Abstract
The Cdc14 dual-specificity phosphatase plays a key role in the mitotic exit of budding yeast cells. Mammals have two homologues, Cdc14a and Cdc14b. Unlike the yeast counterpart, neither Cdc14a nor Cdc14b seems to be essential for mitotic exit. To determine the physiological function of Cdc14b, we generated mice deficient in the phosphatase. The mutant mice were viable and did not display overt abnormalities. However, these mice developed signs of aging at much younger ages than the wild-type mice. At the cellular level, the Cdc14b-deficient mouse embryonic fibroblasts (MEFs) grew more slowly than the controls at later passages as a result of increased rates of senescence. Consistent with these premature-aging phenotypes, Cdc14b-deficient cells accumulated more endogenous DNA damage than the wild-type cells, and more Cdc14b-deficient MEFs entered senescence than control MEFs in response to exogenous DNA damage. However, no deficiencies in DNA damage checkpoint response were detected in Cdc14b mutant cells, suggesting that the function of Cdc14b is required for efficient DNA damage repair.
INTRODUCTION
The yeast Cdc14 phosphatase is required for mitotic exit (3, 20). In mammals, there are two Cdc14 homologues, Cdc14a and Cdc14b (11). Cdc14a was shown to localize in the centrosome and may function in the cycling of that organelle (10, 14). It was also implicated in the maturation of oocytes (17). In contrast, Cdc14b is a nuclear protein with apparent accumulation in the nucleolus, a localization pattern similar to that of its yeast counterpart (5, 10, 14). Studies in cultured cancer cell lines suggested that Cdc14b might function in microtubule bundling and stability (5), centrosome duplication (23), G1 phase length control (16), and the G2/M DNA damage checkpoint (2). However, a role in mitotic exit has never been demonstrated for Cdc14b. Recently, CDC14B was deleted in HCT116 human colorectal carcinoma cells. The mutant cells were viable and free of any noticeable defects in mitosis or other cell cycle events (4). These different and somewhat controversial findings raise questions as to the physiological function of Cdc14b.
Here, we report the generation and analyses of Cdc14b-deficient mice. We found that Cdc14b mutant animals were viable and did not display overt abnormalities. However, the mice developed signs of aging at much younger ages than the wild-type mice. These signs include cataracts and kyphosis. At the cellular level, the Cdc14b-deficient mouse embryonic fibroblasts (MEFs) displayed a slow-growth phenotype at later passages, likely a result of the increased rate of senescence in the mutant cells. Consistent with these premature-aging phenotypes, Cdc14b-deficient cells accumulated more endogenous DNA damage than the wild-type cells, and more Cdc14b-deficient MEFs than control MEFs entered senescence in response to exogenous DNA damage. These results are consistent with the notion that CDC14B is required for efficient DNA damage repair in human and chicken cells (15).
MATERIALS AND METHODS
Generation of Cdc14b-deficient mice.
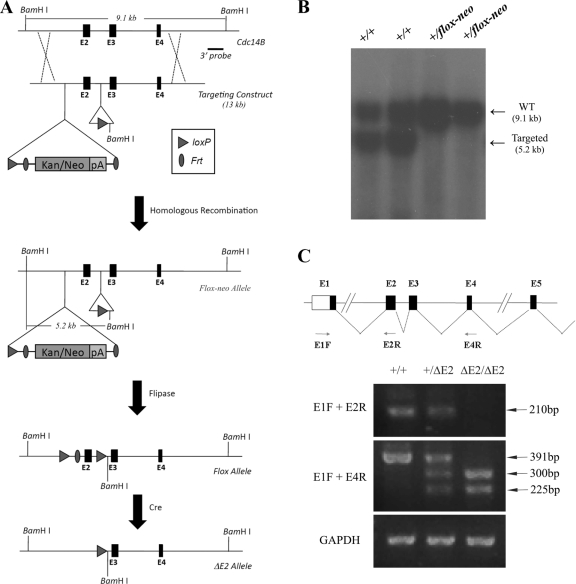
We floxed exon 2 to create a conditional allele of Cdc14b (Fig. 1 A). After germ line transmission, the Cdc14b+/flox-neo mice were first crossed with Flpase mice to remove the Frt-flanked neo cassette, generating Cdc14b+/flox mice that were further crossed with Meox2-Cre mice to delete exon 2 (21). Cdc14b+/ΔE2 mice were back-crossed to a C57BL/6 background for more than five generations. The following primers were used for the PCR-based genotyping: 5′-ACA GCA GAC CAA AGA GTG CAA C and 5′-CAC ACA CAC ACA CCA ATC GTA G for the Cdc14b+ allele; 5′-ATC ACC TAC CAC CAT GGT CC and 5′-AGC ACC CAG CTG CAA GTG TT for the Cdc14bΔE2 allele.
