Abstract
Background
NFATp is one member of a family of transcriptional activators whose nuclear accumulation and hence transcriptional activity is regulated in mammalian cells. Human NFATp exists as a phosphoprotein in the cytoplasm of naive T cells. Upon antigen stimulation, NFATp is dephosphorylated, accumulates in nuclei, and functions to regulate transcription of genes including those encoding cytokines. While the properties of the DNA binding domain of NFATp have been investigated in detail, biochemical studies of the transcriptional activation and regulated association with nuclei have remained unexplored because of a lack of full length, purified recombinant NFATp.
Results
We developed methods for expressing and purifying full length recombinant human NFATp that has all of the properties known to be associated with native NFATp. The recombinant NFATp binds DNA on its own and cooperatively with AP-1 proteins, activates transcription in vitro, is phosphorylated, can be dephosphorylated by calcineurin, and exhibits regulated association with nuclei in vitro. Importantly, activation by recombinant NFATp in a reconstituted transcription system required regions of the protein outside of the central DNA binding domain.
Conclusions
We conclude that NFATp is a bona fide transcriptional activator. Moreover, the reagents and methods that we developed will facilitate future studies on the mechanisms of transcriptional activation and nuclear accumulation by NFATp, a member of an important family of transcriptional regulatory proteins.
Background
NFAT (Nuclear Factor of Activated T cells) is a family of transcriptional activators that stimulate the expression of genes including those encoding immunomodulatory cytokines [1,2,3,4,5]. The transcriptional effects of individual NFAT family members, including NFATp, NFATc, NFAT3, NFAT4/x, and NFAT5, are beginning to emerge. For example, NFATp, NFATc, and NFAT4 participate in the activation of T and B cells [1, 6]. NFATc appears to be critical for proper cardiac muscle-cell differentiation [7], and NFAT3 functions in cardiac hypertrophy [8]. NFAT4/x has been implicated in development of immature thymocytes [5]. NFAT5 is involved in the transcriptional regulation of osmotic stress response genes [9, 10].
All NFAT proteins except NFAT5 exist as phosphoproteins and are maintained in the cytoplasm of resting cells [2, 3, 11, 12]. NFAT nuclear localization is regulated by the action of a specific phosphatase and a number of kinases [11,12,13,14,15,16,17,18,19,20,21]. In the case of T cells, antigen stimulation elicits a calcium-dependent signaling pathway that results in the activation of calcineurin, which directly dephosphorylates NFATp in preparation for nuclear import [11, 22]. Once in the nucleus, NFAT can bind DNA elements in target promoters, often in association with other resident and co-induced nuclear proteins [1].
Sequence comparison of NFAT proteins revealed that the DNA-binding domain and a region referred to as the NFAT homology region (NHR) share sequence similarity, while other regions of the NFAT proteins share little or no sequence similarity (a schematic of NFATp is shown in Figure 1A) [1]. The DNA-binding domains are similar among all NFAT members [4, 5, 10, 23, 24] and allow NFAT proteins to bind DNA with sequence specificity as monomers [4]. NFAT proteins (except NFAT5) contain NHRs located N-terminal to the DNA binding domain that function to regulate nuclear localization in cells [12, 15, 25]. NFAT proteins are highly phosphorylated in the NHR, and the phosphatase calcineurin binds directly to sequences in the NHR [11,12,13, 15, 19, 25,26,27]. Regions outside of the NHR and DNA binding domain are thought to contain transcriptional activation domains. For example, the N- and C-terminal regions (amino acids 1-171 and 727-927, respectively) of murine NFATp function as activation domains when fused to a heterologous DNA binding domain in transient transfection assays in Jurkat cells [28]. The C-terminal regions of NFAT proteins are unique in sequence and may be responsible for functional differences among NFAT family members.
Figure 1.
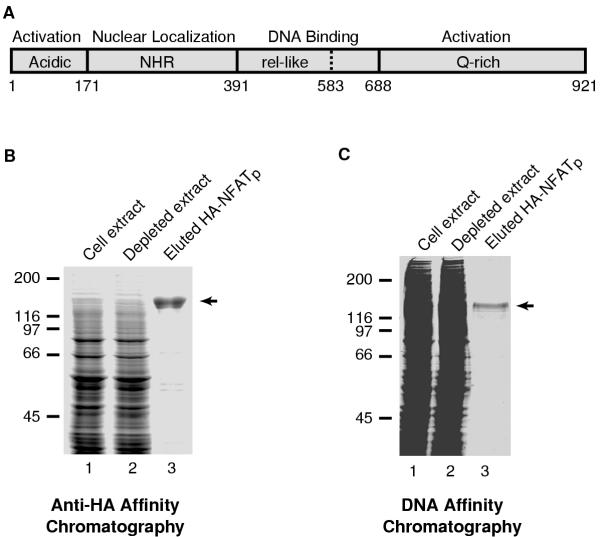
Expression and purification of recombinant HA-NFATp in insect cells. (A) Schematic of the functional domains of NFATp. The DNA binding domain is central to the protein and the minimal DNA binding domain is located between amino acids 391 and 583. The N-terminal region (amino acids 1-390) contains an activation domain rich in acidic amino acids and the region of the protein that binds calcineurin and is involved in regulated nuclear localization. The C-terminal region (amino acids 688-921) is unique to NFATp and contains an activation domain that is rich in glutamines. (B) HA-NFATp was purified by anti-HA affinity chromatography from baculovirus-infected Hi-five cell extracts. Portions of insect cell extract containing over-expressed HA-NFATp (lane 1), depleted extract (lane 2), and the purified/eluted HA-NFATp (lane 3) were resolved by SDS-PAGE and stained with coomassie brilliant blue. The position of full-length HA-NFATp is indicated with an arrow. (C) HA-NFATp was purified by DNA affinity chromatography from baculovirus-infected Hi-five cell extracts. Portions of insect cell extract containing over-expressed HA-NFATp (lane 1), depleted extract (lane 2), and the purified/eluted HA-NFATp (lane 3) were resolved by SDS-PAGE and stained with coomassie brilliant blue. The position of full-length HA-NFATp is indicated with an arrow.
