Abstract
Background
Infectious diseases result in millions of deaths each year. Physical interactions between pathogen and host proteins often form the basis of such infections. While a number of methods have been proposed for predicting protein–protein interactions (PPIs), they have primarily focused on intra-species protein–protein interactions.
Methodology
We present an application of a supervised learning method for predicting physical interactions between host and pathogen proteins, using the human–HIV system. Using a Support Vector Machine with a linear kernel, we explore the use of a number of features including domain profiles, protein sequence k-mers, and properties of human proteins in a human PPI network. We achieve the best cross-validation performance when we use a combination of all three of these features. At a precision value of 70% we obtain recall values greater than 40%, depending on the ratio of positive examples to negative examples used during training. We use a classifier trained using these features to predict new PPIs between human and HIV proteins. We focus our discussion on those predicted interactions that involve human proteins known to be critical for HIV replication and propagation. Examples of predicted interactions with support in the literature include those necessary for viral attachment to the host membrane and subsequent invasion of the host cell.
Significance
Unlike intra-species PPIs, host–pathogen PPIs have not yet been experimentally detected on a large scale, though they are likely to play important roles in pathogenesis and disease outcomes. Computational methods that can robustly and accurately predict host–pathogen PPIs hold the promise of guiding future experiments and gaining insights into potential mechanisms of pathogenesis.
Keywords: Host–pathogen interactions, Protein interaction prediction, Systems biology, Infectious disease
1. Introduction
Infectious diseases cause millions of deaths each year. Despite enormous effort, many mechanisms of infection and pathogenesis still remain poorly understood. A potentially powerful application of protein–protein interaction (PPI) networks lies in using them to obtain insights into the molecular mechanisms underlying infectious diseases, especially since interactions between pathogen proteins and host proteins play key roles in initiating and sustaining infection. In a recent study, we surveyed the landscape of human proteins that interact with viruses and other pathogens (Dyer et al., 2008). We collected host–pathogen PPIs from seven public databases. Apart from strains of HIV and four other viruses, we found that for every other pathogen, at most 100 physical interactions are currently known between proteins in that pathogen and human proteins. Therefore, the severe lack of large-scale datasets detailing interactions between host and pathogen proteins is a significant hurdle to progress in host–pathogen systems biology. Consequently, it becomes imperative to develop computational methods that can robustly and accurately predict host–pathogen PPIs. Such predictors can guide cost effective experimental strategies to detect host–pathogen PPIs, drive research on how pathogens infect host cells, and help identify potential targets for therapeutics.
While a number of methods have been proposed for predicting PPIs, they have primarily focused on intra-species PPIs (Jansen et al., 2003; Ng et al., 2003; Pellegrini et al., 1999; Qi et al., 2006; Sharan et al., 2005; Sprinzak and Margalit, 2001; Yu et al., 2004; Zhang et al., 2004). Applying these methods to host–pathogen systems is made difficult by two factors. First, as we have already noted, experimental studies on most human pathogens have so far detected very small numbers of PPIs, making it difficult to build comprehensive training sets. Second, a number of data types used as features by previous methods, such as gene expression and knockout phenotypes, are not readily available for host–pathogen systems. We are aware of only a few computational methods for predicting host–pathogen PPIs (HP PPIs). Despite these limitations, methods using sequence-signature pairs and homology have been used to predict HP-PPIs (Davis et al., 2007; Dyer et al., 2007; Krishnadev and Srinivasan, 2008; Lee et al., 2008; Qi et al., 2010).
In this paper, we build a supervised predictor for human–HIV PPIs. We have selected this system because (i) HIV is a retrovirus that can lead to a failure of the immune system (AIDS), which kills millions of people yearly and (ii) successful prediction of PPIs for this well-studied host–pathogen system will set the stage for subsequent work on other less-studied systems.
We obtained known human–HIV PPIs from a number of small-scale experiments and from manually curated data. We used these data to train a Support Vector Machine (SVM) classifier using different combinations of features, including domain profiles, frequencies of protein sequence k-mers, and network characteristics of the human interactors in a human PPI network. We compared the performance of an SVM with a linear kernel on different combinations of features. We found that using a combination of protein sequence four-mers, protein domains, and PPI network information achieves the best performance, with precision greater than 70% for recall greater than 40%, depending on the ratio of positive examples to negative examples used during training.
We used this predictor to identify potentially novel viral interacting partners for human proteins. We focused our attention on those human proteins that are known to play an important role in HIV infection (Brass et al., 2008). Many predicted interactions involving these human proteins had considerable support in the literature. These interactions illustrate how the virus has evolved to manipulate host cellular processes to carry out successful pathogenesis. For example, predicted interactions with human cell surface proteins and human nuclear pore proteins are known to play a critical role in the initial invasion of the cell and subsequent movement of viral material across the nuclear membrane. We discuss in depth predicted interactions involving these and other host proteins.
