Abstract
Little is known about how combinations of histone marks are interpreted at the level of nucleosomes. The second PHD finger of human BPTF is known to specifically recognize histone H3 when methylated on lysine 4 (H3K4me2/3); here we examine how additional heterotypic modifications influence BPTF binding. Using peptide surrogates, three acetyllysine ligands are indentified for a PHD-adjacent bromodomain in BPTF via systematic screening and biophysical characterization. Although the bromodomain displays limited discrimination amongst the three possible acetyllysines at the peptide level, marked selectivity is observed for only one of these sites, H4K16ac, in combination with H3K4me3 at the mononucleosome level. In support, these two histone marks constitute a unique trans-histone modification pattern that unambiguously resides within a single nucleosomal unit in human cells, and this module co-localizes with these marks in the genome. Together, our data call attention to nucleosomal patterning of covalent marks in dictating critical chromatin associations.
The mechanism by which covalent modifications of histones and DNA contribute to the chromatin structural states that govern all DNA-templated processes is a central question to understanding genome management and its disregulation in human disease. Advances in understanding the role of chromatin modifications may be divided into two separate veins: enumerating and characterizing chromatin modification-effector pairs (Taverna et al., 2007), and discerning relative modification patterns at the genome-level and correlating these patterns to function (Bernstein et al., 2005; Guenther et al., 2007; Wang et al., 2008). However, the convergence of these two areas– how different chromatin binding modules simultaneously engage these modification patterns to transduce downstream function — remains poorly understood.
We have recently proposed that multivalent engagement of nucleosomal units bearing distinct epigenetic signatures by chromatin-modification complexes may be involved in many chromatin transactions (Ruthenburg et al., 2007b). While compelling tests of the ‘multivalency hypothesis’ have yet to occur, earlier studies have provided hints that this phenomenon may be more general than currently appreciated: i.) greater net binding affinity and substrate specificity beyond the sum of constituent parts may arise in the binding of two proximal acetyllysines in the H4 tail by the bromodomain proteins hTaf1 and Brdt (Jacobson et al., 2000; Moriniere et al., 2009); ii.) multiple contact surfaces distributed over a number of subunits (some of which appear to be histone-modification dependent) are required for Rpd3S histone deacetylase complex binding to a single nucleosome (Li et al., 2007). However, the interplay between discrete histone modification-dependent interactions has not been well studied in a nucleosomal context, nor is there a clear example of a protein complex or single polypeptide that simultaneously engages two or more histone modifications on a nucleosome for which the discrete constituent interactions are clearly defined. Thus, several key questions posed in the histone code hypothesis (Strahl and Allis, 2000) still remain unresolved: how are combinations of histone modifications interpreted at the molecular level, are there units of recognition beyond single tails, and what are the functional consequences?
Here, we sought to address how chromatin modification patterns may be simultaneously engaged on the nucleosome level using a PHD finger and adjacent bromodomain of the NURF chromatin remodelling complex subunit BPTF as a paradigm to provide insights into the above questions. We examine the biochemical, structural and functional properties endowed by a bivalent configuration of these linked effector domains, the simplest case of multivalent histone modification-dependent nucleosomal engagement (Ruthenburg et al., 2007b).
BPTF in the context of the NURF complex is an essential regulator of chromatin structure in development (Badenhorst et al., 2002; Landry et al., 2008; Wysocka et al., 2006), bringing about transcriptional activation or repression in a locus-specific manner (Bai et al., 2007; Kwon et al., 2008) by virtue of the complex’s chromatin remodelling activity (Hamiche et al., 1999; Tsukiyama and Wu, 1995). The second PHD finger of BPTF, implicated in recruitment or stabilization of the NURF complex to active homeotic genes as a consequence of MLL1-mediated H3K4 trimethylation, is followed closely by a bromodomain whose mechanistic role is obscure (Wysocka et al., 2006). The spatial coupling of these two domains is sufficiently tight to permit determination of the structure of the terminal PHD-bromodomain module spanned by an apparently rigid linker α-helix (Li et al., 2006). While the molecular details of H3K4me3 binding by the PHD-finger are known, the ligand for the associated bromodomain remains unclear. Given that the bromodomain is a well-established histone acetyllysine recognition domain (Dhalluin et al., 1999; Mujtaba et al., 2007), we envisioned that together with the PHD-finger, this bivalent structural element may bind two different classes of histone modifications generally associated with euchromatin and transcription initiation (Guenther et al., 2007; Ruthenburg et al., 2007a; Shogren-Knaak et al., 2006). In support, deletion of both the PHD finger and the adjacent bromodomain rescued BPTF knock-down in Xenopus less efficiently than a PHD finger mutation that completely abolishes H3K4me3 interactions (Wysocka et al., 2006). To examine the nature of this putative bivalent nucleosomal recognition by the PHD-bromo module (Figure 1A), we first sought to identify and characterize potential bromodomain binding partners.
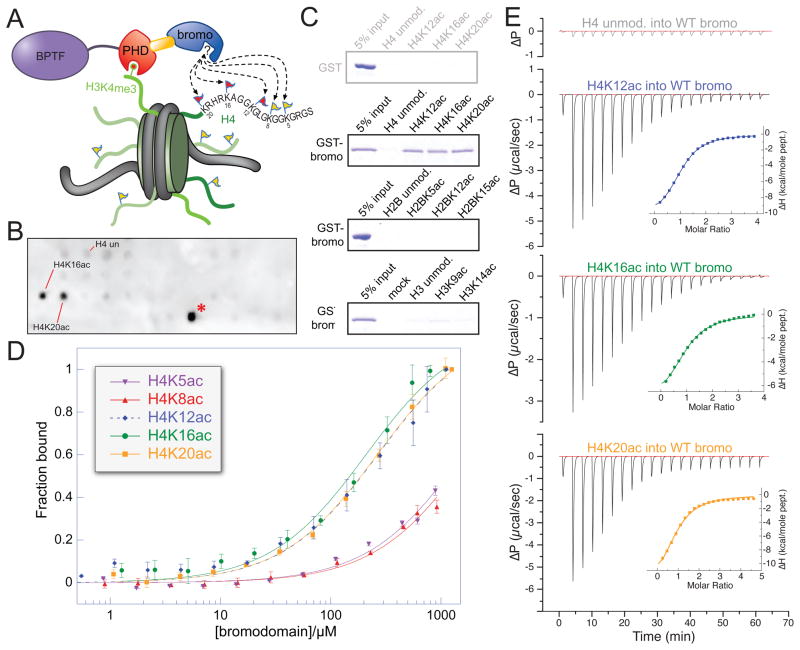
Figure 1. Systematic characterization of preferred acetylated histone ligands for the BPTF bromodomain.
(A) Schematic representation of a putative bivalent nucleosomal interaction with the BPTF PHD-bromo module. The known point of contact (Li et al., 2006; Wysocka et al., 2006) is illustrated between the PHD-finger (red) and H3K4me3 (red circle atop green histone tail), while the bromodomain (blue) interacts with an unknown acetylation site (flags on histone tails). (B) A SPOT blot of an array containing all known human core histone acetylation sites on a modified cellulose scaffold probed with GST-tagged bromodomain. A representative SPOT blot (controls and remaining replicates, Figure S1A–C), displaying reproducible staining for H4 peptides (residues 11–25) acetylated on the ε-amines of lysines 16 and 20, respectively (H4K16ac, H4K20ac). The staining of H2BK85ac (red asterisk) appears to be a peptide-HRP interaction and there is no detectable binding between the bromodomain and this peptide in solution (Figure S1E). (C) Peptide pull-down experiments with GST and GST-BPTF bromodomain (GST-bromo) against three peptide series indicated. Full gels with GST controls are available in Figure S1H. (D) Fluorescence polarization anisotropy-based titration of the BPTF bromodomain against each of the indicated peptides (data are represented as mean ± SD). (E) Isothermal titration calorimetry-based binding curves: the indicated H4 peptides are titrated into a solution of BPTF bromodomain (see Figure S1F for Kd values). See also Figure S1.
