Abstract
Rationale
In early heart development, platelet derived growth factor (PDGF) receptor expression in the heart ventricles is restricted to the epicardium. Previously, we showed that PDGFRβ is required for coronary vascular smooth muscle cell (cVSMC) development, but a role for PDGFRα, has not been identified. Therefore, we investigated the combined and independent roles of these receptors in epicardial development.
Objective
To understand the contribution of PDGF receptors in epicardial development and epicardial derived cell fate determination.
Methods and results
By generating mice with epicardial specific deletion of the PDGF receptors, we found that epicardial EMT was defective. Sox9, an SRY-related transcription factor, was reduced in PDGF receptor-deficient epicardial cells, and overexpression of Sox9 restored epicardial migration, actin reorganization, and EMT gene expression profiles. The failure of epicardial EMT resulted in hearts that lacked epicardial-derived cardiac fibroblasts and cVSMC. Loss of PDGFRα, resulted in a specific disruption of cardiac fibroblast development, while cVSMC development was unperturbed.
Conclusions
Signaling through both PDGF receptors is necessary for epicardial EMT and formation of epicardial mesenchymal derivatives. PDGF receptors also have independent functions in the development of specific epicardial derived cell fates.
Keywords: epicardium, PDGF, cardiac fibroblast, EMT, sox9
Introduction
Cardiac disease is the leading cause of death in the industrial world. While recent stem cell therapies have attempted to regenerate myocardium, there are still many physiological barriers to overcome, including fibrosis, inflammation, and insufficient blood vessel generation. Induction of cardiomyocyte regeneration is one proposed way to improve cardiac function, but it is clear that the non-cardiomyocyte populations in the heart also contribute to the repair process. Non-cardiomyocyte lineages (endothelial cells, vascular smooth muscle cells, and cardiac fibroblasts) are essential for blood vessel formation and matrix organization, and an understanding of the developmental signals that shape these cells may provide insights into disease pathogenesis and better heart injury therapies.
Coronary vascular smooth muscle cells (cVSMC) and cardiac fibroblasts develop from the epicardium in a multi-step process involving cell proliferation, epithelial-to-mesenchymal transition (EMT), and mesenchymal cell fate specification1. Several proteins have been implicated in the development of cVSMC from the epicardium2, but less is known about the epicardial derived cardiac fibroblast population. It is proposed that cardiac fibroblasts are essential for normal cardiac function, and their role in matrix deposition during cardiac injury is well established. Yet, signaling pathways regulating their development are poorly understood.
Platelet derived growth factor (PDGF) receptor tyrosine kinases are important for embryonic development and play essential roles in the forming vasculature3. Previously, we identified a role for PDGFRβ as an important factor regulating epicardial derived cVSMC development4. We and others observed PDGFRα in the epicardium5, 6; however, no data exists regarding the fate of epicardial derived cells (EPDCs) when PDGFRα is disrupted. The receptors are co-expressed in the epicardium until E13.5, but after this time point receptor expression becomes mutually exclusive. These initial findings led us to investigate the role of PDGFRα individually and combined with PDGFRβ during EPDC formation.
Using cre/loxP recombination, we generated animals that lacked PDGFRα, PDGFRβ or both PDGF receptors in the epicardium. Epicardial deletion of both PDGF receptors resulted in failure of epicardial EMT and EPDC formation. Loss of PDGF signaling led to reduced Sox9 expression, and when Sox9 expression was restored in mutant hearts, the EMT defect was rescued. Interestingly, mutants lacking only one of the PDGF receptor genes exhibited a lineage specific requirement for each individual receptor. Loss of PDGFRα resulted in a deficit in cardiac fibroblast formation, while cVSMC development was unperturbed. Conversely, PDGFRβ was required for cVSMC development4 but not cardiac fibroblast development. Combined, our data demonstrate a novel role for PDGF receptors in epicardial EMT and EPDC development.
Methods
Additional methods are available in the supplemental material.
Experimental Animals
Mice were maintained on a mixed C57/Bl6 X 129SV background. The strains in these experiments included PDGFRαGFP7, PDGFRαfl8, PDGFRβfl9, 10, R26RYFP11, R26RtdT12 (Jackson Labs), R26RLacZ13 WT1iCre14, Tie2CreTg/015 and Gata5CreTg/016. All animal protocols and experiments were approved by the UTSW IACUC and conformed to NIH guidelines for care and use of laboratory animals. Gata5Cre transgenic and WT1iCre mice were kindly provided by Dr. Ruiz Pilar-Lozano (Burnham Institute) and Dr. William Pu (Harvard), respectively. WT1iCre/+ animals were induced with tamoxifen at indicated timepoints. Tamoxifen (MP Biomedicals, 0215673894) was dissolved in sunflower seed oil (Sima, S5007) at 20mg/ml. 0.1mg/g body weight of tamoxifen was administered by oral gavage. Controls used in most experiments were Gata5Cre negative littermates. Animals used for controls in fibroblast isolation(s) were PDGFRαfl/+ PDGFRβfl/+ R26RLacZ/+ Gata5Cre+.
Results
Loss of both PDGF receptors causes defects in epicardial cell migration
Both PDGF receptors, α and β, are expressed by the epicardium, and loss of PDGFRβ alone causes a reduction in cVSMC4. To investigate a combined role for PDGF receptor signaling, we deleted PDGF receptors from epicardial cells using mice expressing the Gata5Cre transgene16. PDGFREKO (EKO – epicardial knockout) hearts had regions of epicardial detachment and hemorrhaging (Figure 1A). The detachment progressed temporally from the dorsal to ventral heart surface but resolved by birth (data not shown). Despite this phenotype, the epicardium expressed multiple, established epicardial markers (Online Figure I, A–B). Because PDGF receptor signaling has been associated with proliferation and survival17, we examined the epicardium and EPDCs for BrdU incorporation and cleaved caspase-3 activation. PDGFREKO mutant values were similar to those obtained in controls (Online Figure I, C–D), suggesting that loss of PDGF receptor signaling does not affect cellular proliferation or survival of epicardial cells in vivo.
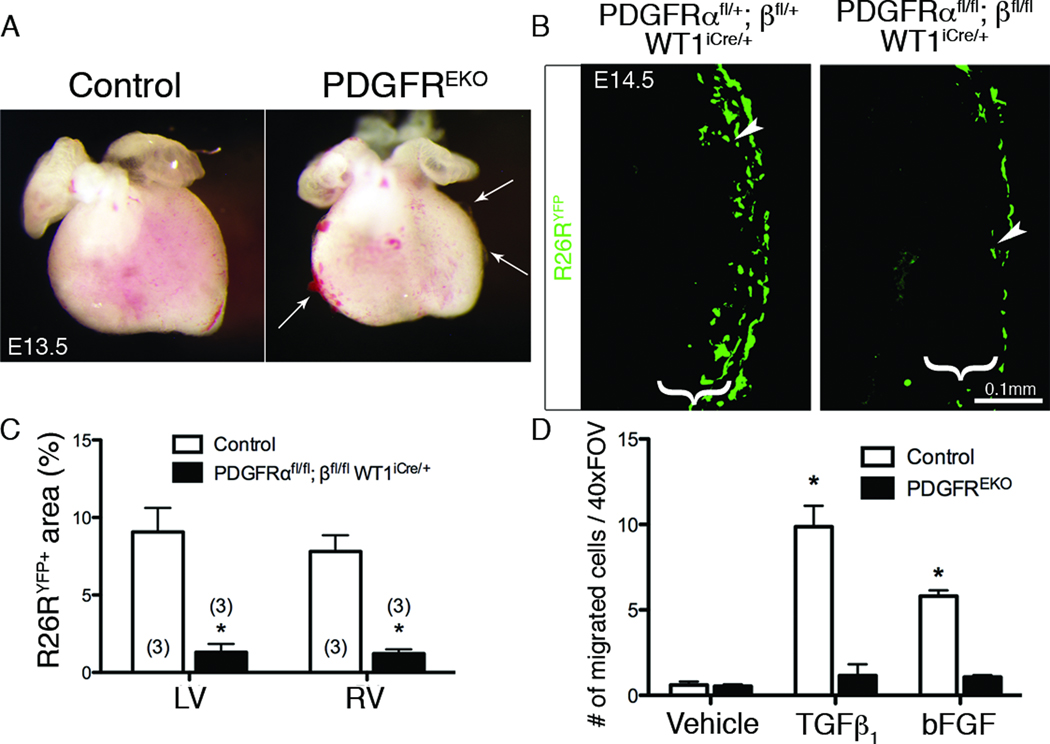
Figure 1. PDGFREKO epicardial cells fail to migrate into the myocardium.
