serotonergic System is Required for Clozapine’s Efficacy. Neuropsychopharmacology. 2010 doi: 10.1038/npp.2010.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleefstra T, et al. Loss-of-function mutations in euchromatin histone methyl transferase 1 (EHMT1) cause the 9q34 subtelomeric deletion syndrome. Am J Hum Genet. 2006;79(2):370–7. doi: 10.1086/505693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fritsch L, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37(1):46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 47.Schaefer A, et al. Control of cognition and adaptive behavior by the GLP/G9a epigenetic suppressor complex. Neuron. 2009;64(5):678–91. doi: 10.1016/j.neuron.2009.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327(5962):213–6. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang Y, et al. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30(21):7152–67. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanacora G, et al. Targeting the glutamatergic system to develop novel, improved therapeutics for mood disorders. Nat Rev Drug Discov. 2008;7(5):426–37. doi: 10.1038/nrd2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ayyanathan K, et al. Regulated recruitment of HP1 to a euchromatic gene induces mitotically heritable, epigenetic gene silencing: a mammalian cell culture model of gene variegation. Genes Dev. 2003;17(15):1855–69. doi: 10.1101/gad.1102803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jakobsson J, et al. KAP1-mediated epigenetic repression in the forebrain modulates behavioral vulnerability to stress. Neuron. 2008;60(5):818–31. doi: 10.1016/j.neuron.2008.09.036. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson MB, et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J Neurosci. 2009;29(24):7820–32. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herz HM, Shilatifard A. The JARID2-PRC2 duality. Genes Dev. 2010;24(9):857–61. doi: 10.1101/gad.1921610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pereira JD, et al. Ezh2, the histone methyltransferase of PRC2, regulates the balance between self-renewal and differentiation in the cerebral cortex. Proc Natl Acad Sci U S A. 2010;107(36):15957–62. doi: 10.1073/pnas.1002530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Burgold T, et al. The histone H3 lysine 27-specific demethylase Jmjd3 is required for neural commitment. PLoS One. 2008;3(8):e3034. doi: 10.1371/journal.pone.0003034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kuzumaki N, et al. Hippocampal epigenetic modification at the brain-derived neurotrophic factor gene induced by an enriched environment. Hippocampus. 2010 doi: 10.1002/hipo.20775. [DOI] [PubMed] [Google Scholar]
- 58.Hunter RG, et al. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106(49):20912–7. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li G, et al. Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 2010;24(4):368–80. doi: 10.1101/gad.1886410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pasini D, et al. JARID2 regulates binding of the Polycomb repressive complex 2 to target genes in ES cells. Nature. 2010;464(7286):306–10. doi: 10.1038/nature08788. [DOI] [PubMed] [Google Scholar]
- 61.Pedrosa E, et al. Positive association of schizophrenia to JARID2 gene. Am J Med Genet B Neuropsychiatr Genet. 2007;144B(1):45–51. doi: 10.1002/ajmg.b.30386. [DOI] [PubMed] [Google Scholar]
- 62.Liu Y, et al. Whole genome association study in a homogenous population in Shandong peninsula of China reveals JARID2 as a susceptibility gene for schizophrenia. J Biomed Biotechnol. 2009;2009:536918. doi: 10.1155/2009/536918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen X, et al. Jumonji modulates polycomb activity and self-renewal versus differentiation of stem cells. Cell. 2009;139(7):1303–14. doi: 10.1016/j.cell.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Peng JC, et al. Jarid2/Jumonji coordinates control of PRC2 enzymatic activity and target gene occupancy in pluripotent cells. Cell. 2009;139(7):1290–302. doi: 10.1016/j.cell.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445(7124):168–76. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 66.Berdasco M, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci U S A. 2009;106(51):21830–5. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–20. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Faber PW, et al. Huntingtin interacts with a family of WW domain proteins. Hum Mol Genet. 1998;7(9):1463–74. doi: 10.1093/hmg/7.9.1463. [DOI] [PubMed] [Google Scholar]
