Abstract
In this report, we have demonstrated that by temporarily removing Kupffer cells (KCs), the transgene expression levels mediated by retrograde intrabiliary infusion (RII) of plasmid DNA, polyethylenimine-DNA, and chitosan nanoparticles were enhanced by 1,927-, 131-, and 23,450-fold, respectively, in comparison with the respective groups without KC removal. KC removal also led to significantly prolonged transgene expression in the liver that received all three carriers. This increased transgene expression was correlated with significantly reduced serum tumor necrosis factor-α level as an indicator for KC activation. These results suggest that KC activation is a significant contributing factor to the lowered transgene expression by polycation-DNA nanoparticles delivered by RII. More importantly, the combination of RII and transient removal of KCs may be adopted as an effective approach to achieving high and persistent transgene expression in the liver mediated by nonviral nanoparticles.
In this study, Dai and colleagues report that by temporarily removing Kupffer cells (KCs), transgene expression levels mediated by retrograde intrabiliary infusion of plasmid DNA, polyethylenimine–DNA, and chitosan nanoparticles were significantly enhanced in comparison with the respective groups without KC removal in rats. They also report that KC removal also led to significantly prolonged transgene expression in the liver that received DNA via all three carriers.
Introduction
For liver-targeted gene delivery, Kupffer cells (KCs) present a major challenge because they can mediate significant sequestration of viral or nonviral nanoparticles. More importantly, KCs as a key component of the innate immune system in the liver also contribute to nonspecific inflammation response following uptake of these nanoparticles or naked plasmid DNA. Several studies have demonstrated that systemic administration of adenoviral (Ad) vectors can activate KCs and lead to rapid clearance of Ad vectors, strong suppression of transgene expression, and liver toxicity (Lieber et al., 1997; Wolff et al., 1997; Alemany et al., 2000; Tao et al., 2001; Y. Zhang et al., 2001). Recently, it has been found that nonviral vectors, such as lipoplexes (liposome/plasmid DNA complexes), can also activate KCs and induce high levels of inflammatory cytokines (Sakurai et al., 2002; Zhang et al., 2005).
Retrograde intrabiliary infusion (RII) has recently been developed as a more effective administration route for the liver-targeted delivery of a variety of vectors, including adenovirus, retrovirus, lipoplexes, and polyplexes, to the liver (Yang et al., 1993; Sullivan et al., 1997; De Godoy et al., 1999; Uehara et al., 1999; Otsuka et al., 2000; X.H. Zhang et al., 2001, 2003; Tominaga et al., 2004; Chen et al., 2005). Compared with two other commonly used administration routes for liver-targeted gene delivery, i.e., intravenous and intraportal infusion, RII provides a more direct access to hepatocytes in liver parenchyma through bile canaliculi (Bircher, 1999). In addition, gene vectors delivered through the bile duct will reduce the nonspecific uptake by KCs, due to the absence of KCs in the biliary system (Bircher, 1999). However, a recent study indicated that the canalicular tight junctions may become permeable to large protein molecules or even 10-nm gold nanoparticles during RII (Hu et al., 2005). Gold nanoparticles infused through bile duct were found also in sinusoidal endothelial cells and KCs. This is likely dependent on the infusion parameters. Similarly, we also observed colocalization of polyethylenimine (PEI)-DNA nanoparticles with KCs after RII in our previous study (Dai et al., 2006). These findings suggest that a fraction of gene delivery vectors administered through RII may cross canalicular tight junctions and the space of Disse. The entry of gene delivery vectors into the blood circulation will possibly lead to the activation of KCs.
It has been demonstrated that temporary and partial removal of KCs with clodronate liposomes can effectively enhance the level and duration of gene expression in the liver after systemic delivery of Ad viruses (Wolff et al., 1997; Schiedner et al., 2003; Van Til et al., 2005). In an effort to understand the role of KCs in nanoparticle-mediated gene delivery through RII, we compared the effect of transient and partial depletion of KCs on the transfection efficiencies of naked DNA, chitosan-DNA nanoparticles, and PEI-DNA nanoparticles when delivered through RII. We also investigated the effect of this treatment on the duration of transgene expression and the serum tumor necrosis factor (TNF)-α level as an indicator of KC activation.