(A) Whole mount images showing regions of epicardial detachment and hemorrhaging (Arrows). (B) R26RYFP IHC was used to examine epicardial cell migration into subepicardial mesenchyme (brackets) from indicated genotypes induced with tamoxifen at E12.5. Arrowheads point to migrated cells within the subepicardial mesenchyme. (C) Quantification of the R26RYFP fluorescent area in (B). N values are indicated in parentheses. (*) p<0.005 (D) Quantification of GFP+ cells within myocardium of E12.5 hearts transduced with an adenovirus expressing GFP and stimulated with hTGFβ1 or bFGF (n=3 for each genotype/condition). Data are represented as mean ± SD. (*) p<0.001 (compared to vehicle treated control) (LV – left ventricle, RV – right ventricle, EKO – epicardial knockout)
Because epicardial formation was unaffected in PDGFREKO mutants, we next assayed hearts for epicardial cell entry into the myocardium. To trace epicardium lacking PDGF receptors, we induced cre-mediated recombination in epicardial cells just prior to EMT (E12.5) using a tamoxifen inducible WT1iCre allele14 and analyzed migration at E14.5. Using R26RYFP reporter activity to follow the epicardial cells, we observed a loss of EPDCs when PDGF receptors were absent (Figure 1B–C). Similar results were obtained when using markers of undifferentiated EPDCs, WT118, 19 or mesenchymal cells, vimentin20. Both markers showed a severe reduction in the region immediately underlying the epicardium in PDGFREKO hearts (Online Figure II and data not shown). Note that vimentin is a broad mesenchymal marker that is also expressed by non-epicardial derived coronary endothelial cells21. Additionally, in an ex vivo migration assay4, fewer cells exited the epicardium in PDGFREKO mutant hearts even when stimulated with EMT-inducing growth factors, hTGFβ122 and bFGF23 (Figure 1D).
PDGF receptor signaling is required for epicardial cell EMT
We surmised that loss of epicardial cell migration was caused by a defect in epicardial EMT. We first examined PDGFREKO embryonic hearts for expression of transcriptional inducers of EMT, Snail24, Slug25, and Sox926, 27. Interestingly, while epicardial genes, such as WT1, Tbx18, were unchanged, we consistently observed a significant reduction in Snail, Slug, and Sox9 transcript levels (Figure 2A).
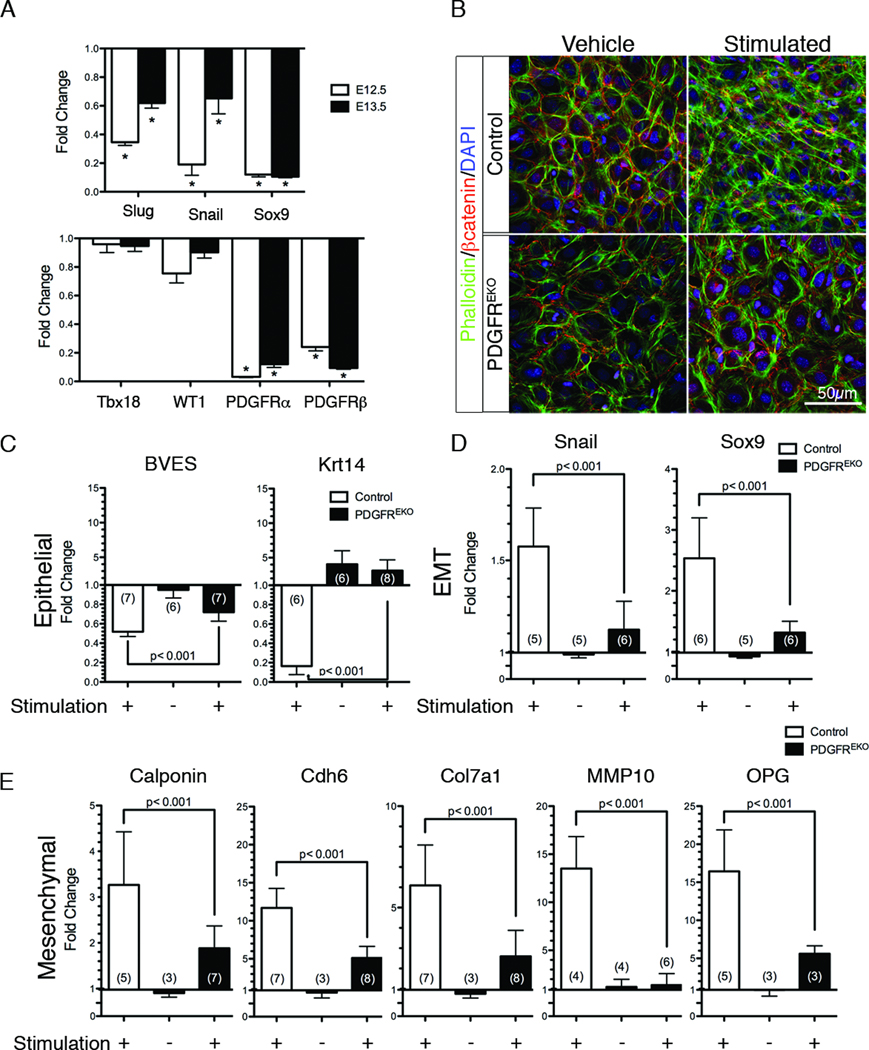
Figure 2. PDGFREKO epicardial cells fail to undergo EMT.
(A) qPCR analysis of gene expression in PDGFREKO whole hearts (atria and conotruncal regions removed) for transcriptional EMT markers (Snail, Slug, Sox9) and epicardial markers (Tbx18, WT1, PDGFRα, PDGFRβ). Data were compared to control littermates and are representative of three independent experiments. (*) p<0.001 (B) βcatenin and phalloidin localization in primary epicardial cultures after 48h of stimulation with hTGFβ1 and PDGFBB. (C–E) qPCR on primary cultures for expression of epithelial, transcriptional, and mesenchymal markers. Data were compared to vehicle treated control epicardial cultures (represented by a baseline of 1.0). Data are represented as mean ± SD. N values are indicated in parentheses.
EMT involves a complex series of events including the loss of epithelial morphology and the acquisition of mesenchymal actin filaments. Therefore, we examined EMT induction in primary epicardial cultures using a cocktail of growth factors. We compared cell morphology by bright field imaging and immunostaining for adherens junctions and filamentous actin organization (βcatenin and phalloidin, respectively). While control cultures lost their epithelial characteristics (junctional βcatenin) and gained mesenchymal cell morphology (cytoplasmic actin stress fibers), PDGFREKO mutant cultures remained epithelial, illustrating a failure to initiate EMT (Figure 2B, Online Figure III, A).
EMT is also associated with changes in gene expression. To evaluate additional EMT markers we performed qPCR analysis. Initial experiments revealed that primary cultures undergo EMT, but many of the genes commonly used to assess EMT were not significantly altered in the stimulated epicardial cultures. For example, we observed no changes in E-cadherin, ZO-1, αSMA and vimentin expression (data not shown). Therefore, to generate an EMT profile specific for primary epicardial cultures, we performed gene expression analysis on E12.5 cultures treated with vehicle or hTGFβ1 (10ng/ml), PDGFBB (20ng/ml), and bFGF (25ng/ml), all three being growth factors known to stimulate EMT28 (GEO Series GSE27181). From these data, we generated a list of candidate genes and verified a subset that correlated with a change from an epithelial to a mesenchymal phenotype. Two epithelial markers that were consistently down-regulated upon EMT induction were Krt1429, 30 and BVES31. We also identified a group of mesenchymal genes that were induced upon EMT induction. This list included Calponin22, Snail32, Sox927, Cdh633, Col7a134, MMP1035, and OPG36–38.
Having established these gene sets, we then investigated their expression during the EMT response in mutant cultures. For every gene examined, we found that expression in the mutant cultures was significantly different from stimulated control cultures (Figure 2C–E). Interestingly, vehicle treated mutant cultures consistently exhibited increased levels of the epithelial gene, Krt14. These data suggest that a defect in the process of EMT was present in PDGFREKO epicardial cells.
Expression of Sox9 in PDGFREKO cells rescues the EMT defect
To determine potential genes that mediate PDGF driven EMT, we screened for gene expression differences using microarray data sets from whole hearts and primary epicardial cultures (GEO Series GSE27181). Comparison of control and PDGFREKO data demonstrated that transcripts of an SRY-related family member, Sox9, were decreased in mutant E12.5 and E13.5 hearts and in primary epicardial cultures (Figure 2A, data not shown).
The correlation of Sox9 transcript levels with PDGF signaling led us to investigate a role for Sox9 in PDGF dependent EMT. In primary epicardial cultures, PDGF stimulation resulted in increased Sox9 expression (Figure 3A). We next determined how Sox9 induced expression impacted these cultures. In the absence of stimulation, Sox9 overexpression had little effect on the cultures, regardless of the genotype. However, AdSox9 transduced cultures stimulated with hTGFβ 1 and PDGFBB changed from an epithelial morphology to a mesenchymal morphology (Figure 3B, Online Figure III, B). These data suggested that additional signaling pathways were required to initiate a Sox9-mediated EMT in our primary epicardial cell cultures, similar to what has been observed in neural crest cells27, 39, but that Sox9 expression could induce EMT even in PDGFREKO epicardial cells.
