Abstract
Surface display is effectively utilized to construct a whole-cell biocatalyst. Codon optimization has been proven to be effective in maximizing production of heterologous proteins in yeast. Here, the cDNA sequence of Rhizopus oryzae lipase (ROL) was optimized and synthesized according to the codon bias of Saccharomyces cerevisiae, and based on the Saccharomyces cerevisiae cell surface display system with α-agglutinin as an anchor, recombinant yeast displaying fully codon-optimized ROL with high activity was successfully constructed. Compared with the wild-type ROL-displaying yeast, the activity of the codon-optimized ROL yeast whole-cell biocatalyst (25 U/g dried cells) was 12.8-fold higher in a hydrolysis reaction using p-nitrophenyl palmitate (pNPP) as the substrate. To our knowledge, this was the first attempt to combine the techniques of yeast surface display and codon optimization for whole-cell biocatalyst construction. Consequently, the yeast whole-cell ROL biocatalyst was constructed with high activity. The optimum pH and temperature for the yeast whole-cell ROL biocatalyst were pH 7.0 and 40 °C. Furthermore, this whole-cell biocatalyst was applied to the hydrolysis of tributyrin and the resulted conversion of butyric acid reached 96.91% after 144 h.
Keywords: Rhizopus oryzae lipase (ROL), Yeast surface display, Codon optimization, Whole-cell biocatalyst
1. Introduction
Lipases (EC 3.1.1.3) catalyze not only hydrolysis of ester bonds under aqueous conditions but also synthesis of ester bonds under microaqueous conditions, being the biocatalysts with broad use (Zaks and Klibanov, 1988; Klibanov, 2001). Despite being widely spread in several species, microbial lipases have more potential for industrial application than those from other sources due to their high productivity, high catalytic specificity, ability to remain active in organic solvents, and ease of genetic manipulation (Hasan et al., 2006). Among the microbial lipases, that from Rhizopus oryzae is one of the most famous and versatile lipases. There are extensive reports on Rhizopus oryzae lipase (ROL) application in bioconversion processes, such as the productions of biodiesel (Matsumoto et al., 2001; Hama et al., 2004; Tamalampudi et al., 2008), 2-monoacylglycerol (2-MAG) (Kaewthong et al., 2005; Esteban et al., 2009) and 1,3-diacylglycerol (1,3-DAG) (Lo et al., 2004; Kristensen et al., 2005). Thus, constructing ROL biocatalyst with high activity could facilitate its industrial application.
Enzyme-displaying yeast, as the application of the yeast cell surface display system, is one of the most attractive biocatalyst systems for bioconversion processes. Enzymes displayed on yeast as whole-cell biocatalysts have many advantages such as simple genetic manipulation and easy production procedure (by cultivation and centrifugation only); thus they are more favorable than immobilized enzymes (Shibasaki et al., 2009). Yeast cell surface display systems have been widely studied, and many heterologous enzymes have been displayed on yeast cell surfaces as whole-cell biocatalysts (Murai et al., 1999; Kondo and Ueda, 2004). It has been proven to be effective to display lipases on yeast cell surfaces for the whole-cell lipase biocatalyst preparation (Hama et al., 2004; Kato et al., 2007).
The hypothesis that codon usage modulates the gene expression has been generally accepted (Das et al., 2009). Differences in codon usages are correlated with the number of isoaccepting tRNA molecules, in both unicellular as well as multicellular organisms (Ikemura, 1985), and in optimizing the growth efficiency of cells (Kurland, 1991). Codon usage was considered a bottleneck of the eventual expression of heterologous protein (Lithwick and Margalit, 2003). Codon optimization has been proven to be effective in maximizing production of heterologous proteins in yeast (Bucarey et al., 2009).
In the present work, we combined the techniques of cell surface display and codon optimization to successfully construct the yeast whole-cell ROL biocatalyst with high activity.
