Abstract
Rice straw is always regarded as a by-product of rice production, but it could be a significant energy source for ruminant animals. Knowledge of the genetic variation and genetic architecture of cell wall traits will facilitate rice breeders by improving relevant traits through selective breeding and genetic engineering. The common wild rice, Oryza rufipogon Griff., which is considered to be the progenitor of Oryza sativa, has been widely utilized for the identification of genes of agronomic importance for rice genetic improvement. In the present study, the mapping of quantitative trait loci (QTLs) for acid detergent fiber (ADF), neutral detergent fiber (NDF), acid detergent lignin (ADL), and ADL/NDF ratio was carried out in two environments using a backcrossed inbred line (BIL) population derived from a cross between the recurrent parent Xieqingzao B (XB) and an accession of Dongxiang wild rice (DWR). The results indicated that all four traits tested were continuously distributed among the BILs, but many BILs showed transgressive segregation. A total of 16 QTLs were identified for the four traits, but no QTLs were in common in two environments, suggesting that environment has dramatic effects on fiber and lignin syntheses. Compared to the QTL positions for grain yield-related traits, there were no unfavorable correlations between grain yield components and cell wall traits in this population. The QTLs identified in this study are useful for the development of dual-purpose rice varieties that are high in grain yield and are also high in straw quality.
Keywords: Rice straw, Acid detergent fiber, Lignin, Quantitative trait loci
1. Introduction
Rice straw is a by-product of rice production, making up 50% of the dry weight of rice plants (Putun et al., 2004). Rice straw used to be burned, and thus, causes pollutions to the environment. However, other applications of rice straw could benefit rice farmers (Putun et al., 2004; Kong et al., 2005). As one of the most important applications, it could be used as a source of carbohydrates for ruminants (Agbagla-Dohnani et al., 2001). Another popular application is that it could be used for the production of cellulosic ethanol. However, as a minor component of the cell wall of plants, lignin has long been recognized for its negative impact on forage quality, paper manufacturing, and, more recently, cellulosic biofuel production (Li et al., 2008). Decreasing the content of lignin to an appropriate level may improve the feed quality and cellulosic biofuel production efficiency (Agbagla-Dohnani et al., 2001; Li et al., 2008). However, too great a decrease may cause crops to become more susceptible to disease and pests, and more likely to suffer from abiotic stress due to the fragility of fibers. Understanding of the genetic basis of cell wall components (CWCs) in rice straw may contribute to breeding and selection activities.
Grass cell walls are primarily composed of glucose bound in cellulose microfibrils that are surrounded by a matrix of hemicellulose (mixed sugar polysaccharide) and lignin. The straw quality for forage utilization can be quantified as acid detergent fiber (ADF), neutral detergent fiber (NDF), and acid detergent lignin (ADL) (van Soest, 1994; Krakowsky et al., 2005). ADF consists largely of cellulose and lignin, NDF consists largely of cellulose, hemicelluloses, and lignin. They are regarded as important and good indicators of forage quality because of their negative relation to digestibility for livestock animals (van Soest, 1994; Agbagla-Dohnani et al., 2001; Kong et al., 2005). These traits also provide good predictions of ethanol yield and production efficiency if straw is used for biofuel production.
Analyses of genetic diversity and genetic control of CWCs have been conducted in the major crops, such as maize (Méchin et al., 2001; Cardinal et al., 2003; Krakowsky et al., 2005; 2006; Barrière et al., 2008; Lorenz et al., 2010; Truntzler et al., 2010), barley (Grando et al., 2005; Abdel-Haleem et al., 2010), ryegrass (Cogan et al., 2005; Xiong et al., 2006), and rice (Xie et al., 2006; Bao et al., 2007; Dong et al., 2008). Due to the importance of fiber quality in cotton and wood, genetic analysis by quantitative trait locus (QTL) mapping has also been conducted in cotton (Chen et al., 2009; Wu et al., 2009) and trees such as Pinus taeda L. (González-Martínez et al., 2007). Most QTLs have small genetic effects, indicating that straw quality traits are primarily controlled by many small-effect genes. The relationships between ADF and NDF are consistently positive in maize (Cardinal et al., 2003; Krakowsky et al., 2005), barley (Grando et al., 2005), and rice (Bao et al., 2007), whereas the relationships between ADF and ADL varied in different studies (Cardinal et al., 2003; Krakowsky et al., 2005; 2006). Cardinal et al. (2003) reported that the relationship between ADF, NDF, and ADL were positive, whereas Krakowsky et al. (2005; 2006) reported a null relationship. In maize, however, it is found that a number of QTLs for ADF, NDF and ADL were located at the same or linked marker loci no matter whether they were correlated or not (Cardinal et al., 2003; Krakowsky et al., 2005; 2006). Some QTLs in the vicinities of the lignin biosynthesis genes, such as caffeic acid-O-methyltransferase, cinnamoyl CoA-reductase, and cinnamyl alcohol dehydrogenase genes, or of the cellulose biosynthesis genes have been identified (Cardinal et al., 2003; Krakowsky et al., 2005; 2006; Truntzler et al., 2010).