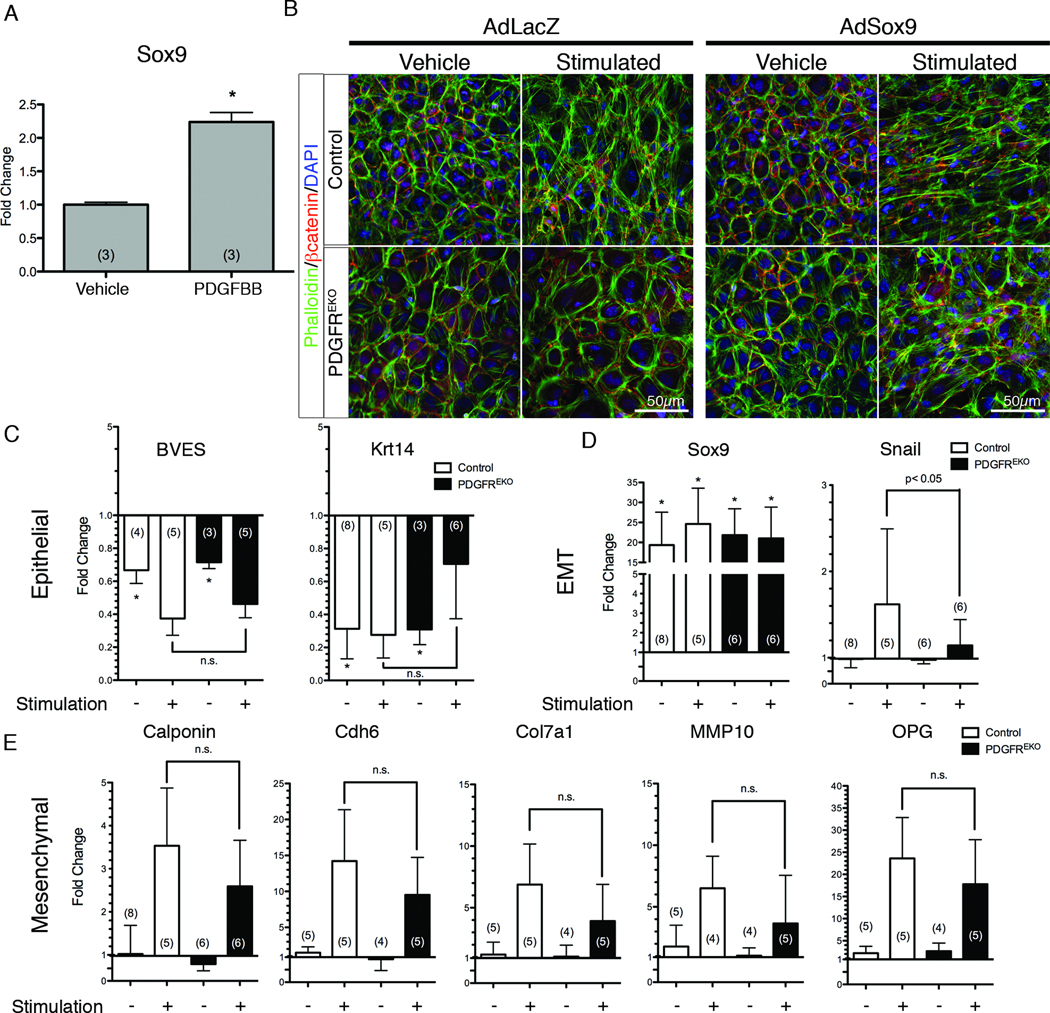
Figure 3. Sox9 rescues PDGF receptor mutant phenotypes.
(A) qPCR for Sox9 expression in primary epicardial cultures stimulated with PDGFBB for 24h. (*) p<0.001 (B) βcatenin and phalloidin localization of control and PDGFREKO cultures stimulated with hTGFβ1 and PDGFBB in the presence of adenoviral LacZ or Sox9. (C–E) qPCR analysis of epithelial, mesenchymal, and EMT transcription factors of primary epicardial cultures transduced with adenoviral Sox9. Data were compared to vehicle treated, GFP adenoviral transduced control epicardial cultures (Online Figure III, D–F) (represented as a baseline of 1.0). Data are represented as mean ± SD. N values are indicated in parentheses. (*) p<0.001 (n.s. – no significant difference)
To examine Sox9’s role in epicardial EMT gene expression, we transduced cultures with AdSox9 (Figure 3C–E). These cultures had reduced epithelial (BVES and Krt14) gene expression, but mesenchymal gene expression remained unchanged. However, similar to the morphological assay, AdSox9 transduced and stimulated (hTGFβ1 and PDGFBB) cultures had both decreased epithelial gene expression and increased mesenchymal gene expression regardless of genotype (Figure 3C–E). Adenoviral transduction alone did not change the gene expression profile of epicardial cells (Online Figure III, D–F).
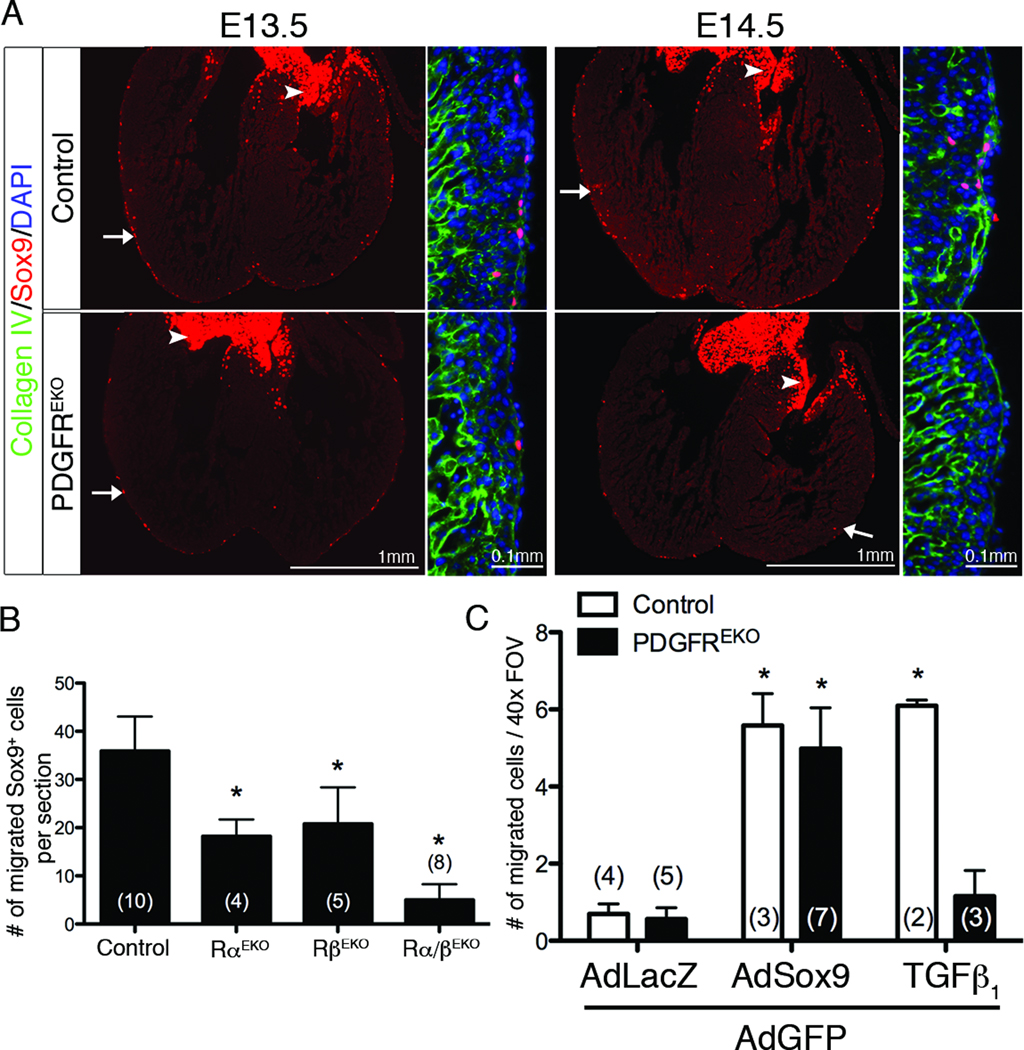
Because Sox9 expression has not been documented in the epicardium previously, we examined hearts for Sox9 protein. Sox9 was present in a subpopulation of epicardial cells at E13.5 (Figure 4A), and a day later, Sox9+ cells were present in both the epicardium and subepicardial mesenchyme (Figure 4A). Using the WT1iCre and R26RYFP alleles, we confirmed that Sox9+ cells are epicardial derived (Online Figure IV, A). In contrast to controls, Sox9 expression in PDGFREKO hearts was significantly reduced at E13.5 and virtually absent at E14.5 (Figure 4A–B). We observed a PDGF receptor gene dosage affect on Sox9 expressing cells that correlated with the number of functional PDGF receptor alleles present (Online Figure IV, B). Individual PDGF receptor epicardial mutants also contained reduced numbers of Sox9+ cells (Figure 4B), suggesting that signaling from either receptor is involved in Sox9 expression.
