Abstract
Soybean (Glycine max (L.) Merrill) is a salt-sensitive crop, and its production is severely affected by saline soils. Therefore, the response of soybean seeds to salt stress during germination was investigated at both physiological and proteomic levels. The salt-tolerant cultivar Lee68 and salt-sensitive cultivar N2899 were exposed to 100 mmol/L NaCl until radicle protrusion from the seed coat. In both cultivars, the final germination percentage was not affected by salt, but the mean germination times of Lee68 and N2899 were delayed by 0.3 and 1.0 d, respectively, compared with controls. In response to salt stress, the abscisic acid content increased, and gibberellic acid (GA1+3) and isopentenyladenosine decreased. Indole-3-acetic acid increased in Lee68, but remained unchanged in N2899. The proteins extracted from germinated seeds were separated using two-dimensional gel electrophoresis (2-DE), followed by Coomassie brilliant blue G-250 staining. About 350 protein spots from 2-DE gels of pH range 3 to 10 and 650 spots from gels of pH range 4 to 7 were reproducibly resolved, of which 18 protein spots showed changes in abundance as a result of salt stress in both cultivars. After matrix-assisted laser desorption ionization-time of flight-mass spectroscopy (MALDI-TOF-MS) analysis of the differentially expressed proteins, the peptide mass fingerprint was searched against the soybean UniGene database and nine proteins were successfully identified. Ferritin and 20S proteasome subunit β-6 were up-regulated in both cultivars. Glyceraldehyde 3-phosphate dehydrogenase, glutathione S-transferase (GST) 9, GST 10, and seed maturation protein PM36 were down-regulated in Lee68 by salt, but still remained at a certain level. However, these proteins were present in lower levels in control N2899 and were up-regulated under salt stress. The results indicate that these proteins might have important roles in defense mechanisms against salt stress during soybean seed germination.
Keywords: Proteomics, Salt stress, Seed germination, Soybean
1. Introduction
Agricultural productivity and the eco-environment are severely affected by soil salinity. It is estimated that about 20% of irrigated land, which yields one-third of the world’s food, is affected by salinity. Moreover, a significant proportion of recently cultivated agricultural land has become saline because of land clearing or irrigation (Munns, 2005). Salinity is one of the major constraints limiting plant growth in some of the most productive agricultural regions of the world (Boyer, 1982). Therefore, the need to develop salt-tolerant cultivars is unavoidable.
However, soybean (Glycine max (L.) Merrill), an important crop that provides fatty acids and proteins for humans and animals, is a salt-sensitive crop (Luo et al., 2005), and its production is severely affected by saline soils. Saline soils significantly alter plant metabolic processes (Levitt, 1980). A high concentration of salt causes ion imbalance, hyperosmotic stress, and oxidative damage (Zhu, 2002). In response to these, plants resort to various pro-survival strategies, most of which are preceded by specific changes in expression levels of proteins whose biological functions are related to salt stress tolerance. The proteomic approach, based on reproducible two-dimensional gel electrophoresis (2-DE) and powerful mass spectrometry (MS) analyses, offers the possibility of identifying those proteins (Gygi and Aebersold, 2000). In fact, the proteomics of soybean in response to abiotic stress have been studied (Zhen et al., 2007; Aghaei et al., 2008; Toorchi et al., 2009; Cheng et al., 2010; Nouri and Komatsu, 2010; Sobhanian et al., 2010). Although the majority of defense proteins are expressed in optimal growth conditions, they have been found to be either overexpressed or underexpressed during stress. These investigations showed that a proteomic approach is useful for analyzing the physiological changes and the functions of stress-induced proteins.
Proteomic analyses also have been performed to analyze soybean seed proteins in mature seeds (Mooney and Thelen, 2004), seed filling (Hajduch et al., 2005; Agrawal et al., 2008), and seed germination (Xu et al., 2006). However, seed germination is one of the most crucial and decisive phases in the growth cycle of plants, because it determines plant establishment and the final yield of the crops. Seeds and young seedlings are frequently confronted with much higher salinities than vigorously growing plants, because germination usually occurs in surface soils, which accumulate soluble salts as a result of evaporation and the capillary rise of water (Almansouri et al., 2001). Limited information is available about salt-response genes in soybean, and the study of protein expression in response to salinity may therefore help identify the related genes and provide a detailed network of salt adaptation mechanisms in this important crop (Aghaei et al., 2008). In this study, proteomic analysis was carried out to identify salt stress-responsive proteins in soybean seed germination in two cultivars, Lee68 (salt-tolerant) and N2899 (salt-sensitive). The application of abiotic stress can result in an altered level of plant growth hormones (Morgan, 1990); therefore, we also investigated the levels of the endogenous hormones indole-3-acetic acid (IAA), gibberellic acid (GA), abscisic acid (ABA), and isopentenyladenosine (iPAs) in seeds germinated under salinity.
2. Materials and methods
2.1. Plant materials and growth condition
Soybean seeds (Glycine max cv. Lee68 and cv. N2899) were obtained from the Chinese National Center for Soybean Improvement. Lee68 is known for its high salt tolerance, while N2899 is a salt-sensitive cultivar identified in our laboratory. For each cultivar, hand-selected seeds of uniform size were sterilized in 1 mg/ml HgCl2 (Genebase) for 2 min and thoroughly washed in distilled water. Seeds were germinated in Petri plates containing Whatman No. 1 filter paper moistened with distilled water or with 100 mmol/L NaCl (Genebase) solution and kept in the dark at 25 °C. A seed was scored as germinated if the primary root reached 2–3 mm length. Seeds were germinated up to 12 d in the dark. Every 12 h, the percentage of germinated seeds (containing abnormal growth) was calculated from three replicates of 300 seeds each. Seeds that had germinated such that the radicle protruded by 2–3 mm from the seed coat were harvested and stored at −80 °C until use (Fig. 1).
Fig. 1.
