Abstract
Sequence-specific DNA binding by transcription factors is central to gene expression regulation. While a number of methods for characterizing DNA-protein interactions are currently available1-6, none have demonstrated both quantitative measurement of affinity and high throughput. To address this challenge, we developed HiTS-FLIP, a technique that couples high-throughput sequencing with direct visualization of in vitro binding to provide quantitative protein-DNA binding affinity measurements at unprecedented depth. HiTS-FLIP analysis of GCN4, the master regulator of the yeast amino acid starvation response7, yielded 440 million binding measurements, enabling determination of context-averaged dissociation constants for all 12mer sequences having submicromolar affinity. These data revealed complex interdependency between motif positions, yielded improved discrimination of in vivo GCN4 binding sites and regulatory targets relative to previous models, and identified sets of genes with distinct GCN4 affinity levels which had distinct functions and expression kinetics. This approach promises to deepen understanding of the interactions that drive transcription.
The precise description of protein-DNA interactions is central to efforts to model and predict gene expression. Methods available for quantitative measurement of DNA-protein interactions in vitro include DNA footprinting, electrophoretic mobility shift assays (EMSA), surface plasmon resonance (SPR), and certain microfluidic methods6, but all have relatively low throughput. One-hybrid systems allow rapid determination of sequence specificity in a yeast (Y1H) or bacterial (B1H) context, but give relatively low resolution of binding sites8. Protein-binding microarrays (PBMs)3 have been widely used9,10, but are typically limited to complete enumeration of binding to motifs of eight nucleotides or fewer. HT-SELEX (High-Throughput Systematic Evolution of Ligands by Exponential Enrichment)5 improves on traditional SELEX, but still requires a resin- or filter-based selection step that may introduce biases. Chromatin immunoprecipitation (ChIP) coupled with microarray analysis (ChIP-Chip2) or with sequencing (ChIP-Seq4) can map in vivo binding sites genome-wide, but often reflect the binding of multi-protein complexes rather than direct binding of the targeted factor. Additionally, ChIP-based methods are limited by antibody availability and specificity for different proteins, conformations and variants, adding to other technical factors to make quantitation difficult11. For these reasons, fully comprehensive and quantitative description of the DNA binding affinity of transcription factors (TFs) has remained elusive. Such a description would be useful for biophysical understanding, for distinguishing direct from indirect binding and isolating intrinsic affinity from effects of chromatin context, and for informing quantitative models of transcription.
Second-generation sequencing instruments can determine one hundred million or more short sequences per run. The Illumina Genome Analyzer (GA) builds millions of distinct clusters on a flow cell, each consisting of several hundred identical DNA molecules. Clusters are sequenced by synthesis in situ, with individual fluorescently tagged nucleotides visualized using a charge-coupled device (CCD) camera to reconstruct the sequence of each cluster12. We reasoned that fluorescently tagged proteins could be added to the flow cell and their binding to each DNA cluster visualized in the same way as fluorophore-tagged nucleotides, and that bound clusters could subsequently be matched to the corresponding sequences based on their position in the flow cell, enabling direct observation of the DNA binding preferences of the tagged protein (Fig. 1a, 1b). The High-Throughput Sequencing – Fluorescent Ligand Interaction Profiling (HiTS-FLIP) procedure is therefore conceptually simple (Fig. 1a): 1) build and sequence ~100 million clusters of genomic or random synthetic DNA; 2) denature and wash away the second strand and rebuild double-stranded DNA; 3) introduce fluorescently-tagged protein to the flow cell; 4) after an optional two-minute wash step, quantify the binding to each cluster by visualizing fluorescence; and 5) map the bound clusters to corresponding sequences and analyze, yielding a comprehensive, quantitative landscape of binding specificity.
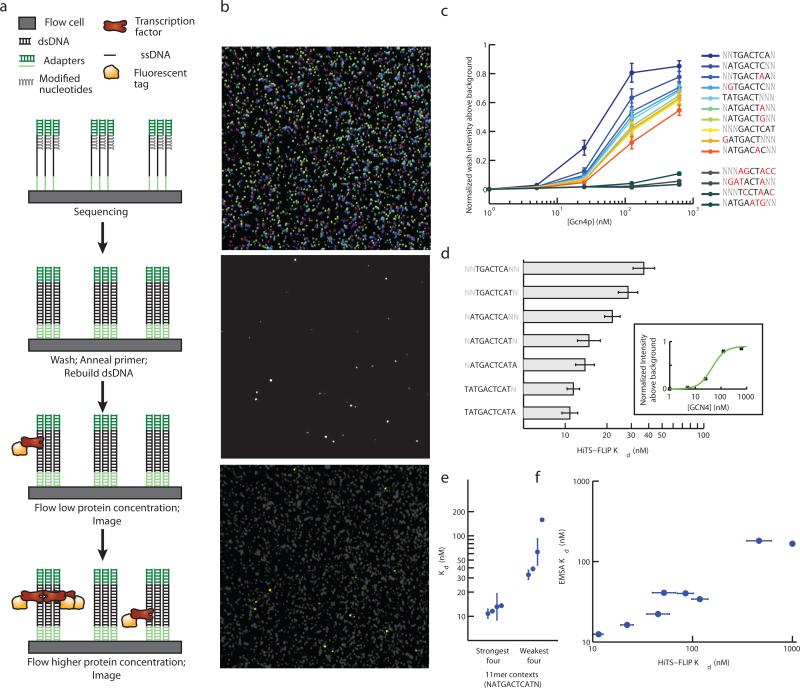
Figure 1. High-throughput sequencing fluorescent ligand interaction profile (HiTS-FLIP) method.