Figure 4. Epicardial expression of Sox9.
(A) Sox9 expression in hearts of indicated genotypes. Images to the right of each frame are higher magnification of left ventricle. Arrows point to examples of Sox9+ cells. Arrowheads indicate Sox9+ cells in the valves. (B) Quantification of Sox9+ cells within the myocardial ventricular wall at E14.5. Valves, epicardium, and septum were excluded from analysis. (C) Quantification of GFP+ cells within myocardium of E12.5 hearts transduced with indicated viruses and/or stimulation with hTGFβ1. Data are represented as mean ± SD. N values are indicated in parentheses. (*) p<0.001 (compared to vehicle treated AdGFP/AdLacZ control)
To determine if Sox9 could rescue the PDGFREKO epicardial migration defect, we transduced control and PDGFREKO hearts with AdGFP and AdSox9 (Figure 4C). AdSox9 transduction was able to induce migration of epicardial cells from both control and PDGFREKO hearts. This result suggested that in whole heart cultures, Sox9 was sufficient for inducing epicardial EMT but not in isolated epicardial cultures. The myocardium may provide additional cues in vivo. Taken together, these results implicate a role for Sox9 in PDGF receptor dependent EMT and demonstrate that Sox9 can partially rescue the EMT defect caused by PDGF receptor deletion in epicardial cells.
Loss of PDGFRα leads to an EMT defect in a subpopulation of epicardial cells
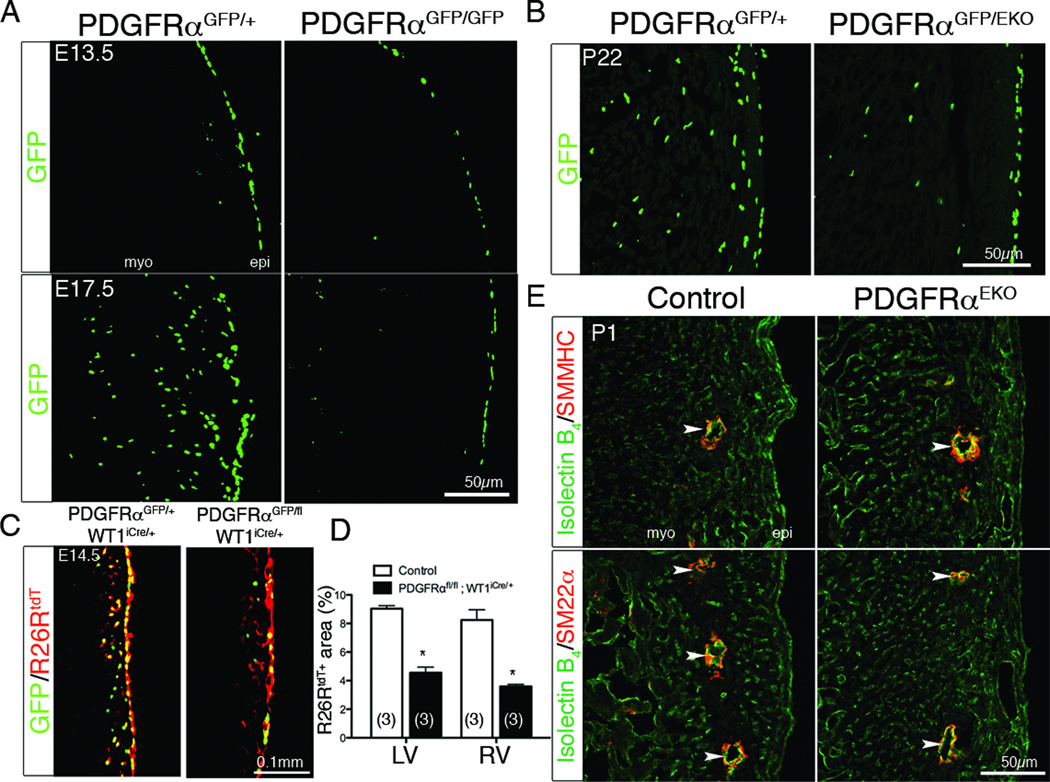
Our data suggested that both PDGF receptors are required for EMT, but results from the PDGFRβ epicardial deletion demonstrated a VSMC lineage defect in epicardial development4. Therefore, we decided to further investigate the individual role of the PDGF receptors during EPDC development. To obtain a more precise view on PDGF receptor expression overlap, we used flow cytometry. We observed that both receptors were initially co-expressed at early embryonic stages (E13.5) but became mutually exclusive at later stages (E16.5) (Online Figure V). Using a GFP knock-in allele7 to follow cells expressing PDGFRα, GFP expression appeared in most epicardial cells at E13.5 (Figure 5A). At E13.5, PDGFRαGFP/GFP mutant hearts exhibited epicardial blistering that was milder than PDGFREKO hearts (data not shown). However, this blistering was less severe than that observed in embryos null for a different PDGFRα allele40. The phenotypic differences described for the PDGFRα null embryos could be caused by differences in genetic background or by the fact that two of these studies (this report and one by Bax et al41) excluded embryos that had retarded growth from analysis. In regards to the number of GFP+ cells in the epicardium, we observed no differences between control and mutant hearts (Figure 5A). However, at E17.5, GFP+ cells were present within the myocardium of heterozygote hearts, but no GFP expressing cells were observed within the myocardium of PDGFRαGFP/GFP hearts. These data suggested that loss of PDGFRα signaling leads to a disruption of a cell population that might arise from the epicardium and is consistent with a recent report showing reduced WT1+ cell migration in PDGFRαGFP/GFP hearts41.
Figure 5. PDGFRα epicardial phenotype.
(A–B) GFP fluorescence was used to follow PDGFRα expressing cells in the indicated genotypes. (C) Confocal images of R26RtdT and PDGFRαGFP fluorescence of the indicated genotypes induced with tamoxifen at E12.5. (D) Quantification of R26RtdT fluorescent area in (C). (*) p<0.001 (E) IHC for coronary endothelial cells (Isolectin B4) and cVSMC (SMMHC and SM22α). (epi – epicardium, myo – myocardium Arrowheads denote coronary vessels).
To determine if the loss of PDGFRα-expressing cells in the myocardium of the null was caused by a failure in PDGFRα dependent cell migration from the epicardium, we deleted PDGFRα in epicardial cells. PDGFRαGFP/EKO hearts showed a reduction of GFP expressing cells within the myocardium, similar to PDGFRαGFP/GFP animals (Figure 5B). Lineage tracing at E12.5 using an inducible, epicardial specific Cre mouse line (WT1iCre)14 and R26RtdT 12 demonstrated that GFP expressing cells were epicardial derived. In addition, when PDGFRα epicardial function was disrupted, migration of PDGFRαGFP positive cells into the heart was reduced (Figure 5C–D).
PDGFRα mutant hearts have a selective loss of cardiac fibroblasts
We next examined if there was a defect in the formation of epicardial derivatives in the absence of PDGF receptor signaling. Because epicardial EMT was disrupted, we expected aberrant cVSMC and cardiac fibroblast development. Surprisingly, the expected Mendelian ratio of PDGFRαEKO, PDGFRβEKO and PDGFREKO mutant animals was recovered at weaning and up to one year after birth. No measurable defects in cardiac size or function were observed (Online Figure VI). Loss of PDGFRβ alone results in an absence of epicardial derived cVSMC, but a secondary population of cVSMC are initially present at the aortic root4, which continues to expand as the animals age (data not shown). This rescue may explain why loss of epicardial cVSMC does not lead to lethality. Examination of the endothelial component of the coronary vasculature suggested that patterning of the vessels in the PDGFREKO hearts was similar to that previously reported for the PDGFRβEKO4, and that endothelial cell presence within the heart was not disrupted by a lack of EPDCs. We determined the consequences of disrupted epicardial EMT by examining hearts for epicardial derivatives. Staining for cVSMC markers, SM22α, smooth muscle myosin heavy chain (SMMHC), α-smooth muscle actin, and PDGFRβ demonstrated that cVSMC content of PDGFRαEKO hearts was unaffected (Figure 5E and data not shown). Because the smooth muscle cell markers that we examined should detect VSMC as well as pericytes, we conclude that loss of PDGFRα does not affect the mural cell lineage. By contrast, PDGFREKO hearts showed a reduction in all of these markers (data not shown) similar to the loss that we reported in PDGFRβ mutant hearts. These data, in combination with the observed loss of GFP+ cells, suggested that PDGFRα might be required for the formation of a distinct EPDC population, cardiac fibroblasts.