2. Results and discussion
Our analysis contained two components. First, we compared SVM results using different feature combinations to identify which subset of features achieved the best performance. Second, we used this set of features to predict new PPIs between human and HIV proteins. The complete list of predicted interactions can be seen in Supplementary Files S1–S3. Each file corresponds to a different ratio of positive examples to negative examples, as further explained below.
2.1. Predictive feature sets
SVMs need both positive and negative examples in the training set. We generated three sets of negative examples (NEs), containing 25, 50, and 100 times the number of pairs of positive examples (PEs). Please see Section 3.4.1 for details. We refer to these datasets using the phrases “1:25 PE:NE ratio,” “1:50 PE:NE ratio,” and “1:100 PE:NE ratio.”
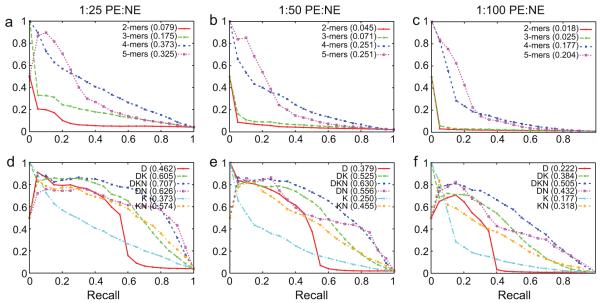
We measured the performance of SVMs trained using different amino acid k-mer sizes using 4-fold cross validation (see Fig. 1(a)–(c)). We computed the area under the precision/recall curve (AUC-PR) in order to compare the performance of different feature sets quantitatively. High AUC scores are characteristic of good predictors. At the 1:25 PE:NE ratio the 4-mer model performed the best with an AUC-PR of 0.373. At the 1:50 PE:NE ratio the 4-mer and 5-mer models had the same AUC-PR score of 0.251. Finally, at the 1:100 PE:NE ratio, the 5-mer model had the best AUC-PR score of 0.204. Subsequent analyses showed that the 4-mer model has the best area under the receiver operation characteristic curves (AUC, data not shown). Since the 4-mer model consistently had the highest or close to the best AUC-PR and AUC in all these tests, we used it in the rest of the analysis.
Fig. 1.
Precision/recall curves for different PE:NE ratios. (a)–(c) Results for four different amino acid k-mer sizes. (d)–(f) Results for combinations of amino acid 4-mers with other features. For each feature set, the AUC-PR score is shown within parentheses.
Fig. 1(d)–(f) displays the precision/recall curves for each of the PE:NE ratios and with different combinations of features: domains (D), protein sequence 4-mers (K), and network properties (N). We performed the analysis for all possible combinations of features, except for the single feature N, since the coverage of this feature was very sparse. As described in we also computed the AUC-PR values to quantitatively compare the performance of the different feature sets. At all three PE:NE ratios, the model trained using domains, amino acid 4-mers, and network properties (DKN) had the highest AUC-PR scores. The scores were 0.707, 0.630, and 0.505 for the 1:25, 1:50, and 1:100 ratios, respectively.
Since we used randomly chosen protein pairs as negative examples, we estimated the robustness of our results to the specific choice of negative examples. We repeated our analysis with ten different randomly generated sets of negative examples. Given our finding that the DKN feature set had the highest AUC-PR score across all three PE:NE ratios, we performed this analysis solely on this feature set. The results show that the variability over different sets of NEs is very small, as can seen by the small error bars in Supplementary Fig. S1 in the precision–recall curves, for values of recall at least 0.1. We concluded that the precise set of randomly selected negative examples did not have much influence on the results.
2.2. Feature importance
Since we used SVMs with linear kernels, we reasoned that the magnitude of the coefficients of the separating hyperplane may allow us to estimate each feature’s importance. Although the separating plane is defined by all features with non-zero coefficients, focusing on features with the largest coefficients yields a qualitative feel for the relative contributions of different features. For each PE:NE ratio, we constructed the SVM model corresponding to the DKN feature set. Several domain pairs appear in the top ten features for all PE:NE ratios. The top ranked feature is the human domain “Four-helical cytokine, core (IPR012351)” and the HIV domain “HIV transactivating regulatory protein Tat (IPR001831)”. Cytokines are a class of proteins that are used extensively in cellular communication and signaling, and in the activation of apoptotic pathways. The viral protein Tat is known to play a critical role in the disruption of normal cell signaling pathways such as the apoptotic pathway (Cossarizza, 2008). Another example is the third ranked domain pair consisting of the human domain “Clathrin adaptor (IPR000804)” and the HIV domain “HIV negative factor Nef (IPR001558)”. The viral Nef protein has been shown to play an important role in disrupting the AP2M1 clathrin adapter pathway by inducing the formation of clathrin-coated pits in the presence of CD4 in an effort to accelerate the rate of endocytosis (Foti et al., 1997; Swigut et al., 2001).
