Abstract
T helper cell differentiation and activation require specific transcriptional programs accompanied by changes in chromatin structure. However, little is known about the chromatin remodeling enzymes responsible. We performed genome-wide analysis to determine the general principles of BRG1 binding, followed by analysis of specific genes to determine whether these general rules were typical of key T cell genes. We found that binding of the remodeling protein BRG1 was programmed by both lineage and activation signals. BRG1 binding positively correlated with gene activity at protein-coding and microRNA (miRNA) genes. BRG1 binding was found at promoters and distal regions, including both novel and previously validated distal regulatory elements. Distal BRG1 binding correlated with expression, and novel distal sites in the Gata3 locus possessed enhancer-like activity, suggesting a general role for BRG1 in long-distance gene regulation. BRG1 recruitment to distal sites in Gata3 was impaired in cells lacking STAT6, a transcription factor that regulates lineage-specific genes. Together, these findings suggest that BRG1 interprets both differentiation and activation signals and plays a causal role in gene regulation, chromatin structure, and cell fate. Our findings suggest that BRG1 binding is a useful marker for identifying active cis-regulatory regions in protein-coding and miRNA genes.
Introduction
Non-antigen-experienced CD4+ T helper cells have the remarkable capacity to differentiate into a variety of effector cell fates, depending on the pathogenic signal or cytokine environment first encountered (24, 54, 55, 83, 84). The fate of each effector is marked by a cytokine signature that protects the organism when appropriately activated but that can contribute to allergy, autoimmunity, and inflammation when not properly regulated. Helper lineages are also marked by key transcriptional regulators. Thus, Th1 cells are characterized by expression of the cytokine gamma interferon (IFN-γ) and the transcription factor T-box 21 (Tbx21 or Tbet), whereas Th2 cells express the signature cytokines interleukin-4 (IL-4), IL-5, and IL-13 and the transcriptional regulator GATA binding protein 3 (Gata3), and Th17 cells are marked by the cytokines IL-17A and IL-F and the transcription factors ROR-γt and ROR-α. CD4+ cells can also become Treg cells, which negatively regulate the immune response and maintain peripheral tolerance.
The signaling events that lead to CD4+ differentiation integrate both activation and lineage signals to trigger cell division and ultimately direct the cytokine and transcription factor signatures (3, 53, 65). For example, antigen activation of naïve Th cells in the presence of the cytokines IL-12 and IFN-γ activates Stat4 and Stat1, resulting in Th1 differentiation and expression of IFN-γ and Tbet. Conversely, IL-4 activates Stat6 and, in the context of TCR stimulation, promotes commitment to the IL-4-producing Th2 lineage and expression of Gata3. Th17 differentiation is directed through exposure to transforming growth factor β1 (TGF-β1), IL-6, and IL-1, whereas IL-17 expression is maintained through IL-23 stimulation, with expression of Ror-γT. Mitogenic signals through the T cell receptor (TCR) mobilize activation-specific transcription factors (including NFATs, EGRs, and NF-κB) that can directly activate transcription of cytokine genes.
Although both the lineage-specific and activation-induced transcription factors are essential for directing subset-specific cytokine gene transcription, it is clear that this process is also strongly influenced by the surrounding chromatin structure. Previous studies have demonstrated permissive histone modifications, such as H3K4me2 and H3K9,14Ac, at the signature-cytokine loci in the appropriate Th lineages and repressive epigenetic (H3K27Me3) features in the opposing Th subsets (2, 4, 8, 9, 19, 36). Recent genome-wide analysis of H3K4Me3 and H3K27Me3 in a variety of Th subsets revealed the expected lineage-restricted modifications at the cytokine loci (85). However, the lineage-restricted transcription factors were marked by both permissive and repressive (“bivalent”) histone modifications in different Th subsets, suggesting that those genes are poised for subsequent activation or silencing. Although histone modifications can follow differentiation signals, some histone modifications are relatively stable during T cell activation (11, 43). Changes in chromatin accessibility at the Th cytokine loci as evidenced by DNase I hypersensitivity (DHS) analysis have revealed both lineage-specific and activation-induced changes in chromatin structures (1, 4, 20, 75). These chromatin markers are associated with regulatory elements, as deletion in situ alters regulation of the associated genes (5, 40, 49, 71). These observations suggest that dynamic changes in chromatin structure in response to mitogenic signals may be best reflected at the level of nuclease accessibility.
While chromatin remodeling is obviously correlated with T helper differentiation, it is less obvious what remodeling enzymes are responsible for these changes and how they recognize their sites of action. One group of remodeling enzymes consists of the ATP-dependent remodeling enzymes. These are multisubunit complexes that contain an ATPase in the SWI/SNF family and that utilize the energy released from ATP hydrolysis to induce changes in chromatin structure. According to the evidence regarding homology outside the ATPase domains, these SWI/SNF ATPase enzymes can be divided into several subfamilies (21). Mammalian SWI/SNF complexes contain 10 to 15 subunits, including either the BRG1 or BRM ATPase (48). It has been suggested that remodeling enzymes are targeted by transcription factors, noncoding RNA, histone modifications, and DNA damage; however, it must also be remembered that remodeling enzymes can function without targeting (12, 15, 29, 38, 77, 79). Different remodeling enzymes can be independently targeted in T cells, which suggests that they use different targeting signals (63).