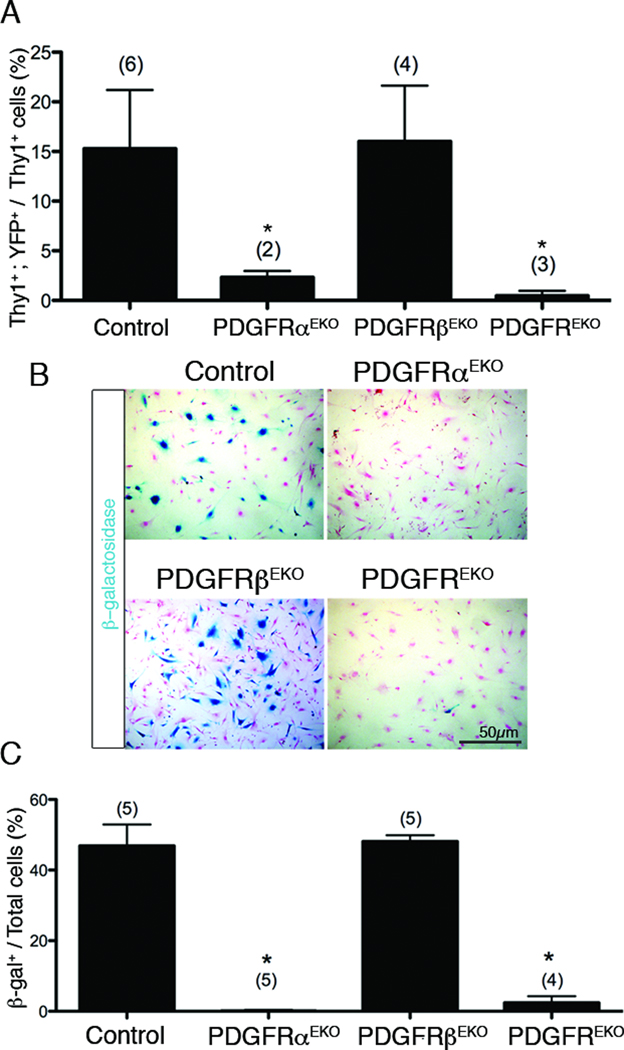
To determine if PDGFRα was required for cardiac fibroblast formation, we analyzed R26RYFP epicardial lineage tagged hearts for a cardiac fibroblast surface marker, Thy142–44. Epicardial derived fibroblasts were defined as YFP+, Thy1+ and CD31− (Figure 6A). Deletion of PDGFRα either individually or in combination with PDGFRβ (PDGFRαEKO and PDGFREKO, respectively) resulted in a loss of epicardial derived cardiac fibroblasts and an absence of YFP+ cells in PDGFREKO hearts. Epicardial derived fibroblast numbers in hearts lacking PDGFRβ (PDGFRβEKO) were similar to controls (Figure 6A). Next, we generated primary cardiac fibroblast cultures and traced the epicardial lineage using Gata5Cre transgene and a R26RLacZ allele to identify EPDCs. Primary cardiac fibroblasts isolated from PDGFREKO and PDGFRαEKO hearts had a paucity of epicardial derived cardiac fibroblasts (β-galactosidase+). By contrast, the number of epicardial derived cardiac fibroblasts observed in controls and PDGFRβEKO hearts were very similar (Figure 6B–C).
Figure 6. PDGFRα is required for epicardial derived cardiac fibroblast formation.
(A) Percentage of epicardially-derived cardiac fibroblasts (Gata5creTg; R26RYFP) from P21–P28 PDGF receptor mutant hearts. There was a subpopulation of Thy1+ CD31+42, 43 cells that were excluded from our analysis. (B) β-galactosidase activity (blue) in passage 1 primary cardiac fibroblast cultures using P21–P28 Gata5creTg; R26RLacZ hearts to follow EPDCs. Nuclear fast red was used as a counterstain. (C) Quantification of blue cells in (B). Data are represented as mean ± SD. N values are indicated in parentheses. (*) p<0.001 (compared to control)
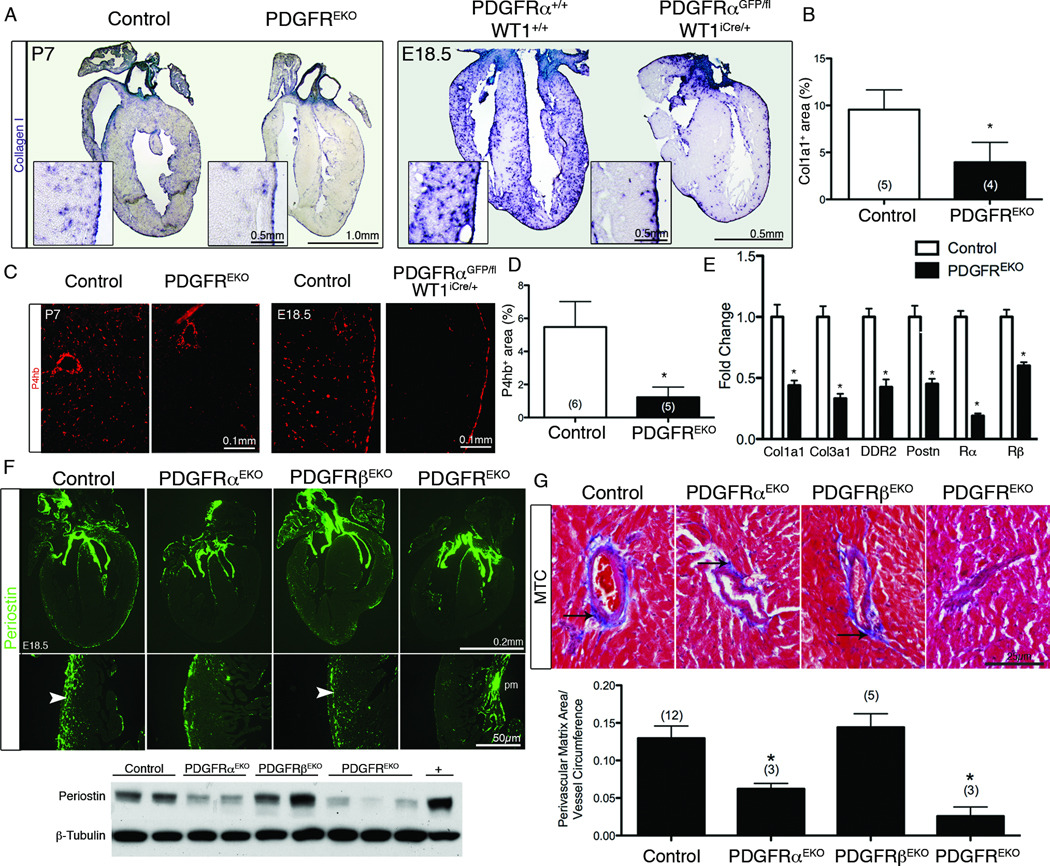
Because a population of cells grew from PDGFREKO and PDGFRαEKO primary fibroblast cultures, we surveyed hearts for overall fibroblast content by detecting transcripts of fibroblast enriched genes. qPCR demonstrated, on average, a 50% reduction in fibroblast gene transcripts in PDGFREKO hearts (Figure 7E). We then used Col1a1 (Figure 7A–B) and prolyl-4-hyroxylase β (P4hb)45 (Figure 7C–D) to identify individual collagen producing cells. To establish the optimal time point to quantify developing fibroblasts, we examined Col1a1 and PDGFRα expression perinatally. Cells expressing these two genes were evident from E18.5 to P7, but after P7 in situ detection of gene expression appeared to taper off (Online Figure VII, A). These data suggested that matrix production by epicardial fibroblasts fell within a very discrete time window. A greater than 50% reduction was observed in the fibroblast population of PDGFREKO at P7 and a reduction in these same fibroblast markers was observed in a PDGFRα epicardial mutant at E18.5 (Figure 7A–D and Online Figure VII, B). These calculations were an over-estimation of remaining fibroblasts as the non-epicardial derived VSMC surrounding the coronary vasculature4, the endocardium, and the epicardium also produced collagen (Figure 7A). To gain insights into the potential source of the cells in primary cardiac fibroblast cultures, we generated fibroblasts from Tie2Cre;R26RLacZ animals, where all cells of endothelial origin should express β-galactosidase, and found that 21±7% (n=4 cultures) of the prolyl-4 hydroxylase and/or αSMA expressing, adherent cells had an endothelial origin as has been previously suggested46.
Figure 7. PDGFRα is required for cardiac fibroblast development.