Germinated soybean seeds of Lee68 and N2899 used in the experiment
N2899+H2O: control N2899; N2899+NaCl: NaCl-treated N2899; Lee68+H2O: control Lee68; Lee68+NaCl: NaCl-treated Lee68
2.2. Extraction and determination of plant hormones
Extraction and immunoassay of IAA were carried out according to Chen et al. (1998b). Extraction and immunoassay of gibberellin A1 & A3 (GA1+3) were carried out as described by Chen et al. (1998a). Extraction and immunoassay of iPAs and ABA were performed as described by Chen et al. (1997). Each assay was replicated three times. The data were analyzed by analysis of variance (ANOVA) and Student’s t-test. All data presented are the mean values.
2.3. Preparation of the protein extract
Total proteins from germinated seeds were extracted according to a modified procedure based upon that of Watson et al. (2003). In brief, a frozen sample was ground in a mortar with liquid nitrogen and incubated with 0.1 g/ml trichloroacetic acid (TCA) (Genebase) and 20 mmol/L dithiothreitol (DTT; Bio-Rad) in acetone (Genebase) at −20 °C for 1 h. The precipitated proteins were pelleted and washed repeatedly with ice-cold acetone containing 20 mmol/L DTT to remove pigments and lipids until the supernatant was colorless. The protein pellet was dried under a vacuum and resuspended in buffer containing 7 mol/L urea (Bio-Rad), 2 mol/L thiourea (Bio-Rad), 0.04 g/ml 3-[(3-cholamidopropyl)-dimethylammonio]-1-propane sulfonate (CHAPS; Bio-Rad), 0.2% (v/v) carrier ampholyte (pH 3–10 or pH 4–7; Bio-Rad), and a cocktail of protease inhibitors (Sigma). Samples were mixed on a vortex mixer for 30 s and ultrasonicated using a VCX600 for 3 min. The insoluble tissue was removed by centrifugation at 15 000×g for 15 min (Beckman). The supernatant was stored at −80 °C. The protein concentration was determined according to Bradford (1976), with bovine serum albumin (BSA; Amresco) as a standard.
2.4. Two-dimensional polyacrylamide gel electrophoresis and image analysis
Immobilized pH gradient (IPG) strips (17 cm, pH 3–10, linear gradient; or 17 cm, pH 4–7, linear gradient; Bio-Rad) were rehydrated at 50 V for 12 h at 20 °C with 350 μl rehydration buffer (7 mol/L urea, 2 mol/L thiourea, 0.04 g/ml CHAPS, 0.2% (v/v) carrier ampholyte, 0.01 g/ml DTT) containing about 1.0 mg (pH 3–10) or 1.5 mg (pH 4–7) of proteins. Focusing was carried out in a Bio-Rad Protean isoelectric focusing (IEF) cell. The voltage setting was 200 V for 30 min, 500 V for 30 min, 1 000 V for 1 h, 2 000V for 1 h, and 8 000 V for 5 h, to a total about 50 000 V∙h. After, IPG strips were equilibrated for 2×10 min in 6 mol/L urea, 30% (v/v) glycerol (Bio-Rad), 0.05 g/ml sodium dodecyl sulfate (SDS; Bio-Rad) in 0.05 mol/L Tris-HCl (pH 6.8; Genebase) containing 0.01 g/ml DTT for the first equilibration step and 0.025 g/ml iodoacetamide (Bio-Rad) for the second equilibration step. The samples were then transferred onto a 0.12 g/ml polyacrylamide gel. Electrophoresis was performed in Tris/glycine/SDS buffer on a Multiphor system (Amersham Pharmacia Biotech) according to the manufacturer’s recommendations. For calibration, low-molecular weight marker proteins (Amersham Biosciences) were applied on the gel via a small piece of filter paper. Gels were stained overnight with Coomassie brilliant blue G-250 (Genebase) according to Neuhoff et al. (1988) and scanned using VersaDoc image system (Bio-Rad). 2-DE gels were processed using PDQuest software V.7.3 (Bio-Rad). Spot quantity normalization occurred through the whole match set, which included all 12 2-DE gels. The gel used as reference in the PDQuest match set corresponded to a co-migration of protein extracts from both Lee68 and N2899 cultivars. Only those with significant (quantitative changes more than two-fold in abundance) and reproducible changes in three replicates were used for further analysis.
2.5. Protein in-gel digestion and matrix-assisted laser desorption ionization-time of flight-mass spectrometry (MALDI-TOF-MS)
Protein spots of interest were excised from the stained gels and in-gel digestion was performed as follows. Gel pieces were washed three times with Milli-Q water, and 50% (v/v) acetonitrile (Sigma) containing 100 mmol/L ammonium bicarbonate (Genebase) was used to remove the dye. Proteins in the gels were reduced in 10 mmol/L DTT dissolved in 50 mmol/L NH4HCO3 (Genebase) solution for at least 1 h at 56 °C, and then incubated with 50 mmol/L iodoacetamide in 50 mmol/L NH4HCO3 at room temperature for 40 min. Gels were then dried by vacuum centrifugation, and incubated for 14 h at 37 °C with 10 μl of 12.5 µg/ml trypsin (modified porcine trypsin, sequencing grade, Promega) in 50 mmol/L NH4HCO3. The resulting tryptic fragments were eluted by diffusion into 50% (v/v) acetonitrile and 0.5% (v/v) TCA, and dried in a speed vacuum. The dried samples were resuspended in 2 μl 0.5% (v/v) trifluoroacetic acid (Genebase). Each sample was mixed with the supernatant of 60% (v/v) acetonitrile saturated with α-cyano-4-hydroxycinnamic acid (Genebase) (1:1, v/v), and then air-dried on the flat surface of a sample plate. The samples were then analyzed with MALDI-TOF-MS (Reflex III, Bruker, Germany) in positive ion reflector mode at an accelerating voltage of 20 kV. Spectra were calibrated using trypsin autolysis products (m/z 842.51 and 2 211.10) as internal standards, and a mixture of standard peptides as external standards.
