Abstract
Hermitamides A and B are lipopeptides isolated from a Papau New Guinea collection of the marine cyanobacterium Lyngbya majuscula. We hypothesized that the hermitamides are ligands for the human voltage-gated sodium channel (hNaV) based on their structural similarity to the jamaicamides. Herein, we describe the nonracemic total synthesis of hermitamides A and B and their epimers. We report the ability of the hermitamides to displace [3H]-BTX at 10 μM more potently than phenytoin, a clinically used sodium channel blocker, a potential binding mode for (S)-hermitamide B in the BTX-binding site, and electrophysiology showing that these compounds are potent blockers of the hNav1.2 voltage-gated sodium channel.
Keywords: Sodium channel blockers, Hermitamide A, Hermitamide B, Lyngbic acid, Lyngbya majuscula
1. Introduction
The human voltage-gated sodium channels (hNav) comprise a family of nine known subtypes of membrane-bound proteins (hNav1.1 to hNav1.9), which play critical physiological roles in the body and are therefore important targets in medicine.1 Their blockade by specific agents has been the basis for the discovery of important anticonvulsants, anesthetics, and antiarrhythmic drugs.2 Despite the success of these agents in the clinic, the sodium channel remains a challenging target for drug discovery.3 In an effort to discover novel ligands for these receptors, we have begun a program to synthesize and evaluate the secondary metabolites of the marine cyanobacterium Lyngbya majuscula for sodium channels inhibition.
Several lipopeptidic secondary metabolites isolated from L. majuscula have shown hNaV activity. For example, the cyclic lipopetides antillatoxin and antillatoxin-B are sodium channel activators with potencies comparable to the brevetoxins.4 The mixed polyketide-peptide kalkitoxin is a potent sodium channel blocking agent.5
Recently, the jamaicamides A, B and C (1–3) (Fig. 1) were isolated from the dark green strain of L. majuscula collected at Hector's Bay in Jamaica.6 These lipopeptides were shown to block sodium channel activity in a cell-based screening assay developed by Manger and co-workers, which involves measuring the end-point of mitochondrial dehydrogenase activity in neuroblastoma cells.7
Figure 1.
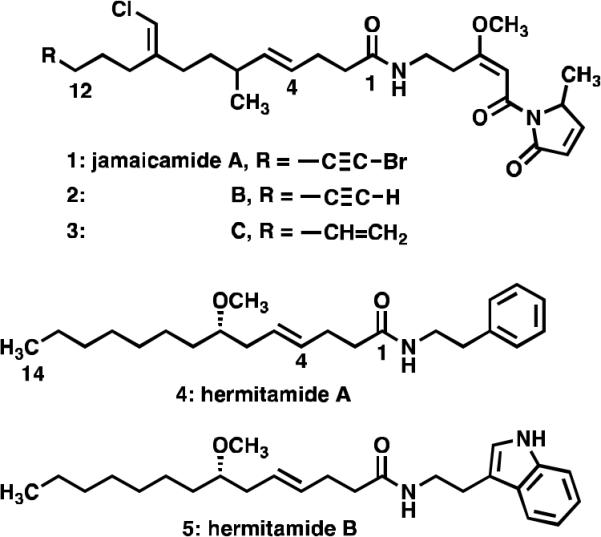
The metabolites jamaicamides A, B, and C (1–3), and the hermitamides A and B (4–5) isolated from L. majuscula.
The hermitamides A (4) and B (5), had been previously reported as metabolites isolated from a Papua New Guinea collection of L. majuscula.8 A careful analysis of the jamaicamide and the hermitamide structures reveal several similarities. For example, each class contains a 14-carbon aliphatic chain, an E olefin between carbon-4 and carbon-5, a 2-carbon chain linker between the amide nitrogen atom and a region of unsaturation (a π-system) (Fig. 2). On the basis of these structural similarities, and on the analysis of two potential modes of binding of the jamaicamides to the sodium channel in relation to that of kalkitoxin (6) (Fig. 3), we hypothesized that hermitamides 4 and 5 might also behave as channel sodium blockers.
Figure 2.
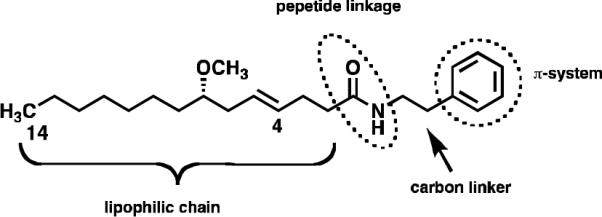
A proposed pharmacophore for jamaicamides A, B, and C (1–3) and the hermitamides A and B (4 and 5) at binding to the voltage-gated sodium channel.
Figure 3.
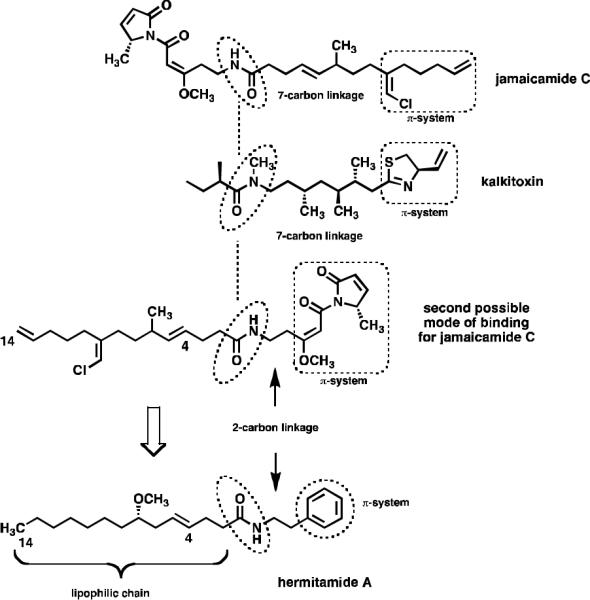
Pharmacophore analysis of jamaicamides, kalkitoxin and the hermitamides.
Herein, we report the first enantioselective total synthesis of both enantiomers of hermitamides 4 and 5, hermitamide mediated displacement of tritiated batrachotoxin ([3H]-BTX) from sodium channels, and functional block of hNav1.2 sodium channel currents in the presence of the hermitamides as measured by patch clamp electrophysiology. A computational homology model of site 2 of the hNaV1.2 channel and a binding model of hermitamide B (5) to this construct is presented.