(A) Col1a1 in situ hybridization of the indicated genotypes and ages. Insets represent higher magnifications of left ventricle. (B) Quantification of interstitial col1a1 expression in (A) as described in online methods. (*) p<0.005 (C) Prolyl-4-hydroxylase β IHC and (D) quantification in the left ventricle of the indicated genotypes. (*) p<0.001. (E) qPCR RNA expression levels in the left ventricle of the indicated genotypes. (n=2 for each genotype) (*) p<0.005 (F) IHC for a fibroblast marker, periostin in E18.5 hearts (upper panel). (pm - papillary muscle) Arrowheads indicate staining present in the ventricular wall. (Lower panel) Western blot of total periostin expression from individual P1 hearts (without aorta, atria, or valves) of indicated genotypes. The positive control is whole cell lysate from differentiated prechondrocyte MC3T3-E1 cells. (G) Masson Trichrome staining (MTC) of coronary arteries in 8–10 week old adult hearts (upper panel). Perivascular collagen deposition is stained blue (arrows). (Lower panel) Quantification of perivascular collagen as seen by MTC. N values are indicated in parentheses. (*) p<0.001 (compared to control)
To determine if a reduction in fibroblasts resulted in any extracellular matrix (ECM) defects, we examined hearts for levels of periostin (Postn), an extracellular matrix molecule secreted by cardiac fibroblasts47. Periostin expression in PDGFRαEKO and PDGFREKO hearts had a marked reduction, while the periostin level in PDGFRβEKO was unaffected (Figure 7F). We also investigated adult mice for generalized defects in matrix deposition. Focusing on perivascular regions, we used Masson Trichrome stain to identify collagen deposition from PDGFRαEKO, PDGFRβEKO, and PDGFREKO hearts. Deletion of PDGFRα either individually or in combination with PDGFRβ (PDGFRαEKO and PDGFREKO, respectively) led to a reduction in collagen, while mice lacking PDGFRβ (PDGFRβEKO) in epicardial cells and their derivatives were similar to controls (Figure 7G).
These results suggested that cardiac fibroblast development is disrupted in PDGFREKO and PDGFRαEKO hearts and that these epicardial derived fibroblasts are required for matrix production in the developing heart. In conclusion, our data show a unique role for PDGF signaling in regulating epicardial EMT and fate specification of EPDCs.
Discussion
Since the discovery of the origin of cVSMC and cardiac fibroblasts over 18 years ago, multiple signaling pathways have been identified that affect the formation, attachment, or EMT of the epicardium. However, few genes have been identified that are essential for cardiac fibroblast formation. Here, we have not only identified a unique requirement for PDGF receptor signaling in regulating epicardial EMT and EPDC formation but also have identified an essential role for, PDGFRα, specifically in cardiac fibroblast formation.
Several growth factor signaling pathways have been implicated in the induction of EMT during development and various pathological states, and PDGF has been linked to the EMT process during cancer progression, organ fibrosis48–50, smooth muscle cell generation from the chicken proepicardium51, and in the regenerating zebrafish heart52. Some suggested mechanisms for PDGF’s role in EMT include stabilization of βcatenin53 or induction of transcriptional activators of EMT, such as ZEB1/2 and Snai254. However, we do not observe these specific effects in epicardial cells after PDGF stimulation. We have identified the transcription factor, Sox9, as a downstream target of PDGF stimulated EMT. Our data show that Sox9 could rescue the EMT defect seen in PDGF receptor mutants and that Sox9 expression was up-regulated upon PDGF stimulation of epicardial cells. These data are consistent with the known role for Sox9 in avian neural crest cell EMT27. Sox9 is a member of the SRY related HMG-box family of transcription factors that is important for the development of many tissues and cell types. Cardiovascular defects have been reported in Sox9 mutants55, 56, but no epicardial phenotype has been described. Our data suggest that Sox9 could be an important component of PDGF receptor signaling during epicardial EMT, but further investigation is necessary to determine the mechanistic link between PDGF and Sox9.
Our data show that loss of both PDGF receptors led to defective EMT and failure to form any epicardial derivatives. Interestingly, individual deletion of the PDGF receptors also led to reduced epicardial EMT and a loss of only a subpopulation of EPDCs. There are two potential scenarios to explain when PDGF receptor function is required. In the first scenario, epicardial cells are heterogeneous, and each epicardial cell would only give rise to a specific lineage of EPDCs, either VSMC or fibroblast. Here, PDGF signaling might regulate a lineage specific EMT. Consistent with this possibility, experiments using limiting amounts of retrovirus to transduce the proepicardial organ57 revealed that tagged cells contributed only to the VSMC lineage. However, there have been no reports suggesting differential gene expression in the epicardium. For example, Tbx18, Tcf21, Raldh2, and both of the PDGF receptors seem to be uniformly expressed in the epicardium prior to EMT. A second possibility for PDGF function is that PDGF signaling by each receptor is redundant in regards to the EMT process, but after EMT, PDGFRα is expressed in fibroblast progenitors, while PDGFRβ is in cVSMC. In this scenario, the most likely role for PDGF receptor signal transduction is expansion and migration of the progenitor population17, 58. These unanswered questions require further investigation using temporal deletion to identify the window of epicardial fate specification.
The role of cardiac fibroblasts in heart pathogenesis is well appreciated, but the function of these cells during development is poorly understood. It has been proposed that cardiac fibroblasts perform a variety of essential duties during heart formation. These include stimulation of cardiomyocyte proliferation43, isolation of the ventricular from the atrial conduction system59, distribution of mechanical forces60, and, of course, deposition and degradation of ECM. Recent estimates are that cardiac fibroblasts comprise about 27% of the cells within the murine heart61, but our data demonstrates that these cells are dispensable for heart development. Under non-pathological conditions, mice without epicardial derived fibroblasts lack adventitial collagen, but heart function is normal.
Because many cell populations have been proposed to contribute to fibrosis formation during pathological circumstances, there is the possibility that another source of fibroblasts fills the void. Proposed origins for this substitute fibroblast population include endothelial cells, fibrocytes, monocytes, and mural cells60. A complete functional substitution by these cells in the absence of epicardial derived fibroblasts is unlikely as mutant hearts that were 8–10 weeks old continued to lack adventitial ECM. Comparison of PDGFRαEKO to PDGFREKO hearts did suggest a partial rescue of the ECM, presumably by the existing cVSMC, although the levels of matrix never appear to reach wild type levels. While our initial examination suggests no major deficits in mutant animals, further studies are warranted to investigate cardiac homeostasis and other functional parameters such as conduction and stress response.
In summary, we demonstrate that cardiac fibroblast and cVSMC development is mediated by a combined role of the PDGF receptors in controlling epicardial EMT. This process appears to be linked to a lineage specific requirement of the receptors. Specifically, PDGFRα, is essential for cardiac fibroblast development. Finally, we establish a novel role for Sox9 as a critical downstream component of PDGF signaling in regulating epicardial EMT.
Novelty and Significance.
What is known?
Cardiac fibroblasts and coronary vascular smooth muscle cells (cVSMC) are epicardial derived cells (EPDC) that arise after an epithelial-to-mesenchymal transition (EMT).
Platelet derived growth factor receptor (PDGFR) β is required for coronary vascular smooth muscle cell development.
PDGF (Platelet derived growth factor) signaling plays a role in coronary vessel remodeling during cardiac zebrafish regeneration.
What new information does this article contribute?
PDGF receptors are required for epicardial EMT and failure of this process leads to animals lacking EPDCs.
Animals lacking EPDCs have no overt phenotype and are viable.
PDGFRα is the first receptor identified to be required for cardiac fibroblast formation, but PDGFRα is dispensable for cVSMC development.
The PDGF receptor genes are required in a lineage specific manner for the formation of the two EPDC cell populations, cVSMC and cardiac fibroblasts.
Cardiac fibrosis is a major consequence of long-term cardiac disease, and the epicardium is the major source of resident cardiac fibroblasts that potentially contribute to this disease. Here, we report that PDGF receptor signaling is required for epicardial EMT. Expression of the transcription factor, Sox9, is reduced in epicardial cells lacking PDGF receptors, and expression of Sox9 rescues EMT in the absence of PDGF receptor signaling. We also report that disruption of the epicardial EMT process leads to the inability to generate cardiac fibroblasts and cVSMC. Additionally, the loss of PDGFRα leads to a defect exclusively in cardiac fibroblast formation. This work is the first example of a lineage specific disruption of epicardial derivatives. Our findings show a previously unidentified role for PDGF receptor signaling in epicardial EMT and EPDC fate specification and provide a novel model to investigate the role of cardiac fibroblasts during embryogenesis as well as in the adult.
Supplementary Material
Acknowledgements
We are grateful to Eric Olson, Ondine Cleaver, and the members of the Tallquist laboratory for critical reading of the manuscript. We also thank Dr. Leon Avery for statistical assistance.
Sources of Funding
This work was supported by NHLBI R01 and U01 grants HL074257 and HL100401, respectively (to M.D.T.); AHA Scientific Development Grant 0330351 (to M.D.T.); AHA predoctoral grant (09PRE2280197, 10PRE3730051) (to C.L.S. and S.T.B, respectively); NIH Ruth Kirschstein predoctoral fellowship F30 (1F30HL096277-01A1) (to C.L.S.); and Institutional Cardiology Training Grant (T32 HL007360-32) (to C.L.S and C.Y.S.).
