Abstract
A hypothetical protein is predicted to be expressed from an open reading frame without known experimental evidence of translation. They constitute a substantial fraction of proteomes. Domain extraction from these hypothetical sequences helps to search for protein coding genes for protein structural and functional annotation. We describe the analysis of prediction data in a sequence dataset of hypothetical protein orthologs of Pongo abelii (orangutan) and Sus scrofa (pig). It should be noted that these orangutan-pig orthologs are also non-homologous to human proteins. These predicted data find application in the genome wide annotation of proteins in poorly understood genomes.
Abbreviations
PDB - Protein Data Bank, DEG - Database of Essential Genes, CDD - Conserved Domain Database, IUCN - International Union for Conservation of Nature.
Keywords: Pongo abelii, Sus scrofa, hypothetical proteins, functional annotation, structure prediction, subcellular localization
Background
The sequencing of some major group of organisms allows comparative analysis of genes, gene families and genomes across phylogenetically divergent genus [1]. The completion of the human genome sequence provides a platform for understanding genetic complexity and elucidating genetic variations contributing to diverse traits and diseases [2]. Orangutans (Pongo abelii) prove very useful in further understanding of Human genetic diseases [7]. The Wild Boar (Sus scrofa) is known to be one of the sources of infection for some human diseases [8]. Orangutans are the only primary arboreal great apes, characterized by strong sexual dimorphism and delayed development of mature male features, a long lifespan and the longest inter-birth interval among mammals [4] with 48 chromosomes [5]. The Orangutans are the most phylogenetically distant great apes from humans, thereby providing an informative perspective on hominid evolution. Structural evolution of the Orangutan genome has proceeded much slower than other great apes, which have played a major role in restructuring other primate genomes [6]. Orangutans also develop cardiovascular disease and spontaneous diabetes like humans. It is found that Orangutan genome might help in developing treatments for such conditions by allowing in-depth analysis of the disease’s evolution in great apes [7]. On the other hand, the pig occupies a unique position amongst mammalian species as a model organism of biomedical importance and commercial value worldwide [9]. It shows similarity in size (at 2.7 Gb) [9], shape and physiology to human and has been used as a major mammalian model for many studies concerning xenotransplantation, cardiovascular diseases, blood dynamics, nutrition, organ-specific toxicity, etc. With the improved knowledge of the structure and function of the pig genome in the last two decades it has been found that this animal shares a high sequence and chromosomal structure homology with humans [2].
We describe the comparative genome analyses of two mammalian species, Pongo abelii and Sus scrofa. The great apes, including Pongo abelii, are highly susceptible to many human diseases, some of which can be fatal while others can cause marked morbidity. There is increasing evidence that diseases can be transmitted from humans to free-living habituated apes, sometimes with serious consequences [3]. Wild boars (Sus scrofa) are indigenous in many countries. These free-living swine populations pose not only ecological concerns but also infectious disease concerns. In addition, recreational hunting of wild boars and consumption of wild boar meat in some regions of the world further provide ample opportunities for direct human contacts with wild boars, and thus created a suitable environment for the transmission of pathogens [8]. Antibodies to a number of zoonotic viruses have been detected in wild boar populations, including Hepatitis E virus (HEV), swine influenza virus and Japanese encephalitis virus. Wild boars infected by these zoonotic viruses have the potential to transmit to humans in close contact [8]. Thus, annotation of hypothetical proteins for these organisms is an imperative necessitate which can further be of assistance in identifying potential targets.
Methodology
Genome dataset
The complete protein sequences of Pongo abelii and Sus scrofa were retrieved from the NCBI database (ftp://ftp.ncbi.nih.gov/genomes/). The sequence dataset shows that 25,862 proteins were function annotated and 3,253 were hypothetical for Pongo abelii. In Sus scrofa, 18,305 were function annotated and 1,875 were hypothetical.
Orthologous hypothetical sequences
The hypothetical sequences of Pongo abelii and Sus scrofa were grouped using CD-HIT (http://weizhong-lab.ucsd.edu/cdhit_suite/) at a certain threshold (60% sequence identity cut off). This eliminated redundant sequences with less than 60% identity. These orthologous sequences were then extracted out manually (47 clusters of 107 proteins).
Human non-homologous proteins
The human protein sequences were retrieved from the NCBI database (ftp://ftp.ncbi.nih.gov/genomes/) with 30,685 function-annotated and 2,925 hypothetical. The analogous hypothetical sequences between Pongo abelii and Sus scrofa (107 proteins) were clustered with the annotated sequences of human using CD-HIT. This resulted in 50 related and 57 divergent sequences.
Functional assignment
The 57 divergent proteins were screened for the presence of enzyme related conserved domains using sequence similarity search with close orthologous family members available in various protein databases using CDD BLAST (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi), INTERPROSCAN (http://www.abi.ac.uk/interpro), PFAM (http://www.pfam.sanger.ac.uk/) and TIGRFAMs (http://blast.jcvi.org/web-hmm/).