While we have not found evidence that these interactions are mediated by the domains, our observations could act as the basis for future mechanistic studies of how HIV proteins interact with human proteins. Supplementary Files S4–S6 contain the lists of all the features used in the three training sets along with the coefficients in the separating hyperplane.
To study the robustness of these results, we performed the following analysis. For each PE:NE ratio, we used the training set to compute the coefficients for each feature. Next we randomly shuffled the labels between the true positive and true negative interactions and repeated the analysis. After performing this step 100 times, we computed the p-value of each feature as the number of random iterations that produced a coefficient at least as large in magnitude as the coefficient computed with the true training set. We observed that none of the top 25+ features had a p greater than 0, i.e., we were not able to generate a random set of data that could generate a feature coefficient at least as large as in the real dataset.
2.3. Literature-based validation of predicted PPIs
Recently Brass et al. (2008) performed a genomic siRNA screen to identify HIV dependency factors (HDFs). By measuring levels of viral protein expression or production of infectious viral particles in human cells after knocking down individual genes, they searched for human genes that are required for HIV to undergo viral replication. Since silencing these genes is not lethal to the cell, HDFs may include many potential host-based therapeutic targets.
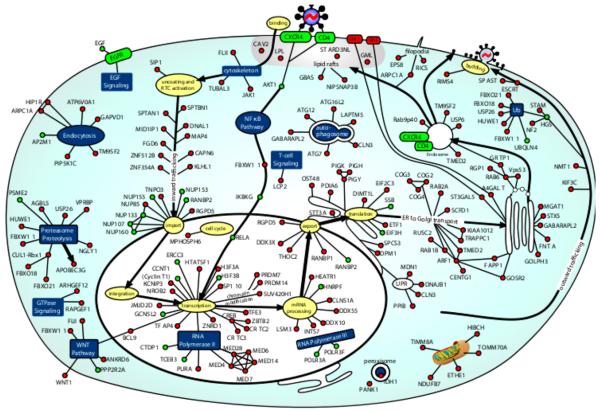
In this section, we focus our discussion on PPIs predicted by our approach where the human protein is an HDF found by Brass et al. (2008). We predicted 46 human–HIV PPIs involving HDFs at the 1:25 PE:NE ratio. We considered a protein pair to be a predicted interaction if the SVM trained on all positive and negative examples assigned that pair a positive score. This score corresponds to a precision of 74.3% and a recall of 65.5%. See Table 1 for a summary of our predictions and Fig. 2 for a visualization of HDFs for which we predict PPIs. Below we discuss predicted interactions involving HDFs that have support in the literature. We did not include the NCBI human–HIV PPI database (Fu et al., 2009) in our set of positive examples. While some of the interactions from our positive examples may be included in this database, we found the NCBI database to be a fertile source for validating our predictions. We were able to find literature support for many of our predictions and we discuss those here.
Table 1.
Summary of the number of predicted PPIs. For each PE:NE ratio, we list the total number of predicted interactions and the number of these that involve HDFs.
| PE:NE ratio |
|||
|---|---|---|---|
| 1:25 | 1:50 | 1:100 | |
| # Predicted PPIs | 1111 | 506 | 182 |
| # Predicted PPIs involving HDFs | 46 | 33 | 16 |
Fig. 2.
Image taken with permission from Brass et al. (2008) and modified. Nodes are HDFs found by Brass et al. (2008). Green nodes are HDFs for which we predict PPIs at the 1:25 PE:NE ratio. Red nodes are HDFs for which we do not predict PPIs. For the sake of clarity, we do not show predicted HIV interactors in the image. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of the article.)
Manipulation of intracellular signaling pathways via cell-surface receptors is a well-established characteristic of HIV infection (Popik and Pitha, 2000). We made several predictions for host proteins found on the cell surface and participating in various signaling pathways. For example, we predicted interactions involving host HDFs Epidermal Growth Factor (EGF) and EGF Receptor (EGFR). These proteins play a critical role in the regulation of cell growth, proliferation, and differentiation. In particular the EGF–Tat and EGFR–Gag interactions have been shown to be critical for promoting cell growth via the host EGF pathway leading to enhanced HIV replication (Nabell et al., 1994; Valiathan and Resh, 2004). We predict both interactions.
