Abstract
Staphylococcus aureus, a versatile human pathogen, is commonly associated with medical device infections. Its capacity to establish and maintain these infections is thought to be related to its ability to form adherent biofilms. In this study, commercially available α-amylase compounds from various biological sources were evaluated for their ability to reduce and prevent biofilm formation of several S. aureus isolates. Our data demonstrates that α-amylase compounds can rapidly detach biofilms of S. aureus, as well as inhibit biofilm formation. Our data also demonstrates that α-amylase compounds have an ability to reduce and disassociate S. aureus cell-aggregates grown in liquid suspension. These findings suggest that commercially available α-amylase compounds could be used in the future to control S. aureus biofilm-related infections.
Keywords: Staphylococcus aureus, α-amylases, Biofilm control, Biofilm-dispersing enzymes.
INTRODUCTION
Staphylococcus aureus is an important human pathogen, particularly in hospital settings, where it is a major source of life threatening bloodstream infections as well as a leading cause of implants and indwelling medical device infections [1-3]. S. aureus is well adapted to the human host and has a large number of factors that enable it to adhere to specific host substrates, evade host defenses, and resist antibiotic therapy [4-7]. One way in which the bacteria become resistant to antibiotics and host defenses is believed to be through biofilm formation [8, 9].
Biofilms are defined as communities of microorganisms that are encased in a self-synthesized extracellular polymeric matrix (EPS) and grow attached to a biotic or abiotic surface [10, 11]. Bacterial biofilms have a significant impact on human health [12, 13], as they frequently exhibit an enhanced pathogenic capability relative to bacteria in solution by virtue of sessile behavior, increased resistance to antimicrobial agents, and the potential for detachment and distal embolization of large biofilm fragments [14].
The increase in biofilm-related infection and the necessity of both preventing and removing microbial biofilms from an infected site has led researchers to examine new ways to control surface attached bacteria. Among the techniques proposed to control biofilms is the use of materials and coatings aimed at inhibiting initial cell adhesion, as well as a variety of treatments designed to reduce pre-existing biofilms, such as heat, cleaning regimens, low-power laser, sonication, chemical treatments, antibiotics, quorum sensing analogs, lectins, and biological control agents such as invertebrates, protozoa, bacteriophages, and predatory bacteria [15-25]. Another biofilm control strategy makes use of enzymes that degrade major structural components of the EPS, resulting in removal of the biofilm. Among the EPS degrading enzymes that showed potential as anti S. aureus biofilm agents are deoxyribonuclease I (DNase I) and dispersin B (DspB) [26-30]. DNase I was shown to degrade extracellular DNA, which functions as a matrix adhesion molecule in biofilms produced by most bacterial species [31], whereas DspB degrades poly-β-1,6-N-acetyl-D-glucosamine (PNAG), an extracellular polysaccharide that also functions in biofilm matrix adhesion in numerous phylogenetically diverse gram-negative and gram-positive bacteria [32-34].
In this study, we examined the potential use of commercially available α-amylase compounds as a method for inhibiting both cell-to-cell and cell-to-surface association of S. aureus, as well as the aptitude of the enzyme to detach pre-existing aggregates and biofilms of S. aureus.
MATERIALS AND METHODS
Bacterial Strains and Culture Conditions
The following S. aureus strains were used in this study: SH1000 (provided by Jeffrey Kaplan, Department of Oral Biology, UMDNJ) [28], SMC3256, a methicillin-resistant conjunctivitis clinical isolate, MU50, a vancomycin intermediate resistance conjunctivitis clinical isolate (both provided by Robert Shanks, Department of Ophthalmology, UPMC) [35], and 6 methicillin-sensitive S. aureus endophalmitis isolates, E253, E417, E424, E425, E427, and E442 (kindly provided by the Campbell Laboratory of Ophthalmic Microbiology, Department of Ophthalmology, UPMC). Staphylococcus epidermidis NJ9709 was also used in the study [26].
Enzymes
The enzymes used in this study were: α-amylase from Aspergillus oryzae (Sigma-Aldrich, St. Louis, MO, Catalog #10065), α-amylase from Bacillus subtilis (Sigma, catalog #10070), α-amylase from human saliva (Sigma, catalog #A1031), and β-amylase from sweet potato (Sigma, catalog #A7005).
Biofilm Formation Assay
Biofilm assay was performed similar to the previously described methods [36]. Overnight cultures of staphylococci were grown in liquid broth. Cultures were diluted, to an A600 of 0.01 (~1x108 CFU/ml), in tryptic soy broth (TSB) supplemented with 0.6% yeast extract, 0.8% glucose, and 0.2% sodium citrate, to give TSB citrate media (TSBC). This medium composition was selected for its ability to stimulate biofilm formation in S. aureus [35, 37]. One hundred microliters of cells were added to individual wells of tissue culture-treated, 96-well polystyrene microtiter dishes (Costar, Corning Inc., Corning, NY). These were incubated in a closed, humidified plastic container for 18 hrs at 37oC before being assayed for biofilm formation. Non-adherent cells were removed by washing and adherent cells were stained with 0.1% crystal violet (CV) as previously described [38]. S. epidermidis biofilms were developed in TSB media [26]. Photographs of the plate were taken with a Canon-scan 4400F digital scanner, then the crystal violet was solubilized using 50% acetic acid for 10 min. Relative biofilm formation was assayed by measuring the absorbance of the crystal violet solution at 600 nm (OD600). In addition to CV, biofilms were also stained and visualized by using 1 mg/ml Congo-red (Sigma-Aldrich, St. Louis, MO), and 1 µl/ml Pico-green dsDNA stain (Invitrogen, Eugene, OR). For biofilm formation on polyurethane, a polymer routinely used in the manufacture of medical devices, a polyurethane tube (Nalgene, Rochester, NY) was cut into 3mm pieces and inserted into a 24-well costar plate. The well was filled with 0.5 ml of TSBC media containing S. aureus SH1000 cells and incubated for 18 hrs to allow biofilm development. For statistical analyses, P values were determined by using a Student t-test performed with Microsoft Excel software. Error bars are shown as one standard deviation.