RESULTS
The BPTF bromodomain binds three different acetylated H4 peptides
Using SPOT blotting (Nady et al., 2008), we screened a spatially arrayed library of all known core histone acetylation marks (Basu et al., 2009). This analysis revealed two acetylation marks that specifically bound recombinant BPTF bromodomain (Figure 1B). Peptides representing histone H4 acetylated lysines 16 and 20 (H4K16ac and H4K20ac) consistently displayed the strongest interaction with the BPTF bromodomain in this assay, whereas the unmodified counterpart displayed no detectable signal above background (Figure 1B, Figure S1C). Affinities for these two peptides and set of control peptides were validated by peptide-pull down, and then examined more carefully via fluorescence polarisation anisotropy (FPA) and isothermal titration calorimetry (ITC) (Figure 1C, D, E). These additional experiments revealed affinity for H4K12ac, perhaps not apparent with SPOT analysis because the array peptide only included three residues N-terminal to the K12ac mark. Quantitative binding measurements lend support to the apparent specificity for these three acetylation sites in the H4 tail whereas other acetylated H4 peptides and unmodified H4 counterparts displayed only weak affinity outside of the experimentally quantifiable range (Figure 1D, E and S1E, F, G, H). These measurements are consistent with the promiscuity and affinity [H4K12ac (Kd, ITC = 69 ± 1μM), H4K16ac (Kd, ITC = 99 ± 7 μM) and H4K20ac (Kd, ITC = 130 ± 10μM)] reported for other bromodomains (Dhalluin et al., 1999; Mujtaba et al., 2007; Zeng et al., 2008).
Properties of BPTF PHD-bromo binding at the peptide versus nucleosome level
With a small set of bromodomain binding-partners characterized, combined with the well-established PHD finger affinity for H3K4me2/3 (Li et al., 2006; Wysocka et al., 2006), we sought to examine the properties of bivalent ligand binding by these two linked binding domains. Simultaneous binding of the BPTF PHD-bromo cassette could have two non-exclusive consequences: allosteric cooperativity wherein the binding of a given peptide in one of the two modules may influence the binding of cognate peptide to the other by a conformational shift (Changeux and Edelstein, 2005); or a multivalent interaction wherein two coupled entities may bind with greater net affinity and specificity than their discrete constituent binding equilibria, largely as an entropic effect (Krishnamurthy et al., 2006; Ruthenburg et al., 2007b). We used peptide-level binding experiments with the PHD-bromo module exploiting the heterotypic ligand binding properties of each domain with one ligand at saturating concentration to query for possible allosteric enhancement. For simplicity, we restricted our initial experiments to H4K16ac in combination with H3K4me3. The binding of the PHD-bromo unit to a fluorescein-labelled H3K4me3 peptide was assessed by FPA in the presence of unlabeled H4K16ac peptide at a concentration 5-fold above its bromodomain binding Kd. This titration did not reveal any significant displacement of the binding curve relative to similar titrations with excess unmodified H4 peptide, or without any H4 peptide (Figure 2A). Moreover, the reciprocal experiment did not detect cooperativity (Figure 2B). Thus we conclude that at the peptide level each binding event is free from detectable allostery.
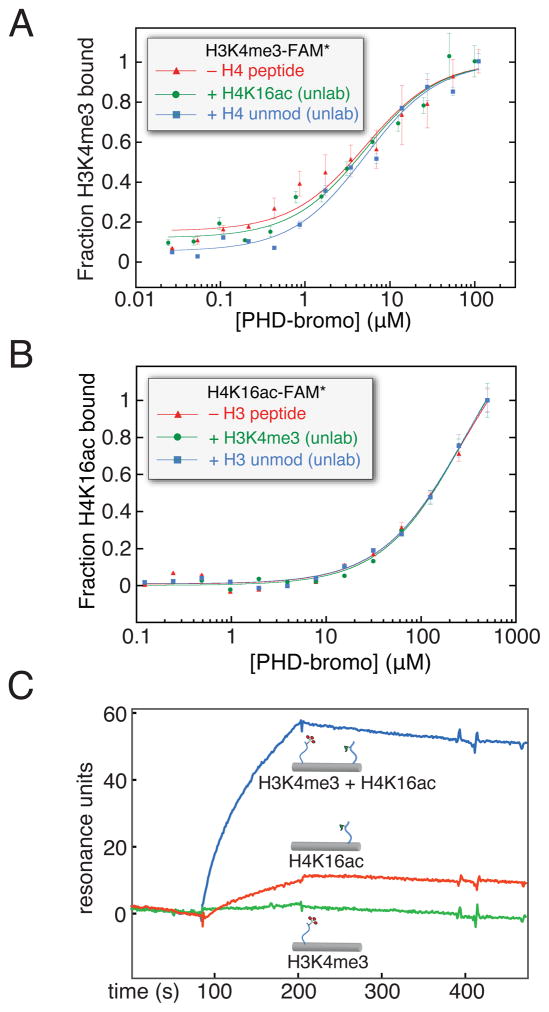
Figure 2. Is there allostery or bivalent cooperativity in simultaneous ligand binding by the BPTF PHD-bromodomain?
(A) The possible allosteric cooperative binding at the peptide level is examined by fluorescence polarization anisotropy using 100 nM fluorescein-labelled H3K4me3 peptide, with either no additional peptide, excess unlabeled H4K16ac peptide (500 μM), or excess unlabeled H4 peptide without acetylation (500 μM). (B) The converse experiment with respect to panel A: with unlabeled H3K4me3 in excess (20 μM, ~10-fold above PHD-H3K4me3 Kd) and fluorescein-H4K16ac (150 nM), protein is titrated and resulting fluorescence polarization anisotropy measured (expressed here as fraction bound). (C) A dsDNA scaffold, selected for rigidity while retaining little predicted bending, was used to covalently install H3K4me3 (H3K4me3[1–8], green), H4K16ac (H4K16ac [12–20], red), or both peptides (blue) at specific positions by disulfide formation with cystamine derivatized convertible dC nucleosides (Figure S2). All of these DNA-protein conjugates were immobilized in different flow channels via a single 3′-biotin linkage to a streptavidin coated surface plasmon resonance chip at low density, untagged PHD-bromo was applied and background binding was subtracted from an empty flow cell. See also Figure S2. (data are represented as mean ± SD)
If the binding of both peptides is effectively bi-molecular, as might be anticipated for the BPTF PHD-bromodomain engaging both marks within a nucleosome, a free energy enhancement in binding might occur due to multivalency (Krishnamurthy et al., 2006; Ruthenburg et al., 2007b). To test this possibility, we developed a novel biophysical assay with histone peptides attached to a rigid DNA duplex to assess the spatial requirements of cooperative and simultaneous histone tail binding. When the spacing between these two short peptides along one face of the B-form DNA duplex is ~71Å (Figure S2A), marked enhancement of PHD-bromodomain binding affinity, relative to DNA-peptide conjugates bearing each single peptide annealed to an unmodified complementary DNA strand, is observed (Figure 2C). Given the distance spanned by the two peptide-binding pockets (~70Å), this result suggests simultaneous binding by both modules. That the H3K4me3 peptide-duplex conjugate displayed little binding affinity was unexpected, because the Kd for the PHD finger binding this peptide is ~100-fold tighter than that of the bromodomain binding H4K16ac (Li et al., 2006). Our interpretation of this result is the H3K4me3 peptide is sufficiently short such that the PHD-finger incurs steric or electrostatic repulsion from the DNA-conjugate upon binding, supported by the observation that this short peptide is bound effectively in the absence of DNA (Figure S2D). Remarkably, this impairment of PHD-finger binding can be partially overcome by distal binding of the bromodomain to the same DNA ruler duplex. Encouraged that simultaneous binding of the PHD-bromodomain modules may provide an affinity enhancement, we sought to study these multivalent interactions on a more physiologically-relevant substrate, the nucleosome.
To assess the consequences of bivalent nucleosome engagement, we sought to construct nucleosomes bearing the desired combinations of posttranslational modifications. To this end, we employed a histone semisynthesis approach of expressed protein ligation (EPL) (Muir, 2003; Shogren-Knaak and Peterson, 2004) to afford homogenously modified histones that could be reconstituted with recombinant human core histones into octamers, then mononucleosomes on a strong positioning sequence (Figures S3 and S4). GST-tagged BPTF PHD-bromodomain was immobilized on a glutathione resin and interrogated for binding to these radiolabeled mononucleosomes. A 2 to 3-fold enhancement of nucleosomal binding affinity was reproducibly observed for nucleosomes bearing both H4K4me3 and H4K16ac over mononucleosomes with only H3K4me3; whereas no binding was observed for the corresponding unmodified species (Figure 3A). Importantly, there was little detectable binding in this assay for nucleosomes acetylated only at H4K16 — as might be anticipated from the ~ 100-fold Kd difference of the two discrete interactions extrapolated to an off-rate dominated binding measurement. To exclude GST-tag dimerization artifacts and surface effects, we performed a reciprocal experiment wherein nucleosomes are immobilized to a solid support and protein without the GST-tag is queried for nucleosome interaction by pull-down, and the results are similar (Figure 3B and S3K,L). We interpret the enhanced binding of doubly-modified mononucleosomes to suggest that both marks may play a role in nucleosome-level binding, yet the PHD-H3K4me3 interaction is dominant.