Fig. 1.
Generation of Cdc14b-deficient mice. (A) Strategy used to generate a conditional allele of Cdc14b. (B) Southern blot analysis. Genomic DNA isolated from tail snips was digested with BamHI, separated on an agarose gel, and blotted. The blot was probed with a 3′ probe shown in panel A. WT, wild type. (C) RT-PCR analysis of the expression of wild-type Cdc14b and Cdc14bΔE2.
Isolation and analyses of MEFs.
MEFs were isolated from embryonic day 12.5 (E12.5) to E13.5 embryos and cultured in Dulbecco's modified Eagle's medium (DMEM) containing 2 mM glutamine, 1% penicillin/streptomycin, and 15% fetal bovine serum (FBS) at 37°C in 5% CO2. The cells were trypsinized and passaged every 3 days, and the 3rd and 7th passages (P3 and P7) of cells were used as early-passage and late-passage MEFs, respectively, unless otherwise indicated.
Immortalized MEFs (iMEFs) were obtained using the classical 3T3 protocol (22). We serum starved (0.5% FBS) the iMEFs for 48 h and then released them into normal culture medium. The serum starvation caused about 50% of the cells to be blocked in G2. Two hours after being released, the cells were exposed to 10-Gy gamma irradiation, and nocodazole (80 ng/ml) was added to the medium immediately after irradiation.
RNA extraction and reverse transcription (RT)-PCR.
Total RNA from MEFs was prepared with Trizol (Invitrogen); 2 μg of total RNA was used to synthesize the first-strand cDNA with Superscript III (Invitrogen). The following pairs of primers were used for PCR after reverse transcription of mouse Cdc14b: E1F (5′-GCC TCC ATG AAG CGT AAG AGC) and E2R (5′-TGC ACT CTT TGG TCT GCT GT) to produce a 210-bp PCR product; E1F and E4R (5′-GCT TTC TCT GAT CAG TGC CAG T) to produce a 391-bp PCR product. Gapdh served as an internal reference (primer pair: Gapdh-F, 5′-CTC CAC TCA CGG CAA ATT CA, and Gapdh-R, 5′-GCA GGA TGC ATT GCT GAC AA).
Western blot assay, immunostaining, and SA–β-Gal staining.
For Western blots, whole-cell lysates were prepared with RIPA buffer, and the protein concentration was measured by the Bradford method. For immunostaining, the cells were cultured on coverslips, fixed in 4% paraformaldehyde (PFA), permeabilized in 0.5% Triton X-100, and blocked with PBS containing 5% normal goat serum. The cells were then incubated with primary antibodies overnight at 4°C or for 2 h at room temperature. After being washed with PBS, the cells were incubated with fluor-conjugated secondary antibodies for 45 min at room temperature. The cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole). Senescence-associated beta-galactosidase (SA–β-Gal) staining was performed as previously described (6).
Behavioral tests.
Mice were tested for fear conditioning according to a 2-day procedure, as we previously described (12).
RESULTS
Generation of Cdc14b-deficient mice.
To elucidate the physiological function of Cdc14b, we generated a conditional-deletion mouse strain. The targeting strategy is depicted in Fig. 1A. We floxed exon 2, deletion of which would result in frameshift. The germ line transmission of the targeted allele was confirmed by Southern blot analysis (Fig. 1B). Cdc14b+/flox-neo mice were first crossed to Meox2-Cre mice (21) to generate systemic deletion of exon 2. Due to the lack of suitable antibodies, we could not confirm the disruption of Cdc14b at the protein level. Therefore, we resorted to RT-PCR analysis with a forward primer located in exon 1 (E1R), a reverse primer in exon 2 (E2R), and another reverse primer in exon 4 (E4R) (Fig. 1C). The primer pair E1F-E1R could amplify only the expected 210-bp fragment from the total RNA prepared from wild-type or E2 deletion heterozygous MEFs, but not from E2 deletion homozygous cells. The deletion of exon 2 resulted in two mRNA species from the E2 deletion allele. Sequencing the two products indicated that one was missing E2 (as expected) and another was missing both E2 and E3. Because of the frameshift, both of these species are not expected to produce functional Cdc14b protein.
