Summary
The number of protein-protein complex structures is nearly 6-times smaller than that of tertiary structures in PDB which limits the power of homology-based approaches to complex structure modeling. We present a new threading-recombination approach, COTH, to boost the protein complex structure library by combining tertiary structure templates with complex alignments. The query sequences are first aligned to complex templates using a modified dynamic programming algorithm, guided by ab initio binding-site predictions. The monomer alignments are then shifted to the multimeric template framework by structural alignments. COTH was tested on 500 non-homologous dimeric proteins, which can successfully detect correct templates for half of the cases after homologous templates are excluded, which significantly outperforms conventional homology modeling algorithms. It also shows a higher accuracy in interface modeling than rigid-body docking of unbound structures from ZDOCK although with lower coverage. These data demonstrate new avenues to model complex structures from non-homologous templates.
Keywords: Protein-protein docking, protein structure prediction, protein complex recognition
INTRODUCTION
Many fundamental cellular processes are mediated by protein-protein interactions. The rate of solving complex structures, which constitutes an important step toward a mechanistic understanding of these processes (Russell et al., 2004), by experimental methods has been slow. By examining the sequence space of protein complexes, Aloy and Russell (Aloy and Russell, 2004) estimated the total number of unique interaction types to be around 10,000. Thus, at the current rate of structure determination of unique protein complexes (~200–300 per year), it would take at least two decades before a complete set of protein complex structures is available. These data highlight the urgent need for developing efficient computational methods for protein complex structure prediction, especially when the structures of homologous proteins are not available.
While rapid progress has been made in protein tertiary structure prediction (Kryshtafovych et al., 2009; Moult et al., 2009; Zhang, 2008), the challenges in generating atomic level protein quaternary structures from amino acid sequence has remained relatively unexplored (Aloy et al., 2005; Lensink and Wodak, 2010a; Russell et al., 2004; Vajda and Camacho, 2004). The effort in complex structure modeling has been mainly focused on rigid-body docking of monomer structures (Gray et al., 2003; Hwang et al., 2010; Katchalski-Katzir et al., 1992; Kozakov et al., 2010; Tovchigrechko and Vakser, 2005), with success often depending on the size and shape complementarity of the interface area, and the hydrophobicity of interface residues (Vajda and Camacho, 2004). One of the major challenges in protein-protein docking is the modeling of binding-induced conformational changes (Lensink and Wodak, 2010a; Mendez et al., 2003; Mendez et al., 2005) in which some progress has recently been made with the development of new docking methods, e.g. SnugDock (Sircar and Gray, 2010), MdockPP (Huang and Zou, 2010), ATTRACT (Zacharias, 2005) and others. Progress in this area was also observed in the recent community-wide docking experiments, CAPRI (Fiorucci and Zacharias, 2010; Janin, 2010; Lensink and Wodak, 2010a; Sircar et al., 2010). However, as an inherent limit, protein-protein docking can be performed only when the structures of the component monomers are known.
The second way of constructing protein-protein complex structures is through homology modeling which has attracted considerable attention in recent years (Aloy et al., 2004; Kundrotas et al., 2008; Lu et al., 2002). Aloy et. al. (Aloy et al., 2004) tried to detect the interaction templates using an evolution based method i.e. a template is identified when both the query and template sequences are in the same Pfam family (Finn et al., 2006). Skolnick and coworkers developed MULTIPROSPECTOR (Lu et al., 2002) which first identifies tertiary templates by the monomer threading program PROSPECTOR (Skolnick et al., 2004). If both query chains hit monomers from the same complex, the complex is assigned as a complex template. Kundrotas et. al. (Kundrotas et al., 2008) recently presented HOMBACOP which used a scheme similar as MULTIPROSPECTOR but with the template of each component identified by sequence profile-profile alignments; it requires the native binding information to assist in the alignment adjustments. One drawback of these monomer based threading algorithms is that the cooperativity of multiple-chain alignments, e.g. binding specificity and burial interactions, cannot be correctly accounted for during the course of threading alignments because the alignment result of one chain is independent from that of another chain.
Here, we present a new method, COTH, for protein-protein complex structure predictions, based on co-threading the sequences of both chains simultaneously through the protein quaternary structure library. To boost the capacity of the protein complex library, a monomer-based threading was performed in parallel through the tertiary structure library with the resultant alignments shifted to complex framework by structural alignments. A new ab initio interface predictor, BSpred, was developed to adjust the complex alignment. The algorithms have been tested on two large-scale bound and unbound benchmarks to examine the strength and weakness in comparison with the conventional rigid-body docking and homology modeling methods, which demonstrated promising new avenues to protein complex structural predictions.
RESULTS
Overall results of COTH on testing proteins
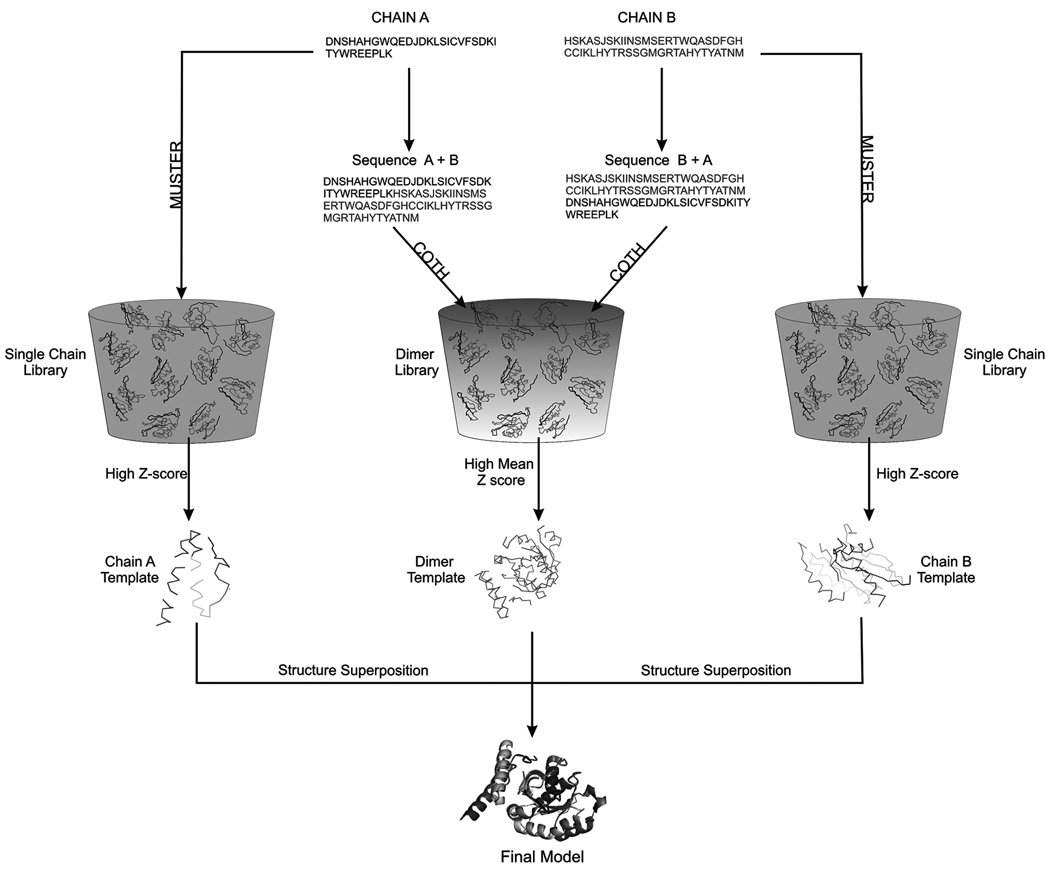
The COTH protocol consists of three consecutive steps: 1) Dimeric threading through multiple-chain complex structure library for chain orientation prediction (called “COTH threading” throughout the article); 2) single-chain threading through tertiary structure library; 3) recombination of tertiary templates and model selection of complex structures (Figure 1). To avoid naming confusion, a list of the programs described in this article is presented in Table 1.
Figure 1. Flowchart of the COTH algorithm for protein complex template identification.
The sequences are first joined in both permutation and threaded against a complex structure library to identify complex templates. Both monomer chains are individually threaded by MUSTER against the tertiary structure library to obtain tertiary structures. The monomer templates are then structurally superposed to the dimer template to generate the final template models. See also Figure S1.
Table 1.
