Abstract
Deregulation of RNA polymerase III (Pol III) transcription enhances cellular tRNAs and 5S rRNA production, leading to an increase in translational capacity to promote cell proliferation, transformation and tumor formation. Phosphorylation of histone H3 (H3ph) is induced by tumor promoters (EGF, UV and TPA) and immediate early genes, such as c-myc, c-jun and c-fos. However, it remains to be determined whether H3ph is involved in RNA Pol III transcription. Here, we report that EGF strongly induced H3ph at serine 28 (H3S28ph). EGF significantly increased transcription of RNA Pol III-dependent genes (Pol III genes), tRNALeu, tRNATyr, 5S rRNA, and 7SL RNA. Inhibition of EGFR, but not PI3K, reduced both H3S28ph and tRNALeu and 5S rRNA transcription. EGF enhanced occupancy of H3S28ph in the promoters of tRNALeu and 5S rRNA. Further analysis indicates that EGF augmented cellular levels of protein and mRNA of TFIIIB subunits, Brf1 and TBP. Brf1 is a specific transcription factor for RNA Pol III genes. EGF enhanced occupancy of H3S28ph in the Brf1 and TBP promoters. Inhibition of H3S28ph by mutant H3S28A repressed Brf1, TBP and tRNALeu and 5S rRNA expression and decreased occupancy of H3S28ph in their promoters. Reduction of Brf1 significantly decreased tRNALeu and 5S rRNA transcription and repressed EGF-induced anchorage-independent growth. Blocking H3S28ph signaling by using mutant H3S28A reduced EGF-induced cell transformation. Together, these results indicate that EGF activates EGFR signaling to induce H3S28ph, which, in turn, upregulates tRNALeu and 5S rRNA transcription through Brf1 and TBP and promotes cell transformation. The studies demonstrate that epigenetic modification of H3S28ph plays a critical role in the activity of Pol III genes.
Keywords: phosphorylation, histone H3, EGFR, Pol III transcription, Brf1, cell transformation
Introduction
RNA polymerase (Pol) III transcribes a variety of untranslated RNAs, including tRNAs, 5S rRNAs, 7SL RNA, 7SK RNA and U6 RNA (Ullu E and Tschudi C, 1984; Dieci et al, 2007, Raha D et al., 2010), while tRNA and 5S rRNA control the translational and growth capacity of cells (Goodfellow et al., 2006; White R, 2004). Oncogenic proteins, such as Ras, c-Jun, and c-Myc, stimulate RNA Pol III-dependent gene (Pol III gene) transcription (Zhong et al., 2004; Johnson and Johnson, 2008); tumor suppressors, such as pRb, p53, PTEN and Maf1 repress transcription of this class of genes (White R, 2004; Johnson et al., 2008a; Woiwode et al., 2008). Studies have indicated that RNA Pol III transcription products are elevated in both transformed and tumor cells suggesting that they play a crucial role in tumorigenesis (White R, 2004; Winter et al., 2000). Consistent with this idea, enhanced Pol III transcription is required for oncogenic transformation (Johnson et al., 2008b, Marshall et al., 2008). The ability of these oncogenic and tumor suppressor proteins to deregulate Pol III transcription results from their capacity to regulate the TFIIIB complex. The TFIIIB complex consists of TATA box-binding protein (TBP) and its associated factors, Brf1 and Bdp1. TFIIIB, together with TFIIIC and RNA Pol III, are required to transcribe tRNA genes, whereas TFIIIB, together with TFIIIA, TFIIIC and RNA Pol III, are required to transcribe 5S rRNA genes. Earlier studies demonstrated that EGF activated EGFR, Ras, and MAP kinases to increase TBP expression, resulting in RNA Pol I and III transcription (Zhong et al., 2004). Further analysis indicated that alteration of cellular levels of TBP affected Bdp1 expression, but did not affect Brf1 expression (Zhong and Johnson, 2009). However, very little is known about whether H3 modifications modulate Pol III transcription. Recent studies have indicated that most active chromatin marks present in Pol II genes are also observed in active Pol III genes (Barski et al., 2010; Oler et al., 2010; Moqtaderi et al., 2010). This implies that epigenetic modifications of histones may regulate activity of Pol III genes.
The maintenance of a repressed or activated status of a gene is often necessary for embryonic development, cellular differentiation, and pathological states (Felsenfeld and Groudine, 2003), and may be regulated by a specific histone code (Jenuwein and Allis, 2001). Post-translational modifications of histone H3 (i.e., phosphorylation, acetylation, methylation and ubiquitination) are known to be involved in chromatin remodeling and transcriptional regulation. Phosphorylation of histone H3 (H3ph) is induced by tumor promoters (EGF, UV, TPA) (Cheung et al., 2000; Zhong, 2001a, b; Kim et al., 2008) and immediate early genes, such as c-myc, c-jun and c-fos (Cheung et al., 2000; Wei et al., 1999). The activation of these oncoproteins promotes cancer development. H3ph at serine 28 (H3S28ph) is associated with mitotic chromosome condensation (Goto et al., 1999) and cell transformation (Kim et al., 2008). Studies have revealed that MAP kinases (i.e., ERK, p38, JNK) and the MAPK downstream component, MSK1, mediated H3S28ph (Zhong et al., 2000, 2001a,b). However, it remains to be determined which pathway upstream of MAPK mediates H3S28ph and whether H3S28ph affects the transcription machinery that regulates the activity of Pol III genes. Our previous study demonstrated that EGF activated EGFR and Ras to enhance TBP expression and RNA Pol III reporter transcription (Zhong et al., 2004). This implies that H3S28ph may mediate Pol III gene transcription. To explore the epigenetic role of H3S28ph on Pol III transcription, we treated mouse epidermal JB6 cells with EGF to examine the effects of H3S28ph on Pol III genes. We have established that Brf1, TBP and tRNALeu and 5S rRNA are new targets of H3S28ph. Our results elucidate a novel role of H3S28ph on the regulation of RNA Pol III transcription machinery. H3S28ph regulates Brf1 expression through EGFR, but not the PI3K pathway to indirectly affect the activity of Pol III genes. In addition, H3S28ph also directly upregulates transcription of tRNALeu and 5S rRNA. These regulatory events serve to modulate cell transformation.