Early-onset aging in Cdc14b-deficient mice.
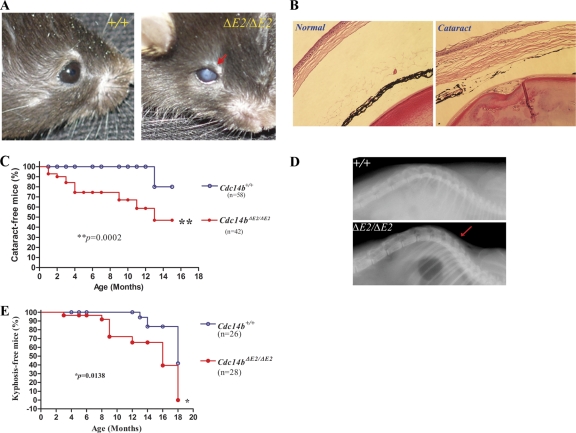
Our initial analysis of the Cdc14b-deficient mice in a mixed genetic background (129 and C57BL/6) did not find any discernible abnormalities. The intercrosses of the heterozygous mice produced homozygous progeny at the Mendelian ratio (25%), indicating that Cdc14b is not required for embryonic development and is unlikely to play a critical role in the control of mitotic exit, as its budding yeast counterpart does. However, after we backcrossed the ΔE2 allele to a C57/B6 background for more than five generations, the homozygous exon 2 deletion mice showed cataracts at young ages while only two wild-type littermates did so in old age (Fig. 2 A and C). Histological analysis of the eyeballs with cataracts showed abnormal corneas and lenses (Fig. 2B). The diseased cornea became loose and thickened, and some surface areas of the ocular lens showed invaginations. Thus, the cataracts in the mutant mice were most likely caused by loss of transparency in the cornea and, to a lesser degree, in the lens. In addition, Cdc14bΔE2/ΔE2 mice also developed kyphosis much earlier than the control animals (Fig. 2D and E), further supporting the idea that the mutant mice were experiencing premature aging.
Fig. 2.
Cdc14b-deficient mice experience premature aging. (A) Eyes of a Cdc14bΔE2/ΔE2 mutant mouse with cataract (arrow) and a wild-type mouse. (B) Hematoxylin and eosin staining of sections of the eyeballs. (C) Cataract-free curve analysis of mice with different genotypes. (D) X-ray analysis of the spine. The arrow indicates the more curved spine (kyphosis) in the mutants. (E) Kyphosis-free curve analysis of the wild-type and mutant mice.
Early-onset senescence in Cdc14b-deficient MEFs.
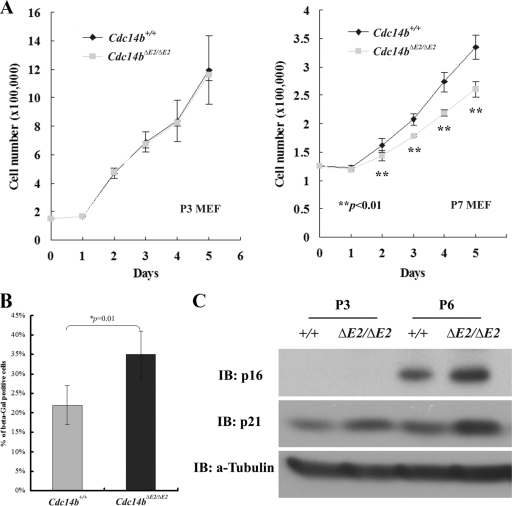
To determine whether the loss of Cdc14b causes any cellular phenotypes, we isolated MEFs from embryos around E13.5. At early passages (less than P6), the mutant cells grew at rates similar to those of the control cells (Fig. 3 A). However, by P7, the mutants showed lower growth rates than control cells (Fig. 3A). Consistent with the reduction in the growth rate, more senescent cells were found in the mutants than in the control, as determined by the SA–β-Gal assay (Fig. 3B). Furthermore, later-passage Cdc14bΔE2/ΔE2 cells expressed more p21Cip1 and p16Ink4a (Fig. 3C), consistent with the observation that more mutant cells entered senescence.