Abbreviations
- PDGF
platelet derived growth factor
- EPDC
epicardial derived cell
- cVSMC
coronary vascular smooth muscle cell
- EMT
epithelial to mesenchymal transition
- myo
myocardium
- epi
epicardium
- LV
left ventricle
- RV
right ventricle
- EKO
epicardial knockout
- pm
papillary muscle
- SEM
subepicardial mesenchyme
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None.
References
- 1.Winter EM, Gittenberger-de Groot AC. Epicardium-derived cells in cardiogenesis and cardiac regeneration. Cell Mol Life Sci. 2007;64:692–703. doi: 10.1007/s00018-007-6522-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olivey HE, Svensson EC. Epicardial-myocardial signaling directing coronary vasculogenesis. Circ Res. 2010;106:818–832. doi: 10.1161/CIRCRESAHA.109.209197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrae J, Gallini R, Betsholtz C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 2008;22:1276–1312. doi: 10.1101/gad.1653708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mellgren AM, Smith CL, Olsen GS, Eskiocak B, Zhou B, Kazi MN, Ruiz FR, Pu WT, Tallquist MD. Platelet-derived growth factor receptor beta signaling is required for efficient epicardial cell migration and development of two distinct coronary vascular smooth muscle cell populations. Circ Res. 2008;103:1393–1401. doi: 10.1161/CIRCRESAHA.108.176768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang J, Gu Y, Li P, Johnson BL, Sucov HM, Thomas PS. Pdgf-a as an epicardial mitogen during heart development. Dev Dyn. 2008;237:692–701. doi: 10.1002/dvdy.21469. [DOI] [PubMed] [Google Scholar]
- 6.Bax NA, Lie-Venema H, Vicente-Steijn R, Bleyl SB, Van Den Akker NM, Maas S, Poelmann RE, Gittenberger-de Groot AC. Platelet-derived growth factor is involved in the differentiation of second heart field-derived cardiac structures in chicken embryos. Dev Dyn. 2009;238:2658–2669. doi: 10.1002/dvdy.22073. [DOI] [PubMed] [Google Scholar]
- 7.Hamilton TG, Klinghoffer RA, Corrin PD, Soriano P. Evolutionary divergence of platelet-derived growth factor alpha receptor signaling mechanisms. Mol Cell Biol. 2003;23:4013–4025. doi: 10.1128/MCB.23.11.4013-4025.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tallquist MD, French WJ, Soriano P. Additive effects of pdgf receptor beta signaling pathways in vascular smooth muscle cell development. PLoS Biol. 2003;1:E52. doi: 10.1371/journal.pbio.0000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Richarte AM, Mead HB, Tallquist MD. Cooperation between the pdgf receptors in cardiac neural crest cell migration. Dev Biol. 2007;306:785–796. doi: 10.1016/j.ydbio.2007.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmahl J, Rizzolo K, Soriano P. The pdgf signaling pathway controls multiple steroid-producing lineages. Genes Dev. 2008;22:3255–3267. doi: 10.1101/gad.1723908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Srinivas S, Watanabe T, Lin CS, William CM, Tanabe Y, Jessell TM, Costantini F. Cre reporter strains produced by targeted insertion of eyfp and ecfp into the rosa26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soriano P. Generalized lacz expression with the rosa26 cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- 14.Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang D, von Gise A, Ikeda S, Chien KR, Pu WT. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kisanuki YY, Hammer RE, Miyazaki J, Williams SC, Richardson JA, Yanagisawa M. Tie2-cre transgenic mice: A new model for endothelial cell-lineage analysis in vivo. Dev Biol. 2001;230:230–242. doi: 10.1006/dbio.2000.0106. [DOI] [PubMed] [Google Scholar]
- 16.Merki E, Zamora M, Raya A, Kawakami Y, Wang J, Zhang X, Burch J, Kubalak SW, Kaliman P, Belmonte JC, Chien KR, Ruiz-Lozano P. Epicardial retinoid x receptor alpha is required for myocardial growth and coronary artery formation. Proc Natl Acad Sci U S A. 2005;102:18455–18460. doi: 10.1073/pnas.0504343102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoch RV, Soriano P. Roles of pdgf in animal development. Development. 2003;130:4769–4784. doi: 10.1242/dev.00721. [DOI] [PubMed] [Google Scholar]
- 18.Zamora M, Manner J, Ruiz-Lozano P. Epicardium-derived progenitor cells require beta-catenin for coronary artery formation. Proc Natl Acad Sci U S A. 2007;104:18109–18114. doi: 10.1073/pnas.0702415104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore AW, McInnes L, Kreidberg J, Hastie ND, Schedl A. Yac complementation shows a requirement for wt1 in the development of epicardium, adrenal gland and throughout nephrogenesis. Development. 1999;126:1845–1857. doi: 10.1242/dev.126.9.1845. [DOI] [PubMed] [Google Scholar]
- 20.Perez-Pomares JM, Macias D, Garcia-Garrido L, Munoz-Chapuli R. Contribution of the primitive epicardium to the subepicardial mesenchyme in hamster and chick embryos. Dev Dyn. 1997;210:96–105. doi: 10.1002/(SICI)1097-0177(199710)210:2<96::AID-AJA3>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 21.Red-Horse K, Ueno H, Weissman IL, Krasnow MA. Coronary arteries form by developmental reprogramming of venous cells. Nature. 2010;464:549–553. doi: 10.1038/nature08873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Compton LA, Potash DA, Mundell NA, Barnett JV. Transforming growth factor-beta induces loss of epithelial character and smooth muscle cell differentiation in epicardial cells. Dev Dyn. 2006;235:82–93. doi: 10.1002/dvdy.20629. [DOI] [PubMed] [Google Scholar]
- 23.Morabito CJ, Dettman RW, Kattan J, Collier JM, Bristow J. Positive and negative regulation of epicardial-mesenchymal transformation during avian heart development. Dev Biol. 2001;234:204–215. doi: 10.1006/dbio.2001.0254. [DOI] [PubMed] [Google Scholar]
- 24.Veltmaat JM, Orelio CC, Ward-Van Oostwaard D, Van Rooijen MA, Mummery CL, Defize LH. Snail is an immediate early target gene of parathyroid hormone related peptide signaling in parietal endoderm formation. Int J Dev Biol. 2000;44:297–307. [PubMed] [Google Scholar]
- 25.Savagner P, Yamada KM, Thiery JP. The zinc-finger protein slug causes desmosome dissociation, an initial and necessary step for growth factor-induced epithelial-mesenchymal transition. J Cell Biol. 1997;137:1403–1419. doi: 10.1083/jcb.137.6.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung M, Chaboissier MC, Mynett A, Hirst E, Schedl A, Briscoe J. The transcriptional control of trunk neural crest induction, survival, and delamination. Dev Cell. 2005;8:179–192. doi: 10.1016/j.devcel.2004.12.010. [DOI] [PubMed] [Google Scholar]
- 27.Sakai D, Suzuki T, Osumi N, Wakamatsu Y. Cooperative action of sox9, snail2 and pka signaling in early neural crest development. Development. 2006;133:1323–1333. doi: 10.1242/dev.02297. [DOI] [PubMed] [Google Scholar]
- 28.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 29.Chamulitrat W, Schmidt R, Chunglok W, Kohl A, Tomakidi P. Epithelium and fibroblast-like phenotypes derived from hpv16 e6/e7-immortalized human gingival keratinocytes following chronic ethanol treatment. Eur J Cell Biol. 2003;82:313–322. doi: 10.1078/0171-9335-00317. [DOI] [PubMed] [Google Scholar]
- 30.Ke XS, Qu Y, Goldfinger N, Rostad K, Hovland R, Akslen LA, Rotter V, Oyan AM, Kalland KH. Epithelial to mesenchymal transition of a primary prostate cell line with switches of cell adhesion modules but without malignant transformation. PLoS One. 2008;3:e3368. doi: 10.1371/journal.pone.0003368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wada AM, Reese DE, Bader DM. Bves: Prototype of a new class of cell adhesion molecules expressed during coronary artery development. Development. 2001;128:2085–2093. doi: 10.1242/dev.128.11.2085. [DOI] [PubMed] [Google Scholar]
- 32.Murray SA, Carver EA, Gridley T. Generation of a snail1 (snai1) conditional null allele. Genesis. 2006;44:7–11. doi: 10.1002/gene.20178. [DOI] [PubMed] [Google Scholar]