a) HiTS-FLIP method schematic. A microfluidic flow cell with anchored single-stranded DNA is sequenced by synthesis. Second-strand DNA is stripped and rebuilt using Klenow and unmodified dNTPs to form dsDNA clusters. Fluorescently labeled Gcn4p is introduced at different concentrations and binding is imaged. b) Partial images from sequencing cycles (false colored for each nucleotide, top), a Gcn4p binding cycle (center), and a merge of the top and center with Gcn4p clusters highlighted in yellow (lower). The images shown represent roughly 0.003% of a flow cell. c) Intensity of binding versus Gcn4p concentration for the top ten 7mers (see text) and four 7mers with expected binding affinity near zero. Mismatches from the consensus sequence are marked in red. Error bars indicate 95% confidence intervals based on estimates from 5 flow cell lanes. d) Dissociation constants (Kd values) calculated by fitting a Hill equation to intensity measurements (inset; for TGACTCA). Top 7mer by Kd as well as selected extensions that had significantly increased affinity are shown. Error bars indicate standard deviation estimated from comparison of 5 flow cell lanes. e) Kd (mean and standard error) for 11mers containing the strongest-binding 9mer sequence, ATGACTCAT. The four 11mers with the strongest Kd (flanked by T,A; T,C; G,A; and C,A) and the four with the weakest Kd (flanked by C,T; A,G; A,C; and C,G) are shown. f) Dissociation constants measured by HiTS-FLIP (mean and standard error of 5 flow cell lanes) compared with those measured by EMSA (mean of duplicate experiments). Nine sequences chosen to have a wide range of expected binding affinity were assayed.
HiTS-FLIP was applied to S. cerevisiae GCN4, a prototypical dimeric basic leucine zipper (bZIP) TF that is the master regulator of the amino acid starvation response7. We chose this factor to test our approach because of the availability of extensive, high-quality binding and activity data suitable for comparison7,13-16, and the interesting roles of this factor in metabolic and morphogenic pathways17. Gcn4p with an mOrange fluorescent protein tag was applied at several concentrations to a flow cell built from a library of randomized 25 bp synthetic DNA. Since dimerization and specific DNA binding involves residues situated at the C-terminus of the protein18, the N-terminal fusion should have minimal effect on DNA binding characteristics. After matching the mOrange-Gcn4p binding intensities to the locations of sequencing clusters (Fig. 1b), we were able to quantify the binding of Gcn4p to more than 88 million DNA clusters corresponding to distinct 25 bp sequences. Selecting the top 0.5% of clusters based on raw fluorescence intensity at a Gcn4p concentration of 125 nM, the known consensus binding heptanucleotide (7mer) TGACTCA19 was enriched 40-fold over its frequency in all clusters. The binding intensity of clusters containing TGACTCA was independent of the 7mer's location within the sequence (Supplementary Fig. 1), indicating that each cluster provides an unbiased measurement of affinity.
Because each 7mer occurred in approximately 100,000 different clusters on the flow cell, we could estimate binding affinities to 7mers based on the 25mer binding data simply by: (i) determining the median binding intensities of clusters containing each 7mer and identifying the 7mer associated with highest binding intensity, (ii) removing clusters containing that 7mer, and (iii) repeating to generate a list of 7mers ranked by binding affinity (alternative algorithms are considered in Supplementary Discussion). The resulting sets of median binding intensity values represent “context-averaged” 7mer affinities since they derive from averages of binding measurements to thousands of 25mers containing a specific 7mer embedded in diverse sequence contexts, analogous to the typical in vivo situation in which TF motifs occur embedded in longer dsDNAs of varying sequence. After normalizing for cluster size and background fluorescence (Supporting Information), binding to all possible oligonucleotides (k-mers) of 7-12 bases in length was quantified using this procedure (Table 1). Oligonucleotides unrelated to the GCN4 consensus exhibited negligible binding, while near-consensus k-mers yielded highly consistent orderings of binding intensity at different Gcn4p concentrations (Fig. 1c). Treating each lane of the flow cell as a technical replicate, the mean binding intensity of k-mers was highly reproducible (confidence intervals, Fig. 1c).
Table 1.
HiTS-FLIP binding statistics.
| k-mer | Mean measurements per k-mer1 | No. of k-mers with KD < 1 μM | Avg. standard error2 of log(KD) |
|---|---|---|---|
| 8mers | 77,485 | 219 | 3.2% |
| 9mers | 18,791 | 1,113 | 3.1% |
| 10mers | 4,528 | 4,546 | 4.0% |
| 11mers | 1,091 | 19,268 | 6.4% |
| 12mers | 475 | 81,852 | 11.4% |
The number of clusters passing our quality filter and the total number of binding measurements were 88,052,302 and 440,261,510, respectively.
Standard error is an average over all k-mers having Kd < 1 μM and was calculated by treating 5 flow cell lanes as technical replicates and fitting a Kd for each.
