Abstract
Cytomegalovirus (CMV) and the human tumor cell share the same objectives: escape the recognition and destruction by the immune system and establish a state of immune tolerance conducive for their development. For early tumor development, the escape of the first lines of defense of the immune surveillance is a critical step which determines survival or destruction. The presence of CMV on the tumor site and its involvement in carcinogenesis as initiator or promoter is increasingly documented. In this article, we highlight the similarity between mechanisms used by tumors and CMV to circumvent the immune defenses and evade from immune surveillance. We suggest that CMV and tumors help one another for their common objective. CMV gets shelter in immunologically poor environment of the tumor cells. In return CMV, by acting directly on the cancer cell and/or on the tumor microenvironment, provides the tumor cell the ways to promote its immune escape and development of immune tolerance.
Keywords: HCMV, tumor, immune escape.
INTRODUCTION
The theory of cancer immunosurveillance proposes that the immune system can detect and eliminate the developing tumors. There were a number of arguments in favor of the immune system’s efficiency to struggle against cancers such as the increased incidence of cancers in immunosuppressed patients, the correlation between intratumoral infiltrating T cells and tumor survival or the discovery of tumor associated antigens recognized by immune cells. Many studies have highlighted the involvement of both innate immunity and adaptive immunity to fight against tumor cells. However, cancer cells could escape recognition and destruction by the immune system and may continue to grow [1, 2]. Thus the formation of a tumor occurs in 3 phases: an initial elimination phase which coincides with the theory of immunosurveillance in which the immune system will recognize and destroy cancer cells. A second equilibrium phase exists between the development of new cancer cells and their destruction. During this phase tumor cells accumulate mutations under the pressure exerted by the immune system that will allow the emergence of less immunogenic variants which will be tolerated by the immune system. The final escape phase during which tumor cell variants, with a phenotype edited by the immune pressure, are able to evade the innate and adaptive defenses and to develop into clinically apparent cancer [1]. At this stage, the inflammatory response generated by immune cells including the release of cytokines, growth factors and pro-angiogenic factors to promote tissue reconstruction and the intake of nutrients, will be diverted by the tumor to ensure its own growth [2].
The human cytomegalovirus (HCMV) is a multifaceted betaherpesvirus that is regarded as asymptomatic, mildly pathogenic virus in immunocompetent host. However it may cause serious in utero infections as well as acute and chronic complications in immunocompromised host [3]. The involvement of HCMV in late inflammatory complications underscores its possible role in inflammatory diseases and cancer. Evidence of this involvement of HCMV in such phenomena is being accumulated (review in: [4-6]). Early in vitro studies suggested that HCMV was able to transform embryonic fibroblasts in culture and to induce chromosomal damages and mutations but HCMV has never been accepted as an oncogenic virus [6]. Later on the concept of "oncomodulation" was proposed to explain the possible contribution of HCMV in tumor progression [7]. The oncomodulation states that HCMV infects the tumor tissue and acts as a cofactor in amplifying mechanisms of carcinogenesis without necessarily initiating tumor. Support for this idea is based on experiments showing that proteins of HCMV (or non-coding RNAs) could influence the genesis and tumor growth acting on the cell cycle, apoptosis, genetic instability, invasiveness, angiogenesis, adhesion and cell migration. These proteins have been the subject of extensive recent reviews [6, 8]. The increased sensitivity of detection of HCMV in tumor tissues (immunohistochemistry, in situ hybridization and polymerase chain reaction (PCR) techniques) originally proposed by Cobbs et al. in 2002 served to highlight the presence of HCMV proteins and DNA in tumor cells but not in adjacent cells of several cancers such as glioma [9], colon cancer [10], prostate cancer [11], and some skin cancers [12]. Interestingly, the presence of HCMV was also highlighted in the pre-cancerous lesions such as colorectal polyps [10], and prostatic intraepithelial neoplasia [11]. The involvement of (virus-induced) inflammation in the initiation of cancer has emerged in the recent years [13, 14]. Chronic HCMV infection triggers biological responses observed in cancer with both stimulation of inflammation and immunosuppression [15]. Indeed, HCMV persistently infects monocytes/ macrophages and induces an unique inflammatory (M1) and immunosuppressive (M2) polarization of macrophages [16, 17]. This atypical M1/M2 phenotype of macrophages is associated with the release of cytokines involved in cancer initiation or promotion such as IL-6, TNFα, and IL-10 [2, 14]. Moreover, HCMV infection activates transcription factors which play a main role in inflammation and carcinogenesis such as NF-κB and the signal transducer and activator of transcription 3 (STAT3) [18, 19].
Despite increasing evidence of the presence of HCMV in tumors and its participation in carcinogenesis [4-6], its involvement in the crucial stage of the tumor immune escape has never really been studied. HCMV is a latent herpesvirus that maintains dynamic relationships with the immune system and we propose that it has the potential to help the tumor cell to evade the first line of host defense and induce a state of immune tolerance in which it can grow. We will discuss side-by-side the means used by HCMV and the tumor cell to escape recognition and destruction by the immune system and to generate a state of immune tolerance (Table 1). Using similar strategies, the HCMV recruited at the inflammatory site generated by the early tumor development and also by infecting the tumor cell is able to offer additional means to escape the immune system and promote its development. Thus we propose a novel involvement of HCMV in carcinogenesis, as a critical cofactor in the final escape phase from immune surveillance resulting in progression to cancer.
Table 1.
Strategies Used by Tumors and CMV for Immune Escape
| Strategy | Tumors | Ref. | CMV | Ref. |
|---|---|---|---|---|
| Impairment in Antigen Processing and Presentation | ||||
| Impairment in surface expression of MHC-I | MHC-I gene mutations, inhibition of MHC-coding genes expression, impairment in antigen binding or peptide transport from the endoplasmic reticulum to the cell surface | [23] | Directly acting on MHC-I molecules or by acting on MHC-I associated proteins, including TAP and tapasin | [25, 27] |
| Escape from Natural Killer Cells | ||||
| NK activating receptors | Immune surveillance of cancer through NKG2D/ NKG2DLs pathway | [32] | Retention of ligands of NKG2D (MICB, ULBP1 and 2) in ER by gpUL16 | [54, 55] |
| Regulation of MICB by cellular miRNA | [64] | Down regulation of MICB by HCMV miR-UL112 | [57] | |
| Intercellular retention of MICA inhibits NK cytotoxicity | [42] | Inhibition and intracellular retention of MICA by UL142 | [56, 59] | |
| Immune surveillance through Natural Cytotoxicity Receptors NKp30, NKp44, NKp46, and NKp80 | [32, 33] | Reduced NKp30-mediated killing by pp65 | [63] | |
| Anti-tumor response by DNAM-1 | [36, 37] | Down-regulation of CD155 expression, a ligand for DNAM-1, by gpUL141 | [60] | |
| NK Inhibitory receptors | Blockade of the inhibitory receptors LIR-1 and NKG2A results in increased NK cell cytotoxicity | [44] | gpUL18 binds with LIR-1 and inhibits LIR-1 + NK cell activity | [47, 48] |
| Expression of “non classical” HLA-E induces a decrease in NK responses through interaction with CD94/NKG2A | [24] | Over expression of HLA-E by gpUL40 | [52, 53] | |
| Blockade of Death Receptors-Mediated Apoptosis | ||||
| Over-expression of antiapoptotic proteins | Over-expression of FLIPL,S which act as caspase 8 inhibitors | [67] | Expression of FLIP by immediate early 2 (IE2) protein | [84] |
| Over-expression of the anti-apoptotic protein Bcl-2 | [70] | Over-expression of Bcl-2 in HCMV infected cells | [10, 85] | |
| Over-expression of other antiapoptotic Bcl-2 family members such as Bcl-x or Mcl-1 | [74, 75] | |||
| Inhibition of pro-apoptotic molecules and death receptors | Decreased expression of Fas Mutations or deletions in genes encoding Fas and TRAIL-R1-R2 |
[76] [77] |
Inhibition of recruitment of pro-caspase 8 to the Death-inducing Signaling Complex (DISC) by product of HCMV UL36 gene | [87] |
| Inhibition of proapoptotic Bcl-2 family members Bax and Bak by product of the HCMV UL37 gene | [86] | |||
| Escape from Complement Attack | ||||
| Complement Regulatory Proteins (CRPs) | Expression of CRPs (CD46, CD55, CD59, and CD35) was noticed in a wide range of cancer cells | [91-93] | HCMV upregulates expression of host-encoded CRPs resulting in protection from complement-dependent lysis | [96, 97] |
| HCMV incorporates host cell-derived CRPs, CD55 and CD59 into its virions | [98] | |||
| Immune Tolerance Establishment | ||||
| Expression of interleukin-10 | Increased release of IL-10 in cancers, conferring a more invasive phenotype | [102-108] | HCMV encodes an IL-10 homolog (cmv-IL-10) that shares human IL-10 immunomodulatory properties | [114- 118] |
| Expression of TGFβ | Overexpression of TGFβ promoted tumor-immune escape and was associated with tumor progression with worse prognosis. | [128-131, 134-136] | HCMV induced transcription and release of TGFβ | [138-140] |
HCMV HELPS TUMORS FOR ESCAPE FROM RECOGNITION AND DESTRUCTION BY THE IMMUNE SYSTEM
Impairment in Antigen Processing and Presentation
The trigger of cellular adaptive immune response depends on antigen processing and presentation to T cells via the major histocompatibility complex class I (MHC-I) and the MHC class II (MHC-II) molecules. Antigen-presenting cells (APCs) present the previously phagocytosed antigens to CD4+ T Cells through MHC-II molecules, which may lead to trigger either an adaptive response or an immune tolerance towards this antigen. MHC-II is formed inside the endoplasmic reticulum (ER) by the assembly of α and β chains associated with a chaperone protein, the invariant chain (Ii), which stabilizes the MHC-αβ complex and guides its migration through the Golgi apparatus until the cell surface. Before reaching the surface, Ii chain is degraded, enabling the MHC complex to bind to the antigenic peptide and to present it to CD4+ T Cells. MHC-I is expressed by every nucleated cell and plays an important role in presenting cytosolic antigenic peptides to CD8+ T Cells. The MHC-I-peptide complex is formed within the endoplasmic reticulum in which the antigenic peptides are translocated by the Transporter Associated with Antigen Processing (TAP).