2. Results and Discussion
2.1 Synthesis of hermitamides A and B
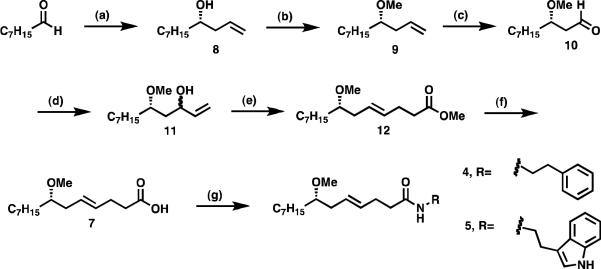
The syntheses of hermitamides 4 and 5 have attracted the interest of several organic synthesis groups. The configuration of the remote stereocenter bearing the methoxy group in compounds 4 and 5 was confirmed by relay synthesis of the respective hermitamides from (S)-lyngbic acid ((S)-7), which was isolated with the hermitamides.8 The preparation of the enantiopure lyngbic acid has been accomplished by ring-opening of a chiral epoxide,9 lipase resolution,10 and asymmetric allylation of the requisite aldehyde.11 A racemic total synthesis of hermitamides 4 and 5 and a formal synthesis of the nonracemic compounds have also been reported.12
Although these methods have proved to be valuable to obtain synthetic intermediates, to the best of our knowledge, completion of the asymmetric total synthesis of hermitamides 4 and 5 has not been reported. We have devised a simpler and efficient synthetic route to hermitamides 4 and 5 based on three key synthetic strategies as follows: i) generation of the asymmetric center by Keck allylation of octanal; ii) sterospecific formation of the E olefin by Johnson-Claisen rearrangement; and iii) condensation of the resulting lyngbic acid with the appropriate amines via carbodiimide chemistry.
The synthesis of nonracemic lyngbic acid (7) depicted in Scheme 1 commenced with the asymmetric allylation of octanal with allyltributylstannane mediated by a titanium-binol complex to give alcohol 8 in 40% yield and greater than 95:5 er as previously reported.13 Treatment of 8 with sodium hydride in dimethylformamide followed by methylation with methyl iodide, afforded the methyl ether 9 in 92% yield. Osmium dihydroxylation of 9 followed by in situ oxidative cleavage of the intermediate with sodium periodate under Johnson-Lemieux conditions yielded the aldehyde 10, which was used in the next step without further purification. Addition of vinylmagnesium bromide in tetrahydrofuran to 10 afforded the corresponding allylic alcohol 11 as a mixture of diastereomers in 51% yield. The Johnson-Claisen rearrangement of 11 was then effected by treatment with trimethylorthoacetate in the presence of a catalytic amount of propionic acid. The mixture was heated at reflux to allow the methanol generated to be distilled off after which the methyl ester of lyngbic acid (12) is obtained in 63% yield. Saponification of 12 with lithium hydroxide in a tetrahydrofuran-water solvent system afforded 7 in 41% yield.14 Finally, the total syntheses of 4 and 5 were accomplished in excellent yields by coupling 7 with phenethylamine (77%) or tryptamine (88%), respectively, via diisopropylcarbodiimide in the presence of 1-hydroxybenzotriazole (Scheme1).
Scheme 1.
Asymmetric synthesis of hermitamides A and B. (a) allyltributylstannane, (R)-BINOL (20 mol %), TiCl2(O-i-Pr)2 (20 mol %), CH2Cl2, 4 Å MS, −20 °C (40%); (b) i. NaH, DMF; ii. MeI (92%); (c) OsO4, NaIO4; (d) vinylmagnesium bromide, THF (51%, 2 steps); (e) CH3C(OCH3)3, n-PrCO2H, reflux (63%); (f) LiOH, H2O-THF (41%); (g) DCC, 1-hydroxybenzotriazole, R-NH2 [R-NH2 = phenethyalamine (77%) or tryptamine (88%). Obs.: EDC in methylene chloride has also been used in a scaled up synthesis of (S)-5 (75%)].
By following the same strategy depicted in Scheme 1, we have also synthesized the racemates of hermitamides 4 and 5 and their corresponding R enantiomers, (R)-4 and (R)-5. The R enantiomers of the hermitamides were synthesized from R-8, which was derived from addition of allyltributylstannane to octanal in the presence of (S)-(−)-1,1'-bi-2-naphthol ((S)-BINOL). The racemic hermitamides were synthesized from racemic 8, which was derived from addition of an ethereal solution of allylmagnesium bromide to octanal. The method also proved to be robust for scale up. Compound 5 was obtained from octanal in seven steps on a four-gram scale using N-(3-dimethylaminopropyl)-N′-ethyl carbodiimide hydrochloride (EDC) in solution of methylene chloride for the last step. All reactions details and yields for the intermediate compounds are reported in the experimental section of this paper.
2.2 Pharmacology
2.2.1 Pharmacophore analysis
The hermitamide structures resemble the jamaicamides in that both are lipopeptidic metabolites that contain a lipophilic side-chain, a peptide linkage, and a carbon linker to an aromatic moiety (the π-system) (Fig. 2). However, the carbon skeletal arrangement of the jamaicamides is much more complicated for both the aliphatic chain and the aromatic moiety, which is a highly unsaturated alkaloid with one stereocenter and a trisubstituted olefin.
In fact, the proposed pharmacophore seems also to be shared with other L. majuscula metabolites, such as the sodium channel blocker kalkitoxin. Examination of possible binding modes of jamaicamide C in relation to kalkitoxin highlights an interesting feature. By aligning each molecule pivoting around the amide bond, a potential second mode of binding for jamaicamide C is revealed. In this conformation the carbon chain linking the π-system to the nitrogen is shortened from 7 to 2 carbon atoms (Fig. 3). If this conformation is active for the binding of jamaicamide C to the sodium channel, then hermitamides 4 and 5 are hypothesized to be potential blocking agents, because they share the following structural features: both present a 14-carbon aliphatic chain bearing an E olefin between carbons C4 and C5, a peptide linkage, and a π-system connected to the nitrogen atom separated by a 2-carbon chain linker (Fig. 3). The hermitamides are structurally simpler than the jamaicamides. If the hermitamides present sodium channel blocking activity, the facility in which these natural products can be synthesized will enable large-scale preparation of this class of sodium channel blockers for further biological testing in vitro and in vivo.
2.2.2 In vitro displacement of [3H]-BTX by hermitamides A and B
In order to determine the binding of hermitamides 4 and 5 to sodium channels, we tested the natural products, the corresponding R enantiomers, and the racemic hermitamides for their ability to displace [3H]-BTX in rat forebrain membranes. Rat sodium channels exhibit a high degree of homology with human sodium channels (>95%),15 and the sodium channel is expressed highly in the forebrain. Batrachotoxin (BTX) is a lipid-soluble neurotoxin that specifically labels site 2 of the voltage-gated sodium channels. This site overlaps with the local anesthetic binding site 9. Ligands that bind to site 9 include anesthetics, anticonvulsants and antiarrthymics. As a positive control, phenytoin, a clinically used sodium channel blocker that is known to displace [3H]-BTX was also included.
