Abstract
Regenerating islet (Reg) proteins are involved in the proliferation and differentiation of diverse cell types. However, whether embryonic stem cells (ESCs) express Reg genes and their corresponding proteins remains unknown. In this study, we probed the expression of Reg family members by mouse ESCs (mESCs). Mouse Reg1 and Reg3γ were detected in undifferentiated stem cells. Furthermore, we tested if gastrin—an inducer of Reg1 expression in committed cells—up-regulates the Reg1 gene in mESCs. Gastrin did not affect the expression of Reg1 either in self-renewing mESCs or under conditions permitting their differentiation. Moreover, overexpression of Reg genes found in various forms of cancer has been linked to dysregulated activation of the canonical Wnt/β-catenin cascade. Given the important roles of Wnt signaling in stem cells, we investigated if activation of Wnt alters the expression of Reg genes in mESCs. Wnt activation led to an increase in Reg1 gene expression with a concomitant increase in the amount of secreted Reg1 protein. Finally, the expression pattern of genes indicative of differentiation was examined in mESCs that were either exposed to soluble Reg1 or overexpressed the Reg1 gene. This is the first account of expression of Reg family members by ESCs. Our results show that the canonical Wnt cascade affects Reg expression and warrants further studies into the potential roles of Reg proteins in stem cell physiology.
Introduction
Regenerating islet (Reg) proteins, which were first discovered in pancreatic stone formation [1], are involved in the proliferation and differentiation of various types of human, rat, and mouse cells [2–4]. The Reg family comprises of 4 subclasses (Reg1, Reg2, Reg3, and Reg4) [5,6] across species with most of the orthologs belonging to the Reg1 and Reg3 groups. The expression of Reg genes is up-regulated in the pancreas after injury and the corresponding proteins promote the regeneration and proliferation of islet cells [7,8] while protecting acinar cells from apoptosis [9]. The Reg2 protein is a potent mitogen of Schwann cells and contributes to the regeneration of motor neurons in mice [10]. Moreover, the generation and maintenance of the villous structure of the small intestine is influenced by Reg1, which is considered a regulator of intestinal cell growth [11]. Despite the intimate link between Reg proteins and the proliferation and/or differentiation of diverse types of cells, no information is available, to date, about the expression and regulation of members of the Reg family in embryonic stem cells (ESCs).
Interestingly enough, overexpression of Reg proteins has been observed in liver tumors [12], pancreatic duct-cell carcinoma [13], testicular cancer [14], and colon cancer [15,16]. Enhanced levels of the human Reg3A (also known as pancreatitis-associated protein (PAP)) and Reg1α were discovered in primary liver tumors with β-catenin mutations suggesting a possible regulation of these Reg genes by the canonical Wnt/β-catenin signaling pathway [17]. A strong association between β-catenin mutations and changes in the expression of Reg genes was also documented in a recent clinical study involving biopsy samples from patients with liver cancer [18]. Dysregulated activation of the canonical Wnt signaling has also been identified in other cancer types (eg, seminoma [19], colon [20]) in which Reg proteins have been shown to be aberrantly overexpressed.
In addition to its role in carcinogenesis, Wnt signaling is important for the maintenance of stem cell pluripotency [21,22] and the expansion of progenitor cells [23]. Canonical Wnt signaling is also involved in the commitment of ESCs toward different phenotypes including neural cells [24], melanocytes [25], hematopoietic cells, and endothelial cells [26]. In the absence of Wnt activation, glycogen synthase kinase-3β (GSK3β) phosphorylates β-catenin, which is subsequently degraded via the ubiquitin-proteosome cascade. Activation of the Wnt/β-catenin pathway by inhibiting the GSK3β with 6-bromoindirubin-3′-oxime (BIO) [27] is sufficient to maintain cultured mouse ESCs (mESCs) and human ESCs (hESCs) in an undifferentiated state [28]. Blocking of GSK3β by BIO or LiCl [29] causes the accumulation and nuclear translocation of β-catenin that acts as a transcriptional cofactor with the T-cell factor/lymphoid enhancer factor (TCF/LEF) activating gene targets of Wnt. The genetic program initiated by canonical Wnt depends on the cellular context [30], and this may explain largely the multitude of effects associated with Wnt signaling. Given the mirror image roles of the canonical Wnt cascade in the biology of stem cells and cancer [31], we hypothesized that if members of the Reg family are expressed in ESCs, such expression may be influenced by Wnt.
In this study, we probed mESCs for the expression of various Reg genes. Only Reg1 and Reg3γ were detected in undifferentiated mESCs. Considering that gastrin induces the expression of Reg1 [32], we explored whether Reg1 is up-regulated in gastrin-treated mESCs. Exposure of self-renewing stem cells to gastrin did not alter their Reg1 profile. In contrast, activation of the canonical Wnt in mESCs boosted the expression of Reg1. This increase was curtailed in the presence of dominant negative TCF (ΔnTCF4). Finally, we investigated if Reg1 stimulates the commitment of mESCs toward a particular lineage. Cells cultured with Reg1 under conditions allowing their differentiation, showed preferential up-regulation of early endoderm markers. Our results show that the expression of Reg1 is modulated by Wnt signaling, which is a key regulator of stem cell fate, and warrant further studies into the role of Reg proteins in ESC physiology.
Materials and Methods
Cell culture
E14Tg2a (Mutant Mouse Regional Resource Centers (MMRRC), University of California, Davis, CA) and R1 (American Type Culture Collection (ATCC), Manassas, VA) mESCs were maintained on feeder cell-free, gelatin-coated (0.1% gelatin in phosphate-buffered saline (PBS); Sigma-Aldrich, St. Louis, MO) tissue culture dishes in 5% CO2/95% air at 37°C as described before [33]. The culture medium consisted of Dulbecco's modified Eagle's medium (DMEM; Sigma-Aldrich) supplemented with 10% ESC-screened FBS (Hyclone, Logan, UT), 2 mM l-glutamine, 0.1 mM nonessential amino acids, 0.055 mM β-mercaptoethanol, penicillin (100 U/mL), streptomycin (100 μg/mL) (all from Invitrogen, Carlsbad, CA), and 1,000 U/mL leukemia inhibitor factor (LIF; Chemicon, Temecula, CA). Medium was replaced every day and the mESCs were subcultured every 2 days. The cells were adapted in defined serum-free medium (DSFM) as described previously [33] and cultured for 3–4 passages prior to their use in the experiments described in this study. The composition of DSFM is as described above except that 10% KnockOut Serum Replacer (Invitrogen) is used instead of FBS. Gastrin (G17) was obtained from EMD Biosciences (San Diego, CA).
To investigate the effects of Reg1 on mESC differentiation, cells were cultured in DSFM without LIF. The culture medium, which was replaced daily, was supplemented with recombinant mouse Reg1 (obtained as a pre-release agent from R&D Systems, Minneapolis, MN). Alternatively, cells were transduced with an adenovirus carrying the Reg1 gene (AdReg1GFP; see “Adenovirus construction and cell transduction” section below).
Mouse 3T3 fibroblasts, human embryonic kidney 293 (HEK293) cells (ATCC), and mouse MIN6 pancreatic β-cells (a kind gift from Prof. J. Miyazaki—University of Tokyo, Japan) were cultured in DMEM with 10% FBS, 2 mM l-glutamine, penicillin (100 U/mL), and streptomycin (100 μg/mL) at 5% CO2/95% air at 37°C. The medium was replaced every 2 days and the cells were passaged every 4 days (6 days for MIN6 cells).
During passaging, the cells were incubated with TrypLE™ (Invitrogen, Carlsbad, CA) in PBS and cell clumps were dissociated by gentle pipetting. The cell suspension was spun down at 200g for 5 min and after removing the supernatant, the cell pellet was resuspended in fresh medium and plated on tissue culture dishes.