Cell Aggregation Assay
Overnight cultures of staphylococci were diluted 1:1000 in TSBC. Five ml of the cell suspension was placed in an 18 mm glass tube and incubated for 18 hr at 37oC on a TC-7 tissue culture roller drum (New Brunswick Sci, New-Brunswick, NJ) set on 30 RPM. To examine the aggregates, the cultures were poured into a 90 mm Petri-dish and visually examined. Fluorescent microscopy was used after 1 μl of Calcofluor-white (Sigma-Aldrich, St. Louis, MO) was added to the tube. To measure the extent of aggregation, the overnight culture tubes containing the aggregates were left to stand for 20 min to allow aggregates to settle to the bottom of the tube. Turbidity of the suspension (optical density of the suspension [ODs]) was measured at 600 nm. The culture was then dispersed by a 10s sonication step, using a VC505 sonicator (Sonics and Materials Inc., Newtown, CT) and the total turbidity was measured (ODt). The percentage of aggregation was estimated as follows: % aggregation =[(ODt – ODs) x 100]/ODt [21, 39]. As for the biofilm assay, TSBC medium was chosen for its ability to stimulate S. aureus cell clustering and aggregation [35, 37].
Inhibition and Reduction Assays
To assess the ability of the enzyme to inhibit biofilm formation or the formation of cell clusters (aggregates), biofilm or aggregation tubes were incubated for 18 hrs with or without (control) the enzyme before being analyzed. To measure the ability of the enzyme to reduce an existing biofilm, cells were first grown in 96-well plates for 18 hrs (pre-formed biofilm). Thereafter, non-adherent cells were removed and fresh TSBC medium, containing the enzyme, was added to the biofilm and incubated at 37oC for the duration of the experiment. To evaluate the ability of the enzyme to disassociate S. aureus aggregates, enzyme was added directly to the tube containing overnight aggregates (pre-formed aggregates). The tubes were returned to incubate on the roller drum following aggregation quantification.
Scanning Electron Microscopy
Experiments were performed as described previously [20, 21]. In brief, S. aureus biofilms were developed on a 12x22 mm PVC plastic cover slip. The cover slips were placed in a 24-well polystyrene cell culture plate and incubated for 18 hrs. Biofilms and enzymatic assays were prepared as described above. S. aureus biofilms were rinsed to remove any planktonic cells before being fixed in 2% glutaraldehyde, 0.1 M sodium cacodylate, and 0.1% ruthenium red. Images were viewed at the air-liquid interface using a Hitachi S-2500 scanning electron microscope (SEM).
RESULTS
Biofilm Reduction and Inhibition Experiments
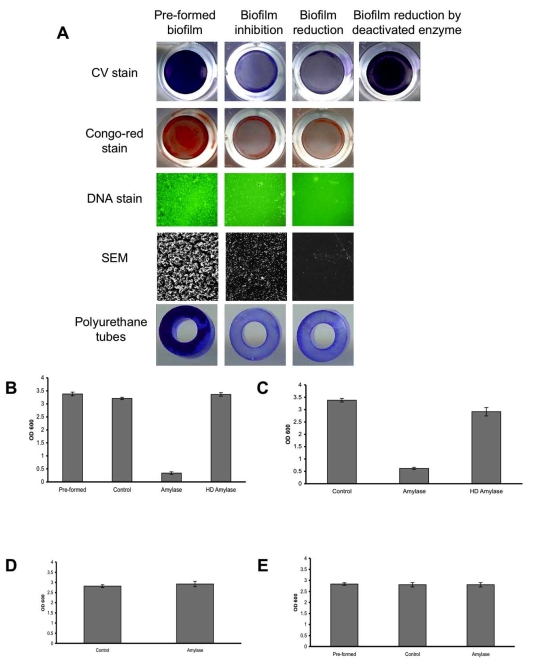
To assess the ability of α-amylase to reduce an existing S. aureus biofilm, S. aureus SH1000 biofilms were developed in 96-well plates. Thereafter, the biofilms were washed and fresh TSBC media containing 100 mg/ml of α-amylase compound from A. oryzae was added to the wells. The plate was incubated for 10 min at 37oC before being analyzed. As seen in Fig. (1A), a clear decrease in CV staining was observed in the amylase treated wells (Biofilm-reduction) compared to the initial biofilm (Pre-formed biofilm). The change in biofilm biomass was further assessed using CV quantification. A significant 90% (P<0.001) reduction in CV staining was measured following a 10 min incubation period with the enzyme (1B, amylase) with a 5% drop in the control sample containing TSBC alone (1B, control). To further validate that the decrease in biofilm biomass is due to the enzyme activity, the enzyme was heat-deactivated (85oC for 120 min). As seen in Fig. (1A and 1B), no reduction in CV staining was observed in the biofilms after incubation with the heat-deactivated enzyme. To assess the effect of the enzyme on a polysaccharide-based component that might be present in the biofilm, parallel wells were stained with Congo-red. A noticeable drop in biofilm Congo-red staining was seen following a 10 min enzyme treatment, compared to the robust staining in the initial biofilm (1A, Pre-formed) and in the control treated sample (data not shown). By using DNA stain and SEM imaging, we were able to further visualize changes in biofilm cell-surface coverage. A clear decrease in biofilm cell density was seen in the enzyme treated samples when compared to the initial biofilm, with the latter exhibiting a substantial biofilm cell population attached to the surface (Fig. 1A).
Fig. (1).
Biofilm reduction and inhibition assay. (A) S. aureus SH1000 biofilms were developed for 18 hrs with (biofilm inhibition) or without (pre-formed biofilm) the addition of 100 mg/ml α-amylase. In another experiment, S. aureus biofilms (pre-formed biofilm) were rinsed and exposed for 10 min to the enzyme (biofilm reduction) or to a heat-deactivated enzyme. Biofilms were rinsed and stained with CV, Congo-red or Pico-green (DNA stain). The experiments were also carried out on S. aureus biofilms developed on polyurethane tubes. For SEM imaging, biofilms were developed on PVC plastic cover slips. DNA stain was visualized under fluorescent microscope at 400X magnification. Scanning electron micrographs were viewed at 500X magnification. Each experiment was carried out at least six times yielding similar results. (B) Quantification of biofilm reduction. Pre-formed overnight biofilms (Pre-formed) were incubated for 10 min with fresh TSBC media (Control), TSBC supplemented with 100 mg/ml enzyme (amylase), or heat deactivated enzyme (HD amylase). The wells were rinsed, stained with CV, and the amount of CV staining was quantified at A600 (OD 600). (C) Quantification of biofilm inhibition. S. aureus cells were cultured in 96-well plates with TSBC media (Control), media supplemented with 100 mg/ml enzyme (amylase), or heat deactivated enzyme (HD amylase). After 18 hrs the plates were rinsed, stained with CV, and the amount of CV staining was quantified. (D) Quantification of S. epidermidis biofilm inhibition. S. epidermidis cells were cultured in 96-well plates with TSB media (Control) and media supplemented with 100 mg/ml enzyme (amylase). After 18 hrs, the plates were rinsed, stained with CV, and the amount of CV staining was quantified. (E) Quantification of S. epidermidis biofilm reduction. Pre-formed overnight biofilms (Pre-formed) were incubated for 4 hrs with fresh TSB media (Control) or TSB supplemented with 100 mg/ml enzyme (amylase). The wells were rinsed, stained with CV, and the amount of CV staining was quantified at A600 (OD 600). Each value represents the mean of 24 wells from one representative experiment. Error bars are shown as one-standard deviation. Each experiment was carried out three times yielding similar results.
