Abstract
Context:
Autoimmune Addison's disease (AD) is the major cause of primary adrenal failure in developed nations. Autoantibodies to 21-hydroxylase (21OH-AA) are associated with increased risk of progression to AD. Highest genetic risk is associated with the Major Histocompatibility region (MHC), specifically human leukocyte antigen (HLA)-DR3 haplotypes (containing HLA-B8) and HLA-DR4.
Objective:
The objective of the study was the further characterization of AD risk associated with MHC alleles.
Design, Setting, and Participants:
MHC genotypes were determined for HLA-DRB1, DQA1, DQB1, MICA, HLA-B, and HLA-A in 168 total individuals with 21OH-AA (85 with AD at referral and 83 with positive 21OH-AA but without AD at referral).
Main Outcome Measure(s):
Genotype was evaluated in 21OH-AA-positive individuals. Outcomes were compared with general population controls and type 1 diabetes patients.
Results:
In HLA-DR4+ individuals, HLA-B15 was found in only one of 55 (2%) with AD vs. 24 of 63 (40%) 21OH-AA-positive nonprogressors (P = 2 × 10−7) and 518 of 1558 (33%) HLA-DR4 patients with type 1 diabetes (P = 1 × 10−8). On prospective follow-up, none of the HLA-B15-positive, 21-hydroxylase-positive individuals progressed to AD vs. 25% non-HLA-B15 autoantibody-positive individuals by life table analysis (P = 0.03). Single nucleotide polymorphism analysis revealed the HLA-DR/DQ region associated with risk and HLA-B15 were separated by multiple intervening single-nucleotide polymorphism haplotypes.
Conclusions:
HLA-B15 is not associated with protection from 21OH-AA formation but is associated with protection from progression to AD in 21OH-AA-positive individuals. To our knowledge, this is one of the most dramatic examples of genetic disease suppression in individuals who already have developed autoantibodies and of novel dominant suppression of an autoimmune disease by a class I HLA allele.
Autoimmune Addison's disease (AD) is an uncommon disorder (1, 2) with insidious disease onset (e.g. weight loss and fatigue) and acute manifestations (in the form of Addisonian crisis) that can be fatal if not properly recognized and treated. The formation of 21-hydroxylase autoantibodies (21OH-AA) precedes the development of AD in the absence of symptoms and is a marker for risk of progression to clinical disease. This is similar to autoantibody formation and disease progression in more common autoimmune diseases such as type 1 diabetes mellitus (T1DM) and thyroiditis. More than 50% of individuals with autoimmune AD have other autoimmune diseases (including type 1 diabetes), making it likely that there is a common pathophysiology (1, 3–5).
Autoimmune polyendocrine syndrome, type 2 (APS-2) often manifests as a combination of AD, T1DM, and/or autoimmune thyroid disease. In contrast to the monogenic autoimmune polyendocrine syndrome, type 1 (APS-1; in which mutations occur in the AIRE gene), APS-2 is a polygenic disorder with the primary susceptibility loci within the major histocompatibility complex (MHC) (6–12). Specific MHC risk for AD (as an isolated disease or as part of APS-2) and T1DM is associated with the class II MHC haplotypes human leukocyte antigen (HLA)-DRB1*0301-DQB1*0201 (HLA-DR3) and HLA-DRB1*04-DQB1*0302 (HLA-DR4) (1, 10). T1DM is more associated with DRB1*0401, whereas AD is more associated with DRB1*0404 (1, 10, 13, 14) in the United States and Norway but not in Italian populations (15). Risk for AD and APS-2 has further been associated with an extended HLA-DR3 haplotype that includes a highly conserved MHC region from HLA-DR3 to HLA-B8 (including the MICA5.1 allele) but less often includes HLA-A1 of the classic extended DR3-A1-B8 haplotype (8–10, 16, 17).
Despite the known prevalence of extended HLA-DR4 haplotypes containing HLA-B15 in T1DM (6, 18), extended haplotypes of HLA-DR4 chromosomes (involving class I alleles) have not been well defined for AD. Here we describe a strikingly different class I association between DR4 haplotypes in individuals with AD vs. individuals with clinically isolated T1DM and individuals with 21OH-AA (many of whom also have T1DM) who have not progressed to clinical AD. We find dominant suppression of progression to AD in 21OH-AA positive, non-Addisonian individuals who have HLA-B15.
Materials and Methods
Study design
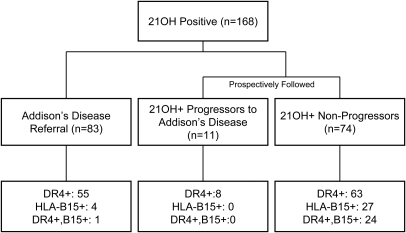
In 1993 we began ongoing HLA genotype analysis of AD referrals as well as 21OH-AA testing of relatives of AD individuals and individuals with T1DM. Follow-up for progression to AD in 21OH-AA+ individuals continues through 2010. Subjects with AD or 21OH-AA positivity were genotyped for HLA-B, HLA-DR, and HLA-DQ. As illustrated in Fig. 1, we have enrolled a total of 168 individuals with positive 21OH-AA status. There were 83 individuals with diagnosed AD upon referral, and 85 individuals without diagnosed AD but with positive 21OH-AA (including 11 who eventually were diagnosed with AD after we detected positive 21OH-AA and 74 who have not progressed). Subjects with 21OH-AA positivity were followed up from the date of first positive antibody detection until disease onset or last follow-up without AD. Of those who progressed, mean time to disease progression was 2.5 yr (confidence interval 95%, 1.5–4.0 yr). Nonprogressors were followed up for a mean of 5.2 yr (confidence interval 95%, 4.4–6.4 yr) from the time of first antibody detection, with the absence of AD typically confirmed via clinical history, ACTH levels, and cortisol levels after Cortrosyn stimulation.