Dongxiang wild rice (Oryza rufipogon Griff., hereafter referred to as DWR) is a common wild rice located at 28°14′ N latitude and 116°30′ E longitude in Dongxiang county, Jiangxi province, China, which is considered to be the northernmost region in China and in the world where O. rufipogon is found (Xie et al., 2010). Wild relatives of cultivated rice are highly diversified and host various genes conferring resistance to biotic and abiotic stresses, thus providing a valuable gene pool for rice genetic improvement (Chen et al., 2010; Xie et al., 2010). Identification of QTL controlling drought tolerance (Zhou et al., 2006), resistance to whitebacked planthopper (Chen et al., 2010), and yield-related components (Tian et al., 2006; Huang et al., 2008) in Chinese DWR have been conducted. Whether DWR carries favorable genes/QTLs for forage quality improvement or for biofuel production is an interesting issue that needs to be addressed.
In this study, a backcrossed inbred line (BIL) population derived from an indica rice Xieqingzao B (hereafter referred to as XB) and a DWR, O. rufipogon Griff. and its genetic linkage map were used to map QTLs controlling the fiber and lignin contents of rice straw. The objective of this study was to dissect the genetic architecture for the fiber and lignin contents in the wild rice, and thus facilitate molecular breeding for the improvement of straw quality with suitable molecular markers.
2. Materials and methods
2.1. Plant materials
The mapping population consists of 202 BILs derived from the interspecific cross XB//XB/DWR (Huang et al., 2008; Chen et al., 2010), in which Oryza sativa ssp. indica var. Xieqingzao B is the maintainer line of the dwarf-abortive cytoplasmic male sterile line Xieqingzao A, and DWR is an accession of O. rufipogon from Dongxiang country, Jiangxi province, China. All of the 202 BILs together with the parents (XB and DWR) were grown at the China National Rice Research Institute, Hangzhou, Zhejiang province, China (hereafter HZ environment) and at the experiment farm of the Rice Research Institute, Jiangxi Academy of Agricultural Sciences, Nanchang, Jiangxi province, China (hereafter NC environment). The field management follows the conventional practices, and all the rice materials were harvested on time when the grains were mature.
2.2. Phenotypic data
Straw was taken only from the stem bearing effective spike of each plant. They were firstly washed to remove soil on the base and then dried in a forced-air oven for 24 h at 60 °C. The sheath and leaves were removed, and only three nodes from the base up were collected. The samples were ground to pass a 1-mm sieve, then they were scanned using a near infrared reflectance spectroscopy (Foss-NIR System, Inc., Silver Spring, MD, USA) to obtain the spectroscopic profiles, and ADF (%), NDF (%) and ADL (%) were calculated with prediction equations developed by Kong et al. (2005). The ADL/NDF ratio denoted the degree of lignification, which is expressed as the percentage of lignin (ADL) in the cell wall (NDF) (Méchin et al., 2001; Barrière et al., 2008).
2.3. Statistical anaysis
Analysis of correlation and analysis of variance (ANOVA) were carried out in Statistical Analysis System (SAS; Version 8.1. SAS Institute Inc., Cary, NC, USA).
2.4. Genetic map construction and QTL detection
Using total DNA extracted from the fresh leaf of a single plant for each of the 202 lines, a linkage map consisting of 149 DNA markers and spanning 1 306.4 cM was constructed in the previous studies (Huang et al., 2008; Chen et al., 2010). QTL analysis was performed using composite interval mapping (CIM) of Windows QTL Cartographer 2.5 (Wang et al., 2005). The presence of a QTL was claimed when a logarithm of the odds (LOD) score was larger than 2.0. Genetic parameters, i.e., additive effects and explained variation of each QTL, were also estimated. QTLs were named following the nomenclature of McCouch et al. (1997).