In another experiment, the ability of the enzyme compound to inhibit biofilm formation was evaluated. S. aureus SH1000 cells were incubated in TSBC media (control) or TSBC media supplemented with 100 mg/ml of α-amylase from A. oryzae. The biofilms were left to develop at 37oC for 18 hrs before the extent of biofilm formation was measured. As before, a robust biofilm developed in the wells incubated with TSBC (1A, Pre-formed). However, a significantly (P<0.001) reduced biofilm formed in the wells which were incubated simultaneously with the enzyme during biofilm development, as seen by CV, Congo-red, DNA, and SEM imaging (1A, biofilm inhibition). CV quantification revealed that biofilm development in the presence of the enzyme was 80% less than the biofilm buildup in either TSBC media alone or TSBC media supplemented by heat-deactivated enzyme (1C, amylase, Control, and HD amylase, respectively). The ability of the enzyme to both reduce and inhibit biofilm formation was consistent on all of the examined surfaces, which included polystyrene (used in the 96-well plates), PVC (used for SEM imaging), and polyurethane (Fig. 1A, polyurethane tubes). Although the α-amylase compound was shown to both inhibit and reduce biofilm buildup of S. aureus, it was not efficient in controlling S. epidermidis biofilms. Similar biofilm accumulation was measured in wells inoculated with S. epidermidis alone and S. epidermidis supplemented with 100 mg/ml α-amylase (Fig. 1D). α-amylase also did not cause S. epidermidis biofilm detachment with biofilm measuring A600=2.8 in wells that were incubated for 4 hrs with α-amylase or TSB control (Fig. 1E).
In order to investigate whether the biofilm reducing and inhibiting effect could be attributed to loss of cell viability, 5x108 CFU/ml S. aureus SH1000 were incubated overnight in PBS buffer containing 100 mg/ml A. oryzae α-amylase compound. No reduction in cell viability was measured after a 24 hr incubation period, with 4x108 CFU/ml remaining in the samples maintained in PBS alone or PBS with amylase. The enzyme also did not exhibit any bactericidal or bacteriostatic effect when incubated for 24 hrs with S. aureus, reaching a cell density of 6.2x109, 6.1x109, and 5.8x109 CFU/ml when incubated in the presence of TSBC, TSBC supplemented with 100 mg/ml amylase, and TSBC containing 200 mg/ml amylase, respectively.
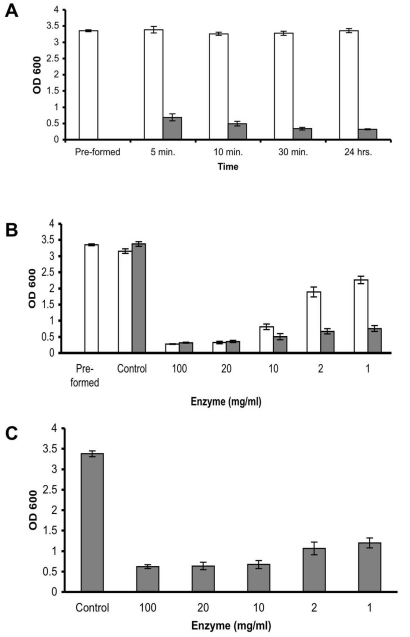
A time point experiment in which S. aureus SH1000 pre-formed biofilms were incubated in the presence of 100 mg/ml enzyme demonstrated that the biofilm reducing activity is extremely rapid, reducing 79% of the existing biofilm within 5 min, and reaching 89% reduction after 30 min of incubation. No significant increase in biofilm reduction was measured following an additional 24 hr incubation period (Fig. 2A). In order to examine the minimal concentration required to effectively remove the biofilm, serial diluted enzyme was added to the pre-formed biofilm. A 90%, 89%, and 72% reduction was observed in the wells which were incubated for 45 min with 100, 20, and 10 mg/ml enzyme, respectively, with a 36% and 23% reduction measured in the wells incubated with 2 and 1 mg/ml (Fig. 2B, white bars). By 24 hrs, a reduction greater than 77% was seen in all of the examined enzyme concentrations (Fig. 2B, gray bars). Incubating S. aureus cells in the presence of 100 to 1 mg/ml enzyme also demonstrated a positive correlation between the enzyme quantity present in the well and biofilm inhibition, with 82% 81%, 80%, 68%, and 64% less biofilm buildup in the wells incubated with 100, 20, 10, 2, and 1 mg/ml enzyme, respectively, as compared to the biofilm in enzyme- free media (Fig. 2C, Control).
Fig. (2).
Enzyme efficacy. (A) Quantification of biofilm reduction at different time points. S. aureus SH1000 biofilms were developed in 96 well plates for 18 hrs (Pre-formed). Thereafter, the wells were rinsed and incubated with fresh TSBC media (white bars) or 100 mg/ml enzyme (gray bars). The wells were than rinsed, stained with CV, and the amount of CV staining was quantified. (B) Quantification of biofilm reduction at different enzyme concentrations. S. aureus biofilms were developed in 96 well plates for 18 hrs (Pre-formed). Thereafter, the wells were rinsed and incubated with fresh TSBC media (Control) or media supplemented with 100 to 1 mg/ml enzyme. CV staining and quantification was assessed after 45 min (white bars) and 24 hrs (gray bars) of incubation. (C) Quantification of biofilm inhibition at different enzyme concentrations. S. aureus cells were cultured in 96-well plates with media (Control) or TSBC supplemented with 100 to 1 mg/ml enzyme. The extent of biofilm formation was analyzed after 18 hrs. Each value represents the mean of 16 wells from one representative experiment. Error bars are shown as one-standard deviation.