The assays were run at 10 μM concentration and recorded as percent displacement of [3H]-BTX. As shown in Table 1, hermitamides 4 and 5 showed potencies equal to or greater than phenytoin. Hermitamide B (5) was the most active natural product, followed by hermitamide A (4), and then lyngbic acid (7). Surprisingly, the remote methoxy stereocenter does not appear to have much effect on the affinity of these compounds.
Table 1.
In vitro displacement of [3H]-BTX by hermitamides A and B and lyngbic acid.
| Compounds | % [3H]-BTX displacement at 10 μMa) | |
|---|---|---|
| (rac)-7 | rac-lyngbic acid | 7.35 |
| (S)-7 | (S)-lyngbic acid | 7.77 |
| (R)-7 | (R)-lyngbic acid | 11.91 |
| (rac)-4 | rac-hermitamide A | 15.71 |
| (S)-4 | (S)-hermitamide A | 15.89 |
| (R)-4 | (R)-hermitamide A | 13.79 |
| (rac)-5 | rac-hermitamide B | 36.05 |
| (S)-5 | (S)-hermitamide B | 29.02 |
| (R)-5 | (R)-hermitamide B | 20.31 |
| 10 | phenytoin | 19.78 |
This screen has an error of approximately 20%
2.2.3 In vitro electrophysiology study via patch clamping
The ability of the hermitamides to displace [3H]-BTX suggests that both bind to the sodium channel at the same site as BTX. In order to test whether these compounds were functionally active as sodium channel blockers, we tested the effects of the hermitamides on human embryonic kidney cells (HEK 293) stably expressing hNav1.2 sodium channels using the twomicroelectrode voltage clamp technique.
As shown in Table 2 and Figure 4, hermitamides 4 and 5 indeed function as sodium channel blockers. Hermitamide A (4) blocks ~50% sodium channel current at 1 μM, whereas hermitamide B (5) was a more potent blocker, eliciting ~80% blockade at 1 μM with a 50% blockage occurring somewhere between 1 μM and 100 nM concentration. Apparently, the aromatic region of these compounds are important for activity, with the indole group of hermitamide B being advantageous over the simple phenyl ring of hermitamide A. Interestingly, the remote methoxy stereocenter does not appear to have much influence on the ability of these compounds to inhibit channel current. This result is in line with our results in Table 1, where hermitamide B displaces [3H]-BTX more efficiently than hermitamide A.
Table 2.
Electrophysiology effects of the hermitamides on hNav1.2.
| Compound | 10 μM | 1 μM | 100 nM |
|---|---|---|---|
| (rac)-4 | 90.2 ± 2.5 (3) | 49.7 ± 3.8 (4) | <10 (3) |
| (S)-4 | 94.4 ± 1.3 (4) | 74.4 ± 2.1 (4) | 15.5 ± 3.7 (4) |
| (R)-4 | 58.7 ± 1.3 (4) | <10 (3) | |
| (rac)-5 | 100 (1) | 88.4 ± 2.9 (4) | 24.3 ± 2.9 (4) |
| (S)-5 | 100 (3) | 87.8 ± 2.2(4) | 17.2 ± 2.4(4) |
| (R)-5 | 80.7 ± 2.2(4) | 28.8 ± 3.0 (4) | |
| Phenytoin | 10 | <10 (3) |
Values represent percentage block (%) ± S.E.M. Values in parenthesis represent n numbers of experiments.
Figure 4.
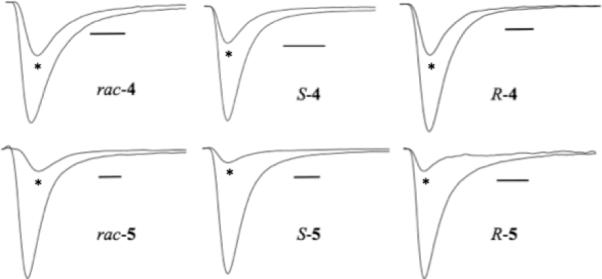
Pharmacological block of sodium currents by hermitamides at hNav1.2 channels expressed in HEK 293 cells. All drugs were tested at 1 μM concentration (scale bar represents 1 ms). *drug response.
2.3 Molecular modeling
Since hermitamide B (5) displaced [3H]-BTX in a competitive binding assay, this suggests that it binds to the sodium channel at site 9. Site 9 overlaps with site 2, which is also known as the local anesthetic (LA) binding site. In order to gain insight into the structural features that might govern the binding of hermitamide B to the hNav receptors, and to use this information for future drug design, a model of the human voltage-gated sodium channel was predicted by homology and hermitamide B was docked to this model.
2.3.1 Homology model of the hNav channel
The sodium channel was modeled using the MthK (PDB: 1lnq) potassium channel as a template. In this model, the S6 helical bends were produced at the serine sites, which correspond to the glycine residues in the MthK structure. A detailed description of how this model was built is found in the experimental section of this paper.
2.3.2 Docking of (S)-hermitamide B to the homology model of the hNav channel
Hermitamide B (5) was docked into our homology model using the program FlexX incorporated in Sybyl 8.0. Comparison with an initially predicted binding model of BTX using the same homology model suggests that the binding mode of hermitamide B differs from BTX at its interactions with the S6 helix residues. In order to provide consistent results, the docked position of hermitamide B was remodeled using a step-by-step manual docking methodology with restrained molecular dynamic (MD) simulations followed by energy minimization. In the restrained MD simulations, the optimum van der Waals and H-bond distance constraints was set between the ligand and the pore forming residues.
The following residues were identified as important for the binding of hermitamide B to the sodium channel: F1283, F1579, L1582, V1583, Y1586 in IVS6; T1279, L1280 in IIIS6; L788, F791, L792, in IIS6; I433, N434, L437 in IS6; and selectivity filter residues D400, E755, K1237 in the domains of I-IV P-loops. Mutation experiments and computational modeling studies further support that several of these residues participate in the BTX and LA binding.16,17,18 As shown in figure 5, binding is driven mainly by a hydrophobic interaction with residue K1237, and H-bonds between the amide group of hermitamide B with N434 and Y1586. Strong hydrophobic contacts were also predicted between hermitamide B and F1283, F1579, L1582, V1583, Y1586 L1280, L788, F791, L792, I433, and L437. The higher affinity of hermitamide B for the sodium channel over hermitamide A may be due to a putative stacking interaction of the indole moiety with F791, and favorable van der Waals contact with L437 and L788. In addition, our predicted model shows no interactions between the sodium channel and the methoxy group. This is in agreement with our biological evaluations showing that the stereochemistry of the methoxy group did not significantly contribute to the activity of the compounds (tables 1 and 2).