Results
EGF-induced H3S28ph modulates transcription of Pol III genes through EGFR pathway
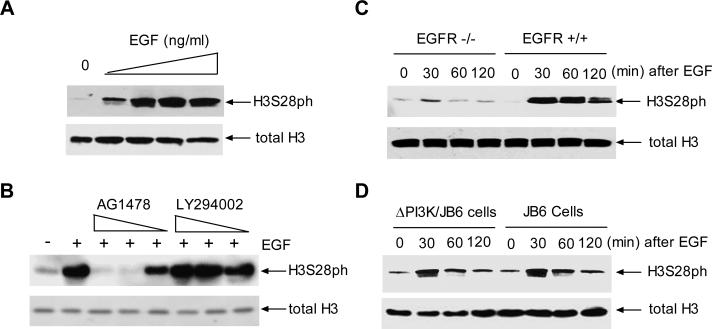
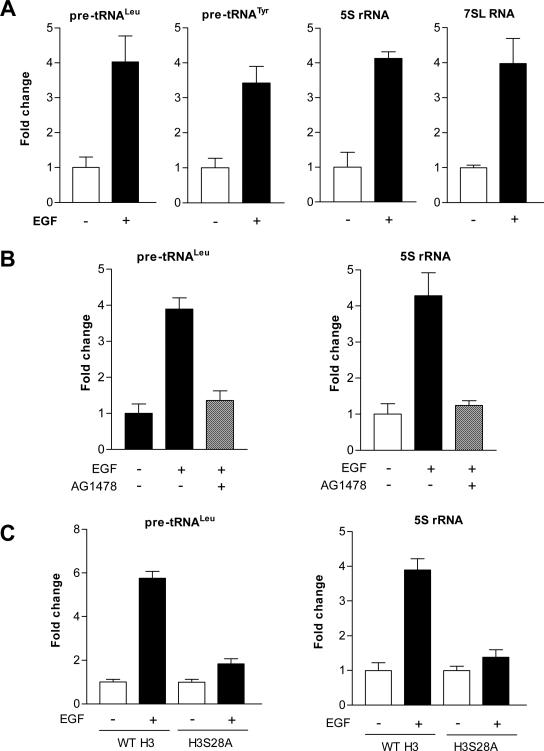
To determine which pathway upstream of MAPK regulates H3S28ph, mouse epidermal JB6 cells were treated with various amounts of EGF for 30 min. As shown in Fig.1A, EGF dramatically induced H3S28ph compared to a control without EGF treatment. Pretreatment of cells with the EGFR inhibitor, AG1478, but not the PI3K inhibitor, LY294002, blocked EGF-induced H3S28ph (Fig. 1B). To further determine whether this event specifically required EGFR, matched mouse embryo fibroblast cells that normally express EGFR were compared to cells that lack EGFR. Immunoblot analysis revealed that deletion of EGFR blocked EGF-induced H3S28ph in EGFR–/– cells compared to the EGFR+/+ cells (Chen et al., 2001) (Fig. 1C). Introduction of the dominant negative mutant of PI3K (ΔPI3K) into JB6 cells did not decrease H3S28ph compared to JB6 cells transfected with empty vector alone (Fig 1D). As shown in Fig. 2A, EGF dramatically increased Pol III genes, tRNALeu, tRNATyr, 5S rRNA, and 7SL RNA transcription. Blocking EGFR using AG1478 reduced the induction of Pol III genes (Fig. 2B). Further analysis indicated that blocking H2S28ph by expressing mutant H3S28A reduced EGF-induced tRNALeu and 5S rRNA transcription (Fig. 2C). Together, these results indicate that activation of EGFR signaling serves to regulate cellular H3S28ph, which modulates transcription of tRNALeu and 5S rRNA.
Fig. 1. EGF-induced H3S28ph requires EGFR, but not PI3K.
(A) EGF induces H3S28ph in JB6 cells. JB6 cells were starved in 0.1% FBS/MEM and treated with 0, 5, 10, 20 or 50ng/ml EGF for 30 min. H3S28ph and total H3 were determined by antibodies as designated. (B) EGFR inhibitor reduces H3S28ph. JB6 cells were pretreated with 2μM AG1478 or 25μM LY294002 for 1h and then treated with 20 ng/ml EGF as indicated. H3S28ph.was detected as in A. (C) EGFR deficiency blocks H3S28ph. EGFR-/- and EGFR+/+ MEFs were treated with 20ng/ml EGF and immunoblot analysis was performed using lysates derived from these MEFs as designated. (D) Dominant negative mutant PI3K (ΔPI3K) did not affect H3S28ph. JB6 cells expressing ΔPI3K or vector were treated with EGF to detect H3S28ph as in C. A representative blot from three independent determinations is shown.
Fig. 2. Blocking H3S28ph represses endogenous Pol III gene transcription.
(A) EGF enhances Pol III gene transcription in JB6 cells. Cells were treated with 20 ng/ml EGF for 60 min and total RNAs were extracted from these cells. The pre-tRNALeu, pre-tRNATyr, 7SL RNA, 5S rRNA, and GAPDH transcripts were measured by RT-qPCR. The fold change was calculated by normalizing to the amount of GAPDH mRNA. (B) Inhibitor of EGFR represses the activity of pre-tRNALeu and 5S rRNA genes. The cells were pretreated with AG1478 for 1h and treated with EGF for 60 min. tRNALeu and 5S rRNA was determined by RT-qPCR. (C) Inhibition of H3S28ph reduces Pol III gene transcription. JB6 cells were transiently transfected with WT H3 and mutant H3S28A expression constructs for 48h and treated with EGF, the amounts of pre-tRNALeu, 5S rRNA, and GAPDH transcripts were measured by RT-qPCR. The bars represent means ± SE of at least three independent determinations.