2.6. Database queries and protein identification
Protein identification was performed by querying peptide mass fingerprinting (PMF) data in the soybean UniGene database (ftp://ftp.ncbi.nih.gov/repository/UniGene/Glycine_max/) using MS-Fit program of Protein Prospector (http://prospector.ucsf.edu). The following parameters were used for database searches with MALDI-TOF peptide mass data: mono-isotopic peak; mass tolerance, 0.2 Da; missed cleavages, 1; and allowed modifications, carbamidomethylation of Cys and oxidation of Met. A positive identification of the protein followed the procedure of Hajduch et al. (2005).
3. Results
3.1. Effects of salinity on seed germination
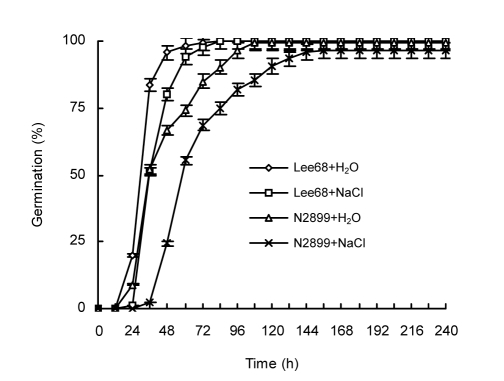
Under optimal conditions, over 98% of Lee68 and N2899 seeds germinated after a mean of 1.5 and 2.1 d, respectively. When exposed to 100 mmol/L NaCl, the final germination percentage in both cultivars was not affected. However, the mean germination times of Lee68 and N2899 were delayed by 0.3 and 1.0 d, respectively, compared to the controls. In addition, control Lee68 and N2899 seeds required 1.2 and 1.5 d, respectively, to reach 50% germination, while seeds germinated in the presence of 100 mmol/L NaCl required 1.5 and 2.5 d, respectively. From imbibition till maximum germination, Lee68 and N2899 took 3.5 and 4.5 d, respectively. Under salt stress this period was 3.5 and 6.5 d, respectively (Fig. 2). We also observed that there were many more abnormally germinated N2899 seeds compared to Lee68. Moreover, germination of N2899 seeds was completely inhibited by exposure to over 150 mmol/L NaCl (data not shown). These observations suggest that 100 mmol/L NaCl affected soybean seed germination, especially in N2899. However, moderate salt stress intensity only delayed germination time, and did not have a severe impact on the final germination percentage.
Fig. 2.
Effects of 100 mmol/L salt concentration on the germination percentages of Lee68 and N2899 seeds
Lee68+H2O: control Lee68; Lee68+NaCl: NaCl-treated Lee68; N2899+H2O: control N2899; N2899+NaCl: NaCl-treated N2899
3.2. Effects of salinity on IAA, GA1+3, ABA, and iPAs levels
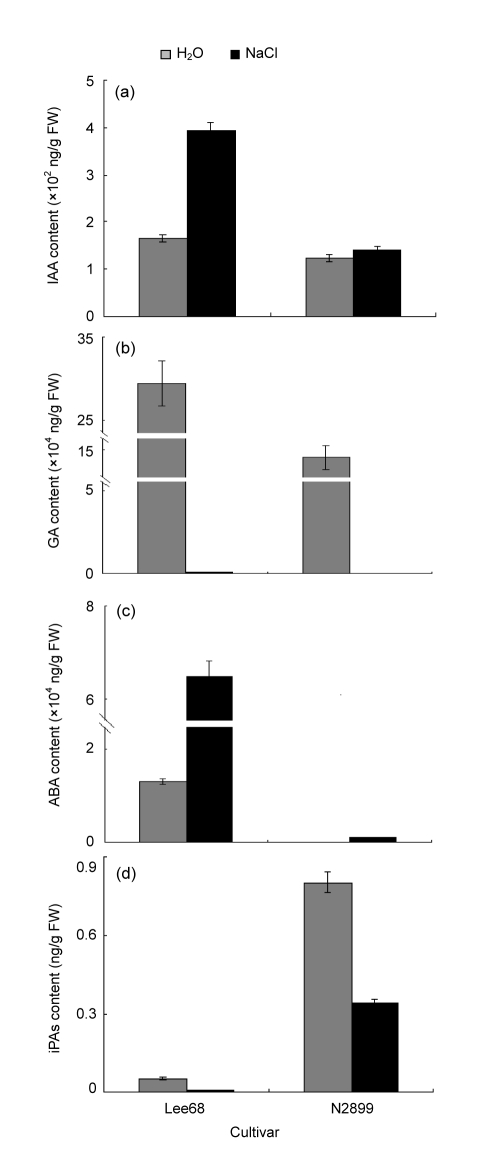
Hormone levels are related to seed germination and stress. Therefore, we detected the endogenous ABA, IAA, GA1+3, and iPAs levels in Lee68 and N2899 seeds germinated in moderate salinity conditions (Fig. 3). The IAA content of salt-treated germinated Lee68 seeds was significantly higher than that of the control (P<0.01). There was no difference in IAA levels among control Lee68, salt-treated N2899, and control N2899 seeds (P>0.05). Salinity had a stronger effect on the GA1+3 level. The GA1+3 content significantly decreased in both cultivars under salt stress (P<0.001). The GA1+3 content in control Lee68 was higher than that in control N2899 (P<0.05). ABA content significantly increased in response to salinity (P<0.001), while the ABA content in salt-treated N2899 was lower than that in control Lee68 (P<0.001). The iPAs content in these four samples was low compared with the other hormones and was decreased by salt in both cultivars (P<0.01). However, the iPAs content in salt-treated N2899 was higher than that in control Lee68 (P<0.01).
Fig. 3.