Figure 5.
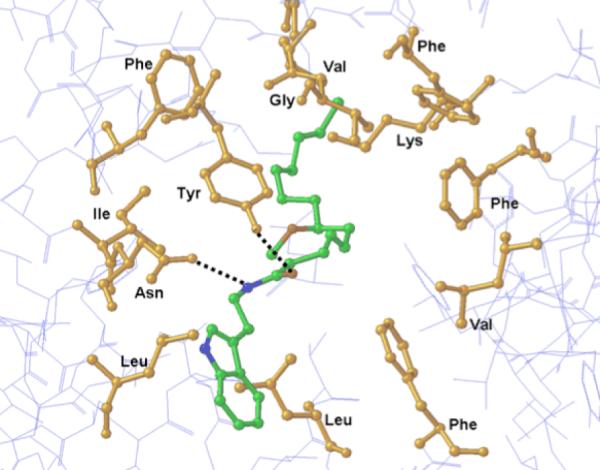
Model showing (S)-hermitamide B docked into the BTX binding site of hNav channel.
3. Conclusion
Blockers of the human voltage-gated sodium channel are widely used in the clinic as anticonvulsants, anesthetics, and antiarrhythmics. However, several challenges remain regarding the sodium channel in drug discovery. For example, selectivity for sodium channel subtypes as well as selectivity for the sodium channels over potassium and calcium channels continue to be a hindrance in further development of drugs for this target. In order to discover new structural classes as a template for future drug discovery efforts, we have synthesized and evaluated the natural products hermitamides A and B isolated from the marine cyanobacteria L. majuscula.
We have reported for the first time the asymmetric total synthesis of these compounds by employing a simple and effective synthetic strategy, i.e. Keck allylation of octanal, sterospecific formation of an E olefin by Johnson-Claisen rearrangement, and coupling of the resulting lyngbic acid with the appropriate amines via carbodiimide chemistry.
We have also demonstrated that hermitamides A and B are inhibitors of the human voltage-gated sodium channel. Our hypothesis for their activity was derived from an envisioned common pharmacophore that is shared with other known sodium channel blockers. By examining two of the potential modes of binding of jamaicamide C in relation to kalkitoxin, both of which are known sodium channel blockers, we reasoned that the hermitamides share a very similar structural framework as the jamaicamides and therefore should also block the sodium channel. Our results show that the hermitamides efficiently displace [3H]-BTX, suggesting that they bind to either site 2 or site 9 of the sodium channel. Hermitamides A and B also proved to be more potent at displacing [3H]-BTX from the sodium channel compared to the clinical agent phenytoin. We then evaluated the functional blocking activity of the hermitamides on hNaV1.2 channels by patch clamp techniques. Indeed both hermitamides A and B were found to be potent inhibitors of sodium channel current in HEK-293 cells stably expressing hNaV1.2.
Initial docking studies of hermitamides A and B to our voltage-gated sodium channel homology model, built on the published MthK potassium channel crystal structure, suggests that the binding is governed mainly by the hydrophobic interaction of the lipophilic side chain of hermitamides with residue K1237, and by the H-bond interaction between the amide group and residues N434 and Y1586. The model also suggests that the slight higher potency of hermitamide B in displacing [3H]-BTX compared to the hermitamide A can be explained by putative stacking interactions of the indole moiety of hermitamide B with residues F791, and favorable van der Waals contact with residues L437 and L788.
Cyanobacteria are a rich source of biologically active secondary metabolites. The hermitamides were isolated from a collection of L. majuscula cyanobacteria isolated in Papua New Guinea. However their function in nature remains unknown. We have shown that these compounds are very potent sodium channel blockers and we present a molecular modeling suggesting a potential binding mode of these natural products to the sodium channel. Caco-2 permeability studies and preliminary in vivo pain studies in our laboratory suggest that optimization of the physicochemical properties of these compounds may allow for translation to potential drugs. Regardless, we believe these natural products provide an interesting new scaffold for drug discovery.
4. Experimental
4.1 Chemistry
4.1.1 General Procedures
Commercially available reagents and solvents were purchased from Sigma–Aldrich, Fisher Scientific, EMD and TCI America. 1H and 13C-NMR spectra were recorded on a Varian 400-NMR model equipment. Optical rotation measurements were performed on a Jasco polarimeter. Flash chromatographic purifications were done on Biotage SP1 chromatographers.
4.1.2 Syntheses
rac-Undec-1-en-4-ol – ((rac)-8)
To a solution of 1.2 mL (7.8 mmol) of octanal in 30 mL of ether was added 15.6 mL of a 1.0 M solution of allylmagnesium bromide in ether. The reaction was allowed to stir at room temperature for 18 h and then quenched with saturated NH4Cl. The product was extracted with ether (2 × 20 mL), dried over Na2SO4 and filtered. The solvent was removed under reduced pressure and the remaining residue was purified by flash chromatography to give 0.79 g (60%) of the allylic alcohol. 1H NMR (400 MHz, CDCl3): δ 5.80-5.70 (m, 1H), 5.04-4.99 (m, 2H), 3.53 (m, 1H), 2.52 (br s, 1H), 2.20-2.05 (m, 2H), 1.38-1.34 (m, 2H), 1.25-1.15 (m, 10H), 0.80 (t, J = 7.1 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 133.6, 116.3, 70.6, 42.5, 37.5, 32.7, 30.6, 30.2, 26.7, 23.7, 15.2.