H3S28ph occupies the Brf1 and TBP promoters to modulate their expression
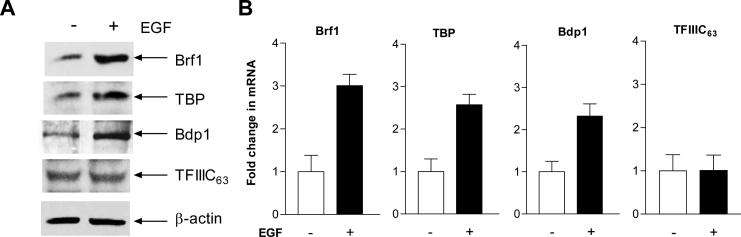
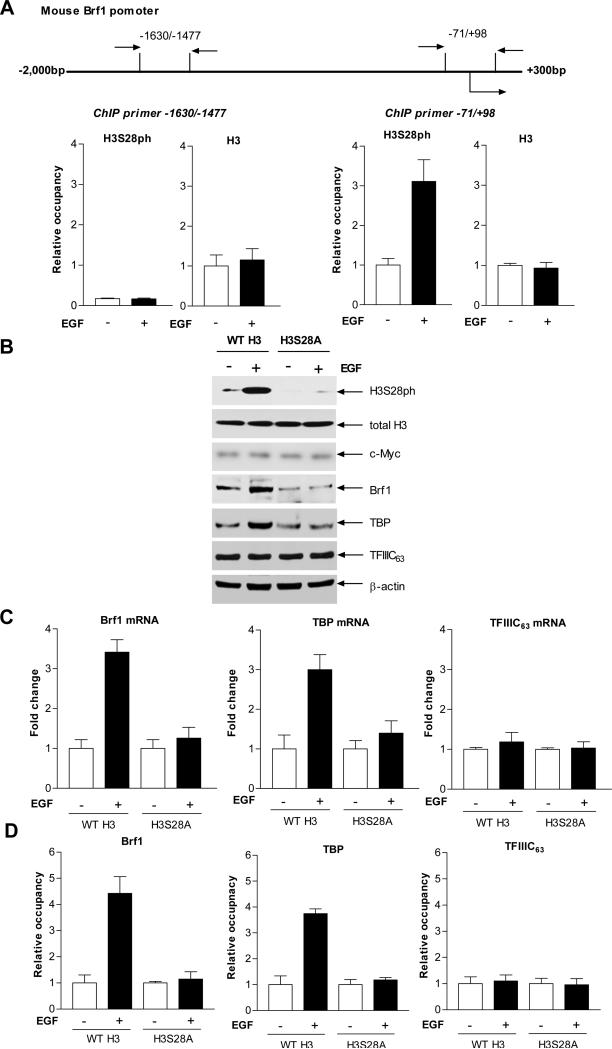
It has previously been shown that EGF increases TBP expression to enhance tRNA reporter transcription (Zhong et al., 2004). It remains to be seen whether EGF affects the expression of the other two TFIIIB subunits, Brf1 and Bdp1. Further analysis indicated that the MAPK, JNK1 positively regulated expression of the TFIIIB subunits TBP, Brf1, and Bdp1. Regulation of Bdp1, but not Brf1, occurred through JNK-mediated alterations in TBP expression (Zhong and Johnson, 2009), suggesting Brf1 and Bdp1 may be regulated independently. As shown in Fig. 3, EGF treatment increased cellular levels of protein and mRNA of Brf1, TBP and Bdp1, but did not affect TFIIIC63. Thus, we further investigated the role of H3S28ph in the regulation of Brf1 and TBP. To determine whether the EGF-mediated increase in H3S28ph required to stimulate Brf1 and TBP expression was due to enhanced occupancy of H3S28ph at the Brf1 and TBP promoters, chromatin immunoprecipition (ChIP) assays were performed. EGF induced a marked increase in the levels of Brf1 and TBP mRNA and occupancy of H3S28ph in the Brf1 and TBP promoters, but not in the TFIIIC63 (Fig. 4A and D). Reduction of H3S28ph by expressing mutant H3S28A decreased its occupancy in the Brf1 and TBP promoters, but did not affect TFIIIC63 (Fig. 4D). We next assessed whether the EGF-induced increase in H3S28ph affected induction of Brf1 and TBP expression. The results indicated that increased H3 expression by the WT H3 expression plasmid in JB6 cells enhanced EGF-mediated Brf1 and TBP expression, but not TFIIIC63 (Fig. 4B and C). In contrast, blocking H3S28ph by mutant H3S28A repressed the induction of Brf1 and TBP expression by EGF (Fig. 4B and C). These results support the idea that EGF mediates an increase in Brf1 and TBP expression primarily through enhanced H3S28ph occupancy of their promoters.
Fig. 3. EGF enhances expression of TFIIIB subunits.
(A) EGF increases cellular levels of TFIIIB subunits in JB6 cells. JB6 cells were treated with EGF. Immunoblot analysis was performed using protein lysates derived from these cells and antibodies against Brf1, TBP, Bdp1, TFIIIC63 and β-actin as designated. A representative blot from three independent determinations is shown. (B) EGF enhances mRNA levels of Brf1, TBP, Bdp1 and TFIIIC63. JB6 cells were treated with EGF. The amounts of Brf1, TBP, Bdp1, TFIIIC63 and GAPDH transcription were measured by RT-qPCR. The values represent means ± SE of three independent determinations.
Fig. 4. EGF induces H3S28ph occupancy in Brf1 and TBP promoters and enhances their expression.