We previously found that the SWI/SNF remodeling enzyme BRG1 is required for Th2 differentiation and transcription of Th2 cytokines (88). BRG1 binding was detected at both the promoters and distal regulatory regions of the IL-4, IL-5, and IL-13 genes. Some of the BRG1 binding sites were specific for Th2 and/or inducible by activation. BRG1 was required for nuclease accessibility at a subset of these binding sites, including the Th2 locus control region (LCR). BRG1 recruitment to the LCR was mediated by lineage-specific and activation-specific transcription factors (Stat6 and NFAT). Histone acetylation at these cytokine genes was also dependent on the activity of BRG1. These results suggest that BRG1 regulates Th2 differentiation by directly regulating Th2 cytokines, perhaps through distal regulatory elements. Related studies have previously found a role for BRG1 in Th1 cells (41, 62, 89). BRG1 also plays an important role in T cell development (13, 14, 32, 82). While those studies of T helper cells identified a functional role for BRG1, they examined only a few genes.
Given that chromatin remodeling serves as an important regulatory mechanism during Th differentiation, we extended our analysis to additional Th subsets in an unbiased manner using genome-wide chromatin immunoprecipitation and sequencing (ChIP-Seq) to generate BRG1 maps of undifferentiated, naïve CD4+ Th cells and effector Th1, Th2, and Th17 cells. We collected a uniform data set for BRG1 binding, employed this resource to ask global questions about BRG1 regulation, and used specific genes to investigate whether these general rules applied to genes that are important for T helper function. BRG1 binding was highly dynamic, responding to activation and the differentiation state. BRG1 binding positively correlated with gene activity. BRG1 was found at regulatory regions for protein-coding and microRNA (miRNA)-coding genes. BRG1 appeared to mark regions of active chromatin with enhancer activity.
MATERIALS AND METHODS
Lymphocyte preparation and culture.
Naïve CD4+ T cells from the spleens and lymph nodes of 4- to 6-week-old BALB/c mice (Taconic) were purified to 95% purity using a CD4+ CD62L+ T cell purification II kit per the instructions of the manufacturer (Miltenyi). Lymphocytes were cultured in RPMI 1640 medium supplemented with 10% fetal calf serum (FCS), 100 U/ml penicillin, 100 μg/ml streptomycin, 1 mM sodium pyruvate, 2 mM l-glutamine, 25 mM HEPES, and 50 μM β-mercaptoethanol. Animal approval was obtained from the National Institute on Aging Animal Care and Use Committee (NIA ACUC) (protocol ASP-365-MJP-Mi), and all experiments conformed to the relevant regulatory standards.
In vitro T helper cell differentiation.
For Th1 and Th2 differentiation, purified CD4+ CD62L+ T cells were plated onto anti-CD3 (1 μg/ml)- and anti-CD28 (2 μg/ml)-coated plates at 1 to 2 × 106 cells/ml in the presence of 10 ng/ml IL-4 and 10 μg/ml anti-IFN-γ (Th2 conditions) or in the presence of 1 ng/ml IL-12 and 10 μg/ml anti-IL-4 (Th1 conditions). IL-2 (100 U/ml) was added 24 h later. Cultures were expanded in IL-2 (100 U/ml) 3 days after initial culture. Th17 cells were cultured using soluble anti-CD3 (4 μg/ml), soluble anti-CD28 (1 μg/ml), 10 μg/ml anti-IL-4, 10 μg/ml anti-IFN-γ, 100 ng/ml IL-6, 10 ng/ml IL-1β, and 1 ng/ml TGF-β as described previously (74). The Th17 cultures were expanded in the presence of 10 ng/ml IL-23 after 3 days. Effector cells were stimulated with phorbol myristate acetate (PMA) (50 ng/ml) and ionomycin (500 ng/ml) for 1.5 h.
Cytokines and antibodies.
The antibodies to CD3 (2C11), IL-4 (11B11), and IFN-γ (R4-6A2) used in Th cell differentiation cultures were obtained from Bio X Cell. Antibody to CD28 was obtained from BD Pharmingen. Recombinant human IL-2 (hIL-2), murine IL-4 (mIL-4), mIL-6, mIL-1β, and mIL-12 were obtained from Peprotech. Recombinant hTGF-β was obtained from R&D. Antibodies used in ChIP experiments were specific to BRG1 (J1; Weidong Wang), H3K9,14 acetylated histones (catalog no. 06-599; Upstate/Millipore), H3K4Me3 (catalog no. 04-745; Millipore), and Stat6 (catalog no. M-200; Santa Cruz Biotech).
mRNA quantitation.
Total RNA was purified using RNeasy columns (Qiagen). cDNA was made using iScript (Bio-Rad) according to the manufacturer's instructions. The mRNA levels of cytokines and transcription factors were determined by real-time quantitative PCR (Q-PCR) using SYBR green (Qiagen) and an ABI 7500 system. Expression levels were normalized to TATA binding protein (TBP) or β-actin as indicated. Oligonucleotide sequences are presented in Table S3 in the supplemental material.