- 33.Inoue T, Inoue YU, Asami J, Izumi H, Nakamura S, Krumlauf R. Analysis of mouse cdh6 gene regulation by transgenesis of modified bacterial artificial chromosomes. Dev Biol. 2008;315:506–520. doi: 10.1016/j.ydbio.2007.12.011. [DOI] [PubMed] [Google Scholar]
- 34.Vindevoghel L, Lechleider RJ, Kon A, de Caestecker MP, Uitto J, Roberts AB, Mauviel A. Smad3/4-dependent transcriptional activation of the human type vii collagen gene (col7a1) promoter by transforming growth factor beta. Proc Natl Acad Sci U S A. 1998;95:14769–14774. doi: 10.1073/pnas.95.25.14769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkins-Port CE, Higgins PJ. Regulation of extracellular matrix remodeling following transforming growth factor-beta1/epidermal growth factor-stimulated epithelial-mesenchymal transition in human premalignant keratinocytes. Cells Tissues Organs. 2007;185:116–122. doi: 10.1159/000101312. [DOI] [PubMed] [Google Scholar]
- 36.Sakata M, Shiba H, Komatsuzawa H, Fujita T, Ohta K, Sugai M, Suginaka H, Kurihara H. Expression of osteoprotegerin (osteoclastogenesis inhibitory factor) in cultures of human dental mesenchymal cells and epithelial cells. J Bone Miner Res. 1999;14:1486–1492. doi: 10.1359/jbmr.1999.14.9.1486. [DOI] [PubMed] [Google Scholar]
- 37.Corallini F, Gonelli A, D'Aurizio F, di Iasio MG, Vaccarezza M. Mesenchymal stem cells-derived vascular smooth muscle cells release abundant levels of osteoprotegerin. Eur J Histochem. 2009;53:19–24. doi: 10.4081/ejh.2009.19. [DOI] [PubMed] [Google Scholar]
- 38.Vidal NO, Brandstrom H, Jonsson KB, Ohlsson C. Osteoprotegerin mrna is expressed in primary human osteoblast-like cells: Down-regulation by glucocorticoids. J Endocrinol. 1998;159:191–195. doi: 10.1677/joe.0.1590191. [DOI] [PubMed] [Google Scholar]
- 39.Cheung M, Briscoe J. Neural crest development is regulated by the transcription factor sox9. Development. 2003;130:5681–5693. doi: 10.1242/dev.00808. [DOI] [PubMed] [Google Scholar]
- 40.Soriano P. The pdgf alpha receptor is required for neural crest cell development and for normal patterning of the somites. Development. 1997;124:2691–2700. doi: 10.1242/dev.124.14.2691. [DOI] [PubMed] [Google Scholar]
- 41.Bax NA, Bleyl VC, Gallini R, Wisse LJ, Hunter J, Van Oorschot AA, Mahtab EA, Lie-Venema H, Goumans MJ, Betsholtz C, Gittenberger-de Groot AC. Cardiac malformations in pdgfralpha mutant embryos are associated with increased expression of wt1 and nkx2.5 in the second heart field. Dev Dyn. 2010;239:2307–2317. doi: 10.1002/dvdy.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hudon-David F, Bouzeghrane F, Couture P, Thibault G. Thy-1 expression by cardiac fibroblasts: Lack of association with myofibroblast contractile markers. J Mol Cell Cardiol. 2007;42:991–1000. doi: 10.1016/j.yjmcc.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Ieda M, Tsuchihashi T, Ivey KN, Ross RS, Hong TT, Shaw RM, Srivastava D. Cardiac fibroblasts regulate myocardial proliferation through beta1 integrin signaling. Dev Cell. 2009;16:233–244. doi: 10.1016/j.devcel.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takeda N, Manabe I, Uchino Y, Eguchi K, Matsumoto S, Nishimura S, Shindo T, Sano M, Otsu KC, Snider P, Conway SJ, Nagai R. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J Clin Invest. 2010;120:254–265. doi: 10.1172/JCI40295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kivirikko KI, Myllyla R, Pihlajaniemi T. Protein hydroxylation: Prolyl 4-hydroxylase, an enzyme with four cosubstrates and a multifunctional subunit. FASEB J. 1989;3:1609–1617. [PubMed] [Google Scholar]
- 46.Zeisberg EM, Tarnavski O, Zeisberg M, Dorfman AL, McMullen JR, Gustafsson E, Chandraker A, Yuan X, Pu WT, Roberts AB, Neilson EG, Sayegh MH, Izumo S, Kalluri R. Endothelial-to-mesenchymal transition contributes to cardiac fibrosis. Nat Med. 2007;13:952–961. doi: 10.1038/nm1613. [DOI] [PubMed] [Google Scholar]
- 47.Katsuragi N, Morishita R, Nakamura N, Ochiai T, Taniyama Y, Hasegawa Y, Kawashima K, Kaneda Y, Ogihara T, Sugimura K. Periostin as a novel factor responsible for ventricular dilation. Circulation. 2004;110:1806–1813. doi: 10.1161/01.CIR.0000142607.33398.54. [DOI] [PubMed] [Google Scholar]
- 48.Fischer AN, Fuchs E, Mikula M, Huber H, Beug H, Mikulits W. Pdgf essentially links tgf-beta signaling to nuclear beta-catenin accumulation in hepatocellular carcinoma progression. Oncogene. 2007;26:3395–3405. doi: 10.1038/sj.onc.1210121. [DOI] [PubMed] [Google Scholar]
- 49.Kong D, Wang Z, Sarkar SH, Li Y, Banerjee S, Saliganan A, Kim HR, Cher ML, Sarkar FH. Platelet-derived growth factor-d overexpression contributes to epithelial-mesenchymal transition of pc3 prostate cancer cells. Stem Cells. 2008;26:1425–1435. doi: 10.1634/stemcells.2007-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ostendorf T, Rong S, Boor P, Wiedemann S, Kunter U, Haubold U, van Roeyen CR, Eitner F, Kawachi H, Starling G, Alvarez E, Smithson G, Floege J. Antagonism of pdgf-d by human antibody cr002 prevents renal scarring in experimental glomerulonephritis. J Am Soc Nephrol. 2006;17:1054–1062. doi: 10.1681/ASN.2005070683. [DOI] [PubMed] [Google Scholar]
- 51.Lu J, Landerholm TE, Wei JS, Dong XR, Wu SP, Liu X, Nagata K, Inagaki M, Majesky MW. Coronary smooth muscle differentiation from proepicardial cells requires rhoa-mediated actin reorganization and p160 rho-kinase activity. Dev Biol. 2001;240:404–418. doi: 10.1006/dbio.2001.0403. [DOI] [PubMed] [Google Scholar]
- 52.Kim J, Wu Q, Zhang Y, Wiens KM, Huang Y, Rubin N, Shimada H, Handin RI, Chao MY, Tuan TL, Starnes VA, Lien CL. Pdgf signaling is required for epicardial function and blood vessel formation in regenerating zebrafish hearts. Proc Natl Acad Sci U S A. 2010;107:17206–17210. doi: 10.1073/pnas.0915016107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang L, Lin C, Liu ZR. P68 rna helicase mediates pdgf-induced epithelial mesenchymal transition by displacing axin from beta-catenin. Cell. 2006;127:139–155. doi: 10.1016/j.cell.2006.08.036. [DOI] [PubMed] [Google Scholar]
- 54.Kong D, Li Y, Wang Z, Banerjee S, Ahmad A, Kim HR, Sarkar FH. Mir-200 regulates pdgf-d-mediated epithelial-mesenchymal transition, adhesion, and invasion of prostate cancer cells. Stem Cells. 2009;27:1712–1721. doi: 10.1002/stem.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bi W, Huang W, Whitworth DJ, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Haploinsufficiency of sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98:6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Akiyama H, Chaboissier MC, Behringer RR, Rowitch DH, Schedl A, Epstein JA, de Crombrugghe B. Essential role of sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci U S A. 2004;101:6502–6507. doi: 10.1073/pnas.0401711101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- 58.Betsholtz C. Biology of platelet-derived growth factors in development. Birth Defects Res C Embryo Today. 2003;69:272–285. doi: 10.1002/bdrc.10030. [DOI] [PubMed] [Google Scholar]
- 59.Kolditz DP, Wijffels MC, Blom NA, van der Laarse A, Hahurij ND, Lie-Venema H, Markwald RR, Poelmann RE, Schalij MJ, Gittenberger-de Groot AC. Epicardium-derived cells in development of annulus fibrosis and persistence of accessory pathways. Circulation. 2008;117:1508–1517. doi: 10.1161/CIRCULATIONAHA.107.726315. [DOI] [PubMed] [Google Scholar]
- 60.Krenning G, Zeisberg EM, Kalluri R. The origin of fibroblasts and mechanism of cardiac fibrosis. J Cell Physiol. 2010;225:631–637. doi: 10.1002/jcp.22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Banerjee I, Fuseler JW, Price RL, Borg TK, Baudino TA. Determination of cell types and numbers during cardiac development in the neonatal and adult rat and mouse. Am J Physiol Heart Circ Physiol. 2007;293:H1883–H1891. doi: 10.1152/ajpheart.00514.2007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.