CD4 positive helper T cells are the primary substrates of HIV. T cells are responsible for activating and directing other immune cells that lack cytotoxic and phagocytic activities, i.e., these cells cannot kill infected cells or pathogens directly. We made several predictions for both the host CD4 proteins, which are supported by the literature. Predicted interacting partners for the host CD4 protein include the viral proteins Vpu and Nef. These interactions have been linked to a depletion of CD4 proteins on the cell surface (Chen et al., 1996). Reduction in the number of CD4+ cells weakens the host’s immune system and makes it more susceptible to infections. Although the direct mechanism is not clear, it has been shown that down regulation of CD4 is required for HIV infection (Tanaka et al., 2003).
We predicted that human HDFs PPP2R2A and PSME2 proteins interact with the HIV Tat protein. Both human proteins are localized to the cytoplasm. PPP2R2A is one of four major Ser/Thr phosphatases that plays a role in the negative control of cell growth and division. During pathogenesis, the interaction between PPP2R2A and Tat has been observed to play a key role in Tat’s ability to act as a transcription factor in the increased production of viral material (Ruediger et al., 1997). Host PSME2 is a subunit of the protein complex responsible for activating the proteosome complex and enhancing the generation of major histocompatability complex class I binding peptides. Viral Tat has been shown to interfere with the antigen presentation via this interaction (Huang et al., 2002; Seeger et al., 1997), leading to a failure of the human immune system to recognize HIV infected cells.
Since viruses lack the machinery needed to replicate their genomes, viral genetic material must first cross the barrier from the cytoplasm into the nucleus in order to make use of the host’s transcriptional machinery. The nuclear pore complex is a large protein complex that spans the nuclear membrane and allows for the transport of molecules across the nuclear envelope including proteins and RNA. We predicted interactions between several viral proteins and host HDFs that are known to be part of the nuclear pore complex including NUP107, NUP133, NUP153, NUP155, and NUP160. One of the predicted interactors is the viral Vpr protein. Viral Vpr has been shown to localize at the nuclear envelope and interact with several nuclear proteins (Le Rouzic et al., 2002). This interaction has been linked with Vpr’s ability to drive the cell into G2 cell cycle arrest resulting in the activation of apoptotic pathways (Andersen et al., 2006). We also predicted interactions of these host HDFs with the viral Tat protein. While no direct interaction has been observed between viral Tat and these nuclear pore proteins, Tat has been shown to possess a Nuclear Localization Sequence (NLS) and is capable of transporting material across the nuclear membrane through the nuclear pore (Efthymiadis et al., 1998). Thus, the predicted interactions involving the viral Tat protein and these host HDFs may be worthy candidates for experimental validation.
Within the nucleus of the host cell, we made several predictions involving host HDF proteins. A goal of these interactions may be to modulate and manipulate host immune response pathways in order to assure continued survival of the virus. One example that highlights this strategy is the predicted interaction between the host HDF RelA and the viral Vpr proteins. RelA is part of the NFκB complex. NFκB is a transcription factor that regulates many biological processes such as inflammation, immunity, and apoptosis. The interaction with Vpr has been shown to inhibit the nuclear translocation of NFκB, thus preventing the host from mounting a successful immune response (Venkatachari et al., 2007).
HIV also makes use of host proteins to drive the expression of its own genetic material. One such example is the host CCNT1 protein, which is a cyclin. Cyclins function as regulators of cyclin dependent kinases, which play an important role in cell cycle progression. Two of the predicted partners of CCNT1 are the viral proteins Tat and Vpr. CCNT1 serves as an essential cofactor for Tat. The interaction between these two has been shown to increase Tat’s affinity for the transactivating response RNA element (TAR) allowing the transcription of viral genes (Bieniasz et al., 1999). The viral Vpr protein has been shown to interact with CCNT1 in tandem with viral Tat to modulate transcription of the viral genome (Sawaya et al., 2000). HIV must also recruit host polymerases to translate viral genetic material. We predict an interaction between the human POLR3A, a DNA-dependent RNA polymerase, and the viral Tat protein. The HIV Tat protein has been shown to upregulate transcription by POLR3A, leading to an increased production of viral proteins (Jang et al., 1992).
3. Materials and methods
We first describe the classifier we used to predict human–HIV HP-PPIs. Next, we present the features we included in this classifier. Finally, we describe our validation protocol.
3.1. Support Vector Machines
The Support Vector Machine (SVM) is a powerful and popular approach in machine learning for classification problems. Given a training set S with each vector in S associated with a label equal to 1 or −1, an SVM classifier computes a hyperplane separating the vectors in S with label 1 from the vectors with label −1, optionally after projecting the vectors to a higher-dimensional feature space. The projection is often represented compactly by a kernel function. An important feature of SVMs is that the separating plane has maximum margin, which is the distance from the separating plane to the closest vector.