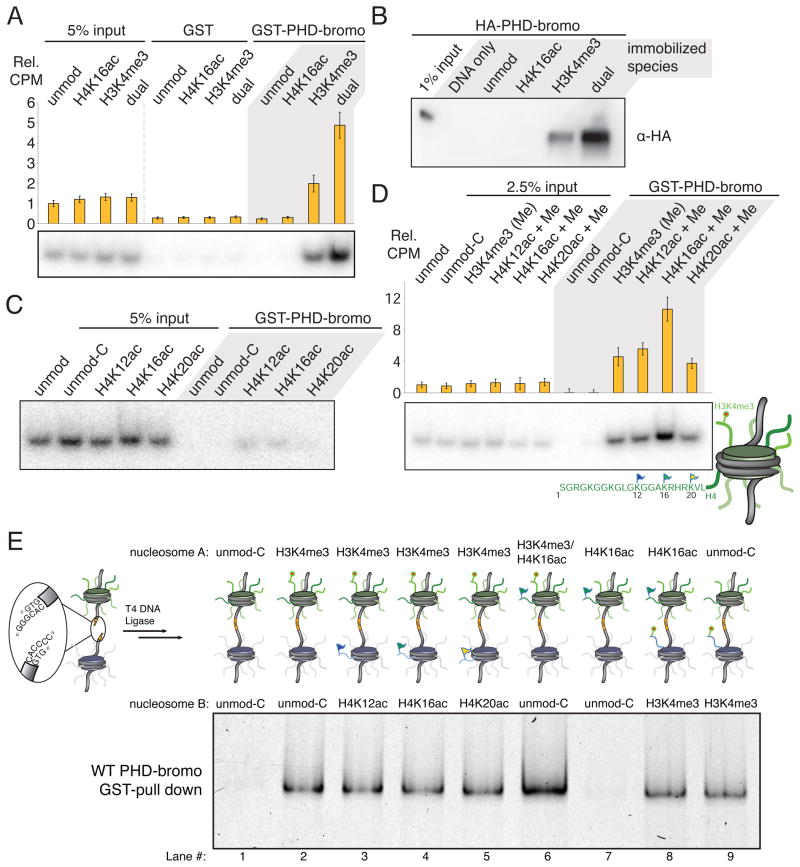
Figure 3. The BPTF PHD-bromo module simultaneously engages two heterotypic trans-histone marks in nucleosomal contexts.
(A) GST pull-down of modified nucleosomes with semisynthetic histones produced by EPL. Nucleosomes (unmod, WT recombinant histones; H4K16ac modified; H3K4me3 modified; and dual, H4K16ac and H3K4me3 modified) pulled-down with resin-bound GST or GST-BPTF PHD-bromo module protein are detected by autoradiography after native gel electrophoresis or scintillation counting normalized to indicated % input (Rel. CPM, yellow bars, represent mean ± SD). (B) The reciprocal experiment relative to Figure 3A -- a Western blot of HA-tagged PHD-bromodomain (without GST-tag) retained on streptavidin immobilized mononucleosomes following extensive washing relative to 1% input. (C) The nucleosomes bearing acetylated H4 alone do not display significant binding in the same GST-pull-down experimental format as described in panel A. An additional control nucleosome species with H3T32C and H4R23C histones (unmod-C) serves as an unmodified nucleosome control that retains the cysteine ligation scars. (D) In this experiment, all three H4 acetylation marks that are bound at the peptide-level (Figure 1) are examined in combination with H3K4me3 at the nucleosome level as in panel A. Data are represented as mean ± SD. (E) GST pull-down of hetero-dinucleosomes composed of two mononucleosomes indicated in positions A and B ligated together; each lane is labelled below numerically. The input dinucleosomes for this experiment are presented in Figure S3N. See also Figure S3.
Using this same assay we revisited the question of specificity amongst the three candidate H4 acetylation marks that were previously identified in peptide-binding assays. Although selectivity between H4K12ac, H4K16ac and H4K20ac peptides is limited, we wondered if there might be additional binding constraints imposed by these marks when presented in a nucleosomal context. While this assay is not sufficiently sensitive to detect bromodomain engagement of the acetylation marks in the absence of H3K4me3 binding (Figure 3C), again we observe a binding enhancement attributable to bivalent engagement only for H3K4me3 in combination with H4K16ac. Surprisingly, the other two acetylation marks (H4K12ac and H4K20ac) when paired with H3K4me3, do not display any binding enhancement beyond that due to H3K4me3 binding alone (Figure 3D). Despite similar peptide-level binding by the bromodomain there is clear binding specificity at the mononucleosome level.
Does the BPTF PHD-bromo module preferentially bind to mononucleosomes or do higher-order chromatin structures present the respective tails in a more productive spatial disposition for engagement? To begin to address this question, we constructed a series of dinucleosomal species via heteromeric DNA ligation of two mononucleosomes (McGinty et al., 2008; Zheng and Hayes, 2004) (see schematic in Figure 3E). Analogous pull-down experiments indicate that the dinucleosome composed of a nucleosome bearing both H3K4me3 and H4K16ac in position A and an unmodified nucleosome in position B is a modestly preferred binding partner of the PHD-bromodomain over the same two marks each in adjacent nucleosomes assorted into either configuration (Lanes 6 versus 4 and 8, Figure 3E, S3N). Further, we do not observe simultaneous binding of H3K4me3 and any if the candidate H4 acetyl marks across two nucleosomes. While this experiment does not exclude the possibility that higher-order nucleosome arrays or different spacing of linker DNA between nucleosomes A and B could produce more favorable tail orientations, our data suggest that the PHD-bromo module engages chromatin in an intra- rather than inter-nucleosomal binding mode under the conditions examined.
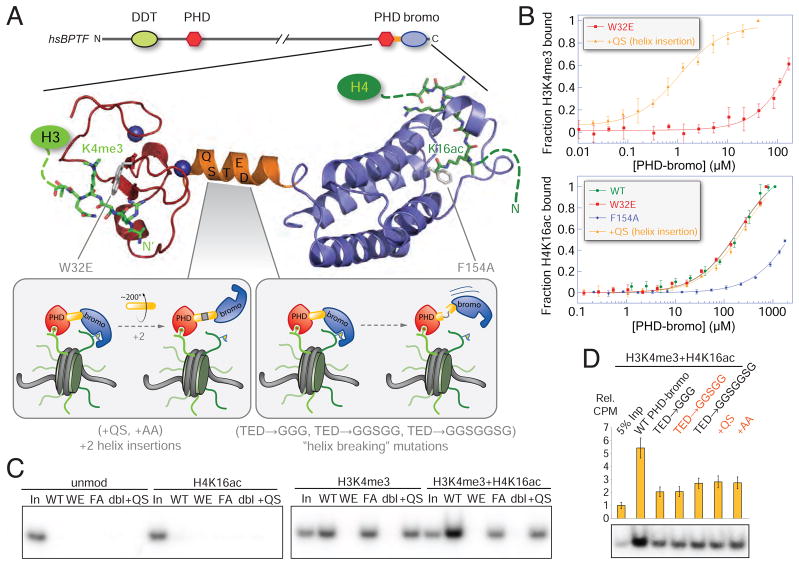
To more precisely examine the intranucleosomal binding properties of the PHD-bromodomain, we next explored a panel of mutations that abrogate the capacity of each domain to bind their respective substrates, as well perturb the spatial apposition of these two domains at the nucleosome level. An insertion of two amino acids (+QS) into the apparently rigid helix that links the PHD and bromodomains should rotate the two domains ~200° out of phase assuming the helix remains intact, changing the relative orientation of histone binding pockets (Figure 4A, inset). Neither perturbation of this helix, nor mutations in the other domain substantially impact a given domain’s intrinsic peptide binding capacity by FPA (Figure 4B). In the GST pull-down experiment, the W32E mutant most severely impaired BPTF association, while the F154A mutant displayed similar binding to the WT protein engaging the H3K4me3-modified nucleosome (Figure 4C). (For compact notation, all amino acid numbering here is relative to the start of the PHD finger at amino acid 2717 in the full protein.) Interestingly, a helix insertion mutant (+QS) is no longer be able to bind doubly-modified nucleosomes with both binding modules concomitantly-- reducing the amount of nucleosome retained to levels commensurate to the WT protein binding nucleosomes bearing only H3K4me3. We further explored the role of this linker with extensive mutagenesis depicted schematically in Figure 4A insets. Perturbations of the linker were designed to extend the helix or introduce flexibility in this linkage with canonical helix-forming or helix breaking residues (Chou and Fasman, 1978). Insertion near or replacement of several residues in the center of the helix uniformly impaired apparent bivalent interaction (Figure 4D). Taken together, these results suggest that the precise relative orientation of the two domains is a critical determinant of bivalent mononucleosome binding potential.