(A) EGF enhances occupancy of H3S28ph in the Brf1 promoter. Schematic of the mouse Brf1 promoter and primers used for ChIP assays are designated relative to putative transcription start sites (TSS) and upstream of TSS (top). JB6 cells were treated with EGF and ChIP assays were performed using antibodies to H3S28ph and H3 and qPCR was used to quantify the amplified DNA. The relative occupancy of the proteins was calculated based on the control (no EGF treatment). (B and C) Inhibiting H3S28ph abrogates EGF-enhanced Brf1 and TBP expression. JB6 cells were transfected with WT H3 or mutant H3S28A expression plasmids for 48 hours and then treated with EGF. H3S28ph, H3, c-Myc, Brf1, TBP, TFIIIC63 and β-actin were determined by immunoblot analysis. A representative blot is shown (B). RT-qPCR was performed on RNA isolated from these cells to measure Brf1, TBP, TFIIIC63 and GAPDH transcripts (C). (D) Expression of mutant H3S28A reduces H3S28ph occupancy in Brf1 and TBP promoters. These cells were treated as indicated in B and C. Chromatin was extracted from these cells to perform ChIP assay with H3S28ph antibody. All values shown are the means ± SEM of at least three independent chromatin preparations.
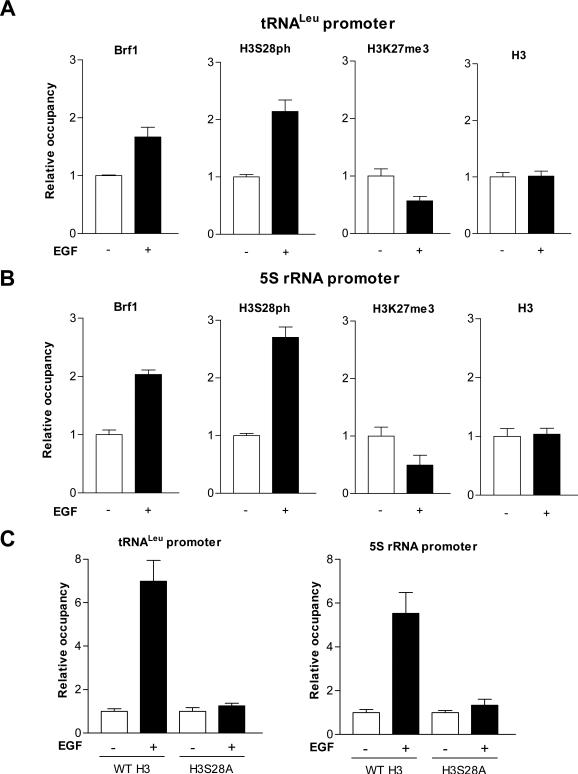
EGF enhanced the occupancy of H3S28ph in tRNA and 5S rRNA promoters
Next, we further determined whether H3S28ph mediated tRNALeu and 5S rRNA transcription. Since Brf1 specifically regulates Pol III gene transcription, we performed ChIP analysis to determine if Brf1 and H3S28ph occupy the Pol III gene promoters. The results revealed that EGF increased the occupancy of tRNALeu and 5S rRNA promoters by Brf1 and H3S28ph (Fig. 5A and B). In contrast, repression of H3S28ph by mutant H3S28A expression decreased occupancy of H3S28ph in the promoters (Fig. 5C). In addition, EGF reduced the H3K27me3 (a marker of gene repression) occupancy of tRNALeu and 5S rRNA promoters (Fig. 5A and B). Recently, by using ChIP-sequence analysis for human CD4+ T cells and human Leukemia K562 cells, Barski and Moqtaderi respectively reported that weak occupancy of H3K27me3 correlates with tRNA expression (Barski et al, 2010; Moqtaderi et al, 2010). Our results of HeK27me3 occupancy in tRNALeu and 5S rRNA promoter are consistent with the analysis of ChIP-sequence. Although the analysis of ChIP-sequencing reveals strong H3K4me1/2/3 and H3Kac and weak H3K27me3 in tRNA genes, it remains to be determined whether H3S28ph modulates the activities of Pol III genes. Our studies demonstrate that H3S28ph may directly function at tRNALeu and 5S rRNA promoters to modulate their transcription.
Fig. 5. EGF increases occupancies of Brf1 and H3S28ph, but decreases H3K27me3 in tRNALeu and 5S rRNA promoters.
(A) EGF-mediated occupancy in the tRNALeu promoter. JB6 cells were treated with or without EGF and ChIP assays were performed using Brf1, H3S28ph, H3K27me3 and H3 antibodies and qPCR with specific tRNALeu primers to quantify the amplified DNA. (B) EGF enhances Brf1, H3S28ph occupancy in the 5S rRNA promoter. ChIP assays were performed as indicated in Fig. 4 and qPCR was used with 5S rRNA primers to quantify the DNA. (C) Expression of mutant H3S28A reduces H3S28ph occupancy in tRNALeu and 5S rRNA promoters. These cells were treated as designed Fig 4B and C. Chromatin was extracted from these cells to perform ChIP assays with H3S28ph antibody. The relative occupancy of the proteins was calculated based on the control (no EGF treatment). All values shown are the means ± SEM of at least three independent chromatin preparations.