Materials and Methods
Preparation of PEI-DNA and chitosan-DNA nanoparticles
Polyethylenimine [branched PEI, average molecular weight (MW) of 25 kDa] was purchased from Sigma–Aldrich (St. Louis, MO). PEI was purified by dialysis against water (dialysis tubing with MWCO 3500; Pierce, Rockford, IL) for 3 days and then lyophilized. Chitosan C390 (MW 390 kDa, deacetylation degree 83.5%) was kindly provided by Vanson Halosource (Redmond, WA). Plasmid pVR1255 is a 6.4-kb pcDNA encoding luciferase driven by human cytomegalovirus promoter (a gift from Dr. Carl J. Wheeler, Vical Inc., San Diego, CA). The plasmid was amplified in Escherichia coli DH5α and purified by a Qiagen Giga plasmid purification kit (endotoxin free; Qiagen, Hilden, Germany). To prepare chitosan-DNA nanoparticles, a chitosan solution (0.02% in 5 mM sodium acetate buffer, pH 5.5) and a DNA solution (100 μg/ml in 5 mM sodium acetate solution) at an N/P ratio of 3 (N/P ratio is defined as the molar ratio of chitosan nitrogen to DNA phosphate) were preheated to 50–55°C separately. An equal volume of both solutions was quickly mixed while vortexing. PEI-DNA nanoparticles were prepared by mixing PEI solution (133 μg/ml in 5% glucose) with an equal volume of DNA solution (100 μg/ml in 5% glucose) at an N/P ratio of 10. Both chitosan-DNA and PEI-DNA nanoparticles were incubated at room temperature for 30 min before characterization and transfection. The particle size and zeta potential of chitosan-DNA and PEI-DNA nanoparticles were measured on a Zetasizer 3000 (Malvern Instruments, Malvern, Worcestershire, UK).
Transient depletion of KCs and RII of PEI-DNA, chitosan-DNA nanoparticles, and naked DNA
Clodronate liposomes were prepared as described (Van Rooijen and Sanders, 1994) and injected intravenously to rats at a dose of 2 μl/g of body weight at 48 hr prior to RII of nanoparticles and naked DNA. Wistar rats (male, 200–250 g) were randomly assigned to groups of 15–17 rats. Animals were laparotomized under general anesthesia, and the liver was isolated from the surrounding tissue. A 33G needle was inserted into the common bile duct, and a tie was used to secure the needle. Nanoparticles and naked DNA were administered at the dose equivalent to 200 μg of plasmid (0.8 mg/kg of body weight) in 4 ml of medium into the common bile duct over 20 min (0.2 ml/min) using a syringe pump. A tie was then placed around the bile duct between the liver and the point of infusion to prevent back flow, and the needle was withdrawn. After 30 min, all ties were removed. The needle hole in the bile duct might require stitches with 10-O nylon suture (Ethicon, Somerville, NJ) to prevent bile leakage, whenever necessary. Rats were kept on normal diet. On days 3, 5, and 7, rats from each group were sacrificed. Major organs (liver, heart, lung, spleen, and kidney) were harvested and stored at −80°C for analysis. Each liver was divided into four sections composed of median, left, right, and caudate lobes. Two milliliters of lysis buffer (0.1% Triton X-100, 2 mM EDTA, and 0.1 M Tris-HCl, pH 7.8) per gram of tissue was used for each sample, and the tissue was homogenized and subjected to two freeze–thaw cycles. The homogenates were centrifuged at 14,000 rpm for 10 min. Luciferase activity in the homogenate was measured for 10 sec on a luminometer and converted to the mass of luciferase expressed per gram of tissue using a standard curve generated in parallel on the same luminometer. Student's t test was used to compare the difference between selected groups. Differences were considered statistically significant when p < 0.05.