In principle, the DNA binding specificity of a protein can be described comprehensively and quantitatively by a complete set of equilibrium binding constants relative to all possible oligonucleotide ligands. Using HiTS-FLIP, conditions can be adjusted and binding re-imaged without resequencing, which we exploited by altering the concentration of Gcn4p on the flow cell to determine equilibrium binding constants. Using 5 different protein concentrations, increasing in 5-fold increments from 1 nM to 625 nM (corresponding to roughly 1 to 1000 Gcn4p molecules per cell nucleus20), we observed a range from no appreciable Gcn4p binding to near-saturating binding for the consensus 7mer, TGACTCA (Fig. 1c). The GA optics are based on total internal reflection illumination of the fluorophores, which excites only those fluorophores situated less than 100nm from the flow cell surface, providing preferential readout of fluorophores attached to the flow cell relative those in solution, alleviating the need for washing12. Although we performed a short (2-minute) wash prior to imaging, similar intensity values and 7-mer rankings were obtained in a pilot experiment that omitted the wash step (Supplementary Discussion). This observation suggests that the wash step had minimal impact on the results and that the low background of the GA's illumination system may make this step unnecessary, which contrasts with the PBM approach, for which the requirement of several wash / dry steps likely prevents measurement of equilibrium values and detection of lower affinity interactions. Fitting a Hill equation to context-averaged binding intensity versus concentration data for each 7mer through 12mer yielded a context-averaged dissociation constant (Kd) value for each oligonucleotide (Fig. 1d, Supplementary Tables 1-5). Four 8mers representing extended versions of the canonical 7mer had significantly higher binding intensities (at 5 nM Gcn4p) than TGACTCA, and the near-palindromic 9mer sequence ATGACTCAT bound more strongly than all 8mers, consistent with previous studies10,19,21.
While in vitro binding preferences of Gcn4p for sequences longer than 9 bp have not been previously described, we had statistical power to observe relatively subtle differences in affinity (Table 1, Fig. 1d), e.g., 11-mer Kd values were estimated within 6.4% on average and 12-mers were estimated within 11.4% (standard error between lanes). Two 10mers, ATGACTCATA and TATGACTCAT, each occurring more than 1000 times on the flow cell, had significantly higher binding intensity than the strongest 9mer (z-test, p < 0.01 at 5nM Gcn4p), and we identified a new, near-palindromic 11-mer consensus, TATGACTCATA, with significantly higher binding intensity than all 10mers (z-tests, p < 0.04, Fig. 1d). No 12mers with significantly higher intensity than the consensus 11mer were found. The relatively small size of the yeast genome (~13 Mbp) implies that individual 11mers will occur only a handful of times on average per haploid genome, which would make it difficult to assess binding preferences of individual 11mers using in vivo methods, illustrating the need for comprehensive in vitro approaches for a full understanding of binding affinity. Even the two nucleotides flanking the strongest-binding 9-mer affected affinity by an order of magnitude (Fig. 1e, Supplementary Fig. 2). We also observed effective binding to sequences with adjacent rather than overlapping half-sites, as in ATGACGTCAT (Supplementary Fig. 3)22.
As an independent validation, dissociation constants were measured for 10 oligonucleotides by EMSA (Supplementary Fig. 4). Dissociation constants determined by HiTS-FLIP exhibited high correlation and a linear relationship with these values and with Kd values reported in the literature for untagged Gcn4p across a range of binding affinities (Fig. 1e, Supplementary Table 6)23, 24. The determination of Kd values for every 11mer and 12mer by HiTS-FLIP, including tens of thousands of values below 1 μM, likely represents the most comprehensive quantitative description of in vitro DNA binding affinity obtained for any protein.
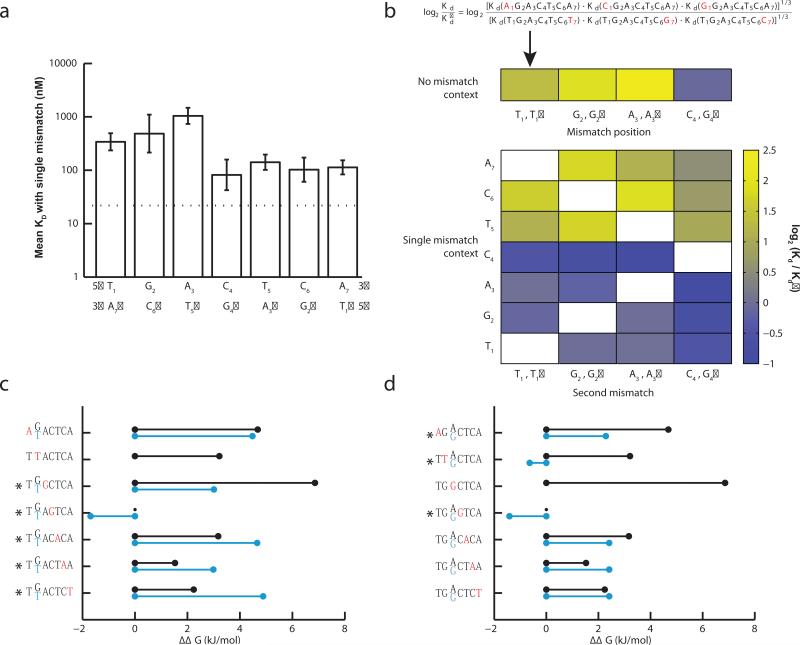
Gcn4p is known to bind as a dimer in which the basic DNA binding domain of each subunit binds optimally to the half-site sequence 5'-TGAC-3'. The preference for C at the 4th position introduces asymmetry in binding to the consensus 7mer, T1G2A3C4T5C6A7, with stronger binding observed to the left than to the right half-site22. Substitutions at positions T1, G2 and A3 resulted in larger increases in Kd (i.e. greater weakening of binding) than at the corresponding positions T1', G2' and A3' of the right half-site, confirming the expected asymmetry (Fig. 2a, 2b).
