Abstract
MicroRNAs (miRNAs) are a newly discovered endogenous class of small noncoding RNAs that play important posttranscriptional regulatory roles by targeting mRNAs for cleavage or translational repression. Accumulating evidence now supports the importance of miRNAs for human embryonic stem cell (hESC) self-renewal, pluripotency, and differentiation. However, with respect to induced pluripotent stem cells (iPSC), in which embryonic-like cells are reprogrammed from adult cells using defined factors, the role of miRNAs during reprogramming has not been well-characterized. Determining the miRNAs that are associated with reprogramming should yield significant insight into the specific miRNA expression patterns that are required for pluripotency. To address this lack of knowledge, we use miRNA microarrays to compare the “microRNA-omes” of human iPSCs, hESCs, and fetal fibroblasts. We confirm the presence of a signature group of miRNAs that is up-regulated in both iPSCs and hESCs, such as the miR-302 and 17–92 clusters. We also highlight differences between the two pluripotent cell types, as in expression of the miR-371/372/373 cluster. In addition to histone modifications, promoter methylation, transcription factors, and other regulatory control elements, we believe these miRNA signatures of pluripotent cells likely represent another layer of regulatory control over cell fate decisions, and should prove important for the cellular reprogramming field.
Introduction
Human embryonic stem cells (hESCs) have gained popularity as a potentially ideal cell candidate for regenerative medicine. First isolated by James Thomson and colleagues in 1998 [1], hESCs are derived from the inner cell mass of the human blastocyte and can be kept in an undifferentiated, self-renewing state indefinitely. In contrast to adult stem cells, hESCs have the advantage of being pluripotent, which endows them with the ability to differentiate into virtually every cell type in the human body. However, the use of human embryos is controversial in the United States, and potential tissue rejection following transplantation in patients remains problematic [2].
One way to circumvent these issues is to generate induced pluripotent stem cells (iPSCs). Mouse and human cells can be reprogrammed to pluripotency through ectopic expression of defined transcription factors [3–11]. The first successful reprogramming of human fibroblast cells into iPSCs was reported independently by Shinya Yamanaka (using OCT4, SOX2, KLF4, c-MYC) [8] and James Thomson (using OCT4, SOX2, NANOG, LIN28) [12]. The main advantage of this approach is that it does not need human embryos or oocytes to generate patient-specific stem cells, and therefore can potentially bypass the ethical and political debates that have surrounded this field for the past decade. Another important benefit is that for the first time, disease-specific stem cells can be created, which will help scientists understand the molecular mechanisms of many common inherited diseases [13].
For a number of reasons, these reprogramming methods have so far been gradual and slow, requiring weeks of cell culture with very low yield of iPSCs [4,11,14,15]. Inefficient delivery of factors to the cells is certainly one obstacle, and this challenge is being actively addressed by many groups. Another obstacle is the general lack of understanding of the molecular changes that underlie reprogramming [16,17]. Understanding the molecular circuitry of reprogramming will greatly benefit the field by providing new targets and pathways that could increase the yield of iPSCs. Efforts to better integrate the genomic and epigenomic networks that control reprogramming have been undertaken [18], but overall the specific mechanisms required for more efficient reprogramming remain elusive.
One potential regulatory mechanism of reprogramming that has so far received little attention is microRNAs (miRNAs). These small, noncoding RNAs play important posttranscriptional regulatory roles by targeting messenger RNAs (mRNAs) for cleavage or translational repression [19], and are key components of an evolutionarily conserved system of RNA-based gene regulation in eukaryotes [20]. Interestingly, hESCs are known to express miRNAs that are often undetectable in adult organs such as miR-371, miR-372, miR-302a, miR-302b, miR-302c, and miR-302d [21–25], whereas Dicer-deficient murine embryonic stem cells (ESCs), which cannot generate miRNAs, have been shown to be defective in differentiation [26,27]. These and other studies suggest that miRNAs likely play key roles in human and murine ESC gene regulation [24,28–32]. A recent study has attempted to incorporate miRNA gene regulation into a model of transcriptional regulatory circuitry of ESCs by generating genome-wide maps of binding sites for key ESC transcription factors such as Oct4, Sox2, and Nanog [33]. These ESC transcription factors were found to bind at many start sites of miRNA transcripts that have been detected in ESCs, such as the miR-302 cluster. Clearly, at least a subset of miRNAs seems to be involved in pluripotency, and these studies have contributed greatly to the understanding of miRNA networks in ESCs.