ChIP assay.
Chromatin immunoprecipitation was performed using methods similar to those described previously (88); details are available on request. Approximately 100 million cells were cross-linked with 1% formaldehyde and quenched with glycine. Cells were lysed with buffer containing 1% sodium dodecyl sulfate (SDS), treated with micrococcal nuclease, sonicated until the average DNA size was approximately 500 bp, and adjusted to concentrations of 0.1% SDS, 1% Triton X-100, and 150 mM NaCl at 5 ml. Sonicates were precleared with protein A Sepharose (Upstate), and IP was performed with BRG1 (J1) or rabbit IgG (catalog no. sc-2027; Santa Cruz). Chromatin was collected with protein A beads without salmon sperm DNA, washed, and eluted with sodium bicarbonate-SDS, and cross-links were reversed, followed by protease treatment and purification of Qiagen columns. Chromatin was quantified by real-time PCR (Q-PCR) using an Applied Biosystems 7500 system with Sybr green detection (Qiagen). For each primer set for each experiment, a six-point standard curve of genomic DNA (0 to 40 ng) was compared to input and immunoprecipitated sample curves, thus correcting for the efficiency of the individual primer sets. Graphs indicate immunoprecipitated chromatin amounts relative to input DNA (percent input). Downstream analysis of ChIP-seq data and data quality is described in detail in Methods in the supplemental material.
DNase I hypersensitivity (DHS) analysis.
Nuclei were purified from effector Th cells as described previously (1). Briefly, nuclei were released by hypotonic lysis in the presence of 0.5% NP-40 and digested with the indicated amounts of DNase I (Worthington) for 3 min at room temperature. The samples were then treated with proteinase K and RNase A, and the DNA was recovered after phenol-chloroform extraction and ethanol precipitation. Stimulated effector Th2 and Th1 cells were enriched with respect to viable cells prior to nuclear isolation using a Dead Cell Removal kit (Miltenyi). DNase I accessibility was assessed by real-time PCR of DNA (88). PCRs were performed using a QuantiTect SYBR green PCR kit (Qiagen) per the manufacturer's instructions and an ABI 7500 system. The DNase I sensitivity is indicated by the percentage of DNA remaining compared to undigested sample results, and DNA content is normalized to a known DNase I-resistant locus (Nfm or Nefm exon 1). Oligonucleotide sequences are presented in Table S2 in the supplemental material.
Episomal reporter assay.
DNA regions surrounding the indicated BRG1 binding sites from the GATA3 locus were cloned using Phusion (Finnzymes), mouse genomic DNA, and the indicated primers. The resulting BamHI fragments were cloned immediately upstream (BglII) or 2.2 kb downstream (BamHI) of the simian virus 40 (SV40) promoter in the pREP4-luc episomal luciferase reporter (44). The reporter constructs were transfected into CEM cells, a human lymphoblastoid T cell line, by the use of an Amaxa Nucleofection V kit (C-16) (Lonza). After 72 h, cells were either left resting or stimulated with PMA-ionomycin for an additional 5 h and harvested for analysis of luciferase activity by the use of a dual-luciferase reporter assay system (Promega). Results were normalized to a pRL-SV40 cotransfected renilla luciferase control plasmid (Promega).
RESULTS
BRG1 binding globally correlates with gene activity in T helper subsets.
We purified naïve T helper cells from BALB/c mice and differentiated these cells in culture according to the scheme presented in Fig. S1A in the supplemental material. We found that the mRNA encoding signature cytokines and transcription factors was restricted to the expected lineages (Figure S1B in the supplemental material and data not shown). We used primary cells rather than cell lines because the chromatin markers are more intense and more dynamic and because the physiology of the cells is better understood.
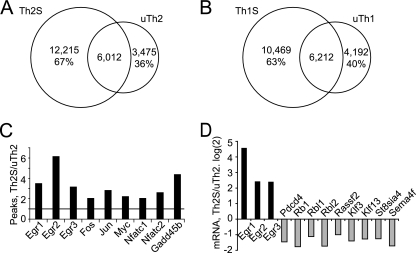
We analyzed six T helper subsets (naïve, unstimulated Th1 [uTh1] unstimulated Th2, stimulated Th1, stimulated Th2, and stimulated Th17) to systematically determine whether BRG1 binding responded to T helper differentiation or activation signals. We used ChIP-seq to perform a genome-wide measurement of the binding of BRG1, a component of the SWI/SNF ATP-dependent remodeling enzyme. We found 7,000 to 19,000 discrete BRG1 binding regions (see Table S1 in the supplemental material). BRG1 binding measured by ChIP-seq was consistent with our previous analysis of binding at Il4, Il5, Il13, Il2, Il3, and Gmcsf by the use of standard ChIP (63, 88) (see Figure S6a in the supplemental material). Differentiated, stimulated cells (Th1S, Th2S, and Th17S) contained the largest number of BRG1 binding regions, naïve cells contained the fewest, and resting differentiated cells (uTh1 and uTh2) were intermediate. The peaks were strongest in differentiated, stimulated cells and weaker in naïve and resting cells. The number of BRG1 binding regions is in good agreement with a previous report of 10,600 BRG1 binding regions in embryonic stem (ES) cells (30). By comparison, these T helper subsets were previously reported to contain 15,000 to 20,000 H3K4me3-modified regions, while H3K27me3 was found at 60,000 to 90,000 regions (85). The BRG1 binding regions we identified were apparently discrete elements with a calculated average length of about 170 bp (see Table S1 in the supplemental material), approximately the size of an enhancer or DNase I-hypersensitive site. This average length was similar to the finding in one previous report of 250-bp BRG1 binding regions in ES cell promoters (34) and much smaller than the 6.2-kb average length determined from genome-wide analysis of BRG1 in ES cells (30). This calculation was made using CisGenome software, which uses a probabilistic model to identify the location, length, and statistical significance of each binding region (33).