A number of activators can interact with NFAT to potentiate synergistic activation of transcription on NFAT-responsive promoters that characteristically consist of multiple NFAT-specific sites. One example is the interleukin-2 (IL-2) promoter, which contains five NFAT binding sites in a region spanning 250 bp upstream of the TATA box [29, 30]. Four of the IL-2 NFAT sites are part of composite elements consisting of NFAT and AP-1 sites that direct the cooperative binding of NFAT proteins and AP-1 components [30]. In biochemical experiments the DNA-binding domains of NFAT proteins (with the exception of NFAT5) are sufficient for cooperative DNA-binding in association with Fos/Jun heterodimers [4, 10, 11]. As supported by structural analysis of the NFAT/cJun/cFos/DNA complex, physical interactions between the DNA binding domain of NFAT and the basic leucine zipper regions of cJun and cFos enhance their interaction with DNA [31].
To date, biochemical analysis of NFATp and other NFAT members has been conducted using only portions and not the full-length versions of these proteins. Recovery of smaller, soluble, truncated forms of NFAT proteins is feasible relative to the more difficult task of isolating active forms of full-length NFAT proteins. Although such domain-based investigations have been constructive in studying the activity and the properties of the NFAT DNA-binding domains, a biochemical analysis of transcriptional activation and regulated nuclear association of NFAT necessitated the characterization of full-length proteins. This is mainly because the domains exhibiting the transactivation properties and those controlling nuclear association are located in regions flanking the central DNA binding domain. We therefore established protocols for expressing and purifying full-length recombinant human NFATp. Utilizing a reconstituted transcription system, we found that NFATp is a bona fide transcriptional activator and that activation requires regions of NFATp outside the DNA binding domain. In addition, we discovered that human NFATp produced in insect cells using a recombinant baculovirus is a phosphoprotein that demonstrates calcineurin-regulated association with nuclei in vitro.
Results and Discussion
Expression and purification of N-terminally tagged NFATp
We chose to begin our analysis of NFAT proteins in vitro by working with human NFATp (B isoform, amino acids 1-921, Figure 1A). A recombinant baculovirus was created for the expression of full length human NFATp with an N-terminal HA tag. HA-NFATp was purified from baculovirus-infected Hi-five cell extracts using anti-HA affinity chromatography followed by elution with an HA-epitope peptide (Figure 1B). The expressed HA-NFATp protein was visible above background in infected Hi-five cell extracts and was efficiently depleted by the anti-HA resin (Figure 1B). This purification resulted in protein that was nearly homogeneous. We also found that HA-NFATp could be purified from cell extracts by DNA affinity chromatography using an immobilized double stranded oligonucleotide containing two high affinity NFAT binding sites, followed by elution with NaCl (Figure 1C).
HA-NFATp binds DNA alone and cooperatively with cJun/cFos
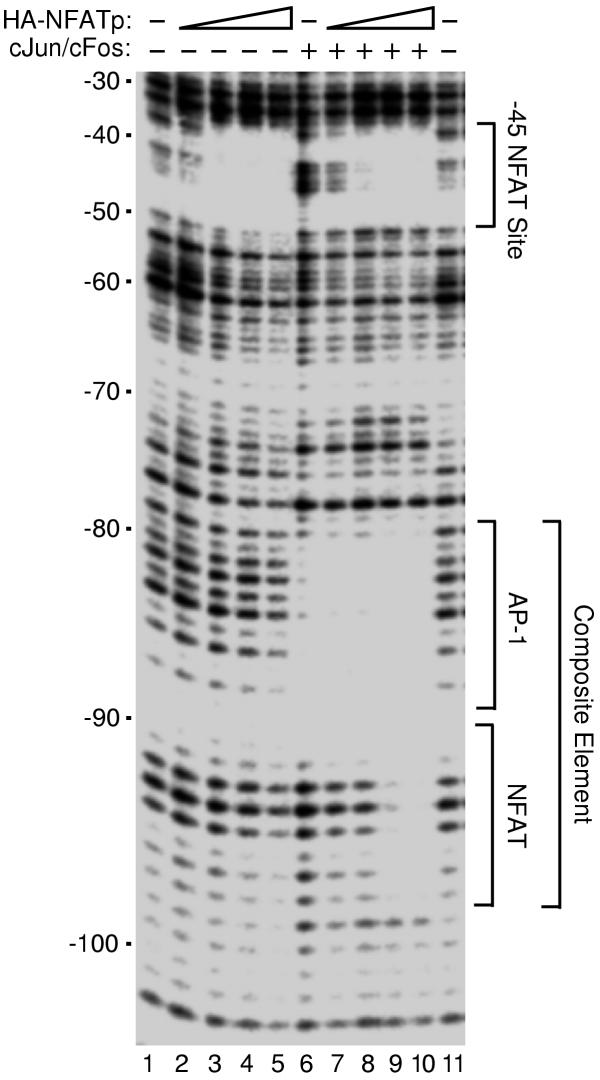
To test the DNA binding activity of recombinant HA-NFATp and the ability to bind DNA cooperatively with cJun/cFos we performed DNase I footprinting. We used a DNA fragment from the human IL-2 promoter that contains a high affinity NFAT site (-45 NFAT site) and a composite element consisting of a low affinity NFAT site and a low affinity AP1 site, to which the DNA binding domain of NFAT has been shown to bind cooperatively with AP-1 proteins [4, 11]. When increasing amounts of HA-NFATp were added to IL-2 DNA in the absence of cJun/cFos, the -45 NFAT site was protected from DNase I digestion, but as expected DNase I digestion of the composite element was unaffected (Figure 2, lanes 1-5). In other studies we found the observed equilibrium dissociation constant (obsKD) of recombinant HA-NFATp for a high affinity NFAT site (mouse IL-4 promoter) to be 1 nM (data not shown). cJun/cFos alone protected the AP-1 site of the composite element (Figure 2, lane 6). When increasing amounts of HA-NFATp were added to IL-2 DNA in the presence of cJun/cFos, clear protection of the entire composite site was observed (Figure 2, lanes 7-10). From DNase I footprinting studies (Figure 2 and data not shown) we conclude that recombinant HA-NFATp binds DNA both on its own and in cooperation with AP-1 proteins (cJun/cFos and cJun/cJun).
Figure 2.