The Effect of α-amylase on S. Aureus Clinical Isolates
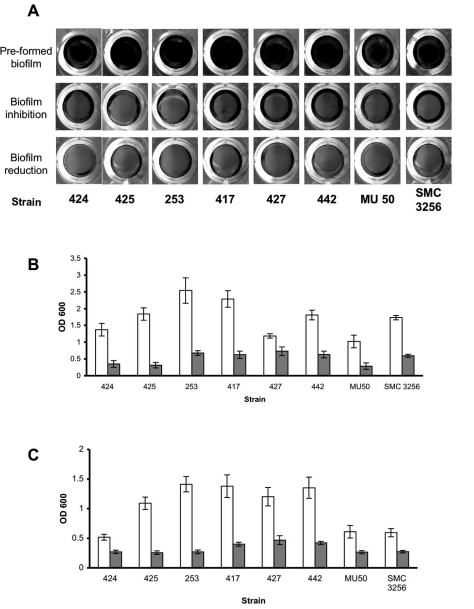
To examine the effect of α-amylase compound on different S. aureus isolates, 8 additional S. aureus clinical isolates were grown in TSBC media with and without the enzyme. As seen in Fig. (3A), the biofilms produced in the presence of the enzyme (3A, biofilm inhibition) were less robust than those grown in enzyme-free media (3A, pre-formed biofilm). These results were also confirmed by CV quantification that demonstrated a decrease of 65% to 83% in biofilm formation (3B, gray bars), with the exception of S. aureus 427, which had 38% less biomass in the treated sample. Treating the pre-formed biofilms with the enzyme for 10 min brought about a measurable decrease in all of the isolates examined, with the most significant decreases of 70%-80% measured for S. aureus isolates 442, 417, 425, and 253, and more modest reductions of 45%-60% measured for isolates 424, 427, MU 50 and SMC 3256 (Fig. 3C).
Fig. (3).
Enzymatic effect on S. aureus clinical isolates. (A) S. aureus clinical isolates were cultured in 96-well plates with TSBC media (Pre-formed biofilm) or TSBC supplemented with 100 mg/ml enzyme (biofilm inhibition). In a parallel experiment, the pre-formed biofilms were incubated for 10 min with the enzyme (biofilm reduction) before being rinsed and stained. Quantification of biofilm inhibition (B) and reduction (C), following exposure to TSBC media alone (white bars) or TSBC containing enzyme (gray bars). Each value represents the mean of 8 wells from one representative experiment. Error bars are shown as one-standard deviation.
Biofilm Inhibition and Reduction by amylases from Different Biological Sources
To compare the biofilm reducing ability of commercially available amylase compounds isolated from different biological sources, S. aureus SH1000 was incubated with 10 mg/ml of the following enzymes: α-amylase compound from B. subtilis, α-amylase compound from human saliva, β-amylase compound from sweet potato, and α-amylase compound from A. oryzae, which was used as a positive control in this experiment. After 10 min of incubation, a substantial reduction in biofilm CV staining was seen only in the wells incubated with α-amylase from A. oryzae (Fig. 4A and 4B, white bars).However, by 3 hrs, a significant reduction (P<0.001) was also observed in the wells incubated with α-amylase from B. subtilis (4B, gray bars). No substantial biofilm reduction was measured for the pre-formed biofilms incubated with β-amylase compound and α-amylase compound from human saliva. In contrast to their biofilm reducing aptitude, all of the α-amylase compounds exhibited some biofilm inhibiting capability when incubated in the presence of S. aureus cells (Fig. 4C), with the most potent inhibiting effect measured for α-amylase compound from A. oryzae (77% inhibition), B. subtilis (88% inhibition), and human saliva (89% inhibition). A more moderate, but measurable, inhibition registered in wells containing β-amylase from sweet potato (49% inhibition).
Fig. (4).
Enzymatic effect of amylases from different biological sources on S. aureus biofilms. (A) S. aureus SH1000 was cultured for 24 hrs in 96-well plates with TSBC media (pre-formed biofilm) or TSBC supplemented with 10 mg/ml of α-amylase (biofilm inhibition) from A. oryzae, B. subtilis, and human saliva and β-amylase from sweet potato. In a parallel experiment, the pre-formed biofilms were incubated for 3 hrs (biofilm reduction) before being rinsed and stained. (B) Quantification of biofilm reduction. Pre-formed biofilms were exposed to the enzymes for 10 min (white bars), 3 hrs (gray bars), and 24 hrs (black bars) before being stained with CV. (C) Quantification of biofilm inhibition. S. aureus was cultured for 24 hrs in 96-well plates with TSBC media (Control) or TSBC supplemented with 10 mg/ml of each enzyme. Each value represents the mean of 6 wells from one representative experiment. Error bars are shown as one-standard deviation.
Since the purity of the commercially acquired compounds is not fully known, biofilm inhibition experiments were conducted in which S. aureus SH1000 was incubated in the presence of α-amylase from A. oryzae with and without the addition of a protease inhibitor (Halttm protease inhibitor cocktail, Thermo Scientific, Rockford, IL). The addition of protease inhibitor did not reduce the biofilm inhibiting effect of the amylase compound, with biofilm buildup reaching A600=0.52, 0.56 and 3.17 in wells inoculated with amylase alone, amylase plus protease inhibitor and amylase free control, respectively. Protease inhibitor cocktail also did not abolish the ability of the amylase compound to remove a preformed biofilm with a 70±1.8% reduction in wells inoculated with amylase alone and 66±4% reduction in wells containing amylase plus protease inhibitor. To determine whether DNase is present in the amylase compound, α-amylase from A. oryzae was prepared and spotted on DifcoTm DNase test agar plates with methyl green (BD, Franklin Lakes, NJ). No DNase activity was seen after incubating the plates for up to 48 hrs (data not shown).