While significant efforts are being applied to ESCs, no study has yet analyzed the miRNA profile of human iPSCs. Determining the miRNAs that are associated with reprogramming may yield significant insight into the specific miRNA expression patterns needed for pluripotency. In this report, we use miRNA microarrays to compare the “microRNA-omes” of human iPSCs, hESCs, and fetal fibroblasts. We confirm the presence of a signature group of miRNAs that are up-regulated in both iPSCs and hESCs, such as the miR-302 and 17–92 clusters, as well as some subtle differences between the two pluripotent cell types, including the miR-371/372/373 cluster. We also note the broad changes in miRNA patterns between pluripotent cells and differentiated fibroblasts. These miRNA profiles are an initial step toward a better understanding of the regulatory networks that govern pluripotency and reprogramming.
Results
Characterization of fibroblasts, iPSCs, and hESCs
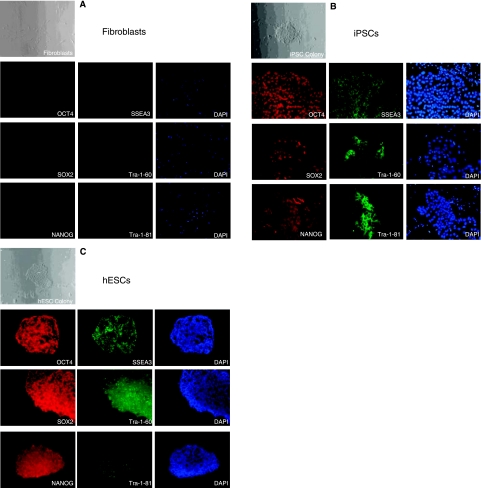
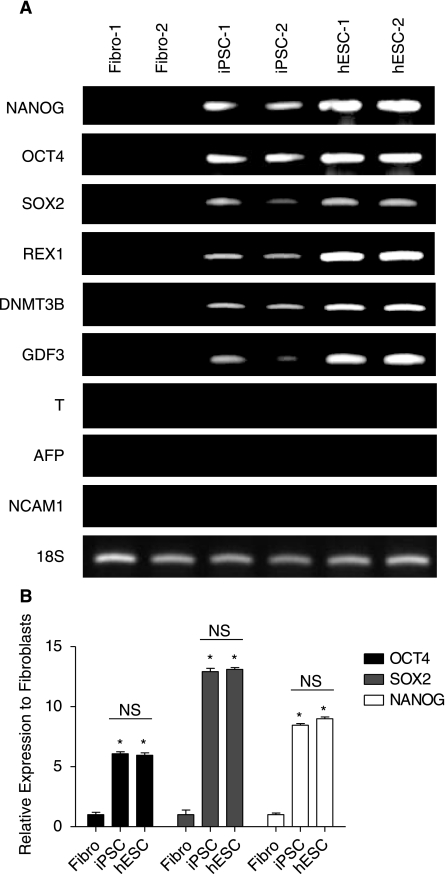
We obtained iPSCs from the James Thomson lab (University of Wisconsin-Madison), which were originally derived from IMR90 fetal fibroblasts (ATCC, Manassas, VA) using the reprogramming factors OCT4, SOX2, NANOG, and LIN28 [10]. hESCs (H7 from Wicell, Madison, WI) and iPSCs were maintained in the undifferentiated state using mTeSR™1 medium (StemCell Technologies, Vancouver, Canada). iPSC colonies exhibited an embryonic-like morphology that was similar to hESCs (Fig. 1). In contrast to IMR90 fibroblasts, iPSCs and hESCs stained positive for a number of well-characterized embryonic markers. We also performed RT-PCR gene expression analysis of selected embryonic and germ layer genes (Fig. 2A). Confirming our immunostaining results, iPSCs and hESCs expressed the same set of embryonic genes (OCT4, SOX2, REX1, DNMT3B, GDF3), but were negative for the differentiation markers Brachyury (T) (mesoderm), AFP (endoderm), and NCAM1 (neuroectoderm). We further verified the expression of OCT4, SOX2, and NANOG in iPSCs and hESCs using quantitative RT-PCR (Fig. 2B). The relative expression levels of these pluripotency genes were similar between the two pluripotent cell types. Taken together, these findings indicate that our iPSCs and hESCs were ostensibly free of spontaneously differentiated contaminants, and expressed appropriate levels of genes associated with pluripotency.
FIG. 1.
Morphology and pluripotent marker staining of human fibroblasts, iPSCs, and hESCs. (A) Fibroblasts exhibit typical morphology of these cells, do not grow in colonies, and do not stain for common markers of embryonic cells. (B) iPSCs exhibit colony formation and show robust staining for embryonic markers. Note that mouse embryonic fibroblasts (MEFs) were co-cultured with iPSCs to maintain them in an undifferentiated state. MEFs stain positive for DAPI, as can be seen in the image panel where they surround the iPSC colony, but are negative for all embryonic markers. (C) Similar to iPSCs, hESCs grow in colonies and stain positive for the same group of embryonic markers.