Effects of salinity on IAA (a), GA (b), ABA (c), and iPAs (d) in Lee68 and N2899 germinated seeds
3.3. 2-DE maps of Lee68 and N2899
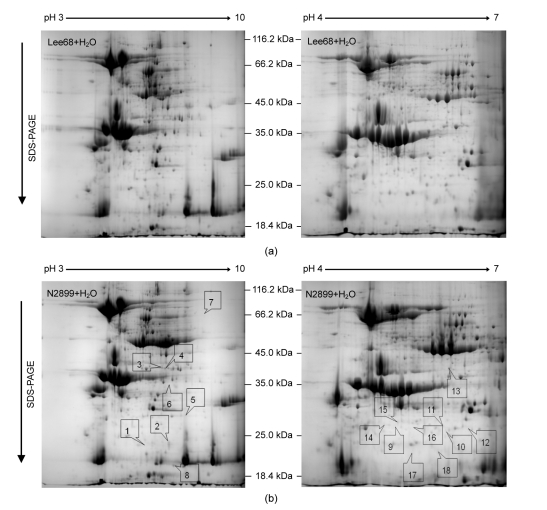
Soybean Lee68 and N2899 displayed different seed vigors and germination times. Therefore, we investigated the proteins expressed in the germinated seeds. Total proteins from Lee68 and N2899 germinated seeds were extracted and separated by 2-DE gels. Initial separations were performed with pH 3–10 IPG strips. The gel maps showed that about 70% of the protein spots were in the region of pH 4–7, as analyzed by PDQuest software (Fig. 4). Therefore, additional analyses with pH 4–7 IPG strips were performed to improve spot resolution. The 2-DE maps of the control Lee68 and N2899 germinated seeds in the pH 3–10 and pH 4–7 ranges are shown in Fig. 4. About 350 protein spots could be resolved in pH 3–10 2-DE gels and 650 protein spots in pH 4–7 2-DE gels stained by Coomassie brilliant blue G-250. In all, there were more than 90 differentially expressed proteins between the 2-DE maps of Lee68 and N2899, although the majority of protein spots were similar. These differentially expressed proteins showed varietal differences and might be associated with resistances, agronomic traits, and qualities between cultivars.
Fig. 4.
2-DE maps of the control Lee68 and N2899 seeds germinated at pH 3–10 and pH 4–7
(a) Lee68+H2O: control Lee68; (b) N2899+H2O: control N2899
3.4. Salt-responsive proteins
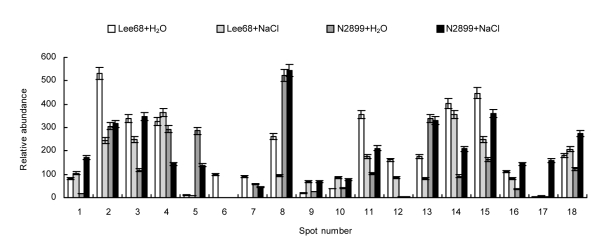
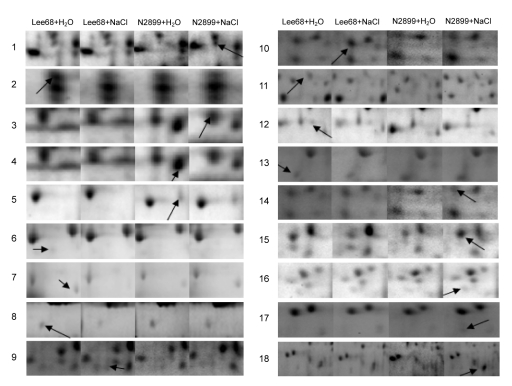
To investigate the response of soybean seed germination to non-lethal concentration salinity levels, Lee68 and N2899 seeds were exposed to 100 mmol/L NaCl until the radicle protruded from the seed coat. Whole proteins from germinated seeds were analyzed by high-resolution 2-DE. Eighteen protein spots showed more than two-fold reproducible differences in abundance as a result of salt stress in both cultivars. The positions of these proteins are indicated by number in Fig. 4. Ten proteins were in the pH 4–7 2-DE gels and eight in the pH 3–10 2-DE gels. The relative abundances of these proteins are shown in Fig. 5. Compared with their controls, 10 proteins were differentially displayed in Lee68 2-DE gels and 12 in N2899 2-DE gels under salt stress. Fig. 6 shows the enlarged maps of the 18 protein spots. These changes were relatively small when compared with the soybean cultivars, but they directly responded to salt stress.
Fig. 5.
Histograms showing the volume changes of 18 differentially expressed spots in Lee68 and N2899 seeds germinated under salt stress
Lee68+H2O: control Lee68; Lee68+NaCl: NaCl-treated Lee68; N2899+H2O: control N2899; N2899+NaCl: NaCl-treated N2899
Fig. 6.
Enlarged maps of the 18 differentially expressed protein spots in Lee68 and N2899 seeds germinated under salt stress Lee68+H2O: control
Lee68; Lee68+NaCl: NaCl-treated Lee68; N2899+H2O: control N2899; N2899+NaCl: NaCl-treated N2899
These differentially expressed proteins were excised from the 2-DE gels and subjected to MALDI-TOF-MS. All proteins obtained PMF. By querying the PMF in the soybean UniGene database, nine proteins were successfully identified (Table 1). The data in Table 1 include assigned protein spot numbers, experimental molecular mass and pI, theoretical molecular mass and pI, number of peptides matched, sequence coverage, number of UniGene accessions, and homologous proteins. The possible functions of these proteins are discussed below.
Table 1.