HA-NFATp binds DNA alone and cooperatively with cJun/cFos. DNase I footprinting assays were performed to investigate the binding of HA-NFATp to a region of the human IL-2 promoter in the absence (lanes 1-5) and presence (lanes 6-10) of recombinant human cJun/cFos (3.2 nM). HA-NFATp was added to reactions at the following final concentrations: 3 nM, lanes 2 and 7; 9 nM, lanes 3 and 8; 27 nM, lanes 4 and 9; and 54 nM, lanes 5 and 10. Footprinting reactions were resolved by denaturing PAGE and analyzed with a Molecular Dynamics PhosphorImager. Positions relative to the transcriptional start site (+1) of the human IL-2 promoter are indicated on the left. Locations of the previously characterized high affinity -45 NFAT site and composite element are indicated on the right.
HA-NFATp activates transcription in a reconstituted in vitro system
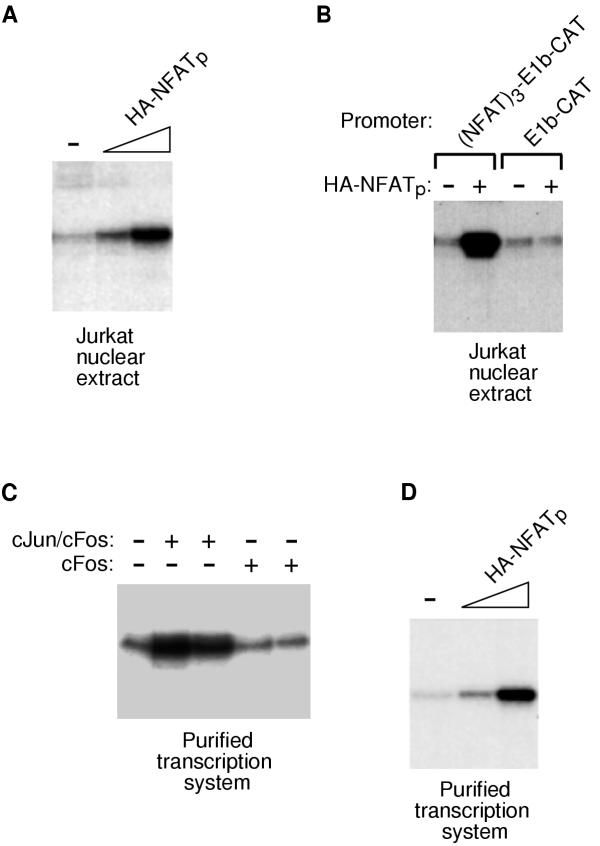
We tested purified HA-NFATp for the ability to activate transcription in vitro. In designing an NFATp-responsive DNA template, we positioned three high-affinity NFAT-binding sites upstream of the adenovirus E1b TATA box and the CAT gene. RNA transcripts produced from the (NFAT)3-E1b-CAT plasmid in vitro can be detected by primer extension. Initial in vitro transcription studies were performed using a nuclear extract from unstimulated Jurkat T cells. As shown in Figure 3A, HA-NFATp activated transcription under these conditions. Activation was dependent on the NFAT sites in the DNA template, since a template lacking NFAT sites did not respond to HA-NFATp (Figure 3B).
Figure 3.
NFATp is a bona fide transcriptional activator. (A) Recombinant HA-NFATp activates transcription in an extract prepared from unstimulated Jurkat cell nuclei. The template used contained three NFAT sites upstream of the E1b TATA box and the CAT gene. Transcripts were analyzed by primer extension using a CAT specific primer followed by denaturing PAGE. The amounts of HA-NFATp used were: 12 ng and 40 ng. (B) Activation by HA-NFATp requires NFAT sites in the DNA template. Basal and NFATp-activated transcription were assessed using unstimulated Jurkat extract and either the (NFAT)3-E1b-CAT template or a template lacking NFAT sites (E1b-CAT). Transcripts were analyzed by primer extension using a CAT specific primer followed by denaturing PAGE. (C) Reconstitution of a purified RNA polymerase II transcription system that is devoid of AP-1 proteins. Transcription was reconstituted from highly purified factors and tested for response to cJun/cFos and cFos alone. The DNA template contained five high affinity AP-1 site upstream of the E1b TATA box and the CAT gene. Transcripts were analyzed by primer extension using a CAT specific primer followed by denaturing PAGE. (D) HA-NFATp activates transcription in a reconstituted transcription system. The DNA template was (NFAT)3-E1b-CAT. The transcription system consisted of highly purified recombinant and native general transcription factors as described in the text.
Although synergistic transcriptional activation presumably resulting from the cooperativity between NFATp and AP-1 has been well established, NFATp transactivation in the absence of AP-1 has not been demonstrated directly. For example, even though the NFAT sites in the DNA template used in the transcription experiments in Figures 3A and 3B were high affinity NFAT binding sites that were not part of a composite element, it remained possible that AP-1 proteins in the nuclear extract contributed to the activation observed. An analysis of the transcriptional properties unique to NFATp required the use of a reconstituted transcription system that is responsive to human NFATp, independent of AP-1 activity. To this end we used an in vitro human RNA polymerase II transcription system reconstituted from highly purified native (n) and recombinant (r) proteins, including (r)TFIIA, (r)TFIIB, (n)TFIID, (r)TFIIE, (r)TFIIF, (n)TFIIH, and (n)RNA polymerase II. Using a promoter containing five high affinity AP-1 sites we found that this transcription system responds well to recombinant cJun/cFos, but does not respond to cFos alone (Figure 3C). This experiment demonstrates that the purified transcription system is largely, if not entirely devoid of AP-1 proteins. When NFATp was added to the reconstituted transcription system it activated transcription to high levels from the DNA template containing three high affinity NFAT sites (Figure 3D). This demonstrates that NFATp is a bona fide transcriptional activator.
Activation domains of NFATp are required for transcriptional activation in vitro
Functional analysis of regions of murine NFATp fused to the yeast GAL4 DNA binding domain using transient transfection assays detected two distinct transactivation domains within the N- and C-terminal regions of NFATp [28]. In this study, however, transcriptional activation by NFATp was not dependent on the identified transactivation domains. The minimal DNA-binding domain of NFATp was sufficient to stimulate reporter gene activation, presumably by recruitment of endogenous AP-1 components through cooperative DNA binding. It is not possible to assess the true transactivation function of NFAT proteins in reporter assays that use a promoter comprised of NFAT/AP-1 composite elements in cells with high levels of endogenous AP-1 proteins. Our finding that recombinant HA-NFATp activates transcription in a reconstituted transcription system provided compelling evidence that NFATp is a bona fide activator. If NFATp acted alone to stimulate transcription in the reconstituted transcription system then the activation we observed would be dependent on the activation domains of NFATp. We therefore applied in vitro biochemical methods for the characterization of NFATp functional domains.