Figure 2. Positional interdependencies in the DNA binding affinity of Gcn4p.
a) Mean dissociation constant (Kd) for the 3 sequences having one mismatch from the canonical 7mer at each position T1 through A7 for the forward strand and T1′ through A7′ for the reverse strand 7-mer. Dotted line represents the Kd for the canonical 7mer. Error bars represent standard error for the 3 sequences. b) Asymmetrical effect of substitutions on binding affinity. Top: 7mers with a single mismatch in the left half (T1 through C4) are compared to the symmetrical substitution in the reverse strand (T1′ through G4′); see equation for example. The log ratio of the geometric mean of binding affinities is shown for each position as a blue-yellow heat map. Bottom: As above, except that each row compares 7mers with an additional mismatch at the position indicated. c) Effect of a representative substitution at each position on free energy (ΔΔG) of binding. More positive values indicate positions where the reference nucleotide was more crucial for binding affinity. Specific substitutions (red) were defined relative to the consensus 7mer (black) or relative to a 7mer having a G2 to T2 substitution (blue). Significant differences between the two ΔΔG values (p < 0.05, z-test) are marked with asterisks. d) As in (c) except that blue bars indicate substitutions relative to a 7mer having an A3 to G3 substitution.
Considering pairwise substitutions relative to the consensus, we observed extensive interdependence. Specifically, the incremental effect on binding of a second mismatch in the same half-site was consistently lower than the effect of the corresponding mismatch in the opposite half-site (Fig. 2b), i.e. two mismatches in the same half-site disrupt binding less than a single mismatch in each half-site. One important consequence of this pattern is that nucleotide positions do not contribute independently to binding, as observed previously for certain transcription factors9,25. Therefore, models that assume independence such as the commonly used position weight matrix (PWM) model cannot accurately capture the DNA binding affinity of Gcn4p. Instead, we advocate the use of the full spectrum of Kd values estimated by HiTS-FLIP for all k-mers of appropriate size (e.g., 8, 10 or 12 bp, depending on the factor and the depth of the data).
Analyzing the effect of individual substitutions on the inferred change in Gibbs free energy of binding for specific second substitutions indicated that presence of a substitution in the same half-site tended to reduce the effects of other substitutions in the same half-site, but exaggerated the effects of substitutions in the other half-site, relative to the effects in a fully consensus background (Fig. 2c, 2d, Supplementary Fig. 5). For example, substitution of position A3 by G destabilized binding by only ~3 kJ/mol in the presence of a 7mer containing a T substitution at the G2 position in the same half-site, compared to a ~7kJ/mol destabilization in the context of the consensus 7mer, with corresponding increases in ΔΔG magnitudes at positions in the other half-site (Fig. 2c). Taken together, the data in Figure 2 suggest a model in which a substitution at one position in a half-site tends to weaken the interaction of the associated Gcn4p monomer with other positions in the same half-site, perhaps through a subtle protein conformational change, making interactions between the other monomer and half-site more critical. Substitution of A3 by G had the surprising effect of converting a G2 → T substitution from strongly destabilizing to weakly stabilizing (Fig. 2d), underscoring the complexity of the affinity landscape.
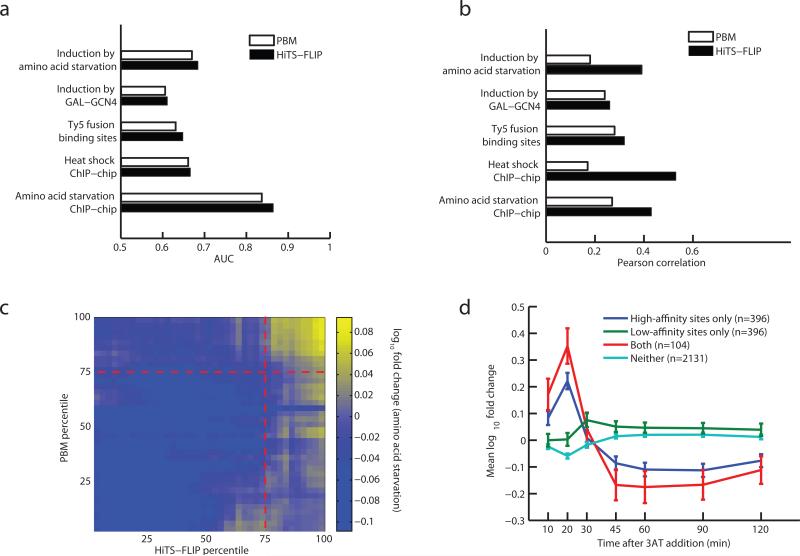
Both HiTS-FLIP and the previously described PBM method26 assess in vitro binding affinity to a wide spectrum of DNA sequences (hundreds of thousands for PBMs, tens of millions for HiTS-FLIP). As a direct comparison, we assessed HiTS-FLIP data relative to available PBM data10. Although we could have used HiTS-FLIP Kd values for all 11- or 12-mers, we restricted our analysis to 8-mers, the size directly addressed by the PBM data, to facilitate a direct comparison of data quality in a manner controlling for data quantity. The two methods agreed on the identities of the top few 8mers, but exhibited substantial differences in magnitudes, particularly for moderate and low affinity 8mers (Supplementary Fig. 6). To assess the relationship to in vivo binding and activity, we predicted the occupancy of promoters using a simple method taking into account the measured Kd values of all 8-mers (Supplementary Methods), or as described previously for the PBM data10 (Supplementary Table 7). Predicted occupancies were compared to data from five published studies reporting: (i) gene expression following amino acid starvation (which is driven largely by GCN4)7; (ii) gene expression following transgenic GCN4 induction14; (iii) Ty5 fusion analysis of Gcn4p binding sites15; and ChIP-chip binding data following (iv) heat shock16 and (v) amino acid starvation13 (both inducers of GCN4 expression). For all five datasets, responsive or bound promoters were more accurately classified using occupancies predicted by HiTS-FLIP (Fig. 3a). Although the improvement in classification assessed by the area under curve (AUC) of the receiver operator characteristic (ROC) curve was fairly modest, the magnitudes of expression induction and Gcn4p binding were much more accurately predicted by HiTS-FLIP, yielding higher Pearson correlations in all cases and substantially higher values in some cases, particularly for comparisons involving amino acid starvation or heat shock (Fig. 3b). This trend was observed independently of the formalism used for converting PBM data into predicted promoter occupancies (Supplementary Discussion).