Naming conventions of methods described in this work.
| Name | Description |
|---|---|
| C-PPA | A multiple-chain threading algorithm with scoring function including profile-profile and secondary structure matches. It is an extension of the PPA algorithm for monomer threading (Wu et al., 2007). |
| C-MUSTER | A multiple-chain threading algorithms with scoring function including similar terms to C-PPA, plus multiple structure-based terms derived for torsion-angle and structural profile matches. It is an extension of the MUSTER algorithm for monomer threading (Wu and Zhang, 2008). |
| COTH threading | A multiple-chain threading algorithm with scoring function including similar terms to C-MUSTER, plus the binding site match. The binding sites for targets are predicted by BSpred. |
| COTH | Models are generated by combining the tertiary templates from MUSTER with the quaternary templates from COTH-threading through structure superposition. |
| COTH-exp | Models are generated by superimposing the experimental unbound monomer structures onto the templates from COTH-threading. |
| COTH-model | Models are generated by superimposing the full-length monomer models onto the templates from COTH-threading. The monomer models were predicted by MUSTER with loops filled by MODELLER. |
| ZDOCK-exp | Models are generated by ZDOCK which docks the experimental unbound monomer structures followed by RDOCK refinement. |
| ZDOCK-model | Models are generated by ZDOCK which docks the full-length monomer models predicted by MUSTER and MODELLER, followed by RDOCK refinement. |
To test COTH, we constructed a non-redundant set of 500 dimeric proteins from the PDB, which is also non-redundant to (below 30% in sequence identity with) the 180 training proteins used in algorithm optimization (Materials and Methods). A list of the testing and training proteins is shown at http://zhanglab.ccmb.med.umich.edu/COTH/proteinlist.html. When COTH is conducted, all homologous templates, which have a sequence identity >30% or are detectable by PSI-BLAST with an E-value<0.5 to the query, are excluded from both dimer and monomer template libraries. These criterions are widely used in protein structure predictions for excluding homologous templates (Simons et al., 2001; Zhang and Skolnick, 2004a).
Evaluation of the global template quality is mainly carried out by TM-score (Zhang and Skolnick, 2004b), complex RMSD, and the alignment coverage. TM-score has been extensively used for quality assessment of protein structure predictions because of its ability in combining alignment accuracy and coverage. TM-score was originally developed for monomer. To calculate the TM-score of dimer models, we first convert the dimer structure into an artificial monomer by connecting the C-terminal of the 1st chain and the N-terminal of the 2nd chain, and then run the TM-score program with the length of the query complex sequence as the normalization scale (see Equation 4 in Method). This definition of complex TM-score has the value in [0, 1] and is sensitive to both the topology of individual chain structures and the relative orientation of two components. In general, either the incorrect component structure or the wrong orientation of the components will result in low TM-score. In other words, a high complex TM-score means the correct modeling of both individual chain structures and the relative orientation (Lorenzen and Zhang, 2007a).
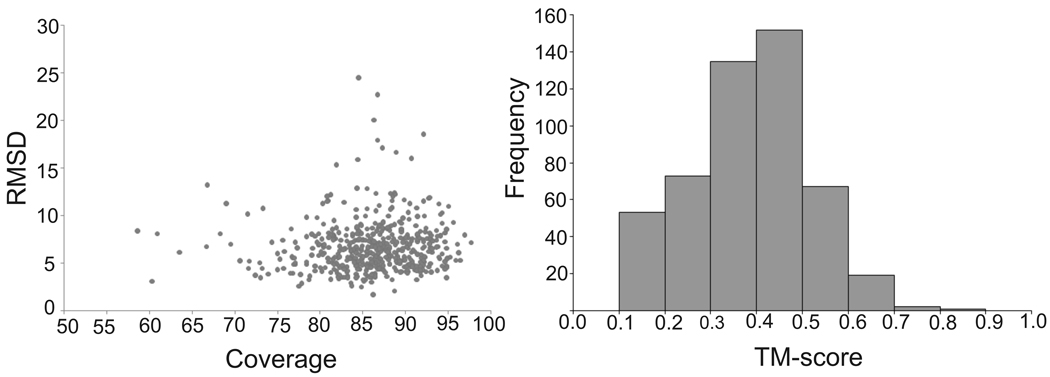
In Figure 2a, we show RMSD versus alignment coverage in the first COTH models. By RMSD here we mean the root-mean-squared-deviation of the threading model and the native structure in the threading aligned region (unless specified, RMSD indicates the global complex RMSD throughout the paper). Even though all homologous templates are excluded, COTH identified notable templates from non-homologous proteins. For example, there are 269 cases (or 293 in the top 10 models) which have the first template with a TM-score >0.4. The average sequence identity between template and query is only 21.2% for the 269 proteins. Despite the low sequence identity, the average alignment coverage is 85.1% and the average RMSD to the native is 5.9 Å in the aligned regions, which demonstrates the ability of COTH to identify non-homologous templates. Alternatively, if we consider templates with an RMSD <6.5Å and the alignment coverage >70% to be reliable, 272 out of 500 targets have reliable templates in the best in top 10 predictions. In Figure 2b we show the distribution of TM-score of the first templates. The majority of targets have templates with a TM-score >0.3, which is significantly higher than the random template selection (TM-score <0.17) (Zhang and Skolnick, 2004b). In the cases where TM-score is in the 0.3–0.4 range, targets often have only the chain orientation correctly predicted but with substantial regions of monomer structures missing or wrongly aligned. This provides opportunities for improvement by further structure refinement based on monomer structure recombination as explored below.
Figure 2. Complex threading results by COTH on 500 non-redundant test proteins.
a) RMSD versus alignment coverage for the best in Top 10 models. b) Histogram of TM-score for the first model.
There are 39 cases, however, which are all hard cases with a significance score of alignments relative to the random, Z-score <2.5 (see Figure S2), where the TM-score of individual ligand and receptor templates are >0.5 but the complex TM-score is <0.4. In these cases, though the quality of the individual chains is good, their predicted orientation is incorrect. The average accuracy for the interface prediction by BSpred is, as expected, poor at only 42.3% with the coverage of 14.9%. We note that among the 39 targets, 21 cases do have templates of correct orientations with a TM-score >0.4 as identified from the complex library by our complex structural alignment algorithm MM-align (Mukherjee and Zhang, 2009) when using the native structure as the probe. Thus, the improvement of the BSpred accuracy in the binding-site predictions is essential to recognize the correct chain orientations for these cases.
Except for the TM-score of the global complex structure, we also assess the modeling quality of protein-protein interface structures, the quality of which is of key importance for the functional annotation of protein complexes. Here, a residue is defined as at the interface if the distance of the Cα atom to any Cα atoms in the counterpart chain is below 10 Å. The interface RMSD, I-RMSD, is the root-mean-squared-deviation of the model and the native structure in the aligned region of the interfaces. The interface coverage, I-cov, is the ratio of the threading aligned interface residues divided by the total number of interface residues in the target. For the 500 targets, the average I-RMSD and I-cov is 12.9 Å and 61.1%, respectively, for the best in the top-5 models (Table 2). This high I-RMSD value is partly due to a few hard cases, which have a very high I-RMSD (>25 Å) because of the completely wrong alignment. If we define a successful threading “hit” as the model which has an I-RMSD ≤ 5Å with at least 50% of the interface residues aligned, there are 186 cases in which COTH generates at least one hit in the top-5 models, despite the exclusion of homologous templates, which represents 37% of the overall sample.
Table 2.
Template identification by different methods on 500 testing proteins.
| Methods | TM-score (first/best in top 5) |
RMSD (coverage)1 | NHit1 | I-RMSD/I-cov1 |
|---|---|---|---|---|
| PSI-BLAST | 0.269/0.293 | 8.19 Å (63.3%) | 124 | 13.7 Å (42.9%) |
| C-PPA | 0.321/0.334 | 5.43 Å (64.8%) | 145 | 13.1 Å (49.3%) |
| C-MUSTER | 0.381/0.412 | 4.51 Å (69.8%) | 161 | 12.8 Å (54.6%) |
| COTH threading | 0.394/0.421 | 4.45 Å (71.0%) | 168 | 12.6 Å (55.8%) |
| COTH | 0.438/0.477 | 4.30 Å (77.6 %) | 186 | 12.9 Å (61.1%) |
Data are shown as the best in top 5 models.
See also Figure S2.
Enzyme-ligand and antigen-antibody are two class of complexes found predominantly in nature. In our testing set there are 236 enzyme-ligand complexes and 169 antigen-antibody complexes. The first COTH templates for enzyme-ligand have an average TM-score 0.441, and an average RMSD 4.1 Å with alignment coverage 86.2%. For the antigen-antibody complexes, the COTH models have an average TM-score 0.410, and an average RMSD 4.6 Å in 86.3% residues. There is a tendency that COTH performs better on enzyme-ligand complexes than antigen-antibody, which somewhat surprisingly coincides with that of the rigid-body docking methods which also performs better on average at docking the enzyme-ligand structures because of the inherent shape complementarity in the complex structures while antigen-antibody interactions have usually larger backbone and side-chain variations in the interfaces (Chen et al., 2003; Comeau et al., 2004; Gray et al., 2003; Katchalski-Katzir et al., 1992; Mendez et al., 2003; Mendez et al., 2005; Tovchigrechko and Vakser, 2006; Vajda and Camacho, 2004). For COTH, however, the higher TM-score is mainly due to the higher conservation of the enzyme-ligand sequence while antigen-antibody complexes can vary greatly in the sequence space. In our test proteins, for example, the average number of sequence homologies as identified by PSI-BLAST from non-redundant sequence databases is 3.12 for enzyme-ligand complexes, which is about two-fold higher than that of antibody-antigen (1.67) and thus allows on average a better construction of sequence profiles for COTH. It should be noted, however, that for both our test proteins and the proteins in the template library, the complexes are represented as dimers although more often than not the antigen-antibody complexes are trimers (the heavy chain and light chain of the antibody and the antigen chain). So, by antigen-antibody complexes here we model only one chain of the antibody (the light chain or the heavy chain) and the antigen chain each time, where the result shown is the average of all antibody chains with the antigen.