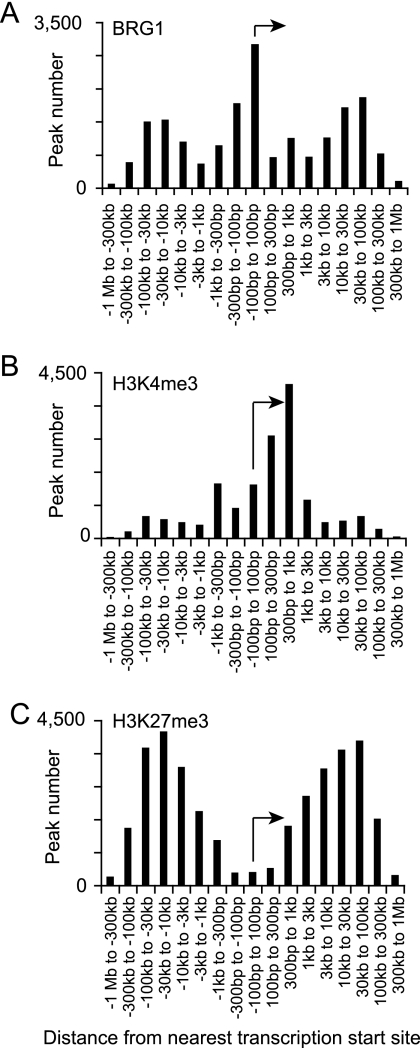
BRG1 has been hypothesized to function at both promoters and distal regulatory regions. For each BRG1 binding region, we calculated the distance to the nearest transcription start site (TSS). In stimulated Th2 cells, BRG1 binding was frequently found at promoters and also at distal regions more than 10 kb upstream and downstream from TSS (Fig. 1A). The results of determinations of the BRG1 binding region distance from the TSS were similar in all cases. Typically, 20% of genes possessed BRG1 binding regions that were at least 10 kb from the nearest promoter (Table 1), suggesting a general role for distal BRG1 binding sites in gene regulation. About 25% of the BRG1 binding sites were located within 1 kb of a transcriptional start site, suggesting a general role for BRG1 at promoters. The distribution of BRG1 binding regions was distinct from the previously reported distribution of H3K4me3, which is a marker for active promoters (85); this suggested that targeting and functions of these sites were independently determined (Fig. 1B and Table 2). BRG1 binding regions were also distributed differently from those of H3K27me3, again suggesting independent targeting and function (Fig. 1C and Table 2); the presence of H3K27me3 is negatively correlated with gene activity (85). BRG1 binding was distinct from these trimethylation markers at specific genes that are important in Th2 cells (see Fig. S2 in the supplemental material).
Fig. 1.
BRG1 binding, H3K4 trimethylation, and H3K27 trimethylation have distinct global distributions relative to transcriptional start sites. BRG1 binding regions and H3K4me3 and H3K27me3 were identified using ChIP-seq analyses and assigned to the nearest transcription start site (TSS) using CisGenome. The distance to the nearest TSS was calculated using Excel. The amount of binding and the number of modified regions over all genes were summed in each window using Excel. The transcription start site and direction are indicated by arrows. BRG1 binding data are from this study; H3K4me3 and H3K27me3 data were published previously (85). BRG1 binding is prominent at promoters and distal regions (10 to 100 kb), and H3K4 trimethylation is predominantly found from the TSS to kb +1, while H3K27 trimethylation is mostly distal from the TSS (3 to 300 kb). (A) BRG1 binding sites with respect to TSS. (B) H3K4 trimethylation with respect to TSS. (C) H3K27 trimethylation with respect to TSS.
Table 1.