In this study, we used an SVM classifier with a linear kernel, i.e., we performed no projection. We also evaluated SVM classifiers with radial basis kernels. We omit these results since the improvement over the linear kernel was marginal. For each host–pathogen protein pair (p, q), we computed a vector of different protein features f(p,q), as explained in the next section. Let S be a training set consisting of (f(p,q), l) pairs, where l ∈ {−1, 1} is the class label of the PPI (p, q). In our case, the labels 1 and −1 corresponded to the classes “PPI” and “non-PPI,” respectively.
3.2. PPI features
We considered three types of protein features in this study: domains (D), protein sequence k-mers (K), and properties in the intra-species human PPI network (N). We explain the rationale for including each of these features below.
3.2.1. Domains (D)
Physical interactions between proteins are often mediated by specific domains (Pawson and Nash, 2003). Previous research has demonstrated the utility of protein-domain information in predicting both intra-species PPIs (Ng et al., 2003; Sprinzak and Margalit, 2001) and host–pathogen PPIs (Dyer et al., 2007).
Let Mp be the set of domains present in a protein p and let M be the set of all domains, over all proteins in our dataset. Our feature vector contained one binary feature for every pair of domains in M × M. For a PPI (p, q), we set the features corresponding to each of the domain pairs in Mp × Mq to be 1 and the remaining features to be 0. We encoded the domain features using pairs of domains since the interaction is often contingent on the presence of the pair. An alternative method that we considered was to compute the probability that a protein pair would contain a pair of domains given that the proteins interacted in the training set. We refrained from using this approach because these probabilities cannot be computed accurately for the currently sparse human–HIV PPI datasets.
3.2.2. Protein sequence k-mers (K)
Since the sequence of a protein determines its structure and consequently its function, it may be possible to predict PPIs using the amino acid sequence of a protein pair. Shen et al. (2007) introduced the “conjoint triad model” for predicting PPIs using only amino acid sequences. Shen et al. (2007) partitioned the twenty amino acids into seven classes based on their electrostatic and hydrophobic properties. For each protein, they counted the number of times each distinct three-mer (set of three consecutive amino acids) occurred in the sequence. To account for protein size, they normalized these counts by linearly transforming them to lie between 0 and 1 (see (Shen et al., 2007) for details). They represented the protein with a 343-element feature vector, where the value of each feature is the normalized count for each of the 343 (73) possible amino acid three-mers. In this paper we explored the use of two-, three-, four-, and five-mers. For each host–pathogen protein pair, we concatenated the feature vectors of the individual proteins. Therefore, each host–pathogen protein pair had a feature vector of length at most 98, 646, 4802, and 33614, in the cases of two-, three-, four-, and five-mers, respectively.
3.2.3. Network properties (N)
Recent studies have suggested that pathogens have evolved to interact with human proteins which are hubs (proteins with many interacting partners) (Dyer et al., 2008; Calderwood et al., 2007) and bottlenecks (proteins that are central to many paths in the network) (Dyer et al., 2008) in the human PPI network. We represented the human PPI network as an undirected graph G= (V,E), where V was the set of human proteins and E was the set of PPIs between them. We defined the degree of a protein in a PPI network as the number of interactions in which it participates, not including self-interactions. We defined the betweenness centrality bc(v) of a protein v as the fraction of shortest paths in G between all protein pairs (u, w) that pass through the protein v. Given u, v, w ∈ V, let σuw denote the number of shortest paths between proteins u and w. Let σuw(v) denote the number of these that pass through v. Then the betweenness centrality of v is
In our analysis, we divided bc(v) by the number of pairs of nodes in G, yielding a quantity between 0 and 1. We used the algorithm devised by Brandes (2001) to compute the betweenness centrality of all nodes in G. For each host–pathogen protein pair, we included two features corresponding to these properties: an integer-valued feature for a human protein’s degree and a real-valued feature for its betweenness centrality.
3.3. Evaluation of performance
We tested the predictive power of six combinations of features: D, DK, DKN, DN, K, and KN using four-fold cross validation. To obtain feature vectors for a particular combination, we simply concatenated the vectors for the individual features. We did not test the predictive power of the N feature set alone because the coverage of these features is small. We used the SVMLight package (Joachims, 1999) for training and testing SVMs. In this package, the parameter C controls the trade-off between maximizing the margin of the separating plane and minimizing the mis-classification error. We systematically varied C, trying alternate powers of 2 between 2−5 and 217 (i.e., 2−5, 2−3, …, 215, 217). For each choice of C, we counted the number of true positives (TP), false positives (FP), true negatives (TN), and false negatives (FN) and computed accuracy (TP + TN/(TP + FP + TN + FN)), precision (TP/(TP + FP)) and recall (TP/(TP + FN)). For each feature combination, we based further analyses and predictions on the value of C that yielded the maximum accuracy for that combination. We plotted precision/recall curves by varying the threshold on the score assigned to protein pairs by the SVM classifier; we considered protein pairs above the threshold to be interacting and those below the threshold as non-interacting.