The effectiveness of the adaptive immune response against tumors depends in part on the ability of tumor cells to present tumors associated antigens to T cells through MHC molecules [20]. To escape from the recognition of its antigens by T Cells, tumor cell has been reported to down regulate both antigen processing and presentation machineries [21, 22]. Moreover, antigenic variations are provided by continuous mutations occurring in the cancer cell. Thereby, even in the presence of an adaptive immune response specifically directed against some of its antigens, tumors generate mutant variants not recognized by the immune system. Repeats of these antigenic mutations play an important role in the formation of the tumor antigenic profile [22]. Importantly, impairment in surface expression of MHC-I peptide has been noticed in many types of cancers and is one of the best-studied mechanisms used by tumor cells to escape from immune recognition [23]. Several mechanisms involved in the alteration of MHC-I expression have been described, including MHC-I gene mutations, inhibition of MHC-coding genes expression, impairment in antigen binding or peptide transport from the endoplasmic reticulum to the cell surface [23, 24].
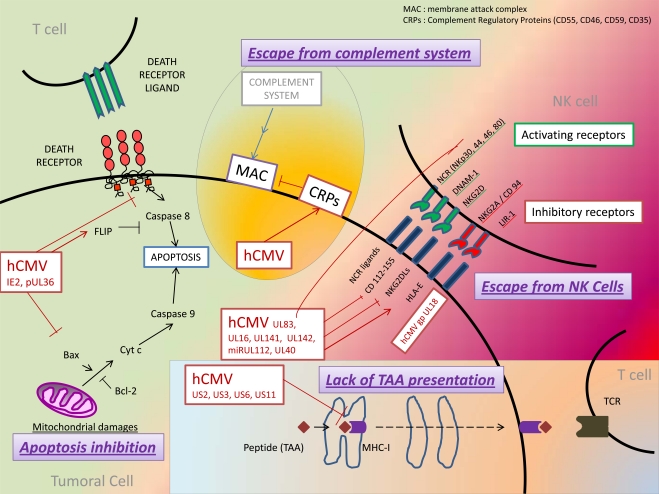
HCMV has developed immune escape strategies by interfering with its antigens processing and presentation through MHC [25, 26] (Fig. 1). Thus, latent HCMV infection of tumor cells might promote the ability of tumor cells to evade immune response. At least four proteins encoded by the Unique Short (US) region of HCMV genome have been involved in the inhibition of MHC-I expression, either by directly acting on MHC-I molecules or by acting on MHC-I associated proteins, including TAP and tapasin which have both chaperone-like and catalytic functions on MHC-I molecules (review in [25, 27]): the protein encoded by US3 gene of HCMV (pUS3) binds to and inhibits tapasin, leading to retention of MHC-I molecules within the ER, whereas proteins pUS2 and pUS11 bind to MHC-I molecules and promote their reverse transport from the ER to the cytosol where they are degraded. Moreover, pUS6 inhibits TAP complex thereby inhibiting peptide translocation from the cytosol to the ER. In addition, at least three proteins encoded by HCMV inhibit the expression of MHC-II [26] : pUS2 protein acts far upstream by specifically binding to the HLA-DR α-chain of MHC-II, leading to its degradation by the proteasome, whereas pUS3 protein affects the MHC-II α β complex by competing with the Ii chain and retaining it in the Golgi. At last, pp65 protein encoded by the gene UL83 (Unique Long 83) acts downstream by mediating an accumulation of MHC-II molecules in perinuclear lysosomes, resulting in degradation of the HLA-DR α-chain. An additional mechanism for MHC-II inhibition is the synthesis of an HCMV interleukin-10 homolog (cmvIL-10). Human IL-10 has been described to inhibit expression of MHC-II to the cell surface [28]. A similar inhibition of MHC-II expression was noticed in peripheral blood mononuclear cells (PBMC) and monocytes treated with cmvIL-10 [29].
Fig. (1).
Model for immune escape in tumors and CMV infection.
Blockade of Cytotoxic Activity from Immune Effectors
Escape from Natural Killer Cells
Natural killer (NK) cells are essential effectors of innate immunity, with both cytotoxicity and cytokine-producing functions [30]. Regulation of NK depends on various stimulatory and inhibitory receptors which respond to the expression of self-molecules such as MHC-I molecules, stress-induced ligands or non-self ligands. Indeed, cells that fail to express MHC-I molecules such as virus-infected or tumor cells, are recognized and eliminated by NK cells, according to the "missing self" hypothesis [31]. Conversely, healthy self-cells which express MHC-I molecules stimulate NK inhibitory receptors and are self-tolerated by the immune system [30].
Some of NK receptors have been particularly involved in the immune surveillance of cancers [32], including four stimulatory receptors which constitute the group of Natural Cytotoxicity Receptor (NCR): NKp30, NKp44, NKp46, NKp80 ; receptors DNAM-1 and NKG2D (Fig. 1). In vitro studies have shown that blockade of some receptors among NKp30, NKp44, NKp46 or NKp80 with monoclonal antibodies inhibited NK cells cytotoxicity against tumor cells [33, 34]. DNAM-1 (also called CD226), an adhesion molecule whose ligands include CD112 and CD155, also seems to play an important antitumor role [35-37]. Its ligands are frequently expressed by tumor cells, causing their lysis in vitro by NK cells. Moreover, DNAM-1-deficient mice were noticed to have an impaired antitumor response and an accelerated tumor growth [38, 39]. NKG2D receptors were extensively studied and found to be expressed by various cells such as NK cells, CD8+ T cells, γδ-T cells and NKT cells [40]. Several NKG2D ligands (NKG2DLs) have been characterized including MICA (MHC class I polypeptide-related sequence A), MICB, ULBP1 (cytomegalovirus UL16-Binding Protein 1) and ULBP 2. These ligands present structural homology with MHC-I molecules but are typically not expressed by healthy-cells. Conversely, expression of NKG2DLs was upregulated in stressed cells with DNA damages [41]. Numerous studies have highlighted the major role played by the NKG2D/NKG2DLs pathway in tumor immune clearance (review in [32]). Conversely, altered expression of NKG2DLs by tumor cell variants conferred a selective advantage in tumor-immune escape [42]. Indeed, in a recent study, tumor cells expressed higher amounts of NKG2DLs in NKG2D-deficient than in wild-type mice, suggesting a selection of tumor cells with weak expression of NKGDLs due to immune pressure mediated by NK cells [43]. To escape from NK-mediated lysis, certain tumor cells with a decrease in “classical” HLA type-I expression (HLA-A, B and C) were demonstrated to express “non classical” HLA type-I molecules including HLA-G and HLA-E. Expression of HLA-E induced a decrease in NK antitumor responses by interacting with the CD94/NKG2A inhibitory receptors [24]. Blockade of the inhibitory receptors LIR-1 and NKG2A also consistently resulted in increased NK cell cytotoxicity against leukemia cells [44].
Because HCMV down-regulates the expression of MHC-I molecules, HCMV-infected cells should be more vulnerable to Natural Killer cell-mediated cytolysis. However, clinical studies in patients with defects in NK functions as well as experimental studies have highlighted that HCMV has developed various mechanisms to evade the recognition and destruction mediated by NK cells [45]. HCMV encodes proteins such as gpUL18 and gpUL40 that impair NK cells responses by stimulating their inhibitory receptors. Glycoprotein gpUL18, which has a highly structural resemblance to MHC-I molecules, forms a trimeric complex by associating with β2-microglobulin and cellular endogenous peptides. For this reason, gpUL18 was the first described viral-MHC homologue with the ability to bind and present peptides [46-48]. gpUL18 was shown to bind with high affinity to the inhibitory receptors LIR-1 expressed by NK cells, monocytes, dendritic cells, B cells and subsets of T cells [48, 49]. Despite initial controversial studies, gpUL18 was recently shown to inhibit LIR-1 + NK cell activity [48, 50, 51]. Moreover, expression of “non classical” MHC-I molecules HLA-E is up-regulated by HCMV- encoded protein gpUL40. This HLA-E over-expression contributes to evade NK-mediated lysis by binding to the NK inhibitory receptors CD94/NKG2A [52, 53].