FIG. 2.
RT-PCR and quantitative RT-PCR analysis of selected genes. mRNA was isolated separately from biological duplicates of IMR90 fetal fibroblasts, iPSCs, and hESCs. (A) As expected, the embryonic genes (OCT4, NANOG, SOX2, REX1, DNMT3B, GDF3) were detectable in iPSCs and hESCs, but not in fetal fibroblasts. To ensure that our cells were in an undifferentiated state and had no differentiated contaminants, we also analyzed markers of mesoderm (Brachyury, T), endoderm (AFP), and neuroectoderm (NCAM1), which were all negative. Human 18S was used as loading control. Primer sequences can be found in Supplementary Materials and Methods. (B) Quantitative RT-PCR analysis of the pluripotency genes OCT4, SOX2, and NANOG confirms similar expression levels of these genes in iPSCs and hESCs. Error bars represent one standard deviation from the mean (*P value < 0.05 vs. fibroblasts; NS = Not Significant).
MiRNA profiling
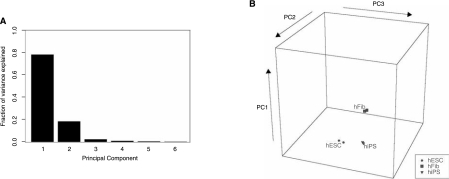
We next wanted to determine the miRNA profiles of all three cell types: IMR90 fibroblast, iPSCs, and hESCs. Using Sanger miRBase Version 10.0 miRNA expression microarrays (LC Sciences, Houston, TX), we analyzed 697 unique miRNAs across biological duplicates of each cell type (see Supplementary Table 1 (Supplementary Table is available online at http://www.liebertpub.com) for full normalized dataset and Supplementary Information for methods). We performed principal component analysis (PCA) on the miRNA profiles to assess the overall similarities and differences in miRNA expression between human iPSCs, hESCs, and fibroblasts. Of the six principal components (PC), the first three explain 78%, 18%, and 2%, respectively, of the total variance in the data (Fig. 3A). All three cell types had similar weights along the first PC, suggesting that this component represents variation in miRNA levels due to factors that are independent of cell type (e.g., promoter strength, pre-miRNA stability) (Fig. 3B). The second and third PCs appear to account for the cell type-specific differences in miRNA expression. The second PC distinguishes fibroblasts from both iPSCs and hESCs, suggesting that it corresponds to the pluripotency state of the cell. Finally, iPSCs and hESCs were separated, though to a lesser degree, along the third PC, indicating that these two cell types are not identical in their miRNA expression profiles. Thus the first three PC indicated that the miRNA expression profile of iPSCs is more similar to that of hESCs than fibroblasts, but still distinct from either.
FIG. 3.
Principal component (PC) analysis of miRNA profiles. (A) Bar plot showing the fraction of total variance in the miRNA data explained by each of the six PCs. The first three components together account for 99% of the total variance. (B) The six miRNA profiles, two each of human iPSCs (hiPS), ESCs (hESC), and fibroblasts (hFib), are plotted along the first three PCs. The coordinates of each point indicate the relative weight of each PC in that profile. As indicated by the scatter, the miRNA profile of iPSCs appears to be more similar to that of hESCs than fibroblasts, but still distinct from either.
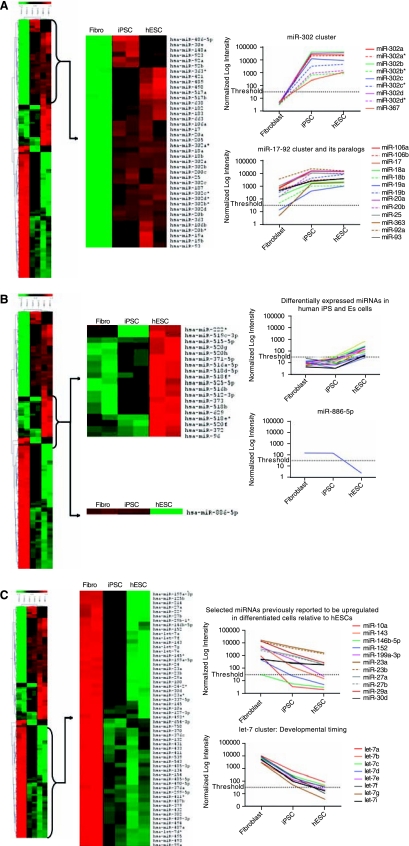
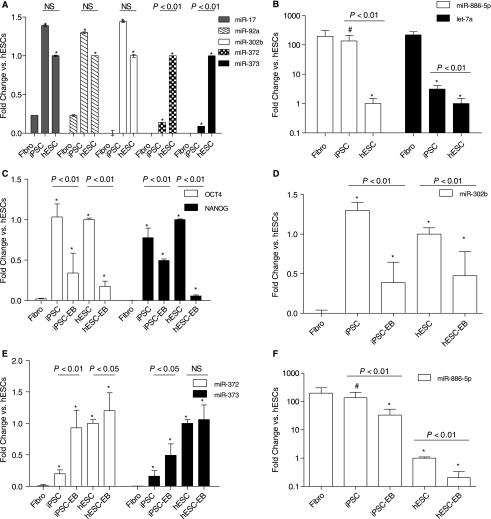
