Abstract
The induction of the granulocytic differentiation of leukemic cells by all-trans retinoic acid (RA) has been a major breakthrough in terms of survival for acute promyelocytic leukemia (APL) patients. Here we highlight the synergism and the underlying novel mechanism between RA and the granulocyte colony-stimulating factor (G-CSF) to restore differentiation of RA-refractory APL blasts. First, we show that in RA-refractory APL cells (UF-1 cell line), PML-RA receptor alpha (RARα) is not released from target promoters in response to RA, resulting in the maintenance of chromatin repression. Consequently, RARα cannot be recruited, and the RA target genes are not activated. We then deciphered how the combination of G-CSF and RA successfully restored the activation of RA target genes to levels achieved in RA-sensitive APL cells. We demonstrate that G-CSF restores RARα recruitment to target gene promoters through the activation of the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway and the subsequent derepression of chromatin. Thus, combinatorial activation of cytokines and RARs potentiates transcriptional activity through epigenetic modifications induced by specific signaling pathways.
INTRODUCTION
The determination of granulopoiesis in pluripotent hematopoietic stem cells results from a multistep process involving a Lin− IL7Ra− Kit+ Sca-1− CD34+ FcγRlo common myeloid progenitor (CMP), Lin− IL7Ra− Kit+ Sca-1− CD34+ FcγRhi granulocyte-monocyte progenitors (GMP), granulocyte CFU (CFU-G), and finally the ultimate maturation steps, which involve myeloblasts, promyelocytes, myelocytes, metamyelocytes, and neutrophils (2, 17, 41). This long process is under close regulation orchestrated by numerous factors, among which cytokines, such as circulating granulocyte colony-stimulating factor (G-CSF) (38) and several transcription factors, such as nuclear retinoic acid (RA) receptors (RARs), play important roles (17, 41).
RARs (α, β, and γ) are ligand-dependent regulators of transcription (for a review see the work of Rochette-Egly and Germain [40]), which as heterodimers with retinoic X receptors (RXRs), bind specific RA response elements (RAREs) located in the promoters of target genes. According to recent studies, in the absence of the ligand, RA, only a small fraction of RAREs are occupied by RXR-RAR heterodimers (6, 34). Upon ligand binding, RARs undergo conformational changes that allow their recruitment to response elements and their interaction with coactivators associated with large complexes with chromatin modifying and remodeling activities that decompact repressive chromatin and pave the way for the recruitment of the transcription machinery.
The importance of RARs in granulopoiesis has been highlighted subsequently by the identification in acute promyelocytic leukemia (APL) of the PML-RARα fusion protein that results from the reciprocal translocation t(15;17) between chromosomes 15 and 17. In the absence of ligand, the fusion protein impedes in a dominant-negative manner the expression of RARα target genes and thus blocks the APL cells at the promyelocytic stage (33, 36) through its ability to occupy RAREs and to interact with complexes encompassing a wide range of epigenetic enzymes with strong repressive activity toward target genes. At pharmacological concentrations, all-trans RA is a highly effective agent that induces terminal differentiation of APL cells both in vitro and in vivo (8). The differentiation process is accompanied by the release of corepressors and the subsequent activation of RARα target genes (33). However, some APL patients present incomplete responsiveness to RA, resulting in patient relapses (13, 28, 43). This RA resistance has been related to the presence of mutations in the ligand-binding domain of the RARα portion of the PML-RARα fusion protein (50). The Arg276Trp mutation, which results in a dramatic decrease in RA binding activity (11, 44), has been found in several patient samples (11) and in the UF-1 cell line (30).
Interestingly, when combined with RA, several signaling pathways potentiate the granulocytic differentiation of APL cells and release RA resistance even in mutated clones (20, 48). In this context, the combination of G-CSF and RA has been shown to potentiate the granulocytic differentiation of APL cells (21) and to achieve the differentiation of several RA-resistant leukemic cells, including the UF-1 cell line (25, 29). However, the molecular mechanism of the relased RA resistance by G-CSF still remains ill defined.
In order to further investigate the cross talk between G-CSF and RA, we compared two APL cell lines, the RA-sensitive NB4 cell line and the RA-refractory UF-1 cell line, which undergoes maximal differentiation when RA is combined with G-CSF. We demonstrate that, when combined with RA, G-CSF restores the epigenetic modifications of histones and the recruitment of RARα to target gene promoters and thus the expression of RA target genes. This functional cascade results from the activation of the extracellular signal-regulated kinase (ERK)/mitogen-activated protein kinase (MAPK) pathway. This study highlights a new concept according to which MAPK signaling might be considered a key pathway for RA-induced granulocytic differentiation.
MATERIALS AND METHODS
Reagents and antibodies.
The human recombinant G-CSF Neupogen Filgrastim (Amgen) was used at a final concentration of 50 ng/ml. Chemical inhibitors UO126, LY294002, and JAK inhibitor I were obtained from Calbiochem. Cells were incubated 30 min with the inhibitor prior to addition of RA and/or G-CSF. The RARα-specific agonist AM580 was purchased from Sigma-Aldrich. Antibodies against CD114, STAT3, phospho-STAT3, AKT, phospho-AKT (Ser473), p38MAPK, phospho-P38MAPK, ERK1/2, phospho-ERK1/2, and phospho-CREB were purchased from Cell Signaling. Antibodies against diacetyl-Lys9/14 histone H3, acetyl-Lys5/8/12/16 histone H4 and against phospho-Ser10 histone H3 were from Millipore. Phycoerythrin-labeled antibodies against CD11b and CD11c were from Pharmingen. Rabbit polyclonal antibodies against the C-terminal F region of RARα, RPα(F), and mouse monoclonal antibodies recognizing the N-terminal A region of RARα [monoclonal antibody 10 (MAb10) α1(A1)] were described previously by Gaub et al. (18). Rabbit polyclonal antibodies against PML (H-238) and SMRT (H-300) were from Santa Cruz Biotechnology.
Cells.
Mononuclear cells from patients' bone marrow samples taken at diagnosis were prepared by Ficoll-Hypaque density gradient purification, and the presence of the PML-RARα fusion was verified by reverse transcription (RT)-PCR as previously described (10). The UF-1 cell line was obtained from Ikeda (Tokyo, Japan) and cultured in RPMI supplemented with 15% stromovascular fraction (SVF) at 37°C in a humidified atmosphere containing 5% CO2. NB4 cells were cultured in RPMI supplemented with 10% fetal bovine serum.