Fig. 3.
Cellular senescence in Cdc14b-deficient MEFs. (A) Growth curve analysis of early- and late-passage MEFs. The error bars indicate standard deviations. (B) Quantitation of SA–β-Gal-positive MEFs at passage 7. (C) Western blot analyses of senescence-related molecular markers. IB, immunoblot.
Proficient DNA damage checkpoint in Cdc14b-deficient cells.
It was previously reported that Cdc14b contributed to the G2/M DNA damage checkpoint (2). A deficiency in the DNA damage checkpoint caused by the loss of Cdc14b function may explain the premature-aging phenotype in Cdc14b mutant mice, given that DNA damage plays a causal role in cellular senescence and aging (1, 8, 9). Therefore, we tested the proficiency of the G2/M DNA damage checkpoint in the absence of Cdc14b.
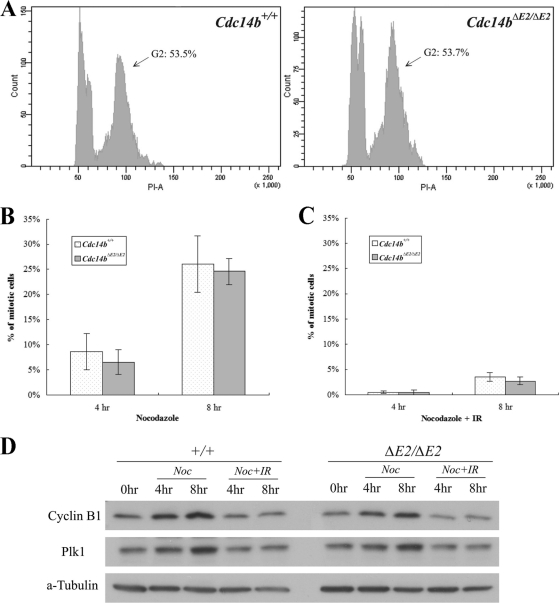
To better synchronize the MEFs for the DNA damage checkpoint analysis, we immortalized the MEFs (iMEFs). There were no significant differences in the efficiency of immortalization between the control and mutant cells. We serum starved the cells for 48 h and then released them into normal medium. The serum starvation caused an apparent G2 block, since 2 h after release, over 50% of the cells were at G2 (Fig. 4 A). To examine the G2/M DNA damage checkpoint, we irradiated the cells 2 h after their release from serum starvation. Immediately after irradiation, nocodazole was added to the medium to block the cells in mitosis. At different time points after irradiation, the cells were harvested for analyses. First, we counted the cells that had entered mitosis (mitotic-index assay). The mock-irradiated wild-type and mutant cells entered mitosis at comparable rates, indicating that the loss of Cdc14b does not impact the normal G2/M transition (Fig. 4B). However, both the wild-type and the mutant cells were blocked from entering mitosis by the irradiation (Fig. 4C). We next analyzed the expression of mitotic regulators. As shown in Fig. 4D, DNA damage inhibited the accumulation of cyclin B1 and Plk1 in both wild-type and mutant cells. Taken together, these results demonstrate that Cdc14b is not required for the G2/M DNA damage checkpoint, in agreement with the conclusion reached with human cells deficient in CDC14B (15).
Fig. 4.
Proficient G2/M DNA damage checkpoint in Cdc14b-deficient MEFs. (A) Fluorescence-activated cell sorting (FACS) analysis of iMEFs 2 h after release from 48-h serum starvation. (B) Mitotic-index analysis of cells without gamma irradiation. Nocodazole (80 ng/ml) was added to the medium of the cells in panel A, and the cells were harvested at 4 and 8 h after the addition of nocodazole (6 and 10 h after release). The error bars indicate standard deviations. (C) Mitotic-index analysis of cells with 10-Gy gamma irradiation. The cells were treated the same as those in panel B but were irradiated immediately before the addition of nocodazole. (D) Western blot analyses of mitotic regulators in the cells from panels B and C. Time zero (0 h) was 2 h after release from serum starvation.
Cdc14b-deficient cells are defective in DNA damage repair.