Using clustered heat maps of the miRNA datasets, we observed many miRNAs that were highly expressed in iPSCs and hESCs but not in fibroblasts (Fig. 4A); a smaller group of miRNAs that exhibited dissimilar expression in iPSCs and hESCs (Fig. 4B); and miRNAs that were up-regulated in fibroblasts but down-regulated in both iPSCs and hESCs (Fig. 4C) (see Supplementary Fig. 1 for full heat maps) (Supplementary Figure is available online at http://www.liebertpub.com) In addition to clustered heat maps, we have also included normalized intensity plots of selected miRNAs that have previously been associated with developmental or stem cell processes. These intensity plots give an indication of the absolute, rather than relative, miRNA expression across the three cell populations. Last, we confirmed the microarray expression patterns of selected miRNAs using quantitative RT-PCR, and also verified that iPSCs could be differentiated into embryoid bodies that express similar patterns of pluripotency genes as hESCs (Fig. 5).
FIG. 4.
Heat maps and signal intensity plots of miRNA expression across fibroblasts, iPSCs, and hESCs. ANOVA analysis demonstrates statistically significant differential miRNA regulation across the three samples. miRNAs with P values below 0.01 were selected for cluster analysis. (A) miRNAs upregulated in both iPSCs and hESCs compared to fibroblasts. Highlighted are the normalized signal intensity plots for the miR-302 cluster and miR-17–92 cluster, including its paralogs miR-106a-92 and miR-106b-25. (B) miRNAs exhibiting opposite expression in iPSCs and hESCs. Note that the normalized signal intensities of the miRNAs are not dramatic, though are statistically significant. In contrast to the group of miRNAs upregulated in hESCs but not in iPSCs (top panel), we observed only one miRNA (miR-886–5p) that was upregulated in iPSCs but not in hESCs (bottom panel). (C) miRNAs upregulated in fibroblasts but down-regulated in iPSCs and hESCs. Highlighted in the top and bottom panels are selected miRNAs known to be important in ESC biology and/or organismal development, such as the let-7 cluster that has a well-established role in late development timing. The clustering was done using hierarchical methods and was performed with average linkage and Euclidean distance metrices. The “threshold” value denotes signal intensities <32, a cutoff used by the microarray service provider, below which quantitation of signal may be inaccurate. Full intensity data can be found in Supplementary Table S1.
FIG. 5.
Quantitative RT-PCR of selected miRNAs confirms expression patterns seen in the microarray data, and also reveals miRNA expression during differentiation. (A) miR-17, miR-92a, and miR-302b show similarly high expression in both pluripotent cell types, but not in fibroblasts. In contrast, miR-372 and miR-373 are expressed at lower levels in iPSCs relative to hESCs, and at even lower levels in fibroblasts. (B) Also confirming the microarray data, miR-886–5p is expressed at statistically significant higher levels in fibroblasts and iPSCs compared to hESCs, and let-7a is more highly expressed in fibroblasts relative to both pluripotent cell types. However, expression of let-7a remains different between iPSCs and hESCs. (C) Differentiation of iPSCs and hESCs to Day 14 embryoid bodies reveals down-regulation of the pluripotency genes OCT4 and NANOG, as expected. (D) miR-302b, part of the miR-302 cluster that is highly expressed in pluripotent cells, is down-regulated in both iPSCs and hESCs at Day 14 of differentiation. (E) miR-372 and miR-373 are moderately up-regulated or unchanged, respectively, with differentiation. (F) miR-886–5p is down-regulated in both pluripotent cell types during differentiation. Error bars represent one standard deviation from the mean of biological duplicate experiments (#P value < 0.05 vs. fibroblasts; *P value < 0.01 vs. fibroblasts; NS = not significant).