- 69.Kirkwood SC, et al. Progression of symptoms in the early and middle stages of Huntington disease. Arch Neurol. 2001;58(2):273–8. doi: 10.1001/archneur.58.2.273. [DOI] [PubMed] [Google Scholar]
- 70.Seong IS, et al. Huntingtin facilitates polycomb repressive complex 2. Hum Mol Genet. 2010;19(4):573–83. doi: 10.1093/hmg/ddp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kim MO, et al. Altered histone monoubiquitylation mediated by mutant huntingtin induces transcriptional dysregulation. J Neurosci. 2008;28(15):3947–57. doi: 10.1523/JNEUROSCI.5667-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ryu H, et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A. 2006;103(50):19176–81. doi: 10.1073/pnas.0606373103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kelly TK, De Carvalho DD, Jones PA. Epigenetic modifications as therapeutic targets. Nat Biotechnol. 2010;28(10):1069–1078. doi: 10.1038/nbt.1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Huang J, Berger SL. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev. 2008;18(2):152–8. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 75.Lall S. Primers on chromatin. Nat Struct Mol Biol. 2007;14(11):1110–5. doi: 10.1038/nsmb1107-1110. [DOI] [PubMed] [Google Scholar]
- 76.Nicodeme E, et al. Suppression of inflammation by a synthetic histone mimic. Nature. 2010 doi: 10.1038/nature09589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Tan J, et al. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–63. doi: 10.1101/gad.1524107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Akbarian S, et al. Chromatin alterations associated with down-regulated metabolic gene expression in the prefrontal cortex of subjects with schizophrenia. Arch Gen Psychiatry. 2005;62(8):829–40. doi: 10.1001/archpsyc.62.8.829. [DOI] [PubMed] [Google Scholar]
- 79.Culhane JC, et al. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J Am Chem Soc. 2010;132(9):3164–76. doi: 10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Black YD, et al. Altered attention and prefrontal cortex gene expression in rats after binge-like exposure to cocaine during adolescence. J Neurosci. 2006;26(38):9656–65. doi: 10.1523/JNEUROSCI.2391-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Porteous DJ, et al. The genetics and biology of DISC1--an emerging role in psychosis and cognition. Biol Psychiatry. 2006;60(2):123–31. doi: 10.1016/j.biopsych.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 82.Green T, et al. Psychiatric disorders and intellectual functioning throughout development in velocardiofacial (22q11.2 deletion) syndrome. J Am Acad Child Adolesc Psychiatry. 2009;48(11):1060–8. doi: 10.1097/CHI.0b013e3181b76683. [DOI] [PubMed] [Google Scholar]
- 83.Kane JM, Correll CU. Pharmacologic treatment of schizophrenia. Dialogues Clin Neurosci. 2010;12(3):345–57. doi: 10.31887/DCNS.2010.12.3/jkane. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Papakostas GI. The efficacy, tolerability, and safety of contemporary antidepressants. J Clin Psychiatry. 2010;71(Suppl E1):e03. doi: 10.4088/JCP.9058se1c.03gry. [DOI] [PubMed] [Google Scholar]
- 85.Bonasio R, Tu S, Reinberg D. Molecular signals of epigenetic states. Science. 2010;330(6004):612–6. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Taverna SD, et al. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14(11):1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29(6):653–63. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 88.Shiio Y, Eisenman RN. Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A. 2003;100(23):13225–30. doi: 10.1073/pnas.1735528100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Justin N, et al. Reading, writing and editing methylated lysines on histone tails: new insights from recent structural studies. Curr Opin Struct Biol. 2010 doi: 10.1016/j.sbi.2010.09.012. [DOI] [PubMed] [Google Scholar]
- 90.Vermeulen M, et al. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell. 2010;142(6):967–80. doi: 10.1016/j.cell.2010.08.020. [DOI] [PubMed] [Google Scholar]
- 91.Taverna SD, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A. 2007;104(7):2086–91. doi: 10.1073/pnas.0610993104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ku M, et al. Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 2008;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]