Abstract
MicroRNA-21 (miR-21) is a key regulator of oncogenic processes. It is significantly elevated in the majority of human tumors and functionally linked to cellular proliferation, survival and migration. In this study, we used two experimental-based strategies to search for novel miR-21 targets. On the one hand, we performed a proteomic approach using two-dimensional differential gel electrophoresis (2D-DIGE) to identify proteins suppressed upon enhanced miR-21 expression in LNCaP human prostate carcinoma cells. The tumor suppressor acidic nuclear phosphoprotein 32 family, member A (ANP32A) (alias pp32 or LANP) emerged as the most strongly downregulated protein. On the other hand, we applied a mathematical approach to select correlated gene sets that are negatively correlated with primary-miR-21 (pri-miR-21) expression in published transcriptome data from 114 B-cell lymphoma cases. Among these candidates, we found tumor suppressor SMARCA4 (alias BRG1) together with the already validated miR-21 target, PDCD4. ANP32A and SMARCA4, which are both involved in chromatin remodeling processes, were confirmed as direct miR-21 targets by immunoblot analysis and reporter gene assays. Furthermore, knock down of ANP32A mimicked the effect of enforced miR-21 expression by enhancing LNCaP cell viability, whereas overexpression of ANP32A in the presence of high miR-21 levels abrogated the miR-21-mediated effect. In A172 glioblastoma cells, enhanced ANP32A expression compensated for the effects of anti-miR-21 treatment on cell viability and apoptosis. In addition, miR-21 expression clearly increased the invasiveness of LNCaP cells, an effect also seen in part upon downregulation of ANP32A. In conclusion, these results suggest that downregulation of ANP32A contributes to the oncogenic function of miR-21.
Keywords: miR-21, ANP32A, SMARCA4, prostate, post-transcriptional regulation
Introduction
MicroRNAs (miRNAs) are short non-coding RNAs involved in post-transcriptional regulation of gene expression. They originate from mRNA-like, long primary transcripts (pri-miRNA) that are processed by two sequential cleavage steps into their mature form. In animals, miRNAs bind to partly complementary sequences located predominantly within the 3′ untranslated regions and mediate translational repression or degradation of their target mRNAs (Bartel, 2004). More than 700 human miRNAs have been identified to date and proteome-wide studies suggested that an individual miRNA can regulate hundreds of targets (Baek et al., 2008; Selbach et al., 2008; Yang et al., 2010). They have been found to play an important role in fundamental cellular processes, including proliferation, apoptosis, migration and differentiation. Hence, aberrant miRNA expression has been correlated and functionally linked to cancerogenesis (Calin et al., 2002; Garzon et al., 2009).
MicroRNA-21 (miR-21) stands out as the only miRNA overexpressed in the vast majority of cancer types analyzed (Krichevsky and Gabriely, 2009). Elevated levels have been demonstrated in solid tumors of, for example, breast, lung, colon, pancreas, prostate, liver, stomach and brain, as well as in hematological cancers, including chronic lymphocytic leukemia, diffuse large B-cell lymphoma (DLBCL) and Hodgkin lymphoma (Chan et al., 2005; Iorio et al., 2005; Volinia et al., 2006; Fulci et al., 2007; Lawrie et al., 2007; Navarro et al., 2008). Direct evidence for the oncogenic potential of miR-21 was revealed by functional studies in several cancer cell lines. Knock down of miR-21 was shown to cause increased apoptosis and reduced invasiveness in glioblastoma (Chan et al., 2005), reduced cell proliferation in breast and cervix cancer (Frankel et al., 2008; Zhu et al., 2008; Yao et al., 2009), as well as decreased cellular invasion and metastasis in colorectal cancer (Asangani et al., 2008). Conversely, miR-21 overexpression resulted in increased tumor cell proliferation, migration and invasion in hepatocellular carcinoma, as well as chemoresistance of cholangiocarcinoma (Meng et al., 2006). Recent work of our group demonstrated that signal transducer and activator of transcription 3 induces pri-miR-21 transcription, and elevation of miR-21 significantly promoted survival of multiple myeloma (Loffler et al., 2007). Therefore, understanding the network of miR-21-induced gene expression changes is of fundamental interest.
To date, experimentally confirmed targets include multiple tumor suppressive components of the p53, TGF-β and mitochondrial apoptosis pathways (Papagiannakopoulos et al., 2008), as well as tumor suppressors PDCD4 (Asangani et al., 2008), PTEN (Meng et al., 2007), RECK (Gabriely et al., 2008), TPM1 (Zhu et al., 2008) and Maspin (Zhu et al., 2008).
The identification of mRNAs targeted by a particular miRNA has been a great challenge because of the only partial complementarity between miRNAs and their targets. Much of the early efforts based on computational algorithms combined continuous pairing of 6–8 bases in the 5′ miRNA seed region and the 3′ untranslated region of mRNAs with phylogenetic conservation of the complementary sequence in the 3′ untranslated region of orthologous genes (Lewis et al., 2003; Kiriakidou et al., 2004; John et al., 2006). However, about 30% of the predicted mRNA target sites cannot be validated experimentally (Martin et al., 2007) and more than one quarter of experimentally validated miRNA targets are not predicted by any of the most commonly used miRNA target prediction programs (Sethupathy et al., 2006). As miRNAs can regulate both mRNA stability and protein synthesis, experimental approaches include gene expression analysis and proteomic screening.
In this study, we searched for novel miR-21 targets applying both strategies. We analyzed LNCaP cells transfected with miR-21-expressing or -control vectors by two-dimensional differential in gel electrophoresis (2D-DIGE). Furthermore, we used the statistical analysis of B-cell lymphoma gene expression data by searching for genes inversely correlated with high pri-miR-21 levels. On the basis of these results, we demonstrate that tumor suppressors ANP32A and SMARCA4 are direct targets of miR-21. Furthermore, we provide experimental evidence that ANP32A is able to contribute to various miR-21-mediated effects.
Results
Identification of putative miR-21 targets by use of 2D-DIGE
In order to identify putative miR-21 targets, a global proteomic approach using DIGE was conducted with samples from cells expressing different miR-21 levels. Recent studies have shown that miR-21 is upregulated in prostate cancer cells (Volinia et al., 2006) and that its expression in prostate cancer cell lines rises with the grade of androgen-independence and malignancy from LNCaP to PC-3 and DU-145 cells (Li et al., 2009). We chose LNCaP cells that express very low levels of miR-21 for transfection with either a pre-miR-21-expressing plasmid or an empty vector and searched for proteins regulated by enhanced miR-21 expression. As measured by quantitative PCR, miR-21 levels were raised to about two- to threefold compared with the control (data not shown). Cells were lysed and proteins were fluorescence-labelled, 24 h after transfection. After co-separating the protein extracts and fluorescence scanning, differentially expressed spots were detected (Figure 1). In summary, 37 protein spots displayed an at least 1.3-fold regulation over three independent experiments. Among the 25 proteins unambiguously identified by mass spectrometry, 5 proteins were represented by two or more spots exhibiting a comparable ratio of expression that were most likely caused by post-translational modification (Supplementary Table 1). Using an at least 1.5-fold differential regulation and a P-value of ⩽0.05 as cut-offs, we found 16 proteins that were all suppressed in the presence of miR-21 (Table 1 and Supplementary Figure 1). This overall downward direction in regulated spots is consistent with the concept of miRNA-mediated translational inhibition.