Fig. 1.
Flow chart illustrating the number of 21OH-AA subjects included in this study.
Study population
The cohort with diagnosed AD has been previously described (17). We have now studied 94 non-APS-1 individuals with AD. There were 40 males and 54 females with the average age of AD onset 26 yr (22 yr in males and 30 yr in females). Thirty-one of 94 individuals also had diabetes, and 36 had other autoimmune diseases including thyroid disease, celiac disease, vitiligo, and lupus. Thirty-nine individuals had no other reported autoimmune disease.
Acquisition of 21OH-AA+ individuals was through screening of T1DM patients (n = 67) or relatives of AD or T1DM individuals (n = 16). Two 21OH-AA+ were referred for autoantibody testing on clinical suspicion of AD but did not progress by clinical testing until more than a year after initial autoantibody positivity. Within the 21OH-AA-positive nonprogressor cohort, 70 (80%) had type 1 diabetes and an additional six individuals had other autoimmune diseases (including autoimmune thyroid disease, celiac disease, and vitiligo). Only 12 individuals who were 21OH-AA positive had no other autoimmune disorders reported; thus, the great majority would be classified as having APS-2 (3). For those who volunteered race information (n = 126), all were Caucasian [Hispanic (n = 4) and white non-Hispanic (n = 122)]. Patients or their parents provided informed consent with institutional review board oversight at the University of Colorado Denver.
Inclusion criteria
AD patients were identified by several methods including referral with the diagnosis of AD from the National Adrenal Disease Foundation, referral of 21OH-AA-positive or symptomatic relatives of Addison's patients, and diabetic patients followed up at the Barbara Davis Center for Childhood Diabetes diagnosed with AD after 21OH-AA screening. All patients in this study were 21OH-AA positive (index >99th percentile of normal). 21OH-AA were measured with a fluid phase radioassay as previously described (10). Patients with the clinical diagnosis of APS-1 (with distinct signs, symptoms, and abnormalities in the AIRE gene) were excluded, and all included patients were interferon-α autoantibody negative (confirming absence of APS-1) (19).
Control data
HLA typing was available for families from the Type 1 Diabetes Genetics Consortium (T1DGC). The T1DGC enrolled 2300 affected sibling pairs with type 1 diabetes and their parents. Completed typing for HLA alleles across the MHC region as well as single-nucleotide polymorphism (SNP) typing more than 2800 SNPs across the MHC is available for all T1DGC individuals (20). Analyses in this paper used a single individual with T1DM per family (n = 2300 individuals).
DR3/4 control individuals from the general population were available from the Diabetes Autoimmunity Study of the Young (DAISY) with HLA typing at birth of approximately 30,000 newborns. Details regarding the DAISY population are provided in the paper from Rewers et al. 1996 (21). There were 271 HLA-DR3/4 (DRB1*0301-DQB1*0201/DRB1*04xx-DQB1*0302)-positive, autoantibody-negative, nondiabetic, unrelated individuals with HLA-B and HLA-A allele typing available. HLA-B frequency determination used only DR3/4-positive individuals, given that this is the highest risk, most common genotype for AD individuals.
Genotyping
We performed HLA-DRB1, HLA-DQB1, HLA-B, and HLA-A typing using hybridization of linear arrays of immobilized, sequence-specific oligonucleotides with amplified exon 2 DNA similar to previously described methodology (22) and direct sequencing of amplified HLA-DRB1 exon 2 to differentiate DRB1*04 subtypes.
MICA genotypes were determined using a fluorescent-based method as reported previously (23–25). Briefly, PCR fragments were generated using primers (MICA forward, 5′-/6-FAM/CCTTTTTTTCAGGGAAAGTGC-3′; MICA reverse, 5′-CCTTACCATCTCCAGAAACTGC-3′) that flank the microsatellite repeat polymorphism in the transmembrane region of the MICA gene (exon 5). Reactions (25 μl) were assembled using FailSafe PCR PreMix J, 2.5 U MasterAmp Taq polymerase (Epicenter, Madison, WI), 10 nmol of each primer, and 100 ng of genomic template. The PCR product was amplified via 35 PCR cycles of 94 C for 30 sec, 57 C for 35 sec, 72 C for 1 min, and a final extension of 72 C for 45 min. Products were diluted 1:50 and were separated by capillary electrophoresis on an ABI3100 Avant genetic analyzer (Applied Biosystems, Foster City, CA). Alleles were identified using GeneMapper version 3.5 (Applied Biosystems). The approximate peak sizes corresponding to each allele are 180 for allele 4, 182.7 for allele 5, 184.1 for allele 5.1, 185.7 for allele 6, and 194.5 for allele 9. The nomenclature used to define the MICA alleles was that of the MICA sequences, 2000 (26).