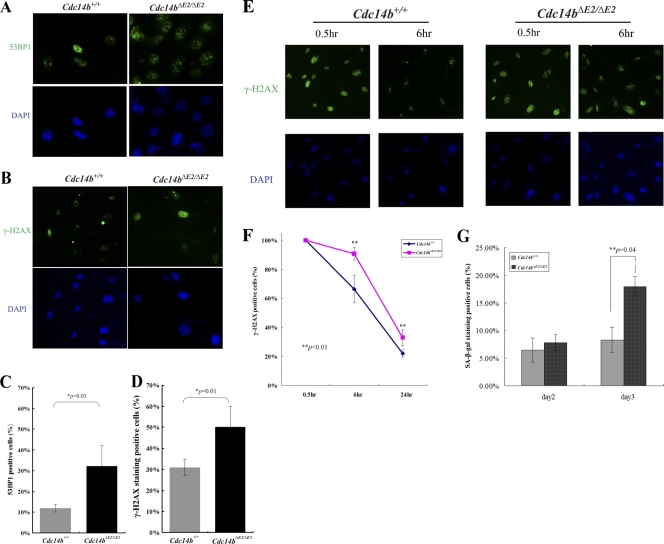
More recently, it was shown in human and chicken cells that Cdc14b was required for efficient DNA damage repair (15). A deficiency in DNA damage repair would be consistent with the premature-aging phenotypes observed in the mutant mice. We therefore first tested if there were elevated levels of endogenous DNA damage. We used two DNA damage markers, γ-H2AX and 53BP1, to gauge the level of DNA damage. Immunostaining of the primary MEFs at late passages showed that Cdc14b mutant MEFs contained significantly more γ-H2AX- and 53BP1-positive cells than wild-type control cells (Fig. 5 A to D). Next, we determined the effect of exogenous DNA damage on Cdc14b mutant MEFs. The asynchronously growing primary MEFs were irradiated; collected at 0.5, 6, and 24 h after irradiation; and processed for immunostaining. As shown in Fig. 5E, at 0.5 h, both wild-type and mutant cells included a large number of damaged cells with no differences between the two cell types. However, with time, while the percentage of γ-H2AX-positive cells decreased in the wild-type MEFs, such decrease was much slower in the mutants (Fig. 5E and F). Consistent with the persistence of DNA damage, a significantly larger fraction of the irradiated mutant MEF population than that of the wild-type population entered senescence, as demonstrated by the SA–β-Gal staining results (Fig. 5G).
Fig. 5.
Defective DNA damage repair in Cdc14b-deficient MEFs. (A and B) Immunofluorescence staining of passage 7 primary MEFs. (C and D) Quantitation of the results in panels A and B. A cell with 5 or more foci of 53BP1 or γ-H2AX staining signals was considered positive. The error bars indicate standard deviations. (E) Immunofluorescence staining of passage 3 primary MEFs after 10-Gy gamma irradiation. (F) Quantitation of the results in panel E. A cell with 20 or more foci of γ-H2AX staining signals was counted as positive in this assay. (G) More Cdc14b mutant MEFs entered senescence after gamma irradiation. At days 2 and 3 after irradiation, the cells were stained for SA–β-Gal.
Cdc14b-deficient mice display defects in other physiological processes.
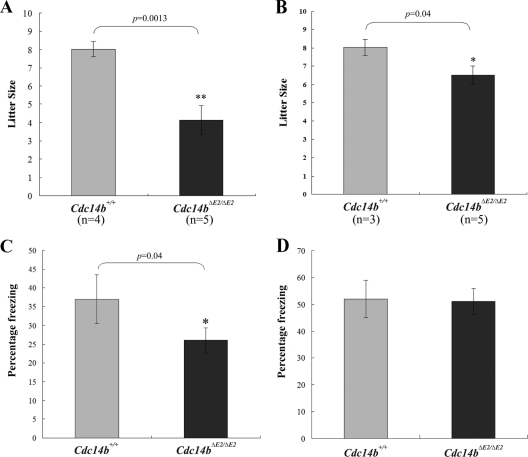
In addition to premature aging, Cdc14b mutant mice were less fertile than the wild-type control, especially females (Fig. 6 A and B). This result is consistent with the report that Cdc14b is involved in the meiosis of mouse oocytes (18). Moreover, since Cdc14b can positively regulate APCCdh1 function (most likely through dephosphorylating Cdh1) (2, 25) and since that APCCdh1 is required for learning and memory (7, 12), we subjected Cdc14b mutant mice to behavior tests to determine whether the lack of this phosphatase impacts neural functions. Similar to Cdh1 heterozygous mice (12), Cdc14b-deficient mice displayed deficits in contextual fear conditioning, but not in cued fear conditioning (Fig. 6C and D).