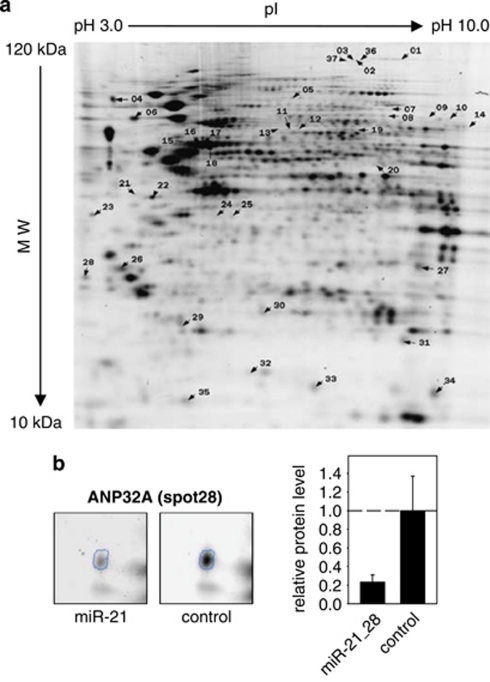
Figure 1.
Identification of putative miR-21 target genes in LNCaP cells by 2D-DIGE. LNCaP cells were transfected with either empty or miR-21 expression vectors, harvested after 24 h, and protein lysates were labeled with Cy3 or Cy5 (miR-21 and control) and Cy2 for the internal standard. (a) Representative gel image obtained after isoelectric focussing at pH 3–10, followed by separation in a 12% SDS–polyacrylamide gel. Numbers indicate spots identified as differentially expressed. (b) Enlarged area maps of spot 28 (corresponding to ANP32A) obtained by separate Cy3 (miR-21) and Cy5 (control) evaluation. The bar chart shows the relative expression level of ANP32A protein in cells transfected with miR-21 or control.
Table 1. Putative targets of miR-21 identified by 2D-DIGE.
| Gene symbola | Protein name | Ratiob | P-value |
|---|---|---|---|
| ANP32A | Acidic nuclear phosphoprotein 32 family, member A | 0.23 | 0.025 |
| ME1 | NADP-dependent malic enzyme | 0.25 | 0.047 |
| NCL | Nucleolin | 0.44 | 0.012 |
| LTA4H | Leukotriene A4 hydrolase | 0.44 | 0.005 |
| PEBP1 | Phosphatidylethanolamine-binding protein 1 | 0.58 | 0.050 |
| SOD1 | Superoxide dismutase 1 | 0.58 | 0.0002 |
| RPSA | 40S ribosomal protein SA | 0.59 | 0.002 |
| EIF5A | Eukaryotic translation initiation factor 5A | 0.59 | 0.035 |
| ATP6V0D1 | Vacuolar proton-ATPase, subunit D | 0.59 | 0.004 |
| GNB2L1 | Guanine nucleotide-binding protein subunit beta-2-like 1 | 0.61 | 0.011 |
| HADHA | Trifunctional enzyme subunit alpha | 0.61 | 0.003 |
| DDX3X | X-linked DEAD box protein 3 | 0.61 | 0.013 |
| EIF3I | Eukaryotic translation initiation factor 3 subunit I | 0.64 | 0.016 |
| CFL1 | Cofilin-1 | 0.65 | 0.016 |
| CSDE1 | Cold shock domain-containing protein E1 | 0.65 | 0.003 |
| HDLBP | High-density lipoprotein-binding protein | 0.65 | 0.023 |
Abbreviations: miR-21, microRNA-21; 2D-DIGE, two-dimensional differential in gel electrophoresis.
Gene symbols following UniProtKB.
Expression level (miR-21 versus control).
For proteins identified by several spots, results with the lowest P-values are shown.
Bioinformatical screening of the 16 candidate mRNAs for miR-21 target sequences by programs requiring a perfect seed match were not successful. By applying the RNA22 tool (Huynh et al., 2006), which allows G:U pairing of the target mRNA to the miRNA seed sequence, we obtained perfect 7-mer seed matches for LTA4H, RPSA, EIF3I, CSDE1 and HDLBP (Supplementary Figure 2a). With regard to cellular function, the majority of proteins listed in Table 1 play a role in biosynthetic and metabolic processes. This is likely because of the fact that many proteins with basal cellular functions show rather high expression levels, which favor their identification by DIGE. Notably, five of them (DDX3X, CSDE1, EIF5A, HDLBP and NCL) are RNA-binding proteins. Among the candidates, tumor suppressor ANP32A showed the strongest downregulation (4.3-fold; Figure 1b).
Identification of putative miR-21 targets by applying statistics on gene expression data
In parallel, we used a mathematical approach based on the analysis of published gene expression data to identify further putative miR-21 targets. We took advantage of the fact that the affymetrix U133A expression array contains a probe set detecting pri-miR-21. The human MIR21 gene overlaps with protein-coding gene TMEM49 (transmembrane protein-49) (Figure 2a), but is independently transcribed from a conserved promoter residing within an intron of TMEM49 (Loffler et al., 2007; Fujita et al., 2008). The pri-miR-21 transcript has been shown to be both capped and polyadenylated (Cai et al., 2004), suggesting its labeling by gene expression profiling using oligo (dT) priming. As shown in Figure 2, the Affymetrix U133A GeneChip exhibits a probe set detecting both TMEM49 and pri-miR-21. For both genes, no other (specific) probe set exists on U133A.
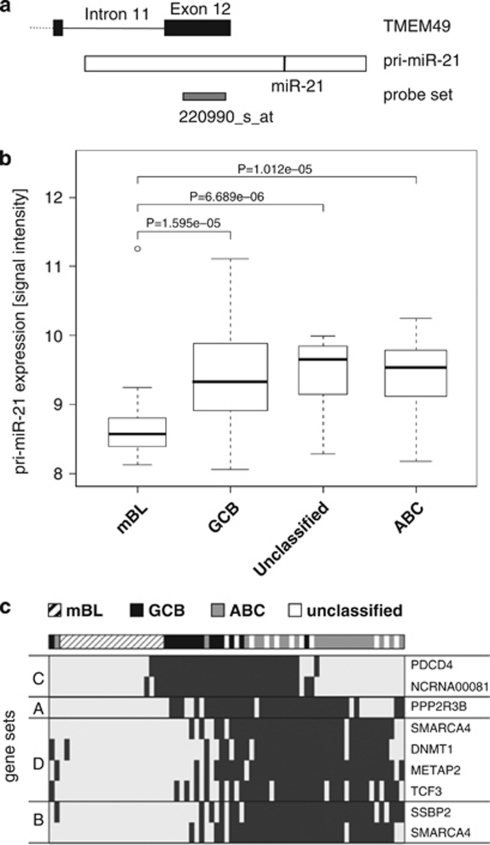
Figure 2.