3.4. Data sets used
We used the Uniprot database (Bairoch et al., 2005) as a source for protein sequence information. We used InterProScan (Quevillon et al., 2005) to determine protein domains. All data used in this study were downloaded in February 2008.
3.4.1. Gold standard datasets
We gathered 1028 human–HIV (isolate HXB2 group M subtype B) PPIs from four public databases: the Biomolecular Interaction Network Database (Gilbert, 2005), the Database of Interacting Proteins (Salwinski et al., 2004), IntAct (Hermjakob et al., 2004), and Reactome (Joshi-Tope et al., 2005). These PPIs formed our positive examples. We also constructed a human intra-species PPI network containing 78,804 PPIs using these four databases along with three additional sources: the Human Protein Reference Database (Mishra et al., 2006), the Molecular INTeraction Database (Zanzoni et al., 2002), and the Munich Information Center for Protein Sequences (Guldener et al., 2006). We used the intra-species network to compute each human protein’s degree and centrality.
Selection of negative examples is a well-recognized challenge for PPI prediction since biological datasets rarely include pairs of proteins that are known not to interact (Ben-Hur and Noble, 2006). The number of truly interacting pairs of human–HIV proteins is likely to be far less than the total set of protein pairs. Therefore, we generated negative examples by randomly pairing human and HIV proteins. In doing so, we ensured that no randomly generated protein pair was already known to interact, i.e., was a positive example. Since we did not know the true number of non-interacting pairs of human–HIV proteins, we tested our prediction methodology with different numbers of negative examples. Specifically, we generated 25, 50, and 100 times as many negative examples as positive examples. Our rationale for trying different PE:NE ratios was that we could observe how the precision and recall of our methodology varies with increasing PE:NE ratio. We used these trends to guide our decisions on which combinations of feature sets achieved the best performance, as explained in Section 3. We note that the true PE:PN ratio is likely to be much smaller than 1:100. As more human–HIV PPIs are detected experimentally, our methods will be able to handle lower PE:PN ratios.
4. Conclusions
We have presented an application of a supervised machine learning method to predict human–pathogen PPIs. Our goal was to predict new physical interactions between human and pathogen proteins that may be critical to pathogenesis. Important aspects of our work include the comparison of different features and their combinations and observing the performance of the predictor for multiple PE:NE ratios. We applied our methodology to the human–HIV system. We found that a model trained using domain-profiles, sequence four-mers, and network characteristics of the human proteins achieved the best performance upon cross validation. When we used this model to predict PPIs involving human proteins known to be critical for HIV infection, we succeeded in predicting many interactions supported by the literature. We expect that other predicted interactions will provide further insights into why these host proteins are critical for HIV. A key extension of this work is to integrate additional types of data (e.g., gene expression) so as to improve the robustness and accuracy of our predictions. It is unclear at this moment how big a host–pathogen interactome will be, especially in the case of RNA viruses such as HIV that have a small number of proteins. As more interactions are identified it will become possible to robustly estimate the size of the host–pathogen interactome in a similar manner to estimates of the sizes of intra-species interactomes (Stumpf et al., 2008). Another important analysis will be to extend this work to other host–pathogen systems.
Supplementary Material
Acknowledgments
The authors wish to thank Abe Brass and Steve Elledge for the use of Fig. 2. This project has been funded in whole or in part with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400035C to Bruno Sobral. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Appendix A. Supplementary data Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.meegid.2011.02.022.