Fig. 6.
Reduced fertility and deficits in learning and memory in Cdc14b-deficient mice. (A) Litter sizes from the crosses between wild-type males and 3- to 4-month-old wild-type or Cdc14b mutant female mice. (B) Litter sizes from crosses between wild-type or Cdc14b mutant males and 3- to 4-month-old wild-type female mice. (C) Contextual fear conditioning test. (D) Cued fear conditioning test. Twelve wild-type and 12 Cdc14bΔE2/ΔE2 3-month-old mice were tested for panels C and D. The error bars indicate standard deviations.
DISCUSSION
Once a cell divides, the daughter cells must return to a ground state (characterized by low cyclin-dependent kinase [CDK] activities) to prepare for another division cycle or, alternatively, to withdraw from the cell cycle. Failure to return to the ground state is associated with unscheduled cell cycle entry and developmental defects in animals. Establishment of this ground state requires not only inhibition of the mitotic kinase CDK1/cyclin B1, but also the dephosphorylation of a large number of CDK1-phosphorylated proteins. The dephosphorylation is carried out primarily by Cdc14 in budding yeasts (3, 20). Despite the presence of two Cdc14 homologues in mammals, Cdc14b was considered much closer to yeast Cdc14 based on their common subcellular localization in the nucleolus. However, a definitive function of Cdc14b in mitotic exit has never been established. In fact, the phosphatase(s) required for mitotic exit in higher eukaryotes seems unrelated to Cdc14. For example, in Xenopus, protein phosphatase 1 (PP1) was shown to catalyze dephosphorylation during mitotic exit (24), and recently, PP2A-B55α was identified as the mitotic exit phosphatase in human cells (19). The lack of mitotic phenotypes in cells with Cdc14a (15) or Cdc14b (reference 4 and this study) deleted unequivocally demonstrates that the mammalian Cdc14 phosphatases do not regulate mitosis. They must have gained other functions during evolution.
Aging (and cellular senescence) is triggered by cellular stresses, including DNA damage (1, 8, 9). The genetic material in a cell is constantly assaulted by endogenous metabolic by-products, such as reactive oxygen species, and by exogenous physical and chemical genotoxic agents. It is estimated that there are about 105 lesions per cell per day in humans (13). In response to such high levels of DNA damage, sophisticated mechanisms have evolved to coordinate cell cycle progression and repair. When the damage is too extensive to repair, the damaged cells are eliminated by apoptosis, as they may impose adverse effects on the well-being of a multicellular organism or enter senescence, a state of permanent arrest. The aging phenotypes of Cdc14b-deficient mice suggest a role of the phosphatase in the DNA damage response. Bassermann et al. (2) reported that Cdc14b contributed to the G2/M DNA damage checkpoint by activating APCCdh1. However, we did not observe any checkpoint defects in the Cdc14b-deficeint MEFs. A similar result was reported recently using HCT116 and DT-40 cells with CDC14B deleted (15). Instead, our results, and those obtained from human and chicken cells indicate that Cdc14b is required for efficient DNA damage repair.
It is unclear how Cdc14b functions in DNA damage repair. It is likely that Cdc14b dephosphorylates substrates involved in DNA damage repair. Given that Cdc14a is also required for efficient DNA damage repair (15), it is plausible that these two homologous phosphatases target the same set of substrates, although neither is sufficient. Identification of the relevant substrates in future studies will be the key to understanding how Cdc14 phosphatases regulate the DNA repair process.
ACKNOWLEDGMENTS
We are indebted to J. Qin, X. Huang, and S. Y. Jung (Baylor College of Medicine) for discussion and support. We thank the transgenic core of BCM for technical help. We also thank other members of the Zhang laboratory for valuable discussions and support.
This work is supported by NIH grants CA116097 and CA122623 to P.Z.
Footnotes
Published ahead of print on 24 January 2011.