Differentiation of APL cells.
The sensitivity of patients' cells to RA-induced differentiation was assessed by morphological criteria and the appearance of burst function (nitroblue tetrazolium [NBT] test) as previously described as a routine test in our laboratory (9). Differentiation of UF-1 and NB4 cells was also assessed by the NBT test and by fluorescence-activated cell sorter (FACS) analysis of CD11b and CD11c surface expression using a FACSCalibur apparatus (Becton Dickinson).
Transactivation.
UF-1 cells were electroporated as previously described (42) with luciferase reporter genes, hRAR2-Luc or RARE3-TK-Luc (5 μg), and treated or not treated with RA and/or G-CSF. All transfections were performed with a galactosidase expression vector (pCH110) as an internal standard. A total of 24 h after transfection, luciferase activity was determined according to standard procedure. Results were expressed as fold induction based on the basal activity of the reporter gene (arbitrarily set at 1) in the absence of any ligand.
Cell extracts and immunoblotting.
Cells were lysed in 20 mM Tris (pH 7.4), 150 mM NaCl, 10 mM EDTA, 10% glycerol, 1% Igepal, 1 mM orthovanadate, and Complete protease inhibitor cocktail (Roche). Proteins were resolved by SDS-PAGE. After electrotransfer, antigen-antibody complexes were revealed by means of peroxidase-labeled secondary antibodies and an enhanced fluoro-chemiluminescence system (ECL plus; Amersham Biosciences).
Nucleofection for siRNA experiments.
Cells were nucleofected using the Amaxa apparatus, the U01 program, and solution T. Small interfering RNAs targeting MEK1 and MEK2 were purchased from Qiagen.
Detection of in vivo phosphorylated RARα and p38MAPK.
Cell extracts were prepared and applied to phosphoprotein affinity purification columns (PhosphoProtein purification system; Qiagen S.A.) according to manufacturer-supplied instructions. After column eluates containing protein peaks were washed, they were concentrated and analyzed by immunoblotting as previously described (6).
Quantitative RT-PCR.
Total RNA was extracted using RNA-Plus (Q-Biogen) reagent according to the manufacturer's instructions. RARα2 gene expression was analyzed as described by Glasow et al. (23), while MEK1 and MEK2 expressions were analyzed using predesigned assays (Assays-on-Demand; Applied Biosystems). Normalization was obtained with ABL gene expression. Results were expressed using the threshold cycle (ΔΔCT) method.
ChIP analysis.
Chromatin immunoprecipitation (ChIP) analysis of CBP/p300 and histone acetylation was performed according to the protocol described by Millipore, as performed by Cras et al. (12), using 106 cells with a Diagenode sonicator, the Bioruptor, for sonication. ChIP analysis of PML-RARα, RARα SMRT, and H3S10p was performed as described by Bruck et al. (6).
RESULTS
G-CSF restores the differentiation of leukemic cells with low RA sensitivity.
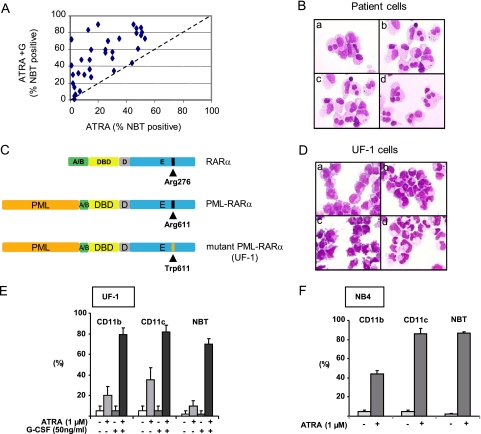
We previously identified a population of APL patients with poorer clinical prognosis, i.e., less than 50% of the patient's APL cells differentiated in response to a low RA concentration (0.1 μM) (9). We tested the efficacy of G-CSF addition to RA in enhancing the differentiation efficiency of RA in APL cells in 110 consecutive samples from APL patients tested at diagnosis; 34 (30%) samples presented with reduced in vitro RA sensitivity. We show that the differentiation efficacy of RA was significantly restored in 28 samples (82%) when G-CSF was added (Fig. 1 A and B) (P < 0.0001, Wilcoxon test), with 18 patient samples depicting more than 50% of differentiated cells (Fig. 1A), i.e., superior to the cutoff prognostic significance (9).
Fig. 1.
G-CSF restores RA-induced differentiation in APL cells with reduced sensitivity to RA. (A) In vitro differentiation of APL blasts at diagnosis from 34 patients with reduced sensitivity to RA. Percentage of NBT-positive cells was analyzed after 3 days of treatment with RA (0.1 μM) ± G-CSF (50 ng/ml). (B) May-Grunwald–Giemsa coloration of patient cells treated for 3 days with medium (a), RA (1 μM) (b), G-CSF (50 ng/ml) (c), or RA (1 μM) and G-CSF (50 ng/ml) (d). (C) Schematic representation (not to scale) of the RARα and PML-RARα proteins with the mutation found in the UF-1 cell line. (D) May-Grunwald–Giemsa coloration of UF-1 cells treated for 3 days with medium (a), RA (1 μM) (b), G-CSF (50 ng/ml) (c), or RA (1 μM) and G-CSF (50 ng/ml) (d). (E) CD11b and CD11c analysis and NBT test with UF-1 cells treated for 3 days with RA (1 μM) and/or G-CSF (50 ng/ml). (F) Same as described in the legend to panel E, with NB4 cells treated for 3 days with RA (1 μM).
As already reported, the UF-1 APL cell line is poorly responsive to RA even at high concentrations (1 μM) due to a mutation in the ligand binding domain (Fig. 1C) and shows enhanced differentiation when RA (1 μM) is combined with G-CSF (50 ng/ml) (25) (Fig. 1D). We show in this study that these morphological and functional changes are correlated with the induced expression of the integrin chains CD11b and CD11c (Fig. 1E). Of note, the addition of G-CSF to RA allowed UF-1 cells to reach differentiation levels similar to that obtained with RA alone in the NB4-sensitive cell line (Fig. 1F). Due to these characteristics, we took advantage of the characteristics of the UF-1 cell line to decipher the molecular mechanism of the RA/G-CSF cross talk which could explain the synergistic effect noted with 80% of AML3 samples that were poor responders to RA.