(S)-1-Undecen-4-ol – ((S)-8)
To a flame dried flask under a N2 atmosphere was added 5.0 g of 4 Å molecular sieves and 100 mL of anhydrous CH2Cl2, followed by 1.9 g (7.8 mmol) of TiCl2(i-PrO)2 and 2.2 g (7.8 mmol) of (R)-(+)-1,1'-bi-2-naphthol. The solution immediately turned red and was stirred at room temperature for 2 h. To this solution was added 17.9 mL (58.3 mmol) of allyltributyltin. The reaction was cooled to −20 °C and 5.0 g (39.0 mmol) of octanal was added. The solution was allowed to stir for 48 h at −20 °C. The solution was filtered through a plug of Celite and the solvent was removed under reduced pressure. The remaining residue was purified by flash chromatography eluting with 1:9 EtOAchexanes to give 2.5 g (38%) of allylic alcohol (S)-8. 1H NMR (400 MHz, CDCl3): δ 5.85-5.75 (m, 1H), 5.11-5.07 (m, 2H), 3.60 (m, 1H), 2.29-2.22 (m, 1H), 2.14-2.07 (m, 1H), 1.91 (br s, 1H), 1.44-1.41 (m, 2H), 1.32-1.22 (m, 10H), 0.85 (t, J = 6.5 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 133.6, 116.8, 70.8, 42.6, 37.6, 32.8, 30.6, 30.3, 26.7, 23.8, 15.4. [α]23D -7.7 (c 1.04, CHCl3); lit.13 [α]25D -6.5 (c 1.04, CHCl3).
(4S)-Methoxy-undec-1-ene - ((S)-9)
To a solution of 2.5 g (14.7 mmol) of alcohol (S)-8 in 50 mL of DMF was added 0.90 g (22.0 mmol) of NaH (60% dispersion in mineral oil) and 1.4 mL (22.0 mmol) of CH3I. The reaction was refluxed for 10 h. The solution was cooled to room temperature, taken up in EtOAc, and washed with saturated LiCl (3×). The organic phase was dried over Na2SO4, filtered, and the solvent was removed under reduced pressure. The resulting residue was purified by flash chromatography eluting with 1:9 EtOAc-hexanes to yield 2.5 g (92%) of the titled compound. 1H NMR (400 MHz, CDCl3): δ 5.85–5.75 (m, 1H), 5.08–5.01 (m, 2H), 3.32 (s, 3H), 3.18 (quin, J = 5.8 Hz, 1H), 2.24 (m, 2H), 1.46–1.40 (m, 2H), 1.30–1.20 (m, 10H), 0.86 (t, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 133.6, 115.7, 80.4, 56.9, 38.6, 34.3, 32.8, 30.8, 30.3, 26.4, 23.8, 15.4. [α]22D -4.0 (c 2.00, CHCl3).
Methyl (4E,7S)-Methoxy-tetradecenoate – ((S)-12)
To a biphasic solution of 2.5 g (13.5 mmol) of alkene (S)-9 in 30 mL of 1:1 Et2O-H2O was added 2.2 mL (5 mol %) of OsO4 (2.5 wt % solution in t-BuOH). At room temperature, 6.4 g (29.7 mmol) of NaIO4 was added dropwise over 30 min. The solution was stirred for an additional 18 h. The reaction was taken up in EtOAc and the aqueous phase was removed. The organic phase was dried over Na2SO4, filtered, and the solvent was removed under reduced pressure to give (3S)-methoxydecanal ((S)-10) as an yellow oil. No further purification was performed.
To a solution of aldehyde (S)-10 in 50 mL of THF was added 27 mL of a 1.0 M solution of vinylmagnesium bromide. The reaction was stirred at room temperature for 3 h and then quenched with saturated NH4Cl. The product was extracted with EtOAc (3 × 30 mL), dried over Na2SO4, filtered, and the solvent was removed under reduced pressure. The resulting residue was purified by flash chromatography eluting with 1:5 EtOAchexanes to afford 1.5 g (51% over two steps) of 3-hydroxy-5S-methoxydodec-1-ene ((S)-11) as a mixture of diastereomers.
To vinyl alcohol (S)-11 in 6 mL of trimethyl orthoacetate was added one drop of propionic acid. The flask was fitted with a distillation head, the solution was heated to 100 °C for 1.5 hours, and the resulting evolution of MeOH was removed by distillation. The solvent was then removed under reduced pressure and the remaining residue was purified by flash chromatography eluting with 1:5 EtOAc-hexanes to afford 1.2 g (63%) of methyl ester (S)-12. 1H NMR (400 MHz, CDCl3): δ 5.47 (m, 2H), 3.66 (s, 3H), 3.32 (s, 3H), 3.12 (quin, J = 5.8 Hz, 1H), 2.24–2.16 (m, 4H), 2.18 (m, 2H), 1.30–1.23 (m, 2H), 1.19–1.09 (m, 10H), 0.87 (t, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 172.5, 130.2, 127.5, 81.6, 58.0, 53.1, 38.2, 36.0, 35.3, 33.8, 31.8, 31.3, 30.0, 27.4, 24.9, 16.5.
(7S, 4E)-Methoxytetradecenoic acid - ((S)-7)
To a solution of 1.1 g (4.1 mmol) of methyl ester (S)-12 in 20 mL of 1:1 THF-H2O was added 0.49 g (20.3 mmol) of LiOH. The solution was allowed to stir at room temperature for 18 h. The solution was then acidified with 1 N HCl and the product was extracted with EtOAc (2 × 30 mL). The organic phase was dried over Na2SO4, filtered, and the solvent was removed under reduced pressure. The remaining residue was purified by flash chromatography eluting with 1:1 EtOAc-hexanes to afford 0.43 g (41%) of carboxylic acid (S)-7. 1H NMR (400 MHz, CDCl3): δ 11.04 (br s, 1H), 5.44 (m, 2H), 3.29 (s, 3H), 3.12 (quin, J = 5.8 Hz, 1H), 2.39–2.29 (m, 4H), 2.15 (m, 2H), 1.42–1.37 (m, 2H), 1.28–1.19 (m, 10H), 0.83 (t, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 176.5, 129.0, 126.4, 80.7, 56.7, 37.1, 34.8, 34.1, 32.7, 30.7, 30.2, 28.7, 26.3, 23.8, 15.4. [α]27D -10.9 (c 1.1, CHCl3); lit.23 [α]26D -11.1 (c 3.9, CHCl3). Anal. Calcd for C15H28O3: C, 70.27; H, 11.01. Found: C, 69.98; H, 11.12.