Results
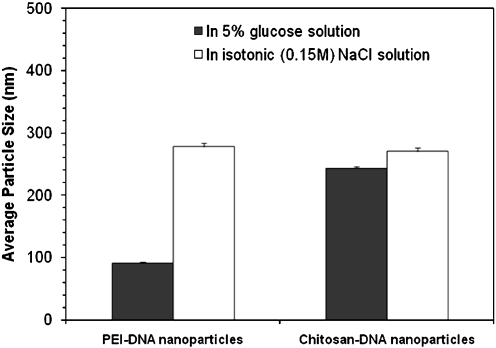
Before RII, we characterized the size of PEI-DNA nanoparticles and chitosan-DNA nanoparticles by dynamic light scattering. In the presence of 5% (w/v) glucose solution, PEI-DNA nanoparticles and chitosan-DNA nanoparticles showed an average particle size of 90.6 ± 2.0 nm and 243.1 ± 2.9 nm (Fig. 1), respectively. After incubation of these nanoparticles with isotonic (0.15 M) NaCl solution for 30 min, the particle size of PEI-DNA nanoparticles increased drastically to 277 ± 6.4 nm. On the other hand, chitosan-DNA nanoparticles appeared more stable in NaCl solution: the average particle size increased only by about 27 nm after 30 min of incubation.
FIG. 1.
Particle size of PEI-DNA nanoparticles (N/P = 10) and chitosan-DNA nanoparticles (N/P = 3) in 5% glucose solution and in isotonic (0.15 M) NaCl solution.
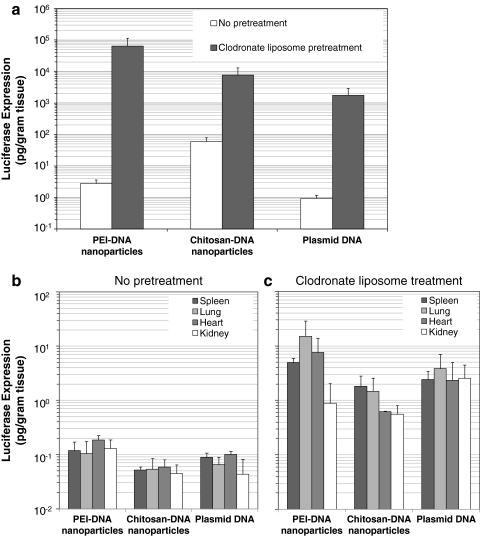
Three days after RII, chitosan-DNA nanoparticles mediated a luciferase expression of 33 pg/g of tissue in the liver of the rats that received no pretreatment. PEI-DNA nanoparticles were less efficient than chitosan-DNA nanoparticles in the liver, and the luciferase expression was about 17-fold lower than that by chitosan-DNA nanoparticles on day 3. It is surprising that naked DNA given by RII also gave a transient luciferase expression (0.62 pg/g of tissue) in the liver. In stark contrast, after clodronate liposome treatment prior to bile duct infusion, luciferase expression in the liver on day 3 mediated by both nanoparticles and naked DNA was significantly increased. As shown in Fig. 2a, the biggest increase (23,477-fold) in transgene expression was observed for PEI-DNA nanoparticles. For chitosan-DNA nanoparticles and naked DNA, the transfection efficiency in the liver was also enhanced by 131- and 1,927-fold, respectively. This significant enhancement in transfection efficiency after partial KC depletion indicated that KCs may be involved in the uptake of polymer-DNA nanoparticles or naked DNA and play an important role in regulating transgene expression mediated by these nanoparticles. It is interesting that, for all three groups, transgene expression in other organs, particularly in the lung and heart, also increased by 20- to 125-fold (Fig. 2c) as a result of KC depletion, even though luciferase activity detected in the liver still accounted for more than 95% of the total expression. These results support the hypothesis that nanoparticles can cross canalicular tight junctions and sinusoidal fenestrae and enter the blood circulation under this infusion condition. Therefore, without KC depletion, uptake of nanoparticles and naked DNA by KCs and KC activation may occur, leading to the reduced transgene expression.
FIG. 2.
Effect of KC depletion on transfection efficiency of different carriers in the liver (a) and in other organs without clodronate liposome treatment (b) and with clodronate liposome treatment following RII (c). Luciferase expression was analyzed on day 3. Each bar represents the mean ± SD (n = 3).