Differentially expressed proteins identified by PMF query
| Spot No. | Theoretical Mr/pI | Experimental Mr/pI | No. of peptides matched | Sequence coverage (%) | Accession No. | Protein name |
| 1 | 28.7/6.3 | 22.7/6.7 | 8 | 31 | Gma.15591 | Unknown protein |
| 3 | 36.9/6.4 | 38.7/7.2 | 7 | 61 | Gma.17053 | Glyceraldehyde 3-phosphate dehydrogenase |
| 7 | 63.8/8.2 | 62.9/8.3 | 19 | 25 | Gma.8130 | Malate synthase |
| 9 | 28.1/5.7 | 26.7/5.3 | 6 | 23 | Gma.18200 | Ferritin |
| 10 | 24.9/6.3 | 26.2/6.1 | 5 | 26 | Gma.448 | 20S proteasome β subunit |
| 11 | 25.4/5.7 | 27.0/6.0 | 7 | 30 | Gma.1917 | Glutathione S-transferase (GST) 10 |
| 14 | 26.1/5.8 | 26.2/6.1 | 5 | 27 | Gma.7612 | Hypothetical 26.0-kDa protein |
| 15 | 27.4/5.9 | 27.0/6.0 | 5 | 19 | Gma.8540 | Seed maturation protein PM36 |
| 16 | 23.6/5.7 | 26.8/5.6 | 4 | 25 | Gma.8517 | Glutathione S-transferase (GST) 9 |
4. Discussion
4.1. Phytohormones
We investigated the effects of salinity on endogenous IAA, GA1+3, ABA, and iPAs in germinated seeds of Lee68 and N2899. The ABA content increased, and GA1+3 and iPAs decreased in response to salinity, while the change of IAA content depended on the cultivar. IAA has a major role in regulating plant growth. Bianco and Defez (2009) reported that the IAA-overproducing RD64 strain showed an increased tolerance to 0.5 mol/L NaCl in Medicagotruncatula. IAA can strengthen the capacity of resistance of the soybean to saline environment (Wei and Chen, 2000). The level of IAA in salt-treated Lee68 was more than two-fold higher than that in the control, but was the same as that in N2899. The increased IAA in salt-tolerant Lee68 might help seed germination under salt stress. The decreased endogenous GA and increased ABA contents have been observed in salt-stressed soybean. GA3 ameliorates the adverse effects of salt stress and restores normal growth and development of soybean (Hamayun et al., 2010b). Germination of stressed seeds was partially restored by the addition of exogenous cytokinin (CTK) (Gidrol et al., 1994). However, Kaur et al. (1998) found that GA3 was more effective than CTK in enhancing the reduced germination and seedling growth of chickpea seeds under salt stress. GA synthesis affects not only alterations in protein expression, but also increases in germination rate, promotes root and shoot length during seed germination, and promotes early seedling development (Gallardo et al., 2002; Kim et al., 2008). In the present study, GA content was significantly reduced and seed germination time was delayed by salt. ABA is involved in responses to environmental stress such as salinity (Jia et al., 2002), and is required by the plant for stress tolerance (Hamayun et al., 2010a). Umezawa et al. (2001) found that the leaf ABA content in Lee (salt-tolerant) increased significantly under salt stress, while that in Enrei (salt-sensitive) showed only a slight increase. It is thus possible that ABA enhances salt tolerance in soybean. Our results also showed that salinity increased the ABA contents of both cultivars. Moreover, the ABA level in Lee68 was much higher than that in N2899, confirming that it might be related to the salt stress tolerance of Lee68. The contents and change ranges of hormones, except iPAs, in Lee68 were higher than those in N2899. This might be one of the reasons why N2899 is more sensitive to salt than Lee68.
4.2. Differentially expressed proteins
In the plant life cycle, seed germination and seedling stages are key developmental stages conditioning the final yield of crops. Both are very sensitive to salt stress. Consequently, researchers have submitted crop seeds to salt treatments and sampled them after varying stress time. However, germination and seedling growth are delayed by salt. Therefore, we studied all soybean seeds that completed the germination process under salt stress in two cultivars with different salt tolerances. We then sampled the germinated seeds whose radicles protruded from the seed coat by 2–3 mm. Eighteen protein spots showed changes in abundance in response to 100 mmol/L NaCl stress in the two cultivars, of which nine proteins were identified.
Spots 9 and 10 were up-regulated by salt in both cultivars. Spot 9 was identified as ferritin. Ferritin, a class of iron-storage proteins, is composed of at least two different subunits and its level decreases gradually during soybean germination (Masuda et al., 2001). However, the abundance of ferritin increased in response to salt stress in both cultivars. A high concentration of salt leads to oxidative damage. Ferritin could be involved in defense mechanisms against iron-mediated oxidative stress (Briat et al., 1999). Proteomic investigations on soybean root also revealed the accumulation ferritin under drought stress (Alam et al., 2010). Therefore, it might have an important role in soybean seed germination and seedlings under stress. Induction of ferritin by ABA has been documented at both transcript and protein levels (Ravet et al., 2009). In maize plantlets, iron overload led to a five-fold increase in ABA concentration in roots and leaves, and ferritin mRNA accumulated in response to exogenous ABA treatment (Lobréaux et al., 1993). In this study, the endogenous ABA and ferritin levels were up-regulated by salt. The result supports the possible involvement of ABA in ferritin gene regulation.
Spot 10 was identified as 20S proteasome subunit β-6. The 20S proteasome is the catalytic core of the 26S proteasome, a proteolytic complex involved in recognizing and catabolizing ubiquitin-protein to remove abnormal proteins (Sassa et al., 2000; Smalle and Vierstra, 2004). The proteasome also operates in the stress response by removing abnormal proteins (Imin et al., 2006). 20S proteasome α subunit A was up-regulated in soybean under osmotic stress (Toorchi et al., 2009). The up-regulation of the 20S proteasome in both cultivars after stress could be associated to the degradation of oxidatively-damaged proteins caused by salt stress. Recent evidence has suggested a role for the ubiquitin-proteasome pathway in CTK, GA, and ABA signalings (Itoh et al., 2003). 26S ubiquitin-proteasome regulatory subunit 4 homolog levels were increased during rice seed germination by GA and ABA (Kim et al., 2008).