Aggregation Reduction and Inhibition Experiments
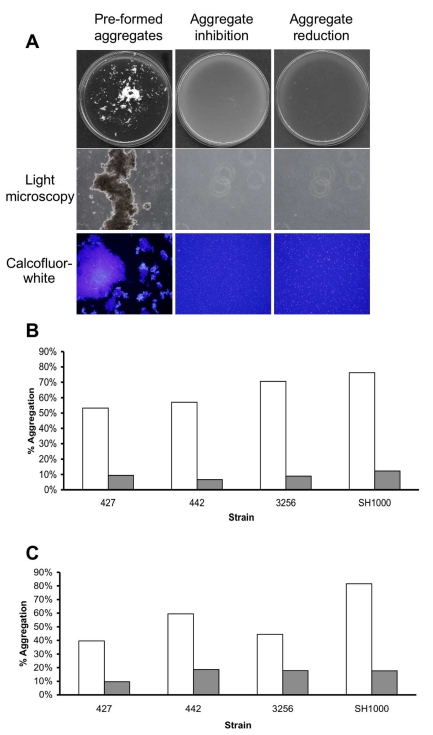
In the previous experiments, emphasis was put on the ability of the enzyme to detach pre-formed biofilms or inhibit the attachment of cells to the examined surface, leading to biofilm reduction and inhibition. In the following experiments, the aim was to investigate whether cell-to-cell associations could also be affected by the presence of α-amylase compound in the surrounding media. To this end, a bioassay was performed in which cells were grown in TSBC media in order to stimulate cell-to-cell association and aggregation. The ability of the enzyme to inhibit the formation of the aggregate was achieved by simultaneously incubating the cells with 50 mg/ml of the enzyme from A. oryzae during growth. As seen in (Fig. 5A) growing S. aureus SH1000 cells in TSBC resulted in the formation of extremely large macro-colonies (>1mm) that were visible by eye (Fig. 5A, Pre-formed aggregates). These macro-colonies, composed of hundreds of cells, were positively stained by the polysaccharide stain Calcofluor-white and visualized by light and fluorescence microscopy (Fig. 5A). Large macro-colonies did not develop in tubes that contained the enzyme (5A, Aggregate inhibition). Light microscopy also confirmed the absence of macro-colonies in the enzyme- treated sample, as only relatively small cell clusters, consisting of 2-10 cells, were formed. In order to quantify the extent of macro-colony inhibition, S. aureus isolates SH1000, 427, 442, and SMC 3256 were grown in the presence of the enzyme and the extent of aggregation was analyzed. These isolates were selected for their enhanced aggregation phenotype in TSBC media. After 18 hrs of incubation, 53% to 76% of the cells were present in aggregates (Fig. 5B, white bars). In contrast, only 6%-12% aggregation was measured in the enzyme- treated tubes (Fig. 5B, gray bars).
Fig. (5).
Aggregation reduction and inhibition experiments. (A) S. aureus SH1000 cells were grown in 18 cm glass tubes containing TSBC media (Pre-formed aggregates) or TSBC supplemented with 100 mg/ml enzyme (Aggregate inhibition) and incubated on a tissue culture roller at 30 RPM. After 18 hrs, the tubes were poured into a 90 mm Petri-dish and visualized by eye and by light microscopy at 400X magnification. Aggregates were also stained for 10 min with 1 µl of Calcofluor-white before being viewed by fluorescent microscopy, magnification set at 400X. To measure the ability of the enzyme to disassociate the aggregates, pre-formed SH1000 aggregates were incubated for 3 hrs with 50 mg/ml enzyme (Aggregate reduction). Quantification of aggregation inhibition (B) and reduction (C) of four S. aureus isolates following exposure to TSBC media alone (white bars) or TSBC containing enzyme (gray bars). Each experiment was carried out four times, yielding similar results. Value represents data from one representative experiment.
In another experiment, the ability of the enzyme to degrade existing macro-colonies was evaluated by adding 100 mg/ml α-amylase compound from A. oryzae into the pre-formed aggregate tube. Within 3 hrs of incubation, a clear reduction in macro-colonies was seen, with aggregates of only 20 cells or less remaining in the enzyme- treated sample (Fig. 5A, aggregate reduction). Quantifying the extent of reduction revealed decreases of 59%, 66%, 74%, and 79% in SH1000 aggregation within 30 min, 1hr, 2hr and 3hr incubation periods, respectively. Aggregation reductions of 77%, 70%, and 65% were measured for isolates 427, 442, and SMC 3256 following a 3 hr incubation period (Fig. 5C).
DISCUSSION
In the last few years, biofilm formation has become an increasing source of concern in industrial, environmental and medical settings. This is especially true in orthopedic medicine, where biofilms pose a significant challenge and can result in some of the most serious complications involving prosthetic and other medical device infections, impacting morbidity, mortality, and medical costs [40]. The necessity of removing the biofilm from the infected site usually requires surgical intervention and radical antimicrobial therapy [41]. A possible therapeutic approach to medical device infections involves the use of EPS-degrading agents that might block, inhibit or reduce cell-to-cell and cell-to-surface associations within the biofilm, bringing about the release of planktonic cells into the environment and allowing the host immune system and antimicrobial drugs to eliminate the infection [42, 43].
In this study, we examined the potential use of commercially available α-amylase compounds as a biofilm control agent against S. aureus. Our work demonstrated that, when grown in the presence of the enzyme, a significant reduction in biofilm buildup is achieved. The biofilm inhibition effect was observed with all of the examined S. aureus isolates and was not influenced by the chemical composition of the surface on which the biofilm developed. A measurable reduction in biofilm buildup was also seen for S. aureus biofilms incubated with α-amylase compounds derived from bacterial, fungal, and human origin. However, a reduced inhibiting effect was registered for β-amylase. The difference in biofilm reduction could be attributed to the fact that β-amylase is an exo-acting carbohydrolase which hydrolyzes the α-1,4-glucosidic linkages of starch only from the non-reducing end of the polysaccharide, as opposed to α-amylase, which can act simultaneously anywhere on the substrate, thus α-amylase tends to be faster-acting than β-amylase [44]. In addition to the enzyme’s capacity to inhibit the formation of S. aureus biofilms, α-amylase also rapidly reduced existing pre-formed biofilms at concentrations as low as 10 mg/ml, with a 1-mg/ml enzyme concentration requiring longer incubation periods to achieve comparable biofilm reduction. As was demonstrated for biofilm inhibition, the biofilm reducing ability was consistent on all of the surfaces and isolates tested as well as with α-amylase compounds derived from A. oryzae and B. subtilis. However, it is interesting to note that α-amylase from human saliva only produced a limited drop in biofilm biomass and that no reduction was observed in the biofilms incubated with β-amylase.
Biofilms in most settings are multicellular structures composed of both cell-to-surface and cell-to-cell associations. In order to examine the effect of the α-amylase compound specifically on cell-to-cell interactions, experiments were conducted in conditions that stimulated cell clustering and aggregation. Here too, α-amylase from A. oryzae had the potency to inhibit the formation of S. aureus aggregates as well as reduce pre-existing aggregates.