3. Results
3.1. Phenotypic variation
The indica parent XB had 36.0%, 60.6%, and 7.7% of ADF, NDF, and ADL contents in HZ environment, and had a slightly higher ADF (36.6%) and NDF (61.1%) contents in NC environment, with a similar ADL content between the two environments. The ADF, NDF, and ADL contents in DWR were 30.8%, 55.8%, and 7.0% in HZ, and 31.3%, 56.6%, and 7.1% in NC, respectively. The ADL/NDF ratio of DWR was slightly smaller than that of XB in both environments (Table 1).
Table 1.
Variation of cell wall traits of parents and BIL population
| Rice | Parameter | ADF (%) |
NDF (%) |
ADL (%) |
ADL/NDF (%) |
||||
| HZ | NC | HZ | NC | HZ | NC | HZ | NC | ||
| DWR | Mean | 30.79 | 31.30 | 55.74 | 56.58 | 7.01 | 7.06 | 12.58 | 12.48 |
| XB | Mean | 35.98 | 36.60 | 60.61 | 61.11 | 7.66 | 7.70 | 12.64 | 12.60 |
| BIL | Mean | 37.69 | 35.63 | 58.81 | 54.76 | 7.16 | 6.14 | 12.14 | 11.93 |
| Range | 26.29–45.76 | 22.07–46.36 | 45.00–75.28 | 43.13–66.55 | 4.14–11.60 | 3.05–8.90 | 7.75–20.18 | 5.01–15.79 | |
| SD | 4.22 | 4.87 | 4.98 | 4.02 | 1.36 | 1.13 | 1.95 | 1.54 | |
| Kurtosis | −0.33 | −0.44 | 0.30 | 0.10 | 0.14 | 0.41 | 1.61 | 2.52 | |
| Skewness | −0.14 | −0.31 | 0.35 | −0.02 | −0.05 | 0.51 | 0.52 | −0.85 | |
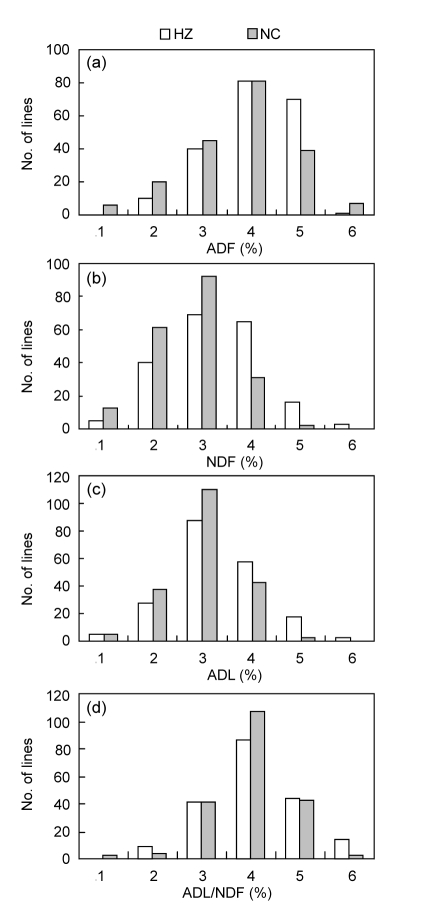
Among the BIL population, wide segregation in the CWCs was found in the two environments (Table 1, Fig. 1). The ADF ranged from 26.3% to 45.8% in HZ and from 22.1% to 46.4% in NC. The NDF content had the mean value of 58.8% in HZ, ranging from 45.0% to 75.3%; while in NC, it had the mean value of 54.8% and ranged from 43.1% to 66.6%. The mean ADL content was 7.2% in HZ and 6.1% in NC, ranging from 4.1% to 11.6% in HZ and from 3.1% to 8.9% in NC. The range of ADL/NDF ratio was from 7.8% to 20.2% with a mean of 12.1% in HZ, and from 5.0% to 15.8% with a mean of 11.9% in NC.
Fig. 1.
Frequency distributions of cell wall traits in BIL population in HZ and NC environments
(a) ADF; (b) NDF; (c) ADL; (d) ADL/NDF ratio
The segregation of these traits was continuously distributed among the whole population in both environments (Fig. 1), but showed significant transgressive distribution. The kurtosis and skewness for ADF, NDF, and ADL were less than 1 indicating their normal distributions (Table 1, Fig. 1). The kurtosis of ADL/NDF ratio was larger than 1, indicating that it displayed the relatively-peaked distribution.