Figure 3. HiTS-FLIP accurately predicts in vivo binding and regulation and reveals impact of lower-affinity binding sites.
a) Predictive power of protein-binding microarrays (PBMs)10 and HiTS-FLIP for in vivo Gcn4p bound regions (ChIP-chip and Ty5 datasets) and Gcn4p-induced genes, as measured by area under the curve (AUC). b) As in (a) except that the correlation between predicted Gcn4p occupancy and in vivo binding intensity or strength of induction is measured. Correlation is for top quintile of predicted bound genes. c) Genes were ranked by predicted occupancy using PBM data or HiTS-FLIP data and induction of genes under amino acid starvation conditions is shown by heat map, with small squares representing bins of genes. Red dotted lines indicate sample cutoffs of the 75th percentile for both methods. d) Gcn4p-sensitive genes are predicted using high-affinity k-mers (Kd < 400nM) or moderate-affinity k-mers (1uM > Kd > 400nM). Mean and standard error of fold-induction by 3-aminotriazole (3AT) treatment, triggering amino acid starvation, is plotted for genes ranking in the top quintile of high-affinity model, moderate-affinity model, both, or neither. Error bars represent ± one standard error.
To further explore the differences in promoter classification between these methods, we analyzed in detail the induction of gene expression following amino acid starvation7. Expression induction was observed not only for genes ranked highly by both methods, but also for a subset of genes ranked highly (> 75th percentile) by HiTS-FLIP but more lowly (< 75th percentile) by PBM, but not for genes ranked highly by PBM only (Fig. 3c). Most (85.4%) of the promoters ranked highly only by HiTS-FLIP contained no 9mer within 1 mismatch of the GCN4 consensus, but many (72.3%) contained one or more 9mers with 2 or 3 mismatches that nevertheless preserved an intact half-site. Such sequences typically score quite poorly by PBM and PWM methods but may have moderate Gcn4p affinity by HiTS-FLIP analysis (an example is shown in Supplementary Fig. 7). Because all of the data in Figure 3 used HiTS-FLIP 8mer Kd values only, the advantages reported relate to data quality rather than quantity and should apply equally to proteins with motifs of 8 bp or shorter.
To assess whether such lower affinity Gcn4p binding sequences play an important role in vivo, we analyzed their association with induction of gene expression. Using all 8mers to score promoters, predicted Gcn4p occupancy and fold induction following amino acid starvation were strongly correlated (Supplementary Fig. 8). But even when scoring only those 8mers identified by HiTS-FLIP as having lower Gcn4p affinity (400nM < Kd < 1μM, n = 143), significant induction of expression was observed for promoters with the highest predicted occupancy (Supplementary Fig. 8), implying that such motifs can have function27. Defining promoters containing high-affinity binding sites (Kd < 400nM, n = 75), lower-affinity sites, both, or neither, we observed distinct responses to GCN4 induction in an amino acid starvation time course28 (Fig. 3d). High-affinity-only promoters exhibited rapid, strong induction, followed by rapid decay, while lower-affinity-only promoters exhibited delayed induction that was more modest in magnitude but more sustained in duration, decaying only gradually. This comparison suggested not only that low-affinity promoters are occupied only at higher Gcn4p concentrations, as expected from standard biophysical models29, but also that the class of genes with high-affinity-only promoters are subject to strong negative feedback regulation. Promoters containing both high- and low-affinity sites were significantly more induced by amino acid starvation than the high-affinity only class, though these groups did not differ in the strength of their high-affinity sites, and similar results were observed following induction of transgenic GCN4 (Supplementary Fig. 9, 10). These observations suggest that lower-affinity sites often augment transcriptional induction as Gcn4p concentration increases, whether paired with strong sites or not. Negative feedback was much more modest following induction of transgenic GCN4 (Supplementary Fig. 10), implicating other components of the amino acid starvation response. Little or no induction was observed for the low-affinity class of genes after amino-acid starvation in GCN4 knockout cells28 (Supplementary Fig. 11), confirming that most or all of the observed effects of these motifs in amino acid starvation are attributable to GCN4.