Analysis of pri-miR-21 expression in B-cell lymphomas. (a) Schematic representation of the human MIR21 gene locus that overlaps with the last two exons of the TMEM49 gene on chromosome 17. Affymetrix probe set 220990_at detects TMEM49 as well as pri-miR-21. The black line within the pri-miR-21 bar indicates the location of mature miR-21 sequence. (b) Box plot showing the expression of pri-miR-21 in gene expression data from 114 lymphomas (Hummel et al., 2006) grouped into molecular Burkitt's lymphoma (mBL) and in DLBCL subgroups GCB (germinal center B cell-like), ABC (activated B cell-like), and in DLBCL samples that cannot be assigned to either subgroup (unclassified). The P-values shown are not adjusted for multiple testing, but remain significant after adjusting with the Bonferroni method for the six possible pairwise tests. (c) In gene expression data from the lymphomas cases above, correlated sets of genes were retrieved and spatially sorted according to a previously published procedure (Lauter et al., 2009). In the heat map, lymphoma cases and the four gene sets negatively correlated to pri-miR-21 expression were arranged that neighboring cases (columns) and neighboring gene sets (rows) are correlated as highly as possible. Data of 43 cases with low spatial information for the circular data representation were excluded. The largest gap between the cases and between the gene sets marks the beginning and the end of the ordered case and gene-set sequence, respectively. The heat map illustrates the expression of the eight putative miR-21 target genes belonging to the four identified gene sets. Light and dark gray indicate high and low expression, respectively.
U133A gene expression data of 114 B-cell lymphomas from Hummel et al. were used for statistical analysis (Hummel et al., 2006). On the basis of their expression signatures, 25 out of 114 cases have been classified as Burkitt's lymphomas. The other cases correspond to the group of DLBCL that can be molecularly subclassified into germinal center B cell-like and activated B cell-like DLBCL types, with some cases remaining unclassified (Rosenwald et al., 2002). Pri-miR-21 showed significant higher expression levels in the DLBCL subgroups than in Burkitt's lymphomas (Figure 2b). Assuming that the amplitude of pri-miR-21 transcription reflects the level of mature miR-21, mRNAs suppressed in the presence of high pri-miR-21 (and TMEM49) expression might represent putative miR-21 targets. To search for genes negatively correlated with the pri-miR-21 probe set, we applied a recently published multivariate procedure for the analysis of gene expression data (Lauter et al., 2009). This algorithm searches for gene sets that show a correlated expression within gene expression studies. Applying such correlated gene sets instead of single genes leads to increased power and stability of statistical analyses. After retrieving all correlated gene sets from the lymphoma expression data, we searched for gene sets exhibiting negative correlation to pri-miR-21. As a result, we obtained four gene sets comprising eight genes on a significance level of 0.01 (Table 2). Remarkably, one of the most frequently validated miR-21 targets, PDCD4, was among the identified genes supporting the reliability of the approach. As expected, opposite to the pri-miR-21 levels, putative miR-21 target genes are expressed higher in Burkitt's lymphomas than in DLBCL (Figure 2c).
Table 2. Putative targets of miR-21 identified by gene set analysis of gene expression data from B-cell lymphomas.
| Gene set | P-value | Gene number | Gene symbol | Gene name |
|---|---|---|---|---|
| A | 0.002 | 1 | PPP2R3B | Protein phosphatase 2 (formerly 2A), regulatory subunit B, β |
| B | 0.004 | 2 | SSBP2 | Single-stranded DNA-binding protein 2 |
| SMARCA4 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 | |||
| C | 0.004 | 2 | PDCD4 | Programmed cell death 4 (neoplastic transformation inhibitor) |
| NCRNA00081 | Non-protein coding RNA 81 | |||
| D | 0.004 | 4 | SMARCA4 | SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin, subfamily a, member 4 |
| DNMT1 | DNA (cytosine-5-)-methyltransferase 1 | |||
| METAP2 | Methionyl aminopeptidase 2 | |||
| TCF3 | Transcription factor 3 |
Abbreviation: miR-21, microRNA-21.
Among these candidates, DNMT1, METAP2 and TCF3 exhibit perfect 7-mer seed matches that include G:U pairing (Supplementary Figure 2b). Five out of eight candidates are involved in transcriptional regulation (SSBP2, SMARCA4, PDCD4, DNMT1 and TCF3). Furthermore, PPP2R3B, SSBP2, SMARCA4 and PDCD4 have been described in the context of tumor suppression, which remarkably agrees with the oncogenic function of miR-21. Tumor suppressor SMARCA4 (also known as BRG1) was found included in two different gene sets (Table 2).
Remarkably, when the four gene sets were used to cluster the cohort of 114 lymphoma cases, not only the Burkitt's lymphomas cases clustered into one group but also the activated B cell-like and germinal center B cell-like subgroups of DLBCL were almost perfectly separated (Figure 2c). Therefore, although the expression of pri-miR-21 appears comparable in activated B cell-like and germinal center B cell-like (see above), its molecular targets might differ between these two DLBCL subgroups. This suggests that the function of miR-21 might depend on the cellular context.
Tumor suppressors ANP32A and SMARCA4 are both downregulated by miR-21
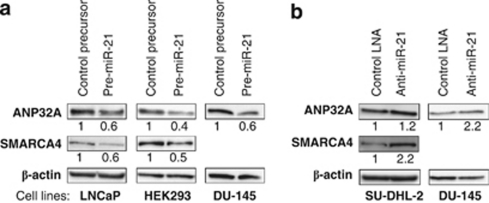
From the two approaches to identify miR-21 targets, ANP32A and SMARCA4 were selected for further confirmation on the basis of their common implication in cancerogenesis and chromatin remodeling. First, we studied their miR-21-mediated negative regulation by immunoblot analysis in cell lines of various origins. Depending on the endogenous miR-21 levels, being either low (LNCaP prostate carcinoma, HEK293 embryonic kidney cells) or high (DU-145 prostate carcinoma, SU-DHL-2 DLBCL), we used miR-21 precursors or anti-miR-21 to enhance or reduce the cellular miR-21 levels, respectively. For DU-145, both strategies were performed. Transfection of miR-21 precursors elevated the miR-21 level about 2.5-fold in DU-145, 10-fold in LNCaP and 20-fold in HEK293 cells. The reduction of the miR-21 levels by anti-miR-21-oligonucleotides was confirmed by a miR-21-sensitive reporter assay resulting in a twofold increase of the luciferase activity (data not shown). As displayed in Figure 3, overexpression (a) and knock down (b) of miR-21 revealed the corresponding vice versa effects on ANP32A protein expression in all four cell lines. The same was true for SMARCA4, with the exception that this protein was not detectably expressed in DU-145. Concluding from these results, ANP32A and SMARCA4 protein levels correlate inversely with miR-21 expression.
Figure 3.