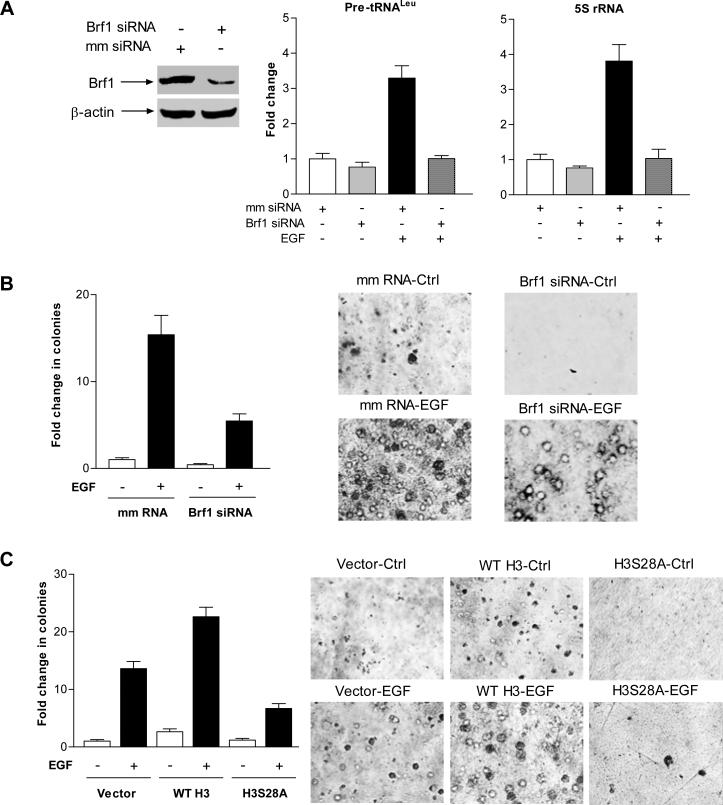
Reduction of H3S28ph decreased transcription of tRNA and 5S rRNA and repressed cell transformation
Previous studies demonstrated that increasing Brf1 expression resulted in enhancement of Pol III gene transcription (Johnson et al., 2008b) and was sufficient for cell transformation (Marshall et al., 2008). Induction of Brf1 expression allowed anchorage-independent colonies to form and promoted tumor formation in mice (Marshall et al., 2008). In the present study we demonstrated that H3S28ph modulated Brf1 expression and tRNA and 5S rRNA transcription (Fig. 2 and 3). Thus, we investigated whether H3S28ph affected EGF-induced cell transformation. The results indicate that inhibiting Brf1 expression with its siRNAs decreased cellular levels of Brf1 protein and mRNA, as well as Pol III gene transcription (Fig. 6A). EGF strongly induced JB6 cell anchorage-independent growth, while reducing Brf1 expression significantly decreased EGF-induced colony formation (Fig. 6B). Furthermore, increasing H3 expression by using stable JB6 cells expressing WT H3 enhanced EGF-induced anchorage-independent growth, compared to stable JB6 cells expressing vector alone. However, blocking H3S28ph by stably expressing mutant H3S28A decreased anchorage-independent growth (Fig. 6C). These results demonstrate that H3S28ph modulates Brf1 expression and enhances tRNA and 5S rRNA transcription, thereby promoting cell transformation.
Fig. 6. Reduction of Brf1 and H3S28ph represses EGF-induced cell transformation.
(A) Repression of Brf1 expression inhibits EGF-induced Pol III gene transcription. JB6 cells were transfected with mismatch (mm) RNA or Brf1 siRNAs for 48 h and treated with EGF. Cell lysates and RNA were isolated from these cells. Immunoblot analysis was performed to determine cellular level of Brf1 protein (left) and pre-tRNALeu (middle), 5S rRNA (right) and GAPDH transcripts were measured by RT-qPCR. (B) Down-regulating Brf1 expression decreases EGF-induced anchorage-independent growth. JB6 cells expressing Brf1 siRNAs were poured in triplicate into 6-well plate with 0.35% agar containing 0ng or 20ng/ml EGF. The cells in A were analyzed for growth in soft agar. (C) Blocking H3S28ph signaling represses EGF-induced cell transformation. JB6 stable cell lines expressing pcDNA3 vector (vector), pcDNA3-wild type H3 (WT H3) or pcDNA3-H3S28A (H3S28A) were poured into 6-well plate with EGF as designated at (B). Cells in (A) and (B) were be incubated at 37°C in 5% CO2 for 1-2 weeks and were fed with fresh complete media with or without EGF twice weekly. Colonies were counted at 1-2 weeks after plating. Values are the means ± SEM (n ≥ 3).
Discussion
This study presents a mechanistic analysis characterizing how the epigenetic modification of histone H3, H3S28ph, modulates endogenous tRNA and 5S rRNA gene transcription. Here, we report that EGF-induced H3S28ph occurs through EGFR, but not the PI3K pathway. Our studies identify the mechanism by which EGF-induced occupancy of H3S28ph in the Brf1 and TBP promoters modulates their expression, which indirectly regulates tRNA and 5S rRNA transcription. In addition, H3S28ph occupies tRNA and 5S rRNA promoters to directly regulate tRNA and 5S rRNA transcription. H3S28ph-mediated Brf1 expression and Pol III gene transcription are critically important to cell transformation. Blocking H3S28ph is sufficient to inhibit tRNA and 5S rRNA transcription and to repress anchorage-independent growth of JB6 cells. These findings support the novel notion that the histone H3 epigenetic modification, H3S28ph, regulates Pol III gene activity.
Previous studies have shown that MAPKs, ERKs, p38 and JNKs are involved in the phosphorylation of histone H3 at serine 10 (H3S10ph) and serine 28 (Zhong et al., 2000, 2001 b). Further analysis indicated that down-stream of MAPKs, MSK1 mediated H3S28ph (Zhong et al., 2001a). In this study, we report that EGFR, but not PI3K, mediated EGF-induced H3S28ph (Fig. 1). Studies have identified H3S10ph and H3S28ph as mitotic markers (Wei et al., 1999; Goho et al., 1999). Phosphorylation of histone H3 at threonine 3 (H3T3ph) has been shown to be necessary for accumulation of the chromosomal passenger complex at the centromere in mitosis (Wang et al., 2010). Phosphorylation of histone H3 at threonine 6 (H3T6ph) mediated androgen receptor (AR)-activated gene expression (Metzger et al., 2010). In addition, H3S10ph and H3S28ph were associated with cell transformation (Kim et al., 2008). These studies suggest that H3ph is critically important to numerous cellular processes. Pol III gene transcription is essential for cell transformation and tumorgenesis (Johnson et al., 2008b; Marshall et al., 2008). However, it is not clear whether H3ph modulates Pol III gene transcription. Our previous study has demonstrated that EGF increased TBP expression and enhanced tRNA reporter transcription through the EGFR-Ras-MAPK pathway (Zhong et al., 2004). Here, we report that EGF induced H3S28ph through the EGFR pathway (Fig. 1) and that H3S28ph indeed modulated endogenous tRNALeu and 5S rRNA transcription.