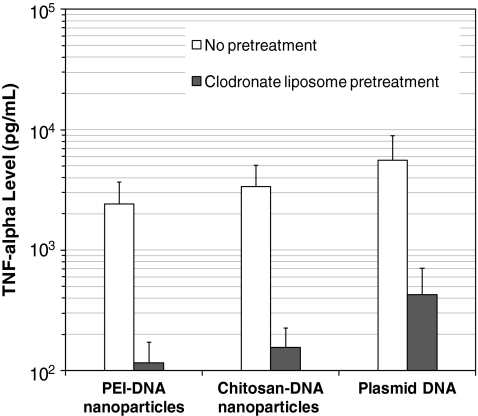
To validate the hypothesis that entry of nanoparticles or plasmid DNA into blood circulation can lead to the activation of KCs, we analyzed the serum TNF-α levels of both KC-depleted rats and normal rats on day 3 after RII of nanoparticles or naked DNA, because the activation of KCs by lipoplexes is usually associated with the production of a high level of TNF-α, one of the major proinflammatory cytokines (Sakurai et al., 2002; Kako et al., 2008). As shown in Fig. 3, both nanoparticles and naked DNA infused through the bile duct induced a high level of TNF-α from 2.4 to 5.6 ng/ml, as compared with a background level of <10 pg/ml. This result indicated that polymer-DNA nanoparticles and naked DNA administered through the bile duct did induce a strong innate immune response. Consistent with this finding, after depletion of KCs, serum TNF-α levels dropped 20-, 21-, and 13-fold in the rats infused with chitosan-DNA nanoparticles, PEI- DNA nanoparticles, and naked DNA, respectively. The significant reduction in the levels of TNF-α after the depletion of KCs suggested that innate immune response after bile duct infusion is very likely to be a result of KC activation by polymer-DNA nanoparticles or naked DNA.
FIG. 3.
Serum level of TNF-α after RII of PEI-DNA nanoparticles, chitosan-DNA nanoparticles, and naked DNA with or without clodronate liposome pretreatment on day 3 (n = 3).
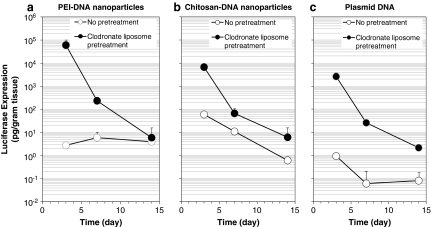
In addition to the effect of boosting transfection efficiency, the depletion of KCs also was found to help prolong transgene expression in the liver. Without pretreatment, PEI-DNA nanoparticles and chitosan-DNA nanoparticles mediated similar levels of transgene expression in the liver (6 and 11 pg/g of tissue, respectively) on day 7. No detectable transgene expression was observed in the liver transfected with plasmid DNA. However, after treatment with clodronate liposomes, significant enhancement of transgene expression was observed in the liver for all three groups (Fig. 4). The biggest boost (428-fold) in transgene expression on day 7 was found in the rats transfected with plasmid DNA. The transgene expression levels in the rats transfected with chitosan-DNA nanoparticles and PEI-DNA nanoparticles were also increased by 6- and 38-fold. On day 14, in untreated rats, PEI-DNA nanoparticles and chitosan-DNA nanoparticles still mediated detectable transgene expression in the liver, but plasmid DNA did not lead to any detectable gene expression. It is interesting that, although the transfection efficiency of chitosan-DNA nanoparticles dropped another 18-fold from day 7 to day 14, the transfection efficiency of PEI-DNA nanoparticles remained almost unchanged. Similar to the results on day 7, treatment with clodronate liposomes led to 10- and 27-fold enhanced transgene expression in the liver transfected with chitosan-DNA nanoparticles and plasmid DNA. However, the transfection efficiency of PEI-DNA nanoparticles was not improved by the treatment on day 14.
FIG. 4.
Transgene expression mediated by PEI-DNA nanoparticles (a), chitosan-DNA nanoparticles (b), and plasmid DNA (c) in the liver with or without clodronate liposome pretreatment. Each data point represents the mean ± SD (n = 3).
