Abstract
Viral Protein Database is an interactive database for three dimensional viral proteins. Our aim is to provide a comprehensive resource to the community of structural virology, with an emphasis on the description of derived data from structural biology. Currently, VPDB includes ˜1,670 viral protein structures from >277 viruses with more than 465 virus strains. The whole database can be easily accessed through the user convenience text search. Interactivity has been enhanced by using Jmol, WebMol and Strap to visualize the viral protein molecular structure.
Availability
The database is available for free at http://www.vpdb.bicpu.edu.in
Keywords: Database, viral proteins, virus, Jmol, STRAP, WebMol
Background
Viruses have always been a major cause for a large number of infectious diseases. Furthermore, recent emergences of new viral diseases have been of concern in normal human activities. Molecular knowledge of viral protein is thus seen to play an important role for developing improved protein-based vaccines, designing novel anti-viral agents and understanding the entry mechanisms of viruses. Although there exists numerous databases and resources for virus structure information, a comprehensive viral protein structural database is unavailable till the date which can provide detailed sequential as well as structural information on each viral protein (see Table 1). ViperDB [1], VIDA [2], VirusMint [3], and PhEVER [4] are the few databases available online for viral proteins. ViperDB archives the icosahedral virus capsid structures, whereas VIDA incorporates open reading frames of animal virus proteins. The VirusMint database contains protein interactions between viral and human proteins from 490 unique viral proteins of more than 110 different strains. PhEVER is a recently developed database that aims to provide accurate evolutionary and phylogenetic information for the analysis of virus-virus and virus-host lateral gene transfer. There is currently no unique resource available till the date for viral protein structure along with their detailed annotated binding interaction information. To address this problem, we have developed VPDB as a comprehensive derived database from the analysis of a large set of viral proteins obtained from PDB [5], PDBsum [6], and UniProtKB [7]. VPDB comprises of derived data from viral proteins designed to serve as a single-stop solution for retrieval and analysis of viral proteins from all the viral groups and species whose 3- dimensional structure has been solved and present in PDB.
Database design
VPDB is based on three layer structure, namely data layer, middle layer and presentation layer. Data layer is consisted of MySQL, whereas middle layer comprises server side PHP and Java scripts. User friendly web based online presentation layer was developed using PHP, Java, HTML and CSS. The database system is implemented in Red Hat (Linux). It employs Apache as a web-server.
Result and Discussion
VPDB is a user friendly database, which allows the user to input their query text and accordingly select the query type ( PDB IDs or Keywords, Author name or Title of the article and Experimental methods) in order to retrieve the related information from the database. Query related hits are displayed in a result page. User can then select the protein of interest by selecting PDB IDs (Figure 1a) for further detailed information.
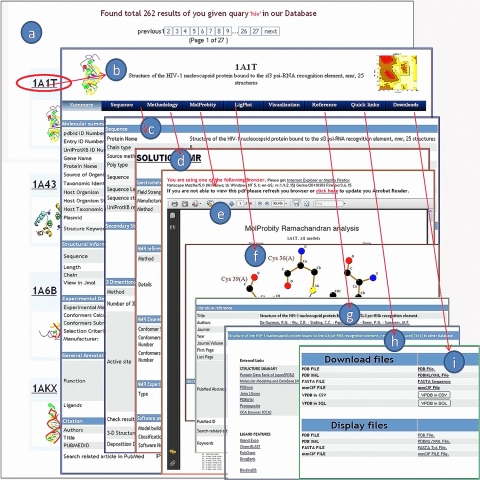
Figure 1.
A snapshot of VPDB web interface, showing detailed results with various tabs and options. (a) the list of the hits as a result of a given query for HIV virus, (b) Summary tab: It contains the detailed information of the selected protein from the list (1A1T), (c) Sequence tab: It contains the sequence information, secondary structure and 3D model, (d) Methodology tab: Parameters for the 3 Dimensional protein building method, (e) MolProbity tab,(f) Ligplot tab, (g) Reference tab: It contains the journal article in detail, (h) Quick links tab, and (i) Download tab.
Database description
Result page has been further classified into nine different tabs. Each tab has its own web page that provides a larger set of data. Summary tab contains the summary of the five sub categories under the five sub headings such as molecular summary, structural summary, experimental details, general annotation and citation (Figure 1b). Sequence tab contains information about the protein sequence, sequence length, chain type, source method, protein sequence in fasta format and UniProtKB reference. Furthermore, secondary structure of protein has been computed using DSSP server [8] for the secondary structural analysis of proteins. Knowledge of the binding pocket is also well known to play an important role for drug designing and existing drug refinement. To make VPDB more comprehensive, the largest binding pocket is identified from the CASTp server based upon the area-volume, although the result shows high but not absolute correlation between pocket volume, and binding affinities. MolProbity tab contains the MolProbity Ramachandran analysis for the three-dimensional protein. It is a web based tool offering quality validation for three-dimensional structure of proteins. The LigPlot tab contains the name of ligand, LigPlot and Nucplot interaction data (Figure 1e). Three java based open-source tools Jmol, WebMol and STRAP has been incorporated in the database for tertiary structure analysis. These tools require a java-enabled browser. Reference tab summarizes the detailed literature information obtained from PubMed. Moreover, Quick links tabs and download tab has been provided for other related database links and to download the database files for viral protein respectively.
Further, for the comparison of viral proteins with the other database, VSE (Virus Search Engines) offers search tools to retrieve the data instantly from source databases. All these information embedded in a single database makes VPDB a better resource for viral protein analysis in comparison to other available virus databases. We believe that VPDB, due to its rich information content will be a useful and novel resource for researchers in the field of structural virology providing an insight into the molecular mechanisms brought about by different viral proteins.
Supplementary material
Acknowledgments
The authors would like to thanks the anonymous reviewers for their valuable comments and Department of Information Technology (DIT), Government of India for their financial support.
Footnotes
Citation:Sharma et al, Bioinformation 6(8): 324-326 (2011)
References
- 1.CM Shepherd, et al. Nucleic Acids Res. 2006;34:D386. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.MM Alba, et al. Nucleic Acids Res. 2001;29:133. doi: 10.1093/nar/29.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A Chatr-aryamontri, et al. Nucleic Acids Res. 2009;37:D669. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.L Palmeira, et al. Nucleic Acids Res. 2011;39:D569. doi: 10.1093/nar/gkq1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. www.pdb.org.
- 6.RA Laskowski. Nucleic Acids Res. 2001;29:221. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.E Boutet, et al. Methods Mol Biol. 2007;406:89. doi: 10.1007/978-1-59745-535-0_4. [DOI] [PubMed] [Google Scholar]
- 8.W Kabsch, C Sander. Biopolymers. 1983;22:2577. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.