In UF-1 cells, RA is inefficient in inducing the recruitment of RARα to target gene promoters.
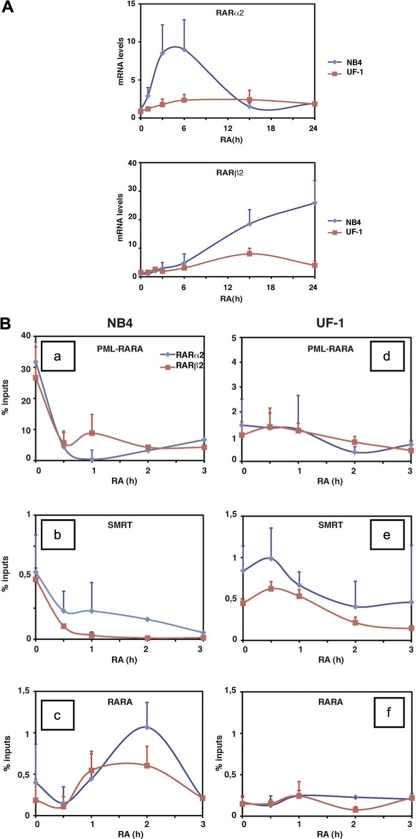
RA-induced granulocytic differentiation is well known to occur through the induction of specific target genes under the control of RAREs, such as RARα2 and RARβ2 genes. The expression of these genes is increased in response to RA (0.1 μM) in the RA-sensitive APL cell line NB4 (Fig. 2 A). On the contrary, they are not activated in the poorly responsive UF-1 cell line (Fig. 2A). Therefore, we compared the two cell lines for PML-RARα occupancy of the RARE-containing promoter regions of the RARα2 and RARβ2 genes in chromatin immunoprecipitation (ChIP) experiments. The specificity of the experimental conditions was checked in the absence of antibodies and with the promoter of the 36B4 gene, which does not contain any RARE (6). As expected, we found that in NB4 cells, both promoters were occupied by PML-RARα (Fig. 2Ba). Interestingly, within 30 min following the addition of pharmacological concentrations of RA (0.1 μM), PML-RARα dissociated from both promoters (Fig. 2Ba). The corepressor SMRT was also bound at the promoters in the absence of RA and dissociated rapidly after RA addition (Fig. 2Bb). Interestingly RARα was hardly detected at the promoters in the absence of RA (Fig. 2Bc). However, after RA addition there was an enrichment of bound RARα at the promoters, which peaked between 1 and 2 h and decreased at 3 h (Fig. 2Bc). Collectively, these results suggest that, in NB4 cells and in the presence of pharmacological doses of RA, the dissociation of PML-RARα and SMRT would allow the recruitment of RARα.
Fig. 2.
APL cells with reduced sensitivity to RA (UF-1) are resistant to RA-induced transcription regulation of target genes. (A) Comparison by quantitative RT (qRT)-PCR analysis of the RARα2 and RARβ2 transcripts in NB4 and UF1 cells treated with 1 μM RA for the indicated times. (B) Kinetic chromatin immunoprecipitation analysis (ChIP) performed with RA-treated NB4 and UF-1 cells and determining the occupancy of the RARα2 and RARβ2 promoters by PML-RARα (a and d), RARα (c and f), and SMRT (b and e). Values (percentage of inputs) are the mean ± SD of duplicates performed with 3 separate chromatin preparations. PML-RARα was immunoprecipitated with anti-PML antibodies, while RARα was specifically immunoprecipitated with monoclonal antibodies raised against the N-terminal region of RARα, which is not present in the PML-RARα fusion protein.
On the contrary, in the poorly responsive UF-1 cell line, similar ChIP experiments revealed that the promoters were less occupied by PML-RARα (Fig. 2Bd). Nevertheless, the corepressor SMRT was still able to bind the promoters (Fig. 2Be). Most importantly, in this cell line, PML-RARα did not dissociate from the promoters after RA addition. SMRT did not dissociate efficiently either (Fig. 2Be). Consequently, we were unable to detect any significant increase in RARα recruitment after RA treatment, even up to 3 h (Fig. 2Bf). All together, these results suggest that in UF-1 cells, RA is unable to induce the recruitment of RARα to the promoters of RA target genes. This might explain the poor transcriptional effect of RA in this cell line.
G-CSF restores the RA-induced expression of RA target genes via restoring the recruitment of RARα at target gene promoters in UF-1 cells.
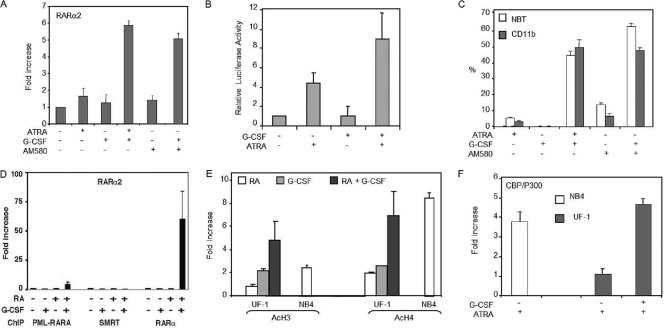
We next investigated whether G-CSF was able to restore the expression of RA target genes in UF-1 cells. G-CSF alone was unable to induce RARα2 and RARβ2 gene expression (Fig. 3A and data not shown). However, when combined with RA, G-CSF increased significantly the expression of both genes to levels observed with NB4 cells treated by RA alone (Fig. 3A and data not shown). Similar results were obtained with an overexpressed luciferase reporter gene under the control of a RARE isolated from the RARβ2 gene promoter (Fig. 3B).
Fig. 3.