3.2. Correlation analysis and analysis of variance
Correlation coefficients of the traits in separate environments and between the two environments are listed in Table 2. In each environment, ADF was positively correlated with NDF (P<0.001), ADL (P<0.001), and ADL/NDF ratio (P<0.001). Positive correlation was also observed between NDF and ADL (P<0.001), which indicated that an increase of one component was associated with an increase in another two components. NDF and ADL/NDF showed a significant relationship in NC environment (P<0.001), but no relationship in HZ environment.
Table 2.
Correlation analysis among the cell wall traits of BIL population
| Traits | Correlation coefficient |
|||
| ADF | NDF | ADL | ADL/NDF | |
| ADF | −0.009 | 0.617*** | 0.799*** | 0.657*** |
| NDF | 0.800*** | 0.099 | 0.504*** | 0.133 |
| ADL | 0.896*** | 0.760*** | 0.006 | 0.909*** |
| ADL/NDF | 0.675*** | 0.360*** | 0.912*** | 0.058 |
The correlation coefficients below diagonal were from data of NC, and those of above diagonal were from data of HZ, and those at diagonal were between data of NC and HZ
Significant at P<0.001
The ADF, NDF, ADL, and ADF/NDF ratio in HZ had no relation with those in NC environment, suggesting that these traits were strongly affected by the environment. ANOVA results indicated that the environment was the major source of variance for all the traits, accounting for 82.0%, 94.7%, 87.8%, and 55.8% of the total variance, respectively (Table 3). The variances of genotype were a little larger than those of genotype×environment interaction.
Table 3.
Mean square data of ANOVA with the two-year data
| Source | df | Mean square |
|||
| ADF | NDF | ADL | ADL/NDF | ||
| Genotype (G) | 201 | 47.5 | 35.7 | 2.6 | 5.3 |
| Environment (E) | 1 | 350.5 | 1 125.6 | 35.4 | 12.1 |
| G(E | 199 | 29.6 | 27.4 | 2.3 | 4.3 |
According to the F-value of ANOVA, all the variances were significant at P<0.001
3.3. QTL analysis
The results of QTLs are summarized in Table 4 and Fig. 2. A total of 16 QTLs were identified by CIM. For ADF, two QTLs at the marker intervals of RM126-RZ56 on chromosome 8 (qADF8) and RM332-RM167 on chromosome 11 (qADF11) were detected with an LOD value larger than 3 in HZ environment. Each QTL explained 6.8% and 8.2% of the total phenotypic variance. In NC, only one QTL located between its flanking markers of RM154 and RG555 on chromosome 2 was identified, explaining 5.5% of the total variance. The alleles of qADF11 and qADF2 of wild rice parent DWR increased the ADF by 1.7% and 2.6%, whereas the DWR allele of qADF8 decreased the ADF by 1.44%.
Table 4.
Locations of QTLs affecting plant cell wall properties detected in BIL population derived from DWR and XB
| Trait | Location | Locus | Chromosome | Marker | LOD | Additive* | R2 |
| ADF | HZ | qADF8 | 8 | RM126-RZ562 | 3.07 | −1.44 | 6.80 |
| qADF11 | 11 | RM332-RM167 | 3.34 | 1.70 | 8.22 | ||
| NC | qADF2 | 2 | RM154-RG555 | 2.64 | 1.32 | 5.51 | |
| NDF | HZ | qNDF7 | 7 | RG678-RM11 | 3.30 | 2.31 | 7.76 |
| qNDF11 | 11 | RM332-RM167 | 2.03 | 1.51 | 4.41 | ||
| NC | qNDF4 | 4 | RM335-RM518 | 2.64 | −1.20 | 5.91 | |
| qNDF6 | 6 | RM253-RM276 | 2.06 | 0.93 | 4.78 | ||
| ADL | HZ | qADL8 | 8 | RM126-RZ562 | 2.27 | −0.38 | 4.71 |
| qADL11-1 | 11 | RM332-RM167 | 4.46 | 0.66 | 12.64 | ||
| NC | qADL6 | 6 | RM253-RM276 | 2.97 | 0.29 | 6.27 | |
| qADL7 | 7 | RM5752-RM6574 | 2.69 | −0.57 | 11.71 | ||
| qADL11-2 | 11 | RM202-RM287 | 2.33 | 0.30 | 4.84 | ||
| ADL/NDF | HZ | qANR11-1 | 11 | RM332-RM167 | 3.19 | 0.77 | 8.09 |
| NC | qANR6 | 6 | RG172-RM340 | 2.33 | −0.52 | 5.81 | |
| qANR7 | 7 | RM5752-RM6574 | 2.62 | −0.86 | 13.75 | ||
| qANR11-2 | 11 | RM202-RM287 | 2.18 | 0.45 | 5.85 |
Additive effect is the effect on the traits if an XB allele was replaced by a DWR allele
Fig. 2.