2. Materials and methods
2.1. Strains, media, and materials
Saccharomyces cerevisiae MS-1 (purchased from Meishan Mauri Yeast Co., Ltd., China) was used as the host strain for cell surface display system. YPD medium was used to cultivate yeast host cells. Yeast transformants were selected by YGCG medium (30 g/L glycerol, 7 g/L yeast nitrogen base without amino acids, 20 g/L casamino acids) containing 200 μg/ml G418 as an antibiotic and 20 g/L galactose as an inducer. For plate medium, 20 g/L agar was added. NheI, SacII, T4 DNA ligase, and DNA polymerase were purchased from TaKaRa (Dalian, China).
2.2. Construction of the fully codon-optimized pro-ROL gene
The wild-type pro-ROL gene was amplified by the polymerase chain reaction (PCR) method using the genomic DNA of Rhizopus oryzae ATCC 34612 (GenBank accession ID: M38352) as a template. To construct the fully codon-optimized pro-ROL gene, the rare codon usage in the pro-ROL sequence was replaced according to the preferred codon usage of Saccharomyces cerevisiae (Andersson and Kurland, 1990), without changing the amino acid sequence (Tables 1 and 2). The optimized pro-ROL-linker gene was synthesized by Sangon Co., Ltd. (Shanghai, China) and the nucleotide sequence was deposited in GenBank (accession ID: GU475119). In addition, a linker sequence consisting of GSSGGSGGSGG SGGSGS was inserted at the C-terminal portion of ROL to keep lipase activity by preserving the conformation of the active site near the C-terminal portion (Washida et al., 2001).
Table 1.
Nucleotide sequence comparison between the non-optimized pro and codon-optimized pro sequences of ROL
| Pro | Sequence | Length (bp) |
| Non-optimized_pro | GTTCCTGTTTCTGGTAAATCTGGATCTTCCAACACCGCCGTCTCTGCATC | 50 |
| Optimized_pro | -----a-----------g-----t-----t-----t--t--t-----t-- | 50 |
| Non-optimized_pro | TGACAATGCTGCCCTCCCTCCTCTCATCTCCAGCCGTTGTGCTCCTCCTT | 100 |
| Optimized_pro | ------c-----tt-g--a--at-g--t--ttcta-a--------a--a- | 100 |
| Non-optimized_pro | CTAACAAGGGAAGTAAAAGCGATCTCCAAGCTGAACCTTACAACATGCAA | 150 |
| Optimized_pro | ----------ttc---gtct--ct-g-----------a------------ | 150 |
| Non-optimized_pro | AAGAATACAGAATGGTATGAGTCCCATGGTGGCAACCTGACATCCATCGG | 200 |
| Optimized_pro | -----c--t--------c--a--t--c-----t---t----t--t--t-- | 200 |
| Non-optimized_pro | AAAGCGTGATGACAACTTGGTTGGTGGCATGACTTTGGACTTACCCAGCG | 250 |
| Optimized_pro | t---a-a--c-----------------t--------------g--atct- | 250 |
| Non-optimized_pro | ATGCTCCTCCTATCAGCCTCTCTAGCTCTACCAACAGCGCC | 291 |
| Optimized_pro | -c-----a--a--ttctt-g---g-a--c--t---tct--t | 291 |
Table 2.