Comparison of different alignment algorithms
To have an objective control of the COTH performance, we conduct experiments with other template alignment algorithms which are implemented in the same template library and with the same sequence identity cutoffs. Despite a number of published template detection algorithms, due to the lack of publicly available web-servers or downloadable programs which are capable of predicting protein complex structures based on homology modeling, here we focus our comparison mainly on PSI-BLAST and several in-house developed programs (see Supporting Information, SI).
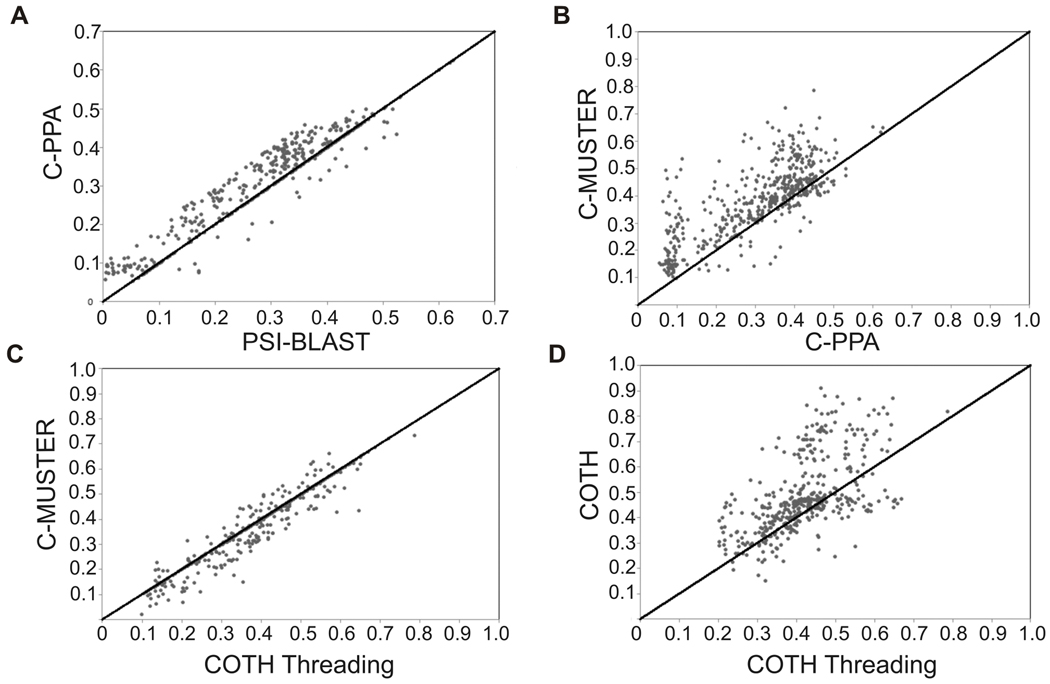
First, PSI-BLAST is a widely-used tool to identify evolutionarily related proteins through iterative sequence-profile alignments (Altschul et al., 1997). Figure 3A shows a comparison of the templates detected by PSI-BLAST and C-PPA, where the latter is a profile-profile alignment method assisted by secondary structure predictions from PSI-PRED (Jones, 1999). In 71% of cases, the C-PPA templates have a higher TM-score than that by PSI-BLAST. The major difference between these two methods is that PSI-BLAST only uses the template sequence while C-PPA uses sequence profile from multiple sequence alignments to represent the templates in the profile-profile alignments, which often contain additional motif conversation signals that aids in the detection of weak evolutionary relationships. Another reason is that C-PPA uses predicted secondary structures (with an accuracy >80%) to assist in adjusting local secondary structure alignments.
Figure 3. Comparison of TM-score of the complex templates as identified by different threading methods.
a) C-PPA versus PSI-BLAST; b) C-MUSTER versus C-PPA; c) C-MUSTER versus COTH threading; d) COTH threading versus COTH.
To test the usefulness of additional structure information in complex template identification, we develop and test C-MUSTER which is a dimeric threading algorithm extended from the monomer threading MUSTER program (Wu and Zhang, 2008). In addition to the profile-profile and secondary structure matches as implemented in C-PPA, C-MUSTER contains multiple structural features predicted from sequences. Figure 3B shows a head-to-head comparison of C-MUSTER and C-PPA. There are obviously more cases (389 versus 92) which are above the diagonal line. The reason for the improvement is that even though sometimes no obvious sequence similarity exists between two proteins, they may share a similar structural framework. Thus, the use of solvent accessibility, torsion angles, structural profile, and hydrophobicity predictions provides insight into the structure of two proteins.
The major difference between C-MUSTER and COTH threading is that COTH threading contains binding site matches from a neural network based prediction algorithm, BSpred. For the 500 testing proteins, the average accuracy of the binding site prediction is 66.8% with the coverage 14.2%. This accuracy is significantly higher than the random prediction (34.2%) with a p-value <10−5. Figure 3C shows the comparison of C-MUSTER versus COTH threading. There are overall 311 cases which have a higher TM-score in the COTH threading alignment than that in C-MUSTER, demonstrating the usefulness of adding the binding-site predictions.
Table 2 summarizes the average TM-score, RMSD, alignment coverage, I-RMSD, I-cov and the number of hits of the template models identified by different methods (PSI-BLAST, C-PPA, C-MUSTER, COTH threading). Compared with PSI-BLAST, C-PPA identifies templates of higher coverage (64.8% versus 63.3%) but with significantly lower RMSD (5.43 Å versus 8.19 Å) which results in a 20% increase in TM-score for the first model. Correspondingly, COTH threading identifies better templates than C-MUSTER and C-PPA in both accuracy and coverage. Overall, the TM-score of COTH threading (0.394) is 46% higher than that by PSI-BLAST (0.269) and there are dominantly more cases with higher TM-score in COTH (427) than in PSI-BLAST. The interface accuracy of the COTH threading is also much higher than PSI-BLAST as indicated by the I-RMSD and I-cov (12.6 Å/55.8% vs. 13.7 Å/42.9%). Again, if we define a hit as I-RMSD < 5 Å with I-cov > 50%, the number of hits in the COTH threading models is 168 which is 35% higher than that by PSI-BLAST (124).