Genomic locations of QTLs for ADF, NDF, ADL and ADL/NDF ratio in HZ and NC environments
Two QTLs were detected in each environment for NDF. The qNDF11 located at the same interval as qADF11, which was detected with a slightly smaller LOD, explaining 7.8% of the total variance. qNDF7 was located at the interval RG678-RM11 on chromosome 7, which was detected with an LOD of 3.3. In NC, qNDF4 at the interval RM335-RM518 on chromosome 4 and qNDF6 at the interval RM253-RM276 on chromosome 6 were detected. They explained 5.9% and 4.8% of the phenotypic variance. The DWR allele of the qNDF4 decreased NDF by 1.2%, whereas the DWR alleles of other three QTLs increased NDF.
Two QTLs for ADL identified in HZ were flanked by the interval RM126-RZ562 on chromosome 8 (qADL8) and RM332-RM167 on chromosome 11 (qADL11-1), respectively. Each QTL explained 4.7% and 12.6% of phenotypic variance. The qADL-11-1 was at the same marker interval as qADF11 and qNDF11. qADL6, qADL7, and qADL11-2 were detected in NC for ADL. The qADL6 and qNDF6 were at the same interval RM253-RM276. The DWR alleles of qADL8 and qADL7 decreased the ADL, while those of other QTLs increased ADL.
For the trait of ADL/NDF ratio, only one QTL was detected in HZ, qADR11-1, which shared the same region as that for qADL11-1. Three QTLs were detected in NC, and two of them on chromosomes 7 and 11 (qADR7 and qADR11-2) shared the common marker intervals as those QTLs for qADL7 and qADL11-2, respectively. Another QTL (qADR6) at the marker interval RG172-RM340 on chromosome 6 was detected with a small LOD, explaining 5.8% of the phenotypic variance. The DWR alleles of qADR6 and qADR7 decreased the ADL/NDF ratio, while those of qADR11-1 and qADR11-2 increased the ADL/NDF ratio.
4. Discussion
The common wild rice, O. rufipogon Griff., the progenitor of cultivated O. sativa, is one of most precious rice germplasms in China (Zhang et al., 2006; Zhou et al., 2006). It is an invaluable gene pool for the genetic improvement of modern rice cultivars (Huang et al., 2008; Chen et al., 2010; Xie et al., 2010). With the mature techniques in genetic linkage map construction and development of the QTL mapping software, the population derived from wild rice makes it a powerful tool for studying the genetics of the economically-important traits from wild rice. Zhou et al. (2006) identified 12 QTLs controlling drought tolerance at the seedling stage from DWR. Using the same population in this study, Huang et al. (2008) identified 23 QTLs for yield-related traits, and found that most of the QTLs are located as clusters, and the enhancing alleles were from XB in 4 clusters and from the DWR in 2 clusters. Chen et al. (2010) detected 3 QTLs for resistance to whitebacked planthopper, and found that the DWR alleles of the 3 QTLs always had the effect for the decrease of seedling mortality. Substantial genetic variation among the BILs for straw quality (Table 1) makes this population a useful resource to study the genetics of cell wall synthesis and composition to facilitate the improvement of straw production and utilization.
A total of 16 QTLs were identified in the two environments in this study (Table 4), 3 for ADF, 4 for NDF, 5 for ADL, and 4 for ADL/NDF ratio (Table 4). Dong et al. (2008) had conducted the QTL mapping for rice straw quality, including the trait of ADF using a recombinant inbred line population derived from two cultivated rice. A total of six QTLs were identified for ADF in the two environments. Among them, one QTL on chromosome 2 could be detected in the two environments, but it was not allelic to the qADF2 in this study when compared their positions in the genetic linkage map. However, the QTL on chromosome 6 (Dong et al., 2008) is allelic to qNDF6 and qADL6 in this study, because they shared the common marker locus RM276. Furthermore, the QTL on chromosome 7 (Dong et al., 2008) is allelic to qNDF7 in this study, because they shared the common marker locus RM11 (Table 4). The QTL on chromosome 4 (Dong et al., 2008) is close to qNDF4 according to their positions in the map. Xie et al. (2010) identified a QTL with minor effect on chromosome 9 for ADF, but this QTL could not be detected in this study. Two QTLs on chromosomes 5 and 9 for ADF and NDF were identified in a doubled haploid population derived from two rice cultivars (Bao et al., 2007), but no common QTLs were found in this study. These facts indicated that the CWC traits were controlled by many QTLs with minor effects that were distributed in different chromosomes and in different rice materials. The present study also found that the environment has a dramatic effect on the CWCs though the genotypic and genotype×environment interaction effects were also significant for all CWC traits (Table 3). This phenomenon is supported by two facts: one is that all the traits tested showed no correlations between HZ and NC environments (Table 2), and the other is that no common QTLs were shared in the two environments. There are six QTLs clustering on chromosome 11, four QTLs for each of four traits detected in HZ, and two QTLs for ADL and ADL/NDF ratio detected in NC; whether they are located at the same region needs to be tested.