Adenovirus construction and cell transduction
The mouse Reg1 was cloned from cDNA prepared from RNA isolated from MIN6 β-cells. The Reg1 cDNA was prepared by PCR using the Phusion high-fidelity DNA polymerase (Finnzymes, Espoo, Finland) and the primers 5′-GGGGGATCCACCATGGCTAGGAACGCCTACTT-3′ and 5′-TTTTGTCGACTCAGCCTTTGAACTTGCAGAC-3′ (start/stop codons are shown in italics) with the BamHI and SalI sites (underlined) inserted to facilitate cloning. The fragment was incorporated in the pCR2.1-TOPO (Invitrogen, Carlsbad, CA) cloning vector by a topoisomerase-catalyzed reaction. Adenoviruses were generated based on a method described previously [34] with modifications. The IRES-eGFP fragment was excised from the pIRES2eGFP (Clontech, Mountain View, CA) vector by digestion with BglII and NotI and inserted into the pShuttle-CMV vector. The resulting plasmid (pShuttle-CMV-IRES-GFP) was digested with BglII and SalI. The cloned Reg1 was excised from pCR2.1-TOPO-mReg1 with BamHI and SalI and ligated with the digested pShuttle-CMV-IRES-GFP resulting in the pShuttle-CMV-mReg1-IRES-GFP. This vector was linearized with PmeI and was transformed into Escherichia coli BJ5183 cells carrying the adenoviral backbone pAdEasy-1 [34]. Clones were selected based on the patterns resulting upon digestion with various restriction endonucleases. A pShuttle-CMV-mReg1-IRES-GFP clone was selected carrying an exact copy of the mouse Reg1 cDNA verified by sequencing. The clone was transformed into XL10-Gold ultracompetent cells (Stratagene, La Jolla, CA), expanded, and prepared for linearization with PacI. The linearized plasmid was transfected into HEK293 cells using Lipofectamine (Invitrogen) according to manufacturer's instructions. Recombinant adenoviruses (AdReg1GFP) were harvested after 1 week and successive rounds of infection of HEK293 cells were performed to increase the virus titer. The adenovirus preparation was purified via filtration (Virabind, Cell Biolabs, San Diego, CA) and its titer was measured using an immunoassay (QuickTiter, Cell Biolabs). Typical titers for AdReg1GFP ranged from 1 to 10 × 1010 plaque-forming units (PFUs)/mL. The expression of the transgene and the corresponding Reg1 protein were confirmed by PCR, western blot, and enzyme-linked immunosorbent assay (ELISA) analyses of samples from cultured cells infected with the AdReg1GFP.
For transduction experiments, 105 cells/cm2 were plated in 24-well tissue culture plates and cultured for 24 h before transfection. Then, virus was added at a multiplicity of infection (MOI) as stated. The MOI was calculated based on the PFU/mL of the virus stock. Six hours postinfection, the medium was replaced with fresh culture medium. Cells infected with an adenovirus carrying the GFP gene downstream of the CMV promoter (AdGFP; Gene Vector Core Laboratory, Baylor College of Medicine, Houston, TX) served as controls for cells transduced with AdReg1GFP.
Cell transfection and reporter assays
Mouse ESCs were plated at 105 cells/cm2 in gelatin-coated 24-well tissue culture plates and cultured for 1 day prior to transfection. Cells were transfected with 0.5 μg DNA using Lipofectamine (Invitrogen, Carlsbad, CA). For each transfection, 300 ng of SuperTOP or SuperFOP [35] (both kind gifts from Dr. R.T. Moon, University of Washington), 100 ng dominant negative TCF4 [36] (ΔnTCF4, kindly provided by Dr. K.W. Kinzler, Johns Hopkins University) or pIRES-eGFP (Clontech, Mountain View, CA), and 100 ng of pCMV-RL (Renilla luciferase; Promega, Madison, WI) were co-transfected. Five hours after transfection, Wnt signaling was activated with the addition of 10 μM BIO (EMD Chemicals, Gibbstown, NJ), 30 mM LiCl (Sigma-Aldrich, St. Louis, MO), or 100 ng/mL Wnt3a (R&D Systems, Minneapolis, MN). Cells were harvested 24 h after Wnt activation and analyzed with the dual luciferase assay (Promega) according to the manufacturer's instructions in a Synergy HT multimode microplate reader (BioTek, Winooski, VT) in luminescence mode. Luciferase signal readings were obtained in relative luminescence units (RLUs) and normalized to the corresponding signal due to the Renilla luciferase activity.
RNA extraction, RT-PCR, and quantitative PCR
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Reverse transcription was performed using the ImProm-II reverse transcriptase (Promega, Madison, WI) with 1 μg of total RNA and 250 ng random primers or oligo(dT)12–18 primers at 42°C for 60 min. PCR runs were carried out with the resulting cDNA for 30–35 cycles at an annealing temperature of 58°C–60°C depending on the primer set. Primer sequences are shown in Table 1.
Table 1.
Primer Sequences (Shown in 5′–3′Orientation)
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Reg1 | TTCAGGCTACCTGGTGTCAGT | CACAGTAGCCACGATTGGAA |
| Reg2 | CTACAGCTTCCAATGTCTGGACT | TCTTCAGGTGACTTTAGGCTCTG |
| Reg3α | CTCAGGACATCTCGTGTCTATTCTT | AGTGACCACGGTTGACAGTAGAG |
| Reg3β | GAATGGAGTAACAATGACGTGATGA | AGTAAATTTGCAGACATAGGGCAAC |
| Reg3γ | GTAACAGTGGCCAATATGTATGGA | TACTCTAGGCCTTGAATTTGCAG |
| Reg3δ | AGAAGCATTTCTCAGGACACCT | CAGTTATAGAAGGTCAGAGGGTCAG |
| Oct3/4A | GGCGTTCTCTTTGGAAAGGTGTTC | CTCGAACCACATCCTTCTCT |
| Nanog | GTGGTTGAAGACTAGCAATGGTC | GAAGTTATGGAGCGGAGCAG |
| Bry | TGCTGCAGTCCCATGATAAC | TGTGCGTCAGTGGTGTGTAA |
| Eomes | GACTTGAATGAACCTTCCAAGACT | ATCTGATGGGATCTAGGGGAAT |
| NeuroD1 | AGATCGTCACTATTCAGAACCTTTT | TCTTCCTCTAGATCCTCATCTTCC |
| Pax6 | AGTCAGACCTCCTCATACTCGTG | ATCACATGCTCTCTCCTTCTCTCT |
| Foxa2 | TGGTCACTGGGGACAAGGGAA | GCAACAACAGCAATAGAGAAC |
| Sox17 | TTTGTGTATAAGCCCGAGATGG | AAGATTGAGAAAACACGCATGAC |
| CCK2R | GATTGCTTATGGTCCCCTACC | GTCTCGCTGTCATTATCACCATC |
| Extl3 | TGTGAGGATATCGCCATGAA | TGCTCATCGTCTCCTCAGAA |
| β-Actin | GCTCTTTTCCAGCCTTCC | GCTCAGGAGGAGCAATGA |
For relative quantification of gene expression, standard real-time polymerase chain reactions were performed on an iCycler quantitative PCR (qPCR) machine (Bio-Rad, Hercules, CA) as described previously [37]. Platinum PCR SuperMix (Invitrogen, Carlsbad, CA) was used with the following reaction conditions: denaturation and polymerase activation at 95°C for 2 min; amplification for 40 cycles at 95°C for 15 s, 58°C or 60°C for 30 s, and 72°C for 1 min. All reactions were run in triplicates on samples from at least 3 experiments. Amplification specificity was verified by a melting curve method. Relative gene expression was calculated by normalizing to the expression of endogenous β-actin, using the ΔΔCT method [38]. The threshold cycle (CT) for β-actin did not vary under different experimental conditions when equal amounts of RNA were used. PCR efficiencies for different primer pairs were determined by a standard curve method on serially diluted templates and found to be ∼1.
Immunocytochemistry
Cells were stained for immunofluorescence as described previously [33]. In brief, cells were washed in PBS and fixed with 4% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) in PBS (pH 7.4) for 20 min at room temperature. After washing with PBS, cells were permeabilized with 0.1% Triton X-100 (Mallinckrodt Baker, Phillipsburg, NJ) in PBS for 15 min and then blocked with 1% normal donkey serum (NDS; Jackson ImmunoResearch Laboratories, West Grove, PA) in PBS for 1 h. Incubation was carried out overnight at 4°C with a primary antibody such as rabbit anti-Oct3/4A, goat anti-Sox17, goat anti-Foxa2 (all from Santa Cruz Biotechnology, Santa Cruz, CA), and mouse anti-β-catenin (BD Biosciences, San Jose, CA). After 3 washes with PBS for 5 min, secondary antibodies were applied for 1 h at room temperature. Donkey anti-rabbit, -goat, and -mouse secondary antibodies conjugated with FITC or Cy3 (Jackson ImmunoResearch Laboratories) were used. Micrographs were acquired with either a Nikon Diaphot microscope equipped with a QImaging Retiga 1300i FAST CCD camera (Burnaby, BC, Canada) or a Zeiss LSM510 confocal laser scanning microscope (Carl Zeiss, Thornwood, NY).
Western blot analysis
Western blot analysis was carried out as described before [33]. In brief, cells were lysed in protein extraction buffer (M-PER; Thermo Fisher Scientific, Rockford, IL) supplemented with a protease inhibitor cocktail (EMD Biosciences, La Jolla, CA) according to the manufacturer's instructions. The total protein content of the samples was determined with the BCA Protein Assay (Thermo Fisher Scientific). Cell lysates were boiled for 5 min and loaded to a sodium dodecyl sulfate-polyacrylamide gel (SDS-PAGE; 4% w/v stacking gel and 12% w/v resolving gel) at 30 μg of total protein per lane. After separation by electrophoresis, sample proteins were transferred onto polyvinylidene difluoride (PVDF) membranes (Thermo Fisher Scientific). The membranes were blocked with 5% (w/v) skim milk in Tris-buffered saline with 0.05% Tween-20 (TBS/T) for 1 h at room temperature and washed several times with TBS/T before overnight incubation at 4°C with primary antibodies against mouse Reg1 (R&D Systems, Minneapolis, MN) and β-actin (Santa Cruz Biotechnology). After incubation with appropriate secondary horseradish peroxidase (HRP)-conjugated antibodies (Jackson ImmunoResearch Laboratories), protein signals were detected with the LumiGLO chemiluminescence reagent (Cell Signaling Technology, Danvers, MA).