Phenethyl (7S,4E)-methoxytetradecenamide - ((S)-4)
To a solution of 0.18 g (0.68 mmol) of carboxylic acid (S)-7 in 10 mL of CH2Cl2 was added 0.12 mL (0.75 mmol) of diisopropylcarbodiimide and 0.10 g (0.75 mmol) of 1-hydroxybenzotriazole. After stirring at room temperature for 10 min, 0.090 mL (0.68 mmol) of phenethylamine was added. The reaction was stirred at room temperature for 15 h, during which a white precipitate formed. The solvent was then removed under reduced pressure and the residue was taken up in EtOAc. The solution was washed with 1 N HCl (3 × 20 mL) followed by saturated NaHCO3 (1 × 20 mL). The organic phase was dried over Na2SO4, filtered, and the solvent was removed under reduced pressure. The remaining residue was purified by flash chromatography eluting with 1:3 EtOAc-hexanes to give 0.19 g (77%) of (S)-4. 1H NMR (400 MHz, CDCl3): δ 7.27–7.13 (m, 5H), 5.82 (br s, 1H), 5.40 (m, 2H), 3.45 (q, J = 6.9 Hz, 2H), 3.26 (s, 3H), 3.09 (quin, J = 5.7 Hz, 1H), 2.76 (t, J = 7.1 Hz, 2H), 2.29–2.25 (m, 2H), 2.17–2.11 (m, 4H), 1.40–1.33 (m, 2H), 1.30–1.22 (m, 10H), 0.83 (t, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 170.2, 137.4, 129.6, 127.4, 127.3, 126.2, 125.2, 80.4, 56.7, 41.3, 37.2, 37.1, 36.5, 34.1, 32.7, 30.7, 30.2, 29.6, 26.3, 23.7, 15.4. [α]23D -8.9 (c 0.45, CHCl3); lit.8 [α]26D -9.3 (c 0.45, CHCl3). Anal. Calcd for C23H37NO2: C, 76.83; H, 10.37; N, 3.90. Found: C, 76.60; H, 10.46; N, 3.89.
2-(1H-indol-3-yl)-ethyl (7S, 4E)-Methoxytetradecenamide -((S)-5)
To a solution of 0.18 g (0.68 mmol) of carboxylic acid (S)-7 in 10 mL of CH2Cl2 was added 0.12 mL (0.75 mmol) of diisopropylcarbodiimide and 0.10 g (0.75 mmol) of 1-hydroxybenzotriazole. After stirring at room temperature for 10 min, 0.11 g (0.68 mmol) of tryptamine was added. The reaction was stirred at room temperature for 15 h. The solvent was then removed under reduced pressure and the residue was taken up in EtOAc. The solution was washed with 1 N HCl (5 × 20 mL) followed by saturated NaHCO3 (1 × 20 mL). The organic phase was dried over Na2SO4, filtered, and the solvent was removed under reduced pressure. The resulting residue was purified by flash chromatography eluting with 1:3 EtOAc-hexanes to give 0.24 g (88%) of (S)-5. 1H NMR (400 MHz, CDCl3): δ 8.89 (br s, 1H), 7.58 (d, J = 7.8 Hz, 1H), 7.35 (d, J = 8.1 Hz, 1H), 7.19 (t, J = 8.1 Hz, 1H), 7.10 (t, J = 7.9 Hz, 1H), 6.97 (s, 1H), 5.80 (br s, 1H), 5.44 (m, 2H), 3.57 (q, J = 6.6 Hz, 2H), 3.32 (s, 3H), 3.15 (quin, J = 5.7 Hz, 1H), 2.95 (t, J = 6.8 Hz, 2H), 2.33–2.28 (m, 2H), 2.18–2.14 (m, 4H), 1.45–1.41 (m, 2H), 1.32–1.22 (m, 10H), 0.89 (t, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 170.4, 135.0, 129.5, 126.2, 126.1, 121.1, 120.8, 118.1, 117.5, 111.6, 110.5, 80.5, 56.7, 40.5, 37.2, 37.1, 34.1, 32.7, 30.7, 30.2, 29.6, 26.3, 26.2, 23.7, 15.4. [α]24D -2.0 (c 2.00, CHCl3); lit.8 [α]26D -4.9 (c 0.15, CHCl3). Anal. Calcd for C25H38N2O2: C, 75.33; H, 9.61; N, 7.03. Found: C, 74.86; H, 9.67; N, 6.92.
(R)-1-Undecen-4-ol - ((R)-8)
Prepared using the same method employed for the preparation of compound (S)-8, with the exception that (S)-BINOL was used as a catalyst.
Yield = 33%. 1H NMR (400 MHz, CDCl3): δ 5.81–5.70 (m, 1H), 5.05–5.00 (m, 2H), 3.55 (m, 1H), 2.54 (br s, 1H), 2.22–2.04 (m, 1H), 2.07 (m, 1H), 1.39–1.35 (m, 2H), 1.27–1.17 (m, 10H), 0.80 (t, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 133.6, 116.4, 70.7, 42.5, 37.5, 32.7, 30.5, 30.2, 26.7, 23.7, 15.3. [α]23D +7.7 (c 1.04, CHCl3); lit.13 [α]25D +6.5 (c 1.04, CHCl3).
(4R)-Methoxy-undec-1-ene - ((R)-9)
Prepared using the same method employed for the preparation of compound (S)-9.
Yield = 80%. 1H NMR (400 MHz, CDCl3): δ 5.71–5.61 (m, 1H), 4.93–4.86 (m, 2H), 3.17 (s, 3H), 3.02 (quin, J = 5.8 Hz, 1H), 2.10 (m, 2H), 1.34–1.30 (m, 2H), 1.19–1.09 (m, 10H), 0.75 (t, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 133.3, 115.3, 80.1, 56.4, 38.4, 34.1, 32.6, 30.6, 30.1, 26.1, 23.6, 15.1.
Methyl (4E,7R)-Methoxy-tetradecenoate - ((R)-12)
Prepared using the same method employed for the preparation of compound (S)-12.
Yield = 51%. 1H NMR (400 MHz, CDCl3): δ 5.33 (m, 2H), 3.51 (s, 3H), 3.17 (s, 3H), 2.99 (quin, J = 5.8 Hz, 1H), 2.25–2.18 (m, 4H), 2.04 (m, 2H), 1.30–1.26 (m, 2H), 1.18–1.09 (m, 10H), 0.74 (t, J = 6.9 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 173.0, 130.1, 127.3, 80.4, 56.1, 51.0, 36.0, 33.6, 33.0, 31.5, 31.8, 29.4, 29.0, 27.6, 24.9, 22.3, 13.7.
(7R, 4E)-Methoxytetradecenoic acid - ((R)-7)
Prepared using the same method employed for the preparation of compound (S)-7.
Yield = 65%. 1H NMR (400 MHz, CDCl3): δ 10.23 (br s, 1H), 5.48 (m, 2H), 3.32 (s, 3H), 3.15 (quin, J = 5.8 Hz, 1H), 2.44–2.31 (m, 4H), 2.18 (m, 2H), 1.44–1.38 (m, 2H), 1.31–1.20 (m, 10H), 0.87 (t, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 176.8 128.9, 126.6, 80.7, 56.9, 37.2, 34.9, 34.3, 32.8, 30.8, 30.3, 28.7, 26.4, 23.9, 15.5. [α]23D +4.2 (c 1.91, CHCl3). Anal. Calcd for C15H28O3: C, 70.27; H, 11.01. Found: C, 70.13; H, 11.21.