The “pluripotent” miRNAs identified in Figure 4A are similar to results from previous studies of hESCs and embryonic tissues [21–23,34]. miRNAs are frequently transcribed together as polycistronic primary transcripts that are then processed into multiple individual mature miRNAs. The genomic organization of these miRNA clusters is often highly conserved, suggesting an important role for coordinated regulation and function. The most dramatic fluctuation in miRNA expression occurred in the miR-302 cluster, which has been consistently associated with ESCs in numerous miRNA profiling and sequencing studies [21–24,35]. Quantitative RT-PCR of miR-302b, one of the miR-302 cluster members, shows that it is similarly down-regulated in both iPSCs and hESCs upon differentiation to day 14 embryoid bodies (Fig. 5d). This group of eight miRNAs, which includes miR-367, is an evolutionarily conserved cluster located in the antisense intron of the protein-coding gene LARP7, a member of the La ribonucleoprotein domain family that is involved in RNA metabolism [36,37]. This poorly understood cluster of miRNAs, along with the LARP7 gene, may represent an important regulatory switch for the transition from embryonic to mature phenotypes in hESCs.
Also apparent from Figure 4A is the up-regulation of the miR-17–92 cluster and its paralogs in iPSCs and hESCs. These miRNAs are components of three paralogous clusters including miR-17–92 at 13q31.3, miR-106a-92 at Xq26.2, and miR-106b-25 at 7q22 with extensive sequence homologies [38]. In the human genome, the miR-17–92 cluster is located within intron 3 of the C13orf25 gene, and encodes six miRNAs (miR-17, miR-18a, miR-19a, miR-20a, miR-19b-1, and miR-92–1) [39,40]. Both the sequences of these mature miRNAs and their organization are highly conserved in all vertebrates, and they have attracted attention due to their possible oncogenicity [40]. This cluster has also been implicated in mammalian development, where loss-of-function of the miR-17–92 cluster resulted in mice that died shortly after birth with lung hypoplasia and cardiac defects [41]. Together with the miR-302 cluster and other up-regulated miRNAs identified in our profiles, the miR-17–92 cluster likely represents an important pluripotency regulatory element.
We were very interested to find any differences in miRNA expression between iPSCs and hESCs, and in Figure 4B we highlight a small group of miRNAs that exhibit dissimilar expression between the two pluripotent cell types. We observed a group of miRNAs that were expressed more highly in hESCs than in iPSCs (top panel), and a single miRNA, miR-886–5p, that was expressed more highly in iPSCs than in hESCs (bottom panel). To confirm these microarray results, we used quantitative RT-PCR to measure the levels of miR-886–5p, miR-372, and miR-373 (Fig. 5A,B); our results confirm that these miRNAs are expressed differently in iPSCs and hESCs. As a follow-up experiment, we also assessed the expression of selected miRNAs after differentiation to day 14 embryoid bodies (Fig. 5E,F). In both pluripotent cell types, the expression patterns of these miRNAs were similar upon differentiation, though we observed two miRNAs, miR-17 and miR-92a, that exhibited significantly higher expression in hESCs after differentiation relative to iPSCs (Supplementary Fig. 2) (Supplementary Figure is available online at http://www.liebertpub.com) Given the lack of annotation of these miRNAs, it is difficult to ascertain their biological significance for reprogramming and pluripotency at present.
Interestingly, except for two miRNAs, miR-629 and miR-96, all of the miRNAs that exhibited higher expression in hESCs relative to iPSCs (listed in the top panel of Fig. 4B) are located together within a 130-kb intergenic region of chromosome 19 (Supplementary Fig. 3) (Supplementary Figure is available online at http://www.liebertpub.com) This enormous meta-region of clustered miRNAs includes the miR-371/372/373 cluster, which has previously been shown to be up-regulated in hESCs [21–24], as well as a larger 54 miRNA cluster spanning 96 kb [42], much of which has also been shown to be up-regulated in hESCs [25]. We were therefore surprised to find that these same chromosome 19 clusters were not expressed at similar levels in iPSCs. Based on this observation, one could hypothesize that robust expression of these clustered miRNAs may not be critical for pluripotency, in contrast to a cluster such as miR-290 (found in mouse) that controls de novo DNA methylation, and is thus critical for cellular pluripotency [31]. In making this hypothesis, however, it is important to note that the normalized signal intensities, a measure of absolute rather than relative expression, show that the overall change in expression among these miRNAs is small. As comparison, the signal intensities for the miR-302 cluster show a much more dramatic fluctuation in expression levels (Fig. 4A). Therefore, though these changes are statistically significant, whether they are biologically significant remains to be determined. It is safe to say that differences, albeit small, do exist between the miRNA profiles of iPSCs and hESCs, and that some of these differences may help define the specific miRNAs required for induction and maintenance of pluripotency.