SNP analysis
Two SNPs were selected to differentiate a common extended DRB1*0404-DQB1*0302-B15 haplotype, with nearly identical SNPs across the MHC, from all other DRB1*04-DQB1*0302-B15 founder chromosomes with variable SNP patterns in the T1DGC data set based on previously described methods (27). The SNPs (rs2242657 C and rs2269475 T) were analyzed in the DR4-B15+, 21OH-AA+, nonprogressor population (n = 19 with usable DNA), available family members, and selected T1DGC controls using Taqman probes (hybridization/extension reaction with a fluorescence detection system developed by Applied Biosystems). To unambiguously assign phase for sentinel SNP typing in 21OH-AA positive nonprogressors, we analyzed family data as well as used homozygosity of the sentinel SNPs.
Statistical analysis
The Fisher's exact test (two sided) was used to calculate P values for association with AD, with α = 0.05. PRISM GraphPad 4 software (GraphPad Inc., San Diego, CA) was used for χ2 (α = 0.05). Life tables were also generated using PRISM GraphPad 4 software, wherein time to progression to AD (or last disease free clinical follow-up) from first detected 21OH-AA were placed in a nonlinear regression model comparing presence or absence of HLA-B15 in various subgroups.
Results
We tested 21OH-AA in individuals referred with AD (AD referrals), the relatives of AD patients, and patients with T1DM only and identified a total of 168 21OH-AA+ individuals (Fig. 1). Autoantibodies to 21-hydroxylase are typically present in 1.5% of non-Addisonian patients with type 1 diabetes and in more than 90% of patients with autoimmune AD (1, 10). The great majority of non-AD, 21OH-AA+ individuals enrolled in the current study had T1DM (80%, n = 68 of 85) as did 33% of the referred patients with existing AD (n = 31 of 94).
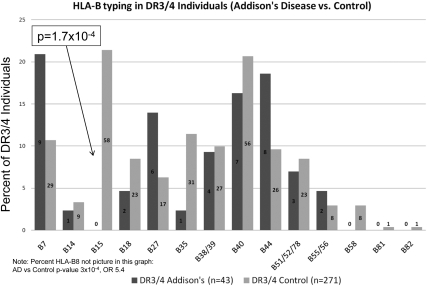
To fix HLA class II genotypes and analyze HLA-B allele association with AD, we initially analyzed DRB1*0301-DQB1*0201/DRB1*04-DQB1*0302 (DR3/4) individuals (the highest risk MHC class II genotype for AD). As illustrated in Fig. 2, in DR3/4 individuals HLA-B15 was the only HLA-B allele significantly decreased in AD (DR3/4 AD = 0 of 43, DR3/4 general population controls = 58 of 271, P = 1.7 × 10−4, odds ratio 0.04). Of note, HLA-B15 is rarely on HLA-DR3 haplotypes but frequently (and most commonly), it is associated with HLA-DR4 haplotypes (18). As we have reported previously, the HLA-B8 allele is positively associated with AD in DR3-positive individuals (not shown in the figure) and was present in 91% (39 of 43) of DR3/4 patients with AD vs. 64% (174 of 271) of DR3/4 DAISY general population controls (P = 3 × 10−4).
Fig. 2.
HLA-DR3/4 AD patients and general population (DAISY) controls were evaluated for HLA-B allele frequency. Within each bar the number indicates the number of individuals with the given HLA-B allele. HLA-B15 was the only HLA-B allele to be significantly decreased in the AD subjects. Corrected P value for HLA-B15 positivity in HLA-DR3/4 is 2.3 × 10−3.
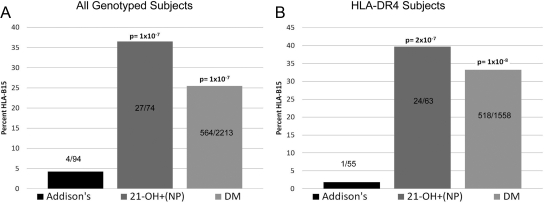
This negative association of HLA-B15 with AD is not limited to DR3/4 subjects. As shown in Fig. 3A, for all AD subjects (n = 94), only 4% had HLA-B15 (including progressors who were followed up to AD and patients with existing AD at referral) compared with 36% of 21OH-AA positive nonprogressors (27 of 74, P = 1 × 10−7) and 25% of patients with T1DM (564 of 2213, P = 1 × 10−7). After stratifying for HLA-DR4+ individuals in these three cohorts (Fig. 3B), the HLA-B15 allele is almost entirely absent in HLA-DR4+ individuals with AD [one of 55 (2%)], but it is common in DR4+, 21OH-AA-positive nonprogressors [24 of 63 (40%), P = 2 × 10−7].
Fig. 3.
HLA-DRB1, HLA-DQB1, and HLA-B were evaluated in AD individuals (including 21-hydroxylase positive progressors), 21-hydroxylase-positive individuals who have not progressed to AD [21-OH+(NP)], and patients with T1DM (DM). A, Percent HLA-B15 in all subjects genotyped for the aforementioned loci. B. Percent HLA-B15 in only the HLA-DR4 positive subjects. All P values listed are relative to AD.