Phenethyl (7R,4E)-methoxytetradecenamide - ((R)-4)
Prepared using the same method employed for the preparation of compound (S)-4.
Yield = 74% for (R)-4. 1H NMR (400 MHz, CDCl3): δ 7.26–7.12 (m, 5H), 5.93 (br s, 1H), 5.40 (m, 2H), 3.42 (q, J = 6.9 Hz, 2H), 3.25 (s, 3H), 3.09 (quin, J = 5.8 Hz, 1H), 2.75 (t, J = 7.1 Hz, 2H), 2.28–2.24 (m, 2H), 2.16–2.10 (m, 4H), 1.43–1.35 (m, 2H), 1.28–1.18 (m, 10H), 0.83 (t, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 170.2, 137.4, 129.5, 127.4, 127.2, 126.1, 125.1, 80.4, 56.7, 41.2, 37.1, 37.0, 36.4, 34.1, 32.6, 30.6, 30.2, 29.6, 26.3, 23.7, 15.3. [α]23D +8.9 (c 0.45, CHCl3). Anal. Calcd for C23H37NO2: C, 76.83; H, 10.37; N, 3.90. Found: C, 76.87; H, 10.57; N, 3.92.
2-(1H-indol-3-yl)-ethyl (7R, 4E)-Methoxytetradecenamide -((R)-5)
Prepared using the same method employed for the preparation of compound (S)-5.
Yield = 96% for (R)-5. 1H NMR (400 MHz, CDCl3): δ 8.97 (br s, 1H), 7.58 (d, J = 7.9 Hz, 1H), 7.35 (d, J = 8.1 Hz, 1H), 7.18 (t, J = 7.9 Hz, 1H), 7.10 (t, J = 7.7 Hz, 1H), 6.97 (s, 1H), 5.84 (br s, 1H), 5.44 (m, 2H), 3.57 (q, J = 6.6 Hz, 2H), 3.33 (s, 3H), 3.16 (quin, J = 5.7 Hz, 1H), 2.95 (t, J = 6.8 Hz, 2H), 2.33–2.29 (m, 2H), 2.18–2.14 (m, 4H), 1.45–1.39 (m, 2H), 1.37–1.29 (m, 10H), 0.90 (t, J = 6.8 Hz, 3H); 13C NMR (100 MHz, CDCl3): δ 170.4, 135.0, 129.5, 126.1, 126.0, 121.1, 120.7, 118.1, 117.5, 111.6, 110.5, 80.5, 56.7, 40.5, 37.2, 37.1, 34.1, 32.7, 30.6, 30.2, 29.6, 26.3, 26.2, 23.7, 15.4. [α]24D +2.0 (c 2.00, CHCl3). Anal. Calcd for C25H38N2O2: C, 75.33; H, 9.61; N, 7.03. Found: C, 74.80; H, 9.65; N, 7.02.
4.2 Modeling of the hNav channel with hermitamide B ((S)-5)
Modeling of the Sodium Channel
Multiple sequence alignment of the S6 transmembrane residues from domains I, III, and IV was carried out using PSI-BLAST and CLUSTALW. Homology modeling of S5, the P-loops, and S6 from all four domains used the MthK channel x-ray structure (PDB: 1lnq) as a template. Since LAs bind to the inactivated form of domains the potassium channel were aligned, i.e. S5 and S6 transmembrane segments based on both homology and secondary structure prediction. Non-homologous regions in the longer P-loops of domains I and III, which correspond to putative glycosylation sites, were deleted. The P-loops, N and C termini were modeled based on homologous segments of the KcsA channel structure (PDB: 1bl8). Sodium channel sequences were aligned versus the MthK channel using ClustalW, and the structure was modeled employing the program Modeler 8.1. To avoid side chain atom contacts, different rotamer states of the residue were considered and the one with minimal contacts, but a favorable interaction was chosen. Local side chain atom minimization was also performed.
Docking of (S)-hermitamide B
Initial docking studies between the ligand and the sodium channel were carried out using the program FlexX. After consistent manual intervention, a final model was arrived. The structure of the Nav-hermitamide B complex was then refined by molecular dynamics simulation using the Amber 9 program suite19 with the PARM98 force-field parameter. The charge and force field parameters of hermitamide B was obtained using the most recent Antechamber module in the Amber 9 program, where hermitamide B was minimized at the MP2/6-31G* level. The SHAKE algorithm20 was used to keep all bonds involving hydrogen atoms rigid. Weak coupling temperature and pressure coupling algorithms21 were used to maintain constant temperature and pressure, respectively. Electrostatic interactions were calculated with the Ewald particle mesh method22 with a dielectric constant at 1Rij and a nonbonded cutoff of 12 Å for thereal part of electrostatic interactions and for van der Waals' interactions. The total charge of the system was neutralized by addition of a chloride ion. The system was solvated in a 14 Å cubic box of water where the TIP3P model8 was used. 3000 steps of minimization of the system were performed in which the sodium channel was constrained by a force constant of 100 kcal/mol/Å2. After minimization, a 10 ps simulation was used to gradually raise the temperature of the system to 298 K while the complex was constrained by a force constant of 20 kcal/mol/Å. Another 20 ps equilibration run was used where only the backbone atoms of the complex were constrained by a force constant of 5 kcal/mol/Å. Final production run of 100 ps was performed with no constraints. When applying constraints, the initial complex structure was used as a reference structure. The PME method10 was used and the time step was 5 fs, and a neighboring pairs list was updated in every 30 steps.
4.3 [3H]-BTX displacement assay
The [3H]-BTX binding assays were performed by Novascreen Biosciences (Hanover, MD). Briefly, rat forebrain membranes (10 mg tissue/well) were incubated with [3H]-BTX (30–60 Ci/mmol). Reactions are carried out in 50 mM HEPES (pH 7.4) containing 130 mM choline chloride at 37 °C for 60 min. The reaction was terminated by rapid vacuum filtration of the reaction contents onto glass fiber filters. Radioactivity trapped onto the filters was determined and compared with control values to ascertain any interactions of the test compound with the Na+channel site 2 binding site. Aconitine (1 μM) was used as a positive control (Sigma Aldrich, Inc., St. Louis, MO).