It was previously suggested that EPS degrading enzymes have a biological role in enabling surface attached cells to dislodge from one site and relocate, for example, Aggregatibacter actinomycetemcomitans biofilm detachment and dispersal is mediated by the production of dispersin B, a poly-β-1,6-N-acetyl-D-glucosamine (PNAG) hydrolyzing enzyme [45-47]. The biological role of dispersin B in A. actinomycetemcomitans and the fact that PNGA is produced in other bacteria such as S. aureus, S. epidermidis, Escherichia coli, Actinobacillus pleuropneumoniae, Yersinia pestis, and Bordetella spp. led to studies aimed at using the enzyme to detach biofilms as well as a means of enhancing the effectiveness of antimicrobial agents [27-29, 47-50]. In addition to dispersin B, other carbohydrate degrading enzymes were shown to have an effect on biofilm EPS components. In Pseudomonas aeruginosa, degradation of the alginate by alginate-lyase enhanced antibiotic biofilm killing [51], whereas the use of cellulase was shown to degrade a carbohydrate-rich component of the matrix. This cellulase-sensitive sugar was also shown to bind Congo-red and is essential for P. aeruginosa pellicle formation and biofilm structure [52, 53]. The role of an amylose-like polysaccharide in biofilm formation was also documented in Salmonella enteritidis [54], Agrobacterium tumefaciens [55], Pseudomonas spp. CE-2 [56], P. putida [57] and Streptomyces coelicolor, in which cellulose-anchored amyloidal fimbriae were shown to mediate biofilm formation [58]. The successful use of proteolytic enzymes with other polysaccharide degrading enzymes, including α-amylase, as means to remove biofilms, was recently demonstrated for bacteria found in the food and brewing industry, including Bacillus cereus, P. fluorescens, Flavimonas oryzihabitans, Lactobacillus brevis, Leuconostoc mesenteroides and Saccharomyces cerevisiae [59-61].
The use of enzymes capable of specifically degrading the constituents of the extracellular S. aureus matrix was the focus of other studies. Chaignon and his colleagues demonstrated that treating S. aureus strain 383 biofilms with proteinase K and trypsin, as well as pancreatine and Pectinex Ultra SP, leads to biofilm detachment, however, biofilm reduction was not seen when dispersin B or sodium periodate was applied [50]. Since both Pectinex Ultra SP and Pancreatin are multi-component enzyme preparations containing protease activity and a wide range of carbohydrolases, it was difficult to attribute the biofilm reducing effect to any single enzyme [62]. The inability to reduce a pre-existing S. aureus biofilm by dispersin B was also concurred by Jeffrey Kaplan’s group. However, their study did demonstrate that dispersin B is effective in inhibiting biofilm formation of the SH1000 strain [28]. Thus, the discrepancy in the ability of an enzyme to reduce S. aureus biofilms at early vs. late stages of biofilm formation is consistent with our finding that some amylase compounds were more competent at inhibiting biofilm buildup than reducing existing biofilms. In addition to the proteolytic and PNAG-modifying enzymes, DNase-I was also shown to be effective in inhibiting and reducing S. aureus SH1000 biofilms [28].
In this study, we have used several commercially available α-amylase compounds. As it is difficult to guarantee the purity of the compounds, we could only speculate that the biofilm controlling aptitude of the compounds resulted from the amylase activity. The finding that several α-amylase compounds had an ability to control S. aureus biofilms, in concert with the fact that the enzyme activity was not inhibited in the presence of protease inhibitors nor did it possess any DNase activity, lessen the possibility that the biofilm removal effect was caused by other enzymes, which might also be present in the acquired compounds. Furthermore, the inability of the compounds to affect S. epidermidis biofilms suggests that the activity of the enzyme is not through the degradation of the PNAG matrix, as this would have affected the stability of the PNAG rich S. epidermidis biofilms [28].
At this point, we can only speculate on the exact origin of the substrate being degraded by the amylase compounds, as the composition of S. aureus biofilms are quite diverse and could vary depending on the strain, the biofilm growth conditions, and the biofilm developmental stage [28, 35, 63-65]. However, based on the research conducted so far, we believe that commercially available polysaccharide-modifying compounds could be used in the future as a preventative measure, to prevent the establishment of a biofilm, or as a method used to eradicate an already existing biofilm infection.
ACKNOWLEDGEMENTS
We would like to thank Dr. Jeffrey Kaplan from UMDNJ, Dr. Robert Shanks, and the Campbell Laboratory of Ophthalmic Microbiology, UPMC, for kindly providing us with the S. aureus isolates. This work was supported by the Foundation of UMDNJ faculty research grant to D.E.K.
REFERENCES
- 1.Raad II, Hanna HA. Intravascular catheter-related infections: new horizons and recent advances. Arch Intern Med. 2002;162:871–8. doi: 10.1001/archinte.162.8.871. [DOI] [PubMed] [Google Scholar]
- 2.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–32. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 3.Donlan RM. Biofilms and device-associated infections. Emerg Infect Dis. 2001;7:277–81. doi: 10.3201/eid0702.010226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park PW, Rosenbloom J, Abrams WR, Mecham RP. Molecular cloning and expression of the gene for elastin-binding protein (ebpS) in Staphylococcus aureus. J Biol Chem. 1996;271:15803–9. doi: 10.1074/jbc.271.26.15803. [DOI] [PubMed] [Google Scholar]
- 5.Hussain M, Haggar A, Heilmann C, Peters G, Flock JI, Herrmann M. Insertional inactivation of Eap in Staphylococcus aureus strain Newman confers reduced staphylococcal binding to fibroblasts. Infect Immun. 2002;70:2933–40. doi: 10.1128/IAI.70.6.2933-2940.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenberg DP, Bayer AS, Cheung AL, Ward JI. Protective efficacy of protein A-specific antibody against bacteremic infection due to Staphylococcus aureus in an infant rat model. Infect Immun. 1989;57:1113–8. doi: 10.1128/iai.57.4.1113-1118.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fedtke I, Gotz F, Peschel A. Bacterial evasion of innate host defenses-the Staphylococcus aureus lesson. Int J Med Microbiol. 2004;294:189–94. doi: 10.1016/j.ijmm.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Stewart PS. Mechanisms of antibiotic resistance in bacterial biofilms. Int J Med Microbiol. 2002;292:107–13. doi: 10.1078/1438-4221-00196. [DOI] [PubMed] [Google Scholar]