Figure 4. Querying the roles of the PHD finger, the helical linker and the bromodomain in bivalent nucleosome binding via mutagenesis.
(A) A ribbon representation of the composite structure of the BPTF PHD-bromodomain module derived from superposition of the previously determined PHD-bromo in complex with H3K4me3 (Li et al., 2006) with the form I BPTF bromodomain H4K16ac complex (Figure 5A). The predicted domain structure of human BPTF shows the position of this module within the whole protein, colored as in Figure 1A and mutations are indicated in grey. Inset panels schematically depict the anticipated consequences of a series of linker helix mutations. (B) Mutations of the PHD-bromo module used to interrogate multivalent binding suggested by the structure assessed by FPA. Upper: a W32E mutation abolishes H3K4me3 binding without disrupting the PHD-finger fold (Li et al., 2006; Pena et al., 2006; Ruthenburg et al., 2007a), while a two amino acid linker helix insertion (labelled +QS), leaves the H3K4me3 binding capacity intact. Lower: a F154A mutation, designed by analogy to previous bromodomain mutagenesis (Dhalluin et al., 1999), abolishes bromodomain binding of H4K16ac; whereas neither the +QS nor the W32E mutants disrupt binding. See Figure S1F for Kd values. (C) Comparison of the WT GST-PHD-bromodomain (WT) to the series of mutant proteins in the same GST pull-down format as Figure 3. Mutants are labelled WE (W32E), FA (F154A), dbl (W32E + F154A), +QS (a QS insertion after S58 in the bridging helix between the PHD) as depicted in panel A, and 5% input is loaded for comparison (In). (D) Mutations designed to break the α-helix (residues 59–61 all mutated to glycine, TED→ GGG; the same residues mutated to glycines in combination with “SG” or SGGS insertions, TED → GGSGG, and TED→ GGSGGSGG, respectively) are compared to mutations intended to extend the helix and thereby rotate the two domains out of phase (+QS or +AA inserted between and S58 and T59) in the same experimental format as panel C. Mutations that effectively insert two amino acids into the linker helix are indicated in red. See also Figure S4. (data are represented as mean ± SD).
Structural analysis reveals two distinct acetyl-histone binding modes
As the mechanism of the composite selectivity in bivalent binding at the nucleosome level remained elusive, we wondered if the molecular basis of H4ac peptide binding might provide meaningful insight. To this end, we solved high resolution crystal structures of two different crystal forms of the BPTF bromodomain in complex with H4K16ac peptide, the preferred ligand of the PHD-bromodomain when combined with H3K4me3; and one structure of PHD-bromodomain in complex with H4K12ac peptide, representing a binding partner that is not selected for in bivalent nucleosome binding as a point of comparison (Figure 5, Table S1). All of these datasets yielded interpretable electron density for acetyllysine and several flanking residues (Figure S5D–F). Beyond the well-documented bromodomain-acetyllysine contacts that are nearly identical in all three structures (Dhalluin et al., 1999; Mujtaba et al., 2007; Zeng et al., 2008), specific contacts are apparent in each structure that account for sequence context specificity.
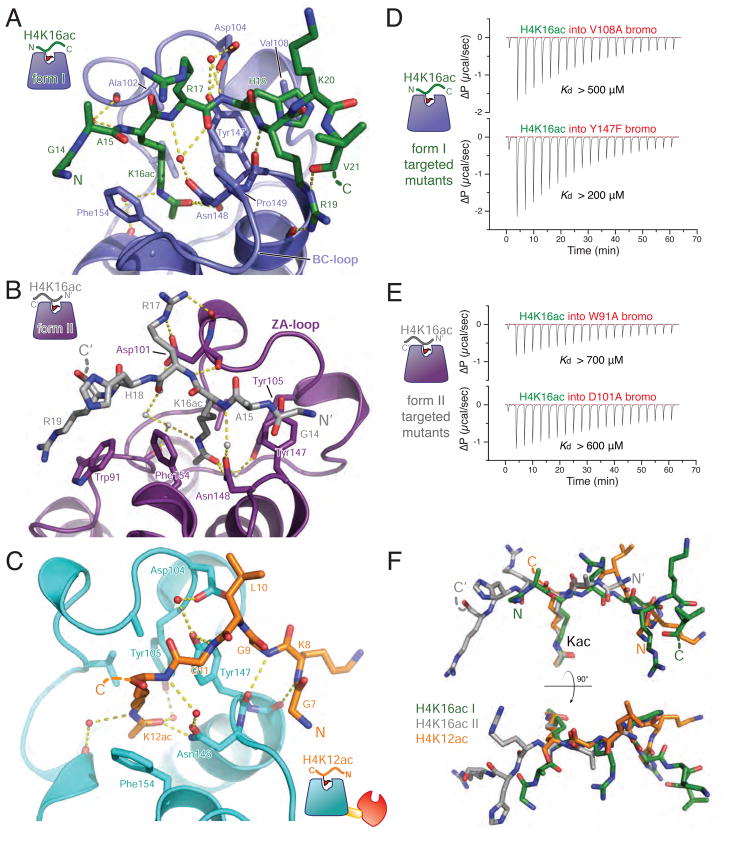
Figure 5. Structural analysis of the BPTF bromodomain peptide complexes.
(A) The model derived from crystal form I (bromodomain in blue) with the apical binding site for the H4K16ac peptide (green) rendered in ribbons and sticks. Hydrogen bonds are displayed as yellow dashed lines. (B) Interactions in crystal form II, the bromodomain is colored purple and the bound H4K16ac peptide is depicted in grey. (C) Interaction of the PHD-bromodomain in complex with H4K12ac peptide (peptide, orange; bromodomain, cyan). (D) ITC with BPTF bromodomain mutants designed to disrupt the form I binding mode (V108A and Y147F). Each of these mutations display modest binding deficits comporting with their modest roles in the form I interface. Dissociation constants were outside of the accurately measurable range, so a lower limit of possible Kd is provided for qualitative comparison. (E) Mutations to disrupt the form II binding interactions while leaving the form II binding mode intact: more substantial mutations, W91A and D101A, produce a more severe loss of affinity. For convenience of comparison, all ΔP scales are identical in scale. (F) Binding conformations of peptides (colored as in previous panels) from each of the structures is compared by Cα-superposition of their respective bromodomains. See also Figure S5.
The crystal form I H4K16ac-bromodomain complex reveals interactions that are analogous to those observed with the GCN5 bromodomain bound to the same mark (Cα r.m.s.d = 0.95 Å, Figure S5C). Remarkably, the H4 peptide orientation (N- to C-terminus) is inverted in our second structure (form II) relative to the first structure (Figure 5A,B,F), despite nearly identical bromodomain conformation (Cα r.m.s.d. = 0.52 Å). A similar reversal of peptide-binding orientation has been previously noted with the PCAF bromodomain, albeit with two different H3 acetylation sites (Zeng et al., 2008). As a point of comparison to the two H4K16ac structures, we examined a PHD-bromodomain complex with ligand that does not contribute to bivalent binding of the PHD-bromodomain in combination with H3K4me3. The structure of the PHD-bromodomain in complex with H4K12ac displays a “reversed” peptide binding orientation-- similar to that observed in the form II complex (Figure 5B, C). However, the ordered region of the H4K12ac peptide is located atop the αB helix, more reminiscent of the peptide positioning in the form I H4K16ac complex (Figure 5A, F; for further details see supplemental information).