Nucleotide sequence comparison between the non-optimized ROL and codon-optimized ROL sequences
| ROL | Sequence | Length (bp) |
| Non-optimized_ROL | TCTGATGGTGGTAAGGTTGTTGCTGCTACTACTGCTCAGATCCAAGAGTT | 50 |
| Optimized_ROL | -----c--------------------------------a--t-----a-- | 50 |
| Non-optimized_ROL | CACCAAGTATGCTGGTATCGCTGCCACTGCCTACTGTCGTTCTGTTGTCC | 100 |
| Optimized_ROL | ---t-----c--------t-----t-----t------a-a--------t- | 100 |
| Non-optimized_ROL | CTGGTAACAAGTGGGATTGTGTCCAATGTCAAAAGTGGGTTCCTGATGGC | 150 |
| Optimized_ROL | -a--------------c-----t--------------------a--c--t | 150 |
| Non-optimized_ROL | AAGATCATCACTACCTTTACCTCCTTGCTTTCCGATACAAATGGTTACGT | 200 |
| Optimized_ROL | -----t--t-----t--c--t--t---t-g--t--c--t--c-------- | 200 |
| Non-optimized_ROL | CTTGAGAAGTGATAAACAAAAGACCATTTATCTTGTTTTCCGTGGTACCA | 250 |
| Optimized_ROL | t------tc---c--g--------t-----ct-g------a-a-----t- | 250 |
| Non-optimized_ROL | ACTCCTTCAGAAGTGCCATCACTGATATCGTCTTCAACTTTTCTGACTAC | 300 |
| Optimized_ROL | ----t------tc---t--t-----c--t--t--------c--------- | 300 |
| Non-optimized_ROL | AAGCCTGTCAAGGGCGCCAAAGTTCATGCTGGTTTCCTTTCCTCTTATGA | 350 |
| Optimized_ROL | -----a--t-----t--t--g-----c---------t-g--t-----c-- | 350 |
| Non-optimized_ROL | GCAAGTTGTCAATGACTATTTCCCTGTCGTCCAAGAACAATTGACCGCCC | 400 |
| Optimized_ROL | a--------t--c-----c-----a--t--t--------------t--t- | 400 |
| Non-optimized_ROL | ACCCTACTTATAAGGTCATCGTTACCGGTCACTCACTCGGTGGTGCACAA | 450 |
| Optimized_ROL | ----a-----c-----t--t-----t--------tt-g--------t--- | 450 |
| Non-optimized_ROL | GCTTTGCTTGCCGGTATGGATCTCTACCAACGTGAACCAAGATTGTCTCC | 500 |
| Optimized_ROL | ------t-g--t--------ct-g------a-a----------------- | 500 |
| Non-optimized_ROL | CAAGAATTTGAGCATCTTCACTGTCGGTGGTCCTCGTGTTGGTAACCCCA | 550 |
| Optimized_ROL | a-----c---tct--t--------t--------aa-a-----------a- | 550 |
| Non-optimized_ROL | CCTTTGCTTACTATGTTGAATCCACCGGTATCCCTTTCCAACGTACCGTT | 600 |
| Optimized_ROL | -t--c--------c--------t--t-----t--a------a-a--t--- | 600 |
| Non-optimized_ROL | CACAAGAGAGATATCGTTCCTCACGTTCCTCCTCAATCCTTCGGATTCCT | 650 |
| Optimized_ROL | -----------c--t-----a--------a--a-----t-----t---t- | 650 |
| Non-optimized_ROL | TCATCCCGGTGTTGAATCTTGGATCAAGTCTGGTACTTCCAACGTTCAAA | 700 |
| Optimized_ROL | g--c--a-----------------t--------------t---------- | 700 |
| Non-optimized_ROL | TCTGTACTTCTGAAATTGAAACCAAGGATTGCAGTAACTCTATCGTTCCT | 750 |
| Optimized_ROL | -t--------------------t-----c--ttc---------t-----a | 750 |
| Non-optimized_ROL | TTCACCTCTATCCTTGACCACTTGAGTTACTTTGATATCAACGAAGGAAG | 800 |
| Optimized_ROL | -----t-----tt-g---------tc------c--c--t--------ttc | 800 |
| Non-optimized_ROL | CTGTTTG | 807 |
| Optimized_ROL | t---c-- | 807 |
2.3. Construction of the expression plasmid
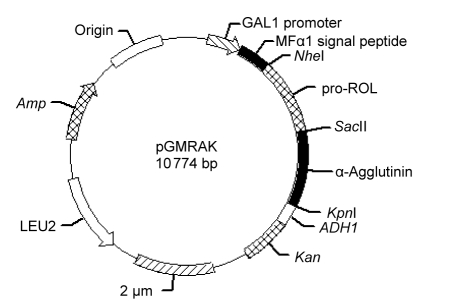