Several HCMV-encoded proteins confer protection from NK lysis by inhibiting NK activating receptors. This immune modulation was described for HCMV-encoded proteins gpUL16, gpUL141, gpUL142, and pp65. Among these proteins, effects of gpUL16 were the most studied. gpUL16 binds to and retains in the ER several ligands of NK activating receptor NKG2D such as MHC I-related molecules MICB and UL16-binding protein family ULBP1 and 2 [54, 55]. This decreased expression of NKGDLs due to sequestration in the ER reduces NK cell recognition of HCMV-infected cells. As mentioned above, NKG2D receptors have been described in other immune cells including dendritic cells and subsets of T cells. The influence of gpUL16 on NKG2D receptors in these cells remains to be assessed. In addition, expression of MICA and MICB was respectively inhibited by the HCMV-encoded MHC-I-related protein gpUL142 and HCMV-microRNA miR-UL112 [56-59]. Moreover, the UL141 gene product, gpUL141, down-regulated the expression of CD155, a ligand for NK cell activating receptors CD226 (DNAM-1) and CD96 (TACTILE) [60]. At last, pp65 (ppUL83), the main tegument protein of HCMV, was shown to impair the activation of NK cells. Relationships between pp65 and the immune system are complex because pp65 is both a major antigenic target for the immune system and an inhibitor of antiviral gene expression in infected cells [61, 62]. pp65 was described to act as a ligand for the NK cell activating receptor NKp30. However, this interaction between pp65 and NKp30 did not activate NK cells but instead dissociated the linked CD3ζ chains from NKp30, leading to a decreased activation signal and a reduced NKp30-mediated killing. Thus, the interaction between pp65 and NKp30 forms an original model by which a viral protein inhibits NK cell-mediated lysis by specifically binding to an activating receptor [63]. On one hand HCMV miRNA-UL112 downregulated MICB, resulting in reduced NK cell mediate lysis [57], while on the other hand several cellular miRNAs have been reported to be upregulated in certain type of cancers and have the ability to regulate MICB [64]. A recent report demonstrates synergistic activity of miR-UL112 with a cellular miRNA to promote immune evasion during HCMV infection [65] but possible synergisms between cellular and HCMV miRNAs for immune evasion of tumor cell need to be further tested.
Blockade of Death Receptor-Mediated Apoptosis in Tumor Cells
Stimulation of the cell membrane death receptor Fas by its ligand FasL results in recruitment of the adapter protein FADD and activation of caspase 8, which unleashes a proteolytic cascade leading to cell death by apoptosis [66]. FasL is expressed by activated Cytotoxic T Lymphocytes (CTL) that, upon activation, specifically recognize a virus-infected cell or a tumor cell over-expressing Fas. Activation of Fas-FasL pathway enables CTL to exert its cytotoxicity. Moreover, apoptosis triggered by Fas-FasL is involved in removal of activated lymphocytes which became unnecessary and might contribute to autoreactivity. Thereby, both Fas and FasL may be expressed by CTL resulting either in their cytotoxicity or in their own apoptosis.
Tumor cell seems to use this dual property of Fas-FasL-mediated apoptosis in lymphocytes (target-cells destruction and stimulation of their own apoptosis) to its advantage. The first strategy used by tumor cells to escape from apoptosis induced by immune effector cells is the over-expression of antiapoptotic proteins including proteins FLIP which act as caspase 8 inhibitors [67] (Fig. 1). Over-expression of FLIP by tumor cells has been described in several cancers such as melanoma [68] and EBV-induced lymphomas [69]. Similarly, over-expression of the anti-apoptotic protein Bcl-2 has been involved in the resistance to both extrinsic and intrinsic apoptosis by tumor cells [70]. Some studies have shown a correlation between over-expression of Bcl-2 in lymphoblastic or leukemic cells and prognosis of survival or response to chemotherapy [71-73]. Moreover, similar results were obtained by studying the expression of other anti-apoptotic members of Bcl-2 family such as Bcl-x or Mcl-1 by tumor cells [74, 75]. A second mechanism used by tumor cells to resist to apoptosis is the inhibition of pro-apoptotic molecules and death receptors. Decreased expression of Fas was shown in tumor cells from hepatocellular carcinoma, colon cancer or melanoma [76]. Moreover, mutations or deletions in genes encoding Fas and TRAIL-R1-R2 were noticed in some cancer cells and involved in some familial forms of cancers [76, 77]. Many studies have suggested that tumor cells could escape from the immune system by expressing FasL and inducing apoptosis in activated Fas+ CTL. This phenomena has been referred to as a "counterattack" from tumor cells to CTL [76, 78, 79]. However, this attractive hypothesis was controversial in other studies which did not highlight FasL expression by tumor cells [80]. In fact, environmental factors and FasL expression levels seem to play an important role to determine which of the CTL or the tumor cell "will kill the other one." Indeed, in the presence of a pro-inflammatory environment, an infiltrate of neutrophils or a high level of FasL expression, CTL would kill tumor cells through the Fas-FasL pathway. Conversely, in the presence of immunosuppressive cytokines such as transforming growth factor-β (TGF-β) or low levels of FasL, CTL express Fas (as observed during last steps of acute inflammatory reactions) and would be the targets of apoptosis triggered by FasL expressed or secreted in a soluble form by the tumor cells [22, 81, 82].
Signaling pathways mediated by tumor necrosis factor (TNF) superfamily have various roles in apoptosis or immune response that impair the survival of latent viruses such as herpesviruses. Thereby, many viruses have developed strategies to escape from the immune system and from the apoptosis of infected cells. Indeed, herpesviruses, including HCMV, inhibit TNF receptors signaling pathways in different points (recent review in [83]). At first, HCMV has been shown to stimulate anti-apoptotic factors which downregulate death receptor-mediated pathways (Fig. 1). Thus, HCMV immediate early 2 (IE2) proteins have been described to activate the expression of FLIP which inhibits death receptor-mediated apoptosis [84]. Similarly, over-expression of Bcl-2 and chemotherapy resistance was reported in cultured cells of neuroblastoma or colon cancer infected by HCMV [10, 85]. Moreover, besides pro-apoptotic factors modulated by HCMV such as p53 [6], HCMV inhibits the recruitment, the activation or the expression of pro-apoptotic actors of death-receptor-mediated pathways. Indeed, the protein encoded by HCMV UL36 gene binds to the pro-caspase 8 and prevents its recruitment to the Death-inducing Signaling Complex (DISC), whereas a product of the HCMV UL37 gene (pUL37x1) inhibits pro-apoptotic Bcl-2 family members Bax and Bak leading to the blockade of mitochondrial membrane permeabilization and release of cytochrome c [86, 87]. As described for tumor cells, HCMV-infected cells were suggested to evade from the immune system by triggering the apoptosis of CTL recruited on infectious site. In favor of this hypothesis, the HCMV Immediate Early protein IE2 was shown to enhance the expression (or secretion) of FasL in HCMV-infected human retinal cells leading to Fas-dependent apoptosis of T lymphocytes [88]. Thus, as suggested with the "attack and counterattack hypothesis" for tumor cells, HCMV-infected cells might have the dual properties of blocking their own death and stimulating Fas-dependent apoptosis of effective T cells.
Escape from Complement Attack
The complement system represents an important effector of the innate defense against pathogens. Activation of complement leads to opsonization and lysis of pathogens and promote an inflammatory response through the production of anaphylatoxins [89]. Through this dual involvement in innate immunity and inflammation, the action of complement might be both favorable and unfavorable towards carcinogenesis [90]. Although the presence and the activation of complement proteins have been noticed in many cancers, their contribution in tumor destruction seems weak. Indeed, tumors cells might escape to complement attack by using various protective mechanisms such as over-expression of membrane-bound Complement Regulatory Proteins (CRPs) and secretion of soluble complement inhibitors [90] (Fig. 1). Expression of CRPs (CD46, CD55, CD59, and CD35) was noticed in a wide range of cancer cells and cell lines including renal tumor cell lines and proximal tubular epithelial cells [91], endometrial malignant cells [92], and colorectal cancers [93]. Furthermore, membrane-associated CRPs on tumor cells were shown to decrease the efficacy of anti-cancer therapy using monoclonal antibodies by impairing both complement-dependent cytotoxicity (CDC) and antibody-dependent cell-mediated cytotoxicity (ADCC). Different models were proposed to enhance monoclonal antibodies treatments by blocking CRPs [90, 94].
To protect the infected cells from complement-mediated lysis, several members of the Herpesviridae and Poxviridae families were shown to encode homologues of Complement Regulatory Proteins (CRPs) [95]. However, no CRPs homologue encoded by HCMV has been identified so far. Nevertheless, HCMV-infected cells were shown to resist to complement attack by several mechanisms: on the one hand, HCMV upregulated the expression of two host-encoded complement regulatory proteins, CD46 and CD55, in fibroblasts and glioblastoma cells, that might in part be explained by the identification of a CMV-responsive element within the CD46 promoter [96, 97]. On the other hand, HCMV incorporates into its virions host cell-derived complement regulatory proteins CD55 and CD59. Blockade of CD55 by antibodies restored complement-mediated effects and reduced viral titers in vitro, indicating an important role of complement inhibition for HCMV replication [98].