Do both of these binding orientations contribute to the net affinity of the bromodomain for H4K16ac peptide? ITC was performed with bromodomain mutants designed to specifically perturb one of the two binding modes while leaving the other binding mode intact (Figure 5D,E). Although the binding of these mutant proteins to H4K16ac peptide was not strong enough to reliably quantify dissociation constants, residual binding affinity is apparent in the solution ITC measurements (compare ITC in Figure 5D,E with the H4 unmodified peptide binding in Figure 1E). Drastic alterations of contacts proper to the form II structure via point mutation of Trp 91 or Asp101 to alanine both severely erode, but do not completely destroy binding (Figure 5E, S5G,H). Selective disruption of interactions found only in the form I structure proved challenging, yielding conservative mutations more modest in their efficacy, V108A and Y147F. Nevertheless, these mutants demonstrate the solution binding relevance of this binding mode (Figure 5D, S5G,I). From these data we conclude that both bromodomain binding modes may play roles in the net affinity for H4K16ac peptide binding in solution. How the distinct molecular interactions in both the H4K12ac and H4K16ac complexes provide a plausible mechanism for nucleosomal H4 acetyl selectivity during bivalent binding of the nucleosome is addressed below (see Discussion).
BPTF co-localizes with doubly-modified nucleosomes in the nucleus
The localization of BPTF to the HOXA9 gene locus contingent upon H3K4 methylation mediated by the MLL complex has previously been established in HEK293 cells (Wysocka et al., 2006). Furthermore, MOF-mediated H4K16ac is highly enriched at the HOXA9 locus in these cells (Dou et al., 2005). Native chromatin immunoprecipitation (ChIP) followed by qPCR on mononucleosome biased fragments recapitulates these trends and affirms more than an order of magnitude signal difference for amplicons along this locus with the H4K12ac and H4K16ac antibodies (Figure 6A). To critically assess the relative import of each element in the BPTF PHD-bromo cassette for the localization of the full-length BPTF polypeptide to the HOXA9 locus, we established HEK293 cell lines with stable and equivalent full BPTF expression (WT and several of the mutants described above, Figure S6A,B). The association of ectopically-tagged BPTF protein with regions of the HOXA9 locus, as assessed by ChIP (Figure 6B), largely recapitulates our in vitro findings within the modest dynamic range of the experiment. All mutations diminish binding, suggesting that a minimally bivalent mode of nucleosomal engagement is important for BPTF recruitment or stabilization at this locus. Importantly, the +QS mutation disrupts localization to HOXA9. Thus, even in the context of full length BPTF and presumably other NURF complex members, the precise orientation of the PHD finger and the bromodomain appears to be crucial for full binding both in vitro and in vivo. Given the cell lines were constructed by stable integration of tagged BPTF into a genome with native BPTF expressed, and that the NURF complex is dimeric in this subunit (Barak et al., 2003), complete loss of localization is not expected. In order bypass this complication, we then restricted our ChIP-seq experiments to the BPTF PHD-bromodomain module.
Figure 6. Bivalent BPTF binding is important for localization of the BPTF PHD-bromodomain and full NURF complex.
(A) Native ChIP comparison of H3K4me3, H4K12ac, and H4K16ac marks. Four primer sets were employed to interrogate the HOXA9 locus (P1–P3, dark and light grey bars represent HOXA9 exons, and an untranslated region, respectively) and a distal intergenic site [(−) control region]. An average of three real-time PCR replicates of a representative experiment is displayed as a function of % input signal, with error bars reflecting PCR product threshold error amongst the replicates. For simplicity of HOXA9 display, the gene structure annotation represents the Crick strand sense of the genome in this as well as panels B and C. Data are represented as mean ± SD. (B) xChIP of HA-tagged BPTF from HEK293 cell lines that exhibit commensurate expression levels of the ectopic tagged constructs at the HOXA9 locus (with primer sets P2–P4). ChIP signal of tagged WT protein expressing cell lines is compared to discrete cell lines bearing the tagged mutant proteins corresponding to mutations depicted in Figure 4. Data are represented as mean ± SD. (C) ChIP-seq data for the HOXA9 locus, with the three histone modification tracks derived from nChIP-sequencing rendered in yellow, green and red as labelled with input tag counts overlayed in grey on the same scale. On the same abscissal scale and register, the xChIP sequencing counts from the 3xFLAG-tagged BPTF PHD-bromodomain are depicted in blue, with attendant input superimposed in grey. All tag counts are normalized by the factor (2×107/total mapped tags per track) and are unique. Below the continuous tag count graph, MACS peak-called regions relative to input for each sequencing track are depicted in rectangles of the same color. (C) Another example of histone modification patterns and PHD-bromodomain binding, displayed as in the previous panel. (E) Plot of average profile of the PHD-bromodomain at peak regions in the other datasets or intersects thereof. The average PHD signal is contoured on the regions that have MACS called peaks for each individual modification (colored as in 6C); or at loci with called peaks within the same 150 bp window for both H3K4me3 and H4K12ac datasets (purple); or H3K4me3 and H4K12ac datasets (blue). See also Figure S6.
Native ChIP followed by Illumina sequencing was performed to fully ascertain the genome-wide distribution of the H3K4me3, H4K12ac and H4K16ac marks in this cell line and correlate them with the PHD-bromodomain localization (Figure 6C–E). The latter dataset was gathered via cross-linking ChIP from a tagged PHD-bromo expressing HEK293 cell-line (Figure S6A). We detect numerous gene-proximal chromatin domains that bear significant peaks for H3K4me3 and each of the two acetylation marks examined; two examples are depicted in Figure 6C and D. Significant overlap between peaks of H3K4me3 and the tagged PHD-bromodomain was observed in a global sense. Although the number of clear peaks for the PHD-bromo is a much smaller set than the H3K4me3, 84% of these PHD-bromo peaks appear within 500 bp of an H3K4me3 peak (Figure S6C). As previously observed (Wang et al., 2008), the acetyl-specific ChIP-seq tracks are qualitatively more diffuse and less peak-like than the H3K4me3 signal and consequently peak-calling relative to input was more challenging. Even so, the average tag-densities differ in a locus specific-manner biased towards euchromatic regions, suggesting that the acetyl histone signal is meaningful (Figure S6D). Given that the PHD-bromo module colocalizes with a subset of H3K4me3 peaks (28%) and the broader distribution of acetyl marks, we would not expect very significant overlap in a global metagene analysis that includes all genes. Indeed, as depicted in Figure S6E, there is some overlap in the PHD-bromo and H3K4me3 average signal plotted for all genes normalized to 3kb, and very modest correlation acetyl mark signal (Shin et al., 2009).
Although the PHD-bromo is not present at every H3K4me3 site, its presence appears to be more correlated with loci that also bear histone H4 acetylation. Plotting the average PHD-bromo tag count over the peak regions in the H4K16ac and H3K4me3 datasets, as well as a where these two peaks intersect (within 150 bp), the apparent tag density is much higher for the PHD-bromo when H3K4me3 is combined with H4K16ac relative to either mark in isolation (Figure 6E). However, this also appears to be the case for the PHD-bromodomain plotted on intervals that are called peaks for both H4K12ac and H3K4me3. Taken together, these data suggest that the PHD-bromodomain tends to co-localize with H3K4me3 in regions that appear to have a reasonably high density of both H4K12 and H4K16 acetylation marks, particularly near TSS elements. The ChIP-seq data are consistent with the proposed role of both H3K4me3 and H4 acetylation playing a role in PHD-bromo recruitment, but there seems to be little distinction between the two H4ac marks when examined by this method. This raises and important question regarding overlapping ChIP-seq peaks—do they actually represent coexistence of two given marks within a single nucleosome?