Indirect enzyme-linked immunosorbent assay
Mouse Reg1 concentrations were determined by an indirect ELISA. On the day of the sampling, supernatants were collected and the cells were harvested by incubation with TrypLE. Cells excluding the Trypan Blue dye (Sigma-Aldrich, St. Louis, MO) were counted in a hemocytometer. Cell counts were used to normalize the total amount of Reg1 protein present in the supernatant. Supernatant samples were transferred to 96-well plates (Nunc Inc., Naperville, IL) at 100 μL/well. Samples of recombinant mouse Reg1 (R&D Systems) were also transferred to the plates and were serially diluted to obtain a standard curve. After overnight incubation at 4°C, the wells were emptied and blocking buffer consisting of 3% bovine serum albumin (BioFX Laboratories, Owings Mills, MD) and 0.1% sodium azide (G-Biosciences, Maryland Heights, MO) in TBS/T was added. The plates were incubated for 1.5 h at 37°C and then rinsed 3 times with TBS/T and a sheep anti-mouse Reg1 antibody (R&D Systems) was added at 0.5 μg/mL to each well. After incubation for 1 h at 37°C, the plates were washed 3 times with TBS/T and a donkey anti-sheep biotinylated secondary antibody (1:5,000; Jackson ImmunoResearch Laboratories) was added to each well for 1 h at room temperature. Subsequently, the wells were again washed 3 times with TBS/T, incubated with alkaline phosphatase-conjugated streptavidin (1:500; Jackson ImmunoResearch Laboratories) for 1 h at room temperature and the TBS/T washing was repeated 3 times. Finally, p-nitrophenyl phosphate (pNPP; Thermo Fisher Scientific) substrate solution was added. Plates were further incubated at 37°C in a humidified chamber for 30–60 min and read on an automated plate reader. The optical density (OD) at 405 nm was correlated to the concentration of Reg1 protein in the samples.
Flow cytometry
Flow cytometry analysis was performed as previously described [33,37]. In brief, cells were incubated with TrypLE (Invitrogen) and collected by centrifugation at 500g for 5 min and fixed for 10 min with 3.7% formaldehyde solution (Sigma-Aldrich). Cells were washed with PBS, blocked with 3% NDS for 30 min, and incubated with the primary antibody goat anti-Sox17 (Santa Cruz Biotechnology) in 1% NDS for 1 h at room temperature. The cells were then washed 3 times with 1% NDS and incubated with a Cy3-conjugated donkey secondary antibody (Jackson ImmunoResearch Laboratories, West Grove, PA) for 30 min at room temperature. After washing with PBS, the cells were analyzed in a FACSCalibur flow cytometer with the CellQuest software (Becton Dickinson, San Jose, CA). Cells were registered as positive if their emitted fluorescence level was higher than 99% of that of samples stained only with the corresponding secondary antibodies.
Statistical analysis
Data are expressed as mean ± SD unless stated otherwise. Statistical analysis including ANOVA was performed using Minitab (Minitab Inc., State College, PA). P values <0.05 were considered as significant.
Results
Reg expression in mESCs
We set out to probe the expression of Reg genes in mESCs by RT-PCR using primers designed specifically for each member of the mouse Reg family (Table 1).For this purpose, we used 2 mESC lines: E14Tg2a and R1. Expression of Reg1 was detected in both lines (Fig. 1A). Reg3γ transcripts were also found in mESCs albeit at lower levels. We did not detect the expression of other Reg members. Pancreatic MIN6 β-cells (mouse insulinoma line) were included in our study since because of their expression of Reg1. It should be noted that Reg proteins are mainly synthesized by pancreatic acinar cells whereas normal β-cells do not express Reg except under pathological conditions (eg, pancreatic injury or cancer) [5]. No expression was evident in 3T3 mouse fibroblasts (negative control). Thus, 2 members of the family of mouse Reg genes, Reg1 and Reg3γ, are expressed in mESCs.
FIG. 1.
Expression of Reg family members in embryonic stem cells (ESCs). (A) Mouse ESC lines E14Tg2a and R1, MIN6 (insulinoma) β-cells, and 3T3 fibroblasts were probed by RT-PCR for the expression of Reg genes. The expression of β-actin is also shown. (B) Western blot analysis of lysates from mESCs, MIN6, and 3T3 cells for their content of Reg1 protein. (C) The amount of Reg1 protein secreted in the medium by mESCs and MIN6 cells was quantified by enzyme-linked immunosorbent assay (ELISA). No Reg1 protein was detectable in supernatants from cultured 3T3 cells. Values are shown as mean ± standard deviation (SD) from 3 experiments. (D) Expression of the putative Reg1 receptor Extl3 in mESCs and MIN6 cells.
Considering the lack of a suitable antibody against the mouse Reg3γ protein, we focused our study mainly on the expression of Reg1 in mESCs. Therefore, we next determined the expression of Reg1 protein. By western blot analysis, mouse Reg1 protein was evident as a ∼16 kDa band (Fig. 1B) in the samples obtained from the mESCs. Given that Reg proteins are secretable, we also examined the content of Reg1 protein in the culture medium by ELISA. Supernatants sampled from mESC cultures after 24 h of incubation contained 13–18 pg Reg1 protein per 106 cells (Fig. 1C). In contrast, <4 pg/106 cells were measured in MIN6 β-cells. In agreement with the absence of Reg1 transcripts in 3T3 cells, there was no detectable Reg1 protein in 3T3 cell culture supernatants.
We also observed the expression of the putative Reg1 receptor [39] (Figs. 1D and 4D), exostosis-like 3 protein (Extl3), which is a member of the multiple exostosis gene family [40]. The Extl3 receptor may mediate signaling triggered by the secreted Reg1 in mESCs as shown for β-cells [39]. Overall, our results show that mESCs express the Reg1 and Reg3γ genes. Moreover, the mESCs synthesize and secrete the Reg1 protein.
FIG. 4.
Reg gene expression is enhanced by activation of the canonical Wnt cascade. (A) The expression of the Reg1 gene was increased in E14Tg2a mESCs treated with 100 ng/mL Wnt3a or 30 mM LiCl compared to untreated (control) cells (their Reg1 gene expression was set to 1). The increase in Reg1 gene expression due to incubation with Wnt3a was curtailed in mESCs transfected with a plasmid encoding ΔnTCF4 (Wnt3a + ΔnTCF4). Cells transfected with an empty vector and treated with Wnt3a (Wnt3a + control vector) were also included in this series of experiments. Results are shown as mean values ± SD from a representative experiment (triplicate samples). *P < 0.005 and **P < 0.001 compared to control (untreated) cells. # P < 0.005 compared to Wnt3a + control vector cells. (B) Wnt activation in mESCs by LiCl or Wnt3a did not lead to statistically significant differences in the expression of the Reg3γ gene in comparison with untreated (control) cells. (C) Other Reg genes were not detected in mESCs even after activation of the Wnt/β-catenin cascade. (D) Expression of the Reg1 receptor Extl3 in mESCs treated with Wnt3a or LiCl. *P < 0.05 compared to untreated (control) mESCs.
Regulation of Reg1 expression in mESCs by gastrin
Many details about the regulation of the Reg1 expression are still unknown. However, Reg1 up-regulation by the gastric hormone gastrin has been documented primarily in rat cells [32,41]. Thus, we first treated MIN6 insulinoma β-cells to see if gastrin has the same effect on the Reg1 gene in mouse cells. Indeed, MIN6 β-cells exposed to 1 nM gastrin for 24 h exhibited a ∼30-fold increase in Reg1 expression compared to untreated MIN6 β-cells (Fig. 2A). Thus, gastrin mediates the up-regulation of Reg1 expression in mouse cells.
FIG. 2.
Induction of Reg1 expression in mouse embryonic stem cells (mESCs) by gastrin. (A) Relative expression of the Reg1 gene in MIN6 cells and mESCs (E14Tg2a) after incubation with 1 nM gastrin for 24 h. Reg1 gene expression is shown as fold-increase compared to untreated cells. Values are shown as mean ± SD. (B) The CCK2R is not detected in undifferentiated mESCs cultured in the presence of leukemia inhibitor factor (LIF) (mESCs). In contrast, MIN6 cells and mESCs grown without LIF (mESCs–LIF) for 4 days express the CCK2R. The expression of the housekeeping gene β-actin is also shown. (C) Mouse ESCs cultivated in the absence of LIF for 4 days were incubated without (−) or with 1 nM gastrin (+) for 24 h. Results are shown compared to the Reg1 expression of E14Tg2a mESCs incubated without gastrin.