BRG1 binding relative to transcriptional start sitesa
| BRG1 binding window | % of all genes |
|---|---|
| Anywhere | 46.9 |
| Promoter | 25 |
| Distal | 19.5 |
| Proximal | 21.7 |
| Promoter only | 13.3 |
| Distal only | 8.2 |
| Proximal only | 9.3 |
BRG1 binding regions in Th2S cells were mapped to the nearest transcription start site (TSS) by the use of CisGenome. The distance to the nearest TSS was calculated using Excel. Regions were assigned to categories based on distance from the TSS. Promoter, bp −300 to +100; proximal, kb −10 to −301 or kb +101 to +10; distal, Mb −1 to bp −10,001 or bp +10,001 to Mb +1. The percentages of all genes with BRG1 binding in these categories were calculated using the Web tool Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Promoter only counts genes that exclusively have promoter category BRG1 binding; promoter counts genes with promoter category BRG1 binding and includes genes that additionally have distal and/or proximal binding.
Table 2.
Correlation of BRG1 binding with H3K4me3 and H3K27me3a
| Comparison | Correlation (R2) |
|---|---|
| BRG1 vs H3K4me3 | 0.11 |
| BRG1 vs H3K27me3 | 0.002 |
For each gene, the amount of BRG1 binding and the level of histone methylation were compared. The number of sequence reads in statistically significant regions was determined using CisGenom and summed using Excel for BRG1, H3K4me3, and H3K27me3 in Th2S cells. BRG1 data were from this study; HeK4me3 and H3K27me3 data were previously published (85). Linear curve fit was determined by least-squares regression. H3K4me3 is a marker for active promoters, while H3K27me3 marks a subset of repressed genes.
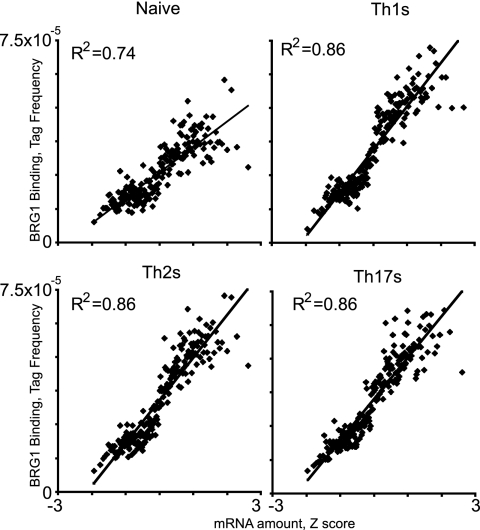
BRG1 has been hypothesized to preferentially support gene activation, though BRG1 is known to activate and repress genes in T cells (14, 41, 88, 89). We found positive global correlation between BRG1 binding and gene activity in all fates tested (Fig. 2). Genes with the most abundant mRNA have more BRG1 binding regions than average, whereas genes with the least abundant mRNA have fewer than average (Fig. 3A). For genes lacking BRG1 binding, the pattern is reversed (Fig. 3B). (Note that the genes with the smallest amount of mRNA were probably silent; this analysis was performed for all genes rather than exclusively for genes defined as active because they were present above an arbitrary threshold.) This suggested that BRG1 primarily facilitated gene activation in these cells. A similar analysis of CD8 T cells revealed a positive correlation of the presence of H3K4me3 with expression, whereas that of H3K27me3 was negatively correlated with expression (6).
Fig. 2.
BRG1 binding positively correlates with gene activity in T helper cells. ChIP-seq and expression data were compared essentially as described previously (6). Genes were sorted based on gene expression values (Z scores) and binned into groups of 100 genes, and the average gene expression value for each bin was calculated. After BRG1 tags in binding regions were assigned to the nearest transcriptional start site, the BRG1 tags contained by the genes in each bin was counted, and the average value was calculated. BRG1 binding in naïve, Th1S, Th2S, and Th17S cells was graphed against gene expression for each bin. All graphs were scaled to the same tag frequency and Z score range to facilitate direct comparison. Gene expression data were published previously (85).
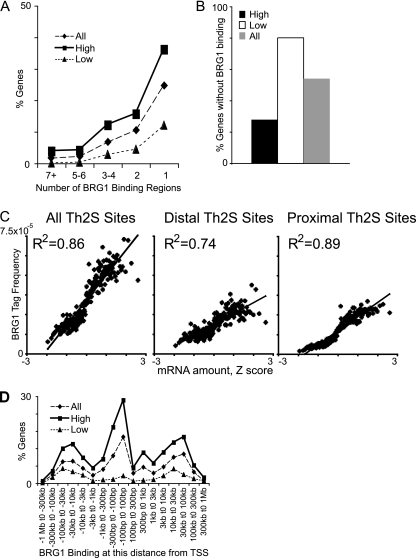
Fig. 3.