To display ROL on the yeast cell surface, the wild-type and optimized pro-ROL-linker genes were inserted into the NheI-SacII section of the expression cassette pGMAK (Guo et al., 2010), respectively, which was constructed in our previous study. The resulting plasmids were named pGMRAK (wild pro-ROL) and pGMRAK (optimized pro-ROL) (Fig. 1). To construct the general expression plasmid pGMAK for cell surface display of Saccharomyces cerevisiae, the GAL1 promoter, the prepro-secretion signal sequence of the mating factor α1 (MFα1 secretion signal sequence), α-agglutinin gene containing the 3′ half of the region encoding 320 amino acids, and the alcohol dehydrogenase terminator (ADH1 terminator) were amplified by PCR using the Saccharomyces cerevisiae MS-1 genomic DNA as the template, and the kanMX gene was amplified from the plasmid pUG6 (Guldener et al., 1996), with recognition sites of relevant restriction enzymes added to the primers in 5′-ends and 3′-ends, respectively. The resulting plasmid was first amplified in Escherichia coli and then extracted and transformed into the Saccharomyces cerevisiae MS-1 by electroporation (Thompson et al., 1998).
Fig. 1.
Construction of the plasmid pGMRAK for ROL-displaying yeast
The plasmid pGMRAK is a multicopy plasmid for expression of the pro-ROL-linker-α-agglutinin fusion gene, containing a secretion signal sequence of MFα1 factor under the control of the GAL1 promoter
2.4. Screening by halo assay
Transformants were spread on YGCG plate-medium containing 0.2% tributyrin as a substrate. Colonies hydrolyzing tributyrin were identified by the formation of clear halos. Saccharomyces cerevisiae MS-1 harboring pGMRAK (wild ROL) and pGMRAK (optimized ROL) and Saccharomyces cerevisiae MS-1 harboring pGMAK (MS-1/pGMAK) as the control were spread on the medium, respectively. The activities of the lipases were examined by the halo formed around the colony.
2.5. Preparation of ROL-displaying yeast
For whole-cell catalyst preparation, the wild-type ROL and codon-optimized ROL-displaying yeasts were cultivated in YGCG medium at 30 °C for 120 h (stationary phase), respectively. After cultivation, the cells were collected by centrifugation (3 000×g, 15 min), washed with distilled water and 50 mmol/L phosphate buffered solution (PBS; pH 7.0), and then lyophilized for 24 h by a freeze-drying system.
2.6. Measurement of lipase activity in yeast
The hydrolysis activities of the codon-optimized ROL and the wild-type ROL displayed on yeast cell surface were measured with p-nitrophenyl palmitate (pNPP) as the substrate (Prim et al., 2000). The assay was conducted in aqueous medium with 1 ml 50 mmol/L PBS (pH 7.0) containing 0.03% (v/v) pNPP, 4 g/L Triton-X, and 1 g/L arabic gum. Reaction was initiated by the addition of 100 μl 1 mg/ml lyophilized ROL-displaying yeast cells and was incubated at 30 °C for 15 min with agitation (150 r/min). After the reaction was complete, 200 μl aliquot of the reaction solution was placed in a 96-well plate to measure the absorbance value of p-nitrophenyl (pNP) released at 410 nm in a microplate reader Multiskan Asent (Labsystems). One unit of hydrolysis activity was defined as the amount of enzyme that released 1 μmol of pNP per minute under the assay conditions described (n=3, relative standard deviation (RSD) was estimated less than 10%).