Discussion
In this study, we administered PEI-DNA nanoparticles, chitosan-DNA nanoparticles, and plasmid DNA through RII, which has recently been shown to be an effective administration route for liver-targeted gene delivery in several animal models, including rats (X.H. Zhang et al., 2001; Dai et al., 2006), pigs (Yang et al., 1993; Otsuka et al., 2000), and primates (Sullivan et al., 1997), using viral vectors (Peeters et al., 1996; De Godoy et al., 1999) and liposomes (Otsuka et al., 2000). The exposure of hepatocytes via RII occurs through bile canaliculi, which measure 0.5–1.25 μm (Bircher, 1999). In addition, the broad distribution of the biliary system provides great access to nearly all hepatocytes in liver parenchyma through bile canaliculi. This method also delivers nanoparticles or DNA preferentially to hepatocytes first, as opposed to KCs lining the luminal surface of the sinusoidal capillaries, when nanoparticles are delivered by intravascular infusion. It is highly likely, though, as our data suggested that, under moderate pressure and infusion volume, there is a pressure-dependent physical opening of canalicular tight junctions, which may lead to the transport of gene carriers into the space of Disse and into the bloodstream through sinusoidal fenestrae. A recent study has shown that large proteins and gold nanoparticles can be transported through canalicular tight junctions during RII infusion (Hu et al., 2005). More importantly, colocalization of gold nanoparticles with KCs was observed. In our previous study, we have also demonstrated that some PEI-DNA nanoparticles, chitosan-DNA nanoparticles, and plasmid DNA were taken up by KCs after RII (Dai et al., 2006).
On the other hand, several studies have revealed that lipoplexes (Dow et al., 1999; Yew et al., 1999; Kako et al., 2008) encapsulating bacterially derived plasmid DNA can induce strong activation of macrophages and subsequent production of high levels of proinflammatory cytokines after intravenous administration. The activation of macrophages is mediated by the strong immunostimulatory effect of unmethylated CpG sequences present at high frequency in bacterially derived plasmid DNA (Krieg, 1999, 2000). To reduce the inflammation response induced by plasmid DNA, depletion of macrophages before systematic administration has been proven to be an effective approach to enhance gene transfection efficiency of lipoplexes (Sakurai et al., 2002). Using the same strategy, we removed KCs with clodronate liposomes by tail-vein injection before RII infusion of nanoparticles or naked DNA to reduce the activation of KCs. The clodronate liposome was chosen for two reasons. First, it effectively removes more than 90% of KCs temporarily for about 6 days, and repopulation of KCs is completed within 2 weeks (Van Rooijen and Sanders, 1994). Second, it preferentially depletes KCs and removes other types of macrophages to a much lesser degree (Van Rooijen and Sanders, 1994; Van Rooijen and Van Kesteren-Hendrikx, 2003). As we expected, depletion of KCs before RII not only significantly enhanced the transfection efficiency in all major organs, but also greatly inhibited the production of TNF-α. These results confirmed that RII can lead to opening of the canalicular tight junction and sinusoidal leakage of the nanoparticles, and this leakage can further result in the uptake of the nanoparticles or DNA and the activation of KCs.
It is also noteworthy that the level of enhancement in transfection efficiency in the liver as a result of KC removal was carrier-dependent (Supplementary Fig. 1, available online at www.liebertonline.com/hum). The biggest boost was found for PEI-DNA nanoparticles. In our previous study, we observed a significantly higher level of colocalization of PEI-DNA nanoparticles with KCs compared with chitosan-DNA nanoparticles and plasmid DNA (Dai et al., 2006). It is also interesting to note that there was a substantially higher number of KCs observed in the rat liver that received PEI-DNA nanoparticles (Dai et al., 2006). Those findings indicated that PEI-DNA nanoparticles may induce a higher level of KC activation, and thus lead to stronger inhibition of transfection efficiency in the liver, compared with the other two groups. In addition, we found that, unlike that in untreated rats, the transfection efficiency of these carriers after KC removal correlated well with that in primary hepatocyte culture (Dai et al., 2006).
The method for transient depletion of KCs is an attractive approach for potential clinical application. Clodronate has been used in Europe and Canada for treating osteoporosis to increase bone mass and reduce fractures. Given by intravenous injection, the effect of clodronate liposome on KC removal can be highly specific (Van Rooijen and Van Kesteren-Hendrikx, 2002). In addition, RII can be achieved in the clinical setting by endoscopic retrograde cholangiopancreatography. Given the drastic improvement of transgene expression by combining RII and transient depletion of KCs, this method may be adopted as a highly effective approach for liver-targeted gene delivery.
Supplementary Material
Acknowledgments
The authors thank Wei-Seng Lim and Xuesong Jiang for their technical assistance. This study was partially supported by grant R01DK068399 (H.Q.M.) from the National Institute of Diabetes and Digestive and Kidney Diseases and a grant from the Agency for Science, Technology and Research of Singapore.