With the known association of HLA-DRB1*0404 in AD and HLA-DRB1*0401 in T1DM as well as the high prevalence of T1DM in our 21OH-AA+ nonprogressor cohort, we examined the frequency of HLA-DRB1*0404 in 21OH-AA+ nonprogressors vs. those with AD. DRB1*0404 in 21OH-AA positive nonprogressors (n = 33 of 63, 52%) and Addison's disease individuals (n = 39 of 56, 67%) were increased relative to patients with T1DM [275 of 1605 (17%), P = 3.7 × 10−17 and 5.7 × 10−10, respectively]. Likewise, DRB1*0401 was present in 20% (11 of 56) of individuals with AD vs. 36% (23 of 63) of 21OH-AA+ nonprogressors (P = 0.05). Comparison of HLA-DR4 positive nonprogressors and individuals with T1DM reveals no significant difference in HLA-B15 frequency [24 of 63 (40%) and 518 of 1558 (34%), respectively, P = 0.34], indicating that HLA-B15 is not associated with decreased development of 21OH-AA. Among non-DR4 patients with 21OH-AA positivity, relatively few individuals had HLA-B15 including three of 39 Addison's patients (8%) compared with three of 12 nonprogressors (25%) (P = 0.13, not significant).
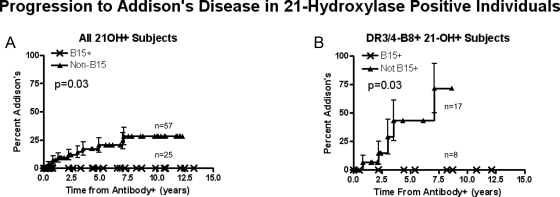
We prospectively follow up individuals who express 21OH-AA for progression to AD (Fig. 1). Despite expressing 21OH-AA, none of the HLA-B15-positive individuals have progressed to AD with up to 13 yr of follow-up. In contrast, nearly 25% of all non-HLA-B15, 21OH-AA-positive individuals progressed by 13 yr of follow-up by life table analysis (Fig. 4A, P = 0.03). Just analyzing the highest-risk HLA genotype (DR3/4 positive, HLA-B8 positive individuals), 75% of those lacking HLA-B15 progressed to AD by 7 yr of follow-up compared with 0% of those with HLA-B15 (P = 0.03).
Fig. 4.
Life table analysis showing protection associated with HLA-B15 in 21OH-AA-positive individuals. A, All genotyped subjects. B, Only DR3-B8/DR4 (the highest risk genotype) subjects.
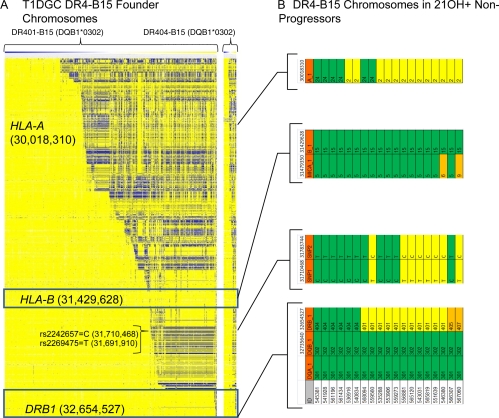
To better define HLA-DR4-B15 extended haplotypes, we analyzed SNPs previously typed across the MHC by the T1DGC consortium. Figure 5 illustrates SNPs across the MHC region in DRB1*0401-B15 and DRB1*0404-B15 founder chromosomes analyzing the large T1DGC diabetes family study (Fig. 5A). A conserved extended DRB1*0401-B15 haplotype was present in approximately two thirds (294 of 446) of the DRB1*0401-B15 chromosomes (all yellow). The conservation of the SNP haplotype begins to break up between HLA-B and HLA-A (with alternative SNPs colored in blue). Among potential variable SNPs, we chose two (rs2242657 and rs2269475) to mark haplotypic variation in the region between HLA-DRB1 and HLA-B in DR4-B15 haplotypes (represented as the 2-SNP haplotypes CT to mark the common DRB1*0404-like conserved haplotype or TC to mark alternative haplotypes). As shown in Fig. 5B, the DR4-B15 haplotypes of 21OH-AA-positive nonprogressors show variation between DRB1 and HLA-B and therefore did not show evidence of a single conserved, long-range MHC haplotype associated with lack of progression.
Fig. 5.
HLA-DR-DQ and HLA-B regions of conservation are noncontiguous (separated by polymorphic regions). A, SNPs across the MHC region. Each column represents a single DR4-B15 founder chromosome (n = 556) in the Type 1 Diabetes Genetics Consortium database of families with phased defined SNP haplotypes. Each row represents a SNP (n = 2828) with the allele in yellow if identical to the allele on the most common conserved DR401-B15 haplotype, in blue if the opposite allele, and in white if the allele could not be defined. The white column separates DRB1*0401 from DRB1*0404 T1DGC chromosomes. B, HLA genotype and sentinel SNP analysis in DR4-B15 chromosomes of 21OH-AA-positive nonprogressors. Alleles similar to the most common conserved DR404-B15 haplotype are seen in green, whereas alleles commonly on DR401-B15 haplotypes are in yellow. Orange represents non-401, non-404 haplotypes. Gene positions (indicated in parentheses in A) are based on National Center for Biotechnology Information Human Genome build 36.
Discussion
For polygenic autoimmune diseases such as T1DM and AD, the MHC region on chromosome 6 is the most important determinant of disease risk (7–10). Within the MHC region, alleles of class II MHC molecules such as DR and DQ are most strongly associated with AD and APS-2 (1, 10, 13, 14, 16, 17). These are immune response genes whose function is to present peptides to T cell receptors of CD4+ T cells. Conversely, the class I MHC molecules (e.g. HLA-B) classically present peptides to cytotoxic T cells (CD8+). An example of HLA class II influence on autoimmune disease is the genotype conferring high risk for AD and T1DM: DR3/4 (DRB1*0301-DQB1*0201/DRB1*04xx-DQB1*0302) (1, 10, 13, 14). In addition to class II HLA alleles, select class I alleles have been reported to increase risk of type 1 diabetes (e.g. HLA-B3906 and HLA-A24) (28–31) and AD (16, 17). Although the HLA-DR3 haplotype association with AD has been well defined, with the highest risk determined by the presence of HLA-B8 on a conserved HLA-DR3 haplotype often without HLA-A1 (17), there are fewer data regarding haplotypes of chromosomes with HLA-DR4.