Although our previous study indicated that EGF increased TBP expression and tRNA reporter transcription in JB6 cells (Zhong et al., 2004), we did not establish whether EGF affected the other two TFIIIB subunits, Brf1 and Bdp1. Our recent analysis further demonstrated that alteration of the cellular level of TBP affected Bdp1 expression, but did not change Brf1 expression (Zhong and Johnson, 2009). In the present study, the results reveal that EGF increased endogenous Pol III gene transcription (Fig. 2) and enhanced cellular levels of TFIIIB subunits, Brf1 and Bdp1 in JB6 cells (Fig. 3). Thus, we investigated whether H3S28ph mediated Brf1 expression, resulting in changes in endogenous Pol III gene transcription. EGF enhanced occupancy of H3S28ph in the promoters of Brf1, TBP, tRNALeu and 5S rRNA, and increased Brf1 and TBP expression as well as tRNALeu and 5S rRNA transcription. In contrast, expression of mutant H3S28A reduced occupancy of H3S28ph in the Brf1, TBP and tRNALeu and 5S rRNA promoters and decreased their expression (Fig. 4 and 5), resulting in repression of cell transformation (Fig. 6). These studies demonstrate that H3S28ph directly and indirectly regulate Pol III genes leading to cell transformation.
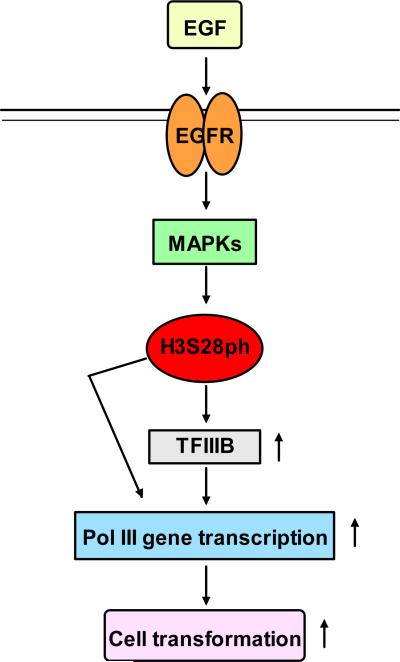
It is possible that a balance of different histone H3 modifications in vivo control gene expression in various physiological and pathological states. The balance between acetylation and deacetylation of core histones is regulated by HATs and HDACs. It has been shown that removal of acetyl groups by HDACs is associated with transcriptional repression (Struhl, 1998; Finnin et al., 1999). The treatment of mammalian cells with inhibitors of HDACs such as TSA or trapoxin was shown to result in increased expression of a variety of genes (Kijima et al., 1993; Yoshida et al., 1995). C-Myc induction enhanced occupancy of tRNALeu and 5S rRNA promoters by acetylated histone H3 and increased the expression of these genes (Kenneth et al., 2007). In addition to increasing acetylation of histone H3, TSA activated MAPKs to induce H3S28ph, whereas H3S28ph, in turn, facilitated acetylation of histone H3 at lysine 9 (Zhong et al., 2003). Immediate early genes, c-myc, c-jun and c-fos, induce H3ph (Cheung et al, 2000; Wei et al, 1999). This implies that c-Myc induction may enhance H3ph to elevate the activity of tRNA and 5S rRNA genes. The role of c-Myc in H3ph is worth further study. H3T6ph prevented demethylation of H3K4 to increase androgen receptor-dependent gene activation (Metzger et al., 2010). H3S10ph was critical for full transcriptional activation of virus-like 30S elements (Brumeir et al., 2010). H3S10ph occupancy of the c-fos promoter mediated gene activity (Shimada et al., 2010). H3K4me1/2/3 is a marker of gene activation, whereas H3K27me3 is a marker of gene repression. Chip-sequence reveals strong H3K4me1/2/3 and H3Kac and weak H3K27me3 in tRNA genes (Barski et al, 2010). Phosphorylation of H3K27me3S28 occurs in response to stress signaling, mitogenic signaling, and retinoic acid (RA)-induced neuronal differentiation. MSK1/2-mediated phosphorylation of H3K27me3S28 activates a subset of polycomb group target genes and modulates the gene expression program which determines cell fate (Gehani et al, 2010). Our results reveal that EGF decreased occupancy of H3K27me3 in tRNALeu and 5S rRNA promoter, while EGF enhanced occupancy of H3S28ph in tRNALeu and 5S rRNA promoters, resulting in upregulation of their transcription. Our studies indicate that epigenetic modifications of histone H3 takes part in the regulation of tRNA and 5S rRNA genes. Together, these analyses support the idea that changes in the balance of different modifications of histone H3 provide a mechanism for the regulation of tRNA and 5S rRNA transcription. H3S28ph not only directly occupies promoters of Pol II genes, such as c-fos and Brf1 and TBP, but also is directly associated with promoters of Pol III genes, tRNALeu and 5S rRNA. It suggests that a common epigenetic regulation of histone H3 mediates both RNA Pol II and RNA Pol III-dependent gene activity. In summary, increased Pol III gene transcription, observed in transformed cells and human tumors, is required for oncogenic transformation. In this present study, we provide evidence that increasing H3S28ph enhances Pol III gene transcription and the rate of cell transformation. The novel findings suggest the possibility that blocking H3S28ph by an inhibitor may be a potential approach to repress cell transformation and tumor development (Fig. 7).
Fig. 7. Schematic illustration of H3S28ph mediating Pol III gene transcription.
EGF induces H3S28ph through the EGFR pathway. H3S28ph increases TFIIIB expression, which in turn regulates Pol III gene transcription. H3S28ph also directly upregulates Pol III gene transcription. Both direct and indirect roles of H3S28ph enhance Pol III gene activity to promote cell transformation.