HCMV, CANCERS AND IMMUNE TOLERANCE ESTABLISHMENT
Defect in anti-tumor immunity against tumor cells and tumor associated antigens is not simply due to a passive process during which the tumor would escape from recognition and destruction by the immune system. An active process of immune tolerance is set up, involving a microenvironment rich in biological tolerogenic agents such as cytokines, immature or dysfunctional antigen-presenting cells (APCs), and regulatory T cells (Treg). Thereby, although tumor associated antigens might be recognized by specific T cells, those will be directed to the ways of tolerance and anergy [22, 99]. Among the biologically active agents released by tumor or stroma cells that may promote immunosuppression and immune escape, we will limit ourselves to the review of interleukin 10 and TGF-β, both in the tumor environment and in the HCMV-infected cell.
Interleukin-10
IL-10 has immunosuppressive properties on several effectors of the immune system. It inhibits Th1 cells activation and proliferation as well as pro-inflammatory cytokines production by acting on Antigen Presenting Cells (in particular macrophages and dendritic cells) [100]. Moreover, IL-10 reduces antigen presentation to T Cells by inhibiting both MHC-I and MHC-II expression [28, 101].
An increased release of IL-10 has been described during cancers. For instance, exposure of dendritic cells to lysates from primary myeloma cells leads to production of IL-10 [102]. Investigation of IL-10 release by melanoma cells themselves has shown a preferential expression of IL-10 by metastasis-derived cells compared to primary-tumor-derived cells that might indicate a more invasive potential of cell variants expressing IL-10 [103]. Moreover, expression of CD1 (involved in primary immune responses to lipids and glycolipids expressed by various tumors) on infiltrating dendritic cells was downregulated in metastatic melanomas through the secretion of IL-10 [104]. Similarly, IL-10 has been involved in the enhancement of liver metastasis and in the impairment of antitumor immunity in murine models of colon cancer [105, 106]. IL-10 was also overproduced and acted as a growth factor for metastatic B-cell lymphoma in rats [107]. Moreover, decreased levels of IL-10 (and TGF-B) induced by low-dose of cyclophosphamide were shown to restore lymphoproliferative immune response in a tumor-bearing rat model [108]. IL-10 is also a critical cytokine for the differentiation of naïve CD4+ T cells into inducible Type 1 T regulatory cells (Tr1) [109]. Expansion of functional T regulatory cells (including FoxP3+ Tregs and Tr1) during cancers, described both in the peripheral blood and in the tumor microenvironment, appears to constitute a crucial means of tumor immune escape [110, 111]. Like other subtypes of Tregs, Tr1 have been involved in the decrease of antitumor immunity and in cancer progression [112, 113].
HCMV encodes an IL-10 homolog, cmvIL-10, that binds to the human IL-10 receptor and competes with cellular IL-10 for binding sites [114, 115]. CmvIL-10 has been described to exert immunosuppressive effects by inhibiting production of pro-inflammatory cytokines by peripheral blood mononuclear cells (PBMC) and dendritic cells and was involved in the immune evasion of HCMV-infected cells. CmvIL-10 was also shown to modulate the expression of MHC-class I and II molecules, and the expression of costimulatory factors and cytokines production by PBMC and dendritic cells [29, 116-118]. Interestingly, cmvIL-10 has been described to activate STAT3 in monocytes and HeLa cells [119, 120]. STAT3 appears to be a central transcription factor in carcinogenesis. Indeed, STAT3 is constitutively activated in many cancers and plays multiple roles in survival, angiogenesis, cell proliferation and metastasis [121]. Furthermore, STAT3 has been involved in restraining antitumor immunity by acting on both innate and adaptative responses and proposed to play a key role in mediating tumor-immune escape [122, 123]. In addition, IL-10 production has been reported during HCMV infection of THP-1 cells [124] and primary macrophages [125].
TGF-β
Transforming growth factor TGF-β is a pleiotropic immunosuppressive cytokine that promotes immune tolerance. TGF-β reduces adaptative immune responses by inhibiting T cells activation, differentiation and proliferation [126]. Moreover, TGF-β inhibits effector functions of cytotoxic T lymphocytes (CTLs) and tumor infiltrating lymphocytes (TILs) by repressing the expression of cytolytic gene products such as perforin, granzymes A and B, FasL or IFN-γ [127, 128]. In a murine model, overexpression of TGF-β in a highly immunogenic tumor was shown to promote tumor- immune escape [129]. Conversely, blockade of TGF-β restored T cells cytotoxicity and tumor clearance in several tumor models [128, 130, 131]. Interestingly, TGF-β might also have a major immunosuppressive and tolerogenic activity by promoting the differentiation of naive T cells into inductible FoxP3 + Cells [126, 132, 133]. In vivo, highly TGF-β plasma levels have been associated with worse prognosis or increased tumor progression in several cancers including breast and prostate cancers [134, 135]. Interestingly, recent studies suggested that immunotherapeutic approaches targeting TGF-β would restore an effective antitumor immune response and reduce tumor growth [136, 137].
In vitro early studies highlighted that HCMV could induce transcription and secretion of TGF-β, in part through HCMV Immediate Early proteins [138, 139]. Moreover, HCMV infection of endothelial cells induced an integrin αvβ6 (a TGF-β activator ) expression, leading to up-regulation of TGF-β [140]. In the same study, over expression of integrin αvβ6 was noticed in diverse human HCMV-infected tissues including lung, placenta, and salivary glands. Two recent studies exploring cytokine production from transplanted patients described both enhanced TGF-β plasma levels and overexpression of TGF-β mRNA in PBMC in patients with acute HCMV infection compared to HCMV uninfected patients [141, 142]. Interestingly, these studies also noticed overexpression of IL-10 mRNA in PBMC and increased soluble FasL plasma levels in recipients with active HCMV. In addition, TGF-β stimulation seems to be beneficial for HCMV. Indeed, TGF-β increased HCMV replication in cultured human lung fibroblasts [143]. Moreover, in cultures of primary murine astrocytes infected with murine cytomegalovirus (MCMV), release of CMV decreased in the presence of monoclonal antibody against TGF-β whereas addition of exogenous TGF-β was followed by an increased release of the virus [144].
CONCLUSION
HCMV has coevolved with the immune system for millions of years and has developed a number of strategies to counter the possible removal by the immune system. In this ongoing tug-of-war, virus gets advantage from immunologically poor environment of tumors to avoid recognition from immune system. Presence of HCMV DNA and proteins in cancerous tissue while absence in adjacent healthy tissue indicates the preference of the virus to persist in tumors where they are less prone to be removed by the immune system. In return, HCMV helps the tumor to avoid immune surveillance through encoding viral proteins (and possibly through non-coding RNAs) and induction of cellular factors having the potential to evade immune response and development of immune tolerance conducive for tumor development.
ACKNOWLEDGEMENTS
Principal Contributions:
Quentin Lepiller and Kashif Aziz Khan wrote the paper. Vincent Di Martino contributed to the design of the manuscript. Georges Herbein supervised the review.
Conflicting interests:
There are no conflicting or financial interests by the authors.
Grant Support:
This work was supported by grants from the University of Franche-Comté to G.H. K.A.K is a recipient of doctoral scholarships from the Higher Education Commission, Pakistan.