We have found that HLA-B15 is associated with dramatic protection from progression to AD in DR4-positive individuals, even in individuals who have already developed 21OH-AA. This is the first report to our knowledge of a class I HLA allele associated with dominant suppression of autoimmune disease in individuals who have already developed autoantibodies. Looking at individuals who have already developed 21OH-AA but who have not progressed to AD, a total of 27 of 74 have HLA-B15, in contrast to patients with AD (four of 94, P = 6 × 10−8, odds ratio 0.07). By life table analysis, risk of progression to AD is greatly diminished in the presence of HLA-B15 (see Fig. 4). When specifically looking at the high-risk genotype DR3/4, none of the Addison's patients had the HLA-B15 allele (n = none of 43), whereas it was present in 35% of 21OH-AA-positive DR3/4 nonprogressors (n = 13 of 37, P = 1 × 10−5), 29% of DR3/4 individuals with T1DM (n = 261 of 910, P = 1.4 × 10−6) and 21% of DR3/4 controls (n = 58 of 271, P = 2 × 10−4).
Given the heterogeneity of autoimmune AD presentation, specifically when it is either isolated or as a part of APS-2 [with T1DM, thyroid autoimmune disease (TAD), or both], it is important to ensure the effect seen with HLA-B15 is not due to association with another comorbid disorder. When looking at the AD and 21OH-AA+ nonprogressor individuals with known comorbidity status, we found that there is still a significant difference in HLA-B15 presence between the two groups when comorbidities are accounted for. In isolated 21OH-AA positivity (without either T1DM or TAD), 30% (three of 10) of individuals had HLA-B15 vs. 5% (two of 39) in the AD group (P = 0.05). Similarly, among those with AD or 21OH-AA nonprogressors with T1DM, TAD, or both, 38% (24 of 63) of nonprogressors vs. 4% (two of 53) of AD had HLA-B15 (P = 4.5 × 10−6).
Because alleles of genes in the MHC are often in long-range linkage disequilibrium (spanning millions of base pairs) (6), it is difficult to determine whether the effect of HLA-B15 is from the specific HLA-B15 allele or from the haplotypes with which it is associated. With known protective effects of DRB1-DQB1 genotypes (including those with DRB1*0403), it is important to assess HLA-B15 associated protection in the context of class II alleles. HLA-B15 is not typically found with DRB1*0403. In the 21OH-AA+ nonprogressor group, only one of 27 individuals with HLA-B15 (4%) also had DRB1*0403 vs. only two of 560 (<1%, P = 0.13) of HLA-B15+T1DGC individuals and none of four AD individuals (P = 1). Looking at our DR3/4 general population controls, only three of 23 individuals with DRB1*0403 had HLA-B15.
Further analysis of sentinel SNPs in the predominantly DRB1*0404 and DRB1*0401 chromosomes indicates that HLA-DR4-B15 haplotypes are polymorphic between DRB1 and HLA-B15. Although long-range association with variation in between these two loci is a formal possibility, our findings suggest that at least two distinct MHC regions contribute to AD (class II and class I), with the simplest hypothesis positing direct influence of HLA-B15 on AD risk.
The paucity of HLA-B15 in AD populations also leads to an interesting observation regarding MICA 5.1, an allele reported to be associated with AD risk (10). MICA is located physically close to HLA-B and is associated with the DR3-B8 haplotype, and the MICA 5.1 allele (which is in strong linkage with HLA-B8) has been associated with increased risk of AD (7, 17, 24). Specifically we reported MICA5.1 homozygosity to be associated with an increased risk of progression to AD in 21OH-AA-positive subjects (10). Part of this association may be explained by MICA5.1's inclusion in the DR3-B8 extended haplotype, but given that DR3-B8 homozygosity is relatively uncommon in AD individuals (six of 94, 6%), increased homozygosity of MICA5.1 (48 of 94, 51%) in AD indicates non-DR3-B8 haplotypes with MICA5.1 is associated risk. Of note, there is strong linkage disequilibrium between HLA-B15 and MICA 5 (not MICA 5.1), which has been independently associated with other immune mediated disorders (e.g. giant cell arteritis and aggressive forms of periodontal disease) (32, 33).
In our series of DR3-B8/DR4 individuals, MICA 5.1/5.1 is found in 28% of 21OH-AA nonprogressors (n = six of 21) vs. 62% of Addison's individuals (n = 24 of 39, P = 0.01). However, when the data are stratified for HLA-B15 (thus removing HLA-B15 positive individuals), almost 50% of 21OH-AA nonprogressors have MICA5.1/5.1, making the difference of MICA 5.1 homozygosity between this group and AD group nonsignificant. Thus, we theorize that the MICA 5.1 homozygosity association with AD may be secondary to linkage disequilibrium of MICA 5.1 with high-risk HLA-B8 haplotypes as well as by the MICA 5 linkage disequilibrium with HLA-B15 low-risk haplotypes associated with decreased risk. To directly test this hypothesis, analysis of larger series and potentially other ethnic groups (with different patterns of linkage disequilibrium) will be needed.