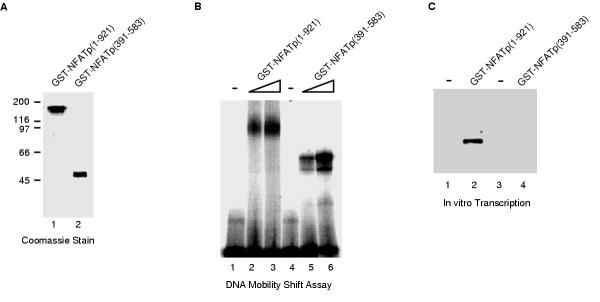
Recombinant baculoviruses were created for the expression of full-length NFATp (amino acids 1-921) and the minimal DNA binding domain of NFATp (amino acids 391-583) fused to GST. GST-NFATp(1-921) and GST-NFATp(391-583) were expressed in insect cells, purified on glutathione sepharose beads, and eluted with glutathione. Figure 4A shows the purity of these two proteins as assessed by SDS-PAGE and staining with coomassie brilliant blue. Both proteins were competent for binding a high affinity NFAT site when tested in electrophoretic mobility shift assays (Figure 4B). The transactivation properties of the two proteins were tested by in vitro transcription using the reconstituted transcription system and a DNA template consisting of three high affinity NFAT sites upstream of the adenovirus major late core promoter and a 200 bp G-less cassette. As shown in Figure 4C, GST-NFATp(1-921) activated transcription, while GST-NFATp(391-583) did not activate transcription. Thus, regions outside of the minimal DNA binding domain were required for transcriptional activation in a reconstituted transcription system. Taken together, our results directly demonstrate that NFATp is a transcriptional activator.
Figure 4.
Transcriptional activation by NFATp in a reconstituted transcription system is dependent on regions outside of the DNA binding domain. (A) GST-fusions to full-length NFATp (amino acids 1-921) and the DNA-binding domain of NFATp (amino acids 391-583) were expressed in insect cells, purified by affinity chromatography, and eluted with glutathione. The purified proteins were analyzed by SDS-PAGE and stained with coomassie brilliant blue. (B) GST-NFATp(1-921) and GST-NFATp(391-583) are active for binding DNA. Proteins were incubated with a double stranded oligonucleotide containing a consensus NFAT-binding site. Protein/DNA complexes and free DNA were resolved on a native 4% polyacrylamide gel, and detected by autoradiography. The final concentrations of GST-NFATp(1-921) and GST-NFATp(391-583) were: 17 nM (lanes 2 and 5) and 35 nM (lanes 3 and 6). (C) GST-NFATp(1-921), but not GST-NFATp(391-583) activates transcription in a reconstituted transcription system. Transcription reactions were performed in a reconstituted RNA polymerase II transcription system using a DNA template consisting of three high affinity NFAT sites upstream of the adenovirus major late core promoter and a 200 bp G-less cassette. 40 ng of either GST-NFATp(1-921) and GST-NFATp(391-583) were added to reactions as indicated. Transcripts were purified, resolved by denaturing PAGE, and visualized with a PhosphorImager.
Recombinant HA-NFATp is phosphorylated and exhibits regulated association with nuclei in vitro
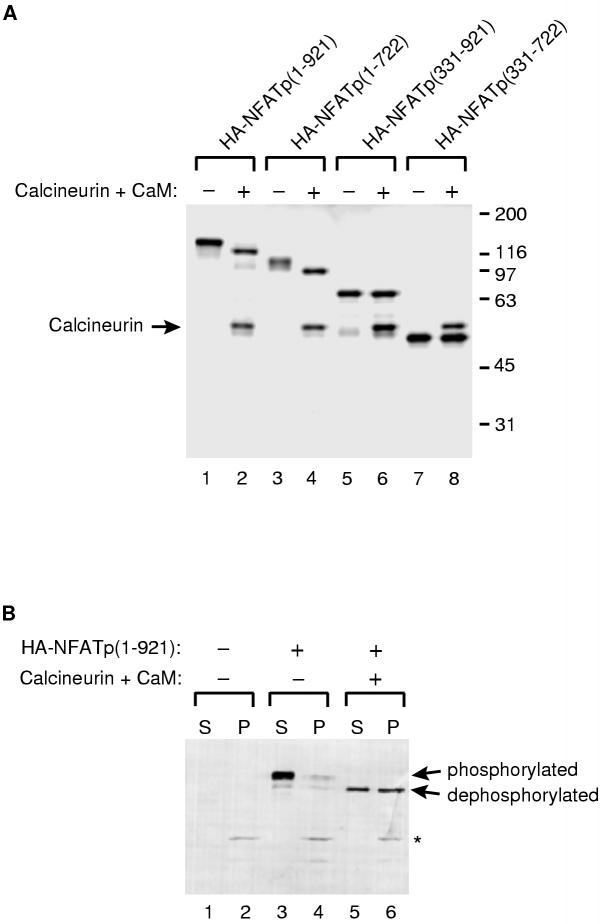
In unstimulated T cells NFATp exists in a highly phosphorylated form that when analyzed by SDS-PAGE has an apparent molecular weight that is significantly larger than its predicted molecular weight [2, 22, 26]. When T cells are stimulated, NFATp is dephosphorylated by calcineurin and it migrates farther on SDS gels than its phosphorylated counterpart. Similarly, when NFATp is purified from unstimulated T cells and subsequently dephosphorylated by calcineurin in vitro, its migration on SDS gels increases (i.e. its apparent molecular weight decreases) [2, 11, 26]. Most of the many phosphorylation sites in NFATp are in the N-terminal region of the protein [15, 27]. Upon purifying recombinant HA-NFATp, we noticed that the protein migrated at an apparent molecular weight similar to the native NFATp in unstimulated T cells (approximately 140 kD when compared to size standards). This led us to hypothesize that the recombinant HA-NFATp purified from baculovirus infected insect cells was phosphorylated. To test this hypothesis and to determine if phosphorylation of a distinct region of HA-NFATp was responsible for aberrant migration in SDS gels, we expressed and purified full-length HA-NFATp and three deletions: HA-NFATp(331-921), HA-NFATp(1-722), and HA-NFATp(331-722). All four proteins were expressed in insect cells from recombinant baculoviruses, purified by anti-HA affinity chromatography, and eluted with HA-epitope peptide. After purification we treated the four proteins with a combination of calcineurin and calmodulin to remove phosphates attached to serines and threonines on the recombinant proteins. The phosphatase treated proteins as well as mock treated controls were resolved by SDS-PAGE and visualized by silver staining. As shown in Figure 5A, the apparent molecular weight of full-length HA-NFATp was significantly decreased by the phosphatase treatment, demonstrating that recombinant HA-NFATp purified from insect cells is phosphorylated. The shift in migration of HA-NFATp was dependent on both calcineurin and calmodulin, and was not efficiently produced with other phosphatases (data not shown). Of the three deletion proteins, only HA-NFATp(1-722) showed a change in migration upon phosphatase treatment; the positions at which both HA-NFATp(331-921) and HA-NFATp(331-722) migrated on SDS gels were not affected by phosphatase treatment. Thus, the N-terminal region of NFATp (amino acids 1-330) was required to observe a phosphorylation-dependent decrease in migration of the recombinant HA-NFATp. This suggested that the major phosphorylation sites on insect cell produced NFATp were in the N-terminal region, as is the case for native NFATp in mammalian T cells [27]. This was confirmed by performing mass spectrometry on peptides recovered from a tryptic digest of recombinant NFATp (W. Clements and J. Goodrich, unpublished data).