Figure 4C shows the miRNAs that are up-regulated in fibroblasts with respect to iPSCs and hESCs. Many of these miRNAs have little annotation and so have poorly defined roles in development and pluripotency, though they can be assumed to be important for maintenance of the differentiated or fetal fibroblast phenotype. The upper half of the magnified heat map in Figure 4C shows miRNAs that are minimally down-regulated in iPSCs (shaded in black), but that are significantly down-regulated in hESCs. A number of these miRNAs have previously been reported to be up-regulated in differentiated cells relative to hESCs [21,23,25], and we have plotted the intensity values for a few of these miRNAs in Figure 4C. Though there is an obvious downward trend in miRNAs when transitioning from the fibroblast to iPSC state, the expression levels of many do not completely decrease to the levels observed in hESCs. One example is let-7a, which showed statistically significant higher expression in iPSCs relative to hESCs as confirmed by quantitative RT-PCR (Fig. 5b). These observations underscore our general findings that there are subtle differences in the miRNA profiles between the two pluripotent cell types.
One group of miRNAs in Figure 4C that has been extensively studied is the let-7 cluster. This cluster is known to be expressed sequentially at specific stages of development to help coordinate developmental timing [20,43]. One recent study has also shown that Lin28, one of the reprogramming factors used by the James Thomson lab to induce pluripotency in fibroblasts, blocks an early transcript of let-7g that is preferentially expressed in adult cells [44]. Furthermore, the promoters of both let-7g and Lin28 are occupied by the embryonic transcription factors Oct4, Sox2, Nanog, and Tcf3 in mice, suggesting that these factors promote the transcription of both primary let-7g and Lin28, which then blocks the maturation of let-7g [33]. Taken together, these studies, along with our new findings, suggest one possible mechanism for the induction of pluripotency in adult cells using ectopic transcription factor expression.
Conclusion
Here we present the first profiling study of the human iPSC miRNA-ome. Our profiles demonstrate a high degree of similarity between iPSCs and hESCs, though with some differences. Because they can regulate numerous genes, often in common pathways, miRNAs may be regulators of cellular processes, akin to transcription factors that control entire programs of cellular differentiation and organogenesis. Therefore, in addition to histone modifications, promoter methylation, transcription factors, and other regulatory control elements, these miRNA signatures likely represent another layer of regulatory control for cell fate decisions, and should prove significant for the cellular reprogramming field.
The idea that miRNAs may contribute to the induction and maintenance of pluripotency arises from their well-established role in development and ESC biology. During development, miRNAs are known to interfere with the expression of mRNAs-encoding factors that control developmental timing, stem cell maintenance, and other developmental and physiological processes [20]. However, though over 450 human miRNA have been described [34], each of which is predicted to target tens if not hundreds of different mRNAs, only a fraction of them have been annotated; the great majority of miRNAs have poorly defined roles in cell fate decisions. This limited annotation, combined with a lack of knowledge of the transcriptional start sites, promoter regions, and downstream mRNA targets of miRNAs, has made it difficult to comprehensively integrate miRNAs into a broader understanding of molecular networks. Unlike other molecular mechanisms that have benefited from extensive research beginning in the latter half of the 20th century, the relatively new field of miRNAs, especially in the case of stem cells, has thus suffered from a lack of molecular context.
Given these issues, attempting to connect iPSC miRNA profiles to the molecular landscapes that underlie reprogramming will raise a number of basic questions. What regulates these small molecules, and what are their effects on downstream gene networks? If noncoding RNAs are regulated similarly to transcription factors, what advantage(s) do they offer over transcription factors in terms of regulation of pluripotency? Could miRNAs, each potentially targeting hundreds of mRNAs, represent nodes of control that dictate a cell's regulatory dynamics? These questions lead to a general observation regarding the fundamental difference between miRNA and mRNA activity. By inhibiting the translation of mRNAs within the cytoplasm, miRNAs directly affect the existing transcriptome of the cell. That is, the pool of mRNA in the cytoplasm—the “old” mRNA pool—is the immediate target of miRNAs. In contrast, transcription factors and other elements are responsible for activation and suppression of genes that will ultimately become the “new” mRNA pool. In the case of cellular reprogramming, as well as stem cell differentiation, it is therefore tempting to hypothesize that miRNAs can help transition the old mRNA pool to the new, with each layer of control required for efficient transformation of the cell. These questions and more will be active areas of investigation in stem cell biology and beyond for the foreseeable future.
Supplementary Material
Acknowledgments
We are grateful to Dr. James Thomson for his laboratory's iPSCs, Dr. Christoph Eicken at LC Sciences (Houston, TX) for help with miRNA microarrays, and Dr. Oscar Abilez for assistance with microscopy.
Research Support
Funding for this work was supplied in part by research grants from California Institute of Regenerative Medicine RS1–00322, Edward Mallinckrodt Jr. Foundation, NIH DP2OD004437, Stanford CVI, and BWF Career Award in Medical Scientist (J.C.W.). K.D.W. is a recipient of a Stanford University Bio-X Graduate Student Fellowship.