Then, we asked if transcription of Reg1 is also increased in mESCs incubated with gastrin. Nevertheless, no significant change in Reg1 expression was observed when mESCs were treated with 1 nM gastrin for 24 h (Fig. 2A). In addition, the expression of Reg1 was not altered even after treating mESCs with higher concentrations (up to 100 nM) of gastrin for 6–48 h (data not shown). Given that gastrin binds to the cholecystokinin 2 receptor (CCK2R) to activate the expression of Reg1, we examined if CCK2R is expressed in MIN6 β-cells and mESCs. Accordingly, strong CCK2R expression was displayed in MIN6 β-cells (Fig. 2B). However, CCK2R transcripts were not detected in mESCs supporting the lack of gastrin-induced activation of Reg1 expression in these cells.
Of note is the fact that mESCs, cultured for 4 days under differentiating conditions with the withdrawal of LIF, expressed CCK2R (Fig. 2B). Yet, when these differentiated cells were incubated with gastrin for 24 h, there was no change in the expression of Reg1 (Fig. 2C). This led us to conclude that gastrin does not induce the expression of Reg1 in self-renewing mESCs and early mESC progeny.
Activation of the canonical Wnt pathway in mESCs
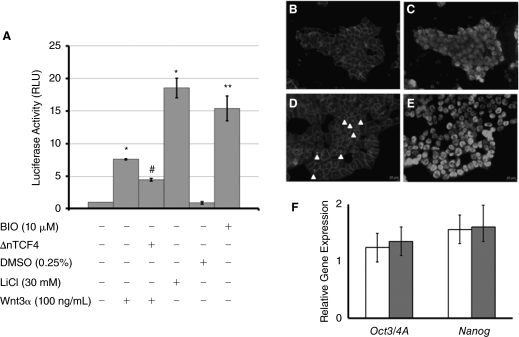
Previous studies indicated a possible link between Wnt/β-catenin signal transduction and the expression of Reg proteins in cancer cells [17,18]. Before examining if such a link exists in mESCs, we defined conditions under which the canonical Wnt pathway can be activated in our mESC lines. Wnt activation was assessed via a reporter assay. Mouse ESCs maintained in the absence of serum [33] were transfected with the SuperTOP plasmid [35] encoding the luciferase gene downstream of a promoter targeted by β-catenin/TCF. Treatment of E14Tg2a mESCs with 30 mM LiCl for 24 h resulted in the most pronounced increase in luciferase signal (18.54 ± 1.49-fold) compared to the baseline signal obtained from untreated mESCs (Fig. 3A). Similarly, the luciferase signal in mESCs incubated with 10 μM BIO was 15.4 ± 1.91-fold higher compared to untreated mESCs. Because dimethyl sulfoxide (DMSO) is used as a solvent for the preparation of BIO stock solutions, cells incubated with 0.25% (v/v) DMSO were also included in these experiments but the corresponding luminescence readings were comparable to those for control mESCs (0.87 ± 0.18-fold). Purified Wnt3a (100 ng/mL) also stimulated Wnt signaling (7.61 ± 0.11-fold) in mESCs. The level of Wnt activation was reduced when the cells were transfected with the ΔnTCF4 construct prior to their exposure to the Wnt3α ligand (4.47 ± 0.22-fold).
FIG. 3.
Activation of the canonical Wnt pathway in mESCs. (A) The activation of Wnt in mESCs transfected with a Wnt/β-catenin activatable promoter driving the expression of luciferase (SuperTOP) was examined under different conditions as shown. The luciferase signal was normalized to that due to Renilla luciferase activity and was expressed in relative luciferase units (RLUs). Cells transfected with SuperFOP (carrying mutated TCF/LEF-binding sites) did not yield signal above the background. *P < 0.005 compared to untreated mESCs. **P < 0.005 compared to mESCs exposed in 0.25% dimethyl sulfoxide (DMSO). # P < 0.005 compared to mESCs treated with Wnt3a only. (B–E) Immunostaining of mESCs for β-catenin and Oct3/4A. Unlike (B) untreated mESCs, cells incubated with Wnt3a (D) displayed nuclear accumulation of β-catenin as indicated by the arrowheads. Co-staining of the cells depicted in (B and D) for Oct3/4A is also shown (C and E). Bars in (B–E): 20 μm. (F) Relative expression of Oct3/4A and Nanog in mESCs treated with 100 ng/mL Wnt3a (white bars) or 30 mM LiCl (dark bars). Results are shown as mean ± SD (n = 3) relative to the expression of the corresponding genes in samples of untreated mESCs.
Activated Wnt/β-catenin signaling is characterized by translocation of the β-catenin to the cell nucleus. This was also evident when R1 mESCs were treated with LiCl or Wnt3a and stained for β-catenin (Fig. 3B). These cells were also positive for Oct3/4A (Fig. 3C and 3E) suggesting that under these experimental conditions the cells remained undifferentiated. This is also corroborated by qPCR results on the expression of the Oct3/4A and Nanog genes by mESCs treated with LiCl or Wnt3a (Fig. 3F).
Effect of Wnt/β-catenin signaling on Reg1 expression in mESCs
We showed that mESCs express Reg1 (and the Reg3γ gene) and established conditions for activation of the canonical Wnt cascade in these cells. We hypothesized that a link exists between the expression of Reg proteins and Wnt/β-catenin signaling in mESCs as suggested for other cell types [17,18]. To test our hypothesis, we implemented the experimental conditions described in the previous paragraph for the activation of the canonical Wnt pathway in mESCs. However, BIO was excluded from these experiments because its stock solution is prepared in DMSO, which may trigger the aberrant differentiation of mESCs thereby perplexing our observations.
Cells treated with 100 ng/mL of Wnt3a displayed 2.63 ± 0.55 times (P < 0.005) higher expression of Reg1 compared to untreated mESCs (Fig. 4A). A more marked up-regulation of Reg1 was noted when cultured mESCs were exposed to 30 mM LiCl (7.5 ± 0.94-fold vs. control mESCs). In contrast, Reg3γ expression did not exhibit statistically significant differences under these conditions (Fig. 4B). Interestingly enough, activation of Wnt/β-catenin signaling did not translate to detectable expression of the other mouse Reg genes, that is, Reg2, Reg3α, Reg3β, and Reg3delta; (Fig. 4C).
We also quantified the gene expression of the putative Reg1 receptor Extl3. Treatment of mESCs with 100 ng/mL Wnt3a did not change the level of mouse extl3 expression (Fig. 4D) despite the increase in Reg1 (Fig. 4A). Nonetheless, cells exposed to LiCl exhibited higher extl3 expression compared to untreated mESCs suggesting that up-regulation of the Reg1 receptor may be associated with the more pronounced Wnt activation by LiCl (compared to Wnt3a) or that an alternative LiCl-dependent mechanism may be in play.
Secretion of Reg1 by mESCs upon activation of the canonical Wnt cascade
Next, we asked if the observed increase in the level of Reg1 gene expression upon activation of the canonical Wnt signaling translated to a commensurate increase in Reg1 protein. To that end, the amount of Reg1 secreted in the culture medium was measured. Indeed, E14Tg2a mESCs incubated with 100 ng/mL of Wnt3a secreted 36 ± 1.12 pg/106 cells compared to 19.98 ± 1.33 pg/106 untreated cells (Fig. 5). Similarly, 35.6 ± 0.87 pg of Reg1 protein per 106 Wnt3a-treated R1 mESCs was measured in the medium versus only 23.09 ± 4.18 pg/106 cells cultured without Wnt3a. Our results support that the increase in the Reg1 mRNA translated to enhanced amounts of secreted protein.
FIG. 5.
Reg1 protein secretion by mESCs. When incubated with 100 ng/mL Wnt3a, E14Tg2a, and R1 mESCs secreted higher amounts of Reg1 protein than control mESCs as enzyme-linked immunosorbent assay (ELISA) analysis revealed. *P < 0.05.
Reg1 and mESC differentiation
Considering the involvement of Reg1 in the differentiation of various progenitor cells, we exposed mESCs to recombinant Reg1 protein and probed the expression of early markers of differentiation toward the 3 germ layers. A previous report [42] indicated that pancreatic ductal and β-cells exposed to exogenous Reg1 showed a mitogenic response whereas cells transfected with a Reg1 expression vector (thereby having elevated intracellular Reg1 protein) showed inhibited growth. Thus, mESCs infected with recombinant adenoviruses carrying the mouse Reg1 and GFP genes (AdReg1GFP) or only the GFP (AdGFP; control) gene were also included in this series of experiments. To explore the potential effects of Reg1 on mESC differentiation, the cells were cultured in the absence of LIF, which is known to prevent the differentiation of mESCs through activation of the STAT3 cascade [43,44].
Differentiating mESCs in the presence of exogenous Reg1 or after infection with AdReg1GFP were probed for the expression of genes characteristic of the 3 germ layers. Mouse ESCs were exposed to various concentrations of Reg1 (results are shown for 100 ng/mL Reg1). Alternatively, the cells were infected with AdReg1GFP (or AdGFP) and after 4 days their gene expression was analyzed. Increased cytotoxicity was observed upon infecting mESCs with either AdReg1GFP or AdGFP at MOIs higher than 100 PFUs/cell. Therefore, results are presented for mESCs infected at a MOI of 100. When mESCs cultured in 24-well plates were infected with AdReg1GFP, an average concentration of 1.66 ± 0.13 ng Reg1 per milliliter of supernatant per 24 h (equivalent to 16.6 ± 1.3 ng/106 cells) was measured by ELISA.