If HLA-B15 itself is protective, its mechanism of action would be of interest because AD is primarily associated with class II HLA alleles. Increased disease risk with class I HLA alleles is typically ascribed to enhanced presentation of autoantigenic peptides to CD8+ T cells. There are reports of class I HLA alleles negatively associated with autoimmune disease (34) along with literature supporting the existence of CD8+ regulatory T cells, which are HLA class I restricted (35–38). This mechanism of pathogenesis, and its possible relation to HLA-related autoimmune disease, warrants further exploration.
Protection mediated by the molecule HLA-B15 likely involves MHC class I structure and the polymorphic amino acids that line the binding pocket. When looking at amino acid sequences in HLA-B*0801 (associated with highest AD risk) and HLA-B*1501, there are 23 total amino acids that differ including seven changes in the binding pocket base and five in the α-helix of the binding groove (39, 40). Sequence differences of multiple alleles will need to be further explored using molecular modeling, pending analysis of larger numbers and multiple populations of individuals with AD.
Even with 21OH-AA and the DR3-B8/DRB1*0404 highest-risk genotype, HLA-B15 (or linked loci) protects from progression to overt AD. This study illustrates the potential of analysis of HLA haplotypes in determining risk in antibody-positive populations. We typically screen patients with T1DM for 21OH-AA, and we annually evaluate adrenal function for those found to be positive. Although AD has classic symptoms and signs, in patients with T1DM initial presentation can be as subtle as increased occurrences of hypoglycemia and decreasing insulin requirement (without hyperpigmentation). If further prospective studies support our findings, the approximate one third of 21OH-AA-positive patients who also have HLA-B15 are at greatly decreased risk of progression to AD.
Acknowledgments
This work was supported by the Type 1 Diabetes Genetics Consortium, a collaborative clinical study sponsored by the National Institute of Diabetes and Digestive and Kidney Diseases, National Institute of Allergy and Infectious Diseases, National Human Genome Research Institute, the Eunice Kennedy Shriver National Institute of Child Health and Human Development, and Juvenile Diabetes Research Foundation International and supported by Grant U01 DK062418. This work was also supported by the National Institutes of Health Grant DK32083, Diabetes Autoimmunity Study in the Young Grant DK32493, Autoimmunity Prevention Center Grant AI050864, Diabetes Endocrine Research Center Grant P30 DK57516, Clinical Research Centers Grants MO1 RR00069 and MO1 RR00051, the Immune Tolerance Network Grant AI15416, the American Diabetes Association, the Juvenile Diabetes Research Foundation Grant 11-2005-15, the Children's Diabetes Foundation, and the Brehm Coalition. P.R.B. is a Fellow of the Pediatric Scientist Development Program. The project described was also supported by Award K12-HD000850 from the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Disclosure Summary: The authors have no conflicts of interest to disclose.
Footnotes
- AD
- Addison's disease
- APS-1
- autoimmune polyendocrine syndrome, type 1
- APS-2
- autoimmune polyendocrine syndrome, type 2
- DAISY
- Diabetes Autoimmunity Study of the Young
- HLA
- human leukocyte antigen
- HLA-DR3
- HLA-DRB1*0301-DQB1*0201
- HLA-DR4
- HLA-DRB1*04-DQB1*0302
- MHC
- major histocompatibility complex
- 21OH-AA
- 21-hydroxylase autoantibodies
- SNP
- single-nucleotide polymorphism
- TAD
- thyroid autoimmune disease
- T1DGC
- Type 1 Diabetes Genetics Consortium
- T1DM
- type 1 diabetes mellitus.
References
- 1. Erichsen MM, Løvås K, Skinningsrud B, Wolff AB, Undlien DE, Svartberg J, Fougner KJ, Berg TJ, Bollerslev J, Mella B, Carlson JA, Erlich H, Husebye ES. 2009. Clinical, immunological, and genetic features of autoimmune primary adrenal insufficiency: observations from a Norwegian registry. J Clin Endocrinol Metab 94:4882–4890 [DOI] [PubMed] [Google Scholar]
- 2. Betterle C, Dal Pra C, Mantero F, Zanchetta R. 2002. Autoimmune adrenal insufficiency and autoimmune polyendocrine syndromes: autoantibodies, autoantigens, and their applicability in diagnosis and disease prediction. Endocr Rev 23:327–364 [DOI] [PubMed] [Google Scholar]
- 3. Eisenbarth GS, Gottlieb PA. 2004. Autoimmune polyendocrine syndromes. N Engl J Med 350:2068–2079 [DOI] [PubMed] [Google Scholar]
- 4. Falorni A, Laureti S, De Bellis A, Zanchetta R, Tiberti C, Arnaldi G, Bini V, Beck-Peccoz P, Bizzarro A, Dotta F, Mantero F, Bellastella A, Betterle C, Santeusanio F. 2004. Italian addison network study: update of diagnostic criteria for the etiological classification of primary adrenal insufficiency. J Clin Endocrinol Metab 89:1598–1604 [DOI] [PubMed] [Google Scholar]