G-CSF restores the transcriptional potential of RA in APL cells (UF-1) with reduced sensitivity to RA. (A) RARα2 expression is induced in UF-1 cells treated for 24 h by G-CSF combined with RA or AM580 as assessed by quantitative real-time RT-PCR. (B) G-CSF enhanced RA-induced transactivation of a RARE element in UF-1 cells. (C) AM580, a selective RARα agonist, reproduced the differentiation obtained with RA alone or in combination with G-CSF in UF-1 cells. (D) ChIP analysis of PML-RARα, SMRT, and RARα recruitment at the RARα gene promoter in UF-1 cells treated for 1 h by RA and/or G-CSF. (E) ChIP analysis of histone H3 and H4 acetylation at the RARα gene promoter in UF-1 and NB4 cells treated for 1 h by RA and/or G-CSF. (F) ChIP experiments performed with NB4 and UF-1 cells treated for 1 h with RA and/or G-CSF and determining the recruitment of CBP/P300 to the RARα gene promoter. In all panels, values are the mean ± SD of duplicates performed with at least two separate experiments.
Interestingly, when combined with AM580, a selective RARα agonist, G-CSF also restored both the activation of the RA target genes (Fig. 3A) and the expression of the differentiation markers (Fig. 3C), suggesting that the potentiating effect of G-CSF would involve RARα.
To gain further mechanistic insights into the effects of G-CSF, ChIP experiments were performed. Our results indicate that G-CSF both alone and in combination with RA did not affect significantly the occupancy of the RARα2 and RARβ2 promoters by either PML-RARα or SMRT and thus did not restore their dissociation from the promoters (Fig. 3D and data not shown). However, the combination of G-CSF and RA restored RARα recruitment to levels similar to those observed with NB4 cells (Fig. 3D and data not shown). All together these results indicate that G-CSF restores the expression of RA target genes in UF-1 cells via the restoration of RARα recruitment to the promoters.
G-CSF enhances the permissiveness of RA target gene promoters in UF-1 cells.
We investigated whether G-CSF increases promoters' accessibility and/or chromatin decompaction through histone modifications, a hallmark of gene transcription activation (24). ChIP assays performed with specific antibodies showed that in UF-1 cells treated with the RA–G-CSF combination for 1 h, histones H3 and H4 were more acetylated (5 to 7 times more) at the RARα2 promoter than in cells treated with RA or G-CSF alone (Fig. 3E). Moreover there was a 5-fold enrichment of the histone acetyltransferase CBP/P300 bound to this promoter (Fig. 3F). Interestingly, the increase in histone acetylation and CBP/p300 recruitment induced by the G-CSF/RA combination in UF-1 cells was similar to that observed with NB4 cells treated with RA alone (Fig. 3E and F). All together these results suggest that in poorly responsive UF-1 cells, G-CSF would restore the expression of RA target genes through histone modifications involved in chromatin permissiveness.
The ERK pathway is involved in the synergistic effect of G-CSF on UF-1 cell differentiation.
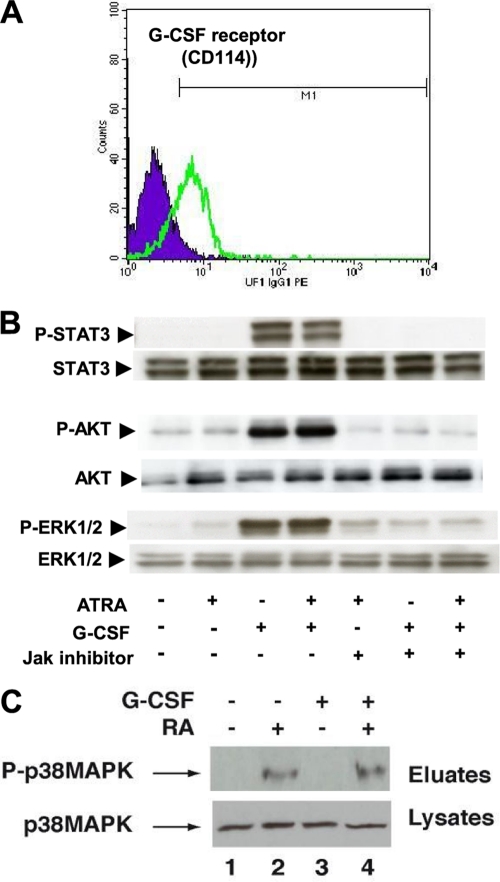
G-CSF activates several signaling cascades through binding to CD114, a membrane-anchored receptor (3). In UF-1 cells, CD114 is expressed (Fig. 4A), and G-CSF induced the rapid activation of the JAK pathway as assessed by the phosphorylation of the downstream STAT3 protein (Fig. 4B). G-CSF also induced the activation of several other downstream signaling cascades, such as AKT and extracellular signal-regulated kinase 1/2 (ERK1/2) MAPK pathways (Fig. 4B). Of note is that RA had no effect on these pathways and did not modulate the effects of G-CSF (Fig. 4B). In contrast, G-CSF had no effect on p38MAPK activity and did not affect the ability of RA to activate this pathway (Fig. 4C).
Fig. 4.
Comparison of the kinases and phosphorylation pathways in NB4 and UF-1 cells. (A) UF-1 cells express the G-CSF receptor (CD114) as analyzed by flow cytometry. (B) Signaling pathways activated by G-CSF and RA in UF-1 cells. Their inhibition by a Jak inhibitor is also shown. (C) G-CSF signaling does not affect the RA-induced activation of p38MAPK.