There was no pronounced difference in the level of expression of the mesoderm genes brachyury [45] and eomes [46] (Fig. 6A) and the ectoderm markers pax6 [47] and neuroD1 [48] among all samples in these experiments. In contrast, we noticed that the expression of sox17 and foxa2 genes, which are characteristic of endoderm progeny [49,50], gradually increased in mESCs treated with exogenous Reg1 or infected with the AdReg1GFP (Fig. 6B). After 4 days of exposure to recombinant Reg1 protein, the cells exhibited 14.6 ± 0.1-fold and 6.6 ± 0.8-fold higher sox17 and foxa2 expression, respectively, when compared to mESC controls (ie, not cultured with LIF and Reg1). Mouse ESCs infected AdReg1GFP also showed a 5.3 ± 0.8-fold and 10.1 ± 1.5-fold increase in sox17 and foxa2 transcripts, respectively, compared to mESCs cultivated without LIF but infected with a control adenoviral vector (AdGFP). Other endoderm genes were also displayed by mESCs treated with Reg1 or infected with AdReg1GFP (Supplementary data available online at www.liebertonline.com/scd).
FIG. 6.
Gene expression in mESCs exposed to exogenous Reg1 or transduced with AdReg1GFP. (A) Quantitative PCR results for the expression of the differentiation markers brachyury (Bry) and eomes [mesoderm], neuroD1 and pax6 [ectoderm]. White bars: mESCs (−LIF) with 100 ng/mL Reg1 (control: mESCs without leukemia inhibitor factor [LIF]). Dark bars: mESCs (−LIF) infected with AdReg1GFP at MOI of 100 (control: mESCs without LIF but infected with AdGFP at a MOI of 100). The cells were probed after 4 days of incubation with Reg1 or adenoviral infection. (B) The endoderm genes foxa2 and sox17 displayed a more pronounced up-regulation in mESCs that were either treated with exogenous Reg1 (white bars) or overexpressed Reg1 after infection with AdReg1GFP (dark bars). Results are shown after 2 and 4 days of exposure to Reg1 or infection with AdReg1GFP. Gene expression from the control samples was set to unity. #P < 0.001 and *P < 0.05 compared to controls. (C) Foxa2 (red) and (D) Sox17 (red) staining (nuclear DNA counterstaining with DAPI) of mESCs exposed to 100 ng/mL Reg1. Undifferentiated E14Tg2a cells stained for (E) Foxa2 and (F) Sox17 (and DAPI) are also shown. Bars: 50 μm. (G) The fraction of Sox17+cells in cultures without LIF (−LIF) and containing 100 ng/mL Reg1 (−LIF + Reg1) was assessed by flow cytometry (# P < 0.001). Results are shown from 3 independent experiments. Color images available online at www.liebertonline.com/scd.
Of note, there was no statistically significant difference in the expression of the genes we examined in uninfected mESCs and AdGFP-infected mESCs (both cultured without LIF; data not shown). In addition, we did not observe a difference in the growth of cells exposed to Reg1 compared to that of cells infected with AdReg1GFP (ie, higher endogenous levels of Reg1) unlike previous findings [42]. Similarly, there was no noticeable difference in the number of cells treated with Reg1 or infected with AdReg1GFP and their respective control cultures suggesting that Reg1 does not impact the survival of differentiating mESCs, at least for the period examined here.
The expression of foxa2 and sox17 in mESCs treated with Reg1 was corroborated by immunocytochemistry results (Fig. 6C–6F). The cells were also probed by flow cytometry for the expression of Sox17 (Fig. 6G). In the absence of LIF, 11.49% ± 0.53% of the differentiating mESCs treated with 100 ng/mL Reg1 for 4 days were Sox17+ compared to only 5.76% ± 0.32% of control mESCs.
Our results show the increased expression of endoderm markers in mESCs in the presence of Reg1 under conditions allowing their differentiation (ie, without LIF). These findings warrant further studies into the potential of Reg1 as a signal coaxing stem cells along particular lineages.
Discussion
Regenerating islet (Reg) proteins have been identified in various types of regenerating tissues [10,11,51,52] and cancer cells [13,14,16] and the expression patterns of Reg genes have been under intense scrutiny. To date, however, there is no information regarding the expression of members of the Reg family in ESCs and their immediate progeny. In this study, we probed the expression of Reg genes in mESCs and detected transcripts of 2 Reg members: Reg1 and Reg3γ. Moreover, we did not observe changes in the Reg1 expression in self-renewing and differentiating mESCs during treatment with gastrin, which is known to induce Reg [32]. To our knowledge, this is also the first time that direct evidence is presented of the effect of the canonical Wnt/β-catenin signaling on the expression of Reg1. We found that activation of Wnt in mESCs is linked to increased expression and secretion of Reg1 whereas there was no measurable effect on the expression of other Reg members. Furthermore, exposure of mESCs to Reg1 under conditions allowing cell differentiation led to a marked up-regulation in genes characteristic of early endoderm. This is the first account of Reg expression by ESCs, its modulation via activation of the canonical Wnt pathway, and its effect on ESC commitment.
Recent studies have identified Reg genes as targets of gastrin in gastric tumors and cancer cell lines [32,41]. Gastrin targets Reg1 through its CCK2R [53]. A C-rich element (C−74CCCTCCC−67 relative to the transcriptional start site) has been identified in the rat Reg1 gene promoter as being responsible for the induction of Reg1 expression by gastrin. Upon examining the mouse Reg1 gene promoter, we found a similar C-rich region (C−76TCCTCCC−69) suggesting a similar mode of regulation. In line with this finding, our results clearly demonstrate an increase in Reg1 expression in mouse MIN6 insulinoma β-cells treated with gastrin. Others [53] observed an elevated expression of Reg in the pancreatic acini of transgenic mice carrying the human CCK2R gene. Nonetheless, gastrin did not affect the expression of Reg1 in mESCs. This may be largely due to the lack of CCK2R in undifferentiated mESCs. However, differentiating mESCs (ie, growing without LIF) treated with gastrin also did not display enhanced levels of Reg1 albeit their expression of CCK2R. It is possible that downstream mediators of gastrin signaling relayed by CCK2R [54] may be unavailable in mESCs and their immediate committed progeny.
A link between the canonical Wnt signaling and Reg gene/protein expression has been suggested previously. Cavard et al. [17] observed the overexpression of REG1A and REG3A in human liver tumors with mutations in the β-catenin gene. In fact, treatment of Huh7 human hepatoma cells with LiCl, an activator of Wnt/β-catenin signaling, led to induction of REG3A expression, which was abolished by siRNA targeting β-catenin. Others have also observed a strong association between the enhanced expression of REG1A and PAP (REG3A) and mutations in the β-catenin in human hepatocarcinomas [18]. Overexpression of REG1A was also found in colon adenomas in which the inactivated adenomatous polyposis coli (APC) promotes the nuclear accumulation of β-catenin [55]—a hallmark of canonical Wnt signaling.
Our findings show a link between the activation of the canonical Wnt cascade and the expression of Reg1 in mESCs. Expression and secretion of Reg1 were elevated when mESCs were treated with Wnt3a or LiCl. This increase was curtailed in cells transfected with a ΔnTCF4 construct suggesting that a mechanism involving TCF/LEF mediates the observed up-regulation of Reg1. In contrast, expression of the Reg3γ gene that is expressed at low levels in mESCs did not change when the activity of Wnt was modulated. Furthermore, other Reg transcripts were not detected in mESCs even after Wnt activation. To that end, there was no noticeable increase in the expression of mouse Reg3a (an ortholog of human PAP) in mESCs with activated Wnt. This is in contrast to previous findings suggestive of the enhanced expression of REG3A due to Wnt signaling in human liver cancer cells [17,18]. This discrepancy may be attributed to species differences and potentially distinct regulatory mechanisms pertaining to the expression of various Reg3 (PAP) orthologs.
Moreover, we hypothesized that mESCs may up-regulate Extl3 to accommodate the increase in Reg1 upon treatment with Wnt3a or LiCl. However, the expression of the putative Reg1 receptor Extl3 was not altered with Wnt activation despite the up-regulation of Reg1. Kobayashi et al. [39] also showed that mouse islets do not increase their expression of the Reg receptor during pancreas regeneration, which stimulates the production of Reg1. These authors noted that the regeneration and proliferation of islet cells (mainly β-cells) are primarily regulated by the expression of the Reg proteins. The effects of Reg in mESCs may also exhibit a similar dependency on the expression of Reg1 rather than its receptor Extl3.