REFERENCES
- 1. Adams P. D. 2009. Healing and hurting: molecular mechanisms, functions, and pathologies of cellular senescence. Mol. Cell 36:2–14 [DOI] [PubMed] [Google Scholar]
- 2. Bassermann F., et al. 2008. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell 134:256–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bembenek J., Yu H. 2003. Regulation of CDC14: pathways and checkpoints of mitotic exit. Front. Biosci. 8:d1275–d1287 [DOI] [PubMed] [Google Scholar]
- 4. Berdougo E., Nachury M. V., Jackson P. K., Jallepalli P. V. 2008. The nucleolar phosphatase Cdc14B is dispensable for chromosome segregation and mitotic exit in human cells. Cell Cycle 7:1184–1190 [DOI] [PubMed] [Google Scholar]
- 5. Cho H. P., et al. 2005. The Dual-specificity phosphatase CDC14B bundles and stabilizes microtubules. Mol. Cell. Biol. 25:4541–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dimri G. P., et al. 1995. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 92:9363–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. García-Higuera I., et al. 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 10:802–811 [DOI] [PubMed] [Google Scholar]
- 8. Hoeijmakers J. H. J. 2009. DNA damage, aging, and cancer. N. Engl. J. Med. 361:1475–1485 [DOI] [PubMed] [Google Scholar]
- 9. Jackson S. P., Bartek J. 2009. The DNA-damage response in human biology and disease. Nature 461:1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kaiser B. K., Zimmerman Z. A., Charbonneau H., Jackson P. K. 2002. Disruption of centrosome structure, chromosome segregation, and cytokinesis by misexpression of human Cdc14A phosphatase. Mol. Biol. Cell 13:2289–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li L., Ernsting B. R., Wishart M. J., Lohse D. L., Dixon J. E. 1997. A family of putative tumor suppressors is structurally and functionally conserved in humans and yeast. J. Biol. Chem. 272:29403–29406 [DOI] [PubMed] [Google Scholar]
- 12. Li M., et al. 2008. The adaptor protein of the anaphase promoting complex Cdh1 is essential in maintaining replicative lifespan and in learning and memory. Nat. Cell Biol. 10:1083–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lindahl T. 1993. Instability and decay of the primary structure of DNA. Nature 362:709–715 [DOI] [PubMed] [Google Scholar]
- 14. Mailand N., et al. 2002. Deregulated human Cdc14A phosphatase disrupts centrosome separation and chromosome segregation. Nat. Cell Biol. 4:317–322 [DOI] [PubMed] [Google Scholar]
- 15. Mocciaro A., et al. 2010. Vertebrate cells genetically deficient for Cdc14A or Cdc14B retain DNA damage checkpoint proficiency but are impaired in DNA repair. J. Cell Biol. 189:631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rodier G., Coulombe P., Tanguay P.-L., Boutonnet C., Meloche S. 2008. Phosphorylation of Skp2 regulated by CDK2 and Cdc14B protects it from degradation by APC-Cdh1 in G1 phase. EMBO J. 27:679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schindler K., Schultz R. M. 2009. The CDC14A phosphatase regulates oocyte maturation in mouse. Cell Cycle 8:1090–1098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Schindler K., Schultz R. M. 2009. CDC14B acts through FZR1 (CDH1) to prevent meiotic maturation of mouse oocytes. Biol. Reprod. 80:795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schmitz M. H. A., et al. 2010. Live-cell imaging RNAi screen identifies PP2A –B55α and importin-β1 as key mitotic exit regulators in human cells. Nat. Cell Biol. 12:886–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 [DOI] [PubMed] [Google Scholar]
- 21. Tallquist M., Soriano P. 2000. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis 26:113–115 [DOI] [PubMed] [Google Scholar]
- 22. Todaro G. J., Green H. 1963. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17:299–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wu J., et al. 2008. Cdc14B depletion leads to centriole amplification, and its overexpression prevents unscheduled centriole duplication. J. Cell Biol. 181:475–483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wu J., et al. 2009. PP1-mediated dephosphorylation of phosphoproteins at mitotic exit is controlled by inhibitor-1 and PP1 phosphorylation. Nat. Cell Biol. 11:644–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang L., et al. 2010. Proteolysis of Rad17 by Cdh1/APC regulates checkpoint termination and recovery from genotoxic stress. EMBO J. 29:1726–1737 [DOI] [PMC free article] [PubMed] [Google Scholar]