- 9.Akiyama H, Kanzaki H, Tada J, Arata J. Staphylococcus aureus infection on cut wounds in the mouse skin: experimental staphylococcal botryomycosis. J Dermatol Sci. 1996;11:234–8. doi: 10.1016/0923-1811(95)00448-3. [DOI] [PubMed] [Google Scholar]
- 10.Gotz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–78. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 11.Fitzpatrick F, Humphreys H, O'Gara JP. The genetics of staphylococcal biofilm formation-will a greater understanding of pathogenesis lead to better management of device-related infection? Clin Microbiol Infect. 2005;11:967–73. doi: 10.1111/j.1469-0691.2005.01274.x. [DOI] [PubMed] [Google Scholar]
- 12.Zegans ME, Shanks RM, O'Toole GA. Bacterial biofilms and ocular infections. Ocul Surf. 2005;3:73–80. doi: 10.1016/s1542-0124(12)70155-6. [DOI] [PubMed] [Google Scholar]
- 13.Fux CA, Costerton JW, Stewart PS, Stoodley P. Survival strategies of infectious biofilms. Trends Microbiol. 2005;13:34–40. doi: 10.1016/j.tim.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 14.Hall-Stoodley L, Costerton JW, Stoodley P. Bacterial biofilms: from the natural environment to infectious diseases. Nat Rev Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- 15.Parkar SG, Flint SH, Brooks JD. Evaluation of the effect of cleaning regimes on biofilms of thermophilic bacilli on stainless steel. J Appl Microbiol. 2004;96:110–16. doi: 10.1046/j.1365-2672.2003.02136.x. [DOI] [PubMed] [Google Scholar]
- 16.Parry JD. Protozoan grazing of freshwater biofilms. Adv Appl Microbiol. 2004;54:167–96. doi: 10.1016/S0065-2164(04)54007-8. [DOI] [PubMed] [Google Scholar]
- 17.Oulahal-Lagsir N, Martial-Gros A, Bonneau M, Blum LJ. "Escherichia coli-milk" biofilm removal from stainless steel surfaces: synergism between ultrasonic waves and enzymes. Biofouling. 2003;19:159–68. doi: 10.1080/08927014.2003.10382978. [DOI] [PubMed] [Google Scholar]
- 18.Nandakumar K, Obika H, Utsumi A, Toshihiko O, Yano T. Recolonization of laser-ablated bacterial biofilm. Biotechnol Bioeng. 2004;85:185–9. doi: 10.1002/bit.10833. [DOI] [PubMed] [Google Scholar]
- 19.Lawrence JR, Scharf B, Packroff G, Neu TR. Microscale evaluation of the effects of grazing by invertebrates with contrasting feeding modes on river biofilm architecture and composition. Microb Ecol. 2002;44:199–207. doi: 10.1007/s00248-001-1064-y. [DOI] [PubMed] [Google Scholar]
- 20.Kadouri D, O'Toole GA. Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl Environ Microbiol. 2005;71:4044–51. doi: 10.1128/AEM.71.7.4044-4051.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kadouri D, Venzon NC, O'Toole GA. Vulnerability of pathogenic biofilms to Micavibrio aeruginosavorus. Appl Environ Microbiol. 2007;73:605–14. doi: 10.1128/AEM.01893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hammer BK, Bassler BL. Quorum sensing controls biofilm formation in Vibrio cholerae. Mol Microbiol. 2003;50:101–04. doi: 10.1046/j.1365-2958.2003.03688.x. [DOI] [PubMed] [Google Scholar]
- 23.Hanlon GW, Denyer SP, Olliff CJ, Ibrahim LJ. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2001;67:2746–53. doi: 10.1128/AEM.67.6.2746-2753.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughes KA, Sutherland IW, Jones MV. Biofilm susceptibility to bacteriophage attack: the role of phage-borne polysaccharide depolymerase. Microbiology. 1998;144:3039–47. doi: 10.1099/00221287-144-11-3039. [DOI] [PubMed] [Google Scholar]
- 25.Alem MA, Douglas LJ. Effects of aspirin and other nonsteroidal anti-inflammatory drugs on biofilms and planktonic cells of Candida albicans. Antimicrob Agents Chemother. 2004;48:41–47. doi: 10.1128/AAC.48.1.41-47.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother. 2004;48:2633–36. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Izano EA, Wang H, Ragunath C, Ramasubbu N, Kaplan JB. Detachment and killing of Aggregatibacter actinomycetemcomitans biofilms by dispersin B and SDS. J Dent Res. 2007;86:618–22. doi: 10.1177/154405910708600707. [DOI] [PubMed] [Google Scholar]
- 28.Izano EA, Amarante MA, Kher WB, Kaplan JB. Differential roles of poly-N-acetylglucosamine surface polysaccharide and extracellular DNA in Staphylococcus aureus and Staphylococcus epidermidis biofilms. Appl Environ Microbiol. 2008;74:470–76. doi: 10.1128/AEM.02073-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Donelli G, Francolini I, Romoli D, et al. Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob Agents Chemother. 2007;51:2733–40. doi: 10.1128/AAC.01249-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allesen-Holm M, Barken KB, Yang L, et al. A characterization of DNA release in Pseudomonas aeruginosa cultures and biofilms. Mol Microbiol. 2006;59:1114–28. doi: 10.1111/j.1365-2958.2005.05008.x. [DOI] [PubMed] [Google Scholar]
- 31.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487–8. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 32.Kaplan JB, Velliyagounder K, Ragunath C, et al. Genes involved in the synthesis and degradation of matrix polysaccharide in Actinobacillus actinomycetemcomitans and Actinobacillus pleuropneumoniae biofilms. J Bacteriol. 2004;186:8213–20. doi: 10.1128/JB.186.24.8213-8220.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Izano EA, Sadovskaya I, Vinogradov E, et al. Poly-N-acetylglucosamine mediates biofilm formation and antibiotic resistance in Actinobacillus pleuropneumoniae. Microb Pathog. 2007;43:1–9. doi: 10.1016/j.micpath.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi AH, Slamti L, Avci FY, Pier GB, Maira-Litran T. The pgaABCD locus of Acinetobacter baumannii encodes the production of poly-{beta}-1-6-N-acetyl glucosamine PNAG that is critical for biofilm formation. J Bacteriol. 2009;191:5953–63. doi: 10.1128/JB.00647-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanks RM, Meehl MA, Brothers KM, et al. Genetic evidence for an alternative citrate-dependent biofilm formation pathway in Staphylococcus aureus that is dependent on fibronectin binding proteins and the GraRS two-component regulatory system. Infect Immun. 2008;76:2469–77. doi: 10.1128/IAI.01370-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shanks RM, Sargent JL, Martinez RM, Graber ML, O'Toole GA. Catheter lock solutions influence staphylococcal biofilm formation on abiotic surfaces. Nephrol Dial Transplant. 2006;21:2247–55. doi: 10.1093/ndt/gfl170. [DOI] [PubMed] [Google Scholar]