A question remains of whether the Reg1 gene is targeted directly by the canonical Wnt pathway. The transcriptional activity of genes targeted by the Wnt/β-catenin pathway is mediated by the TCF/LEF complex. We noted that transfection with ΔnTCF4 reduced the expression of Reg1 in mESCs treated with Wnt3α or LiCl. Still, analysis of the Reg1 gene promoter up to 2 kb from the transcription starting site (data not shown) did not reveal clusters of TCF/LEF-binding sites (but only isolated sites). Conceivably then, another protein that synergizes with the TCF/LEF may activate directly the transcription of Reg1. Thus, the exact mechanism by which canonical Wnt modulates Reg expression in ESCs requires further investigation. Current efforts in our laboratory are directed toward elucidating this mechanism.
One could argue that a cross talk between the Wnt and gastrin pathways may affect the expression of Reg1. This is because gastrin expression is targeted by canonical Wnt signaling. Overexpression of activated β-catenin stimulates the human gastrin promoter 2- to 3-fold through a putative TCF-binding site [56] and a similar Wnt-dependent regulation has been shown for the mouse gastrin gene promoter [57]. Conceivably, signaling through the Wnt/β-catenin cascade up-regulates gastrin, which then causes an increase in Reg gene expression. Although activation of Wnt led to up-regulation of Reg1 in undifferentiated mESCs, we did not observe induction of Reg1 by gastrin and these cells lacked the CCK2R. Even differentiating mESCs, which expressed CCK2R, did not boost their expression of Reg1 during treatment with gastrin. Based on our findings, we consider unlikely that Wnt targeting of the gastrin gene may cause subsequent activation of Reg1 expression in self-renewing and differentiating ESCs.
Despite mounting evidence about the important roles of Reg proteins in the proliferation and differentiation of cells in regenerating tissues and cancer cells, there is no information on the role of these proteins in stem cell physiology. Here, we showed that self-renewing mESCs (ie, in the presence of LIF) express Reg1, Reg3γ (albeit at a lower level), and the putative Reg receptor Extl3. Although the role of Reg in undifferentiated ESCs is unclear, Reg signaling may be involved in stem cell proliferation considering the mitogenic effects of Reg proteins on various types of committed cells [10,11,58].
We also began to investigate the potential of Reg1 as a differentiation stimulus for mESCs. Exposure to Reg1 or overexpression of the protein under conditions permitting differentiation (ie, without LIF) led to the up-regulation of early endoderm genes in mESCs. Such up-regulation was observed at higher concentrations of Reg1 than the baseline production of Reg1 by undifferentiated mESCs (∼13–18 pg/[(106 cells)(24 h)]). Then, an increase in the production of Reg1 may be necessary for Reg-induced differentiation to transpire. We showed that Wnt activation enhances Reg1 in undifferentiated mESCs. Canonical Wnt signaling is involved in various facets of embryo development and stem cell specification [59–62]. Hence, this signal may be acting in vivo to regulate the expression of Reg genes/proteins during development.
Yet, such induction of Reg by Wnt (and possibly by other signals) may further stem cell differentiation. We found that mESCs exposed to recombinant Reg1 or after overexpressing the Reg1 gene exhibit increased levels of endoderm markers. To that end, Reg genes/proteins exist primarily in endoderm-derived tissues although their presence in other tissues has been reported as well [10,63]. In a recent report [64], Reg1 was shown to promote the differentiation of pancreatic acinar cells whereas inhibition of Reg1 led to β-cell and possibly ductal cell phenotypes. The Reg1 mRNA exhibits a 20-fold increase in the acinar pancreas after 16 weeks of gestation [65,66] in human embryos. The expression of PAP genes has also been detected in fetal pancreas, stomach, jejunum, and colon [67]. During mouse development, the presence of Reg1 mRNA is detected on E9 just before pancreatic organogenesis [68] and on E13 in the fetal intestine [11]. To our knowledge, there is no information available on the expression of Reg genes at earlier times of embryonic development. Hence, whether Reg proteins contribute to cell differentiation in vivo during early embryogenesis is still an open question.
It should be noted that Reg1−/− knockout mice develop normally although they exhibit diminished pancreatic β-cell hyperplasia in response to appropriate stimuli [52] and differences in the small intestine compared to wild-type mice [11]. The lack of embryonic phenotype of Reg1−/− mice suggests that Reg1 may not be necessary during the early stages of development. Then, our results on the Reg1-associated up-regulation of endoderm genes in differentiating mESCs point to a possible compensatory or complementary role of Reg1 in progenitor cell differentiation.
To assess the effect of Reg1 on mESC differentiation, recombinant Reg1 protein was added to the cultures of mESCs in the absence of LIF or Reg1 was overexpressed by infecting cells with a recombinant adenovirus carrying the Reg1 gene (AdReg1GFP). It was reported previously that Reg1 can inhibit the growth of pancreatic cells when overexpressed within cells or in very high extracellular levels (over 100 nM) [42]. In our study, the concentrations of exogenous Reg1 and expressed Reg1 after infection with AdReg1GFP were well below 100 nM. To that end, we did not observe pronounced differences in the growth of mESCs cultivated without LIF and either exposed to exogenous Reg1 protein or overexpressing Reg1 after adenoviral transduction. It will be interesting to examine if elevated Reg1 affects the survival and proliferation of stem cells subjected to directed differentiation with physiologically relevant factors (eg, activin and Wnt ligands [33,37,69]) toward endoderm cells.
Lastly, aberrant overexpression of Reg proteins has been involved in various types of cancer, for example in the colon, testes, and liver [14,16–18,70,71]. The dysregulated activation of the canonical Wnt cascade has also been associated with tumorigenesis [20,30,72,73]. More recently, the mirror image roles of Wnt/β-catenin signaling in the biology of stem cells and cancer have been at the epicenter of intense research [31,74]. Here, we demonstrated the enhancement in Reg expression by activation of the canonical Wnt pathway in stem cells. Our findings warrant further investigations into novel mechanisms of stem cell carcinogenesis involving members of the Reg family.
Supplementary Material
Acknowledgments
The authors would like to thank Drs. K.W. Kinzler (Johns Hopkins University) and R.T. Moon (University of Washington) for providing the ΔnTCF4 and SuperTOP/SuperFOP constructs, respectively. We thank Li (Grace) Lu for her excellent technical assistance. This work was supported by grants from the New York State Stem Cell Science Trust (NYSTEM contract # C024355) and the National Institutes of Health (HL092398) to E.S.T.
Author Disclosure Statement
The authors declare no conflict of interest.
References
- 1.De Caro A. Lohse J. Sarles H. Characterization of a protein isolated from pancreatic calculi of men suffering from chronic calcifying pancreatitis. Biochem Biophys Res Commun. 1979;87:1176–1182. doi: 10.1016/s0006-291x(79)80031-5. [DOI] [PubMed] [Google Scholar]
- 2.Terazono K. Yamamoto H. Takasawa S. Shiga K. Yonemura Y. Tochino Y. Okamoto H. A novel gene activated in regenerating islets. J Biol Chem. 1988;263:2111–2114. [PubMed] [Google Scholar]
- 3.Lechene de la Porte P. de Caro A. Lafont H. Sarles H. Immunocytochemical localization of pancreatic stone protein in the human digestive tract. Pancreas. 1986;1:301–308. doi: 10.1097/00006676-198607000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Katsumata N. Chakraborty C. Myal Y. Schroedter IC. Murphy LJ. Shiu RP. Friesen HG. Molecular cloning and expression of peptide 23, a growth hormone-releasing hormone-inducible pituitary protein. Endocrinology. 1995;136:1332–1339. doi: 10.1210/endo.136.4.7895644. [DOI] [PubMed] [Google Scholar]
- 5.Graf R. Schiesser M. Reding T. Appenzeller P. Sun LK. Fortunato F. Perren A. Bimmler D. Exocrine meets endocrine: pancreatic stone protein and regenerating protein—two sides of the same coin. J Surg Res. 2006;133:113–120. doi: 10.1016/j.jss.2005.09.030. [DOI] [PubMed] [Google Scholar]