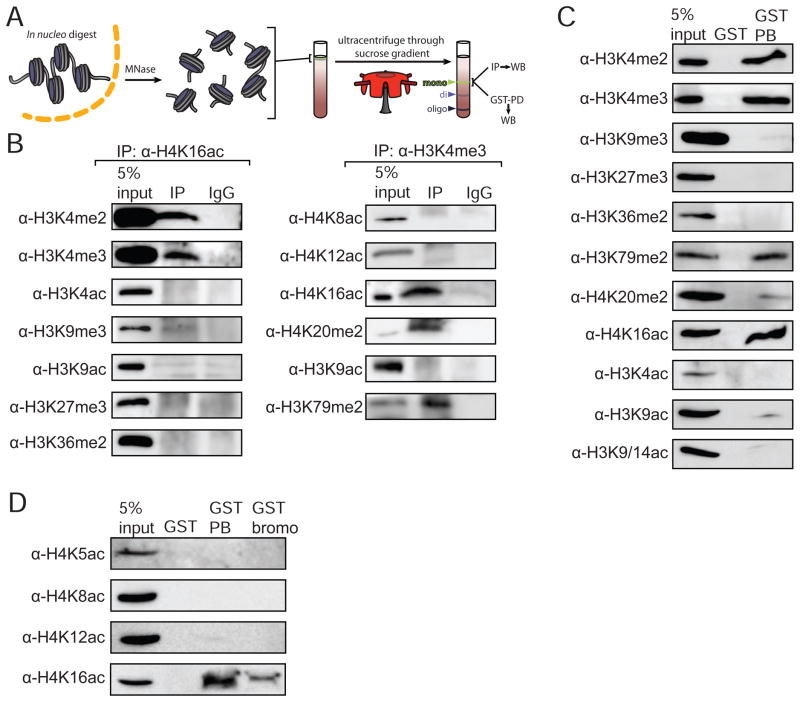
In order for the simultaneous bivalent binding of NURF described in vitro to be meaningful, mononucleosomes bearing both H3K4me3 and H4K16ac marks must exist in cells. To address this issue, we isolated high-purity mononucleosomes by sucrose gradient fractionation of MNase fragmented chromatin derived from HEK293 nuclei (Figure 7A and Figure S7A–F) (Mizzen et al., 1999). We then examined these purified nucleosomes for the coexistence of H3K4me3 with H4K16ac marks by co-immunoprecipitation relative to additional modifications and variants. There is a substantial pool of mononucleosomes bearing both modifications implicated in BPTF binding, H3K4me2/3 and H4K16ac (Figure 7B), while neither of these marks were found to reside in the same mononucleosomes as canonical repressive marks (H3K9me3, H3K27me3). To exclude the possibility of off-target antibody recognition bias, we performed the reciprocal immunoprecipitation experiments—employing an α-H3K4me3 antibody for IP followed by staining for H4K16ac (Figure 7B, right panel). H3K4me3 antibodies did not robustly IP H4K12ac— another mark that the bromodomain is capable of binding at the peptide level (Figure 1)-- or any other H4 acetylation mark. This finding suggests another possible source of selectivity: H4K12ac/H3K4me3 doubly-modified mononucleosomes do not appear to be present in a detectable population (antibodies are not available to H4K20ac, the other histone mark bound effectively by the BPTF bromodomain at the peptide level; see Figure 1). However, we observed substantial co-occupancy of H3K4me3 and H4K20me2, consistent with the observation that >80% of H4K20 is dimethylated at any given time while the corresponding acetylation is not very abundant (Pesavento et al., 2008).
Figure 7. The BPTF PHD-bromodomain preferentially engages a native population of H3K4me3 and H4K16ac doubly-modified nucleosomes.
(A) Schematic representation of the experiments: mononucleosome pools were isolated from in nucleo MNase digests and sucrose gradient ultracentrifugation (See Figure S5 for details). (B) Coimmunoprecipitation and western blotting of highly purified mononucleosomes with the well-validated modification and sequence-specific histone antibodies indicated. (C) Recombinantly produced GST-PHD-bromodomain (GST-PB) was used to pull-down native mononucleosomes and the associated material is compared to 5% of the input mononucleosome pool and GST alone for the ability to bind native nucleosomes contingent upon modification patterns. (D) An additional pull-down with the GST tagged bromodomain alone (GST-bromo) was performed alongside GST and GST-PB, and the bound material was probed by Western Blot against the four acetyl marks on the H4 tail for which there are antibodies available. See also Figure S7.
The analogous pull-down from the same highly purified mononucleosomal pools employing GST-tagged BPTF in place of antibodies again demonstrates preferential binding of the PHD-bromo module to H3K4me3 and H4K16ac bearing nucleosomes (Figure 7C,D). Additional coexisting marks resembled the pattern observed in the mononucleosomal IPs. Consistent with our semisynthetic nucleosome experiments, the PHD-bromo module bound a greater quantity of H4K16ac mononucleosomes as compared to the bromodomain alone (Figure 7D), with similar resin loading levels (Figure S7F). We did not detect significant BPTF binding to H4K12ac modified mononucleosomes under these conditions-- this could reflect lower abundance of intranucleosomal H3K4me3, but may also be a function of reduced BPTF binding. In order to distinguish these two possibilities, we cultured cells in trichostatin A, a potent HDAC inhibitor that serves to enrich otherwise transient acetylation marks like H4K12ac (Pesavento et al., 2006), and despite substantial enrichment of the mark, we observed minimal binding to the PHD-bromo module (Figure S7H).
DISCUSSION
One explicit prediction of the histone code hypothesis is the combinatorial readout of multiple histone marks (Strahl and Allis, 2000), although experimental support for this prediction is lacking. Here we present evidence for recognition of multiple heterotypic histone modifications simultaneously in a binding event that spans two different histone tails to establish nucleosome-level engagement contingent upon two discrete modifications. Of the three acetylated peptides with measureable bromodomain affinity, H4K16ac is an intuitively attractive BPTF binding partner. This mark is installed by the MOF acetyltransferase that may reside within the same MLL1 complex (Dou et al., 2005) that methylates H3K4 (Milne et al., 2002), the preferred binding partner of the proximal PHD finger (Li et al., 2007; Wysocka et al., 2006). The aggregate affinity of the PHD-bromo module for H3K4me3 and H4K16ac doubly-modified nucleosomes is greater than that of the PHD and bromodomains alone, yet more modest in magnitude than perhaps one would anticipate from discrete module-mark dissociation constants. Importantly, the bivalent nature of this interaction appears to effectively enhance the specificity of the bromodomain. Tethering of the PHD-finger to nucleosomal H3K4me3 appears to constrain the bromodomain to bind only H4K16ac, although at the peptide level there is not much distinction in binding affinities for this mark relative to the two flanking H4 acetyl marks.
What is the molecular mechanism for this composite specificity? Notwithstanding the molecular basis of each domain’s interactions with cognate peptides and the clear importance of the linker in permitting bivalent interactions, the precise molecular mechanism of this selectivity remains elusive. Our combined structural and mutagenesis studies with the BPTF bromodomain provide one potential explanation—the H4K16ac peptide has access to two alternate binding modes, while the H4K12ac may have access to only one of these peptide-binding orientations. Modelling suggests that only the peptide orientation in the form I H4K16ac bromodomain complex structure will accommodate a reasonable H3K4me3 approach angle in a nucleosome (Figure S5J), whereas the binding mode in the crystal form II structure does not seem to be compatible with bivalent binding (Figure S5K). Using similar constraints we are unable to model the PHD-bromodomain, spanning H4K12ac and H3K4me3 that is consistent with binding orientations observed in the structures. Literal interpretation of this bivalent model brings the BPTF PHD-bromo module in close contact with DNA. Yet, inspection of protein surface electrostatics suggest that the association may not be this intimate-- H3 tail flexibility could enable bivalent binding without engendering significant electrostatic repulsion from the nucleosomal DNA (Figure S5J, red arrow displays potential rigid body movement of PHD-bromo module away from the nucleosome). This model is consistent with the hydroxyl-radical footprint of the Drosophila NURF complex bound to a strongly positioned nucleosome -- the region of DNA near the pseudo-dyad at the duplex entrance/exit where the N-terminal H3 and H4 tails emerge from the octamer core is protected from cleavage (Schwanbeck et al., 2004). However, unambiguous delineation of the mechanism of H4 acetyl mark discrimination awaits the structural elucidation of the doubly-modified mononucleosome in complex with the PHD-bromodomain.
How might this modest affinity gain be functionally important for the full NURF complex? It appears that that every element of the bivalent nucleosome-binding interface described here is requisite for proper localization of the NURF complex to developmentally important loci in human cells. Yet in Drosophila, genetic deletions of a large portion of the C-terminus of BPTF (including the PHD-bromodomain) present no developmental defects outside of gametogenesis (Kwon et al., 2009), and there are known DNA-sequence specific factors involved in NURF recruitment (Badenhorst et al., 2005; Tsukiyama and Wu, 1995; Xiao et al., 2001). In vertebrates there are no analogous factors known to be involved in NURF recruitment, nor have clear DNA sequence elements related to NURF recruitment been identified. Here the role of histone modifications in the recruitment or stabilization of NURF complex at target loci appear to play a more important role, as removal of PHD-bromo module results in severe homeotic, hematopoietic, and gut abnormalities not found in flies (Wysocka et al., 2006).
It is likely the interplay of histone-modification specific interactions combined with other chromatin contacts determines the ultimate energetics and specificity in binding that culminates in genomic localization. Indeed, the bivalent interaction described here could be tetravalent in the context of the NURF complex as there are two copies of BPTF in the NURF complex (Barak et al., 2003). Beyond the established nucleosomal contacts made by other NURF complex subunits-- SNF2L likely has at least three DNA contacts (by analogy to ISWI) (Grune et al., 2003; Schwanbeck et al., 2004) and RbAp46/48 binds the first helix of H4 (Verreault et al., 1998)-- Drosophila BPTF (NURF301) bears three distinct nucleosomal interaction regions, only one of which may be attributed to the PHD-bromo module (Xiao et al., 2001). Specific interactions with the H4 tail, proximal to K16 and K20 seem to be essential for remodelling activity of NURF and other ISWI family complexes (Clapier et al., 2001; Hamiche et al., 2001; Shogren-Knaak et al., 2006).