ANP32A and SMARCA4 are regulated by miR-21 in various cell lines. Prostate cancer (LNCaP, DU-145), DLBCL (SU-DHL2), or embryonic kidney (HEK293) cells were transfected with miR-21 or control precursor (a) or with anti-miR-21 or control LNA (b). Cell lysates were prepared and immunoblot analysis of ANP32A and SMARCA4 was performed 72 h post transfection. Numbers indicate quantification relative to the corresponding β-actin loading control.
ANP32A and SMARCA4 are direct targets of miR-21
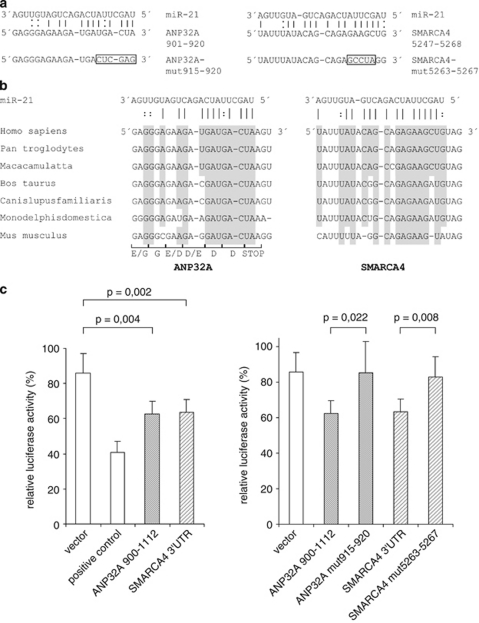
To test whether ANP32A and SMARCA4 are directly regulated by miR-21, we searched both mRNA sequences for predicted miR-21-binding sites using RNA22. By allowing mismatch pairing of the target mRNA to the miRNA seed sequence compensated by 3′ matches, a putative miR-21 complementary region in the ANP32A sequence (nt901–920 of NM_006305) and SMARCA4 (nt5247–5268 of NM_001128849) were found (Figure 4a). These target sites show a high interspecies homology in mammals (Figure 4b). For ANP32A, whose target site overlaps the open reading frame, this conservation includes the wobble bases putatively forming the miR-21–mRNA interaction. We constructed reporter genes containing a 213 bp sequence of ANP32A starting 22 nucleotides 5′ of the stop codon and the entire 3′ untranslated region of SMARCA4 downstream of the luciferase gene. A reporter construct containing three successive miR-21-binding sites was used as positive control. The reporter plasmids were transfected into LNCaP cells along with pre-miR-21 or a precursor control, and luciferase activity was determined. Enhanced expression of miR-21 significantly reduced reporter activity of both constructs to about 70% compared with the control (Figure 4c, left panel). We next mutated the reporter constructs by altering bases in the predicted miR-21 target sequences (Figure 4a). These mutations abolished the suppressive effect of pre-miR-21 on the luciferase activity of the ANP32A and SMARCA4 constructs (Figure 4c, right panel), verifying the functionality of the putative binding sites. Taken together, our results indicate that ANP32A and SMARCA4 are direct targets of miR-21.
Figure 4.
ANP32A and SMARCA4 are direct targets of miR-21. (a) Putative miR-21-binding sites within ANP32A (NM_006305) and SMARCA4 (NM_001128849) genes. Perfect matches are indicated by a line; G:U pairs by a colon. The frame marks nucleotides mutated for the reporter gene assays. (b) Comparison of nucleotides between the miR-21 sequence and its targets in various species. For the ANP32A site overlapping the open reading frame, amino acids are indicated. (c) Cloned ANP32A and SMARCA4sequences are targeted by miR-21 (left panel), and mutation of the putative binding sites abolishes this effect to wild type level (right panel). pMIR-REPORT vectors containing ANP32A-900-1112, SMARCA4-3′ untranslated region (3′UTR) and their mutated variants were co-transfected with miR-21 or control precursor into LNCaP cells. A vector containing three miR-21-binding sites was used as a positive control. Luciferase activity was assayed after 48 h. Data are normalized to the corresponding control sample. Values are the means±s.d. from six independent experiments. The P-values were determined using a two-tailed t-test.
ANP32A contributes to the transformation promoting effects of miR-21
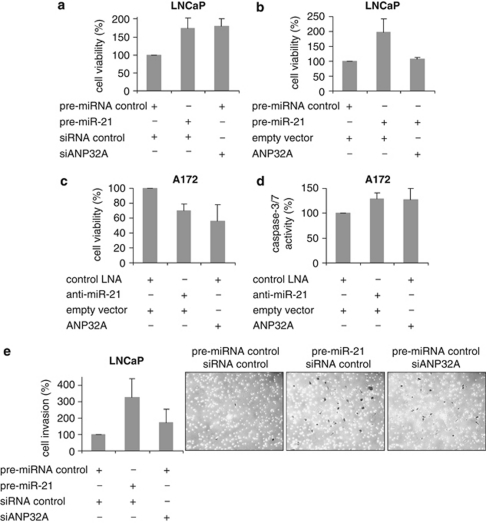
miR-21 has been demonstrated to support tumor cell growth and invasion. Therefore, we next investigated the proliferative effect of miR-21 in LNCaP prostate cancer cells. As determined after 5 days, enhanced miR-21 expression almost doubles the cell number compared with cells transfected with the control precursor (Figures 5a and b). To investigate the biological importance of ANP32A in the miR-21 target network, we studied whether ANP32A influences the proliferation of LNCaP cells. For this purpose, we performed a knock down of ANP32A protein (Supplementary Figure 3) and determined the cellular metabolic activity reflecting the number of viable cells. Lowering the ANP32A protein level by RNA interference raised the LNCaP cell number to the same level as obtained by enhanced miR-21 expression (Figure 5a). Conversely, a high ANP32A protein level abrogated the miR-21-mediated effect on cell viability (Figure 5b). Furthermore, we analyzed whether ANP32A could also contribute to the miR-21-induced effects on cells of different origins exhibiting high miR-21 levels. For the glioblastoma cell line A172 it has been shown that reduction of endogenous miR-21 level causes a significant drop in cell number (Chan et al., 2005). In our hands, anti-miR-21-oligonucleotides reduced cell viability to about 70% and increased the pro-apoptotic caspase activity to 130%. Enhanced ANP32A expression mimicked these miR-21-induced effects (Figures 5c and d). To study the effect of miR-21 on the migration potential of LNCAP cells, we used the Matrigel Invasion Assay. Cells were transfected with pre-miR-21 or ANP32A-specific small-interfering RNA and the corresponding controls. After 48 h, cells were seeded in the invasion chambers for 20 h. Afterwards, LNCaP cells that passed the matrigel were stained and counted. As shown in Figure 5e, enhanced miR-21 expression increased the number of migrated cells more than threefold. ANP32A knockdown by RNA interference also led to a significant higher cell invasion, but did not reach the miR-21-induced level. Taken together, these results suggest that ANP32A accounts for the various biological effects of miR-21 in different cell types.
Figure 5.