- 6.Liu JL. Cui W. Li B. Lu Y. Possible roles of reg family proteins in pancreatic islet cell growth. Endocr Metab Immune Disord Drug Targets. 2008;8:1–10. doi: 10.2174/187153008783928361. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T. Yonemura Y. Yonekura H. Suzuki Y. Miyashita H. Sugiyama K. Moriizumi S. Unno M. Tanaka O. Kondo H. Pancreatic beta-cell replication and amelioration of surgical diabetes by Reg protein. Proc Natl Acad Sci USA. 1994;91:3589–3592. doi: 10.1073/pnas.91.9.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zenilman ME. Magnuson TH. Swinson K. Egan J. Perfetti R. Shuldiner AR. Pancreatic thread protein is mitogenic to pancreatic-derived cells in culture. Gastroenterology. 1996;110:1208–1214. doi: 10.1053/gast.1996.v110.pm8613011. [DOI] [PubMed] [Google Scholar]
- 9.Ortiz EM. Dusetti NJ. Vasseur S. Malka D. Bödeker H. Dagorn JC. Iovanna JL. The pancreatitis-associated protein is induced by free radicals in AR4-2J cells and confers cell resistance to apoptosis. Gastroenterology. 1998;114:808–816. doi: 10.1016/s0016-5085(98)70595-5. [DOI] [PubMed] [Google Scholar]
- 10.Livesey FJ. O'Brien JA. Li M. Smith AG. Murphy LJ. Hunt SP. A Schwann cell mitogen accompanying regeneration of motor neurons. Nature. 1997;390:614–618. doi: 10.1038/37615. [DOI] [PubMed] [Google Scholar]
- 11.Ose T. Kadowaki Y. Fukuhara H. Kazumori H. Ishihara S. Udagawa J. Otani H. Takasawa S. Okamoto H. Kinoshita Y. Reg I-knockout mice reveal its role in regulation of cell growth that is required in generation and maintenance of the villous structure of small intestine. Oncogene. 2007;26:349–359. doi: 10.1038/sj.onc.1209799. [DOI] [PubMed] [Google Scholar]
- 12.Lasserre C. Christa L. Simon MT. Vernier P. Bréchot C. A novel gene (HIP) activated in human primary liver cancer. Cancer Res. 1992;52:5089–5095. [PubMed] [Google Scholar]
- 13.Kimura N. Yonekura H. Okamoto H. Nagura H. Expression of human regenerating gene mRNA and its product in normal and neoplastic human pancreas. Cancer. 1992;70:1857–1863. doi: 10.1002/1097-0142(19921001)70:7<1857::aid-cncr2820700708>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 14.Mauro V. Carette D. Chevallier D. Michiels JF. Segretain D. Pointis G. Sénégas-Balas F. Reg I protein in healthy and seminoma human testis. Histol Histopathol. 2008;23:1195–1203. doi: 10.14670/HH-23.1195. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe T. Yonekura H. Terazono K. Yamamoto H. Okamoto H. Complete nucleotide sequence of human reg gene and its expression in normal and tumoral tissues. The reg protein, pancreatic stone protein, and pancreatic thread protein are one and the same product of the gene. J Biol Chem. 1990;265:7432–7439. [PubMed] [Google Scholar]
- 16.Zenilman ME. Kim S. Levine BA. Lee C. Steinberg JJ. Ectopic expression of reg protein: A marker of colorectal mucosa at risk for neoplasia. J Gastrointest Surg. 1997;1:194–201. doi: 10.1016/s1091-255x(97)80109-6. discussion 201. [DOI] [PubMed] [Google Scholar]
- 17.Cavard C. Terris B. Grimber G. Christa L. Audard V. Radenen-Bussiere B. Simon MT. Renard CA. Buendia MA. Perret C. Overexpression of regenerating islet-derived 1 alpha and 3 alpha genes in human primary liver tumors with beta-catenin mutations. Oncogene. 2006;25:599–608. doi: 10.1038/sj.onc.1208860. [DOI] [PubMed] [Google Scholar]
- 18.Yuan RH. Jeng YM. Chen HL. Hsieh FJ. Yang CY. Lee PH. Hsu HC. Opposite roles of human pancreatitis-associated protein and REG1A expression in hepatocellular carcinoma: association of pancreatitis-associated protein expression with low-stage hepatocellular carcinoma, beta-catenin mutation, and favorable prognosis. Clin Cancer Res. 2005;11:2568–2575. doi: 10.1158/1078-0432.CCR-04-2039. [DOI] [PubMed] [Google Scholar]
- 19.Chang H. Guillou F. Taketo MM. Behringer RR. Overactive beta-catenin signaling causes testicular sertoli cell tumor development in the mouse. Biol Reprod. 2009;81:842–849. doi: 10.1095/biolreprod.109.077446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korinek V. Barker N. Morin PJ. van Wichen D. de Weger R. Kinzler KW. Vogelstein B. Clevers H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 21.Willert K. Brown JD. Danenberg E. Duncan AW. Weissman IL. Reya T. Yates JR., III III Nusse R. Wnt proteins are lipid-modified and can act as stem cell growth factors. Nature. 2003;423:448–452. doi: 10.1038/nature01611. [DOI] [PubMed] [Google Scholar]
- 22.Haegele L. Ingold B. Naumann H. Tabatabai G. Ledermann B. Brandner S. Wnt signalling inhibits neural differentiation of embryonic stem cells by controlling bone morphogenetic protein expression. Mol Cell Neurosci. 2003;24:696–708. doi: 10.1016/s1044-7431(03)00232-x. [DOI] [PubMed] [Google Scholar]
- 23.Chenn A. Walsh CA. Regulation of cerebral cortical size by control of cell cycle exit in neural precursors. Science. 2002;297:365–369. doi: 10.1126/science.1074192. [DOI] [PubMed] [Google Scholar]
- 24.Davidson KC. Jamshidi P. Daly R. Hearn MT. Pera MF. Dottori M. Wnt3a regulates survival, expansion, and maintenance of neural progenitors derived from human embryonic stem cells. Mol Cell Neurosci. 2007;36:408–415. doi: 10.1016/j.mcn.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 25.Fang D. Leishear K. Nguyen TK. Finko R. Cai K. Fukunaga M. Li L. Brafford PA. Kulp AN. Xu X. Smalley KS. Herlyn M. Defining the conditions for the generation of melanocytes from human embryonic stem cells. Stem Cells. 2006;24:1668–1677. doi: 10.1634/stemcells.2005-0414. [DOI] [PubMed] [Google Scholar]
- 26.Woll PS. Morris JK. Painschab MS. Marcus RK. Kohn AD. Biechele TL. Moon RT. Kaufman DS. Wnt signaling promotes hematoendothelial cell development from human embryonic stem cells. Blood. 2008;111:122–131. doi: 10.1182/blood-2007-04-084186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meijer L. Skaltsounis AL. Magiatis P. Polychronopoulos P. Knockaert M. Leost M. Ryan XP. Vonica CA. Brivanlou A. Dajani R. Crovace C. Tarricone C. Musacchio A. Roe SM. Pearl L. Greengard P. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 28.Sato N. Meijer L. Skaltsounis L. Greengard P. Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 29.Klein PS. Melton DA. A molecular mechanism for the effect of lithium on development. Proc Natl Acad Sci USA. 1996;93:8455–8459. doi: 10.1073/pnas.93.16.8455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giles RH. van Es JH. Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta. 2003;1653:1–24. doi: 10.1016/s0304-419x(03)00005-2. [DOI] [PubMed] [Google Scholar]
- 31.Reya T. Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434:843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 32.Ashcroft FJ. Varro A. Dimaline R. Dockray GJ. Control of expression of the lectin-like protein Reg-1 by gastrin: role of the Rho family GTPase RhoA and a C-rich promoter element. Biochem J. 2004;381(Pt 2):397–403. doi: 10.1042/BJ20031793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kehoe DE. Lock LT. Parikh A. Tzanakakis ES. Propagation of embryonic stem cells in stirred suspension without serum. Biotechnol Prog. 2008;24:1342–1352. doi: 10.1002/btpr.57. [DOI] [PubMed] [Google Scholar]
- 34.He TC. Zhou S. da Costa LT. Yu J. Kinzler KW. Vogelstein B. A simplified system for generating recombinant adenoviruses. Proc Natl Acad Sci USA. 1998;95:2509–2514. doi: 10.1073/pnas.95.5.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Veeman MT. Slusarski DC. Kaykas A. Louie SH. Moon RT. Zebrafish prickle, a modulator of noncanonical Wnt/Fz signaling, regulates gastrulation movements. Curr Biol. 2003;13:680–685. doi: 10.1016/s0960-9822(03)00240-9. [DOI] [PubMed] [Google Scholar]
- 36.He TC. Sparks AB. Rago C. Hermeking H. Zawel L. da Costa LT. Morin PJ. Vogelstein B. Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 37.Lock LT. Tzanakakis ES. Expansion and differentiation of human embryonic stem cells to endoderm progeny in a microcarrier stirred-suspension culture. Tissue Eng Part A. 2009;15:2051–2063. doi: 10.1089/ten.tea.2008.0455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livak KJ. Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 39.Kobayashi S. Akiyama T. Nata K. Abe M. Tajima M. Shervani NJ. Unno M. Matsuno S. Sasaki H. Takasawa S. Okamoto H. Identification of a receptor for reg (regenerating gene) protein, a pancreatic beta-cell regeneration factor. J Biol Chem. 2000;275:10723–10726. doi: 10.1074/jbc.275.15.10723. [DOI] [PubMed] [Google Scholar]