Figure 5.
Recombinant HA-NFATp is phosphorylated and displays regulated nuclear association in vitro. (A) HA-NFATp is phosphorylated in its N-terminal region and can be dephosphorylated by calcineurin in vitro. Full length NFATp (lanes 1 and 2) and three deletion mutants (lanes 3-8) were expressed with N-terminal HA-tags using recombinant baculoviruses. All four proteins were purified by anti-HA affinity chromatography and eluted with HA-epitope peptide. The proteins were individually incubated with calcineurin and calmodulin (CaM, even lanes) or mock treated in reactions lacking calcineurin and calmodulin (odd lanes). Proteins were resolved by SDS-PAGE and visualized by silver staining. The position of the calcineurin in the SDS gel is indicated. Note the shift in migration of HA-NFATp(1-921) and HA-NFATp(1-722) upon treatment with phosphatase. (B) HA-NFATp associates with nuclei in vitro only after treatment with calcineurin. HA-NFATp that was pre-treated with calcineurin and CaM (lanes 5 and 6) or mock treated (lanes 3 and 4) was mixed with HeLa nuclei. A control reaction was performed without added HA-NFATp (lanes 1 and 2). After incubation, the nuclei were pelleted by centrifugation and washed. Protein in the pelleted nuclei fractions (P) or supernatant fractions (S) were resolved by SDS-PAGE and analyzed by protein immunoblotting with anti-HA antibody. The positions of phosphorylated HA-NFATp and dephosphorylated HA-NFATp are indicated. The position of a non-specific protein in HeLa nuclei that is recognized by the anti-HA antibody is indicated with an asterisk.
The phosphorylation state of NFATp controls its nuclear localization in cells. Specifically, phosphorylated NFATp is cytoplasmic, and upon dephosphorylation NFATp accumulates in the nucleus [1]. Upon discovering that recombinant HA-NFATp was phosphorylated in its N-terminal region we asked if the phosphorylation state of HA-NFATp would influence its ability to associate with nuclei in an in vitro assay. Nuclei were prepared from HeLa cells. HA-NFATp was either treated with calcineurin and calmodulin or mock treated, and the two proteins were then mixed with the nuclei. After incubation, nuclei were pelleted by centrifugation and thereby separated from unbound protein, which remained in the supernatant. The nuclei were washed, and protein associated with the nuclei as well as protein in the supernatants was solubilized in SDS sample buffer, resolved by SDS-PAGE, and analyzed by protein immunoblotting with anti-HA antibody. As shown in Figure 5B, the majority of phosphorylated HA-NFATp was found in the supernatant and not in the pellet, while approximately 50% of the dephosphorylated NFATp was found associated with nuclei. Moreover, when HA-NFATp(331-722), which was not highly phosphorylated, was tested in the nuclei-association assay it was primarily found in the pellet with the nuclei, regardless of whether it was treated with calcineurin or not (data not shown). Taken together these data lead us to conclude that the phosphorylation state of recombinant HA-NFATp can control its association with nuclei in vitro in a manner that is very similar to the regulation of nuclear association observed for NFAT proteins in cells.
Conclusions
We have found that human NFATp is a bona fide transcriptional activator and that regions outside of the central DNA binding domain are required for NFATp to activate transcription. Our experiments in a reconstituted transcription system lacking contaminating AP-1 proteins and using DNA templates with high affinity NFAT sites demonstrate that NFATp can function as a transcriptional activator on its own. We have also shown that recombinant NFATp purified from insect cells is a phosphoprotein that exhibits association with nuclei in vitro only after dephosphorylation by calcineurin. The recombinant full length and deletion mutants of human NFATp used in the studies described here as well as the methods that we have developed will be valuable for future biochemical studies of the function of NFATp in DNA binding, transcriptional activation, phosphorylation/dephosphorylation, and nuclear association.
Materials and Methods
Plasmids and baculoviruses
A plasmid pREP-NFATp was provided by Tim Hoey (Tularik, Inc.) [4]. pBS-KS+-NFATp was generated by digesting pREP-NFATp with BssH II and Dra I, filling in the ends with Klenow, and ligating the resulting fragment into the Sma I site of pBS-KS+. pVL1392-HAX-NFATp was generated by subcloning a fragment excised from pBS-KS+-NFATp with Spe I and EcoR I into pVL1392-HAX (gift of S. Ruppert and R. Tjian) digested with Spe I and EcoR I. pBS-KS+-NFATp(NdeI), which has an Nde I site, containing the NFATp translational start site ATG, was generated in two steps. A region of NFATp spanning the translational start site ATG and an internal Mro I restriction site was amplified by PCR, creating a product with an Nde I site at the translational start site ATG as well as an upstream Xba I site. The PCR product was digested with Xba I and Mro I and ligated into pBS-KS+-NFATp that had been digested with Xba I and Mro I. pVL1392-HAX-NFATp(1-722) was made by subcloning a Nde I - Msc I fragment from pBS-KS+-NFATp(NdeI) into the Nde I and Sma I sites of pVL1392-HAX. pVL1393-HAX-NFATp(331-921) was made by digesting pBS-KS+-NFATp with SgrA I and filling in the overhang by treatment with Klenow. After further digestion with EcoR I, the NFAT DNA fragment was isolated and ligated into the Sma I and EcoR I sites of pVL1393HAX. A pVL1392-HAX-NFATp(331-722) was made by subcloning a Nde I - Sma I fragment from pGEX-NFATp(331-722) (J. Goodrich, unpublished) into the Nde I and Sma I sites of pVL1392-HAX.