- 5. Badenhoop K, Walfish PG, Rau H, Fischer S, Nicolay A, Bogner U, Schleusener H, Usadel KH. 1995. Susceptibility and resistance alleles of human leukocyte antigen (HLA) DQA1 and HLA DQB1 are shared in endocrine autoimmune disease. J Clin Endocrinol Metab 80:2112–2117 [DOI] [PubMed] [Google Scholar]
- 6. Alper CA, Larsen CE, Dubey DP, Awdeh ZL, Fici DA, Yunis EJ. 2006. The haplotype structure of the human major histocompatibility complex. Hum Immunol 67:73–84 [DOI] [PubMed] [Google Scholar]
- 7. Gambelunghe G, Falorni A, Ghaderi M, Laureti S, Tortoioli C, Santeusanio F, Brunetti P, Sanjeevi CB. 1999. Microsatellite polymorphism of the MHC class I chain-related (MIC-A and MIC-B) genes marks the risk for autoimmune Addison's disease. J Clin Endocrinol Metab 84:3701–3707 [DOI] [PubMed] [Google Scholar]
- 8. Gombos Z, Hermann R, Kiviniemi M, Nejentsev S, Reimand K, Fadeyev V, Peterson P, Uibo R, Ilonen J. 2007. Analysis of extended human leukocyte antigen haplotype association with Addison's disease in three populations. Eur J Endocrinol 157:757–761 [DOI] [PubMed] [Google Scholar]
- 9. Weetman AP, Zhang L, Tandon N, Edwards OM. 1991. HLA associations with autoimmune Addison's disease. Tissue Antigens 38:31–33 [DOI] [PubMed] [Google Scholar]
- 10. Yu L, Brewer KW, Gates S, Wu A, Wang T, Babu SR, Gottlieb PA, Freed BM, Noble J, Erlich HA, Rewers MJ, Eisenbarth GS. 1999. DRB1*04 and DQ alleles: expression of 21-hydroxylase autoantibodies and risk of progression to Addison's disease. J Clin Endocrinol Metab 84:328–335 [DOI] [PubMed] [Google Scholar]
- 11. Neufeld M, Maclaren NK, Blizzard RM. 1981. Two types of autoimmune Addison's disease associated with different polyglandular autoimmune (PGA) syndromes. Medicine (Baltimore) 60:355–362 [DOI] [PubMed] [Google Scholar]
- 12. Bilbao JR, Martín-Pagola A, Pérez De Nanclares G, Calvo B, Vitoria JC, Vázquez F, Castaño L. 2003. HLA-DRB1 and MICA in autoimmunity: common associated alleles in autoimmune disorders. Ann NY Acad Sci 1005:314–318 [DOI] [PubMed] [Google Scholar]
- 13. Aly TA, Ide A, Jahromi MM, Barker JM, Fernando MS, Babu SR, Yu L, Miao D, Erlich HA, Fain PR, Barriga KJ, Norris JM, Rewers MJ, Eisenbarth GS. 2006. Extreme genetic risk for type 1a diabetes. Proc Natl Acad Sci USA 103:14074–14079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Erlich H, Valdes AM, Noble J, Carlson JA, Varney M, Concannon P, Mychaleckyj JC, Todd JA, Bonella P, Fear AL, Lavant E, Louey A, Moonsamy P. 2008. HLA DR-DQ haplotypes and genotypes and type 1 diabetes risk: analysis of the Type 1 Diabetes Genetics Consortium Families. Diabetes 57:1084–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gambelunghe G, Kockum I, Bini V, De Giorgi G, Celi F, Betterle C, Giordano R, Libè R, Falorni A. 2005. Retrovirus-like long-terminal repeat DQ-LTR13 and genetic susceptibility to type 1 diabetes and autoimmune Addison's disease. Diabetes 54:900–905 [DOI] [PubMed] [Google Scholar]
- 16. Eisenbarth G, Wilson P, Ward F, Lebovitz HE. 1978. HLA type and occurrence of disease in familial polyglandular failure. N Engl J Med 298:92–94 [DOI] [PubMed] [Google Scholar]
- 17. Baker PR, Baschal EE, Fain PR, Triolo TM, Nanduri P, Siebert JC, Armstrong TK, Babu SR, Rewers MJ, Gottlieb PA, Barker JM, Eisenbarth GS. 2010. Haplotype analysis discriminates genetic risk for DR3-associated endocrine autoimmunity and helps define extreme risk for Addison's disease. J Clin Endocrinol Metab 95:E263–E270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Baschal EE, Aly TA, Jasinski JM, Steck AK, Noble JA, Erlich HA, Eisenbarth GS. 2009. Defining multiple common “completely” conserved major histocompatibility complex SNP haplotypes. Clin Immunol 132:203–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Meager A, Visvalingam K, Peterson P, Möll K, Murumägi A, Krohn K, Eskelin P, Perheentupa J, Husebye E, Kadota Y, Willcox N. 2006. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med 3:e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brown WM, Pierce J, Hilner JE, Perdue LH, Lohman K, Li L, Venkatesh RB, Hunt S, Mychaleckyj JC, Deloukas P; Type 1 Diabetes Genetics Consortium 2009. Overview of the MHC fine mapping data. Diabetes Obes Metab 11(Suppl 1):2–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rewers M, Bugawan TL, Norris JM, Blair A, Beaty B, Hoffman M, McDuffie RS, Jr, Hamman RF, Klingensmith G, Eisenbarth GS, Erlich HA. 1996. Newborn screening for HLA markers associated with IDDM: diabetes autoimmunity study in the young (DAISY). Diabetologia 39:807–812 [DOI] [PubMed] [Google Scholar]