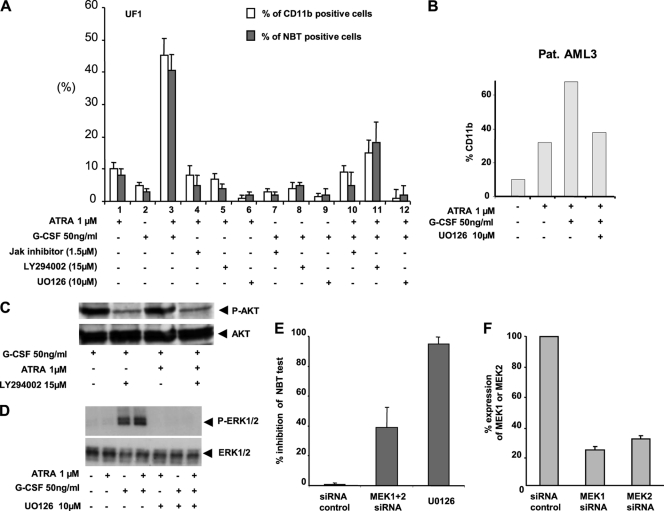
We then investigated whether inhibition of the JAK, Akt, or ERK/MAPK pathway would abrogate the G-CSF–RA synergy. Preincubation of UF-1 cells with a pan-JAK kinase inhibitor, at a dose that does not alter cell viability, inhibited the effects of G-CSF on all the above kinase pathways (Fig. 4B). It also partially abrogated the synergy between G-CSF and RA for granulocytic differentiation (Fig. 5A). In contrast, inhibition of the downstream PI3K/Akt pathway with LY294002 (Fig. 5C) inhibited partially the synergy (Fig. 5A). The most efficient inhibitor to completely abrogate the granulocytic differentiation induced by the G-CSF–RA combination was UO126, an inhibitor of the MEK/ERK1/2 pathway (Fig. 5A, B, and D). Similar results were obtained after the knockdown of MEK1 and MEK2 gene expression using specific siRNA (Fig. 5E and F). Collectively, these results highlight a major role for the MEK/ERK1/2 pathway in the synergistic effect of G-CSF on the RA-induced differentiation of RA-resistant APL UF-1 cells.
Fig. 5.
ERKs are involved in the potentiating effect of the G-CSF–RA combination on the differentiation of UF-1 cells. (A) UF-1 cells were incubated for 30 min with the indicated inhibitor prior to addition of RA and/or G-CSF. Differentiation was monitored after 3 days by analysis of the surface expression of the CD11b antigen or the NBT test. (B) Fresh patient cells (Pat. AML3) were incubated for 30 min with UO126 prior to addition of RA and/or G-CSF. Differentiation was monitored after 3 days by analysis of the surface expression of the CD11b antigen. (C and D) Western blot analysis of P-Akt and P-Erk inhibition by LY294002 and UO126, respectively, in UF-1 cells treated for 10 minutes by RA with or without G-CSF. (E) siRNA directed against MEK1 and MEK2 inhibited the differentiation of UF-1 cells induced by the association RA with G-CSF. (F) qRT-PCR analysis of MEK1 and MEK2 gene expression 48 h after transfection with the corresponding siRNAs. In all panels, values are the mean ± SD of duplicates performed with at least two separate experiments.
The G-CSF-induced MEK/ERK1/2 pathway restores RARα recruitment at RA-target gene promoters through chromatin remodeling.
As MAPKs and their downstream kinases have been shown to have an essential function in phosphorylating transcription factors and/or preparing promoter chromatin for gene activation (6, 45), we next investigated whether, in UF-1 cells, this pathway plays a role in the potentiating effect of G-CSF on the permissiveness of RA target genes and the subsequent expression of these genes.
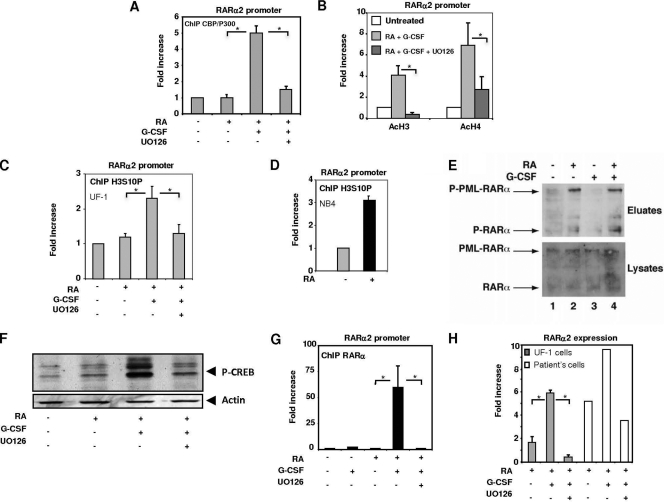
Most interestingly, on UF-1 cells treated by the combination of RA and G-CSF, restoration of histone H3 and H4 acetylation and of histone acetyl transferase CBP/P300 recruitment at the RARα2 gene promoter was noted and abrogated by the MEK inhibitor UO126, as assessed by ChIP assays (Fig. 6A and B). As histone H3 acetylation is known to be controlled by phosphorylation at S10 according to the histone code (39), we analyzed H3S10 phosphorylation in ChIP assays. As shown in Fig. 6C, in UF-1 cells, histone H3 was not phosphorylated in response to RA alone, but the addition of G-CSF restored this effect to a level similar to that observed with NB4 cells (Fig. 6D). Given that in response to RA in sensitive cells, RARα is rapidly phosphorylated and that this phosphorylation process has been shown to control RARα recruitment to the promoters of RA target genes (6), we investigated whether RARα and PML-RARα had a phosphorylation defect in the poorly responsive UF-1 cell line. In fact, the amount of phosphorylated RARα increased rapidly in response to RA, and the addition of G-CSF did not alter this phosphorylation pattern (Fig. 6E). PML-RARα was also phosphorylated in response to RA alone, and the pattern was not modified by the addition of G-CSF (Fig. 6E). Collectively, these results indicate that the synergy between RA and G-CSF does not involve the phosphorylation status of the transcriptional proteins RARα and PML-RARα. However, another transcription factor, CREB, was found to be phosphorylated in UF-1 cells after treatment by RA and G-CSF (Fig. 6F).
Fig. 6.
G-CSF-activated ERKs restore RA target gene transcription through histone phosphorylation. (A and B) ChIP experiments showing that the MEK inhibitor UO126 reduces the recruitment of CBP/p300 (A) and the acetylation of histones H3 and H4 (B) to the RARα2 promoter, induced by the RA–G-CSF combination. (C and D) ChIP analysis of histone H3 phosphorylation in NB4 and UF-1 cells treated with RA and/or G-CSF. (E) G-CSF does not affect the RA-induced phosphorylation of RARα and PML-RARα in UF-1 cells as assessed by immunoblotting after phosphoprotein affinity purification. (F) G-CSF induces the phosphorylation of CREB through ERKs. (G) ChIP analysis of RARα recruitment to the RARα2 promoter in UF-1 cells treated with the RA–G-CSF combination in the absence or presence of UO126. (H) qRT-PCR analysis of the RARα2 gene expression in UF-1 cells or fresh patient cells treated with the RA–G-CSF combination in the presence or absence of the MEK inhibitor UO126. In all panels, values are the mean ± SD of duplicates performed with at least two separate experiments. * indicates a P value of <0.05 as determined by two-tailed Student's t test.