pVL-GST-NFATp was created by subcloning a Nde I - EcoR I fragment containing the NFATp cDNA from pBS-KS+-NFATp(NdeI) into the Nde I and EcoR I sites of pVL1392-GST (gift of S. Ruppert and R. Tjian). pVL-GST-NFATp(391-583) was generated in two steps. First, pBS-NFATp(391-583) was created using single-stranded pBS-KS+-NFATp(NdeI) for site-directed mutagenesis with two oligonucleotides: a unique in-frame ATG codon contained in an Nde I site was created in place of the codon for amino acid 390 using an oligonucleotide of sequence 5'-CATCTGCAGCATCCATATGACTGCATCCCTC-3' and a stop codon was created in the NFATp cDNA after amino acid 583 using an oligonucleotide of sequence 5'-GTTGAAAGACAATAAACAGAATTCTGCCTGGTCTATG-3'. In the second step, the Nde I EcoR I fragment from pBS-NFATp(391-583) was ligated into the Nde I and EcoR I sites of pVL1392-GST.
p(NFAT)3-E1b-CAT, used as template DNA in the in vitro transcription assays, was created by inserting three direct copies of the murine IL-4 high affinity NFAT site (region -82 to -64 of the murine IL-4 promoter) into the Xba I site of plasmid E1b-CAT. p(NFAT)3-MLP-G-less consists of three direct copies of the murine IL-4 high affinity NFAT site (region -82 to -64 of the murine IL-4 promoter) upstream of an adenovirus major late core promoter (-53 to +10) and a 200 bp G-less cassette. To create p(NFAT)3-MLP-G-less a DNA fragment containing the three NFAT sites was recovered from the digestion of the p(NFAT)3-E1b-CAT with Xba I and ligated into the Xba I of pΔML200 (gift of J. Parvin) [32, 33]. p(AP-1)5-E1b-CAT has been described previously [34].
Baculovirus stocks were produced according to standard procedures by individually cotransfecting parental plasmids (described above) along with Baculogold DNA (PharmMingen) into Sf-9 insect cells.
Expression and purification of recombinant proteins
For the preparation of HA-NFATp, Hi-five cells were infected with the HA-NFATp virus for 44 hr at 27°C. Cell were harvested by low-speed centrifugation and resuspended in lysis buffer (20 mM Tris (pH 7.9), 20% (v/v) glycerol, 1 mM EDTA, 0.5 M NaCl, 0.1% NP-40, 0.2 mM PMSF, 1 μg/ml leupeptin, 1.4 μg/ml pepstatin A, and 1 mM DTT). Sonication was performed at 4°C for 5 min before centrifugation at 14,000 rpm for 20-30 min. The recombinant protein was purified on anti-HA-conjugated beads and washed two times in TGEMD buffer (20 mM Tris (pH 7.9), 20% (v/v) glycerol, 1 mM EDTA, 5 mM MgCl2, 0.1% NP-40, 0.2 mM PMSF, 1 μg/ml leupeptin, 1.4 μg/ml pepstatin A, and 1 mM DTT) containing 1 M NaCl and an additional two times in TGEMD buffer containing 0.2 M NaCl. HA-NFATp was subsequently eluted with 1 mg/ml HA peptide resuspended in elution buffer (20 mM Tris (pH 7.9), 20% (v/v) glycerol, 1 mM EDTA, 5 mM MgCl2, 0.2 M NaCl, 0.1% NP-40, 0.2 mM PMSF, 1 μg/ml leupeptin, 1.4 μg/ml pepstatin A, and 1 mM DTT). The eluate was spun through a Millex-GV4 filter, frozen in liquid nitrogen, and stored at -80°C. A similar protocol was used for the expression and purification of HA-NFATp(1-722), HA-NFATp(331-921), and HA-NFATp(331-722).
HA-NFATp was purified from insect cell extracts by DNA affinity chromatography. Insect cell extracts were passed over a column consisting of a double stranded DNA containing two high affinity NFAT sites immobilized on streptavidin sepharose via a biotin at one 5' end. Protein was step-eluted off of the resin with TGEMD buffer containing increasing concentrations of NaCl: 0.2 M, 0.5 M and 1.0 M. Most of the HA-NFATp was found in the 1.0 M NaCl fraction and was highly purified (shown in Figure 1C).
Recombinant GST-NFAT proteins were purified from Hi-five cell extracts (48 hr post-baculovirus infection) by affinity chromatography using glutathione-conjugated sepharose beads. Cell lysates (prepared as described above) were incubated with glutathione sepharose beads for 2 hours at 4°C. This was followed by extensive washes in TGEMD (1.0 M and 0.2 M NaCl). The purified proteins were eluted with reduced glutathione at 1 mg/ml in elution buffer (100 mM Tris (pH 7.9), 120 mM NaCl). The eluted fractions were spun through 0.22 μM Millex filters (Millipore), dialyzed for 6 hours at 4°C in dialysis buffer (20 mM Tris (pH 7.9), 100 mM KCl, 20% (v/v) glycerol, and 1 mM DTT), frozen in liquid nitrogen, and stored at -80°C. The expression and purification of recombinant human cJun/cFos (full length proteins) will be described elsewhere (H. Ferguson and J. Goodrich, manuscript in preparation).