Sustained amino acid starvation, which is associated with stronger GCN4 induction, presumably requires activation of overlapping but distinct pathways than from transient amino acid starvation, a conclusion that was consistent with Gene Ontology analyses (Supplementary Table 8). For example, amino acid biosynthetic functions were very strongly enriched in genes with high-affinity sites in their promoters generally. However, the lower affinity class of genes was not enriched for these functions, but instead was more modestly enriched for other functions including carbohydrate metabolism (Supplementary Table 8), which may represent functions needed only in the presence of sustained amino acid starvation. Together, our analyses identify a function for specific lower-affinity GCN4-binding motifs (often not detected by PWM models) that is reminiscent of the role of binding site affinity in tuning developmental expression in certain systems30.
The HiTS-FLIP method enables any researcher to convert a sequencing instrument into a powerful tool for studying DNA-protein interactions, providing several unique advantages relative to other methods. First, it provides tens to hundreds of millions of binding measurements, far more than is possible using any other technology, enabling analysis of complex interdependencies between positions and analysis of longer or more complex binding motifs. Methods like HT-Selex5 use second generation sequencing to identify bound sequences, but the binding affinities are inferred by using counts of recovered sequences rather than generating thousands to hundreds of thousands of direct, independent measurements of affinities to every possible k-mer, as in HiTS-FLIP, and require selection, washing and PCR steps between binding and sequencing, which may introduce biases. The ability to measure multiple fluorescent wavelengths could allow hetero- and homo-dimeric forms bound to the flow cell to be measured simultaneously in the same experiment using distinct fluorescent tags on individual proteins. Finally, HiTS-FLIP can be multiplexed, by adding up to 8 different proteins to the 8 lanes of the flow cell prior to sequencing, or adding different proteins sequentially to an entire flow cell after a single sequencing run, which could reduce cost per protein by at least several-fold. We expect that the availability of comprehensive catalogs of DNA binding affinities will deepen our understanding of the mechanisms underlying gene expression.
Supplementary Material
Acknowledgements
The authors thank Gerhard Braus for providing the GCN4 containing plasmids, Neil Clarke for sharing amino acid starvation microarray timecourse data, Amy Keating and Aaron Reinke for advice on EMSA experiments, Thom Theara, Stuart Levine and the Biomicro Center at MIT for help with Illumina sequencing technology and initial experiments, and Laurie Boyer, Neil Clarke, Wendy Gilbert, Phillip Sharp and members of the Burge lab for helpful discussion and comments on the manuscript. This work was supported by a Human Frontiers Science Program long-term fellowship (R. N.), a Computational Science Graduate Fellowship from the Office of Science in the Department of Energy under contract DE-FG02-97ER25308 (R.C.F.), by a major equipment grant from the NSF (no. 0821391) and by grants from the NIH (C. B. B.).
Methods
Protein expression and purification
A carrier vector was used to build the GCN4 fused to mOrange sequence. The GFPGCN4 sequence was PCR amplified from the vector pME2126 provided by the G. H. Braus lab and inserted in the carrier vector between Bgl2 and Not1 restriction sites, with a Spe1 site between the GFP and GCN4 coding sequences. Consequently mOrange missing the stop codon (Clontech) was introduced in place of GFP. The whole mOrange-GCN4 sequence was then cloned into the pET151/D-TOPO vector (Invitrogen) according to the manufacturer's instructions. The final construct generated a 6xHis-mOrange-GCN4 fusion gene that was verified by sequencing and transformed into BL21Star bacteria (Invitrogen). Protein production was induced by 1mM IPTG for 4 hours at 37°C. The protein was purified using a Ni-NTA Fast Start kit (Qiagen) following the manufacturer's protocol. The purity of the protein was verified on a NuPAGE 10% Bis-Tris Urea gel (Invitrogen).
HiTS-FLIP experiment
All steps were performed using standard Illumina protocols, except where noted.
The following oligonucleotides were used:
The library pChip_N25: 5’ P-TAAGN25TAGA 3’
pChip_bot_R: 5’ GAACCGCTCTTCCGATCTA 3’
pChip_top_R: 5’ P-TCGGAAGAGCGGTTCAG 3’
pChip_bot_L: 5’ P-CTTAGATCGGAAGAGCGTCGT 3’
pChip_top_L: 5’ ACACGACGCTCTTCCGATC 3’
mOrange-Gcn4p was quantified using Nanodrop and was diluted to the desired concentrations (1, 5, 25, 125, and 625 nM) in 1x PBS/0.01% Tween/BSA buffer:
| 1x PBS/0.01% Tween | 1940ul |
| BSA (NEB, 10mg/ml) | 60ul |
| Total | 2000ul |
1. Library construction
1.1 pChip_bot_R and pChip_top_R were annealed to form adaptor R; and pChip_bot_L and pChip_top_R were annealed to form adaptor L:
| 10x NEB buffer 2 | 5ul |
| Oligo 1 (100uM) | 12.5ul |
| Oligo 2 (100uM) | 12.5ul |
| H2O | 20ul |
| Total | 50ul |
The samples were heated up at 95°C in a heat block for 5 min, and then the heat block was left to cool down to room temperature.