References
- Andersen JL, DeHart JL, Zimmerman ES, Ardon O, Kim B, et al. HIV-1 Vpr-induced apoptosis is cell cycle dependent and requires Bax but not ANT. PLoS Pathog. 2006;2:e127. doi: 10.1371/journal.ppat.0020127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, et al. The universal protein resource (UniProt) Nucleic Acids Res. 2005;33:D154–159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Hur A, Noble WS. Choosing negative examples for the prediction of protein–protein interactions. BMC Bioinform. 2006;7(Suppl. 1):S2. doi: 10.1186/1471-2105-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD, Grdina TA, Bogerd HP, Cullen BR. Recruitment of cyclin T1/P-TEFb to an HIV type 1 long terminal repeat promoter proximal RNA target is both necessary and sufficient for full activation of transcription. Proc. Natl. Acad. Sci. U.S.A. 1999;96:7791–7796. doi: 10.1073/pnas.96.14.7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes U. A faster algorithm for betweenness centrality. Math. Sociol. 2001;25:163–177. [Google Scholar]
- Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, et al. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921–926. doi: 10.1126/science.1152725. [DOI] [PubMed] [Google Scholar]
- Calderwood MA, Venkatesan K, Xing L, Chase MR, Vazquez A, et al. Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 2007;104:7606–7611. doi: 10.1073/pnas.0702332104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BK, Gandhi RT, Baltimore D. CD4 down-modulation during infection of human T cells with human immunodeficiency virus type 1 involves independent activities of vpu, env, and nef. J. Virol. 1996;70:6044–6053. doi: 10.1128/jvi.70.9.6044-6053.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A. Apoptosis and HIV infection: about molecules and genes. Curr. Pharm. Des. 2008;14:237–244. doi: 10.2174/138161208783413293. [DOI] [PubMed] [Google Scholar]
- Davis FP, Barkan DT, Eswar N, McKerrow JH, Sali A. Host pathogen protein interactions predicted by comparative modeling. Protein Sci. 2007;16:2585–2596. doi: 10.1110/ps.073228407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer MD, Murali TM, Sobral BW. Computational prediction of host–pathogen protein protein interactions. Bioinformatics. 2007;23:i159–i166. doi: 10.1093/bioinformatics/btm208. [DOI] [PubMed] [Google Scholar]
- Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4:e32. doi: 10.1371/journal.ppat.0040032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efthymiadis A, Briggs LJ, Jans DA. The HIV-1 Tat nuclear localization sequence confers novel nuclear import properties. J. Biol. Chem. 1998;273:1623–1628. doi: 10.1074/jbc.273.3.1623. [DOI] [PubMed] [Google Scholar]
- Foti M, Mangasarian A, Piguet V, Lew DP, Krause KH, et al. Nef-mediated clathrin-coated pit formation. J. Cell. Biol. 1997;139:37–47. doi: 10.1083/jcb.139.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, et al. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37:D417–422. doi: 10.1093/nar/gkn708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert D. Biomolecular interaction network database. Brief Bioinform. 2005;6:194–198. doi: 10.1093/bib/6.2.194. [DOI] [PubMed] [Google Scholar]
- Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, et al. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, et al. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32:D452–455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Seifert U, Salzmann U, Henklein P, Preissner R, et al. The RTP site shared by the HIV-1 Tat protein and the 11S regulator subunit alpha is crucial for their effects on proteasome function including antigen processing. J. Mol. Biol. 2002;323:771–782. doi: 10.1016/s0022-2836(02)00998-1. [DOI] [PubMed] [Google Scholar]
- Jang KL, Collins MK, Latchman DS. The human immunodeficiency virus tat protein increases the transcription of human Alu repeated sequences by increasing the activity of the cellular transcription factor TFIIIC. J. Acquir. Immune Defic. Syndr. 1992;5:1142–1147. [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, et al. A Bayesian networks approach for predicting protein–protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Joachims T. Advances in Kernel Methods – Support Vector Learning. MIT-Press; 1999. Making Large-Scale SVM Learning Practical. [Google Scholar]
- Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, et al. REACTOME: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428–432. doi: 10.1093/nar/gki072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnadev O, Srinivasan N. A data integration approach to predict host–pathogen protein–protein interactions: application to recognize protein interactions between human and a malarial parasite. In Silico Biol. 2008;8:235–250. [PubMed] [Google Scholar]
- Rouzic E. Le, Mousnier A, Rustum C, Stutz F, Hallberg E, et al. Docking of HIV-1 Vpr to the nuclear envelope is mediated by the interaction with the nucleoporin hCG1. J. Biol. Chem. 2002;277:45091–45098. doi: 10.1074/jbc.M207439200. [DOI] [PubMed] [Google Scholar]