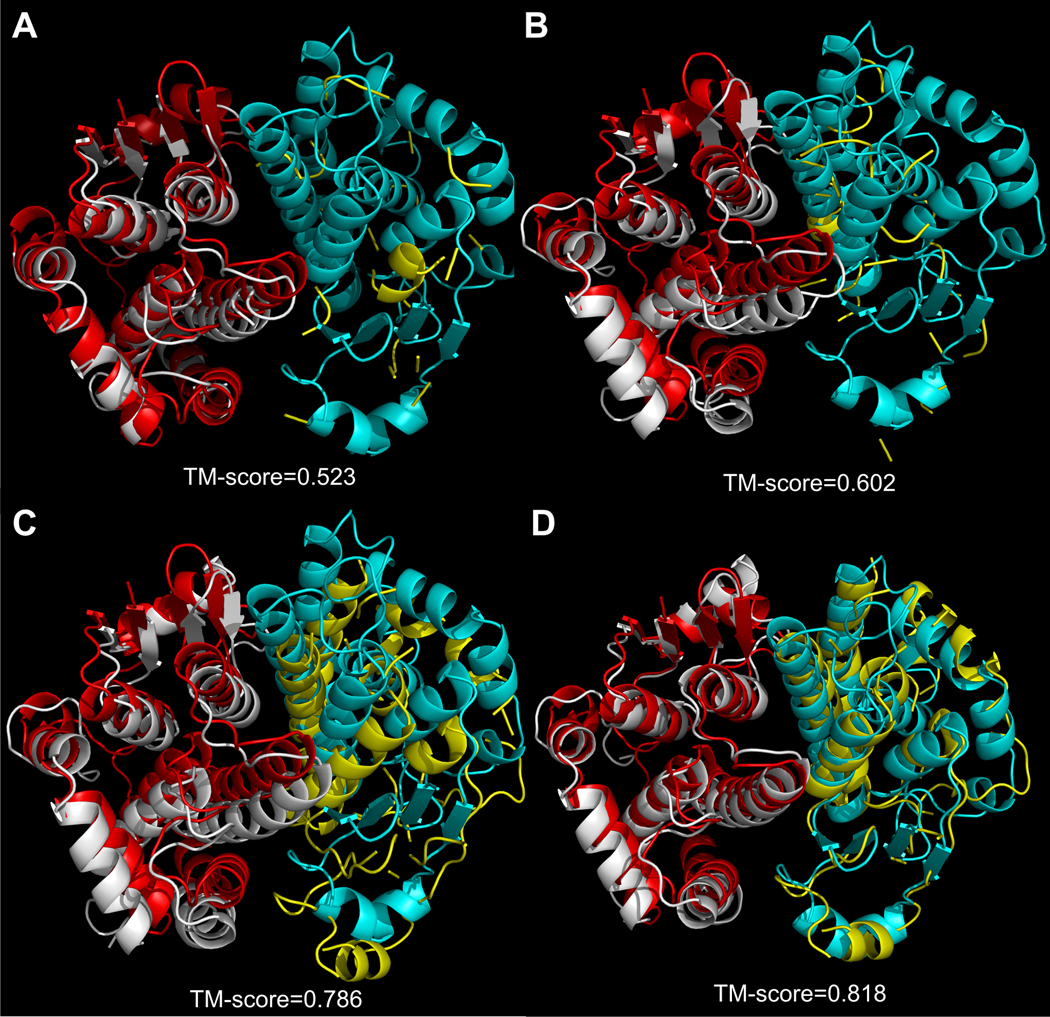
Figure 4 is a typical example of dimer structure (PDB ID: 16gsA0–16gsB0), which reflects the difference of alignments identified by the different methods. First, both PSI-BLAST and C-PPA identify 2c8uA0–2c8uB0 as the best template but C-PPA produces a more accurate alignment and an increased coverage (57.2% for PSI-BLAST and 65.4% for C-PPA) which accounts for the rise in TM-score from 0.523 to 0.602. C-MUSTER identifies 1k3oA0–1k3oB0 as the top template with a sequence identity 25% to the query sequence 16gsA0–16gsB0, which leads to an overall higher coverage 89.9% and a much improved TM-score 0.786. COTH threading, on the other hand, chooses a different protein 1gtaA1–1gtaA2 as the highest scoring template with alignment coverage 94%; the resulting template has the maximum TM-score at 0.818. This better template selection is mainly due to the BSpred binding-site prediction which has an accuracy of 79.4%. The orientation of 1gtaA1–gtaA2 is more similar to the query protein than 1k3oA0–1k3oB0 as identified by the BSpred prediction, which predicts 31 interface residues of the query 16gsA0–16gsB0 and leads to a better alignment reflecting the orientation of the chains correctly.
Figure 4. Examples of improvement over controls.
Template structures produced for the human pi class glutathione transferase by four different threading methods from (a) PSI-BLAST (b) C-PPA (c) C-MUSTER (d) COTH-threading, which have been superimposed onto the experimental structure of the query protein. The experimental structure of 16gsA0–16gsB0 is shown in red for chain 1 and cyan for chain 2 while the models from the threading algorithms are represented in silver for chain1 and yellow for chain 2.
Structure combination of threading templates
Template complexes of similar structures are essential for the COTH threading. However, the algorithm can be constrained due to the limited number of available structures in the complex structure library (currently 6,118 structures at 70% sequence identify cutoff in the PDB. Please refer to Methods section for details). The tertiary structure library, on the other hand, is much larger (38,884 structures at the same cutoff) and hence monomer threading has a much greater scope to identify homologous or analogous structures. In fact, Zhang and Skolnick (Zhang and Skolnick, 2005) demonstrated that the current PDB library is sufficiently complete to solve in principle the protein structure prediction problem for single-domain proteins, i.e. for any single-domain protein there is at least one protein in the PDB which is close to the target protein so that a full-length model of correct topology can be constructed by the template-based modeling methods. Thus, we believe that the tertiary structure of the component chains may be predicted with a better quality by the monomer threading algorithm through tertiary structure library and the quaternary structure prediction should benefit if tertiary templates are combined with the COTH threading frames.
In Figure 3D, we present a head-to-head comparison of the templates by COTH threading versus that by COTH threading followed by monomer structure recombination (called “COTH” instead of “COTH threading” throughout the paper, see naming convention in Table 1). In the latter case, we first identify monomer templates by MUSTER (Wu and Zhang, 2008) using monomer sequence as the query, and identify dimer templates by COTH threading using dimer sequences as the query. In the second step, we superpose the monomer templates on the COTH threading templates by TM-score program (Zhang and Skolnick, 2004b) to obtain the final complex models by combining the monomer and dimer alignments, where all structures in the chain of longer alignment with a steric clash with another chain during structure combination are excluded. For the 1,000 (500×2) testing monomers, the MUSTER templates have a higher TM-score than that from the COTH threading in 893 cases. When combining the MUSTER templates with the COTH threading, in almost all the cases, this structure recombination results in an increase in alignment coverage, while in 399 out of 500 cases, the global RMSD of the complexes decreases despite the increase in alignment coverage. Overall, the TM-score of the final COTH model is higher than the original COTH threading template in 443 cases. The average TM-score of the first COTH model is 0.438, 11% higher than that of the COTH threading templates (Table 2).
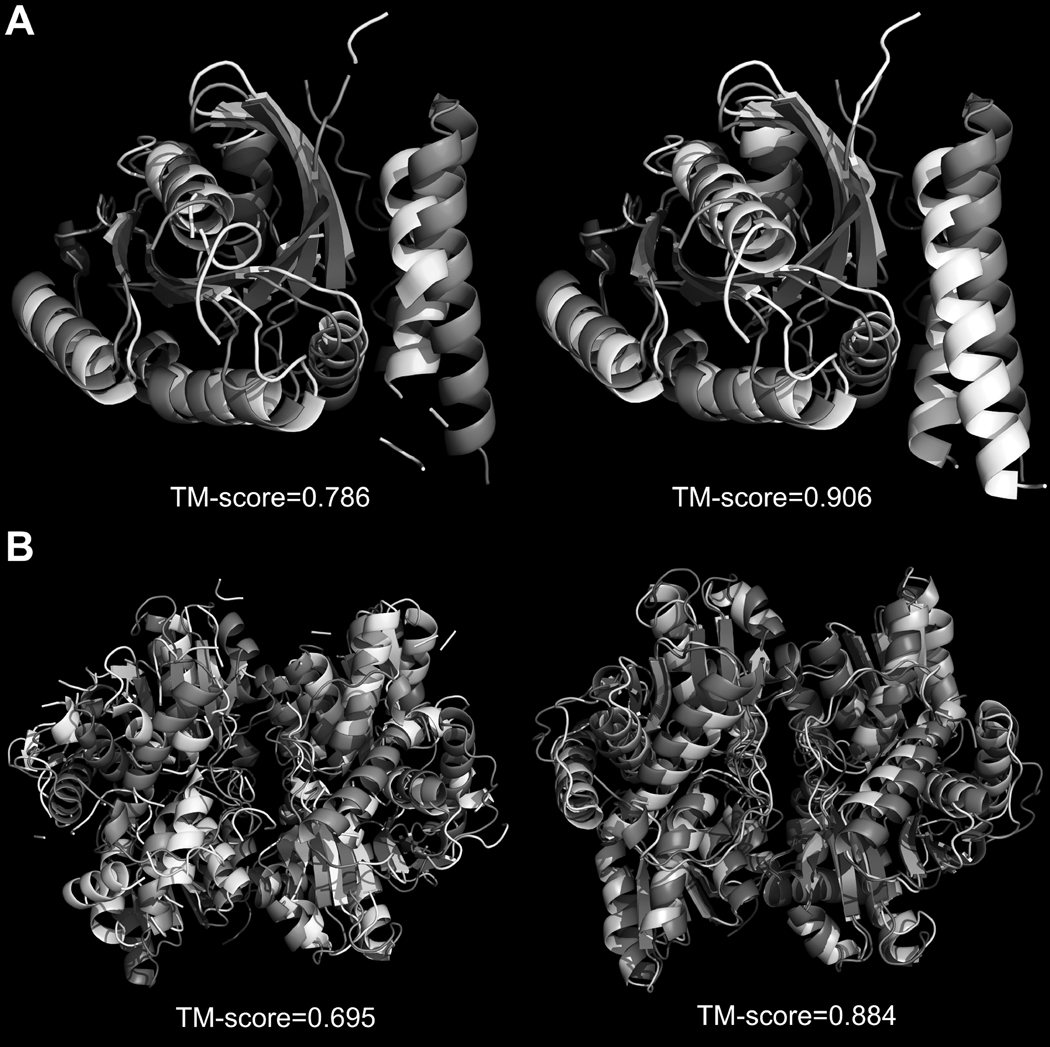
In Figure 5, we cite two typical examples to illustrate the improvement of structure recombination, one is a heterodimer and another is a homodimer. Figure 5A is an example of a near-native heterodimeric structure identified by threading for 1z0kA–1z0kB. The figure on the left shows the first template identified by COTH threading superimposed on the native structure which has a TM-score 0.786 and a RMSD/coverage 2.16Å/86.9%. Despite the correct chain orientation of the template, the alignments of some loops in Chain A and considerable portion of Chain B are missed. The figure on the right is the final template model predicted by COTH. The majority of missed regions in original COTH threading alignment are recuperated through MUSTER alignments with the structural coverage increased from 86.9% to 94.7%; the alignment accuracy is also slightly improved with the RMSD decreased from 2.16 Å to 2.01 Å. This results in an overall TM-score increase from 0.786 to 0.906.