Interestingly, this ERK-dependent activation of chromatin modifier actors (histone phosphorylation, histone acetyltransferase [CBP/p300], and histone acetylation) in the presence of RA and G-CSF resulted in the recruitment of RARα onto its promoter (Fig. 6G) and the induction of RARα2 expression (Fig. 6H). This recruitment and gene expression were completely abrogated by a preincubation of UF-1 cells or fresh patient cells with the MEK inhibitor UO126 (Fig. 6G and H).
All these results converge to lead to the conclusion that, in UF-1 cells, the G-CSF-induced MEK/ERK1/2 pathway restores the activation of RA target genes through the increased permissiveness of the RAR promoter via phosphorylation of histones and the subsequent recruitment of the HAT protein, histone acetylation, and recruitment and phosphorylation of transcription factors.
DISCUSSION
In this study, we outlined a new mechanism underlying the synergism between RA and G-CSF for the differentiation of RA-resistant APL cells. Indeed, we found that, when combined to RA, G-CSF restores the epigenetic modifications of histones and the recruitment of RARα at target gene promoters through the activation of the ERK/MAPK pathway. To our knowledge, this is the first report of synergy between RA and cytokines occurring at promoters of RA target genes.
With 110 fresh samples from APL patients enrolled in two multicenter APL trials, we confirmed that G-CSF and RA cooperate for the granulocytic differentiation of sensitive (21) and RA-resistant primary APL cells (23, 25, 29). Interestingly, the G-CSF–RA combination significantly restored the differentiation of more than 80% of APL patient samples with reduced in vitro RA sensitivity. This enhanced response in the presence of G-CSF in poorly responsive APL samples was found to be similar to that previously reported for the UF-1 cell line, harboring Arg276Trp in the PML-RARα gene. As no inherent mechanism of the efficacy of this combination has yet been reported, we endeavored to decipher the underlying mechanism of the restored granulocytic differentiation in the UF-1 cells and to correlate it with that in APL patient samples.
The effects of RA on APL cells are linked to transcription processes and gene expression. In line with this, RA target genes, such as RARα2 and RARβ2, are strongly activated during the differentiation of RA-sensitive APL cells, such as fresh APL patient cells and the NB4 cell line. Interestingly we found that these genes were not induced in the RA-resistant APL UF-1 cell line. However, their expression was restored by the G-CSF–RA combination in parallel with the granulocytic differentiation of the cells. It thus appeared that the enhanced effect in the presence of G-CSF could be attributed to the transcriptional control of RA target genes.
First, we demonstrated that in RA-responsive NB4 cells, the promoters of the RARα2 and RARβ2 genes are occupied by PML-RARα and SMRT, in agreement with the findings of other studies (33). After RA addition, PML-RARα dissociated rapidly from the promoters with SMRT. This dissociation was followed by chromatin modifications (histone phosphorylation and acetylation) and the recruitment of RARα. It has been proposed that the global changes in the repressive marks would not be triggered by the release of the corepressor complexes from PML-RARα but from the rapid loss of the fusion protein at the DNA binding sites (33).
In RA-poorly responsive UF-1 cells, PML-RARα occupies the promoters of RA target genes in the absence of ligand, even if present in amounts smaller than those in NB4 cells. The corepressor SMRT also occupies the promoters. Most importantly, in response to RA, neither PML-RARα nor SMRT dissociated from the promoters, most probably due to the inability of PML-RARα, which is mutated at R276, to bind retinoic acid. Indeed, R276 is located in helix H5 and participates to the architecture of the ligand-binding pocket through hydrogen bonds and salt bridges (37). It also guides the entrance of the ligand into the pocket. Mutation of this residue disrupts all this network, and the receptor can neither bind RA nor undergo the conformational changes that are required for dissociation from promoters and corepressors and for coactivator recruitment (11, 44). Thus, the chromatin of RA target genes in the UF-1 cell line remains in a repressive state and cannot undergo phosphorylation and acetylation processes according to the histone code. Consequently, RARα cannot be recruited to the promoters, RARα is not expressed, and granulocytic differentiation cannot occur.
The most important clue of the present study is, however, that, in UF-1 cells, RARα recruitment is restored when RA is combined with G-CSF. This previously unsuspected effect of G-CSF raised the question of how G-CSF contributes to the restoration of RARα recruitment and therefore to the transcription of RA target genes.
G-CSF, as most cytokines are, is known to activate the JAK/STAT pathway and the downstream PI3K/Akt and MAPK signaling pathways (3, 14, 27) in normal and leukemic cell lines. We confirmed that these pathways are also activated in UF-1 cells, suggesting that they might cooperate with RA for the restoration of transcription and differentiation. Most interestingly, we found that among these pathways, ERKs were critical for both the overall differentiating effect of the G-CSF–RA combination and for the restoration of RA target gene transcription, as inhibition of ERKs abrogated all these effects. The PI3K/Akt pathway seems to be less involved, as its inhibition did not abrogate efficiently the restoration of the RA effects. In line with this, one must note that Akt has been shown to antagonize the activity of RARs and of most of its targets (22, 32).
ERKs are well known to phosphorylate several transcription factors and coregulators as well as histones and thus to regulate epigenetics and transcription dynamics (15, 16). According to our results, G-CSF-activated ERKs did not affect the phosphorylation of PML-RARα, which remained stalled on the promoters with SMRT. Thus, ERKs did not restore the dissociation of PML-RARα and SMRT from the promoters. ERKs did not affect either the phosphorylation of RARα, which is normally phosphorylated in these cells by the p38MAPK pathway. Interestingly, we found that, in UF-1 cells, G-CSF-activated ERKs restored the phosphorylation of histone H3, allowing the initiation of the other observed modifications, such as acetylation according to the histone code. They also induced the phosphorylation of transcription factors such as CREB, which is a well known target of these kinases (1), and cross-talks with RARs at RA target gene promoters (31). Therefore, it is a good candidate to cooperate with histone modelers and modifiers to decompact chromatin in response to the RA–G-CSF combination. Thus, all these phosphorylation processes mediated by ERKs appear to be at the basis of the derepressive effects of G-CSF on chromatin. In other words, G-CSF-activated ERKs bypass the repressive state of chromatin due to mutated PML-RARα, firmly stalled on the promoters, through the initiation and/or the coordination of several phosphorylation events according to the histone code to make the promoters available for RARα recruitment (39, 46).