From the additive effects of each QTL, we found that some alleles of wild rice could decrease, while others increase the target traits. For example, the DWR alleles of qADF8 and qADL8 decreased ADF and ADL in HZ environment, while the alleles of qADF11, qNDF11, qADL11, and qADR11 increased the four traits in HZ environment. The same situation could be found for the QTLs identified in NC environment (Table 4). This fact indicated that wild rice carries both favorable and unfavorable alleles for straw quality improvement. The breeder could choose the favorable alleles for straw quality improvement using marker-assisted selection method.
According to the relative positions of markers on the reference genetic maps (http://www.gramene.org/), candidate genes or transcription factors involving in lignin and cellulose biosyntheses (Barrière et al., 2009) were searched. A gene similar to cinnamoyl CoA reductase (Os02g0180700) was found nearby the qADF2 on chromosome 2. One myeloblastosis (MYB) transcription factor (Os04g0110300) was close to qNDF4, and another one (Os11g0180900) was located at the qADF11, qADL11-1, or qANR11-1 region. Three putative cellulose synthases were found close to or located at the regions of qANR6 (Os06g0601600), qADL7 (Os07g0208500), and qADL8 (Os08g01 60500). The regions of qANR6 also included two putative genes encoding 4-coumarate-CoA ligase (Os06g0656500) and cinnamoyl-CoA reductase (Os06g0623600). However, whether the QTLs were true genes involved in lignin or cellulose biosynthesis awaits further fine mapping of these traits.
Huang et al. (2008) detected a total of 23 QTLs for grain yield-related traits using the same genetic population. Comparing the map position of the QTLs for grain yield and for CWCs, only two loci are shared between two studies. The qADF2 in this study was located at the same marker interval as the QTL for grain yield per plant and spikelet fertility. Unfortunately, the allele of wild rice of qADF2 increased the ADF, but decreased the grain yield per plant and spikelet fertility (Huang et al., 2008). The qNDF7 was located at the same interval as the QTL for the number of filled grains per panicle and the total number of spikelets per panicle. The wild allele of this locus could increase not only the NDF, but also the number of filled grains per panicle and the total number of spikelets per panicle, which is considered as a favorable QTL for cell wall fiber and grain yield. No unfavorable correlations between the straw quality and grain yield suggest dual purpose of rice breeding be conducted on quality and biomass yield for biofeedstock production, while not at the expense of grain yield.