Author Disclosure Statement
No competing financial interests exist.
References
- Alemany R. Suzuki K. Curiel D.T. Blood clearance rates of adenovirus type 5 in mice. J. Gen. Virol. 2000;81:2605–2609. doi: 10.1099/0022-1317-81-11-2605. [DOI] [PubMed] [Google Scholar]
- Bircher J. Oxford Textbook of Clinical Hepatology. Oxford University Press; NY: 1999. pp. 13–21. [Google Scholar]
- Chen C.Y. Liu H.S. Lin X.Z. Hydrodynamics-based gene delivery to the liver by bile duct injection of plasmid DNA—the impact of lasting biliary obstruction and injection volume. Hepatogastroenterology. 2005;52:25–28. [PubMed] [Google Scholar]
- Dai H. Jiang X. Tan G.C., et al. Chitosan-DNA nanoparticles delivered by intrabiliary infusion enhance liver-targeted gene delivery. Int. J. Nanomedicine. 2006;1:507–522. doi: 10.2147/nano.2006.1.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Godoy J.L. Malafosse R. Fabre M., et al. In vivo hepatocyte retrovirus-mediated gene transfer through the rat biliary tract. Hum. Gene Ther. 1999;10:249–257. doi: 10.1089/10430349950019039. [DOI] [PubMed] [Google Scholar]
- Dow S.W. Fradkin L.G. Liggitt D.H., et al. Lipid-DNA complexes induce potent activation of innate immune responses and antitumor activity when administered intravenously. J. Immunol. 1999;163:1552–1561. [PubMed] [Google Scholar]
- Hu J. Zhang X. Dong X., et al. A remarkable permeability of canalicular tight junctions might facilitate retrograde, non-viral gene delivery to the liver via the bile duct. Gut. 2005;54:1473–1479. doi: 10.1136/gut.2005.070904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kako K. Nishikawa M. Yoshida H. Takakura Y. Effects of inflammatory response on in vivo transgene expression by plasmid DNA in mice. J. Pharm. Sci. 2008;97:3074–3083. doi: 10.1002/jps.21254. [DOI] [PubMed] [Google Scholar]
- Krieg A.M. Direct immunologic activities of CpG DNA and implications for gene therapy. J. Gene Med. 1999;1:56–63. doi: 10.1002/(sici)1521-2254(199901/02)1:1<56::aid-jgm5>3.3.co;2-y. [DOI] [PubMed] [Google Scholar]
- Krieg A.M. The role of CpG motifs in innate immunity. Curr. Opin. Immunol. 2000;12:35–43. doi: 10.1016/s0952-7915(99)00048-5. [DOI] [PubMed] [Google Scholar]
- Lieber A. He C.Y. Meuse L., et al. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J. Virol. 1997;71:8798–8807. doi: 10.1128/jvi.71.11.8798-8807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsuka M. Baru M. Delriviere L., et al. In vivo liver-directed gene transfer in rats and pigs with large anionic multilamellar liposomes: routes of administration and effects of surgical manipulations on transfection efficiency. J. Drug Target. 2000;8:267–279. doi: 10.3109/10611860008997905. [DOI] [PubMed] [Google Scholar]
- Peeters M.J. Patijn G.A. Lieber A., et al. Adenovirus-mediated hepatic gene transfer in mice: comparison of intravascular and biliary administration. Hum. Gene Ther. 1996;7:1693–1699. doi: 10.1089/hum.1996.7.14-1693. [DOI] [PubMed] [Google Scholar]
- Sakurai F. Terada T. Yasuda K., et al. The role of tissue macrophages in the induction of proinflammatory cytokine production following intravenous injection of lipoplexes. Gene Ther. 2002;9:1120–1126. doi: 10.1038/sj.gt.3301784. [DOI] [PubMed] [Google Scholar]
- Schiedner G. Hertel S. Johnston M., et al. Selective depletion or blockade of Kupffer cells leads to enhanced and prolonged hepatic transgene expression using high-capacity adenoviral vectors. Mol. Ther. 2003;7:35–43. doi: 10.1016/s1525-0016(02)00017-5. [DOI] [PubMed] [Google Scholar]