4.4 Sodium hNav1.2 channel electrophysiology
Human embryonic kidney cells (HEK) cells stably expressing human Nav1.2 were a kind gift from Dr. H.A. Hartmann (University of Baltimore, Maryland, USA) and were grown in DMEM/F12 media (Invitrogen, Corp, CA, USA) supplemented with 10% fetal bovine serum, penicillin (100 U/ml), streptomycin (100 μg/ml) and G418 (500 μg/ml; Sigma, MO, USA). Cells were grown in a humidified atmosphere of 5% CO2 and 95% air at 37 °C.
Sodium currents were recorded using the whole-cell configuration of the patch clamp recording technique with an Axopatch 200 amplifier (Molecular Devices). All voltage protocols were applied using pCLAMP 9 software and a Digidata 1322A (Molecular Devices). Currents were amplified and low pass filtered (2 kHz) and sampled at 33 kHz. Borosilicate glass pipettes were pulled using a Brown-Flaming puller (model P87, Sutter Instruments Co, Novato, CA) and heat polished to produce electrode resistances of 0.5–1.5 MΩ when filled with the following electrode solution (in mM); CsCl 130, MgCl2 1, MgATP 5, BAPTA 10, HEPES 5 (pH adjusted to 7.4 with CsOH). Cells were plated on glass coverslips and superfused with solution containing the following composition; (in mM) NaCl 130, KCl 4, CaCl2 1, MgCl2 5, HEPES 5, and glucose 5 (pH adjusted to 7.4 with NaOH).
Compounds were prepared as 100 mM stock solutions in DMSO and diluted to the desired concentration in perfusion solution. The maximum DMSO concentration used was 0.1% and had no effect on current amplitude. All experiments were performed at room temperature (20–22 °C). After establishing whole-cell, a minimum series resistance compensation of 75% was applied. Sodium currents were elicited by a depolarizing step from a holding potential of −60 mV to +10 mV for a duration of 25 ms at 15 s intervals. Compounds were applied after a 3 min control period and continued until a steady state current amplitude was observed. All data represent percentage mean block ± standard error of the mean (SEM).
Acknowledgments
We thank Georgetown University Drug Discovery Program and the Lombardi Cancer Center for funding this research.
References and notes
- 1.Zuliani V, Patel MK, Fantini M, Rivara M. Curr. Top. Med. Chem. 2009;9:396–415. doi: 10.2174/156802609788317856. [DOI] [PubMed] [Google Scholar]
- 2.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. PNAS. 1996;93:9270–9275. doi: 10.1073/pnas.93.17.9270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anger T, Madge DJ, Mulla M, Riddall D. J. Med. Chem. 2001;44:115–137. doi: 10.1021/jm000155h. [DOI] [PubMed] [Google Scholar]
- 4.Li WI, Berman FW, Okino T, Yokokawa F, Shioiri T, Gerwick WH, Murray TF. PNAS. 2001;98:7599–7604. doi: 10.1073/pnas.121085898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LePage KT, Goeger D, Yokokawa F, Asano T, Shioiri T, Gerwick WH, Murray TF. Toxicol. Lett. 2005;158:133–139. doi: 10.1016/j.toxlet.2005.03.007. [DOI] [PubMed] [Google Scholar]
- 6.Edwards DJ, Marquez BL, Nogle LM, McPhail K, Goeger DE, Roberts MA, Gerwick WH. Chem. Biol. 2004;11:817–833. doi: 10.1016/j.chembiol.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 7.Manger RL, Leja LS, Lee SY, Hungerford JM, Hokama Y, Dickey RW, Granade HR, Lewis R, Yasumoto T, Wekell MM. J. AOAC Int. 1995;78:521–527. [PubMed] [Google Scholar]
- 8.Tan LT, Okino T, Gerwick WH. J. Nat. Prod. 2000;63:952–955. doi: 10.1021/np000037x. [DOI] [PubMed] [Google Scholar]
- 9.Mueller C, Voss G, Gerlach H. Liebigs Ann. Chem. 1995;4:673–676. [Google Scholar]
- 10.Sankaranarayanan S, Sharma A, Chattopadhyay S. Tetrahedron Asymmetry. 1996;7:2639–2643. [Google Scholar]
- 11.Li Y, Chen J, Cao X-P. Synthesis. 2006;2:320–324. [Google Scholar]
- 12.Virolleaud M-A, Menant C, Fenet B, Piva O. Tetrahedron Lett. 2006;47:5127–5130. [Google Scholar]
- 13.Costa AL, Piazza MG, Tagliavini E, Trombini C, Umani-Ronchi A. J. Am. Chem. Soc. 1993;115:7001–7002. [Google Scholar]
- 14.During the preparation of this report, a nearly identical route to a lynbic acid analog with an extended carbon chain was published. Feng J-P, Shi Z-F, Li Y, Zhang J-T, Qi X-L, Chen J, Cao X-P. J. Org. Chem. 2008;73:6873–6876. doi: 10.1021/jo800876u..
- 15.Cummins TR, Xia Y, Haddad GG. J. Neurophysiol. 1994;71:1052–1064. doi: 10.1152/jn.1994.71.3.1052. [DOI] [PubMed] [Google Scholar]
- 16.Ragsdale DS, McPhee JC, Scheuer T, Catterall WA. Science. 1994;265:1724–1728. doi: 10.1126/science.8085162. [DOI] [PubMed] [Google Scholar]
- 17.Yarov-Yarovoy V, Brown J, Sharp EM, Clare JJ, Scheuer T, Catterall WA. J. Biol. Chem. 2001;276:20–27. doi: 10.1074/jbc.M006992200. [DOI] [PubMed] [Google Scholar]
- 18.Yarov-Yarovoy V, McPhee JC, Idsvoog D, Pate C, Scheuer T, Catterall WA. J. Biol. Chem. 2002;277:35393–35401. doi: 10.1074/jbc.M206126200. [DOI] [PubMed] [Google Scholar]
- 19.Case DA, Darden TA, Cheatham TE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. AMBER 9. University of California; San Francisco: 2006. [Google Scholar]
- 20.Hanson RN, Lee CY, Friel CJ, Dilis R, Hughes A, DeSombre ER. J. Med. Chem. 2003;46:2865–2876. doi: 10.1021/jm0205806. [DOI] [PubMed] [Google Scholar]
- 21.Berendsen HJC, Postma JPM, vanGunsteren WF, DiNola A, Haak JR. J. Chem. Phys. 1984;81:3684–3690. [Google Scholar]
- 22.Darden T, York D, Pedersen L. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 23.Cardellina JH, II, Dalietrios D, Marner F-J, Mynderse JS, Moore RE. Phytochemistry. 1978;17:2091–2095. [Google Scholar]