ANP32A contributes to the oncogenic potential of miR-21. MiR-21 precursor, ANP32A-specific small-interfering RNA (siRNA) and an ANP32A-expressing plasmid as well as their corresponding controls were introduced into LNCaP and A172 cells, as indicated. Afterwards, cell proliferation (a–c), caspase-3/7-activity (d), and Matrigel invasion (e) were assayed. For the latter, representative pictures of the invaded cells (black colored) on the assay membrane were taken. Bar graphs represent mean values±s.d. from three independent experiments.
With regard to SMARCA4, knockdown experiments in LNCaP and SUDHL-2 cells did not reveal an increase of cellular viability (data not shown).
Discussion
miRNAs have emerged as important post-transcriptional regulators of gene expression. By influencing the expression of multiple target genes simultaneously, miRNAs function as dimmer switches by fine-tuning the gene expression patterns and comprise the potential of complex changes in cellular physiology. Among them, miR-21 has been shown to be a key regulator of oncogenic processes (Selcuklu et al., 2009).
We applied two experimental-based strategies to identify novel miR-21 targets. With a proteomic approach, we identified 16 proteins either directly or indirectly suppressed by enhanced miR-21 expression in LNCaP prostate cancer cells. None of these novel candidates was found by target site prediction algorithms requiring the presence of a perfect miRNA seed match. This finding is consistent with the results obtained by Yang et al. on miR-21 targets in breast cancer cells, finding that less than 10% proteins identified by a proteomic approach were predicted by Pictar, Targetscan and miRanda (Yang et al., 2009). Nevertheless, five of our candidates possess a perfect 7-mer seed match when G:U pairing is allowed. Furthermore, with the exception of Cofilin-1 (no hit), all putative targets show between 1 and 3 hits out of ten different algorithms searching for miRNA target sites that are currently available by miRecords (mirecords.biolead.org) (Xiao et al., 2009). For comparison, among the 27 validated miR-21 targets listed in the miRecords database (december 2010), the number of hits by these ten algorithms ranges from 1 to 6. Furthermore, our candidate DDX3X has also been identified by Yang et al. (2009). These observations indicate that a majority of direct candidates likely possess imperfect seed base pairing supported by 3′ compensatory target sites (Brennecke et al., 2005), as shown for ANP32A and SMARCA4 in this study.
Applying a multivariate procedure for searching correlated gene sets in expression data from lymphoma patients, we obtained a rather short list of eight genes that show a significantly lower expression in the presence of a high pri-miR-21 level. This statistical strategy acts on the assumption that finding gene sets with correlated expression patterns increases the probability of a biological significant context. There have been two concerns in using probe set 220990_s_at for a miR-21 target gene search. The probe set does not only detect pri-miR-21 but also the overlapping gene TMEM49, and miR-21 levels might undergo a post-transcriptional regulation. Nevertheless, the presence of PDCD4, which was bioinformatically predicted and experimentally confirmed as a miR-21 target in several cell types (Asangani et al., 2008; Frankel et al., 2008; Lu et al., 2008; Zhu et al., 2008; Yao et al., 2009), strongly supports the reliability of our approach.
We did not observe an overlap of both candidate lists, which might result from the different nature of the underlying mechanisms (mRNA destabilization or translational inhibition) and from the different cell types analyzed. A cell-type-specific target gene knockdown would, for example, be the outcome of a synergistic action of two or more differentially expressed miRNAs. Moreover, the DIGE approach is limited to proteins of intermediate to high abundance and favours the display of proteins that separate apart from the bulk of other proteins. In particular, SMARCA4 could not be separated by DIGE because of its high molecular weight of about 200 kDa.
The direct regulation of ANP32A (from 2D-DIGE) and SMARCA4 (from gene expression analysis) was confirmed by reporter gene assays. Both proteins were found to be negatively correlated with miR-21 levels, for example, downregulated by miR-21 overexpression and upregulated by miR-21 inhibition in several cell lines. In addition to RNA22, miR-21 target sequences in ANP32A and SMARCA4 were also suggested by two other prediction programs offered by miRecords (miRanda, RNAhybrid for ANP32A, and PITA, RNAhybrid for SMARCA4). The putative miR-21 target sites show an explicit interspecies conservation and mediate a reduced reporter activity upon miR-21 overexpression. Moreover, they were functionally characterized by site-directed mutagenesis. Notably, the miR-21-binding element in ANP32A overlaps the stop codon.
ANP32A has been identified in various molecular contexts accounting for its numerous aliases: leucin-rich acidic nuclear protein (Matsuoka et al., 1994), putative HLA class II-associated protein I (Vaesen et al., 1994), inhibitor-1 of protein phophatase-2A (Li et al., 1995, 1996), phosphoprotein 32 (Chen et al., 1996) and Mapmodulin (Ulitzur et al., 1997). Hence, ANP32A is a multifunctional protein and involved in diverse biological events. Its tumor suppressive function is based on both cancer pathology analysis (Brody et al., 2007; Hoffarth et al., 2008) and functional studies demonstrating that overexpression of ANP32A inhibits oncogene-mediated transformation (Chen et al., 1996; Brody et al., 1999; Bai et al., 2001). Functions of ANP32A that might be related to its tumor suppressive activity are the inhibition of protein phosphatase 2A (Li et al., 1996) and histone acetyltransferases (Seo et al., 2001), as well as the stimulation of apoptosis by caspase activation (Pan et al., 2009). By being a part of the inhibitor of histone acetyltransferases complex, ANP32A is involved in modulating chromatin structure and transcriptional regulation (Seo et al., 2001). Interestingly, in addition to ANP32A, inhibitor of histone acetyltransferases contains isoforms A and B of the SET protein that belongs to the extended candidate list of putative miR-21 targets obtained by 2D-DIGE (Supplementary Table S1) and also possesses miR-21-binding sites as predicted by miRecords. Therefore, our results indicate that miR-21 suppresses the function of the inhibitor of histone acetyltransferases complex.
SMARCA4 is the catalytical component of the SWI/SNF chromatin-remodeling complex that regulates gene expression by disrupting histone–DNA contacts in an ATP-dependent manner (Khavari et al., 1993). It acts in association with many transcription factors by inducing an open state of the nucleosome (Reisman et al., 2009). There are some observations proposing SMARCA4 as a tumor suppressor. SMARCA4-heterozygous mice have an increased predisposition to tumor development (Bultman et al., 2000). Furthermore, the SMARCA4 gene is frequently silenced in cancer (Reisman et al., 2009; Rodriguez-Nieto and Sanchez-Cespedes, 2009) and its ectopic expression induces tumor cell growth arrest (Dunaief et al., 1994).
miR-21 promotes several transformation parameters in tumor cells derived from various origins (Meng et al., 2007; Frankel et al., 2008; Yao et al., 2009). In agreement with published data, we found that high miR-21 levels increase tumor cell viability and migration as well as inhibit pro-apoptotic caspase activity. We show that these effects can be mimicked by the respective manipulation of ANP32A expression. These data suggest that ANP32A is part of the network of miR-21 target genes that increase the transformational potential of cells. As for SMARCA4, the vast majority of experiments demonstrating its tumor suppressing activity were performed in cells lacking SMARCA4 expression (Strober et al., 1996; Wong et al., 2000). In agreement with our results, neither enhanced proliferation nor decreased apoptosis have been demonstrated upon application of SMARCA4-specific small-interfering RNA in the literature to date. In contrast, in normal mammary epithelial cells, SMARCA4 knockdown even inhibited cell proliferation (Cohet et al., 2010). Given the fundamental function of SMARCA4 in cellular transcription, the precise cellular context leading to its tumor suppressive effect remains to be elucidated.