References
- 1.Thomson JA. Itskovitz-Eldor J. Shapiro SS. Waknitz MA. Swiergiel JJ. Marshall VS. Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 2008;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Swijnenburg R-J. Schrepfer S. Govaert JA. Cao F. Ransohoff K. Sheikh AY. Haddad M. Connolly AJ. Davis MM. Robbins RC. Wu JC. Immunosuppressive therapy mitigates immunological rejection of human embryonic stem cell xenografts. Proc Natl Acad Sci USA. 2008;105:12991–12996. doi: 10.1073/pnas.0805802105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aoi T. Yae K. Nakagawa M. Ichisaka T. Okita K. Takahashi K. Chiba T. Yamanaka S. Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- 4.Maherali N. Sridharan R. Xie W. Utikal J. Eminli S. Arnold K. Stadtfeld M. Yachechko R. Tchieu J. Jaenisch R. Plath K. Hochedlinger K. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2008;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Nakagawa M. Koyanagi M. Tanabe K. Takahashi K. Ichisaka T. Aoi T. Okita K. Mochiduki Y. Takizawa N. Yamanaka S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- 6.Okita K. Ichisaka T. Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2008;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 7.Park I-H. Zhao R. West JA. Yabuuchi A. Huo H. Ince TA. Lerou PH. Lensch MW. Daley GQ. Reprogramming of human somatic cells to pluripotency with defined factors. Nature. 2008;451:141–146. doi: 10.1038/nature06534. [DOI] [PubMed] [Google Scholar]
- 8.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2008;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 9.Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 10.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V. Stewart R. Slukvin II. Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2008;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Wernig M. Meissner A. Foreman R. Brambrink T. Ku M. Hochedlinger K. Bernstein BE. Jaenisch R. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2008;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 12.Yu J. Vodyanik MA. Smuga-Otto K. Antosiewicz-Bourget J. Frane JL. Tian S. Nie J. Jonsdottir GA. Ruotti V. Stewart R. Slukvin Thomson JA. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–1920. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 13.Park IH. Arora N. Huo H. Maherali N. Ahfeldt T. Shimamura A. Lensch MW. Cowan C. Hochedlinger K. Daley GQ. Disease-specific induced pluripotent stem cells. Cell. 2008. [Epub ahead of print]. [DOI] [PMC free article] [PubMed]
- 14.Brambrink T. Foreman R. Welstead GG. Lengner CJ. Wernig M. Suh H. Jaenisch R. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell. 2008;2:151–159. doi: 10.1016/j.stem.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadtfeld M. Maherali N. Breault DT. Hochedlinger K. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell. 2008;2:230–240. doi: 10.1016/j.stem.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaenisch R. Young R. Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell. 2008;132:567–582. doi: 10.1016/j.cell.2008.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamanaka S. Strategies and new developments in the generation of patient-specific pluripotent stem cells. Cell Stem Cell. 2007;1:39–49. doi: 10.1016/j.stem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 18.Mikkelsen TS. Hanna J. Zhang X. Ku M. Wernig M. Schorderet P. Bernstein BE. Jaenisch R. Lander ES. Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 20.Carrington JC. Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 21.Lakshmipathy U. Love B. Goff LA. Jörnsten R. Graichen R. Hart RP. Chesnut JD. MicroRNA expression pattern of undifferentiated and differentiated human embryonic stem cells. Stem Cells Dev. 2008;16:1003–1016. doi: 10.1089/scd.2007.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suh M-R. Lee Y. Kim JY. Kim S-K. Moon S-H. Lee JY. Cha K-Y. Chung HM. Yoon HS. Moon SY. Kim VN. Kim K-S. Human embryonic stem cells express a unique set of microRNAs. Dev Biol. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 23.Morin RD. O'Connor MD. Griffith M. Kuchenbauer F. Delaney A. Prabhu A-L. Zhao Y. McDonald H. Zeng T. Hirst M. Eaves CJ. Marra MA. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res. 2008;18:610–621. doi: 10.1101/gr.7179508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laurent LC. Chen J. Ulitsky I. Mueller F-J. Lu C. Shamir R. Fan J-B. Loring JF. Comprehensive microRNA profiling reveals a unique human embryonic stem cell signature dominated by a single seed sequence. Stem Cells. 2008;26:1506–1516. doi: 10.1634/stemcells.2007-1081. [DOI] [PubMed] [Google Scholar]