Footnotes
Project supported by the National Basic Research Program (973) of China (No. 2002CCC00800), the Jiangxi Provincial Inviting Tender Project for Principal Research Topic (No. 20068), and the Ministry of Agriculture of China (Nos. 200803034 and 201103007)
References
- 1.Abdel-Haleem H, Bowman J, Giroux M, Kanazin V, Talbert H, Surber L, Blake T. Quantitative trait loci of acid detergent fiber and grain chemical composition in hulled×hull-less barley population. Euphytica. 2010;172(3):405–418. doi: 10.1007/s10681-009-0066-6. [DOI] [Google Scholar]
- 2.Agbagla-Dohnani A, Noziere P, Clement G, Doreau M. In sacco degradability, chemical and morphological composition of 15 varieties of European rice straw. Anim Feed Sci Technol. 2001;94(1-2):15–27. doi: 10.1016/S0377-8401(01)00296-6. [DOI] [Google Scholar]
- 3.Bao JS, Jin L, Shen Y, Xie JK. Genetic mapping of quantitative trait loci associated with fiber and lignin content in rice. Cereal Res Commun. 2007;35(1):23–30. doi: 10.1556/CRC.35.2007.1.4. [DOI] [Google Scholar]
- 4.Barrière Y, Thomas J, Denoue D. QTL mapping for lignin content, lignin monomeric composition, p-hydroxycinnamate content, and cell wall digestibility in the maize recombinant inbred line progeny F838×F286. Plant Sci. 2008;175(4):585–595. doi: 10.1016/j.plantsci.2008.06.009. [DOI] [Google Scholar]
- 5.Barrière Y, Méchin V, Lafarguette F, Manicacci D, Guillon F, Wang H, Lauressergues D, Pichon M, Bosio M, Tatout C. Toward the discovery of maize cell wall genes involved in silage quality and capacity to biofuel production. Maydica. 2009;54(2/3):161–198. [Google Scholar]
- 6.Cardinal AJ, Lee M, Moore KJ. Genetic mapping and analysis of quantitative trait loci affecting fiber and lignin content in maize. Theor Appl Genet. 2003;106(5):866–874. doi: 10.1007/s00122-002-1136-5. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Qian N, Guo W, Song Q, Li B, Deng F, Dong C, Zhang T. Using three overlapped RILs to dissect genetically clustered QTL for fiber strength on Chro. D8 in upland cotton. Theor Appl Genet. 2009;119(4):605–612. doi: 10.1007/s00122-009-1070-x. [DOI] [PubMed] [Google Scholar]
- 8.Chen J, Huang DR, Wang L, Liu GJ, Zhuang JY. Identification of quantitative trait loci for resistance to whitebacked planthopper, Sogatella furcifera, from an interspecific cross Oryza sativa×O. rufipogon . Breed Sci. 2010;60(2):153–159. doi: 10.1270/jsbbs.60.153. [DOI] [Google Scholar]
- 9.Cogan NOI, Smith KF, Yamada T, Francki MG, Vecchies AC, Jones ES, Spangenberg GC, Forster JW. QTL analysis and comparative genomics of herbage quality traits in perennial ryegrass (Lolium perenne L.) Theor Appl Genet. 2005;110(2):364–380. doi: 10.1007/s00122-004-1848-9. [DOI] [PubMed] [Google Scholar]
- 10.Dong CF, Cai QS, Wang CL, Harada J, Nemoto K, Shen YX. QTL analysis for traits associated with feeding value of straw in rice (Oryza sativa L.) Rice Sci. 2008;15(3):195–200. doi: 10.1016/S1672-6308(08)60042-6. [DOI] [Google Scholar]
- 11.González-Martínez SC, Wheeler NC, Ersoz E, Nelson CD, Neale DB. Association genetics in Pinus taeda L. I. Wood property traits. Genetics. 2007;175(1):399–409. doi: 10.1534/genetics.106.061127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grando S, Baum M, Ceccarelli S, Goodchild A, El-Haramein FL, Jahoor A, Backes G. QTL for straw quality characteristics identified in recombinant inbred lines of a Hordeum vulgare×H. spontaneum cross in a Mediterranean environment. Theor Appl Genet. 2005;110(4):688–695. doi: 10.1007/s00122-004-1894-3. [DOI] [PubMed] [Google Scholar]
- 13.Huang DR, Chen J, Hou LJ, Fan YY, Zhuang JY. Identification of QTLs for yield traits in the BC1F5 population of Xieqingzao B//Xieqingzao B/Dongxiang wild rice. J Agric Biotech. 2008;16:977–982. [Google Scholar]
- 14.Kong XL, Xie JK, Wu XL, Huang YJ, Bao JS. Rapid prediction of acid detergent fiber, neutral detergent fiber, and acid detergent lignin of rice materials by near-infrared spectroscopy. J Agric Food Chem. 2005;53(8):2843–2848. doi: 10.1021/jf047924g. [DOI] [PubMed] [Google Scholar]