DNase I footprinting
DNase I footprints were performed with a 187 bp DNA fragment containing the -111 to +42 region of the human IL-2 promoter and some surrounding vector DNA. The DNA fragment was generated by PCR and was 32P-labeled on the 5'-end of the nontemplate strand. Reactions contained poly(dG-dC) • poly(dG-dC) as a nonspecific competitor (final concentration of 2 μg/ml). HA-NFATp and cJun/cFos (amounts indicated in Figure 2 legend) were incubated with promoter DNA (final concentration 0.2 nM) for 15 min at 30°C under buffer conditions that were identical to those used for transcription in a reaction volume of 20 μl. 2 μl of a solution containing 0.06 Units/μl DNase I (Promega) and 10 mM CaCl2 was added to each reaction. After a 30 sec incubation at 30°C, reactions were stopped with 40 μl of stop solution containing 25 mM EDTA, 125 mM KCl, and 10 μg of carrier yeast RNA. SDS was added to each reaction to a final concentration of 0.5%. Reactions were incubated at 65°C for 15 min and then placed on ice for 10 min. After a 10 min spin at 14,000 rpm in a microcentrifuge, the supernatants were transferred to new tubes. DNA was ethanol precipitated and resuspended in formamide loading buffer. Products were resolved by 8% denaturing PAGE.
In vitro transcription
Transcription reactions using the reconstituted transcription system were performed as previously described [34] with the following modifications. Each reaction contained 50 ng of either p(NFAT)3-E1b-CAT, or p(NFAT)3-MLP-G-less. Where indicated recombinant NFATp proteins were pre-incubated with promoter DNA for five min at 30°C prior to the addition of the remaining general transcription factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH) and RNA polymerase II. After the addition of general transcription factors, reactions were incubated 30°C for 20 min. Nucleoside triphosphates were added and RNA synthesis was allowed to proceed for 20 min at 30°C. Transcription reactions were stopped, RNA transcripts were processed, and where indicated primer extension was performed as previously described [34].
Electrophoretic mobility shift assays
The binding reactions were performed in a total volume of 10 μl in a buffer consisting of 10 mM Tris (pH 7.9), 50 mM KCl, 10% glycerol, 50 μg/ml BSA, 1 mM DTT, 10 mM Hepes, 5 mM MgCl2, 10 μg/ml calf thymus DNA, and 1.25 fmoles of double-stranded oligonucleotide bearing a high-affinity NFAT site (murine IL-4 promoter from -82 to -64). GST-NFATp(1-921) and GST-NFATp(391-583) were incubated with the template for 20 min at 30°C. Protein/DNA complexes and free DNA were resolved on 4% native polyacrylamide gels in 0.5X TBE for 4 hours at 4°C. Gels were dried on 3 mm chromatography paper (Whatman) and subjected to PhosphorImager analysis.
Dephosphorylation of recombinant NFATp proteins
Recombinant HA-tagged NFATp proteins (approximately 500 ng) were incubated with calcineurin (500 ng, Sigma) and calmodulin (200 ng, Sigma) in 21 μl of buffer that had the following final concentrations of components: 7 mM Tris-HCl, 5 mM Hepes (pH 7.6), 90 mM NaCl, 15 mM betamercaptoethanol, 0.3 mM DTT, 1 mM EDTA, 30% (v/v) glycerol, 3 mM MgCl2, and 1.5 mM MnCl2. Control reactions were performed with NFATp proteins in the absence of added calcineurin and calmodulin. All reactions were incubated at 30°C for 20 min. Products were resolved on and 8% SDS gel and visualized by silver staining.
Nuclei association assays
Nuclei were prepared from HeLa cells by standard procedures [35] and resuspended in 10 packed nuclei volumes of buffer A (10 mM Hepes (pH 7.9) 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 0.5 mM PMSF). For each binding reaction, 5 μl of NFATp dephosphorylation reaction (or a mock treated control reaction) was added to 10 μl of nuclei slurry and allowed to incubate with gentle mixing for 10 min at room temperature. After incubation, nuclei were pelleted by centrifugation in a microfuge at 3000 rpm for 2 min. Supernatants were transferred to new tubes and nuclei were subsequently washed two times with 50 μl of buffer A. Protein in the supernatant and pellet fractions were dissolved in SDS sample buffer, resolved by 8% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-HA antibody.
Acknowledgments
Acknowledgments
We thank Tim Hoey for the NFATp cDNA. J.A.G. is grateful to Robert Tjian and Tom Cech for their generous support, especially during the early stages of this work. This research was supported by a Public Health Service grant GM-55235 from the National Institutes of Health. J.A.G. is currently a Pew Scholar in the Biomedical Sciences and was a Special Fellow of the Leukemia Society of America during the early stages of this work. L.J.K. was supported in part by a NIH Predoctoral Training Grant T32 GM08345 and Beverly Sears Dean's Small Grants (University of Colorado).
Contributor Information
Loree J Kim, Email: loree.kim@u.washington.edu.
Heather A Ferguson, Email: heather.ferguson@colorado.edu.
Anita G Seto, Email: anita.seto@colorado.edu.
James A Goodrich, Email: james.goodrich@colorado.edu.