- 25.Bar M. Wyman SK. Fritz BR. Qi J. Garg KS. Parkin RK. Kroh EM. Bendoraite A. Mitchell PS. Nelson AM. Ruzzo WL. Ware C. Radich JP. Gentleman R. Ruohola-Baker H. Tewari M. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26:2496–2505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanellopoulou C. Muljo SA. Kung AL. Ganesan S. Drapkin R. Jenuwein T. Livingston DM. Rajewsky K. Dicerdeficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2008;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murchison EP. Partridge JF. Tam OH. Cheloufi S. Hannon GJ. Characterization of Dicer-deficient murine embryonic stem cells. Proc Natl Acad Sci USA. 2008;102:12135–12140. doi: 10.1073/pnas.0505479102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen C. Ridzon D. Lee C-T. Blake J. Sun Y. Strauss WM. Defining embryonic stem cell identity using differentiation-related microRNAs and their potential targets. Mamm Genome. 2008;18:316–327. doi: 10.1007/s00335-007-9032-6. [DOI] [PubMed] [Google Scholar]
- 29.Ivey KN. Muth A. Arnold J. King FW. Yeh R-F. Fish JE. Hsiao EC. Schwartz RJ. Conklin BR. Bernstein HS. Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang F. Kaneda M. O'Carroll Dn. Hajkova P. Barton SC. Sun YA. Lee C. Tarakhovsky A. Lao K. Surani MA. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2008;21:644–648. doi: 10.1101/gad.418707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinkkonen L. Hugenschmidt T. Berninger P. Gaidatzis D. Mohn F. Artus-Revel CG. Zavolan M. Svoboda P. Filipowicz W. MicroRNAs control de novo DNA methylation through regulation of transcriptional repressors in mouse embryonic stem cells. Nat Struct Mol Biol. 2008;15:259–267. doi: 10.1038/nsmb.1391. [DOI] [PubMed] [Google Scholar]
- 32.Zovoilis A. Nolte J. Drusenheimer N. Zechner U. Hada H. Guan K. Hasenfuss G. Nayernia K. Engel W. Multipotent adult germline stem cells and embryonic stem cells have similar microRNA profiles. Mol Hum Reprod. 2008;14:521–529. doi: 10.1093/molehr/gan044. [DOI] [PubMed] [Google Scholar]
- 33.Marson A. Levine SS. Cole MF. Frampton GM. Brambrink T. Johnstone S. Guenther MG. Johnston WK. Wernig M. Newman J. Calabrese JM. Dennis LM. Volkert TL. Gupta S. Love J. Hannett N. Sharp PA. Bartel DP. Jaenisch R. Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Landgraf P. Rusu M. Sheridan R. Sewer A. Iovino N. Aravin A. Pfeffer Sb. Rice A. Kamphorst AO. Landthaler M. Lin C. Socci ND. Hermida L. Fulci V. Chiaretti S. Foà R. Schliwka J. Fuchs U. Novosel A. Müller R-U. Schermer B. Bissels U. Inman J. Phan Q. Chien M. Weir DB. Choksi R. De Vita G. Frezzetti D. Trompeter H-I. Hornung V. Teng G. Hartmann G. Palkovits M. Di Lauro R. Wernet P. Macino G. Rogler CE. Nagle JW. Ju J. Papavasiliou FN. Benzing T. Lichter P. Tam W. Brownstein MJ. Bosio A. Borkhardt A. Russo JJ. Sander C. Zavolan M. Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2008;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bar M. Wyman SK. Fritz BR. Qi J. Garg KS. Parkin RK. Kroh EM. Bendoraite A. Mitchell PS. Nelson AM. Ruzzo WL. Ware C. Radich JP. Gentleman R. Ruohola-Baker H. Tewari M. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells. 2008;26(10):2496–505. doi: 10.1634/stemcells.2008-0356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ge X. Wu Q. Wang SM. SAGE detects microRNA precursors. BMC Genomics. 2008;7:285–285. doi: 10.1186/1471-2164-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Krueger BJ. Jeronimo Cl. Roy BB. Bouchard A. Barrandon C. Byers SA. Searcey CE. Cooper JJ. Bensaude O. Cohen EA. Coulombe B. Price DH. LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 2008;36:2219–2229. doi: 10.1093/nar/gkn061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hayashita Y. Osada H. Tatematsu Y. Yamada H. Yanagisawa K. Tomida S. Yatabe Y. Kawahara K. Sekido Y. Takahashi T. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2008;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 39.Croce CM. Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 40.Mendell JT. miRiad roles for the miR-17–92 cluster in development and disease. Cell. 2008;133:217–222. doi: 10.1016/j.cell.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ventura A. Young AG. Winslow MM. Lintault L. Meissner A. Erkeland SJ. Newman J. Bronson RT. Crowley D. Stone JR. Jaenisch R. Sharp PA. Jacks T. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bentwich I. Avniel A. Karov Y. Aharonov R. Gilad S. Barad O. Barzilai A. Einat P. Einav U. Meiri E. Sharon E. Spector Y. Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2008;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 43.Reinhart BJ. Slack FJ. Basson M. Pasquinelli AE. Bettinger JC. Rougvie AE. Horvitz HR. Ruvkun G. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2008;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 44.Viswanathan SR. Daley GQ. Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320:97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.