2.7. Optimum temperature and pH
The optimum temperature for the codon-optimized ROL on yeast cell surface was determined by measuring the hydrolysis activity at pH 7.0 in a water bath at temperatures ranging from 30 to 50 °C. The optimum pH was determined at 30 °C with pH buffer solutions ranging from 6.0 to 8.0. The hydrolysis activity was measured at 30 °C and pH 7.0, using pNPP as substrate.
2.8. Thermostability assay
The thermostability of the codon-optimized ROL on yeast cell surface was measured as follows: the whole-cell biocatalyst was washed twice with distilled water after centrifugation and resuspended in 50 mmol/L PBS (pH 7.0). The suspension was then incubated at 40, 50 and 60 °C for 2–6 h, respectively. Aliquots of the cell suspensions were taken out and chilled immediately on ice at regular intervals. The residual hydrolysis activity was measured at 30 °C and pH 7.0, using pNPP as substrate.
2.9. Application of the whole-cell biocatalyst
The codon-optimized ROL-displaying yeast was cultivated in 150 ml YGCG broth medium containing 20 g/L galactose as the inducer and 1% (v/v) tributyrin as the substrate. Aliquots of 1 ml were taken from the reaction mixture every 8 h. The amount of butyric acid produced by hydrolysis reaction was measured by high-performance liquid chromatography (HPLC), in an Agilent 1200 series Rapid Resolution LC System (Agilent Technologies, Palo Alto, CA). The analyses were performed isocratically at 1.2 ml/min and 40 °C with a rapid resolution high throughput (RRHT) column (Zorbax SB-C18, 6×107 Pa, 4.6 mm×100 mm, 1.8 µm particle size) equipped with a 0.2-µm pore size inline filter. The mobile phase was: 80% of solvent A consisting of 25 mmol/L NaH2PO4 with 0.05% (v/v) trifluoroacetic acid (TFA) and 1.5% (v/v) methanol, adjusted to pH 2.31, plus 20% of solvent B consisting of methanol. The retention time for butyric acid was 5.85 min.
3. Results
3.1. Comparison of wild-type ROL and codon-optimized ROL genes
As the rare codons were replaced, a fully codon-optimized pro-ROL gene has been constructed. A sequence comparison between the codon-optimized sequence and wild-type one was shown in Tables 1 and 2. Compared to the non-optimized sequence, the fully codon-optimized gene had 217 nucleotide variations in all 1 098 nucleotides, resulting in 178 changes out of 366 codons. Further study demonstrated that the overall G+C content of codon-optimized pro-ROL sequence was reduced to 41.53% from 46.81% of the non-optimized sequence.
3.2. Expression of the recombinant gene
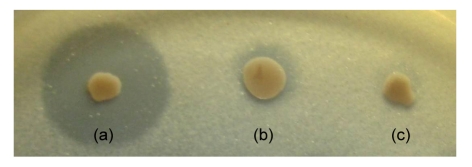
A clear halo was observed around the colonies of codon-optimized ROL-displaying yeast on plate medium containing tributyrin as substrate, while the wild-type ROL-displaying yeast showed a smaller one, but none for the control yeast (Fig. 2). The result indicates that the ROL-α-agglutinin fusion protein was expressed in an active form, and the hydrolysis activity of ROL was significantly improved by codon-optimization.
Fig. 2.
Halo formation of ROL-displaying yeasts cultivated for 144 h
Transformants were spread on YGCG solid medium containing 2 g/L tributyrin as substrate. (a) MS-1/pGMRAK (codon-optimized ROL); (b) MS-1/pGMRAK (wild ROL); (c) MS-1/pGMAK (control)
3.3. Lipase activity and characteristics
According to the time curve of biomass of the recombinant MS-1/pGMRAK, the cells reached stationary phase after 120 h cultivation (data not shown). The hydrolysis activity of the codon-optimized ROL displayed on the yeast cell surface was 25 U/g dried cells in the stationary phase, while that of the non-optimized one was 1.96 U/g dried cells. In addition, the activity of the cell-free supernatant was negligible throughout the entire cultivation. These results confirmed that the ROL-α-agglutinin fusion protein was not secreted into the culture medium but immobilized on the yeast cells firmly as a whole-cell biocatalyst.