Functional categorization
Analysis of hypothetical proteins using function prediction tools have shown variable results for conserved domain search. Hence, rough confidence levels such as 100% (all four methods concur), 75% (three methods concur), 50% (2 methods concur) and 25% (others) have been assigned (data not shown).
Protein structure prediction
The structures of the hypothetical protein sequences were predicted using the PS2 Protein Structure Prediction Server (http://www.ps2.life.nctu.edu.tw/) which accepts protein sequences in FASTA format and uses the strategies of pair wise and multiple alignment by combining the powers of the programs PSI-BLAST, IMPALA and T-COFFEE for constructing the protein 3D structures using the best scored orthologous templates. The structures which were not predicted using PS2 Server were then assigned using BlastP against the PDB database.
Essential proteins and sub-cellular localization
The protein sequences were subjected to DEG (http://tubic.tju.edu.cn/deg/) for essential function assignment. Sub-cellular localization tools such as Balanced Sub-cellular Localization Predictor − BaCelLo (http://gpcr.biocomp.unibo.it/bacello/pred.htm), CELLO (http://cello.life.nctu.edu.tw/) and WOLF PSORT (http://wolfpsort.org/) were subsequently used for assigned as shown in Figure 1.
Figure 1.
Flowchart describing identification and analysis of hypothetical orthologs in pig and orangutan.
Discussion
The orthologous hypothetical proteins in the genomes of Pongo abelii and Sus scrofa were described in this study. Computational prediction of protein function, structure and sub cellular localization is a key for genome annotation. We identified 57 hypothetical proteins orthologous between pig and orangutan with human non-homology using the procedure illustrated in Figure 1. This exercise identified enzyme related conserved domains in 36 hypothetical proteins using four web based prediction tools. The remaining 21 hypothetical proteins show no putative conserved domains. Each protein was then classified into a family based on the presence of specific domain in the sequence.
PS2 server predicted the three dimensional structures for 15 sequences using homology search criteria. It should be noted that the remaining 42 hypothetical proteins lacked sufficient templates for homology building. Weak assignments were then made for these sequences using BLASTP search against the PDB database. The dataset is also subjected to the assignment of essential functions (Figure 2) in addition to sub-cellular localization predictions as given in Figure 3 Figure 4and Figure 5. Thus, the role of pig-orangutan orthologous hypothetical proteins as enzymes, essential proteins and localization function is unclearly realized. It should also be noted that a consensus is not seen among the different prediction tools for this dataset. Thus, the predicted data should be cautiously interpreted. Nonetheless, these predicted data provide a framework for understanding genomes through iterative function assignments and annotations.
Figure 2.
Distribution of essential and non-essential hypothetical proteins.
Figure 3.
Sub-cellular localization prediction using BaCelLo.
Figure 4.
Sub-cellular localization prediction using CELLO
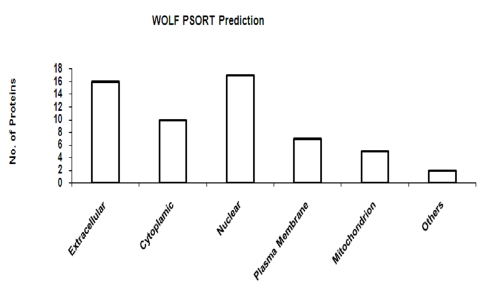
Figure 5.
Subcellular localization prediction using WOLF PSORT.
Conclusion
Prediction of protein function, structure, essentiality and sub-cellular localization for hypothetical proteins is an important component of protein description in genome annotation.
Footnotes
Citation:Jitendra et al, Bioinformation 6(8): 297-299 (2011)
References
- 1.AS Reddy, IS Day. BMC Genomics. 2001;2:2. [Google Scholar]
- 2.N Verma, et al. Proteomics. 2011;11:776. [Google Scholar]
- 3.MH Woodford, et al. Oryx. 2002;36:153. [Google Scholar]
- 4.MA van Noordwijk, CP van Schaik. Am J Phys Anthropol. 2005;127:79. doi: 10.1002/ajpa.10426. [DOI] [PubMed] [Google Scholar]
- 5.Y Fan, et al. Genome Res. 2002;12:1651. doi: 10.1101/gr.337602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DP Locke, et al. Nature. 2011;469:529. [Google Scholar]
- 7. http://www.nature.com/news/2011/110126/full/news.2011.50.html.
- 8.XJ Meng, et al. Philos Trans R Soc Lond B Biol Sci. 2009;364:2697. doi: 10.1098/rstb.2009.0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.AE Hart, et al. Genome Biol. 2007;8:R168. doi: 10.1186/gb-2007-8-8-r168. [DOI] [PMC free article] [PubMed] [Google Scholar]