Taken together, previous reports along with the identification of ANP32A and SMARCA4 as novel target genes indicate that miR-21 acts as key oncomiR by favouring neoplastic transformation on multiple levels. Understanding these gene networks could allow the development of new approaches for cancer diagnosis and therapy.
Materials and methods
Cell culture and reagents
Prostate carcinoma cell lines LNCaP and DU-145 (both from DSMZ, Braunschweig, Germany) and DLBCL cell line SUDHL-2 (from R Dalla-Favera, New York, NY, USA) were maintained in RPMI 1640. HEK293 and glioblastoma A172 cells were cultivated in Dulbecco′s modified Eagle′s medium. All media were supplemented with 10% fetal calf serum (FCS) and 100 U/ml of penicillin and streptomycin (Invitrogen, Karlsruhe, Germany). Antibodies against ANP32A (sc-5652) and SMARCA4 (sc-17796) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-β-actin (Clone AC-74) antibody was from Sigma (Taufkirchen, Germany). Precursor molecules for miR-21 and controls were purchased from Ambion (Applied Biosystems, Darmstadt, Germany), antisense and control oligonucleotides (miRCURY LNA knockdown) for miR-21 knockdown were from Exiqon (Vedbaek, Denmark). Validated small-interfering RNA against ANP32A and control (AllStars Negative Control siRNA) was taken from Qiagen (Hilden, Germany).
2D-DIGE and protein identification
The pre-miR-21 expressing vector (pSuper-miR21) has been described previously (Loffler et al., 2007). Empty control vector or pSuper-miR21 was introduced into LNCaP cells using Amaxa Nucleofector II Device with Kit R and program T-09, according to the manufacturer's protocol (Lonza, Cologne, Germany). Transfection efficiency over 80% was monitored by EGFP coexpression. After 24 h, cells were washed with phosphate buffered saline and 10 m Tris (pH 8.0) containing 5 m magnesium acetate. Subsequently, cells were harvested in cell lysis buffer (20 m Hepes pH 7.2, 10% glycerol, 1% Triton X-100, 1 m EDTA, 0.5% protease inhibitor cocktail (Sigma P8340), 1.25% Benzonase (Merck, Darmstadt, Germany)).
Labeling of the samples was performed following manufacturer's instructions (GE Healthcare, Buckinghamshire, UK). Briefly, cellular proteins were precipitated, resuspended in labeling buffer and aliquots stained by different fluorescent cyanine dyes. Samples were pooled, mixed with rehydration solution and applied to a rehydration tray. Separation in a two-dimensional gel and further processing was performed as described (Morbt et al., 2009). In short, immobilized pH gradient (IPG) strips were rehydrated overnight, after focusing equilibrated, and subsequently alkylated. Strips were then placed on acrylamide gels. Directly after the run, gels were scanned and dried between cellophane sheets. Images of the DIGE gels were quantitatively analyzed using Delta 2D software (Decodon GmbH, Greifswald, Germany). Differentially expressed proteins were identified using the following parameters: expression ratio lower than 0.75 or higher than 1.35 and a P-value of P<0.05, as obtained by the software's integrated Student's t-test. Proteins of interest were cut from dried gels and identified by mass spectrometry. Tryptic digestion was carried out with porcine trypsin as described by Santos et al. (2007). Identification of the extracted peptides was done by using a Bruker Ultraflex III (Bruker Daltonik, Bremen Daltonik, Bremen, Germany) or by reversed-phase nano-LC and a tandem mass spectrometry mass spectrometer (Jehmlich et al., 2008). Database searches were carried out using the MS/MS ion search (MASCOT, http://www.matrixscience.com) against all entries of the Swiss-Prot database (http://www.expasy.org). Proteins were specified as unambiguously identified, if the Mowse score was higher than 100 and at least two different peptides (P<0.05) were used for identification. Molecular weight and pI of the identified protein were cross-checked with the gel position of the excised spot.
Statistical analysis of gene expression
Statistical analysis was performed on a published U133A gene expression data set of 114 lymphoma cases (‘training data set') (Hummel et al., 2006). To avoid repeated gene nominations, the given 22277 probe sets were condensed to 9580 different genes. For the pairwise tests for differential expression of the pri-miR-21 probe set 220990_s_at, we used the Mann–Whitney U-test. To find genes negatively correlated with 220990_s_at, the procedure for searching essential sets of variables in high-dimensional data described by Lauter et al. (2009) was applied. For our particular application, the following program parameters were set: The sum of deviation squares of a gene must be greater than 35, yielding 1284 genes remaining in the analysis. Spearman rank correlation was used. Gene sets were constructed under the condition that the rank correlation between a source gene and a partner gene must be greater than √0.55≈0.74. Resampling was carried out with 500 random permutations of the data. The multiple level of significance was α=0.01. Negative, one-sided significance test was applied. The β-sums statistic of a gene set was calculated only from the three highest β-values. Data clustering and visualization was performed as described (Lauter et al., 2009).
RNA isolation and quantitative PCR
To confirm the transfection efficiency of the pre-miR-21-expressing plasmid or miR-21 precursor oligonucleotides, RNA was isolated and miR-21 expression levels were determined as described (Loffler et al., 2007).
Western blot analysis
DU-145 and HEK293 were transfected with 2 μ oligonucleotides using the MicroPorator MP-100, according to manufacturer's instructions (Invitrogen). Electroporation of SUDHL-2 and LNCaP was as above except that, for SUDHL-2 cells, program O-17 was used. Cells were harvested 72 h post transfection in RIPA buffer. Aliquots (30 μg) were separated on a 10% SDS–polyacrylamide gel electrophoresis and transferred to a Immobilon-P Transfer membrane (Millipore, Schwalbach, Germany). Immunodetection was performed using chemiluminescence (SuperSignal West Dura Extended Duration Substrate, Thermo Scientific, Rockfort, IL, USA).
Construction of expression vectors
For reporter gene assays, sequences of ANP32A and SMARCA4 containing the potential miR-21-binding sites were PCR-amplified from LNCaP total cDNA and cloned into the pMIR-Report vector (Ambion) using the forward primers 5′-TTTACTAGTGAGGGAGAAGATGATGAC-3′ and 5′-GCCCCGACATTCCAGTC-3′, and the reverse primers 5′-TTTAAGCTTTTTTATTCCACCCCCACC-3′ and 5′-GCGTTTTGTTGTTGGTTTAATT-3′, respectively. Mutations into the seed region of the putative miR-21-binding site of pMIR-Report-3′SMARCA4 were introduced using the QuikChange site-directed mutagenesis kit (Agilent Technologies, Böblingen, Germany) using primer 5′- CCATATTTATACAGCAGAGCCTAGGTAGGACTGTTTGTG-3′. Mutagenesis of ANP32A was performed by an inverse PCR approach with primers mut929-34_fwd 5′-GAGTTTTGAAAAATTCCTATTGTG-3′ and mut929-34_rev 5′-GAGATTCCACTTAGTCATCATCT-3′ as well as mut915-20_fwd 5′-GAGAGTGGAATAACCTATTTTGA-3′ and mut915-20_rev: 5′-GAGTCATCTTCTCCCTC-3′. Mutations are underlined.