1.2 The library pChip_N25 was ligated (room temperature; 20min) to the adaptors R and L:
| 2x T4 DNA ligation buffer | 10ul |
| pChip_N25 (25uM) | 2ul |
| adaptor R (25uM) | 2ul |
| adaptor L (25uM) | 2ul |
| T4 DNA ligase | 2ul |
| H2O | 2ul |
| Total | 20ul |
1.3 The library was PCR amplified (12 cycles):
| Ligation mix from step 2 | 1ul |
| 5x phusion buffer | 10ul |
| PE1.0 | 1ul |
| PE2.0 | 1ul |
| 25mM dNTP | 0.5ul |
| Phusion | 0.5ul |
| H2O | 36ul |
| Total | 50ul |
| 98°C 30 sec | |
| 98°C 10 sec | |
| 65°C 30 sec | |
| 72°C 30 sec | |
| 72°C 5 min | |
| 4°C ∞ |
1.4 The PCR product was purified on a 6% TBE PAGE gel. The ~135bp band was eluted out from the gel, ethanol precipitated and quantified by Bioanalyzer.
2. Cluster generation, linearization, blocking and primer hybridization
2.1 The clusters were grown using the Illumina standard protocol, starting from ~3-4pM template to give a density of ~160K/tile.
2.2 The clusters were linearized and blocked using standard protocol.
2.3 The sequencing primer was hybridized using standard protocol.
3. Sequencing
36 cycles of sequencing were performed using standard protocol. At the end of sequencing a final cleavage step was added.
Note: it is imperative not to take out or move the flow cell between the sequencing and protein binding experiments.
4. Double stranded DNA generation
4.1 To avoid the delivery of scan mix before protein imaging, the ImageCyclePump.xml config file (usually within C:\Illumina\SCS2.6\DataCollection\bin\Config) was modified as follows: “<ImageCyclePump On=“true” AutoDispense=“false”>” was changed to “<ImageCyclePump On=“false” AutoDispense=“false”>”.
4.2 1uM of the primer used for dsDNA generation (5’ /5Alex647N/-ACACTCTTTCCCTACACGACGCTCTTCCGATCT 3’) was manually hybridized using receipt “GA2_Manual_ReHyb_v7.xml.”
4.3 The reagent on location 2 of GA was replaced with 1X NEB buffer 2/0.01% Tween buffer:
| 10x NEB buffer 2 | 100ul |
| 10% Tween | 1ul |
| H2O | 899ul |
| Total | 1000ul |
4.4 100ul of 1X NEB buffer 2/0.01% Tween buffer were manually delivered at a rate of 100ul/min from location 2 of GA.
4.5 The reagent on location 2 of GA was replaced with with Resynthesis Mix:
| 10X NEB buffer 2 | 100ul |
| 25mM dNTP mix | 10ul |
| Klenow enzymev(NEB, 5U/ul) | 20ul |
| 10% TWEEN | 1ul |
| H2O | 869ul |
| Total | 1000ul |
4.6 100ul of Resynthesis Mix were manually delivered at a rate of 100ul/min from location 2 of GA.
4.7 The Peltier temperature controller of GA was set to 37C via the manual control tab, and kept at 37C for 30 minutes.
4.8 The Peltier temperature controller was set to 20C via the manual control tab.
5. Protein binding
5.1 The reagent on location 2 of GA was replaced with with 1X PBS/0.01% Tween buffer:
| 1x PBS buffer (Invitrogen) | 9990ul |
| 10% Tween | 10ul |
| Total | 10000ul |
5.2 100ul of 1X PBS/0.01% Tween buffer were manually delivered at a rate of 100ul/min from location 2 of GA.
5.3 The reagent on location 2 of GA was replaced with 1X PBS/0.01% Tween/BSA buffer:
| 1x PBS/0.01% Tween | 1940ul |
| BSA (NEB, 10mg/ml) | 60ul |
| Total | 2000ul |
5.4 100ul of 1X PBS/0.01% Tween/BSA were manually delivered at a rate of 100ul/min from location 2 of GA.
5.5 The reagent on location 2 of GA was replaced with protein solution, starting with the lowest concentration.
5.6 100ul of protein solution were manually at a rate of 50ul/min from location 2 of GA, and incubated at 20°C for 10 mins.
5.7 The reagent on location 2 of GA was replaced with 1X PBS/0.01% Tween buffer.
5.8 100ul of 1X PBS/0.01% Tween buffer were delivered at a rate of 50ul/min from location 2 of GA.
5.9 The flowcell was imaged using recipe “GA2_1Cycle_Protein_Imaging_No Calibration_Findedge_AC-200ms_GT-400ms_v7.xml”.
5.10 Steps 5.1-5.9 were repeated for binding with each protein concentration, from the lowest to the highest.
6. Image processing and mapping
Image analysis and base-calling was performed using version 1.5.1 of the Genome Analyzer software pipeline. The 36 cycles of sequencing were analyzed using the Firecrest and Bustard pipeline components with parameters estimated from a control lane of phiX174 phage DNA using the --control-lane option. Only 25 bases of sequence varied, since nucleotides 1 through 3 and 29 through 36 matched to the adapter sequences. Gcn4p binding cycles were analyzed using Firecrest by appending the images to the end of the sequencing runs, i.e. analyzing 36 sequencing cycles plus 5 protein binding cycles, making 41 total imaging cycles. Coordinates and raw fluorescent intensities for each cluster were extracted from Firecrest output. Fluorescent intensities were matched to sequences for clusters having the same coordinates. Bustard was only run on the 36 sequencing cycles alone, not the protein binding cycles. Only clusters passing default quality filters for all nucleotides (all 36 base calls were made) and mapping clusters in all five Gcn4p concentrations were used for further analysis.