Genome-level ChIP experiments have yielded a wealth of information about the localization of the H3K4me3 and H4K16ac histone marks—there are domains of significant overlap, particularly at active homeotic gene clusters. However, there is little data available that suggest what the absolute modification densities are at a given locus, so it is unclear to what extent spatially overlapping modification patterns described by ChIP-seq represent modifications that synchronously reside within a given nucleosome. By restricting the native chromatin queried to highly purified mononucleosomes, we provide compelling evidence for robust H4K4me3 and H4K16ac coexisting within a single nucleosome. In contrast, only modest coexistence of H3K4me3 and H4K12ac is detected by this method. This underscores a potential pitfall in interpreting ChIP-seq data: significant overlap of H3K4me3 with both of these acetyl marks on average may be observed across large genomic regions. Thus, apparent co-localization by this measure does not necessarily mean that two such marks actually co-exist in the same mononucleosome.
The discovery of a significant pool of doubly modified H3K4me3/H4K16ac mononucleosomes is consistent with the biochemical identification of both enzymes responsible for installing these marks associating within the MLL1 complex in one preparation (Dou et al., 2005; Milne et al., 2002). The coexistence of these marks at MLL1-regulated loci by ChIP is also consistent with the notion that this distinct population of doubly-modified mononucleosomes reside there. The clear coexistence of H3K4me3 and H3K79me2 marks in mononucleosomes was unanticipated, although others have noted this overlap by ChIP-seq (Wang et al., 2008). Interestingly, both H3K4me3 and H3K79me2 marks are downstream of H2B ubiquitylation (Briggs et al., 2002), and recent biochemical studies suggest direct stimulation of each responsible human methyltransferases by this ubiquitylation (Kim et al., 2009; McGinty et al., 2008).
Here we have provided evidence not only for simultaneous recognition of two heterotypic histone marks in a binding event that spans two histone tails as they project from the nucleosome, but also provide potential resolution of a purported weakness in the histone code hypothesis (Strahl and Allis, 2000). If more than one module is capable of binding a given mark, and each of these discrete modules resides in a different complex that transduces different downstream functional consequences, how can that mark itself have any unique information-encoding potential (Becker, 2006)? For example, the ‘spit personality’ of the H3K4me3 mark may be engaged by Taf3 of the TFIID complex to increase transcription (Vermeulen et al., 2007), or the ING2-bearing mSin3a complex to silence certain genes upon DNA damage (Shi et al., 2006). How can complex localization in these diametrically opposed processes be governed by the same mark? Our work suggests that both the PHD finger and bromodomain binding modules, as well as their relative orientation, are important for the full BPTF binding and Hox gene localization of the NURF complex. In this case, it is not the information content of interpreting a single mark that matters; rather it is the combination of engaging a pattern of marks and perhaps other local chromatin features that ultimately dictates cellular localization. Our findings call attention to the histone code being more complex than the unique interpretation of single mark and provide support for multivalent recognition of the chromatin polymer.
In addition to BPTF, there are 22 other polypeptides in the human proteome that have display linked PHD fingers and bromodomains, some of which are remarkably similar to the BPTF-PHD module studied here (Ruthenburg et al., 2007b). Initial work with several of these proteins suggests that each domain may be important for chromatin association (Eberharter et al., 2004; Ragvin et al., 2004; Tsai et al., 2010; Zhou and Grummt, 2005)— investigation of whether similar bivalent interactions play a role in their nuclear function will be of interest. More generally, we anticipate other examples of such combinatorial patterns being recognized by multivalent contacts at the level of single nucleosomes, oligonucleosomes and chromatin territories will be important for numerous genomic transactions.
EXPERIMENTAL PROCEDURES
SPOT blots, peptide pull-downs and fluorescence polarization anisotropy and ITC were performed essentially as described (Li et al., 2006; Nady et al., 2008; Wysocka et al., 2006). Details of recombinant protein production, X-ray crystallography, DNA-ruler assays, histone semisynthesis, and nucleosome reconstitution are available in the Extended Experimental Procedures. In brief, recombinant histones were reconstituted into octamers with semisynthetic histones prepared via expressed protein ligation as well as complementary recombinant histones (Muir, 2003; Shogren-Knaak and Peterson, 2004); then nucleosomes on a [32P] PNK end-labelled strong positioning sequence. Each nucleosome type was incubated with glutathione resin-immobilized GST-BPTF and washed five times over 45 minutes—the retained nucleosomal DNA was eluted from the resin, imaged by autoradiography of samples applied to native gels and/or quantified by scintillation counting. In nucleo MNase digestion was performed as a hybrid of previous conditions (Brand et al., 2008; Mizzen et al., 1999; O’Neill and Turner, 2003) with some modifications to the buffer conditions, and IPs were performed using conventional protocols. ChIP was performed according to established protocols or adaptations thereof, see Supplemental Experimental Procedures for details.
Supplementary Material
Acknowledgments
We would like to thank the staff at beamline 24ID-C of the Advanced Photon Source at the Argonne National Laboratory, and the staff at beamline X29 of the National Synchrotron Light Source at Brookhaven National Laboratory, supported by the US Department of Energy, for assistance with data collection. This work is based upon research conducted at the Northeastern Collaborative Access Team (NE-CAT) beamlines of the Advanced Photon Source and the Macromolecular Crystallography Research Resource (PXRR) at the National Synchrotron Light Source, which are supported by the National Center for Research Resources at the National Institutes of Health. We would like to thank L. Liang for assistance in protein production and crystallization of BPTF H4 peptide complexes; H. A. Zebrowski for assistance in the preparation of the SPOT membrane; S.S. Yi of the Microchemistry and Proteomics Core at Memorial-Sloan Kettering Cancer Center, for synthesis of peptides; Y. Wei of the Rockefeller Chemical Biology Spectroscopy center for use of their Biacore instrument; S. Yokoyama and H. Kurumizaka for an expression construct bearing codon optimized histone H4; K. Chiang for his 3_601_3_x32 repeat plasmid; M. Vila-Perello for HF cleavage of Boc-peptides; H. Dormann for HP1-chromodomain recombinantly produced protein; H. Yu of the Rockefeller Proteomics Core for MS assistance; C. Wu for the human BPTF cDNA; and L. Baker, F. Casadio, J. Denu, P.W. Lewis, K-M. Noh, R.G. Roeder, R. Sadeh, D. Shechter, T. Swigut, G. G. Wang and J. Wysocka for valuable discussions and scientific input. A.J.R is supported by Irvington Institute Fellowship Program of the Cancer Research Institute, R.K.M. is supported by an MSTP grant. This work was supported by a MERIT grant from the NIH and funds from The Rockefeller University to C.D.A., as well as funds from the Leukemia and Lymphoma Society and Starr Foundation to C.D.A and D.J.P., D.J.P. is supported by funds from the Abby Rockefeller Mauze Trust, the Dewitt Wallace and Maloris Foundations and T.W.M. is supported by an award from the NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badenhorst P, Voas M, Rebay I, Wu C. Biological functions of the ISWI chromatin remodeling complex NURF. Genes Dev. 2002;16:3186–3198. doi: 10.1101/gad.1032202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai X, Larschan E, Kwon SY, Badenhorst P, Kuroda MI. Regional control of chromatin organization by noncoding roX RNAs and the NURF remodeling complex in Drosophila melanogaster. Genetics. 2007;176:1491–1499. doi: 10.1534/genetics.107.071571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. Embo J. 2003;22:6089–6100. doi: 10.1093/emboj/cdg582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, et al. Proteome-wide prediction of acetylation substrates. Proc Natl Acad Sci U S A. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker PB. Gene regulation: a finger on the mark. Nature. 2006;442:31–32. doi: 10.1038/442031a. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, 3rd, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Brand M, Rampalli S, Chaturvedi CP, Dilworth FJ. Analysis of epigenetic modifications of chromatin at specific gene loci by native chromatin immunoprecipitation of nucleosomes isolated using hydroxyapatite chromatography. Nat Protoc. 2008;3:398–409. doi: 10.1038/nprot.2008.8. [DOI] [PubMed] [Google Scholar]