- 37.Shanks RM, Donegan NP, Graber ML, et al. Heparin stimulates Staphylococcus aureus biofilm formation. Infect Immun. 2005;73:4596–606. doi: 10.1128/IAI.73.8.4596-4606.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coico R, Kowalik T, Quarles J, Stevenson B, Taylor R, editors. Current protocols in microbiolgy. New York: Wiley; 2005. [Google Scholar]
- 39.Kadouri D, Jurkevitch E, Okon Y. Involvement of the reserve material poly-beta-hydroxybutyrate in Azospirillum brasilense stress endurance and root colonization. Appl Environ Microbiol. 2003;69:3244–50. doi: 10.1128/AEM.69.6.3244-3250.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Campoccia D, Montanaro L, Arciola CR. The significance of infection related to orthopedic devices and issues of antibiotic resistance. Biomaterials. 2006;27:2331–39. doi: 10.1016/j.biomaterials.2005.11.044. [DOI] [PubMed] [Google Scholar]
- 41.Trampuz A, Zimmerli W. New strategies for the treatment of infections associated with prosthetic joints. Curr Opin Investig Drugs. 2005;6:185–190. [PubMed] [Google Scholar]
- 42.Kaplan JB. Therapeutic potential of biofilm-dispersing enzymes. Int J Artif Organs. 2009;32:545–54. doi: 10.1177/039139880903200903. [DOI] [PubMed] [Google Scholar]
- 43.Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89:205–18. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toda H, Nitta Y, Asanami S, Kim JP, Sakiyama F. Sweet potato beta-amylase. Primary structure and identification of the active-site glutamyl residue. Eur J Biochem. 1993;216:25–38. doi: 10.1111/j.1432-1033.1993.tb18112.x. [DOI] [PubMed] [Google Scholar]
- 45.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous beta-hexosaminidase activity. J Bacteriol. 2003;185:4693–98. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaplan JB, Meyenhofer MF, Fine DH. Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J Bacteriol. 2003;185:1399–404. doi: 10.1128/JB.185.4.1399-1404.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Izano EA, Sadovskaya I, Wang H, et al. Poly-N-acetylglucosamine mediates biofilm formation and detergent resistance in Aggregatibacter actinomycetemcomitans. Microb Pathog. 2008;44:52–60. doi: 10.1016/j.micpath.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Venketaraman V, Lin AK, Le A, Kachlany SC, Connell ND, Kaplan JB. Both leukotoxin and poly-N-acetylglucosamine surface polysaccharide protect Aggregatibacter actinomycetemcomitans cells from macrophage killing. Microb Pathog. 2008;45:173–80. doi: 10.1016/j.micpath.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Izano EA, Shah SM, Kaplan JB. Intercellular adhesion and biocide resistance in nontypeable Haemophilus influenzae biofilms. Microb Pathog. 2009;46:207–13. doi: 10.1016/j.micpath.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, Jabbouri S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Appl Microbiol Biotechnol. 2007;75:125–32. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- 51.Alkawash MA, Soothill JS, Schiller NL. Alginate lyase enhances antibiotic killing of mucoid Pseudomonas aeruginosa in biofilms. APMIS. 2006;114:131–38. doi: 10.1111/j.1600-0463.2006.apm_356.x. [DOI] [PubMed] [Google Scholar]
- 52.Friedman L, Kolter R. Genes involved in matrix formation in Pseudomonas aeruginosa PA14 biofilms. Mol Microbiol. 2004;51:675–90. doi: 10.1046/j.1365-2958.2003.03877.x. [DOI] [PubMed] [Google Scholar]
- 53.Friedman L, Kolter R. Two genetic loci produce distinct carbohydrate-rich structural components of the Pseudomonas aeruginosa biofilm matrix. J Bacteriol. 2004;186:4457–65. doi: 10.1128/JB.186.14.4457-4465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Solano C, Garcia B, Valle J, et al. Genetic analysis of Salmonella enteritidis biofilm formation: critical role of cellulose. Mol Microbiol. 2002;43:793–808. doi: 10.1046/j.1365-2958.2002.02802.x. [DOI] [PubMed] [Google Scholar]
- 55.Matthysse AG, Marry M, Krall L, et al. The effect of cellulose overproduction on binding and biofilm formation on roots by Agrobacterium tumefaciens. Mol Plant Microbe Interact. 2005;18:1002–10. doi: 10.1094/MPMI-18-1002. [DOI] [PubMed] [Google Scholar]
- 56.Jain A, Bhosle NB. Role of beta 1-4 linked polymers in the biofilm structure of marine Pseudomonas sp. CE-2 on 304 stainless steel coupons. Biofouling. 2008;24:283–90. doi: 10.1080/08927010802140857. [DOI] [PubMed] [Google Scholar]
- 57.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol Microbiol. 2010;75:815–26. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 58.de Jong W, Wosten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Mol Microbiol. 2009;73:1128–40. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- 59.Lequette Y, Boels G, Clarisse M, Faille C. Using enzymes to remove biofilms of bacterial isolates sampled in the food-industry. Biofouling. 2010;26:421–31. doi: 10.1080/08927011003699535. [DOI] [PubMed] [Google Scholar]
- 60.Molobela P, Cloete TE, Beukes M. Protease and amylase enzymes for biofilm removal and degradation of extracellular polymeric substances (EPS) produced by Pseudomonas fluorescens bacteria. African J Microbiol Res. 2010;4:1515–24. [Google Scholar]
- 61.Walker SL, Fourgialakis M, Cerezo B, Livens S. Removal of microbial biofilms from dispense equipment: The effect of enzymatic pre-digestion and detergent treatment. J Inst Brew. 2007;113:61–6. [Google Scholar]
- 62.Johansen C, Falholt P, Gram L. Enzymatic removal and disinfection of bacterial biofilms. Appl Environ Microbiol. 1997;63:3724–8. doi: 10.1128/aem.63.9.3724-3728.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sutherland I. Biofilm exopolysaccharides: a strong and sticky framework. Microbiology. 2001;147:3–9. doi: 10.1099/00221287-147-1-3. [DOI] [PubMed] [Google Scholar]
- 64.Sadovskaya I, Chaignon P, Kogan G, Chokr A, Vinogradov E, Jabbouri S. Carbohydrate-containing components of biofilms produced in vitro by some staphylococcal strains related to orthopaedic prosthesis infections. FEMS Immunol Med Microbiol. 2006;47:75–82. doi: 10.1111/j.1574-695X.2006.00068.x. [DOI] [PubMed] [Google Scholar]
- 65.Kogan G, Sadovskaya I, Chaignon P, Chokr A, Jabbouri S. Biofilms of clinical strains of Staphylococcus that do not contain polysaccharide intercellular adhesin. FEMS Microbiol Lett. 2006;255:11–6. doi: 10.1111/j.1574-6968.2005.00043.x. [DOI] [PubMed] [Google Scholar]