Figure 5. Structure superposition improves template quality.
Superposition of the native structure (darker shade) with the template structures generated by COTH threading (lighter shade, left) and COTH threading plus recombination (lighter shade, right). a) GTP-Bound Rab4Q67L GTPase (PDB ID: 1z0kA0–1z0kB0). b) 1-aminocyclopropane-1-carboxylate deaminase (PDB ID: 1f2dA0–1f2dB0).
The second example is from the homodimer 1f2dA0–1f2dB0 shown in Figure 5B. The dimeric template identified by the COTH threading is extracted from the homodimer 1wdwB0–1wdwD0 which shares a sequence identity 14.5%. The TM-score of this template to native is 0.696 and the RMSD/coverage is 4.02Å/90.7%. MUSTER, on the other hand, identifies 1j0aA from the tertiary structure library as template for both component chains. After the superposition and combination of the MUSTER templates, the TM-score of the complex model increases to 0.884. Again, the MUSTER templates improve both the alignment coverage and the alignment accuracy of COTH, with RMSD/coverage changed to 2.42Å/93.5%.
Here, although COTH uses monomer threading from MUSTER, it is essentially different from the separate monomer-based alignments in many of the former methods (Aloy et al., 2004; Kundrotas et al., 2008; Lu et al., 2002). In these former methods, the single-chain threading is on the monomers extracted from the complex structure library and both monomer and dimer structures are dictated by the dimer structure library. But in COTH, the single-chain threading of MUSTER is through the independent tertiary structure library, which are then recombined with the dimer alignments. Overall, the chain orientation is eventually decided by the dimer threading while the MUSTER single-chain threading serves to improve the quality of monomers and the alignment coverage of the complexes by the use of a nearly 6-fold more complete tertiary structure library.
Comparison of COTH with docking algorithms
Docking and threading-recombination are different approaches to the modeling of protein-protein complex structures. While the goal of the docking algorithms is to find the correct orientation and binding sites of the components given the bound/unbound monomer structures, COTH is designed to generate complex structures from sequences with the aid of template identifications. Nevertheless, it is of interest to examine the overall modeling results of COTH and the well-established rigid-body docking algorithms with the purpose for understanding where the two methods stand in a head to head comparison.
We select ZDOCK (Chen et al., 2003; Li et al., 2003; Wiehe et al., 2007) as a representative example of the rigid-body docking algorithms partly due to its continuing good performances in the CAPRI experiments. The ZDOCK package is also publically downloadable at http://zdock.bu.edu. Because the threading-based methods have only part of the chain with structure predictions while docking is usually performed on full-length structures, to have fair comparisons, we design 4 additional experiments which are all on full-length structures. First, we run ZDOCK on the unbound experimental structures, i.e. running the first step rigid body docking using ZDOCK followed by refinement with RDOCK, which is called “ZDOCK-exp” in Tables 1 and 3. In the second experiment, we constructed full-length models for each individual chain by MUSTER (Wu and Zhang, 2008) and MODELLER (Sali and Blundell, 1993) and then use ZDOCK to dock the full-length models, called “ZDOCK-model” in Tables 1 and 3. In the third experiment, we construct complex structures by superposing the unbound experimental structures of individual chains to the template frame from COTH-threading, called “COTH-exp”. In the fourth experiments, we superpose the full-length model of individual chains modeled by MUSTER and MODELLER onto the COTH-threading template frame, called “COTH-model” in Tables 1 and 3. There were no further refinements conducted in the latter two COTH-based modeling.
Table 3.
Summary of the best in top 10 models on 77 ZDOCK benchmark proteins.
| Methods | Interface-Accuracy (Coverage)1 |
Contacts-Accuracy (Coverage)2 |
NHit3 | Median I- RMSD |
|---|---|---|---|---|
| COTH | 59.8% (31.7%) | 34.2% (33.4%) | 28 | 6.37 Å |
| COTH-exp | 70.2% (39.8%) | 47.4% (42.3%) | 23 | 7.76 Å |
| COTH-model | 63.6% (38.7%) | 40.5% (40.3%) | 21 | 7.92 Å |
| ZDOCK-exp | 67.7% (64.5%) | 46.6% (48.8%) | 26 | 8.29 Å |
| ZDOCK-model | 56.4% (49.7%) | 30.1% (40.4%) | 20 | 9.78 Å |
Accuracy (coverage) of the predicted interface residues.
Accuracy (coverage) of the predicted inter-chain contacts.
Number of hits which have an I-RMSD ≤ 5Å to the native.
See also Tables S1, S2, and S3.
It should be mentioned that the models generated by COTH (and all other threading methods) are Cα only which were copied from the template proteins. But for COTH-exp and COTH-model, since the monomer structures are full-atomic, the final combined models are full-atomic as well (similar to the ZDOCK models).
Table 3 summarizes results (the best in top ten models) of the five methods on 77 dimeric complexes in the ZDOCK Benchmark Set 3.0 (Hwang et al., 2008) (the rest of complexes are higher order oligomers and were thus omitted from this study). Since the unbound monomer structures in docking studies are usually similar to the native, instead of examining TM-score and RMSD of the global structure, here we assess the model quality mainly by the interface structure predictions, in a similar way as the CAPRI experiments (Lensink and Wodak, 2010a; Mendez et al., 2003; Mendez et al., 2005).
Interface residue prediction
For the assessment of the interface residue predictions, we define the Accuracy and Coverage of interface residues as
| (1) |
| (2) |
where an “interface residue” is defined as the residue whose Cα atom lies within 10Å of any Cα atoms of any residues in the opposite chain. Since models constructed from threading are Cα only, we do not use the full-atom definition of interface residue as used in CAPRI (Lensink and Wodak, 2010b). However, since our definition is consistent for all the methods compared here, it should allow for an objective assessment of our method. It is found that COTH-based approaches generally have higher binding-site prediction accuracy, but with lower coverage, than the models by ZDOCK, no matter if we use the experimental unbound structures (70.2% vs. 67.7% accuracy and 39.8% and 64.5% coverage) or the MODELLER models (63.3% vs. 56.4% accuracy and 38.7% and 49.7% coverage) for docking. For the 12 “hard” targets as classified in the ZDOCK benchmark dataset (most are antigen-antibody complexes), for example, the average accuracy of the predicted interface residues is 44.8% with the coverage of 42.6% in the ZDOCK models, while the models constructed by superposition of unbound structures to the COTH templates have an average interface accuracy of 60.3% with the coverage of 30.3%. Of the 12 cases, the ZDOCK models have an accuracy higher than 50% in 4 cases while 7 of the COTH models have the accuracy over 50%.
Interface contact prediction
Since the binding-site prediction accuracy only counts for the total number of the correctly predicted residues in the interface area which nevertheless may interact with incorrect residues of the cross chain in the model, in Column 3 of Table 3 we list the accuracy of the interface contacts predicted for the best in the top 10 models. Similarly, the accuracy of interface contact predictions is defined as the number of the correctly predicted contacts across two chains divided by the total number of cross-chain contacts in the model; the coverage is the number of correctly predicted interface contacts divided by the observed cross-chain contacts in the native structure.
Since threading alignments provide only Cα traces, we defined the inter-chain residue contacts based on amino acid specific 20×20 Cα distance and standard deviation matrices, which were calculated from 6,118 non-redundant dimer structures in our library (see Tables S1 and S2). In the calculations, since the experimental complex structures are full-atomic, we defined the inter-chain residue pairs as contact if the distance of any heavy atoms is below 5 Å. Interestingly, the mean distance of Cα atoms is generally smaller between the same amino acids than that between different amino acid types (Table S1), which indicates that the similar amino acids tend to be packed tighter than the different amino acid pairs. Two residues are predicted to be in contact if the distance between their C-alpha atoms is ≤ (di,j + sdi,j) where di,j is the mean C-alpha distance between residue i and residues j taken from Table S1 and sdi,j is the standard deviation taken from Table S2.