In conclusion, the present study demonstrated that granulocytic differentiation of APL cells is a finely tuned process with crucial steps at which specific transcription factors and signaling pathways are required. The correct RA-induced differentiation program necessitates the correct expression of target genes (47), with correct activation of epigenetic modifications (33). Here we highlighted that RA resistance can be reversed upon addition of cytokines which restore gene transcription through MAPK signaling-mediated reactivation of histone modifications. Such results corroborate that epigenetic modifications are ideal targets for therapeutic intervention (49). In conjunction with HDAC inhibitors, cytokine-activated signaling pathways such as MAPK/ERKs provide an additional level of therapeutical differentiation of not only RA-resistant APLs but all other AML subtypes.
ACKNOWLEDGMENTS
This work was supported by grants from the Fondation de France (2006 008197), Institut National du Cancer (RO6066HH, RO9101HH, PL06-095, and PL09-194), and Fondation pour la Recherche Médicale (DEQ20090515423). A.C. was a recipient of a Poste d'accueil fellowship by INSERM. The Ministère de la Recherche et de l'Enseignement Supérieur and the Association pour la Recherche sur le Cancer (ARC) supported C.F. The Ligue Nationale contre le Cancer and ARC supported N.B. ARC supported V.D.
We thank Nicolas Ferre for excellent technical assistance and Bernard Boursin for excellent help in the realization of figures.
Footnotes
Published ahead of print on 24 January 2011.
REFERENCES
- 1. Aggarwal S., et al. 2006. Nonclassical action of retinoic acid on the activation of the cAMP response element-binding protein in normal human bronchial epithelial cells. Mol. Biol. Cell 17:566–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Akashi K., Traver D., Miyamoto T., Weissman I. L. 2000. A clonogenic common myeloid progenitor that gives rise to all myeloid lineages. Nature 404:193–197 [DOI] [PubMed] [Google Scholar]
- 3. Avalos B. R. 1996. Molecular analysis of the granulocyte colony-stimulating factor receptor. Blood 88:761–777 [PubMed] [Google Scholar]
- 4. Reference deleted.
- 5. Reference deleted.
- 6. Bruck N., et al. 2009. A coordinated phosphorylation cascade initiated by p38MAPK/MSK1 directs RARalpha to target promoters. EMBO J. 28:34–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Reference deleted.
- 8. Cassinat B., Chomienne C. 2001. Biological features of primary APL blasts: their relevance to the understanding of granulopoiesis, leukemogenesis and patient management. Oncogene 20:7154–7160 [DOI] [PubMed] [Google Scholar]
- 9. Cassinat B., et al. 2001. In vitro all-trans retinoic acid sensitivity of acute promyelocytic leukemia blasts: a novel indicator of poor patient outcome. Blood 98:2862–2864 [DOI] [PubMed] [Google Scholar]
- 10. Castaigne S., et al. 1990. All-trans retinoic acid as a differentiation therapy for acute promyelocytic leukemia. I. Clinical results. Blood 76:1704–1709 [PubMed] [Google Scholar]
- 11. Côté S., et al. 2002. Response to histone deacetylase inhibition of novel PML/RARalpha mutants detected in retinoic acid-resistant APL cells. Blood 100:2586–2596 [DOI] [PubMed] [Google Scholar]
- 12. Cras A., et al. 2007. Epigenetic patterns of the retinoic acid receptor beta2 promoter in retinoic acid-resistant thyroid cancer cells. Oncogene 26:4018–4024 [DOI] [PubMed] [Google Scholar]
- 13. Ding W., et al. 1998. Leukemic cellular retinoic acid resistance and missense mutations in the PML-RARalpha fusion gene after relapse of acute promyelocytic leukemia from treatment with all-trans retinoic acid and intensive chemotherapy. Blood 92:1172–1183 [PubMed] [Google Scholar]
- 14. Dong F., Larner A. C. 2000. Activation of Akt kinase by granulocyte colony-stimulating factor (G-CSF): evidence for the role of a tyrosine kinase activity distinct from the Janus kinases. Blood 95:1656–1662 [PubMed] [Google Scholar]
- 15. Edmunds J. W., Mahadevan L. C. 2006. Cell signaling. Protein kinases seek close encounters with active genes. Science 313:449–451 [DOI] [PubMed] [Google Scholar]
- 16. Foulds C. E., Nelson M. L., Blaszczak A. G., Graves B. J. 2004. Ras/mitogen-activated protein kinase signaling activates Ets-1 and Ets-2 by CBP/p300 recruitment. Mol. Cell. Biol. 24:10954–10964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Friedman A. D. 2007. Transcriptional control of granulocyte and monocyte development. Oncogene 26:6816–6828 [DOI] [PubMed] [Google Scholar]
- 18. Gaub M. P., et al. 1992. Immunodetection of multiple species of retinoic acid receptor alpha: evidence for phosphorylation. Exp. Cell Res. 201:335–346 [DOI] [PubMed] [Google Scholar]
- 19. Reference deleted.
- 20. Giafis N., et al. 2006. Role of the p38 mitogen-activated protein kinase pathway in the generation of arsenic trioxide-dependent cellular responses. Cancer Res. 66:6763–6771 [DOI] [PubMed] [Google Scholar]
- 21. Gianní M., et al. 1994. Retinoic acid and granulocyte colony-stimulating factor synergistically induce leukocyte alkaline phosphatase in acute promyelocytic leukemia cells. Blood 83:1909–1921 [PubMed] [Google Scholar]
- 22. Gianni M., et al. 2002. Down-regulation of the phosphatidylinositol 3-kinase/Akt pathway is involved in retinoic acid-induced phosphorylation, degradation, and transcriptional activity of retinoic acid receptor gamma 2. J. Biol. Chem. 277:24859–24862 [DOI] [PubMed] [Google Scholar]
- 23. Glasow A., Prodromou N., Xu K., von Lindern M., Zelent A. 2005. Retinoids and myelomonocytic growth factors cooperatively activate RARA and induce human myeloid leukemia cell differentiation via MAP kinase pathways. Blood 105:341–349 [DOI] [PubMed] [Google Scholar]
- 24. Glass C. K., Rosenfeld M. G. 2000. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 14:121–141 [PubMed] [Google Scholar]
- 25. Higuchi T., Kizaki M., Omine M. 2004. Induction of differentiation of retinoic acid-resistant acute promyelocytic leukemia cells by the combination of all-trans retinoic acid and granulocyte colony-stimulating factor. Leuk. Res. 28:525–532 [DOI] [PubMed] [Google Scholar]
- 26. Reference deleted.