Spots 1, 3, 14, and 16 were up-regulated by salt in N2899 and were unaffected in Lee68. Although Spots 1 and 14 were matched with UniGenes, providing evidence for the existence of the proteins, they were annotated as unknown proteins. Glyceraldehyde 3-phosphate dehydrogenase (GPD) (Spot 3) is a ubiquitous enzyme involved in glycolysis and gluconeogenesis (Duée et al., 1996). Basic metabolism change is a general response to stress. GPD is one of target genes regulating the response of cells to salt stress and may aid in the development of new salt-tolerant cultivars in soybean (Nouri et al., 2011). Jeong et al. (2001) transferred GPD gene to potato to improve salt tolerance in transgenic potato plants. Spot 16 was identified as glutathione S-transferase (GST) 9. GSTs are encoded by a large and diverse gene family in plants and there are 25 identified accurate full-length sequence clones in soybean. It is clear that GST activity levels frequently increase in response to stimuli that cause oxidative damage, but the mechanisms involved in protection are unclear (McGonigle et al., 2000). Overexpression of GSTs and glutathione peroxidases enhanced the growth of transgenic tobacco seedlings during chilling and salt stress (Roxas et al., 1997). GSTs have an important role in the response of plants to changing environmental conditions. Therefore, gene engineering strategies involving GSTs could improve soybean salt tolerance. Furthermore, they might have a role in hormonal regulation (Marrs, 1996). Kim et al. (2008) reported that GST was modulated by GA and ABA.
Spots 11 and 15 were down-regulated in Lee68, but up-regulated in N2899 under salt stress. Interestingly, Spot 11 was identified as GST 10. This result indicates that one protein changes its expression differently between cultivars and that different members of a family are differentially regulated in the same cultivar under salt stress. Spot 15 was identified as seed maturation protein PM36. It is synthesized at late embryogenesis and is degraded rapidly at the early stage of seed germination (Blackman et al., 1991; Hsing et al., 1998). In contrast to most seed maturation proteins, protein PM36 contains a TENA_THI-4 conservative domain. It was predicted that PM36, similarly to seed maturation protein PM4, acted as a storage form of biotin to support seedling growth during germination (Hsing et al., 1998). In the present study, PM36 was up-regulated by salt in N2899. We infer that PM36 might be similar to other late embryogenesis abundant (LEA) proteins and has a protective role in the plant cell under stress conditions (Skriver and Mundy, 1990).
Spot 7 was down-regulated by salt in Lee68, but was unaffected in N2899. Malate synthase (Spot 7) is a characteristic enzyme of the glyoxylate cycle. In germinating oilseeds, the glyoxylate cycle has a key role in converting acetyl-coenzyme A produced by fatty acid β-oxidation into oxaloacetate, and subsequently into sugar (Smith, 2002). It was confirmed that malate synthase was activated in the prolonged anaerobic environment (Ying et al., 2005). The significance of the diminished levels of malate synthase in Lee68 requires further research.
In summary, the proteins we identified are related to salt stress. GPD, GST 9, GST 10, and seed maturation protein PM36 have protective roles in stress tolerance. In control Lee68, these proteins were maintained a high level and, although they were down-regulated by salt stress, they still remained at a certain level. However, in control N2899, their levels were the lowest and were up-regulated by salt stress. It can be hypothesized that these proteins help seed germination under salt stress and Lee68 itself has a stronger resistance mechanism. NaCl at 100 mmol/L might represent low salinity to Lee68, but relatively high salinity to N2899. We could also infer this interpretation from the delayed mean germination time and increased duration of germination. Therefore, N2899 was injured more than Lee68 under a non-lethal concentration of NaCl (100 mmol/L). To achieve the process of germination, these defensive proteins were up-regulated.
The expression and regulation of genes under salt stress is a complicated process, and is affected by experimental materials, salt concentration, treatment methods, and stress time. Our research contributes to the understanding of the mechanisms of seed germination in response to salt stress. Future studies will be directed towards the function of the identified proteins and the regulatory network of hormones in the salt response.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30800692), the National Basic Research Program (973) of China (Nos. 2010CB125906 and 2009CB118400), and the National High-Tech R & D Program (863) of China (No. 2006AA10Z1C1)
References
- 1.Aghaei K, Ehsanpour AA, Shah AH, Komatsu S. Proteome analysis of soybean hypocotyl and root under salt stress. Amino Acids. 2008;36(1):91–98. doi: 10.1007/s00726-008-0036-7. [DOI] [PubMed] [Google Scholar]
- 2.Agrawal GK, Hajduch M, Graham K, Thelen JJ. In-depth investigation of soybean seed-filling proteome and comparison with a parallel study of rapeseed. Plant Physiol. 2008;148(1):504–518. doi: 10.1104/pp.108.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alam I, Sharmin SA, Kim KH, Yang JK, Choi MS, Lee BH. Proteome analysis of soybean roots subjected to short-term drought stress. Plant Soil. 2010;333(1-2):491–505. doi: 10.1007/s11104-010-0365-7. [DOI] [Google Scholar]
- 4.Almansouri M, Kinet JM, Lutts S. Effect of salt and osmotic stresses on germination in durum wheat (Triticum durum Desf.) Plant Soil. 2001;231(2):243–254. doi: 10.1023/A:1010378409663. [DOI] [Google Scholar]