- 40.Wise CA. Clines GA. Massa H. Trask BJ. Lovett M. Identification and localization of the gene for EXTL, a third member of the multiple exostoses gene family. Genome Res. 1997;7:10–16. doi: 10.1101/gr.7.1.10. [DOI] [PubMed] [Google Scholar]
- 41.Higham AD. Bishop LA. Dimaline R. Blackmore CG. Dobbins AC. Varro A. Thompson DG. Dockray GJ. Mutations of RegIalpha are associated with enterochromaffin-like cell tumor development in patients with hypergastrinemia. Gastroenterology. 1999;116:1310–1318. doi: 10.1016/s0016-5085(99)70495-6. [DOI] [PubMed] [Google Scholar]
- 42.Mueller CM. Zhang H. Zenilman ME. Pancreatic reg I binds MKP-1 and regulates cyclin D in pancreatic-derived cells. J Surg Res. 2008;150:137–143. doi: 10.1016/j.jss.2008.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Williams RL. Hilton DJ. Pease S. Willson TA. Stewart CL. Gearing DP. Wagner EF. Metcalf D. Nicola NA. Gough NM. Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature. 1988;336:684–687. doi: 10.1038/336684a0. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda T. Nakamura T. Nakao K. Arai T. Katsuki M. Heike T. Yokota T. STAT3 activation is sufficient to maintain an undifferentiated state of mouse embryonic stem cells. EMBO J. 1999;18:4261–4269. doi: 10.1093/emboj/18.15.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herrmann BG. Labeit S. Poustka A. King TR. Lehrach H. Cloning of the T gene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- 46.Ciruna BG. Rossant J. Expression of the T-box gene Eomesodermin during early mouse development. Mech Dev. 1999;81:199–203. doi: 10.1016/s0925-4773(98)00243-3. [DOI] [PubMed] [Google Scholar]
- 47.Walther C. Gruss P. Pax-6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]
- 48.Lee JE. Hollenberg SM. Snider L. Turner DL. Lipnick N. Weintraub H. Conversion of Xenopus ectoderm into neurons by NeuroD, a basic helix-loop-helix protein. Science. 1995;268:836–844. doi: 10.1126/science.7754368. [DOI] [PubMed] [Google Scholar]
- 49.Kanai-Azuma M. Kanai Y. Gad JM. Tajima Y. Taya C. Kurohmaru M. Sanai Y. Yonekawa H. Yazaki K. Tam PP. Hayashi Y. Depletion of definitive gut endoderm in Sox17-null mutant mice. Development. 2002;129:2367–2379. doi: 10.1242/dev.129.10.2367. [DOI] [PubMed] [Google Scholar]
- 50.Perea-Gómez A. Shawlot W. Sasaki H. Behringer RR. Ang S. HNF3beta and Lim1 interact in the visceral endoderm to regulate primitive streak formation and anterior-posterior polarity in the mouse embryo. Development. 1999;126:4499–4511. doi: 10.1242/dev.126.20.4499. [DOI] [PubMed] [Google Scholar]
- 51.Terazono K. Uchiyama Y. Ide M. Watanabe T. Yonekura H. Yamamoto H. Okamoto H. Expression of reg protein in rat regenerating islets and its co-localization with insulin in the beta cell secretory granules. Diabetologia. 1990;33:250–252. doi: 10.1007/BF00404804. [DOI] [PubMed] [Google Scholar]
- 52.Unno M. Nata K. Noguchi N. Narushima Y. Akiyama T. Ikeda T. Nakagawa K. Takasawa S. Okamoto H. Production and characterization of Reg knockout mice: reduced proliferation of pancreatic beta-cells in Reg knockout mice. Diabetes. 2002;51(Suppl 3):S478–S483. doi: 10.2337/diabetes.51.2007.s478. [DOI] [PubMed] [Google Scholar]
- 53.Gigoux V. Clerc P. Sanchez D. Coll MG. Corominola H. Leung-Theung-Long S. Pénicaud L. Gomis R. Seva C. Fourmy D. Dufresne M. Reg genes are CCK2 receptor targets in ElasCCK2 mice pancreas. Regul Pept. 2008;146:88–98. doi: 10.1016/j.regpep.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 54.Dufresne M. Seva C. Fourmy D. Cholecystokinin and gastrin receptors. Physiol Rev. 2006;86:805–847. doi: 10.1152/physrev.00014.2005. [DOI] [PubMed] [Google Scholar]
- 55.Buckhaults P. Rago C. St Croix B. Romans KE. Saha S. Zhang L. Vogelstein B. Kinzler KW. Secreted and cell surface genes expressed in benign and malignant colorectal tumors. Cancer Res. 2001;61:6996–7001. [PubMed] [Google Scholar]
- 56.Koh TJ. Bulitta CJ. Fleming JV. Dockray GJ. Varro A. Wang TC. Gastrin is a target of the beta-catenin/TCF-4 growth-signaling pathway in a model of intestinal polyposis. J Clin Invest. 2000;106:533–539. doi: 10.1172/JCI9476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lei S. Dubeykovskiy A. Chakladar A. Wojtukiewicz L. Wang TC. The murine gastrin promoter is synergistically activated by transforming growth factor-beta/Smad and Wnt signaling pathways. J Biol Chem. 2004;279:42492–42502. doi: 10.1074/jbc.M404025200. [DOI] [PubMed] [Google Scholar]
- 58.Okamoto H. The Reg gene family and Reg proteins: with special attention to the regeneration of pancreatic beta-cells. J Hepatobiliary Pancreat Surg. 1999;6:254–262. doi: 10.1007/s005340050115. [DOI] [PubMed] [Google Scholar]
- 59.Chen Q. Zhang Y. Lu J. Wang Q. Wang S. Cao Y. Wang H. Duan E. Embryo-uterine cross-talk during implantation: the role of Wnt signaling. Mol Hum Reprod. 2009;15:215–221. doi: 10.1093/molehr/gap009. [DOI] [PubMed] [Google Scholar]
- 60.van Amerongen R. Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 61.Moon RT. Brown JD. Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 62.Kléber M. Sommer L. Wnt signaling and the regulation of stem cell function. Curr Opin Cell Biol. 2004;16:681–687. doi: 10.1016/j.ceb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 63.de la Monte SM. Ozturk M. Wands JR. Enhanced expression of an exocrine pancreatic protein in Alzheimer's disease and the developing human brain. J Clin Invest. 1990;86:1004–1013. doi: 10.1172/JCI114762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sanchez D. Mueller CM. Zenilman ME. Pancreatic regenerating gene I and acinar cell differentiation: influence on cellular lineage. Pancreas. 2009;38:572–577. doi: 10.1097/mpa.0b013e3181a1d9f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mally MI. Otonkoski T. Lopez AD. Hayek A. Developmental gene expression in the human fetal pancreas. Pediatr Res. 1994;36:537–544. doi: 10.1203/00006450-199410000-00022. [DOI] [PubMed] [Google Scholar]
- 66.Baeza N. Moriscot C. Figarella C. Guy-Crotte O. Vialettes B. Reg protein: a potential beta-cell-specific growth factor? Diabetes Metab. 1996;22:229–234. [PubMed] [Google Scholar]
- 67.Bartoli C. Baeza N. Figarella C. Pellegrini I. Figarella-Branger D. Expression of peptide-23/pancreatitis-associated protein and Reg genes in human pituitary and adenomas: comparison with other fetal and adult human tissues. J Clin Endocrinol Metab. 1998;83:4041–4046. doi: 10.1210/jcem.83.11.5217. [DOI] [PubMed] [Google Scholar]
- 68.Perfetti R. Raygada M. Wang Y. Zenilman ME. Egan JM. Denno KM. Sadler TW. Shuldiner AR. Regenerating (reg) and insulin genes are expressed in prepancreatic mouse embryos. J Mol Endocrinol. 1996;17:79–88. doi: 10.1677/jme.0.0170079. [DOI] [PubMed] [Google Scholar]
- 69.D'Amour KA. Agulnick AD. Eliazer S. Kelly OG. Kroon E. Baetge EE. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol. 2005;23:1534–1541. doi: 10.1038/nbt1163. [DOI] [PubMed] [Google Scholar]
- 70.Bernard-Perrone FR. Renaud WP. Guy-Crotte OM. Bernard P. Figarella CG. Okamoto H. Balas DC. Senegas-Balas FO. Expression of REG protein during cell growth and differentiation of two human colon carcinoma cell lines. J Histochem Cytochem. 1999;47:863–870. doi: 10.1177/002215549904700703. [DOI] [PubMed] [Google Scholar]
- 71.Macadam RC. Sarela AI. Farmery SM. Robinson PA. Markham AF. Guillou PJ. Death from early colorectal cancer is predicted by the presence of transcripts of the REG gene family. Br J Cancer. 2000;83:188–195. doi: 10.1054/bjoc.2000.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jamieson CH. Ailles LE. Dylla SJ. Muijtjens M. Jones C. Zehnder JL. Gotlib J. Li K. Manz MG. Keating A. Sawyers CL. Weissman IL. Granulocyte-macrophage progenitors as candidate leukemic stem cells in blast-crisis CML. N Engl J Med. 2004;351:657–667. doi: 10.1056/NEJMoa040258. [DOI] [PubMed] [Google Scholar]
- 73.Chan EF. Gat U. McNiff JM. Fuchs E. A common human skin tumour is caused by activating mutations in beta-catenin. Nat Genet. 1999;21:410–413. doi: 10.1038/7747. [DOI] [PubMed] [Google Scholar]
- 74.Malanchi I. Huelsken J. Cancer stem cells: never Wnt away from the niche. Curr Opin Oncol. 2009;21:41–46. doi: 10.1097/CCO.0b013e32831d1faf. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.