Materials and methods
Reagents and antibodies
EGF was from Sigma-Aldrich. Cell culture medium (MEM) and DMEM, G418, Lipofectin reagent, Lipofectamine 2000, TRIzol reagent and OPTI-MEM were from Invitrogen. Antibodies against TBP, Bdp1, β-actin, TFIIIC63 and c-Myc probe were obtained from San Cruz. Histone H3 and phospho-H3S28 antibodies were from Cell Signaling. Brf1 antibody was from Bethyl laboratories Inc. Inhibitors of AG1478 and LY294002 were from A.G. Scientific, Inc. The sequences of primers and Brf1 siRNAs and primers were listed in supplements. Expression plasmids of histone H3 were kindly provided by Dr. Masaki Inagaki (Aichi, Japan).
RT-qPCR and transfection assays
Total RNA was isolated from mouse epidermal JB6 cells or engineered JB6 cells using single step extraction method with TRIzol reagent (Invitrogen). Precursor of tRNALeu and tRNATyr, 7SL RNA and 5S rRNA transcripts were measured as described previously (Crighton et al., 2003). For transient transfection assays, JB6 cells were transfected with plasmids or siRNAs as described previously (Zhong et al., 2004). Serum-free medium was added to each dish with Lipofectin-DNA or Lipofectamine2000-siRNA complexes, and cells were further incubated for 4h. The medium was changed with 5% FBS/MEM and cells were incubated for 48h before harvesting. Protein concentrations of the resultant lysates were measured by the Bradford method.
Stable transfection and cell Anchorage-independent growth
Stable transfections were conducted using the Lipofectin reagent (Invitrogen) and pCMV-Tag3 vector (Vector), pCMV-Tag3-wild-type histone H3 (WT H3) or pCMV-Tag3-mutant histone H3S28A (H3S28A). The stable transfections were carried out as described in the protocol from Invitrogen. In brief, for each 6 cm dish of JB6 cells to be transfected, 4 μg DNA vector, WT H3 or H3S28A into 100 μl OPTI-MEM media and 4 μl lipofectin reagent in 100 μl OPTI-MEM media were incubated for 5 min at room temperature, respectively. Diluted DNA and lipofectin reagent were mixed and incubated for 30 min at room temperature to form the DNA-lipofectin complexes. DNA-lipofectin complex was added into each dish and mixed. The cells were incubated at 37 °C for 4 h and then media were changed to fresh growth media for 48 h. Selective media containing 400μg/ml G-418 were added to each dish and the surviving G418-resistant cell populations were pooled. Early passage number of selected cells were used and maintained in the presence of G-418 (200μg/ml). G418 selected cells were tested for c-Myc epitope-tagged histone H3 by immunoblot analysis with a c-Myc-tag antibody.
JB6 cells transfected with mismatch RNA and mouse Brf1 siRNAs (Table S1). The Brf1 siRNAs are pooled and correspond to nucleotide positions 723-741, 1623-1641 and 2415-2435 within exon 4, 12 and 18 of mouse Brf1, respectively. The transiently transfected JB6 cells or JB6 stable cell lines expressing vector, WT H3 or H3S28A (2 × 104 cells/well in 6-well plate) were suspended in 0.35% (w/v) agar in 10% FBS/MEM with or without EGF (20ng/ml), over a bottom layer of media with 0.5% (w/v) agar. Cells were fed fresh complete media with EGF twice weekly. Colonies were counted 1-2 weeks after plating.
Immunoblot analysis
Immunoblot analysis was carried out as previously described (Zhong and Johnson, 2009). Cells were grown to 85% confluency in 5% FBS/MEM and then serum deprived using 0.1% FBS/MEM for 3 h. Where noted, cells were pretreated with 2 μM of the EGFR inhibitor, AG 1478 or 25μM PI3K inhibitor, LY 294002 for 1 hr and then incubated with or without 20 ng/ml of EGF. Lysates (50 μg of protein) were subjected to immunoblot analysis. Membranes were probed with specific antibodies as indicated. Hybond-P membrane was used for protein transfer. Bound primary antibody was visualized using horseradish peroxidase-conjugated secondary antibody (Vector Laboratories) and enhanced chemiluminescence reagents (Amersham).
Supplementary Material
Acknowledgements
We want to thank D.L., Johnson and M. R. Stallcup (University of Southern California) for scientific discussions. We would like to thank Z. Dong (University of Minnesota) and C. Huang (University of New York) who provided cell lines. This work was supported by National Institutes of Health grants AA017288 to S.Z and DK025836-26 to D.L.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, et al. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat Struct Mol Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunmeir R, Lagger S, Simboeck E, Sawicka A, Egger G, Hagelkruys A, et al. Epigenetic regulation of a murine retrotransposon by a dual histone modification mark. PLoS Genet. 2010;6:e1000927. doi: 10.1371/journal.pgen.1000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Ma WY, She QB, Wu E, Liu G, Bode AM, Dong Z. Transactivation of the epidermal growth factor receptor is involved in 12-O-tetradecanoylphorbol-13-acetate-induced signal transduction. J Biol Chem. 2001;276:46722–28. doi: 10.1074/jbc.M107156200. [DOI] [PubMed] [Google Scholar]
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell. 2000;5:905–15. doi: 10.1016/s1097-2765(00)80256-7. [DOI] [PubMed] [Google Scholar]
- Crighton D, Woiwode A, Zhang C, Mandavia N, Morton JP, Warnock LJ, et al. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO. J. 2003;22:2810–20. doi: 10.1093/emboj/cdg265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–22. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- Felsenfeld G, Groudine M. Controlling the double helix. Nature. 2003;421:448–453. doi: 10.1038/nature01411. [DOI] [PubMed] [Google Scholar]
- Finnin MS, Donigian JR, Cohen A, Richon VM, Rifkind RA, Marks PA, et al. Structures of a histone deacetylase homologue bound to the TSA and SAHA inhibitors. Nature. 1999;401:188–193. doi: 10.1038/43710. [DOI] [PubMed] [Google Scholar]