- Lee SA, Chan CH, Tsai CH, Lai JM, Wang FS, et al. Ortholog-based protein–protein interaction prediction and its application to inter-species interactions. BMC Bioinform. 2008;9(Suppl. 12):S11. doi: 10.1186/1471-2105-9-S12-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, et al. Human protein reference database – 2006 update. Nucleic Acids Res. 2006;34:D411–414. doi: 10.1093/nar/gkj141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabell LM, Raja RH, Sayeski PP, Paterson AJ, Kudlow JE. Human immunodeficiency virus 1 Tat stimulates transcription of the transforming growth factor alpha gene in an epidermal growth factor-dependent manner. Cell Growth Differ. 1994;5:87–93. [PubMed] [Google Scholar]
- Ng SK, Zhang Z, Tan SH. Integrative approach for computationally inferring protein domain interactions. Bioinformatics. 2003;19:923–929. doi: 10.1093/bioinformatics/btg118. [DOI] [PubMed] [Google Scholar]
- Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- Pellegrini M, Marcotte EM, Thompson MJ, Eisenberg D, Yeates TO. Assigning protein functions by comparative genome analysis: protein phylogenetic profiles. Proc. Natl. Acad. Sci. U.S.A. 1999;96:4285–4288. doi: 10.1073/pnas.96.8.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popik W, Pitha PM. Exploitation of cellular signaling by HIV-1: unwelcome guests with master keys that signal their entry. Virology. 2000;276:1–6. doi: 10.1006/viro.2000.0581. [DOI] [PubMed] [Google Scholar]
- Qi Y, Bar-Joseph Z, Klein-Seetharaman J. Evaluation of different biological data and computational classification methods for use in protein interaction prediction. Proteins. 2006;63:490–500. doi: 10.1002/prot.20865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Tastan O, Carbonell JG, Klein-Seetharaman J, Weston J. Semi-supervised multi-task learning for predicting interactions between HIV-1 and human proteins. Bioinformatics. 2010;26:i645–i652. doi: 10.1093/bioinformatics/btq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, et al. Inter-ProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruediger R, Brewis N, Ohst K, Walter G. Increasing the ratio of PP2A core enzyme to holoenzyme inhibits Tat-stimulated HIV-1 transcription and virus production. Virology. 1997;238:432–443. doi: 10.1006/viro.1997.8873. [DOI] [PubMed] [Google Scholar]
- Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, et al. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya BE, Khalili K, Gordon J, Taube R, Amini S. Cooperative interaction between HIV-1 regulatory proteins Tat and Vpr modulates transcription of the viral genome. J. Biol. Chem. 2000;275:35209–35214. doi: 10.1074/jbc.M005197200. [DOI] [PubMed] [Google Scholar]
- Seeger M, Ferrell K, Frank R, Dubiel W. HIV-1 tat inhibits the 20 S proteasome and its 11 S regulator-mediated activation. J. Biol. Chem. 1997;272:8145–8148. doi: 10.1074/jbc.272.13.8145. [DOI] [PubMed] [Google Scholar]
- Sharan R, Suthram S, Kelley RM, Kuhn T, McCuine S, et al. Conserved patterns of protein interaction in multiple species. Proc. Natl. Acad. Sci. U.S.A. 2005;102:1974–1979. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J, Zhang J, Luo X, Zhu W, Yu K, et al. Predicting protein–protein interactions based only on sequences information. Proc. Natl. Acad. Sci. U.S.A. 2007;104:4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprinzak E, Margalit H. Correlated sequence-signatures as markers of protein–protein interaction. J. Mol. Biol. 2001;311:681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, et al. Estimating the size of the human interactome. Proc. Natl. Acad. Sci. U.S.A. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigut T, Shohdy N, Skowronski J. Mechanism for down-regulation of CD28 by Nef. EMBO J. 2001;20:1593–1604. doi: 10.1093/emboj/20.7.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M, Ueno T, Nakahara T, Sasaki K, Ishimoto A, et al. Down-regulation of CD4 is required for maintenance of viral infectivity of HIV-1. Virology. 2003;311:316–325. doi: 10.1016/s0042-6822(03)00126-0. [DOI] [PubMed] [Google Scholar]
- Valiathan RR, Resh MD. Expression of human immunodeficiency virus type 1 gag modulates ligand-induced downregulation of EGF receptor. J. Virol. 2004;78:12386–12394. doi: 10.1128/JVI.78.22.12386-12394.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatachari NJ, Majumder B, Ayyavoo V. Human immunodeficiency virus (HIV) type 1 Vpr induces differential regulation of T cell costimulatory molecules: direct effect of Vpr on T cell activation and immune function. Virology. 2007;358:347–356. doi: 10.1016/j.virol.2006.08.030. [DOI] [PubMed] [Google Scholar]
- Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, et al. Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanzoni A, Montecchi-Palazzi L, Quondam M, Ausiello G, Helmer-Citterich M, et al. MINT: a molecular INTeraction database. FEBS Lett. 2002;513:135–140. doi: 10.1016/s0014-5793(01)03293-8. [DOI] [PubMed] [Google Scholar]
- Zhang LV, Wong SL, King OD, Roth FP. Predicting co-complexed protein pairs using genomic and proteomic data integration. BMC Bioinform. 2004;5:38. doi: 10.1186/1471-2105-5-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.