- 5.Bianco C, Defez R. Medicago truncatula improves salt tolerance when nodulated by an indole-3-acetic acid-overproducing Sinorhizobium meliloti strain. J Exp Bot. 2009;60(11):3097–3107. doi: 10.1093/jxb/erp140. [DOI] [PubMed] [Google Scholar]
- 6.Blackman SA, Wettlaufer SH, Obendorf RL, Leopold AC. Maturation proteins associated with desiccation tolerance in soybean. Plant Physiol. 1991;96(3):868–874. doi: 10.1104/pp.96.3.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer JS. Plant productivity and environment. Science. 1982;218(4571):443–448. doi: 10.1126/science.218.4571.443. [DOI] [PubMed] [Google Scholar]
- 8.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72(1-2):248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 9.Briat JF, Lobréaux S, Grignon N. Regulation of plant ferritin synthesis: how and why. Cell Mol Life Sci. 1999;56(1-2):155–166. doi: 10.1007/s000180050014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen JG, Du XM, Zhou X, Zhao HY. Levels of cytokinins in the ovules of cotton mutants with altered fiber development. J Plant Growth Regul. 1997;16(3):181–185. doi: 10.1007/PL00006994. [DOI] [Google Scholar]
- 11.Chen JG, Zhou X, Zhang YZ. Gibberellin responding and non-responding dwarf mutants in foxtail millet. Plant Growth Regul. 1998;26(1):19–24. doi: 10.1023/A:1006091601256. [DOI] [Google Scholar]
- 12.Chen JG, Cheng SH, Cao WX, Zhou X. Involvement of endogenous plant hormones in the effect of mixed nitrogen source on growth and tillering of wheat. J Plant Nutr. 1998;21(1):87–97. doi: 10.1080/01904169809365385. [DOI] [Google Scholar]
- 13.Cheng L, Gao X, Li S, Shi M, Javeed H, Jing X, Yang G, He G. Proteomic analysis of soybean [Glycine max (L.) Meer.] seeds during imbibition at chilling temperature. Mol Breeding. 2010;26(1):1–17. doi: 10.1007/s11032-009-9371-y. [DOI] [Google Scholar]
- 14.Duée E, Olivier-Deyris L, Fanchon E, Corbier C, Branlant G, Dideberg O. Comparison of the structures of wild-type and a N313T mutant of Escherichia coli glyceraldehyde 3-phosphate dehydrogenases: implication for NAD binding and cooperativity. J Mol Biol. 1996;257(4):814–838. doi: 10.1006/jmbi.1996.0204. [DOI] [PubMed] [Google Scholar]
- 15.Gallardo K, Job C, Groot SP, Puype M, Demol H, Vandekerckhove J, Job D. Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 2002;129(2):823–837. doi: 10.1104/pp.002816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gidrol X, Lin WS, Dégousée N, Yip SF, Kush A. Accumulation of reactive oxygen species and oxidation of cytokinin in germinating soybean seeds. Eur J Biochem. 1994;224(1):21–28. doi: 10.1111/j.1432-1033.1994.tb19990.x. [DOI] [PubMed] [Google Scholar]
- 17.Gygi SP, Aebersold R. Mass spectrometry and proteomics. Curr Opin Chem Biol. 2000;4(5):489–494. doi: 10.1016/S1367-5931(00)00121-6. [DOI] [PubMed] [Google Scholar]
- 18.Hajduch M, Ganapathy A, Stein JW. A systematic proteomic study of seed filling in soybean. Establishment of high-resolution two-dimensional reference maps, expression profiles, and an interactive proteome database. Plant Physiol. 2005;137(4):1397–1419. doi: 10.1104/pp.104.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hamayun M, Khan SA, Shinwari ZK, Khan AL, Ahmad N, Lee IJ. Effect of salt stress on growth attributes and endogenous growth hormones of soybean cultivar Hwangkeumkong. Pak J Bot. 2010;42(5):3103–3112. [Google Scholar]
- 20.Hamayun M, Khan SA, Khan AL, Shin JH, Ahmad B, Shin DH, Lee IJ. Exogenous gibberellic acid reprograms soybean to higher growth and salt stress tolerance. J Agric Food Chem. 2010;58(12):7226–7232. doi: 10.1021/jf101221t. [DOI] [PubMed] [Google Scholar]
- 21.Hsing YC, Tsou CH, Hsu TF, Chen ZY, Hsieh KL, Hsieh JS, Chow TY. Tissue- and stage-specific expression of a soybean (Glycine max L.) seed maturation biotinylated protein. Plant Mol Biol. 1998;38(3):481–490. doi: 10.1023/A1006079926339. [DOI] [PubMed] [Google Scholar]
- 22.Imin N, Kerim T, Weinman JJ, Rolfe BG. Low temperature treatment at the young microspore stage induces protein changes in rice anthers. Mol Cell Proteom. 2006;5(2):274–292. doi: 10.1074/mcp.M500242-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Itoh H, Matsuoka M, Camille M. A role for the ubiquitin-26S proteasome pathway in gibberellin signaling. Trends Plant Sci. 2003;8(10):492–497. doi: 10.1016/j.tplants.2003.08.002. [DOI] [PubMed] [Google Scholar]
- 24.Jeong MJ, Park SC, Byun MO. Improvement of salt tolerance in transgenic potato plants by glyceraldehyde-3-phosphate dehydrogenase gene transfer. Mol Cell. 2001;12(2):185–189. [PubMed] [Google Scholar]
- 25.Jia GX, Zhu ZQ, Chang FQ, Li YX. Transformation of tomato with the BADH gene from Atriplex improves salt tolerance. Plant Cell Rep. 2002;21(2):141–146. doi: 10.1007/s00299-002-0489-1. [DOI] [Google Scholar]
- 26.Kaur S, Gupta AK, Kaur N. Gibberellin A3 reverses the effect of salt stress in chickpea (Cicerarietinum L.) seedlings by enhancing amylase activity and mobilization of starch in cotyledons. Plant Growth Regul. 1998;26(2):85–90. doi: 10.1023/A:1006008711079. [DOI] [Google Scholar]
- 27.Kim ST, Kang SY, Wang Y, Kim SG, Hwang DH, Kang KY. Analysis of embryonic proteome modulation by GA and ABA from germinating rice seeds. Proteomics. 2008;8(17):3577–3587. doi: 10.1002/pmic.200800183. [DOI] [PubMed] [Google Scholar]
- 28.Levitt J. Responses of Plants to Environmental Stresses: Water, Radiation, Salt and Other Stresses. Vol. 2. New York: Academic Press; 1980. pp. 365–402. [Google Scholar]