In general ZDOCK, generates models of comparable contact accuracy and coverage as COTH when experimental unbound structures are used for docking and for structure superposition, i.e. 0.466 vs 0.474 for accuracy and 48.8% vs 42.3% for coverage, by ZDOCK and COTH respectively. When the predicted full-length models (by MUSTER + MODELLER) are used, however, the contact accuracy by COTH-model (0.405) is higher by 35% than ZDOCK-model (0.301), where the coverage of the contact predictions by the two methods is similar (40.3% vs. 40.4%). Interestingly, the accuracy of COTH-model, which combines full-length models to the COTH templates, is also better than COTH itself that combines MUSTER threading templates (34.2%). This is mainly due to around 1/3 test cases where the MUSTER threading has substantial gaps in the interface area which reduce the accuracy and coverage of the contact predictions. When the full-length models are constructed, the gapped regions were filled and the overall accuracy and coverage of contacts are increased.
Even using the experimental unbound structures, COTH slightly outperforms ZDOCK in the hard cases when conformational changes are involved in protein-ligand binding (Hwang et al., 2008). In the 12 hard cases, for example, the ZDOCK models have a contact accuracy >50% in 4 cases (2nz8A:B, 2ot3A:B, 1r8sA:E, 2c0lA:B) while the COTH models have an accuracy higher than 50% in 5 cases (1iraY:X, 2ot3A:B, 2c0lA:B, 1ibrA:B, 1pxvA:C). Of the 5 COTH winning cases, only two (2ot3A:B and 2c0lA:B) has the ZDOCK models with a contact accuracy >50%; for the other 2 cases where ZDOCK has an accuracy >50% both the COTH models have a contact accuracy below 50%, which demonstrates that the two methods are essentially complementary to each other in terms of predicting the structure of protein complexes. Again, in all the contact predictions, ZDOCK has generally a higher coverage than COTH.
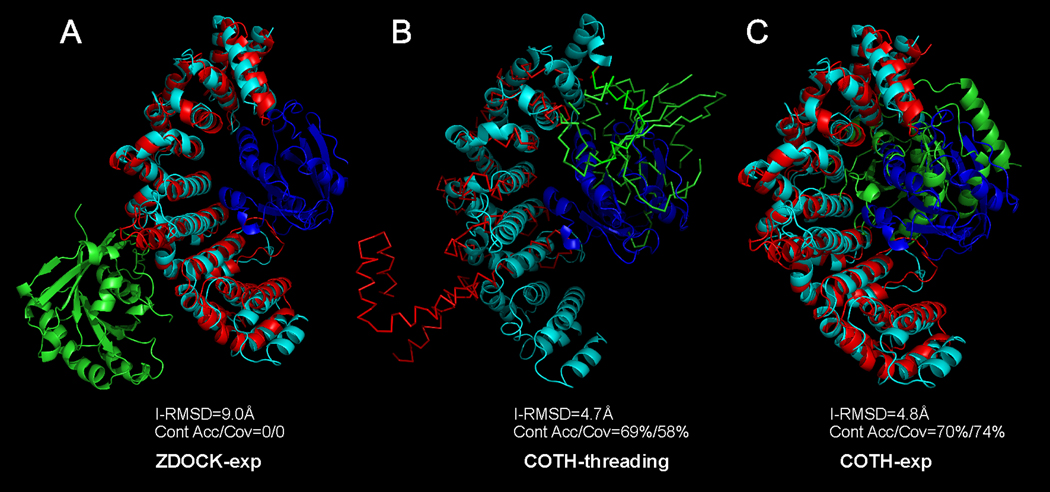
In Figure 6, we show one example of the hard targets from the Ran-Importin beta complex (PDB ID 1ibrA:B). ZDOCK (the best in top 10 models, ranked 5 in this case) put the Ran chain on the convex site of the crescent structure of the Importin beta chain but in the native structure Ran actually binds on the concave site, which resulted in a high I-RMSD (9 Å) with the interface contact accuracy and coverage as 0% (Figure 6A). On the other hand, the COTH-threading (the best in top 10 models, ranked 2 in this case) detected the template of mDIA1-RhoC complex (PDB ID: 1z2c) with a sequence identity 12.4% to the target which has 79.4% of residues aligned. Despite the wrong topology of the C-terminal of the template on the Importin beta chain, the Ran chain was aligned at an approximately correct location of the concave site, which has an I-RMSD=4.7 Å with an interface contact accuracy 68.6% and coverage 57.5% (Figure 6B). When we superposed the experimental unbound structure to the template, we got a complex model of the I-RMSD=4.8 Å, with an interface contact accuracy 70.1% and coverage 74.2%. Because the unbound experimental structures have a closer topology to the target than the COTH-threading template, after the COTH superposition, the global topology of the complex structure is also markedly improved with the overall TM-score increasing from 0.435 to 0.692 and the RMSD decreasing from 5.4 Å to 3.85 Å (Figure 6C).
Figure 6. Modeling result of ZDOCK and COTH on the Ran-Importin beta complex.
The native complex is represented in Cyan (larger chain) and Blue (smaller chain) while the predicted models represented as Red (larger chain) and Green (smaller chain) respectively. (A) ZDOCK-exp; (B) COTH-threading; (C) COTH-exp with unbound experimental structures superimposed on the COTH-threading template.
In general, the ZDOCK model has a higher coverage in the interface and contact predictions. One reason for the difference is that ZDOCK tries to geometrically match the ligand and receptor structures and the contact area of two chains in ZDOCK is usually maximized, while in COTH, the threading alignment is designed to identify the best global structure and chain-orientation match. When the unbound experimental structures or predicted single-chain models are combined with the threading templates, they were simply shifted through superposition to the complex frame without attempt to maximize the geometric contact area of the interface. Therefore, even though the orientation of the monomer chains is correctly modeled in COTH, the coverage of interface contact predictions is usually lower. Further docking refinement simulations, e.g. by backbone displacement and side-chain optimization as done in ROTAFIT (Lorenzen and Zhang, 2007b), may be used to fine-tune the complex structure and improve the interface coverage and contact accuracy. Another factor for the coverage reduction is the alignment gaps in COTH threading which may appear in the interface regions and reduce the residue coverage. This has been partly amended in COTH-exp and COTH-model when full-length structures were used.
Accuracy of interface structure
The accuracy of the interface structure is assessed by the interface RMSD, I-RMSD. A full list of the I-RMSD values by the five methods, COTH, COTH-exp, COTH-model, ZDOCK-exp, ZDOCK-model, is given in Table S3. For all such analysis reported here, the best in top 10 (according to rank) models for each method has been used. The average I-RMSD by different methods is almost randomly distributed due to the large fluctuations of a few high I-RMSD targets. In Column 4 of Table 3, we counted the number of hits in the 77 targets where a hit is defined as a target with I-RMSD<5 Å. For COTH, since gap may involve in the interface area, we request that a hit should have at least 50% of the interface residues aligned. Overall, the number of hits by the four methods with full-length models is similar, ranging from 20 to 26, where ZDOCK is slightly better on experimental unbound structures and COTH has only one more hit on predicted models. The COTH models have the highest number of hits (28) which is partly due to the lower alignment coverage. Again, the COTH-based methods are highly complementary to the docking-based methods. For example, there are only 12 targets commonly hit by both COTH-exp and ZDOCK-exp methods. If we take the top 5 models (according to rank) from each of the methods, the number of hits in the top 10 models will increase from 26 to 33. Meanwhile, there are only 9 targets commonly hit by both COTH-model and ZDOCK-model methods. If we take the top 5 models from each of these two methods, the number of hits in the top 10 models will increases from 21 to 28. In Column 5, we also present the median I-RMSD of the models by different methods, where the COTH based models have generally a lower median I-RMSD than the ZDOCK models.
DISCUSSIONS
We developed a new algorithm for protein complex structure modeling by threading-based template identification and the monomer-dimer alignment combination. The algorithm takes the advantage of the well-established threading alignment methods in protein structure prediction and the complement of tertiary and quaternary structure libraries. The ab initio binding site prediction is further exploited to assist the chain orientation selections.
The COTH method has been tested on two independent sets of protein-protein complexes. In the first test on 500 non-homologous complexes, COTH produces predictions with a TM-score >0.4 (or RMSD<6.5 Å with alignment coverage >70%) for nearly half of the cases when all homologous templates with a sequence identity >30% or detectable by PSI-BLAST with E-value <0.5 are excluded. Detailed comparisons of four different alignment methods show COTH threading with ab initio binding site predictions outperforms C-MUSTER, a direct extension of the tertiary threading algorithm combining multiple structural information; C-MUSTER in turn performs better than the profile-profile based alignments methods, which outperforms the sequence-profile alignment by PSI-BLAST. Overall, the COTH threading, combining the advantages of the profile-profile alignment and multiple-resource structure information, outperforms PSI-BLAST by 46% in TM-score. When combining the tertiary threading alignments, the improvement over PSI-BLAST increase to 63%. Another observed trend in COTH is that the threading-based methods tend to be more reliable for enzyme-ligand complexes as compared to antibody-antigen complexes due to the conservation in sequence profiles in the former.