- Briggs SD, Xiao T, Sun ZW, Caldwell JA, Shabanowitz J, Hunt DF, Allis CD, Strahl BD. Gene silencing: trans-histone regulatory pathway in chromatin. Nature. 2002;418:498. doi: 10.1038/nature00970. [DOI] [PubMed] [Google Scholar]
- Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- Chou PY, Fasman GD. Empirical predictions of protein conformation. Annu Rev Biochem. 1978;47:251–276. doi: 10.1146/annurev.bi.47.070178.001343. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Tackett AJ, Smith ER, Fukuda A, Wysocka J, Allis CD, Chait BT, Hess JL, Roeder RG. Physical association and coordinate function of the H3 K4 methyltransferase MLL1 and the H4 K16 acetyltransferase MOF. Cell. 2005;121:873–885. doi: 10.1016/j.cell.2005.04.031. [DOI] [PubMed] [Google Scholar]
- Eberharter A, Vetter I, Ferreira R, Becker PB. ACF1 improves the effectiveness of nucleosome mobilization by ISWI through PHD-histone contacts. EMBO J. 2004;23:4029–4039. doi: 10.1038/sj.emboj.7600382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell. 2007;130:77–88. doi: 10.1016/j.cell.2007.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Kang JG, Dennis C, Xiao H, Wu C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc Natl Acad Sci U S A. 2001;98:14316–14321. doi: 10.1073/pnas.251421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell. 2009;137:459–471. doi: 10.1016/j.cell.2009.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy VM, Estroff LM, Whitesides GM. Multivalency in Ligand Design. In: Jahnke W, Erlanson DA, editors. Fragment-based Approaches in Drug Discovery. Weinheim: Wiley-VCH; 2006. pp. 11–53. [Google Scholar]
- Kwon SY, Xiao H, Glover BP, Tjian R, Wu C, Badenhorst P. The nucleosome remodeling factor (NURF) regulates genes involved in Drosophila innate immunity. Dev Biol. 2008;316:538–547. doi: 10.1016/j.ydbio.2008.01.033. [DOI] [PubMed] [Google Scholar]
- Kwon SY, Xiao H, Wu C, Badenhorst P. Alternative splicing of NURF301 generates distinct NURF chromatin remodeling complexes with altered modified histone binding specificities. PLoS Genet. 2009;5:e1000574. doi: 10.1371/journal.pgen.1000574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E, Matta J, Tessarollo L, Zhang YE, Ko MS, et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS Genet. 2008;4:e1000241. doi: 10.1371/journal.pgen.1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ilin S, Wang W-K, Duncan E, Wysocka J, Allis CD, Patel DJ. Molecular basis for the site-specific readout of histone H3 K4 trimethylation by the BPTF PHD finger of NURF. Nature. 2006;442:91–95. doi: 10.1038/nature04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Gogol M, Carey M, Lee D, Seidel C, Workman JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–1054. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDot1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne TA, Briggs SD, Brock HW, Martin ME, Gibbs D, Allis CD, Hess JL. MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell. 2002;10:1107–1117. doi: 10.1016/s1097-2765(02)00741-4. [DOI] [PubMed] [Google Scholar]
- Mizzen CA, Brownell JE, Cook RG, Allis CD. Histone acetyltransferases: preparation of substrates and assay procedures. Methods Enzymol. 1999;304:675–696. doi: 10.1016/s0076-6879(99)04041-0. [DOI] [PubMed] [Google Scholar]
- Moriniere J, Rousseaux S, Steuerwald U, Soler-Lopez M, Curtet S, Vitte AL, Govin J, Gaucher J, Sadoul K, Hart DJ, et al. Cooperative binding of two acetylation marks on a histone tail by a single bromodomain. Nature. 2009;461:664–668. doi: 10.1038/nature08397. [DOI] [PubMed] [Google Scholar]
- Muir TW. Semisynthesis of proteins by expressed protein ligation. Annu Rev Biochem. 2003;72:249–289. doi: 10.1146/annurev.biochem.72.121801.161900. [DOI] [PubMed] [Google Scholar]
- Mujtaba S, Zeng L, Zhou MM. Structure and acetyl-lysine recognition of the bromodomain. Oncogene. 2007;26:5521–5527. doi: 10.1038/sj.onc.1210618. [DOI] [PubMed] [Google Scholar]
- Nady N, Min J, Kareta MS, Chedin F, Arrowsmith CH. A SPOT on the chromatin landscape? Histone peptide arrays as a tool for epigenetic research. Trends Biochem Sci. 2008;33:305–313. doi: 10.1016/j.tibs.2008.04.014. [DOI] [PubMed] [Google Scholar]
- O’Neill LP, Turner BM. Immunoprecipitation of native chromatin: NChIP. Methods. 2003;31:76–82. doi: 10.1016/s1046-2023(03)00090-2. [DOI] [PubMed] [Google Scholar]
- Pena PV, Davrazou F, Shi X, Walter KL, Verkhusha VV, Gozani O, Zhao R, Kutateladze TG. Molecular mechanism of histone H3K4me3 recognition by plant homeodomain of ING2. Nature. 2006;442:100–103. doi: 10.1038/nature04814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Bullock CR, LeDuc RD, Mizzen CA, Kelleher NL. Combinatorial modification of human histone H4 quantitated by two-dimensional liquid chromatography coupled with top down mass spectrometry. J Biol Chem. 2008;283:14927–14937. doi: 10.1074/jbc.M709796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- Ragvin A, Valvatne H, Erdal S, Arskog V, Tufteland KR, Breen K, AMOY, Eberharter A, Gibson TJ, Becker PB, et al. Nucleosome binding by the bromodomain and PHD finger of the transcriptional cofactor p300. J Mol Biol. 2004;337:773–788. doi: 10.1016/j.jmb.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007a;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Li H, Patel DJ, Allis CD. Multivalent engagement of chromatin modifications by linked binding modules. Nat Rev Mol Cell Biol. 2007b;8:983–994. doi: 10.1038/nrm2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Liu T, Manrai AK, Liu XS. CEAS: cis-regulatory element annotation system. Bioinformatics. 2009;25:2605–2606. doi: 10.1093/bioinformatics/btp479. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Li H, Ruthenburg AJ, Allis CD, Patel DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–1040. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai WW, Wang Z, Yiu TT, Akdemir KC, Xia W, Winter S, Tsai CY, Shi X, Schwarzer D, Plunkett W, et al. TRIM24 links a non-canonical histone ignature to breast cancer. Nature. 2010;468:927–932. doi: 10.1038/nature09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukiyama T, Wu C. Purification and properties of an ATP-dependent nucleosome remodeling factor. Cell. 1995;83:1011–1020. doi: 10.1016/0092-8674(95)90216-3. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Mulder KW, Denissov S, Pijnappel WW, van Schaik FM, Varier RA, Baltissen MP, Stunnenberg HG, Mann M, Timmers HT. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell. 2007;131:58–69. doi: 10.1016/j.cell.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosomal DNA regulates the core-histone-binding subunit of the human Hat1 acetyltransferase. Curr Biol. 1998;8:96–108. doi: 10.1016/s0960-9822(98)70040-5. [DOI] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocka J, Swigut T, Xiao H, Milne TA, Kwon SY, Landry J, Kauer M, Tackett AJ, Chait BT, Badenhorst P, et al. A PHD finger of NURF couples histone H3 lysine 4 trimethylation with chromatin remodelling. Nature. 2006;442:86–90. doi: 10.1038/nature04815. [DOI] [PubMed] [Google Scholar]
- Xiao H, Sandaltzopoulos R, Wang HM, Hamiche A, Ranallo R, Lee KM, Fu D, Wu C. Dual functions of largest NURF subunit NURF301 in nucleosome sliding and transcription factor interactions. Mol Cell. 2001;8:531–543. doi: 10.1016/s1097-2765(01)00345-8. [DOI] [PubMed] [Google Scholar]
- Zeng L, Zhang Q, Gerona-Navarro G, Moshkina N, Zhou MM. Structural basis of site-specific histone recognition by the bromodomains of human coactivators PCAF and CBP/p300. Structure. 2008;16:643–652. doi: 10.1016/j.str.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C, Hayes JJ. Probing core histone tail-DNA interactions in a model dinucleosome system. Methods Enzymol. 2004;375:179–193. doi: 10.1016/s0076-6879(03)75012-5. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Grummt I. The PHD finger/bromodomain of NoRC interacts with acetylated histone H4K16 and is sufficient for rDNA silencing. Curr Biol. 2005;15:1434–1438. doi: 10.1016/j.cub.2005.06.057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.