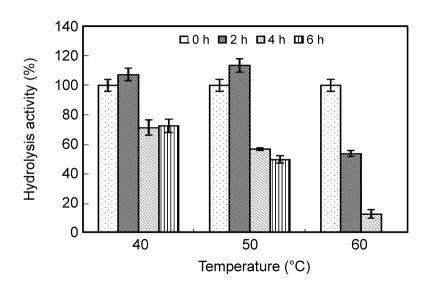
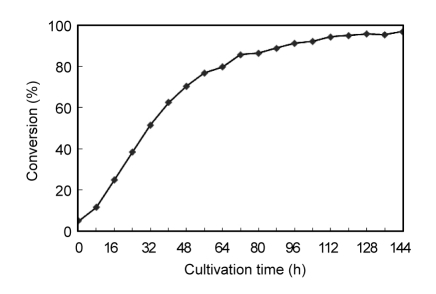
According to the measurement conducted, the optimum pH and temperature for the fully codon-optimized whole-cell biocatalyst were pH 7.0 and 40 °C. As shown in Fig. 3, after 2-h incubation at 40 and 50 °C, the residual activity increased to 107% and 113%, respectively. After 6-h incubation at 40 and 50 °C, the residual activity still remained 72% and 50%, respectively. However, it reduced to 13% after 4-h incubation at 60 °C. After 6-h incubation at 60 °C, cell debris was observed in the sample, implicating that yeast cells were fractured after the heating process. As shown in Fig. 4, the fully codon-optimized whole-cell biocatalyst was applied to tributyrin hydrolysis reaction and the conversion of butyric acid reached 96.91% after 144 h.
Fig. 3.
Thermostability of MS-1/pGMRAK
The cells suspension was incubated at 40, 50 and 60 °C for 2–6 h, the residual hydrolysis activity of the lipase was measured by using pNPP as substrate at 30 °C and pH 7.0. Residual activity was denoted by taking the initial value at 30 °C as 100% (n=3)
Fig. 4.
Time courses of butyric acid content in the tributyrin hydrolysis reaction
The amount of butyric acid produced by hydrolysis reaction was measured by HPLC. The conversion of butyric acid reached 96.91% after 144 h
4. Discussion
Codon usage could play a key role in regulating gene expression, because rare codons may interrupt mRNA stability and its translation rate; moreover, high G+C content may decrease the translational efficiency or even result in failed expression (Sinclair and Choy, 2002). In this study, the satisfying expression of the modified ROL gene in Saccharomyces cerevisiae confirmed that the codon optimization according to the favored codon usage of Saccharomyces cerevisiae had a positive impact on lipase heterologous expression. As shown in the results, the hydrolysis activity of the displayed codon-optimized ROL was 25 U/g dried cells, 12.8-fold higher than that of the wild-type one, which indicates that ROL displayed in high efficiency via codon optimization. To our knowledge, this was the first example to combine the techniques of yeast surface display and codon optimization, and consequently the fully codon-optimized ROL-displaying yeast as a whole-cell biocatalyst was constructed, with much higher enzymatic activity in the Saccharomyces cerevisiae cell surface display systems using α-agglutinin as an anchor. We could infer that the high activity of the yeast whole-cell ROL biocatalyst is attributed to the combination of the two techniques, thus leading to highly efficient ROL display on cell surface, because the codon usage is a key factor for the mRNA translation rate and so for protein synthesis efficiency in cells (Proshkin et al., 2010).
In conclusion, the strategy of combining the techniques of cell surface display and codon optimization is successfully developed, thus the yeast whole-cell ROL biocatalyst with high activity was constructed, which could be of great potential in bioconversion processes.