In the second test of 77 protein complexes from ZDOCK benchmark 3.0, we compared COTH with ZDOCK, which constructs complex structures by docking unbound experimental structures (or predicted full-length monomer models). It is found that COTH performs favorably with a higher accuracy than ZDOCK in predicting the binding-site interface residues; however, the number of interface residues in the COTH prediction is lower. For the interface contact prediction and the accuracy of interface structure represented by interface RMSD, COTH shows a complementary performance with ZDOCK, especially for the hard cases when binding-induced conformational changes are involved. A method to fine-tuning the local position of the COTH threading templates is under construction, which is expected to improve interface match of the complex structures and increase the interface coverage and contact prediction accuracy.
Since COTH has benefited from recombination of monomer threading templates from MUSTER, the algorithm can be further improved by exploiting the meta-server threading approaches. A recent experiment showed that combining templates from multiple threading programs results in at least 7% TM-score increase compared to the best single threading methods (Wu and Zhang, 2007). The COTH method currently takes on average 30 minutes for a medium sized dimer protein of about 400 amino acids on 2.6GHz AMD processors. This efficiency in CPU cost ensures the feasibility of accommodating increasingly larger structure libraries as well as including more single-chain based meta-server threading approaches. It represents also a favor in the speed of calculation compared to the docking methods which usually cost several hours for docking one pair structures.
The COTH algorithm is expected to be used to produce templates for the logical next step of constructing full-length models of protein complexes by building the unaligned gapped regions and refining the complex structures, the development of which is under progress. Thus, COTH not only represents one of the first, fast and reliable methods for predicting template structures of protein complexes from the sequence information, it also has the potential to be used for full-length protein complex structure reassembly by the extension of the tertiary structure assemble method of I-TASSER (Wu et al., 2007; Zhang, 2009). The COTH on-line server is publicly accessible at http://zhanglab.ccmb.med.umich.edu/COTH.
MATERIALS AND METHODS
COTH is a hierarchical threading approach to fold-recognition and structural recombination of protein-protein complexes. For a given complex protein, COTH takes only the amino acid sequences of both chains (i.e. Chain A and B) as the input. It proceeds by joining the chains in both orders, i.e. ChainA-ChainB and ChainB-ChainA, to represent the dimer sequence for template identification. The joined dimeric sequences are then threaded through a representative complex library of the PDB by a process called “COTH threading”, to identify complex templates of similar quaternary structure to the target. Meanwhile, the individual chains of the complex are threaded separately through a representative tertiary structure library by the monomer threading algorithm MUSTER, to identify the monomer templates of similar tertiary structure to the individual target chains. Finally, the top monomer template structures from MUSTER are superimposed onto the top complex templates from COTH-threading, to generate complex structure models which are the output of the COTH pipeline (Figure 1). A detailed description of the procedures is given in Supplementary Information. Here, we briefly explain some of the key steps.
Template libraries
Two libraries were created for COTH. The first is a representative monomer structure library collected from the PDB at the pair-wise sequence identity <70%. Obsolete structures and theoretical models are removed. For multiple domain proteins, both individual domains and the whole proteins are used as the template entries. The second is a non-redundant dimeric structure library screened from DOCKGROUND (Douguet et al., 2006) with the pair-wise sequence identity cutoff at 70% after an initial filtering to remove irregular structures, transmembrane complexes and the complexes with alternate binding modes. Complexes with less than 30 interface residues or with a buried surface area ≤250 Å2 are ignored to rule out possible crystallization artifacts. However, if a new structure has an overall sequence identity >70% to an old structure existing in the library but has one chain sharing less than 70% sequence identity to the corresponding chain of the old structure, the new structure is also included in the library. This helps account for the targets which have big common receptor structures but with different small ligand proteins (often with different orientation). Higher-order complexes are split into dimers by taking all possible dimeric combinations. As of February, 2010, the libraries consist of 38,884 monomer and 6,118 dimer structures.
Single-chain monomeric threading
The single-chain threading is carried out by an extension of the MUSTER algorithm (Wu and Zhang, 2008) through the tertiary structure library. The scoring function of MUSTER is based on the close and remote sequence profile-profile alignments, assisted by the secondary structure predictions, structural profiles accounting for residue depth in the structure, solvent accessibility, torsion angle prediction, and hydrophobic scale.
BSpred
BSpred is a new neural network (NN) program for protein-protein binding residue predictions. It was trained on a set of non-homologous protein complexes that are non-homologous to the testing proteins of this work. The training was conducted in 3 layers with 50 hidden neurons by the standard Back-Propagation algorithm. On a window size of 21 residues, the input training features of BSpred consists of the PSI-BLAST position specific scoring matrix (PSSM), the secondary structure prediction, the solvent accessibility, and the distinctive hydrophobicity of amino acids at interfaces. Based on the observation that interface residues are often sequentially clustered (Ofran and Rost, 2003), a post-process smoothening procedure is introduced, i.e. a residue with NN score>−0.1 is considered as an interface residue only if at least 6 other neighboring residues (from i−3 to i+3) are also predicted to be interface residues. Furthermore, any predicted interface residues, which were not predicted to be solvent exposed by solvent accessibility prediction, are eliminated from the final interface residue list. The BSpred program can be freely downloaded at http://zhanglab.ccmb.med.umich.edu/BSpred.
COTH threading
The alignment of the query and template complexes is generated by a modified dynamic programming algorithm that is designed to avoid unphysical cross alignments (see Figure S1). The scoring function of aligning the ith residue of the query and the jth residue of the template is given by
| (1) |
where ‘q’ stands for the query and ‘t’ for the template. The first term in Eq. 1 represents the sequence-derived profiles where Pcq(i, k) is the frequency of the kth amino acid at the ith position of the multiple sequence alignment by PSI-BLAST at an E-value cutoff of 0.001; Pdq(i, k) is the “remote homology” frequency matrix by PSI-BLAST with E-value<1.0; Lt(j, k) is the PSSM log-odds profile of the template. The second term denotes the secondary structure match and δ(sq(i), st(j)) equals 1 when the secondary structures of i and j are the same and −1 when the secondary structures are different. The third term counts the depth of the aligned residues where Pst(j, k) is the depth dependent structure profile and Lq(i, k) is the PSSM profile of the query. The fourth, fifth and sixth terms compute the match between the solvent accessibility, phi angle and psi angle of the query and the template, respectively. The seventh term counts the hydrophobic match of the residues based on the hydrophobic scoring matrix. The eighth term computes the match between the predicted interface residues of the query by BSpred and the interface residues of the template, where Iq(i) is the interface index of ith query residue (0 or 1) and It(j) is that for jth residue on the template. The last parameter of c8 is introduced to avoid the alignment of unrelated residues in the local regions.
Thus, the COTH threading has 10 free parameters (8 weights in Eq. 1, a gap opening (Go) and a gap extension penalty (Ge)). To determine the parameters, we construct a 10-dimensional parameter space and run COTH on 180 randomly selected non-homologous proteins from DOCKGROUND that are also non-homologous to the test proteins, with parameters taken from each of the grid lattice in the 10-dimension system. The optimal parameters are selected when the highest average TM-score for the 180 training proteins is achieved. As a result, the optimized parameters are: c1=0.80, c2=0.34, c3=1.7, c4=0.29, c5=0.29, c6=0.37, c7=0.20, c8=−4.90, Go=10.11, Ge=0.95.
TM-score of complex structures
TM-score for complex structure prediction is an extension of that for monomers (Zhang and Skolnick, 2004b). To calculate the TM-score of complex structures, the component chains are first tandem connected into artificial single chains. This treatment of complex structures as rigid single-chain structures (rather than two separated chains) will help assess the relative orientation of the chains in the TM-score because the superposition of all chains in this treatment uses the same rotation matrix, while calculating TM-scores by individual chains will result in using different rotation matrices for different chains and thus cannot measure the relative chain orientation. The best structural superposition between the artificial single-chain structures is then identified by maximizing
| (2) |
where Lcomplex is the total length of all chains in the target complex and Lali is the number of the aligned residue pairs. di is the distance of ith pair of Cα atoms after the superposition of model and native structures. . max[…] indicates the optimal superposition to maximize the overall TM-score value.
HIGHLIGHTS.