- 15.Krakowsky MD, Lee M, Coors JG. Quantitative trait loci for cell-wall components in recombinant inbred lines of maize (Zea mays L.). I: stalk tissue. Theor Appl Genet. 2005;111(2):337–346. doi: 10.1007/s00122-005-2026-4. [DOI] [PubMed] [Google Scholar]
- 16.Krakowsky MD, Lee M, Coors JG. Quantitative trait loci for cell wall components in recombinant inbred lines of maize (Zea mays L.). II: leaf sheath tissue. Theor Appl Genet. 2006;112(4):717–726. doi: 10.1007/s00122-005-0175-0. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Weng JK, Chapple C. Improvement of biomass through lignin modification. Plant J. 2008;54(4):569–581. doi: 10.1111/j.1365-313X.2008.03457.x. [DOI] [PubMed] [Google Scholar]
- 18.Lorenz AJ, Coors JG, Hansey CN, Kaeppler SM, de Leon N. Genetic analysis of cell wall traits relevant to cellulosic ethanol production in maize (Zea mays L.) Crop Sci. 2010;50(3):842–852. doi: 10.2135/cropsci2009.04.0168. [DOI] [Google Scholar]
- 19.McCouch SR, Cho YG, Yano M, Paul E, Blinstrub M. Report on QTL nomenclature. Rice Genet Newslett. 1997;14:11–13. [Google Scholar]
- 20.Méchin V, Argillier O, Hebert Y, Guingo E, Moreau L, Charcosset A, Barriere Y. Genetic analysis and QTL mapping of cell wall digestibility and lignification in silage maize. Crop Sci. 2001;41(3):690–697. doi: 10.2135/cropsci2001.413690x. [DOI] [Google Scholar]
- 21.Putun AE, Apaydin E, Putun E. Rice straw as a bio-oil source via pyrolysis and steam pyrolysis. Energy. 2004;29(12-15):2171–2180. doi: 10.1016/j.energy.2004.03.020. [DOI] [Google Scholar]
- 22.Tian F, Li DJ, Fu Q, Zhu ZF, Fu YC, Wang XK, Sun CQ. Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (O. sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor Appl Genet. 2006;112(3):570–580. doi: 10.1007/s00122-005-0165-2. [DOI] [PubMed] [Google Scholar]
- 23.Truntzler M, Barrière Y, Sawkins MC, Lespinasse D, Betran J, Charcosset A, Moreau L. Meta-analysis of QTL involved in silage quality of maize and comparison with the position of candidate genes. Theor Appl Genet. 2010;121(8):1465–1482. doi: 10.1007/s00122-010-1402-x. [DOI] [PubMed] [Google Scholar]
- 24.van Soest PJ. Nutritional Ecology of the Ruminant. 2nd Ed. Ithaca, NY: Cornell University Press; 1994. pp. 108–121. [Google Scholar]
- 25.Wang S, Basten CJ, Zeng ZB. Windows QTL Cartographer 2.5. Raleigh, NC, USA: Department of Statistics, North Carolina State University; 2005. [June 30, 2009]. Available from http://statgen.ncsu.edu/qtlcart/WQTLCart.htm. [Google Scholar]
- 26.Wu J, Gutierrez OA, Jenkins JN, McCarty JC, Zhu J. Quantitative analysis and QTL mapping for agronomic and fiber traits in an RI population of upland cotton. Euphytica. 2009;165(2):231–245. doi: 10.1007/s10681-008-9748-8. [DOI] [Google Scholar]
- 27.Xie JK, Wu XL, Jin L, Wan Y, Huang YJ, Bao JS. Identification of simple sequence repeat (SSR) markers for acid detergent fiber in rice straw by bulked segregant analysis. J Agric Food Chem. 2006;54(20):7616–7620. doi: 10.1021/jf061432h. [DOI] [PubMed] [Google Scholar]
- 28.Xie J, Agrama HA, Kong D, Zhuang J, Hu B, Wan Y, Yan W. Genetic diversity associated with conservation of endangered Dongxiang wild rice (Oryza rufipogon) Genet Resour Crop Evol. 2010;57(4):597–609. doi: 10.1007/s10722-009-9498-z. [DOI] [Google Scholar]
- 29.Xiong Y, Fei SZ, Brummer EC, Moore KJ, Barker RE, Jung G, Curley J, Warnke SE. QTL analyses of fiber components and crude protein in an annual×perennial ryegrass interspecific hybrid population. Mol Breed. 2006;18(4):327–340. doi: 10.1007/s11032-006-9034-1. [DOI] [Google Scholar]
- 30.Zhang X, Zhou SX, Fu YC, Su Z, Wang XK, Sun CQ. Identification of a drought tolerant introgression line derived from dongxiang common wild rice (O. rufipogon Griff.) Plant Mol Biol. 2006;62(1-2):247–259. doi: 10.1007/s11103-006-9018-x. [DOI] [PubMed] [Google Scholar]
- 31.Zhou SX, Tian F, Zhou ZF, Fu YC, Wang XK, Sun CQ. Identification of quantitative trait loci controlling drought tolerance at seedling stage in Chinese Dongxiang common wild rice (Oryza rufipogon Griff.) Acta Genet Sin. 2006;33(6):551–558. doi: 10.1016/S0379-4172(06)60084-X. [DOI] [PubMed] [Google Scholar]