- Sullivan D.E. Dash S. Du H., et al. Liver-directed gene transfer in non-human primates. Hum. Gene Ther. 1997;8:1195–1206. doi: 10.1089/hum.1997.8.10-1195. [DOI] [PubMed] [Google Scholar]
- Tao N.J. Gao G.P. Parr M., et al. Sequestration of adenoviral vector by Kupffer cells leads to a nonlinear dose response of transduction in liver. Mol. Ther. 2001;3:28–35. doi: 10.1006/mthe.2000.0227. [DOI] [PubMed] [Google Scholar]
- Tominaga K. Kuriyama S. Yoshiji H., et al. Repeated adenoviral administration into the biliary tract can induce repeated expression of the original gene construct in rat livers without immunosuppressive strategies. Gut. 2004;53:1167–1173. doi: 10.1136/gut.2003.013748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T. Honda K. Hatano E., et al. Gene transfer to the rat biliary tract with the HVJ-cationic liposome method. J. Hepatol. 1999;30:836–842. doi: 10.1016/s0168-8278(99)80137-8. [DOI] [PubMed] [Google Scholar]
- Van Rooijen N. Sanders A. Liposome mediated depletion of macrophages: mechanism of action, preparation of liposomes and applications. J. Immunol. Methods. 1994;174:83–93. doi: 10.1016/0022-1759(94)90012-4. [DOI] [PubMed] [Google Scholar]
- Van Rooijen N. Van Kesteren-Hendrikx E. Clodronate liposomes: perspectives in research and therapeutics. J. Liposome Res. 2002;12:81–94. doi: 10.1081/lpr-120004780. [DOI] [PubMed] [Google Scholar]
- Van Rooijen N. Van Kesteren-Hendrikx E. “In vivo” depletion of macrophages by liposome-mediated “suicide”. Methods Enzymol. 2003;373:3–16. doi: 10.1016/s0076-6879(03)73001-8. [DOI] [PubMed] [Google Scholar]
- Van Til N.P. Markusic D.M. Van Der Rijt R., et al. Kupffer cells and not liver sinusoidal endothelial cells prevent lentiviral transduction of hepatocytes. Mol. Ther. 2005;11:26–34. doi: 10.1016/j.ymthe.2004.09.012. [DOI] [PubMed] [Google Scholar]
- Wolff G. Worgall S. Van Rooijen N., et al. Enhancement of in vivo adenovirus-mediated gene transfer and expression by prior depletion of tissue macrophages in the target organ. J. Virol. 1997;71:624–629. doi: 10.1128/jvi.71.1.624-629.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.P. Raper S.E. Cohn J.A., et al. An approach for treating the hepatobiliary disease of cystic-fibrosis by somatic gene-transfer. Proc. Natl. Acad. Sci. U.S.A. 1993;90:4601–4605. doi: 10.1073/pnas.90.10.4601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yew N.S. Wang K.X. Przybylska M., et al. Contribution of plasmid DNA to inflammation in the lung after administration of cationic lipid:pDNA complexes. Hum. Gene Ther. 1999;10:223–234. doi: 10.1089/10430349950019011. [DOI] [PubMed] [Google Scholar]
- Zhang J.S. Liu F. Huang L. Implications of pharmacokinetic behavior of lipoplex for its inflammatory toxicity. Adv. Drug Deliver. Rev. 2005;57:689–698. doi: 10.1016/j.addr.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Zhang X.H. Collins L. Sawyer G.J., et al. In vivo gene delivery via portal vein and bile duct to individual lobes of the rat liver using a polylysine-based nonviral DNA vector in combination with chloroquine. Hum. Gene Ther. 2001;12:2179–2190. doi: 10.1089/10430340152710522. [DOI] [PubMed] [Google Scholar]
- Zhang X.H. Sawyer G.J. Dong X.B., et al. The in vivo use of chloroquine to promote non-viral gene delivery to the liver via the portal vein and bite duct. J. Gene Med. 2003;5:209–218. doi: 10.1002/jgm.340. [DOI] [PubMed] [Google Scholar]
- Zhang Y. Chirmule N. Gao G.P., et al. Acute cytokine response to systemic adenoviral vectors in mice is mediated by dendritic cells and macrophages. Mol. Ther. 2001;3:697–707. doi: 10.1006/mthe.2001.0329. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.