References
- Rao A, Luo C, Hogan PG. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- McCaffrey PG, Luo C, Kerppola TK, Jain J, Badalian TM, Ho AM, Burgeon E, Lane WS, Lambert JN, Curran T, Verdine GL, Rao A, Hogan PG. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- Northrop JP, Ho SN, Chen L, Thomas DJ, Timmerman LA, Nolan GP, Admon A, Crabtree GR. NF-AT components define a family of transcription factors targeted in T-cell activation. Nature. 1994;369:497–502. doi: 10.1038/369497a0. [DOI] [PubMed] [Google Scholar]
- Hoey T, Sun YL, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- Masuda ES, Naito Y, Tokumitsu H, Campbell D, Saito F, Hannum C, Arai K, Arai N. NFATx, a novel member of the nuclear factor of activated T cells family that is expressed predominantly in the thymus. Mol Cell Biol. 1995;15:2697–2706. doi: 10.1128/mcb.15.5.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakh L, Ghosh P, Rice NR. Expression of NFAT-family proteins in normal human T cells. Mol Cell Biol. 1997;17:2475–2484. doi: 10.1128/mcb.17.5.2475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. The transcription factor NF-ATc is essential for cardiac valve formation. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. A calcineurin-dependent transcriptional pathway for cardiac hypertrophy. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyakawa H, Woo SK, Dahl SC, Handler JS, Kwon HM. Tonicity-responsive enhancer binding protein, a rel-like protein that stimulates transcription in response to hypertonicity. Proc Natl Acad Sci USA. 1999;96:2538–2542. doi: 10.1073/pnas.96.5.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rodriguez C, Aramburu J, Rakeman AS, Rao A. NFAT5, a constitutively nuclear NFAT protein that does not cooperate with Fos and Jun. Proc Natl Acad Sci USA. 1999;96:7214–7219. doi: 10.1073/pnas.96.13.7214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J, McCaffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T cell transcription factor NFATp is a substrate for calcineurin and interacts with the DNA-binding domains of Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- Masuda ES, Liu J, Imamura R, Imai SI, Arai KI, Arai N. Control of NFATx1 nuclear translocation by a calcineurin-regulated inhibitory domain. Mol Cell Biol. 1997;17:2066–2075. doi: 10.1128/mcb.17.4.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Beals CR, Sheridan CM, Turck CW, Gardner P, Crabtree GR. Nuclear export of NF-ATc enhanced by glycogen synthase kinase-3. Science. 1997;275:1930–1934. doi: 10.1126/science.275.5308.1930. [DOI] [PubMed] [Google Scholar]
- Beals CR, Clipstone NA, Ho SN, Crabtree GR. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin- sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- Turner H, Gomez M, McKenzie E, Kirchem A, Lennard A, Cantrell DA. Rac-1 regulates nuclear factor of activated T cells (NFAT) C1 nuclear translocation in response to Fcepsilon receptor type 1 stimulation of mast cells. J Exp Med. 1998;188:527–537. doi: 10.1084/jem.188.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow CW, Rincon M, Cavanagh J, Dickens M, Davis RJ. Nuclear accumulation of NFAT4 opposed by the JNK signal transduction pathway. Science. 1997;278:1638–1641. doi: 10.1126/science.278.5343.1638. [DOI] [PubMed] [Google Scholar]
- Loh C, Carew JA, Kim J, Hogan PG, Rao A. T-cell receptor stimulation elicits an early phase of activation and a later phase of deactivation of the transcription factor NFAT1. Mol Cell Biol. 1996;16:3945–3954. doi: 10.1128/mcb.16.7.3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Burgeon E, Carew JA, McCaffrey PG, Badalian TM, Lane WS, Hogan PG, Rao A. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol Cell Biol. 1996;16:3955–3966. doi: 10.1128/mcb.16.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Shibasaki F, Price R, Guillemot J-C, Yano T, Dotsch V, Wagner G, Ferrara P, McKeon F. Intramolecular masking of nuclear import signal on NFAT4 by Casein Kinase I and MEKK1. Cell. 1998;93:851–861. doi: 10.1016/s0092-8674(00)81445-2. [DOI] [PubMed] [Google Scholar]
- Porter CM, Havens MA, Clipstone NA. Identification of amino acid residues and protein kinases involved in the regulation of NFATc subcellular localization. J Biol Chem. 2000;275:3543–3551. doi: 10.1074/jbc.275.5.3543. [DOI] [PubMed] [Google Scholar]
- Shaw KT, Ho AM, Raghavan A, Kim J, Jain J, Park J, Sharma S, Rao A, Hogan PG. Immunosuppressive drugs prevent a rapid dephosphorylation of transcription factor NFAT1 in stimulated immune cells. Proc Natl Acad Sci USA. 1995;92:11205–11209. doi: 10.1073/pnas.92.24.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain J, Burgeon E, Badalian TM, Hogan PG, Rao A. A similar DNA-binding motif in NFAT family proteins and the Rel homology region. J Biol Chem. 1995;270:4138–4145. doi: 10.1074/jbc.270.8.4138. [DOI] [PubMed] [Google Scholar]
- Ho S, Timmerman L, Northrop J, Crabtree GR. Cloning and characterization of NF-ATc and NF-ATp: the cytoplasmic components of NF-AT. Adv Exp Med Biol. 1994;365:167–173. doi: 10.1007/978-1-4899-0987-9_17. [DOI] [PubMed] [Google Scholar]
- Luo C, Shaw KT-Y, Raghavan A, Aramburu J, Garcia-Cozar F, Perrino BA, Hogan PG, Rao A. Interaction of calcineurin with a domain of the transcription factor NFAT1 that controls nuclear import. Proc Natl Acad Sci USA. 1996;93:8907–8912. doi: 10.1073/pnas.93.17.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaffrey PG, Perrino BA, Soderling TR, Rao A. NF-ATp, a T lymphocyte DNA-binding protein that is a target for calcineurin and immunosuppressive drugs. J Biol Chem. 1993;268:3747–3752. [PubMed] [Google Scholar]
- Okamura H, Aramburu J, Garcia-Rodriguez C, Viola JP, Raghavan A, Tahiliani M, Zhang X, Qin J, Hogan PG, Rao A. Concerted dephosphorylation of the transcription factor NFAT1 induces a conformational switch that regulates transcriptional activity. Mol Cell. 2000;6:539–550. doi: 10.1016/s1097-2765(00)00053-8. [DOI] [PubMed] [Google Scholar]
- Luo C, Burgeon E, Rao A. Mechanisms of transactivation by nuclear factor of activated T cells-1. J Exp Med. 1996;184:141–147. doi: 10.1084/jem.184.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand DB, Shaw J-P, Bush MR, Replogle RE, Belagaje R, Crabtree GR. Characterization of antigen receptor response elements within the interleukin-2 enhancer. Mol Cell Biol. 1988;8:1715–1724. doi: 10.1128/mcb.8.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney JW, Sun YL, Glimcher LH, Hoey T. Novel NFAT sites that mediate activation of the interleukin-2 promoter in response to T-cell receptor stimulation. Mol Cell Biol. 1995;15:6299–6310. doi: 10.1128/mcb.15.11.6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Parvin JD, Timmers HTM, Sharp PA. Promoter specificity of basal transcription factors. Cell. 1992;68:1135–1144. doi: 10.1016/0092-8674(92)90084-p. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Hahn S, Sharp PA, Guarente L. Function of a yeast TATA element binding protein in a mammalian transcription system. Nature. 1988;334:37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- Galasinski SK, Lively TN, Grebe de Barron A, Goodrich JA. Acetyl-CoA stimulates RNA polymerase II transcription and promoter binding by TFIID in the absence of histones. Mol Cell Biol. 2000;20:1923–1930. doi: 10.1128/MCB.20.6.1923-1930.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam JD, Martin PL, Shastry BS, Roeder RG. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]