REFERENCES
- 1.Dunn GP, Old LJ, Schreiber RD. The three Es of cancer immunoediting. Annu Rev Immunol. 2004;22:329–60. doi: 10.1146/annurev.immunol.22.012703.104803. [DOI] [PubMed] [Google Scholar]
- 2.Mantovani A, Romero P, Palucka AK, Marincola FM. Tumour immunity: effector response to tumour and role of the microenvironment. Lancet. 2008;371:771–83. doi: 10.1016/S0140-6736(08)60241-X. [DOI] [PubMed] [Google Scholar]
- 3.Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–70. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 4.Soderberg-Naucler C. Does cytomegalovirus play a causative role in the development of various inflammatory diseases and cancer? J Intern Med. 2006;259:219–46. doi: 10.1111/j.1365-2796.2006.01618.x. [DOI] [PubMed] [Google Scholar]
- 5.Soderberg-Naucler C. HCMV microinfections in inflammatory diseases and cancer. J Clin Virol. 2008;41:218–23. doi: 10.1016/j.jcv.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 6.Michaelis M, Doerr HW, Cinatl J. The story of human cytomegalovirus and cancer: increasing evidence and open questions. Neoplasia. 2009;11:1–9. doi: 10.1593/neo.81178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cinatl J Jr, Cinatl J, Vogel JU, Rabenau H, Kornhuber B, Doerr HW. Modulatory effects of human cytomegalovirus infection on malignant properties of cancer cells. Intervirology. 1996;39:259–69. doi: 10.1159/000150527. [DOI] [PubMed] [Google Scholar]
- 8.Bain M, Sinclair J. The S phase of the cell cycle and its perturbation by human cytomegalovirus. Rev Med Virol. 2007;17:423–34. doi: 10.1002/rmv.551. [DOI] [PubMed] [Google Scholar]
- 9.Cobbs CS, Harkins L, Samanta M, et al. Human cytomegalovirus infection and expression in human malignant glioma. Cancer Res. 2002;62:3347–50. [PubMed] [Google Scholar]
- 10.Harkins L, Volk AL, Samanta M, et al. Specific localisation of human cytomegalovirus nucleic acids and proteins in human colorectal cancer. Lancet. 2002;360:1557–63. doi: 10.1016/S0140-6736(02)11524-8. [DOI] [PubMed] [Google Scholar]
- 11.Samanta M, Harkins L, Klemm K, Britt WJ, Cobbs CS. High prevalence of human cytomegalovirus in prostatic intraepithelial neoplasia and prostatic carcinoma. J Urol. 2003;170:998–1002. doi: 10.1097/01.ju.0000080263.46164.97. [DOI] [PubMed] [Google Scholar]
- 12.Zafiropoulos A, Tsentelierou E, Billiri K, Spandidos DA. Human herpes viruses in non-melanoma skin cancers. Cancer Lett. 2003;198:77–81. doi: 10.1016/s0304-3835(03)00269-6. [DOI] [PubMed] [Google Scholar]
- 13.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–44. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 14.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–99. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soroceanu L, Cobbs CS. Is HCMV a tumor promoter? Virus Res. 2010 doi: 10.1016/j.virusres.2010.10.026. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan G, Bivins-Smith ER, Smith MS, Smith PM, Yurochko AD. Transcriptome analysis reveals human cytomegalovirus reprograms monocyte differentiation toward an M1 macrophage. J Immunol. 2008;181:698–711. doi: 10.4049/jimmunol.181.1.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan G, Bivins-Smith ER, Smith MS, Yurochko AD. NF-kappaB and phosphatidylinositol 3-kinase activity mediates the HCMV-induced atypical M1/M2 polarization of monocytes. Virus Res. 2009;144:329–33. doi: 10.1016/j.virusres.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yurochko AD, Kowalik TF, Huong SM, Huang ES. Human cytomegalovirus upregulates NF-kappa B activity by transactivating the NF-kappa B p105/p50 and p65 promoters. J Virol. 1995;69:5391–400. doi: 10.1128/jvi.69.9.5391-5400.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Slinger E, Maussang D, Schreiber A, et al. HCMV-encoded chemokine receptor US28 mediates proliferative signaling through the IL-6-STAT3 axis. Sci Signal. 2010;3:ra58. doi: 10.1126/scisignal.2001180. [DOI] [PubMed] [Google Scholar]
- 20.Novellino L, Castelli C, Parmiani G. A listing of human tumor antigens recognized by T cells: March 2004 update. Cancer Immunol Immunother. 2005;54:187–207. doi: 10.1007/s00262-004-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Croci DO, Zacarias Fluck MF, Rico MJ, Matar P, Rabinovich GA, Scharovsky OG. Dynamic cross-talk between tumor and immune cells in orchestrating the immunosuppressive network at the tumor microenvironment. Cancer Immunol Immunother. 2007;56:1687–700. doi: 10.1007/s00262-007-0343-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rabinovich GA, Gabrilovich D, Sotomayor EM. Immunosuppressive strategies that are mediated by tumor cells. Annu Rev Immunol. 2007;25:267–96. doi: 10.1146/annurev.immunol.25.022106.141609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garcia-Lora A, Algarra I, Garrido F. MHC class I antigens, immune surveillance, and tumor immune escape. J Cell Physiol. 2003;195:346–55. doi: 10.1002/jcp.10290. [DOI] [PubMed] [Google Scholar]
- 24.Algarra I, Garcia-Lora A, Cabrera T, Ruiz-Cabello F, Garrido F. The selection of tumor variants with altered expression of classical and nonclassical MHC class I molecules: implications for tumor immune escape. Cancer Immunol Immunother. 2004;53:904–10. doi: 10.1007/s00262-004-0517-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hansen TH, Bouvier M. MHC class I antigen presentation: learning from viral evasion strategies. Nat Rev Immunol. 2009;9:503–13. doi: 10.1038/nri2575. [DOI] [PubMed] [Google Scholar]
- 26.Wiertz EJ, Devlin R, Collins HL, Ressing ME. Herpesvirus interference with major histocompatibility complex class II-restricted T-cell activation. J Virol. 2007;81:4389–96. doi: 10.1128/JVI.01525-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller-Kittrell M, Sparer TE. Feeling manipulated: cytomegalovirus immune manipulation. Virol J. 2009;6:4. doi: 10.1186/1743-422X-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koppelman B, Neefjes JJ, de Vries JE, de Waal Malefyt R. Interleukin-10 down-regulates MHC class II alphabeta peptide complexes at the plasma membrane of monocytes by affecting arrival and recycling. Immunity. 1997;7:861–71. doi: 10.1016/s1074-7613(00)80404-5. [DOI] [PubMed] [Google Scholar]
- 29.Spencer JV, Lockridge KM, Barry PA, et al. Potent immunosuppressive activities of cytomegalovirus-encoded interleukin-10. J Virol. 2002;76:1285–92. doi: 10.1128/JVI.76.3.1285-1292.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vivier E, Tomasello E, Baratin M, Walzer T, Ugolini S. Functions of natural killer cells. Nat Immunol. 2008;9:503–10. doi: 10.1038/ni1582. [DOI] [PubMed] [Google Scholar]
- 31.Ljunggren HG, Karre K. In search of the 'missing self': MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–44. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 32.Raulet DH, Guerra N. Oncogenic stress sensed by the immune system: role of natural killer cell receptors. Nat Rev Immunol. 2009;9:568–80. doi: 10.1038/nri2604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pende D, Parolini S, Pessino A, et al. Identification and molecular characterization of NKp30, a novel triggering receptor involved in natural cytotoxicity mediated by human natural killer cells. J Exp Med. 1999;190:1505–16. doi: 10.1084/jem.190.10.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pessino A, Sivori S, Bottino C, et al. Molecular cloning of NKp46: a novel member of the immunoglobulin superfamily involved in triggering of natural cytotoxicity. J Exp Med. 1998;188:953–60. doi: 10.1084/jem.188.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bottino C, Castriconi R, Pende D, et al. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J Exp Med. 2003;198:557–67. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlsten M, Bjorkstrom NK, Norell H, et al. DNAX accessory molecule-1 mediated recognition of freshly isolated ovarian carcinoma by resting natural killer cells. Cancer Res. 2007;67:1317–25. doi: 10.1158/0008-5472.CAN-06-2264. [DOI] [PubMed] [Google Scholar]
- 37.El-Sherbiny YM, Meade JL, Holmes TD, et al. The requirement for DNAM-1, NKG2D, and NKp46 in the natural killer cell-mediated killing of myeloma cells. Cancer Res. 2007;67:8444–9. doi: 10.1158/0008-5472.CAN-06-4230. [DOI] [PubMed] [Google Scholar]