- Goodfellow SJ, Innes F, Derblay LE, MacLellan WR, Scott PH, White RJ. Regulation of RNA polymerase III transcription during hypertrophic growth. EMBO J. 2006;25:1522–33. doi: 10.1038/sj.emboj.7601040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H, Tomono Y, Ajiro K, Kosako H, Fujita M, Sakurai M, et al. Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J. Biol. Chem. 1999;274:25543–25549. doi: 10.1074/jbc.274.36.25543. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Johnson DL, Johnson SA. Cell biology. RNA metabolism and oncogenesis. Science. 2008a;320:461–462. doi: 10.1126/science.1158680. [DOI] [PubMed] [Google Scholar]
- Johnson SA, Dubeau L, Johnson DL. Enhanced RNA polymerase III-dependent transcription is required for oncogenic transformation. J Biol Chem. 2008b;283:19184–19191. doi: 10.1074/jbc.M802872200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SA, Zhang C, Fromm J, Willis IM, Johnson DL. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol Cell. 2007;26:367–379. doi: 10.1016/j.molcel.2007.03.021. [DOI] [PubMed] [Google Scholar]
- Kenneth NS, Ramsbottom BA, Gomez-Roman N, Marshall L, Cole PA, White RJ. TRRAP and GCN5 are used by c-Myc to activate RNA polymerase III transcription. Proc Natl Acad Sci USA. 2007;104:14917–14922. doi: 10.1073/pnas.0702909104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kijima M, Yoshida M, Sugita K, Horinouchi S, Beppu T. Trapoxin, an antitumor cyclic tetrapeptide, is an irreversible inhibitor of mammalian histone deacetylase. J Biol Chem. 1993;268:22429–22435. [PubMed] [Google Scholar]
- Kim HG, Lee KW, Cho YY, Kang NJ, Oh SM, Bode AM, et al. Mitogen- and stress-activated kinase 1-mediated histone H3 phosphorylation is crucial for cell transformation. Cancer Res. 2008;68:2538–2547. doi: 10.1158/0008-5472.CAN-07-6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall L, Kenneth N, White R. Elevated tRNA(iMet) synthesis can drive cell proliferation and oncogenic transformation. Cell. 2008;133:78–89. doi: 10.1016/j.cell.2008.02.035. [DOI] [PubMed] [Google Scholar]
- Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, et al. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792–796. doi: 10.1038/nature08839. [DOI] [PubMed] [Google Scholar]
- Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, et al. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat Struct Mol Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat Struct Mol Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, et al. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc Natl Acad Sci U S A. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Nakadai T, Fukuda A, Hisatake K. cAMP-response element-binding protein (CREB) controls MSK1-mediated phosphorylation of histone H3 at the c-fos promoter in vitro. J Biol Chem. 2010;285:9390–9401. doi: 10.1074/jbc.M109.057745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K, Moqtaderi Z. The TAFs in the HAT. Cell. 1998;94:1–4. doi: 10.1016/s0092-8674(00)81213-1. [DOI] [PubMed] [Google Scholar]
- Ullu E, Tschudi C. Alu sequences are processed 7SL RNA genes. Nature. 1984;312:171–72. doi: 10.1038/312171a0. [DOI] [PubMed] [Google Scholar]
- Wang F, Dai J, Daum JR, Niedzialkowska E, Banerjee B, Stukenberg PT, et al. Histone H3 Thr-3 phosphorylation by haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky M, Allis CD. Phosphorylation of histone H3 Is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- White RJ. RNA polymerase III transcription and cancer. Oncogene. 2004;23:3208–3216. doi: 10.1038/sj.onc.1207547. [DOI] [PubMed] [Google Scholar]
- Winter A, Sourvinos G, Allison S, Tosh K, Scott P, Spandidos D, et al. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc Natl Acad Sci U S A. 2007;97:12619–12624. doi: 10.1073/pnas.230224097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woiwode A, Johnson SA, Zhong S, Zhang C, Roeder RG, Teichmann M, et al. PTEN represses RNA polymerase III-dependent transcription by targeting the TFIIIB complex. Mol Cell Biol. 2008;28:4204–4214. doi: 10.1128/MCB.01912-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Ma WY, Dong Z. ERKs and p38 kinases directly mediate UVB-induced phosphorylation of H3 at serine 10. J Biol Chem. 2000;275:20980–20984. doi: 10.1074/jbc.M909934199. [DOI] [PubMed] [Google Scholar]
- Zhong S, Goto H, Inagaki M, Dong Z. Phosphorylation of histone H3 at serine 28 and acetylation of histone H3 at lysine 9 induced by TSA. Oncogene. 2003;22:5291–5297. doi: 10.1038/sj.onc.1206507. [DOI] [PubMed] [Google Scholar]
- Zhong S, Jansen C, Goto H, Inagaki M, Dong Z. Ultraviolet B-induced phosphorylation of histone H3 at serine 28 mediated by MSK1. J Biol Chem. 2001a;276:33213–33219. doi: 10.1074/jbc.M103973200. [DOI] [PubMed] [Google Scholar]
- Zhong S, Johnson DL. The JNKs differentially regulate RNA polymerase III transcription by coordinately modulating the expression of all TFIIIB subunits. Proc Natl Acad Sci U S A. 2009;106:12682–12687. doi: 10.1073/pnas.0904843106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S, Zhang Y, Goto H, Inagaki M, Dong Z. MAP kinases mediated UVB-induced phosphorylation of histone H3 at serine 28. J Biol Chem. 2001b;276:12932–12937. doi: 10.1074/jbc.M010931200. [DOI] [PubMed] [Google Scholar]
- Zhong S, Zheng C, Johnson DL. Epidermal Growth Factor enhances cellular TBP levels and induces RNA polymerase I- and III-dependent gene activity. Mol. Cell. Biol. 2004;24:5119–5129. doi: 10.1128/MCB.24.12.5119-5129.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida M, Horinouchi S. Trichostatin and leptomycin. Inhibition of histone deacetylation and signal-dependent nuclear export. Ann N Y Acad Sci. 1999;886:23–36. doi: 10.1111/j.1749-6632.1999.tb09397.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.