Distal and proximal BRG1 binding sites positively correlate with gene activity. (A) Highly expressed genes have more BRG1 binding regions. The number of BRG1 binding regions was counted for each gene. The percentage of genes that have the indicated number of binding regions was graphed for all genes (19,050 genes), “high” genes (the 2,032 genes with the most abundant mRNA), and “low” genes (the 1,208 genes with the least abundant mRNA). A total of 3.9% of the high genes had 7 or more BRG1 binding regions versus 0.08% of the low genes. (B) Genes with the lowest level of expression are more likely to lack BRG1 binding regions. A total of 80% of the low genes had no BRG1 binding versus 28% of the high genes. (C) Distal BRG1 binding and proximal BRG1 binding positively correlate with gene activity. Genes were sorted based on gene expression values (Z scores) and binned into groups of 100 genes, and the average gene expression level for each bin was calculated. BRG1 tags in binding regions were assigned to the nearest transcriptional start site, except for the proximal sites, for which only BRG1 binding from kb −10 to kb +10 with respect to the TSS was counted, while for distal sites only BRG1 binding from kb −100 to kb −10 and kb +10 to kb +100 kb was counted. BRG1 binding was graphed against gene expression for each bin. All graphs were scaled to the same tag frequency and Z score range to facilitate direct comparison. Gene expression data were published previously (85). (D) BRG1 binding is enriched in highly expressed genes at all distances relative to the TSS. The percentages of genes in each category with a BRG1 binding region at the indicated interval from the TSS are graphed. At every interval, the high genes are overrepresented and the low genes are underrepresented relative to all genes.
We identified a large number of BRG1 binding sites that were distal to known TSSs; these sites could play a role in gene activation or, alternatively, could have another function or represent system noise. We asked whether the proximal and distal BRG1 binding regions correlated with gene activity. We compared gene activity for (i) all BRG1 sites, (ii) proximal sites (within 10 kb of a TSS), and (iii) distal sites (10 to 100 kb from a TSS). All of these classes of BRG1 binding sites positively correlated with gene activity (Fig. 3C). BRG1 binding at all distances from TSSs was enriched in highly expressed genes (Fig. 3D). Together, these data support the hypothesis that BRG1 regulates gene expression through both distal and proximal sites.
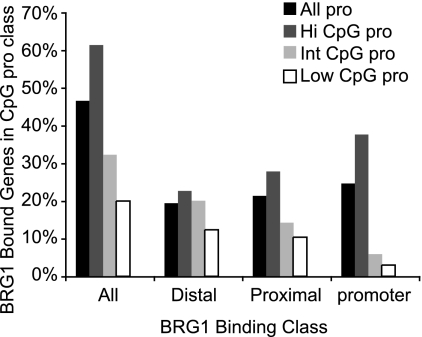
Recently, a unifying model was proposed in which promoters in CpG islands might be independent of SWI/SNF function and non-CpG-island promoters might require SWI/SNF function because of low nucleosome stability at the CpG promoters (64). In contrast, others found that human regulatory regions (including promoters) in T cells have high nucleosomal occupancy, which is correlated with high GC content (80). Among all of the genes examined, we found that BRG1 binding was modestly enriched in genes with high levels of CpG content promoters and underrepresented in genes with intermediate or low levels of CpG content promoters in Th2S cells (Fig. 4). BRG1 sites at the corresponding promoter were most sensitive to promoter CpG content, while distal BRG1 binding sites were less sensitive, suggesting that the function of distal BRG1 sites might be independent of promoter CpG type (Fig. 4). Among the genes reported to be regulated by BRG1 in T helper cells (41, 88, 89), there was one high-level CpG promoter (Il12rβ2), there were four intermediate-level CpG promoters (Ifnγ, Il4, Il5, and Il10), and there was one low-level CpG promoter (Il13).
Fig. 4.
Promoter CpG content and BRG1 binding in Th2S cells. Genes were grouped by promoter type as high CpG promoter (Hi CpG Pro), intermediate CpG promoter (Int CpG pro), or low CpG promoter (Low CpG pro) according to a previously published promoter CpG content list (47). Genes not found in that list are not shown as a separate type; “All pro” includes all 4 classes. Genes were also grouped by BRG1 binding class; “all” includes genes with at least 1 BRG1 binding region anywhere; “distal” includes genes that have at least one BRG1 binding region in the interval Mb −1 to kb −10 or kb +10 to Mb +1; “proximal” includes genes that have at least one BRG1 binding region in the interval kb −10 to kb −300 or kb +100 to kb 10; “promoter” includes genes that have at least one BRG1 binding region in the interval kb −300 to +100. For each BRG1 binding class, the height of a promoter class bar indicates the percentage of promoters of that type with BRG1 binding of that class. As shown in Table 1, BRG1 binding to genes as promoter sites and BRG1 binding to genes as distal sites are not mutually exclusive. High CpG promoter genes included those encoding Tbx21, Gata3, c-Maf, Ltβ, Rad50, Stat1, Stat3, Stat5a, Stat6, IL-12r-β2, and Ahr; intermediate CpG promoter genes included those encoding IFN-γ, IL-4, IL-5, IL-17a, IL-10, IL-2, FoxP3, TNF, Stat4, and Stat5b; low CpG promoter genes included Rorc, Il17f, Il13, Il21, Il3, Gmcsf, Ccr2, Ccr3, and Ccr5.
BRG1 binding changes during T cell activation.
Previously, we found that binding of another remodeling ATPase, SNF2H, was not strongly dependent on activation (63). Histone modifications were also reported to be relatively stable during T cell activation (11, 43). In contrast, we found that stimulation of Th1 and Th2 cells rapidly changed BRG1 binding at a subset of BRG1 sites in a small number of genes (63, 88). This change occurred after differentiation was complete and was dependent on the activation-specific transcription factor NFAT at the LCR in the Th2 cluster.