- 27. Hunter M. G., Avalos B. R. 1998. Phosphatidylinositol 3′-kinase and SH2-containing inositol phosphatase (SHIP) are recruited by distinct positive and negative growth-regulatory domains in the granulocyte colony-stimulating factor receptor. J. Immunol. 160:4979–4987 [PubMed] [Google Scholar]
- 28. Imaizumi M., et al. 1998. Mutations in the E-domain of RAR portion of the PML/RAR chimeric gene may confer clinical resistance to all-trans retinoic acid in acute promyelocytic leukemia. Blood 92:374–382 [PubMed] [Google Scholar]
- 29. Jansen J. H., et al. 1999. Complete remission of t(11;17) positive acute promyelocytic leukemia induced by all-trans retinoic acid and granulocyte colony-stimulating factor. Blood 94:39–45 [PubMed] [Google Scholar]
- 30. Kizaki M., et al. 1996. Establishment and characterization of a novel acute promyelocytic leukemia cell line (UF-1) with retinoic acid-resistant features. Blood 88:1824–1833 [PubMed] [Google Scholar]
- 31. Kruyt F. A., Folkers G., van den Brink C. E., van der Saag P. T. 1992. A cyclic AMP response element is involved in retinoic acid-dependent RAR beta 2 promoter activation. Nucleic Acids Res. 20:6393–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lefebvre B., Brand C., Flajollet S., Lefebvre P. 2006. Down-regulation of the tumor suppressor gene retinoic acid receptor beta2 through the phosphoinositide 3-kinase/Akt signaling pathway. Mol. Endocrinol. 20:2109–2121 [DOI] [PubMed] [Google Scholar]
- 33. Martens J. H., et al. 2010. PML-RARalpha/RXR alters the epigenetic landscape in acute promyelocytic leukemia. Cancer Cell 17:173–185 [DOI] [PubMed] [Google Scholar]
- 34. Martens J. H., Rao N. A., Stunnenberg H. G. 20 October 2010. Genome-wide interplay of nuclear receptors with the epigenome. Biochim. Biophys. Acta [Epub ahead of print.] doi: 10.1016/j.bbadis.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 35. Reference deleted.
- 36. Morey L., et al. 2008. MBD3, a component of the NuRD complex, facilitates chromatin alteration and deposition of epigenetic marks. Mol. Cell. Biol. 28:5912–5923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Renaud J. P., et al. 1995. Crystal structure of the RAR-gamma ligand-binding domain bound to all-trans retinoic acid. Nature 378:681–689 [DOI] [PubMed] [Google Scholar]
- 38. Robb L. 2007. Cytokine receptors and hematopoietic differentiation. Oncogene 26:6715–6723 [DOI] [PubMed] [Google Scholar]
- 39. Rochette-Egly C. 2005. Dynamic combinatorial networks in nuclear receptor-mediated transcription. J. Biol. Chem. 280:32565–32568 [DOI] [PubMed] [Google Scholar]
- 40. Rochette-Egly C., Germain P. 2009. Dynamic and combinatorial control of gene expression by nuclear retinoic acid receptors (RARs). Nucl. Recept. Signal. 7:e005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rosenbauer F., Tenen D. G. 2007. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 7:105–117 [DOI] [PubMed] [Google Scholar]
- 42. Rousselot P., et al. 1994. The PML-RAR alpha gene product of the t(15;17) translocation inhibits retinoic acid-induced granulocytic differentiation and mediated transactivation in human myeloid cells. Oncogene 9:545–551 [PubMed] [Google Scholar]
- 43. Schachter-Tokarz E., et al. 2010. PML-RARalpha ligand-binding domain deletion mutations associated with reduced disease control and outcome after first relapse of APL. Leukemia 24:473–476 [DOI] [PubMed] [Google Scholar]
- 44. Takayama N., Kizaki M., Hida T., Kinjo K., Ikeda Y. 2001. Novel mutation in the PML/RARalpha chimeric gene exhibits dramatically decreased ligand-binding activity and confers acquired resistance to retinoic acid in acute promyelocytic leukemia. Exp. Hematol. 29:864–872 [DOI] [PubMed] [Google Scholar]
- 45. Vicent G. P., et al. 2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell 24:367–381 [DOI] [PubMed] [Google Scholar]
- 46. Vicent G. P., Zaurin R., Ballaré C., Nacht A. S., Beato M. 2009. Erk signaling and chromatin remodeling in MMTV promoter activation by progestins. Nucl. Recept. Signal. 7:e008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang S., et al. 2009. RARalpha2 expression is associated with disease progression and plays a crucial role in efficacy of ATRA treatment in myeloma. Blood 114:600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Witcher M., Shiu H. Y., Guo Q., Miller W. H., Jr 2004. Combination of retinoic acid and tumor necrosis factor overcomes the maturation block in a variety of retinoic acid-resistant acute promyelocytic leukemia cells. Blood 104:3335–3342 [DOI] [PubMed] [Google Scholar]
- 49. Yoo C. B., Jones P. A. 2006. Epigenetic therapy of cancer: past, present and future. Nat. Rev. Drug Discov. 5:37–50 [DOI] [PubMed] [Google Scholar]
- 50. Zhou D. C., et al. 2002. Frequent mutations in the ligand-binding domain of PML-RARalpha after multiple relapses of acute promyelocytic leukemia: analysis for functional relationship to response to all-trans retinoic acid and histone deacetylase inhibitors in vitro and in vivo. Blood 99:1356–1363 [DOI] [PubMed] [Google Scholar]