- 22. Bugawan TL, Erlich HA. 1991. Rapid typing of HLA-DQB1 DNA polymorphism using nonradioactive oligonucleotide probes and amplified DNA. Immunogenetics 33:163–170 [DOI] [PubMed] [Google Scholar]
- 23. Zake LN, Ghaderi M, Park YS, Babu S, Eisenbarth G, Sanjeevi CB. 2002. MHC class I chain-related gene alleles 5 and 5.1 are transmitted more frequently to type 1 diabetes offspring in HBDI families. Ann NY Acad Sci 958:309–311 [DOI] [PubMed] [Google Scholar]
- 24. Park YS, Sanjeevi CB, Robles D, Yu L, Rewers M, Gottlieb PA, Fain P, Eisenbarth GS. 2002. Additional association of intra-MHC genes, MICA and D6S273, with Addison's disease. Tissue Antigens 60:155–163 [DOI] [PubMed] [Google Scholar]
- 25. Triolo TM, Baschal EE, Armstrong TK, Toews CS, Fain PR, Rewers MJ, Yu L, Miao D, Eisenbarth GS, Gottlieb PA, Barker JM. 2009. Homozygosity of the polymorphism MICA5.1 identifies extreme risk of progression to overt adrenal insufficiency among 21-hydroxylase antibody-positive patients with type 1 diabetes. J Clin Endocrinol Metab 94:4517–4523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson J, Pérez-Rodríguez M, Waller MJ, Cuillerier B, Bahram S, Yao Z, Albert ED, Madrigal JA, Marsh SG. 2001. MICA sequences 2000. Immunogenetics 53:150–169 [DOI] [PubMed] [Google Scholar]
- 27. Barker JM, Triolo TM, Aly TA, Baschal EE, Babu SR, Kretowski A, Rewers MJ, Eisenbarth GS. 2008. Two single nucleotide polymorphisms identify the highest-risk diabetes HLA genotype: potential for rapid screening. Diabetes 57:3152–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Noble JA, Valdes AM, Bugawan TL, Apple RJ, Thomson G, Erlich HA. 2002. The HLA class I A locus affects susceptibility to type 1 diabetes. Hum Immunol 63:657–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Valdes AM, Erlich HA, Noble JA. 2005. Human leukocyte antigen class I B and C loci contribute to type 1 diabetes (T1D) susceptibility and age T1D onset. Hum Immunol 66:301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nejentsev S, Howson JM, Walker NM, Szeszko J, Field SF, Stevens HE, Reynolds P, Hardy M, King E, Masters J, Hulme J, Maier LM, Smyth D, Bailey R, Cooper JD, Ribas G, Campbell RD, Clayton DG, Todd JA. 2007. Localization of type 1 diabetes susceptibility to the MHC class I genes HLA-B and HLA-A. Nature 450:887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Howson JM, Walker NM, Clayton D, Todd JA. 2009. Confirmation of HLA class II independent type 1 diabetes associations in the major histocompatibility complex including HLA-B and HLA-A. Diabetes Obes Metab 11(Suppl 1):31–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gonzalez-Gay MA, Rueda B, Vilchez JR, Lopez-Nevot MA, Robledo G, Ruiz MP, Fernández O, Garcia-Porrua C, Gonzalez-Escribano MF, Martín J. 2007. Contribution of MHC class I region to genetic susceptibility for giant cell arteritis. Rheumatology (Oxford) 46:431–434 [DOI] [PubMed] [Google Scholar]
- 33. Roshna T, Thomas R, Nandakumar K, Banerjee M. 2006. A case-control study on the association of human leukocyte antigen-A*9 and -B*15 alleles with generalized aggressive periodontitis in an Indian population. J Periodontol 77:1954–1963 [DOI] [PubMed] [Google Scholar]
- 34. Sia C, Weinem M. 2005. The role of HLA class I gene variation in autoimmune diabetes. Rev Diabet Stud 2:97–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bisikirska B, Colgan J, Luban J, Bluestone JA, Herold KC. 2005. TCR stimulation with modified anti-CD3 mAb expands CD8 T cell population and induces CD8CD25 Tregs. J Clin Invest 115:2904–2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Issazadeh S, Kjellén P, Olsson T, Mustafa M, Holmdahl R. 1997. Major histocompatibility complex-controlled protective influences on experimental autoimmune encephalomyelitis are peptide specific. Eur J Immunol 27:1584–1587 [DOI] [PubMed] [Google Scholar]
- 37. Schmidt D, Verdaguer J, Averill N, Santamaria P. 1997. A mechanism for the major histocompatibility complex-linked resistance to autoimmunity. J Exp Med 186:1059–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Madakamutil LT, Maricic I, Sercarz E, Kumar V. 2003. Regulatory T cells control autoimmunity in vivo by inducing apoptotic depletion of activated pathogenic lymphocytes. J Immunol 170:2985–2992 [DOI] [PubMed] [Google Scholar]
- 39. Kaas Q, Ruiz M, Lefranc MP. 2004. IMGT/3Dstructure-DB and IMGT/StructuralQuery, a database and a tool for immunoglobulin, T cell receptor and MHC structural data. Nucleic Acids Res 32:D208–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ehrenmann F, Kaas Q, Lefranc MP. 2010. IMGT/3Dstructure-DB and IMGT/DomainGapAlign: a database and a tool for immunoglobulins or antibodies, T cell receptors, MHC, IgSF and MhcSF. Nucleic Acids Res 38:D301–D307 [DOI] [PMC free article] [PubMed] [Google Scholar]