Global comparison of unstimulated versus stimulated cells following differentiation revealed redistribution of BRG1 binding (Fig. 5A and B). This was at the same time that cytokine mRNA expression was also strongly induced by stimulation (see Fig. S1B in the supplemental material). Thousands of BRG1 binding regions were lost and gained in both the Th1 and Th2 fates following brief (1.5-h) stimulation. In each lineage, unstimulated and stimulated cells shared approximately 6,000 BRG1 binding regions. We identified immediate-early genes as a members of a gene class that were induced by T cell activation in a lineage-independent manner; these genes had more BRG1 binding regions in stimulated cells than in unstimulated cells (Fig. 5C). Examination of activation-inducible cytokines genes revealed similar features.
Fig. 5.
Global activation-specific BRG1 binding. (A and B) Global redistribution of BRG1 binding. BRG1 binding regions were compared in Th2 cells (A) and Th1 cells (B). The numbers of BRG1 binding regions that are unique to stimulated cells, unique to resting cells, and shared are shown. The percentage for the regions unique to stimulated cells (Th2S and Th1S) represents the number of BRG1 binding regions unique for stimulated cells divided by the total number of BRG1 binding regions in stimulated cells. The percentage for regions unique to resting cells (uTh2 and uTh1) represents the number of BRG1 binding regions unique for resting cells divided by the total number of BRG1 binding regions in resting cells. (C) For each immediate-early gene, the ratio of BRG1 binding regions in stimulated versus resting Th2 cells was calculated; the horizontal line at 1 on the y axis indicates the position at which unchanged genes would be located. (D) Regulation of mRNA amounts by activation of Th2 cells. mRNA was prepared from resting and activated Th2 cells, synthesized with cDNA, and quantified using Q-RT-PCR. Each mRNA ratios represents the log2 amount of mRNA in activated cells divided by the amount in resting cells, so values greater than zero represent activation-induced genes (black bars) whereas genes expressed at higher levels in resting cells have values less than zero (light bars).
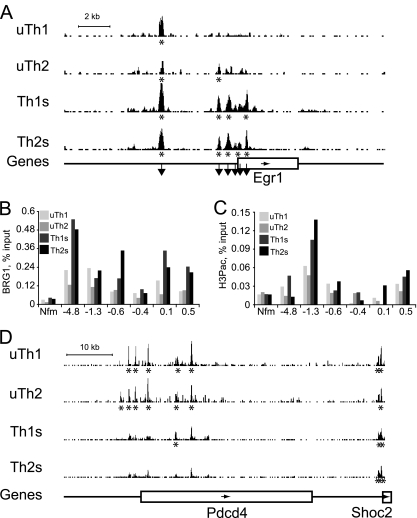
We asked whether these global patterns were typical of specific genes regulated by T cell activation. BRG1 binding regions in the immediate-early transcription factor EGR1 were more abundant and stronger following stimulation (Fig. 6A). We confirmed these findings using standard ChIP and independent samples (Fig. 6B). BRG1 binding did not correlate with a histone modification frequently found at immediate-early genes (H3S10PK14ac), suggesting independent targeting for BRG1 binding and this histone marker (Fig. 6C). Several of the BRG1 binding regions associated with c-myc are very distal upstream regions implicated in human cancers, and one distal BRG1 binding region we found was adjacent to a recently identified distal myc human enhancer (72, 87). Together, these findings suggest a role for BRG1 in establishing activation-induced gene expression.
Fig. 6.
Activation-specific BRG1 binding at specific genes. (A and D) ChIP-seq profiles for resting (uTh1 and uTh2) and activated (Th1S and Th2S) cells are shown. The BRG1 occupancy values (y axis) are identical for all graphs to allow direct comparison (minimum tag frequency of 0, maximum tag frequency of 1.14 × 10−5). Asterisks indicate statistically significant BRG1 binding regions identified using CisGenome. Genes are indicated as boxes below the profiles, and the arrows inside the boxes indicate the direction of transcription. (A) For the Egr1 locus, Chr18, nt 35010000 to 35030000 (MM9) are shown. A scale bar indicates 2 kb along the x axis. Downward pointing arrows indicate regions analyzed by standard ChIP as shown in panels B and C. (D) For the Pdcd4 locus, Chr19, nt 53950000 to 54020000 (MM9) are shown. A scale bar indicates 10 kb along the x axis. (B) BRG1 binding was measured by standard ChIP, and the results were normalized to the input samples. Control IgG ChIP results were similar to those obtained with Nfm, a negative-control locus. The y axis labels indicate distances from the Egr1 TSS in kilobases. This experiment was a biological replicate of the ChIP-seq experiment whose results are depicted in Fig. 6A (but performed with different mice, with cells harvested on different days, etc.). (C) H3S10pK14ac levels were measured by standard ChIP and normalized to the input samples. Control IgG IP results were similar to those obtained with Nfm (Nefm exon 1), a negative-control locus. The locus position is indicated with respect to distance from the Egr1 TSS in kilobases. This experiment was performed using the same chromatin used for the BRG1 ChIP whose results are shown in panel B.