- 38.Gilfillan S, Chan CJ, Cella M, et al. DNAM-1 promotes activation of cytotoxic lymphocytes by nonprofessional antigen-presenting cells and tumors. J Exp Med. 2008;205:2965–73. doi: 10.1084/jem.20081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iguchi-Manaka A, Kai H, Yamashita Y, et al. Accelerated tumor growth in mice deficient in DNAM-1 receptor. J Exp Med. 2008;205:2959–64. doi: 10.1084/jem.20081611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bauer S, Groh V, Wu J, et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–9. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- 41.Gasser S, Orsulic S, Brown EJ, Raulet DH. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 2005;436:1186–90. doi: 10.1038/nature03884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fuertes MB, Girart MV, Molinero LL, et al. Intracellular retention of the NKG2D ligand MHC class I chain-related gene A in human melanomas confers immune privilege and prevents NK cell-mediated cytotoxicity. J Immunol. 2008;180:4606–14. doi: 10.4049/jimmunol.180.7.4606. [DOI] [PubMed] [Google Scholar]
- 43.Guerra N, Tan YX, Joncker NT, et al. NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity. 2008;28:571–80. doi: 10.1016/j.immuni.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Godal R, Bachanova V, Gleason M, et al. Natural killer cell killing of acute myelogenous leukemia and acute lymphoblastic leukemia blasts by killer cell immunoglobulin-like receptor-negative natural killer cells after NKG2A and LIR-1 blockade. Biol Blood Marrow Transplant. 2010;16:612–21. doi: 10.1016/j.bbmt.2010.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilkinson GW, Tomasec P, Stanton RJ, et al. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–12. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beck S, Barrell BG. Human cytomegalovirus encodes a glycoprotein homologous to MHC class-I antigens. Nature. 1988;331:269–72. doi: 10.1038/331269a0. [DOI] [PubMed] [Google Scholar]
- 47.Wagner CS, Ljunggren HG, Achour A. Immune modulation by the human cytomegalovirus-encoded molecule UL18, a mystery yet to be solved. J Immunol. 2008;180:19–24. doi: 10.4049/jimmunol.180.1.19. [DOI] [PubMed] [Google Scholar]
- 48.Yang Z, Bjorkman PJ. Structure of UL18, a peptide-binding viral MHC mimic, bound to a host inhibitory receptor. Proc Natl Acad Sci USA. 2008;105:10095–100. doi: 10.1073/pnas.0804551105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603–13. doi: 10.1016/s1074-7613(00)80135-1. [DOI] [PubMed] [Google Scholar]
- 50.Prod'homme V, Griffin C, Aicheler RJ, et al. The human cytomegalovirus MHC class I homolog UL18 inhibits LIR-1+ but activates LIR-1- NK cells. J Immunol. 2007;178:4473–81. doi: 10.4049/jimmunol.178.7.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wagner CS, Walther-Jallow L, Buentke E, Ljunggren HG, Achour A, Chambers BJ. Human cytomegalovirus-derived protein UL18 alters the phenotype and function of monocyte-derived dendritic cells. J Leukoc Biol. 2008;81:56–63. doi: 10.1189/jlb.0307181. [DOI] [PubMed] [Google Scholar]
- 52.Tomasec P, Braud VM, Rickards C, et al. Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science. 2000;287:1031. doi: 10.1126/science.287.5455.1031. [DOI] [PubMed] [Google Scholar]
- 53.Ulbrecht M, Martinozzi S, Grzeschik M, et al. Cutting edge: the human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J Immunol. 2000;164:5019–22. doi: 10.4049/jimmunol.164.10.5019. [DOI] [PubMed] [Google Scholar]
- 54.Dunn C, Chalupny NJ, Sutherland CL, et al. Human cytomegalovirus glycoprotein UL16 causes intracellular sequestration of NKG2D ligands, protecting against natural killer cell cytotoxicity. J Exp Med. 2003;197:1427–39. doi: 10.1084/jem.20022059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu J, Chalupny NJ, Manley TJ, Riddell SR, Cosman D, Spies T. Intracellular retention of the MHC class I-related chain B ligand of NKG2D by the human cytomegalovirus UL16 glycoprotein. J Immunol. 2003;170:4196–200. doi: 10.4049/jimmunol.170.8.4196. [DOI] [PubMed] [Google Scholar]
- 56.Chalupny NJ, Rein-Weston A, Dosch S, Cosman D. Down-regulation of the NKG2D ligand MICA by the human cytomegalovirus glycoprotein UL142. Biochem Biophys Res Commun. 2006;346:175–81. doi: 10.1016/j.bbrc.2006.05.092. [DOI] [PubMed] [Google Scholar]
- 57.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–81. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wills MR, Ashiru O, Reeves MB, et al. Human cytomegalovirus encodes an MHC class I-like molecule (UL142) that functions to inhibit NK cell lysis. J Immunol. 2005;175:7457–65. doi: 10.4049/jimmunol.175.11.7457. [DOI] [PubMed] [Google Scholar]
- 59.Ashiru O, Bennett NJ, Boyle LH, Thomas M, Trowsdale J, Wills MR. NKG2D ligand MICA is retained in the cis-Golgi apparatus by human cytomegalovirus protein UL142. J Virol. 2009;83:12345–54. doi: 10.1128/JVI.01175-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tomasec P, Wang EC, Davison AJ, et al. Downregulation of natural killer cell-activating ligand CD155 by human cytomegalovirus UL141. Nat Immunol. 2005;6:181–8. doi: 10.1038/ni1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Browne EP, Shenk T. Human cytomegalovirus UL83-coded pp65 virion protein inhibits antiviral gene expression in infected cells. Proc Natl Acad Sci USA. 2003;100:11439–44. doi: 10.1073/pnas.1534570100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kalejta RF. Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev. 2008;72:249–65. doi: 10.1128/MMBR.00040-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Arnon TI, Achdout H, Levi O, et al. Inhibition of the NKp30 activating receptor by pp65 of human cytomegalovirus. Nat Immunol. 2005;6:515–23. doi: 10.1038/ni1190. [DOI] [PubMed] [Google Scholar]
- 64.Stern-Ginossar N, Gur C, Biton M, et al. Human microRNAs regulate stress-induced immune responses mediated by the receptor NKG2D. Nat Immunol. 2008;9:1065–73. doi: 10.1038/ni.1642. [DOI] [PubMed] [Google Scholar]
- 65.Nachmani D, Lankry D, Wolf DG, Mandelboim O. The human cytomegalovirus microRNA miR-UL112 acts synergistically with a cellular microRNA to escape immune elimination. Nat Immunol. 2010;11:806–13. doi: 10.1038/ni.1916. [DOI] [PubMed] [Google Scholar]
- 66.Strasser A, Jost PJ, Nagata S. The many roles of FAS receptor signaling in the immune system. Immunity. 2009;30:180–92. doi: 10.1016/j.immuni.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Medema JP, de Jong J, van Hall T, Melief CJ, Offringa R. Immune escape of tumors in vivo by expression of cellular FLICE-inhibitory protein. J Exp Med. 1999;190:1033–8. doi: 10.1084/jem.190.7.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Griffith TS, Chin WA, Jackson GC, Lynch DH, Kubin MZ. Intracellular regulation of TRAIL-induced apoptosis in human melanoma cells. J Immunol. 1998;161:2833–40. [PubMed] [Google Scholar]
- 69.Tepper CG, Seldin MF. Modulation of caspase-8 and FLICE-inhibitory protein expression as a potential mechanism of Epstein-Barr virus tumorigenesis in Burkitt's lymphoma. Blood. 1999;94:1727–37. [PubMed] [Google Scholar]
- 70.Weller M, Malipiero U, Aguzzi A, Reed JC, Fontana A. Protooncogene bcl-2 gene transfer abrogates Fas/APO-1 antibody-mediated apoptosis of human malignant glioma cells and confers resistance to chemotherapeutic drugs and therapeutic irradiation. J Clin Invest. 1995;95:2633–43. doi: 10.1172/JCI117965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Campos L, Rouault JP, Sabido O, et al. High expression of bcl-2 protein in acute myeloid leukemia cells is associated with poor response to chemotherapy. Blood. 1993;81:3091–6. [PubMed] [Google Scholar]
- 72.Coustan-Smith E, Kitanaka A, Pui CH, et al. Clinical relevance of BCL-2 overexpression in childhood acute lymphoblastic leukemia. Blood. 1996;87:1140–6. [PubMed] [Google Scholar]
- 73.Hermine O, Haioun C, Lepage E, et al. Prognostic significance of bcl-2 protein expression in aggressive non-Hodgkin's lymphoma. Groupe d'Etude des Lymphomes de l'Adulte (GELA) Blood. 1996;87:265–72. [PubMed] [Google Scholar]
- 74.Schulze-Bergkamen H, Ehrenberg R, Hickmann L, et al. Bcl-x(L) and Myeloid cell leukaemia-1 contribute to apoptosis resistance of colorectal cancer cells. World J Gastroenterol. 2008;14:3829–40. doi: 10.3748/wjg.14.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Violette S, Poulain L, Dussaulx E, et al. Resistance of colon cancer cells to long-term 5-fluorouracil exposure is correlated to the relative level of Bcl-2 and Bcl-X(L) in addition to Bax and p53 status. Int J Cancer. 2002;98:498–504. doi: 10.1002/ijc.10146. [DOI] [PubMed] [Google Scholar]
- 76.Igney FH, Krammer PH. Immune escape of tumors: apoptosis resistance and tumor counterattack. J Leukoc Biol. 2002;71:907–20. [PubMed] [Google Scholar]
- 77.Straus SE, Jaffe ES, Puck JM, et al. The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood. 2001;98:194–200. doi: 10.1182/blood.v98.1.194. [DOI] [PubMed] [Google Scholar]