Software
Software and scripts to analyze a set of images from an Illumina sequencing instrument, run the Illumina software pipeline to map protein-binding images to sequencing images, and extract binding intensities and sequences is available at: http://genes.mit.edu/burgelab/hitsflip/
Other information
Details of image normalization, correction for photobleaching, estimation of dissociation constants, EMSA assays and bioinformatic analysis of yeast expression and DNA binding data are described in Supplementary Methods.
Footnotes
Competing Interest Statement
Other than the fact that S.L., I.K., D.S., R.L., L.Z. and G.P.S. are employees of Illumina, Inc., the authors do not have any conflicts of interest with respect to this work.
Supporting Online Material
Supporting Methods and Discussion
Figures S1 – S11
Tables S1 – S8
References
- 1.Klug SJ, Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20:97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- 2.Ren B, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 3.Mukherjee S, et al. Rapid analysis of the DNA-binding specificities of transcription factors with DNA microarrays. Nat Genet. 2004;36:1331–1339. doi: 10.1038/ng1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fordyce PM, et al. De novo identification and biophysical characterization of transcription-factor binding sites with microfluidic affinity analysis. Nat Biotechnol. 2010;28:970–975. doi: 10.1038/nbt.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Natarajan K, et al. Transcriptional profiling shows that Gcn4p is a master regulator of gene expression during amino acid starvation in yeast. Mol Cell Biol. 2001;21:4347–4368. doi: 10.1128/MCB.21.13.4347-4368.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noyes MB, et al. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu C, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottardo R. Modeling and analysis of ChIP-chip experiments. Methods Mol Biol. 2009;567:133–143. doi: 10.1007/978-1-60327-414-2_9. [DOI] [PubMed] [Google Scholar]
- 12.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harbison CT, et al. Transcriptional regulatory code of a eukaryotic genome. Nature. 2004;431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chua G, et al. Identifying transcription factor functions and targets by phenotypic activation. Proc Natl Acad Sci U S A. 2006;103:12045–12050. doi: 10.1073/pnas.0605140103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang H, Johnston M, Mitra RD. Calling cards for DNA-binding proteins. Genome Res. 2007;17:1202–1209. doi: 10.1101/gr.6510207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y, Klutstein M, Simon I, Mitchell T, Bar-Joseph Z. A combined expression-interaction model for inferring the temporal activity of transcription factors. J Comput Biol. 2009;16:1035–1049. doi: 10.1089/cmb.2009.0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Herzog B, Streckfuss-Bomeke K, Braus GH. A Feedback Circuit between Transcriptional Activation and Self-Destruction of Gcn4 Separates Its Metabolic and Morphogenic Response in Diploid Yeasts. J Mol Biol. doi: 10.1016/j.jmb.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 18.Hope IA, Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986;46:885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- 19.Oliphant AR, Brandl CJ, Struhl K. Defining the sequence specificity of DNA-binding proteins by selecting binding sites from random-sequence oligonucleotides: analysis of yeast GCN4 protein. Mol Cell Biol. 1989;9:2944–2949. doi: 10.1128/mcb.9.7.2944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jorgensen P, et al. The size of the nucleus increases as yeast cells grow. Mol Biol Cell. 2007;18:3523–3532. doi: 10.1091/mbc.E06-10-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hill DE, Hope IA, Macke JP, Struhl K. Saturation mutagenesis of the yeast his3 regulatory site: requirements for transcriptional induction and for binding by GCN4 activator protein. Science. 1986;234:451–457. doi: 10.1126/science.3532321. [DOI] [PubMed] [Google Scholar]
- 22.Sellers JW, Vincent AC, Struhl K. Mutations that define the optimal half-site for binding yeast GCN4 activator protein and identify an ATF/CREB-like repressor that recognizes similar DNA sites. Mol Cell Biol. 1990;10:5077–5086. doi: 10.1128/mcb.10.10.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hollenbeck JJ, Oakley MG. GCN4 binds with high affinity to DNA sequences containing a single consensus half-site. Biochemistry. 2000;39:6380–6389. doi: 10.1021/bi992705n. [DOI] [PubMed] [Google Scholar]
- 24.Cranz S, Berger C, Baici A, Jelesarov I, Bosshard HR. Monomeric and dimeric bZIP transcription factor GCN4 bind at the same rate to their target DNA site. Biochemistry. 2004;43:718–727. doi: 10.1021/bi0355793. [DOI] [PubMed] [Google Scholar]
- 25.Man TK, Stormo GD. Non-independence of Mnt repressor-operator interaction determined by a new quantitative multiple fluorescence relative affinity (QuMFRA) assay. Nucleic Acids Res. 2001;29:2471–2478. doi: 10.1093/nar/29.12.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bulyk ML. Analysis of sequence specificities of DNA-binding proteins with protein binding microarrays. Methods Enzymol. 2006;410:279–299. doi: 10.1016/S0076-6879(06)10013-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanay A. Extensive low-affinity transcriptional interactions in the yeast genome. Genome Res. 2006;16:962–972. doi: 10.1101/gr.5113606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prill RJ, et al. Towards a Rigorous Assessment of Systems Biology Models: The DREAM3 Challenges. PloS One. 2010 doi: 10.1371/journal.pone.0009202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gertz J, Siggia ED, Cohen BA. Analysis of combinatorial cis-regulation in synthetic and genomic promoters. Nature. 2009;457:215–218. doi: 10.1038/nature07521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gaudet J, Mango SE. Regulation of organogenesis by the Caenorhabditis elegans FoxA protein PHA-4. Science. 2002;295:821–825. doi: 10.1126/science.1065175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.