Footnotes
Project supported by the National High-Tech R & D Program (863) of China (No. 2006AA10Z308), the National Science Foundation of China (No. 20776130), the Zhejiang Provincial Natural Science Foundation of China (No. Y4090309), and the Zhejiang Provincial Science and Technology Program of China (No. 2009C32009)
References
- 1.Andersson SGE, Kurland CG. Codon preferences in free-living microorganisms. Microbiol Rev. 1990;54(2):198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bucarey SA, Noriega J, Reyes P, Tapia C, Saenz L, Zuniga A, Tobar JA. The optimized capsid gene of porcine circovirus type 2 expressed in yeast forms virus-like particles and elicits antibody responses in mice fed with recombinant yeast extracts. Vaccine. 2009;27(42):5781–5790. doi: 10.1016/j.vaccine.2009.07.061. [DOI] [PubMed] [Google Scholar]
- 3.Das S, Roymondal U, Sahoo S. Analyzing gene expression from relative codon usage bias in yeast genome: a statistical significance and biological relevance. Gene. 2009;443(1-2):121–131. doi: 10.1016/j.gene.2009.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Esteban L, Munio MD, Robles A, Hita E, Jimenez MJ, Gonzalez PA, Camacho B, Molina E. Synthesis of 2-monoacylglycerols (2-MAG) by enzymatic alcoholysis of fish oils using different reactor types. Biochem Eng J. 2009;44(2-3):271–279. doi: 10.1016/j.bej.2009.01.004. [DOI] [Google Scholar]
- 5.Guldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24(13):2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo Q, Zhang W, Ma LL, Chen QH, Chen JC, Zhang HB, Ruan H, He GQ. A food-grade industrial arming yeast expressing β-1,3-1,4-glucanase with enhanced thermal stability. J Zhejiang Univ-Sci B (Biomed & Biotechnol) 2010;11(1):41–51. doi: 10.1631/jzus.B0900185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hama S, Yamaji H, Kaieda M, Oda M, Kondo A, Fukuda H. Effect of fatty acid membrane composition on whole-cell biocatalysts for biodiesel-fuel production. Biochem Eng J. 2004;21(2):155–160. doi: 10.1016/j.bej.2004.05.009. [DOI] [Google Scholar]
- 8.Hasan F, Shah AA, Hameed A. Industrial applications of microbial lipases. Enzyme Microb Technol. 2006;39(2):235–251. doi: 10.1016/j.enzmictec.2005.10.016. [DOI] [Google Scholar]
- 9.Ikemura T. Codon usage and transfer-RNA content in unicellular and multicellular organisms. Mol Biol Evol. 1985;2(1):13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 10.Kaewthong W, Sirisansaneeyakul S, Prasertsan P, H-Kittikun A. Continuous production of monoacylglycerols by glycerolysis of palm olein with immobilized lipase. Process Biochem. 2005;40(5):1525–1530. doi: 10.1016/j.procbio.2003.12.002. [DOI] [Google Scholar]
- 11.Kato M, Fuchimoto J, Tanino T, Kondo A, Fukuda H, Ueda M. Preparation of a whole-cell biocatalyst of mutated Candida antaretica lipase B (mCALB) by a yeast molecular display system and its practical properties. Appl Microbiol Biotechnol. 2007;75(3):549–555. doi: 10.1007/s00253-006-0835-2. [DOI] [PubMed] [Google Scholar]
- 12.Klibanov AM. Improving enzymes by using them in organic solvents. Nature. 2001;409(6817):241–246. doi: 10.1038/35051719. [DOI] [PubMed] [Google Scholar]
- 13.Kondo A, Ueda M. Yeast cell-surface display—applications of molecular display. Appl Microbiol Biotechnol. 2004;64(1):28–40. doi: 10.1007/s00253-003-1492-3. [DOI] [PubMed] [Google Scholar]