A novel algorithm for template-based protein-protein complex structure prediction
Significant ability to recognize non-homologous complex templates
Boost protein quaternary structure library by tertiary structure recombination
Complementary to rigid-body protein-protein docking methods
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Dr. Sitao Wu for help in training BSpred, Dr. Thom Verven and Dr. Zhiping Weng for stimulating discussions. This work was supported in part the National Science Foundation (Career Award 1027394); and the National Institute of General Medical Sciences (GM083107, GM084222).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, Gavin AC, Bork P, Superti-Furga G, Serrano L, Russell RB. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- Aloy P, Pichaud M, Russell RB. Protein complexes: structure prediction challenges for the 21st century. Curr Opin Struct Biol. 2005;15:15–22. doi: 10.1016/j.sbi.2005.01.012. [DOI] [PubMed] [Google Scholar]
- Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nat Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Li L, Weng Z. ZDOCK: an initial-stage protein-docking algorithm. Proteins. 2003;52:80–87. doi: 10.1002/prot.10389. [DOI] [PubMed] [Google Scholar]
- Comeau SR, Gatchell DW, Vajda S, Camacho CJ. ClusPro: a fully automated algorithm for protein-protein docking. Nucleic Acids Res. 2004;32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douguet D, Chen HC, Tovchigrechko A, Vakser IA. DOCKGROUND resource for studying protein-protein interfaces. Bioinformatics. 2006;22:2612–2618. doi: 10.1093/bioinformatics/btl447. [DOI] [PubMed] [Google Scholar]
- Finn R, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam:clans, webtools and services. Nucleic Acids Research. 2006:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiorucci S, Zacharias M. Binding site prediction and improved scoring during flexible protein-protein docking with ATTRACT. Proteins. 2010;78:3131–3139. doi: 10.1002/prot.22808. [DOI] [PubMed] [Google Scholar]
- Gray JJ, Moughon S, Wang C, Schueler-Furman O, Kuhlman B, Rohl CA, Baker D. Protein-protein docking with simultaneous optimization of rigid-body displacement and side-chain conformations. J Mol Biol. 2003;331:281–299. doi: 10.1016/s0022-2836(03)00670-3. [DOI] [PubMed] [Google Scholar]
- Huang SY, Zou X. MDockPP: A hierarchical approach for protein-protein docking and its application to CAPRI rounds 15–19. Proteins. 2010;78:3096–3103. doi: 10.1002/prot.22797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Pierce B, Mintseris J, Janin J, Weng Z. Protein-protein docking benchmark version 3.0. Proteins. 2008;73:705–709. doi: 10.1002/prot.22106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang H, Vreven T, Pierce BG, Hung JH, Weng Z. Performance of ZDOCK and ZRANK in CAPRI rounds 13–19. Proteins. 2010;78:3104–3110. doi: 10.1002/prot.22764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janin J. The targets of CAPRI Rounds 13–19. Proteins. 2010;78:3067–3072. doi: 10.1002/prot.22774. [DOI] [PubMed] [Google Scholar]
- Jones DT. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- Katchalski-Katzir E, Shariv I, Eisenstein M, Friesem AA, Aflalo C, Vakser IA. Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques. Proc Natl Acad Sci U S A. 1992;89:2195–2199. doi: 10.1073/pnas.89.6.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D, Hall DR, Beglov D, Brenke R, Comeau SR, Shen Y, Li K, Zheng J, Vakili P, Paschalidis I, Vajda S. Achieving reliability and high accuracy in automated protein docking: ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins. 2010;78:3124–3130. doi: 10.1002/prot.22835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryshtafovych A, Fidelis K, Moult J. CASP8 results in context of previous experiments. Proteins. 2009;77 Suppl 9:217–228. doi: 10.1002/prot.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundrotas P, Lensink M, Alexov E. Homology based modelling of 3D structures of protein complexes using alignments of modified sequence profiles. International Journal of Biological Macromolecules. 2008;43:198–208. doi: 10.1016/j.ijbiomac.2008.05.004. [DOI] [PubMed] [Google Scholar]
- Lensink M, Wodak S. Docking and scoring protein interactions: CAPRI 2009. Proteins. 2010a doi: 10.1002/prot.22818. [DOI] [PubMed] [Google Scholar]
- Lensink MF, Wodak SJ. Blind predictions of protein interfaces by docking calculations in CAPRI. Proteins. 2010b;78:3085–3095. doi: 10.1002/prot.22850. [DOI] [PubMed] [Google Scholar]
- Li L, Chen R, Weng Z. RDOCK: refinement of rigid-body protein docking predictions. Proteins. 2003;53:693–707. doi: 10.1002/prot.10460. [DOI] [PubMed] [Google Scholar]
- Lorenzen S, Zhang Y. Identification of near-native structures by clustering protein docking conformations. Proteins. 2007a;68:187–194. doi: 10.1002/prot.21442. [DOI] [PubMed] [Google Scholar]
- Lorenzen S, Zhang Y. Monte Carlo refinement of rigid-body protein docking structures with backbone displacement and side-chain optimization. Protein Sci. 2007b;16:2716–2725. doi: 10.1110/ps.072847207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L, Lu H, Skolnick J. MULTIPROSPECTOR: An algotihm for the predictio of protein-protein interactions by multimeric threading. Proteins: Structure, Function and Genetics. 2002;49:350–364. doi: 10.1002/prot.10222. [DOI] [PubMed] [Google Scholar]
- Mendez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- Mendez R, Leplae R, Lensink MF, Wodak SJ. Assessment of CAPRI predictions in rounds 3–5 shows progress in docking procedures. Proteins. 2005;60:150–169. doi: 10.1002/prot.20551. [DOI] [PubMed] [Google Scholar]
- Moult J, Fidelis K, Kryshtafovych A, Rost B, Tramontano A. Critical assessment of methods of protein structure prediction-Round VIII. Proteins-Structure Function and Bioinformatics. 2009;77:1–4. doi: 10.1002/prot.22589. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Zhang Y. MM-align: a quick algorithm for aligning multiple-chain protein complex structures using iterative dynamic programming. Nucleic Acids Res. 2009;37:e83. doi: 10.1093/nar/gkp318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ofran Y, Rost B. Predicted protein-protein interaction sites from local sequence information. FEBS Lett. 2003;544:236–239. doi: 10.1016/s0014-5793(03)00456-3. [DOI] [PubMed] [Google Scholar]
- Russell RB, Alber F, Aloy P, Davis FP, Korkin D, Pichaud M, Topf M, Sali A. A structural perspective on protein-protein interactions. Curr Opin Struct Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Simons KT, Strauss C, Baker D. Prospects for ab initio protein structural genomics. J. Mol. Biol. 2001;306:1191–1199. doi: 10.1006/jmbi.2000.4459. [DOI] [PubMed] [Google Scholar]
- Sircar A, Chaudhury S, Kilambi KP, Berrondo M, Gray JJ. A generalized approach to sampling backbone conformations with RosettaDock for CAPRI rounds 13–19. Proteins. 2010;78:3115–3123. doi: 10.1002/prot.22765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sircar A, Gray JJ. SnugDock: paratope structural optimization during antibody-antigen docking compensates for errors in antibody homology models. PLoS Comput Biol. 2010;6 doi: 10.1371/journal.pcbi.1000644. e1000644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skolnick J, Kihara D, Zhang Y. Development and large scale benchmark testing of the PROSPECTOR 3.0 threading algorithm. Protein. 2004;56:502–518. doi: 10.1002/prot.20106. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. Development and testing of an automated approach to protein docking. Proteins. 2005;60:296–301. doi: 10.1002/prot.20573. [DOI] [PubMed] [Google Scholar]
- Tovchigrechko A, Vakser IA. GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 2006;34:W310–W314. doi: 10.1093/nar/gkl206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vajda S, Camacho CJ. Protein-protein docking: is the glass half-full or half-empty? Trends Biotechnol. 2004;22:110–116. doi: 10.1016/j.tibtech.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Wiehe K, Pierce B, Tong WW, Hwang H, Mintseris J, Weng Z. The performance of ZDOCK and ZRANK in rounds 6–11 of CAPRI. Proteins. 2007;69:719–725. doi: 10.1002/prot.21747. [DOI] [PubMed] [Google Scholar]
- Wu S, Skolnick J, Zhang Y. Ab initio modeling of small proteins by iterative TASSER simulations. BMC Biol. 2007;5:17. doi: 10.1186/1741-7007-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zhang Y. MUSTER: Improving protein sequence profile-profile alignments by using multiple sources of structure information. Proteins. 2008;72:547–556. doi: 10.1002/prot.21945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu ST, Zhang Y. LOMETS: A local meta-threading-server for protein structure prediction. Nucl. Acids. Res. 2007;35:3375–3382. doi: 10.1093/nar/gkm251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias M. ATTRACT: protein-protein docking in CAPRI using a reduced protein model. Proteins. 2005;60:252–256. doi: 10.1002/prot.20566. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Progress and challenges in protein structure prediction. Curr. Opin. Struct. Biol. 2008;18:342–348. doi: 10.1016/j.sbi.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER: Fully automated protein structure prediction in CASP8. Proteins. 2009;77:100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Automated structure prediction of weakly homologous proteins on a genomic scale. Proc. Natl. Acad. Sci. USA. 2004a;101:7594–7599. doi: 10.1073/pnas.0305695101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. Scoring function for automated assessment of protein structure template quality. Proteins. 2004b;57:702–710. doi: 10.1002/prot.20264. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Skolnick J. The protein structure prediction problem could be solved using the current PDB library. Proc. Natl. Acad. Sci. USA. 2005;102:1029–1034. doi: 10.1073/pnas.0407152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.