To construct a vector expressing ANP32A, a DNA fragment covering the ANP32A coding region was amplified from LNCaP cDNA by PCR using the forward primer 5′-GCCGCCATGGAGATGGGCAGACGG-3′ and reverse primer 5′-TTAGTCATCATCTTCTCC-3′. After subcloning the PCR fragment in pCR-Blunt II-Topo (Invitrogen), sequence of ANP32A was released with XbaI/BamHI and inserted into pCDNA3.1(+) opened with NheI/BamHI.
miRNA target reporter assay
LNCaP cells were co-transfected with pMIR-Report vector containing the sequences of ANP32A or SMARCA4 or their mutated constructs, β-galactosidase vector for normalization (0.5 μg each) and miR-21 precursor or control (2 μ) using MicroPorator MP-100. The cells were lysed and reporter activity was measured 48 h post transfection using Luciferase Assay System (Promega, Mannheim, Germany) and chemiluminescent β-Gal Reporter Gene Assay (Roche Diagnostics, Mannheim, Germany).
Cell viability and caspase 3/7 assays
3000 LNCaP or 1000 A172 cells were seeded in 96-well plates and transfected the following day using Lipofectamine 2000 (Invitrogen) or FuGENE HD Transfection Reagent (Roche Diagnostics), respectively. Oligonucleotides were used with a final concentration of 100 n. For overexpression of ANP32A, 0.4 μg (LNCaP) or 0.2 μg (A172) plasmid were transfected per well. Five days post transfection, the CellTiter-Glo Luminescent Cell Viability Assay or Caspase-Glo 3/7 Assay (Promega) was performed.
Invasion assay
LNCaP cells were co-transfected with miR-21 precursor, siANP32A or control (2 μ) using MicroPorator MP-100. After 2 days of cultivation, cells were transferred into invasion chambers (BD Biosciences, Heidelberg, Germany) containing a matrigel-coated membrane of 8 μm pore size. 10% FCS as chemoattractant was added only to the lower compartment. After incubation for 20 h, the non-invaded cells were removed from the upper surface of the membrane by a cotton swab. The invaded cells were fixed using methanol, stained by Toluidine blue and counted per membrane.
Acknowledgments
We thank Riccardo Dalla-Favera (New York) for kindly providing SUDHL-2 cells. The work was partly supported by the DFG grant BR2892/3-1 and a junior research grant by the Medical Faculty, University of Leipzig.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
Supplementary Material
References
- Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai J, Brody JR, Kadkol SS, Pasternack GR. Tumor suppression and potentiation by manipulation of pp32 expression. Oncogene. 2001;20:2153–2160. doi: 10.1038/sj.onc.1204294. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. doi: 10.1371/journal.pbio.0030085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brody JR, Kadkol SS, Mahmoud MA, Rebel JM, Pasternack GR. Identification of sequences required for inhibition of oncogene-mediated transformation by pp32. J Biol Chem. 1999;274:20053–20055. doi: 10.1074/jbc.274.29.20053. [DOI] [PubMed] [Google Scholar]
- Brody JR, Witkiewicz A, Williams TK, Kadkol SS, Cozzitorto J, Durkan B, et al. Reduction of pp32 expression in poorly differentiated pancreatic ductal adenocarcinomas and intraductal papillary mucinous neoplasms with moderate dysplasia. Mod Pathol. 2007;20:1238–1244. doi: 10.1038/modpathol.3800974. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La MC, Nicholson J, Gilliam A, et al. A Brg1 null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- Chen TH, Brody JR, Romantsev FE, Yu JG, Kayler AE, Voneiff E, et al. Structure of pp32, an acidic nuclear protein which inhibits oncogene-induced formation of transformed foci. Mol Biol Cell. 1996;7:2045–2056. doi: 10.1091/mbc.7.12.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohet N, Stewart KM, Mudhasani R, Asirvatham AJ, Mallappa C, Imbalzano KM, et al. SWI/SNF chromatin remodeling enzyme ATPases promote cell proliferation in normal mammary epithelial cells. J Cell Physiol. 2010;223:667–678. doi: 10.1002/jcp.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaief JL, Strober BE, Guha S, Khavari PA, Alin K, Luban J, et al. The retinoblastoma protein and BRG1 form a complex and cooperate to induce cell cycle arrest. Cell. 1994;79:119–130. doi: 10.1016/0092-8674(94)90405-7. [DOI] [PubMed] [Google Scholar]
- Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2008;283:1026–1033. doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- Fujita S, Ito T, Mizutani T, Minoguchi S, Yamamichi N, Sakurai K, et al. miR-21 Gene expression triggered by AP-1 is sustained through a double-negative feedback mechanism. J Mol Biol. 2008;378:492–504. doi: 10.1016/j.jmb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Fulci V, Chiaretti S, Goldoni M, Azzalin G, Carucci N, Tavolaro S, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- Gabriely G, Wurdinger T, Kesari S, Esau CC, Burchard J, Linsley PS, et al. MicroRNA 21 promotes glioma invasion by targeting matrix metalloproteinase regulators. Mol Cell Biol. 2008;28:5369–5380. doi: 10.1128/MCB.00479-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Calin GA, Croce CM. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–179. doi: 10.1146/annurev.med.59.053006.104707. [DOI] [PubMed] [Google Scholar]
- Hoffarth S, Zitzer A, Wiewrodt R, Hahnel PS, Beyer V, Kreft A, et al. pp32/PHAPI determines the apoptosis response of non-small-cell lung cancer. Cell Death Differ. 2008;15:161–170. doi: 10.1038/sj.cdd.4402256. [DOI] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, et al. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- Huynh T, Miranda K, Tay Y, Ang Y-S, Tam W-L, Thomson AM, et al. A pattern-based method for the identification of microRNA-target sites and their corresponding RNA/RNA complexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- Jehmlich N, Schmidt F, von BM, Richnow HH, Vogt C. Protein-based stable isotope probing (Protein-SIP) reveals active species within anoxic mixed cultures. ISME J. 2008;2:1122–1133. doi: 10.1038/ismej.2008.64. [DOI] [PubMed] [Google Scholar]
- John B, Sander C, Marks DS. Prediction of human microRNA targets. Methods Mol Biol. 2006;342:101–113. doi: 10.1385/1-59745-123-1:101. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Kiriakidou M, Nelson PT, Kouranov A, Fitziev P, Bouyioukos C, Mourelatos Z, et al. A combined computational-experimental approach predicts human microRNA targets. Genes Dev. 2004;18:1165–1178. doi: 10.1101/gad.1184704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krichevsky AM, Gabriely G. miR-21: a small multi-faceted RNA. J Cell Mol Med. 2009;13:39–53. doi: 10.1111/j.1582-4934.2008.00556.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauter J, Horn F, Rosolowski M, Glimm E. High-dimensional data analysis: selection of variables, data compression and graphics—application to gene expression. Biom J. 2009;51:235–251. doi: 10.1002/bimj.200800207. [DOI] [PubMed] [Google Scholar]