- 14.Kristensen JB, Xu XB, Mu HL. Diacylglycerol synthesis by enzymatic glycerolysis: screening of commercially available lipases. J Am Oil Chem Soc. 2005;82(5):329–334. doi: 10.1007/s11746-005-1074-5. [DOI] [Google Scholar]
- 15.Kurland CG. Codon bias and gene-expression. FEBS Lett. 1991;285(2):165–169. doi: 10.1016/0014-5793(91)80797-7. [DOI] [PubMed] [Google Scholar]
- 16.Lithwick G, Margalit H. Hierarchy of sequence-dependent features associated with prokaryotic translation. Genome Res. 2003;13(12):2665–2673. doi: 10.1101/gr.1485203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo SK, Baharin BS, Tan CP, Lai OM. Lipase-catalysed production and chemical composition of diacylglycerols from soybean oil deodoriser distillate. Eur J Lipid Sci Technol. 2004;106(4):218–224. doi: 10.1002/ejlt.200300888. [DOI] [Google Scholar]
- 18.Matsumoto T, Takahashi S, Kaieda M, Ueda M, Tanaka A, Fukuda H, Kondo A. Yeast whole-cell biocatalyst constructed by intracellular overproduction of Rhizopus oryzae lipase is applicable to biodiesel fuel production. Appl Microbiol Biotechnol. 2001;57(4):515–520. doi: 10.1007/s002530100733. [DOI] [PubMed] [Google Scholar]
- 19.Murai T, Ueda M, Shibasaki Y, Kamasawa N, Osumi M, Imanaka T, Tanaka A. Development of an arming yeast strain for efficient utilization of starch by co-display of sequential amylolytic enzymes on the cell surface. Appl Microbiol Biotechnol. 1999;51(1):65–70. doi: 10.1007/s002530051364. [DOI] [PubMed] [Google Scholar]
- 20.Prim N, Blanco A, Martinez J, Pastor FIJ, Diaz P. estA, a gene coding for a cell-bound esterase from Paenibacillus sp. BP-23, is a new member of the bacterial subclass of type B carboxylesterases. Res Microbiol. 2000;151(4):303–312. doi: 10.1016/S0923-2508(00)00150-9. [DOI] [PubMed] [Google Scholar]
- 21.Proshkin S, Rahmouni AR, Mironov A, Nudler E. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science. 2010;328(5977):504–508. doi: 10.1126/science.1184939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shibasaki S, Maema H, Ueda M. Molecular display technology using yeast-arming technology. Anal Sci. 2009;25(1):41–49. doi: 10.2116/analsci.25.41. [DOI] [PubMed] [Google Scholar]
- 23.Sinclair G, Choy FYM. Synonymous codon usage bias and the expression of human glucocerebrosidase in the methylotrophic yeast, Pichia pastoris . Protein Expr Purif. 2002;26(1):96–105. doi: 10.1016/S1046-5928(02)00526-0. [DOI] [PubMed] [Google Scholar]
- 24.Tamalampudi S, Talukder MR, Hama S, Numata T, Kondo A, Fukuda H. Enzymatic production of biodiesel from Jatropha oil: a comparative study of immobilized-whole cell and commercial lipases as a biocatalyst. Biochem Eng J. 2008;39(1):185–189. doi: 10.1016/j.bej.2007.09.002. [DOI] [Google Scholar]
- 25.Thompson JR, Register E, Curotto J, Kurtz M, Kelly R. An improved protocol for the preparation of yeast cells for transformation by electroporation. Yeast. 1998;14(6):565–571. doi: 10.1002/(SICI)1097-0061(19980430)14:6<565::AID-YEA251>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 26.Washida M, Takahashi S, Ueda M, Tanaka A. Spacer-mediated display of active lipase on the yeast cell surface. Appl Microbiol Biotechnol. 2001;56(5-6):681–686. doi: 10.1007/s002530100718. [DOI] [PubMed] [Google Scholar]
- 27.Zaks A, Klibanov AM. Enzymatic catalysis in nonaqueous solvents. J Biol Chem. 1988;263(7):3194–3201. [PubMed] [Google Scholar]