- 78.Hahne M, Rimoldi D, Schroter M, et al. Melanoma cell expression of Fas(Apo-1/CD95) ligand: implications for tumor immune escape. Science. 1996;274:1363–6. doi: 10.1126/science.274.5291.1363. [DOI] [PubMed] [Google Scholar]
- 79.Igney FH, Krammer PH. Tumor counterattack: fact or fiction? Cancer Immunol Immunother. 2005;54:1127–36. doi: 10.1007/s00262-005-0680-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chappell DB, Zaks TZ, Rosenberg SA, Restifo NP. Human melanoma cells do not express Fas (Apo-1/CD95) ligand. Cancer Res. 1999;59:59–62. [PMC free article] [PubMed] [Google Scholar]
- 81.Andreola G, Rivoltini L, Castelli C, et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J Exp Med. 2002;195:1303–16. doi: 10.1084/jem.20011624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen JJ, Sun Y, Nabel GJ. Regulation of the proinflammatory effects of Fas ligand (CD95L) Science. 1998;282:1714–7. doi: 10.1126/science.282.5394.1714. [DOI] [PubMed] [Google Scholar]
- 83.Sedy JR, Spear PG, Ware CF. Cross-regulation between herpesviruses and the TNF superfamily members. Nat Rev Immunol. 2008;8:861–73. doi: 10.1038/nri2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chiou SH, Yang YP, Lin JC, et al. The immediate early 2 protein of human cytomegalovirus (HCMV) mediates the apoptotic control in HCMV retinitis through up-regulation of the cellular FLICE-inhibitory protein expression. J Immunol. 2006;177:6199–206. doi: 10.4049/jimmunol.177.9.6199. [DOI] [PubMed] [Google Scholar]
- 85.Cinatl J Jr, Cinatl J, Vogel JU, et al. Persistent human cytomegalovirus infection induces drug resistance and alteration of programmed cell death in human neuroblastoma cells. Cancer Res. 1998;58:367–72. [PubMed] [Google Scholar]
- 86.Norris KL, Youle RJ. Cytomegalovirus proteins vMIA and m38.5 link mitochondrial morphogenesis to Bcl-2 family proteins. J Virol. 2008;82:6232–43. doi: 10.1128/JVI.02710-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Goldmacher VS. Cell death suppression by cytomegaloviruses. Apoptosis. 2005;10:251–65. doi: 10.1007/s10495-005-0800-z. [DOI] [PubMed] [Google Scholar]
- 88.Chiou SH, Liu JH, Hsu WM, et al. Up-regulation of Fas ligand expression by human cytomegalovirus immediate-early gene product 2: a novel mechanism in cytomegalovirus-induced apoptosis in human retina. J Immunol. 2001;167:4098–103. doi: 10.4049/jimmunol.167.7.4098. [DOI] [PubMed] [Google Scholar]
- 89.Dunkelberger JR, Song WC. Complement and its role in innate and adaptive immune responses. Cell Res. 2010;20:34–50. doi: 10.1038/cr.2009.139. [DOI] [PubMed] [Google Scholar]
- 90.Markiewski MM, Lambris JD. Is complement good or bad for cancer patients? A new perspective on an old dilemma. Trends Immunol. 2009;30:286–92. doi: 10.1016/j.it.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gorter A, Blok VT, Haasnoot WH, Ensink NG, Daha MR, Fleuren GJ. Expression of CD46, CD55, and CD59 on renal tumor cell lines and their role in preventing complement-mediated tumor cell lysis. Lab Invest. 1996;74:1039–49. [PubMed] [Google Scholar]
- 92.Murray KP, Mathure S, Kaul R, et al. Expression of complement regulatory proteins-CD 35, CD 46, CD 55, and CD 59-in benign and malignant endometrial tissue. Gynecol Oncol. 2000;76:176–82. doi: 10.1006/gyno.1999.5614. [DOI] [PubMed] [Google Scholar]
- 93.Watson NF, Durrant LG, Madjd Z, Ellis IO, Scholefield JH, Spendlove I. Expression of the membrane complement regulatory protein CD59 (protectin) is associated with reduced survival in colorectal cancer patients. Cancer Immunol Immunother. 2006;55:973–80. doi: 10.1007/s00262-005-0055-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Macor P, Tedesco F. Complement as effector system in cancer immunotherapy. Immunol Lett. 2007;111:6–13. doi: 10.1016/j.imlet.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 95.Lee SH, Jung JU, Means RE. 'Complementing' viral infection: mechanisms for evading innate immunity. Trends Microbiol. 2003;11:449–52. doi: 10.1016/j.tim.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 96.Nomura M, Kurita-Taniguchi M, Kondo K, et al. Mechanism of host cell protection from complement in murine cytomegalovirus (CMV) infection: identification of a CMV-responsive element in the CD46 promoter region. Eur J Immunol. 2002;32:2954–64. doi: 10.1002/1521-4141(2002010)32:10<2954::AID-IMMU2954>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 97.Spiller OB, Morgan BP, Tufaro F, Devine DV. Altered expression of host-encoded complement regulators on human cytomegalovirus-infected cells. Eur J Immunol. 1996;26:1532–8. doi: 10.1002/eji.1830260719. [DOI] [PubMed] [Google Scholar]
- 98.Spear GT, Lurain NS, Parker CJ, Ghassemi M, Payne GH, Saifuddin M. Host cell-derived complement control proteins CD55 and CD59 are incorporated into the virions of two unrelated enveloped viruses. Human T cell leukemia/lymphoma virus type I (HTLV-I) and human cytomegalovirus (HCMV) J Immunol. 1995;155:4376–81. [PubMed] [Google Scholar]
- 99.Zou W. Immunosuppressive networks in the tumour environment and their therapeutic relevance. Nat Rev Cancer. 2005;5:263–74. doi: 10.1038/nrc1586. [DOI] [PubMed] [Google Scholar]
- 100.Maynard CL, Weaver CT. Diversity in the contribution of interleukin-10 to T-cell-mediated immune regulation. Immunol Rev. 2008;226:219–33. doi: 10.1111/j.1600-065X.2008.00711.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kurte M, Lopez M, Aguirre A, et al. A synthetic peptide homologous to functional domain of human IL-10 down-regulates expression of MHC class I and Transporter associated with Antigen Processing 1/2 in human melanoma cells. J Immunol. 2004;173:1731–7. doi: 10.4049/jimmunol.173.3.1731. [DOI] [PubMed] [Google Scholar]
- 102.Fiore F, Nuschak B, Peola S, et al. Exposure to myeloma cell lysates affects the immune competence of dendritic cells and favors the induction of Tr1-like regulatory T cells. Eur J Immunol. 2005;35:1155–63. doi: 10.1002/eji.200425093. [DOI] [PubMed] [Google Scholar]
- 103.Dummer W, Bastian BC, Ernst N, Schanzle C, Schwaaf A, Brocker EB. Interleukin-10 production in malignant melanoma: preferential detection of IL-10-secreting tumor cells in metastatic lesions. Int J Cancer. 1996;66:607–10. doi: 10.1002/(SICI)1097-0215(19960529)66:5<607::AID-IJC4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 104.Gerlini G, Tun-Kyi A, Dudli C, Burg G, Pimpinelli N, Nestle FO. Metastatic melanoma secreted IL-10 down-regulates CD1 molecules on dendritic cells in metastatic tumor lesions. Am J Pathol. 2004;165:1853–63. doi: 10.1016/S0002-9440(10)63238-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jessup JM, Samara R, Battle P, Laguinge LM. Carcinoembryonic antigen promotes tumor cell survival in liver through an IL-10-dependent pathway. Clin Exp Metastasis. 2004;21:709–17. doi: 10.1007/s10585-004-7705-z. [DOI] [PubMed] [Google Scholar]
- 106.Kawamura K, Bahar R, Natsume W, Sakiyama S, Tagawa M. Secretion of interleukin-10 from murine colon carcinoma cells suppresses systemic antitumor immunity and impairs protective immunity induced against the tumors. Cancer Gene Ther. 2002;9:109–15. doi: 10.1038/sj.cgt.7700418. [DOI] [PubMed] [Google Scholar]
- 107.Rico MJ, Matar P, Gervasoni SI, Bonfil RD, Calcaterra N, Scharovsky OG. The transition to the metastatic phenotype of rat lymphoma cells involves up-regulation of IL-10 receptor expression and IL-10 secretion. Clin Exp Metastasis. 2005;22:127–35. doi: 10.1007/s10585-005-5140-4. [DOI] [PubMed] [Google Scholar]
- 108.Matar P, Rozados VR, Gervasoni SI, Scharovsky OG. Down regulation of T-cell-derived IL-10 production by low-dose cyclophosphamide treatment in tumor-bearing rats restores in vitro normal lymphoproliferative response. Int Immunopharmacol. 2001;1:307–19. doi: 10.1016/s1567-5769(00)00028-x. [DOI] [PubMed] [Google Scholar]
- 109.Levings MK, Sangregorio R, Galbiati F, Squadrone S, de Waal Malefyt R, Roncarolo MG. IFN-alpha and IL-10 induce the differentiation of human type 1 T regulatory cells. J Immunol. 2001;166:5530–9. doi: 10.4049/jimmunol.166.9.5530. [DOI] [PubMed] [Google Scholar]
- 110.Zou W. Regulatory T cells, tumour immunity and immunotherapy. Nat Rev Immunol. 2006;6:295–307. doi: 10.1038/nri1806. [DOI] [PubMed] [Google Scholar]
- 111.Allan SE, Broady R, Gregori S, et al. CD4+ T-regulatory cells: toward therapy for human diseases. Immunol Rev. 2008;223:391–421. doi: 10.1111/j.1600-065X.2008.00634.x. [DOI] [PubMed] [Google Scholar]