- 29.Lobréaux S, Hardy T, Briat JF. Abscisic acid is involved in the iron-induced synthesis of maize ferritin. EMBO J. 1993;12(2):651–657. doi: 10.1002/j.1460-2075.1993.tb05698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo QY, Yu BJ, Liu YL. Differential sensitivity to chloride and sodium ions in seedlings of Glycine max and G. soja under NaCl stress. J Plant Physiol. 2005;162(9):1003–1012. doi: 10.1016/j.jplph.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 31.Marrs KA. The functions and regulation of glutathione S-transferases in plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47(1):127–158. doi: 10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- 32.Masuda T, Goto F, Yoshihara T. A novel plant ferritin subunit from soybean that is related to a mechanism in iron release. J Biol Chem. 2001;276(22):19575–19579. doi: 10.1074/jbc.M011399200. [DOI] [PubMed] [Google Scholar]
- 33.McGonigle B, Keeler SJ, Lau SMC. A genomics approach to the comprehensive analysis of the glutathione S-transferase gene family in soybean and maize. Plant Physiol. 2000;124(3):1105–1120. doi: 10.1104/pp.124.3.1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mooney BP, Thelen JJ. High-throughput peptide mass fingerprinting of soybean seed proteins: automated workflow and utility of UniGene expressed sequence tag databases for protein identification. Phytochemistry. 2004;65(3):1733–1744. doi: 10.1016/j.phytochem.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 35.Morgan PW. Effects of Abiotic Stresses on Plant Hormone Systems. In: Alscher RG, Cumming JR, editors. Stress Responses in Plants: Adaptation and Acclimation Mechanism. New York: Wiley-Liss; 1990. [Google Scholar]
- 36.Munns R. Genes and salt tolerance: bringing them together. New Phytol. 2005;167(3):645–660. doi: 10.1111/j.1469-8137.2005.01487.x. [DOI] [PubMed] [Google Scholar]
- 37.Neuhoff V, Arold N, Taube D, Ehrhardt W. Improved staining of proteins in polyacrylamide gels including isoelectric focusing gels with clear background at nanogram sensitivity using Coomassie Brilliant Blue G-250 and R-250. Electrophoresis. 1988;9(6):205–262. doi: 10.1002/elps.1150090603. [DOI] [PubMed] [Google Scholar]
- 38.Nouri MZ, Komatsu S. Comparative analysis of soybean plasma membrane proteins under osmotic stress using gel-based and LC MS/MS-based proteomics approaches. Proteomics. 2010;10(10):1930–1945. doi: 10.1002/pmic.200900632. [DOI] [PubMed] [Google Scholar]
- 39.Nouri MZ, Toorchi M, Komatsu S. Proteomics Approach for Identifying Abiotic Stress Responsive Proteins in Soybean. In: Sudaric A, editor. Soybean Molecular Aspects of Breeding. Croatia: InTech; 2011. pp. 187–214. [Google Scholar]
- 40.Ravet K, Touraine B, Boucherez J, Briat JF, Gaymard F, Cellier F. Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis . Plant J. 2009;57(3):400–412. doi: 10.1111/j.1365-313X.2008.03698.x. [DOI] [PubMed] [Google Scholar]
- 41.Roxas VP, Smith RK, Allen ER. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat Biotechnol. 1997;15(10):988–991. doi: 10.1038/nbt1097-988. [DOI] [PubMed] [Google Scholar]
- 42.Sassa H, Oguchi S, Inoue T. Primary structural features of the 20S proteasome subunits of rice (Oryza sativa) Gene. 2000;250(1-2):61–66. doi: 10.1016/S0378-1119(00)00190-6. [DOI] [PubMed] [Google Scholar]
- 43.Skriver K, Mundy J. Gene expression in response to abscisic acid and osmotic stress. Plant Cell. 1990;2(6):503–512. doi: 10.1105/tpc.2.6.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smalle J, Vierstra RD. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev Plant Biol. 2004;55(1):555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 45.Smith SM. Does the glyoxylate cycle have an anaplerotic function in plants? Trends Plant Sci. 2002;7(1):12–13. doi: 10.1016/S1360-1385(01)02189-6. [DOI] [PubMed] [Google Scholar]
- 46.Sobhanian H, Razavizadeh R, Nanjo Y, Ehsanpour AA, RastgarJazii F, Motamed N, Komatsu S. Proteome analysis of soybean leaves, hypocotyls and roots under salt stress. Proteome Sci. 2010;8(1):19. doi: 10.1186/1477-5956-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toorchi M, Yukawa K, Nouri MZ, Komatsu S. Proteomics approach for identifying osmotic-stress related proteins in soybean roots. Peptides. 2009;30(12):2108–2117. doi: 10.1016/j.peptides.2009.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Umezawa T, Shimizu K, Kato M, Ueda T. Effects of non-stomatal components on photosynthesis in soybean under salt stress. Jpn J Trop Agric. 2001;45(1):57–63. [Google Scholar]
- 49.Watson BS, Asirvatham VS, Wang LJ, Sumner LW. Mapping the proteome of Barrel Medic (Medicagotruncatula) Plant Physiol. 2003;131(3):1104–1123. doi: 10.1104/pp.102.019034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wei AL, Chen YZ. Effect of IAA on soybean seedling’s membrance injury and salt resistance. Acta Bot Boreal Occident Sin. 2000;20(3):410–414. (in Chinese) [Google Scholar]
- 51.Xu XY, Zheng R, Li CM, Gai JY, Yu DY. Differential proteomic analysis of seed germination in soybean. Prog Biochem Biophys. 2006;33(11):1106–1112. (in Chinese) [Google Scholar]
- 52.Ying LU, Wu YR, Han B. Anaerobic induction of isocitrate lyase and malate synthase in submerged rice seedlings indicates the important metabolic role of the glyoxylate cycle. Acta Biochem Biophys Sin. 2005;37(6):406–414. doi: 10.1111/j.1745-7270.2005.00060.x. [DOI] [PubMed] [Google Scholar]
- 53.Zhen Y, Qi JL, Wang SS, Su J, Xu GH, Zhang MS, Miao L, Peng XX, Tian D, Yang YH. Comparative proteome analysis of differentially expressed proteins induced by Al toxicity in soybean. Physiol Plant. 2007;131(4):542–554. doi: 10.1111/j.1399-3054.2007.00979.x. [DOI] [PubMed] [Google Scholar]
- 54.Zhu JK. Salt and drought stress signals transduction in plants. Annu Rev Plant Biol. 2002;53(1):247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]