- Lawrie CH, Soneji S, Marafioti T, Cooper CD, Palazzo S, Paterson JC, et al. MicroRNA expression distinguishes between germinal center B cell-like and activated B cell-like subtypes of diffuse large B cell lymphoma. Int J Cancer. 2007;121:1156–1161. doi: 10.1002/ijc.22800. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Li M, Guo H, Damuni Z. Purification and characterization of two potent heat-stable protein inhibitors of protein phosphatase 2A from bovine kidney. Biochemistry. 1995;34:1988–1996. doi: 10.1021/bi00006a020. [DOI] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- Li T, Li D, Sha J, Sun P, Huang Y. MicroRNA-21 directly targets MARCKS and promotes apoptosis resistance and invasion in prostate cancer cells. Biochem Biophys Res Commun. 2009;383:280–285. doi: 10.1016/j.bbrc.2009.03.077. [DOI] [PubMed] [Google Scholar]
- Loffler D, Brocke-Heidrich K, Pfeifer G, Stocsits C, Hackermuller J, Kretzschmar AK, et al. Interleukin-6 dependent survival of multiple myeloma cells involves the Stat3-mediated induction of microRNA-21 through a highly conserved enhancer. Blood. 2007;110:1330–1333. doi: 10.1182/blood-2007-03-081133. [DOI] [PubMed] [Google Scholar]
- Lu Z, Liu M, Stribinskis V, Klinge CM, Ramos KS, Colburn NH, et al. MicroRNA-21 promotes cell transformation by targeting the programmed cell death 4 gene. Oncogene. 2008;27:4373–4379. doi: 10.1038/onc.2008.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Schouest K, Kovvuru P, Spillane C. Prediction and validation of microRNA targets in animal genomes. J Biosci. 2007;32:1049–1052. doi: 10.1007/s12038-007-0106-0. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Taoka M, Satozawa N, Nakayama H, Ichimura T, Takahashi N, et al. A nuclear factor containing the leucine-rich repeats expressed in murine cerebellar neurons. Proc Natl Acad Sci USA. 1994;91:9670–9674. doi: 10.1073/pnas.91.21.9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Henson R, Lang M, Wehbe H, Maheshwari S, Mendell JT, et al. Involvement of human micro-RNA in growth and response to chemotherapy in human cholangiocarcinoma cell lines. Gastroenterology. 2006;130:2113–2129. doi: 10.1053/j.gastro.2006.02.057. [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morbt N, Mogel I, Kalkhof S, Feltens R, Roder-Stolinski C, Zheng J, et al. Proteome changes in human bronchoalveolar cells following styrene exposure indicate involvement of oxidative stress in the molecular-response mechanism. Proteomics. 2009;9:4920–4933. doi: 10.1002/pmic.200800836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro A, Gaya A, Martinez A, Urbano-Ispizua A, Pons A, Balague O, et al. MicroRNA expression profiling in classic Hodgkin lymphoma. Blood. 2008;111:2825–2832. doi: 10.1182/blood-2007-06-096784. [DOI] [PubMed] [Google Scholar]
- Pan W, da Graca LS, Shao Y, Yin Q, Wu H, Jiang X. PHAPI/pp32 suppresses tumorigenesis by stimulating apoptosis. J Biol Chem. 2009;284:6946–6954. doi: 10.1074/jbc.M805801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Shapiro A, Kosik KS. MicroRNA-21 targets a network of key tumor-suppressive pathways in glioblastoma cells. Cancer Res. 2008;68:8164–8172. doi: 10.1158/0008-5472.CAN-08-1305. [DOI] [PubMed] [Google Scholar]
- Reisman D, Glaros S, Thompson EA. The SWI/SNF complex and cancer. Oncogene. 2009;28:1653–1668. doi: 10.1038/onc.2009.4. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Nieto S, Sanchez-Cespedes M. BRG1 and LKB1: tales of two tumor suppressor genes on chromosome 19p and lung cancer. Carcinogenesis. 2009;30:547–554. doi: 10.1093/carcin/bgp035. [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- Santos PM, Roma V, Benndorf D, von BM, Harms H, Sa-Correia I. Mechanistic insights into the global response to phenol in the phenol-biodegrading strain Pseudomonas sp. M1 revealed by quantitative proteomics. OMICS. 2007;11:233–251. doi: 10.1089/omi.2007.0009. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Selcuklu SD, Donoghue MT, Spillane C. miR-21 as a key regulator of oncogenic processes. Biochem Soc Trans. 2009;37:918–925. doi: 10.1042/BST0370918. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Strober BE, Dunaief JL, Guha S, Goff SP. Functional interactions between the hBRM/hBRG1 transcriptional activators and the pRB family of proteins. Mol Cell Biol. 1996;16:1576–1583. doi: 10.1128/mcb.16.4.1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitzur N, Humbert M, Pfeffer SR. Mapmodulin: a possible modulator of the interaction of microtubule-associated proteins with microtubules. Proc Natl Acad Sci USA. 1997;94:5084–5089. doi: 10.1073/pnas.94.10.5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaesen M, Barnikol-Watanabe S, Gotz H, Awni LA, Cole T, Zimmermann B, et al. Purification and characterization of two putative HLA class II associated proteins: PHAPI and PHAPII. Biol Chem Hoppe Seyler. 1994;375:113–126. doi: 10.1515/bchm3.1994.375.2.113. [DOI] [PubMed] [Google Scholar]
- Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Shanahan F, Chen Y, Lian L, Ha P, Hendricks K, et al. BRG1, a component of the SWI-SNF complex, is mutated in multiple human tumor cell lines. Cancer Res. 2000;60:6171–6177. [PubMed] [Google Scholar]
- Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chaerkady R, Beer MA, Mendell JT, Pandey A. Identification of miR-21 targets in breast cancer cells using a quantitative proteomic approach. Proteomics. 2009;9:1374–1384. doi: 10.1002/pmic.200800551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Chaerkady R, Kandasamy K, Huang TC, Selvan LD, Dwivedi SB, et al. Identifying targets of miR-143 using a SILAC-based proteomic approach. Mol Biosyst. 2010;6:1873–1882. doi: 10.1039/c004401f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Q, Xu H, Zhang QQ, Zhou H, Qu LH. MicroRNA-21 promotes cell proliferation and down-regulates the expression of programmed cell death 4 (PDCD4) in HeLa cervical carcinoma cells. Biochem Biophys Res Commun. 2009;388:539–542. doi: 10.1016/j.bbrc.2009.08.044. [DOI] [PubMed] [Google Scholar]
- Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.