- 112.Bergmann C, Strauss L, Wang Y, et al. T regulatory type 1 cells in squamous cell carcinoma of the head and neck: mechanisms of suppression and expansion in advanced disease. Clin Cancer Res. 2008;14:3706–15. doi: 10.1158/1078-0432.CCR-07-5126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang X, Huang H, Yuan J, et al. CD4-8- dendritic cells prime CD4+ T regulatory 1 cells to suppress antitumor immunity. J Immunol. 2005;175:2931–7. doi: 10.4049/jimmunol.175.5.2931. [DOI] [PubMed] [Google Scholar]
- 114.Kotenko SV, Saccani S, Izotova LS, Mirochnitchenko OV, Pestka S. Human cytomegalovirus harbors its own unique IL-10 homolog (cmvIL-10) Proc Natl Acad Sci USA. 2000;97:1695–700. doi: 10.1073/pnas.97.4.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Redpath S, Ghazal P, Gascoigne NR. Hijacking and exploitation of IL-10 by intracellular pathogens. Trends Microbiol. 2001;9:86–92. doi: 10.1016/s0966-842x(00)01919-3. [DOI] [PubMed] [Google Scholar]
- 116.Raftery MJ, Wieland D, Gronewald S, Kraus AA, Giese T, Schonrich G. Shaping phenotype, function, and survival of dendritic cells by cytomegalovirus-encoded IL-10. J Immunol. 2004;173:3383–91. doi: 10.4049/jimmunol.173.5.3383. [DOI] [PubMed] [Google Scholar]
- 117.Chang WL, Baumgarth N, Yu D, Barry PA. Human cytomegalovirus-encoded interleukin-10 homolog inhibits maturation of dendritic cells and alters their functionality. J Virol. 2004;78:8720–31. doi: 10.1128/JVI.78.16.8720-8731.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Slobedman B, Barry PA, Spencer JV, Avdic S, Abendroth A. Virus-encoded homologs of cellular interleukin-10 and their control of host immune function. J Virol. 2009;83:9618–29. doi: 10.1128/JVI.01098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Spencer JV. The cytomegalovirus homolog of interleukin-10 requires phosphatidylinositol 3-kinase activity for inhibition of cytokine synthesis in monocytes. J Virol. 2007;81:2083–6. doi: 10.1128/JVI.01655-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Gruber SG, Gloria Luciani M, Grundtner P, Zdanov A, Gasche C. Differential signaling of cmvIL-10 through common variants of the IL-10 receptor 1. Eur J Immunol. 2008;38:3365–75. doi: 10.1002/eji.200837718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Aggarwal BB, Kunnumakkara AB, Harikumar KB, et al. Signal transducer and activator of transcription-3, inflammation, and cancer: how intimate is the relationship? Ann N Y Acad Sci. 2009;1171:59–76. doi: 10.1111/j.1749-6632.2009.04911.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kortylewski M, Kujawski M, Wang T, et al. Inhibiting Stat3 signaling in the hematopoietic system elicits multicomponent antitumor immunity. Nat Med. 2005;11:1314–21. doi: 10.1038/nm1325. [DOI] [PubMed] [Google Scholar]
- 123.Wang T, Niu G, Kortylewski M, et al. Regulation of the innate and adaptive immune responses by Stat-3 signaling in tumor cells. Nat Med. 2004;10:48–54. doi: 10.1038/nm976. [DOI] [PubMed] [Google Scholar]
- 124.Nordoy I, Rollag H, Lien E, et al. Cytomegalovirus infection induces production of human interleukin-10 in macrophages. Eur J Clin Microbiol Infect Dis. 2003;22:737–41. doi: 10.1007/s10096-003-1028-x. [DOI] [PubMed] [Google Scholar]
- 125.Khan KA, Coaquette A, Davrinche C, Herbein G. Bcl-3-regulated transcription from major immediate-early promoter of human cytomegalovirus in monocyte-derived macrophages. J Immunol. 2009;182:7784–94. doi: 10.4049/jimmunol.0803800. [DOI] [PubMed] [Google Scholar]
- 126.Li MO, Wan YY, Sanjabi S, Robertson AK, Flavell RA. Transforming growth factor-beta regulation of immune responses. Annu Rev Immunol. 2006;24:99–146. doi: 10.1146/annurev.immunol.24.021605.090737. [DOI] [PubMed] [Google Scholar]
- 127.Ahmadzadeh M, Rosenberg SA. TGF-beta 1 attenuates the acquisition and expression of effector function by tumor antigen-specific human memory CD8 T cells. J Immunol. 2005;174:5215–23. doi: 10.4049/jimmunol.174.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Thomas DA, Massague J. TGF-beta directly targets cytotoxic T cell functions during tumor evasion of immune surveillance. Cancer Cell. 2005;8:369–80. doi: 10.1016/j.ccr.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 129.Torre-Amione G, Beauchamp RD, Koeppen H, et al. A highly immunogenic tumor transfected with a murine transforming growth factor type beta 1 cDNA escapes immune surveillance. Proc Natl Acad Sci USA. 1990;87:1486–90. doi: 10.1073/pnas.87.4.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Won J, Kim H, Park EJ, Hong Y, Kim SJ, Yun Y. Tumorigenicity of mouse thymoma is suppressed by soluble type II transforming growth factor beta receptor therapy. Cancer Res. 1999;59:1273–7. [PubMed] [Google Scholar]
- 131.Gorelik L, Flavell RA. Immune-mediated eradication of tumors through the blockade of transforming growth factor-beta signaling in T cells. Nat Med. 2001;7:1118–22. doi: 10.1038/nm1001-1118. [DOI] [PubMed] [Google Scholar]
- 132.Fantini MC, Becker C, Monteleone G, Pallone F, Galle PR, Neurath MF. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25- T cells through Foxp3 induction and down-regulation of Smad7. J Immunol. 2004;172:5149–53. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 133.Huehn J, Polansky JK, Hamann A. Epigenetic control of FOXP3 expression: the key to a stable regulatory T-cell lineage? Nat Rev Immunol. 2009;9:83–9. doi: 10.1038/nri2474. [DOI] [PubMed] [Google Scholar]
- 134.Shariat SF, Shalev M, Menesses-Diaz A, et al. Preoperative plasma levels of transforming growth factor beta(1) (TGF-beta(1)) strongly predict progression in patients undergoing radical prostatectomy. J Clin Oncol. 2001;19:2856–64. doi: 10.1200/JCO.2001.19.11.2856. [DOI] [PubMed] [Google Scholar]
- 135.Chod J, Zavadova E, Halaska MJ, Strnad P, Fucikova T, Rob L. Preoperative transforming growth factor-beta 1 (TGF-beta 1) plasma levels in operable breast cancer patients. Eur J Gynaecol Oncol. 2008;29:613–6. [PubMed] [Google Scholar]
- 136.Fujita T, Teramoto K, Ozaki Y, et al. Inhibition of transforming growth factor-beta-mediated immunosuppression in tumor-draining lymph nodes augments antitumor responses by various immunologic cell types. Cancer Res. 2009;69:5142–50. doi: 10.1158/0008-5472.CAN-08-2499. [DOI] [PubMed] [Google Scholar]
- 137.Mazzocca A, Fransvea E, Lavezzari G, Antonaci S, Giannelli G. Inhibition of transforming growth factor beta receptor I kinase blocks hepatocellular carcinoma growth through neo-angiogenesis regulation. Hepatology. 2009;50:1140–51. doi: 10.1002/hep.23118. [DOI] [PubMed] [Google Scholar]
- 138.Michelson S, Alcami J, Kim SJ, et al. Human cytomegalovirus infection induces transcription and secretion of transforming growth factor beta 1. J Virol. 1994;68:5730–7. doi: 10.1128/jvi.68.9.5730-5737.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Yoo YD, Chiou CJ, Choi KS, et al. The IE2 regulatory protein of human cytomegalovirus induces expression of the human transforming growth factor beta1 gene through an Egr-1 binding site. J Virol. 1996;70:7062–70. doi: 10.1128/jvi.70.10.7062-7070.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Tabata T, Kawakatsu H, Maidji E, et al. Induction of an epithelial integrin alphavbeta6 in human cytomegalovirus-infected endothelial cells leads to activation of transforming growth factor-beta1 and increased collagen production. Am J Pathol. 2008;172:1127–40. doi: 10.2353/ajpath.2008.070448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Y, Liu Y, Zhang Y, et al. The role of the CD95, CD38 and TGFbeta1 during active human cytomegalovirus infection in liver transplantation. Cytokine. 2006;35:193–9. doi: 10.1016/j.cyto.2006.08.001. [DOI] [PubMed] [Google Scholar]
- 142.Zhang Y, Wang YL, Liu YW, et al. Change of peripheral blood mononuclear cells IFN-gamma, IL-10, and TGF-beta1 mRNA expression levels with active human cytomegalovirus infection in orthotopic liver transplantation. Transplant Proc. 2009;41:1767–9. doi: 10.1016/j.transproceed.2009.03.064. [DOI] [PubMed] [Google Scholar]
- 143.Alcami J, Paya CV, Virelizier JL, Michelson S. Antagonistic modulation of human cytomegalovirus replication by transforming growth factor beta and basic fibroblastic growth factor. J Gen Virol. 1993;74:269–74. doi: 10.1099/0022-1317-74-2-269. [DOI] [PubMed] [Google Scholar]
- 144.Kossmann T, Morganti-Kossmann MC, Orenstein JM, Britt WJ, Wahl SM, Smith PD. Cytomegalovirus production by infected astrocytes correlates with transforming growth factor-beta release. J Infect Dis. 2003;187:534–41. doi: 10.1086/373995. [DOI] [PubMed] [Google Scholar]