We identified genes that were repressed in both Th1 and Th2 cells following stimulation (Fig. 5D). At Pdcd4, St8sia4, Rb1, Rbl1, Rbl2, Klf3, Embigin, and Sema4f, fewer BRG1 binding regions were found in stimulated Th1 and Th2 cells than in resting cells (Fig. 6D and data not shown). BRG1 binding again positively correlated with gene activity, suggesting a role for BRG1 in maintaining expression of these genes in resting cells.
Thus, brief stimulation of T helper cells strongly altered the pattern of BRG1 binding, both globally and at specific genes. This finding contrasts with our previous finding that SNF2H binding is relatively insensitive to T cell activation (63) and with reports that histone modifications are relatively stable during T cell activation (11, 43). These findings suggest that there is a tighter temporal link between BRG1 binding and gene activity than exists for SNF2H or histone modifications and that BRG1 binding positively correlated with the activity of activation-induced and activation-repressed genes.
BRG1 binding changes during T helper differentiation.
We asked whether the global pattern of BRG1 binding changed during differentiation of naïve cells to the Th1, Th2, and Th17 fates in response to lineage-programming signals. Previously, BRG1 was found to have a functional role in Th1 and Th2 differentiation (41, 88, 89). Approximately one-fourth of the BRG1 binding regions found in each fate were absent in the other fates (Table 3). There was also a conserved core of 6,000 BRG1 binding regions shared by differentiated T helper cells (Th1S, Th2S, and Th17S), suggesting significant functional similarity. In contrast, fewer shared binding regions were found when the naïve subset was also included. As is consistent with these numbers, BRG1 binding in Th2S cells correlated best with Th1S cells, correlated well with resting Th2 cells, correlated less with naïve T helper cells, and correlated least with ES cells (Table 4). Thus, global comparison of naïve cells to differentiated cells revealed widespread redistribution of BRG1 binding in response to lineage signals, which is consistent with a role for BRG1 in programming the fates of those cells.
Table 3.
BRG1 redistribution during T cell differentiationa
| BRG1 binding region(s) | No. of peaks | % total |
|---|---|---|
| N only | 1,603 | 23.10 |
| 1S only | 2,573 | 17.10 |
| 2S only | 4,171 | 24.10 |
| 17S only | 5,521 | 33.60 |
| 1S/2S/17S | 6,504 | 40.00 |
| N/1S/2S/17S | 2,420 | 14.90 |
The percent total value for the N-only number of BRG1 binding regions represents the number of N BRG1 binding regions not found in 1S, 2S, or 17S divided by the total number of N BRG1 binding regions. 1S, 2S, and 17S percentages were calculated similarly. The percent value for the 1S/2S/17S number of BRG1 binding regions represents the number of BRG1 binding regions shared by 1S/2S/17S divided by the average number of binding regions in Th1S, Th2S, and Th17S. The percent value for the N/1S/2S/17S number of BRG1 binding regions represents the number of BRG1 binding regions shared by N/1S/2S/17S divided by the average number of binding regions in Th1S, Th2S, and Th17S.
Table 4.
Correlation of BRG1 binding in different cell fatesa
| Comparison | Correlation (R2) |
|---|---|
| Th1S vs Th2S | 0.85 |
| uTh2 vs Th2S | 0.65 |
| Naïve vs Th2S | 0.21 |
| ES vs Th2S | 0.01 |
BRG1 binding levels in each gene were compared for two cell fates. The number of sequence reads in statistically significant regions in Th2S, Th1S, uTh2, naïve, and ES cells was determined using CisGenome and summed using Excel for BRG1. Th2S, Th1S, uTh2, and naïve data were from this study; ES data were previously published (30). Linear curve fit was determined by least-squares regression.
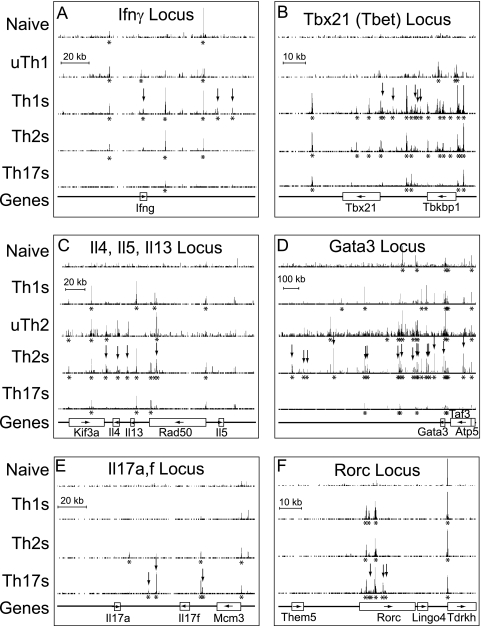
We asked whether these genome-wide averages were representative of the remodeling landscape at specific genes with known fate-specific roles in T helper differentiation and function. For each differentiated cell fate, we analyzed one lineage-specific cytokine and one lineage-specific transcription factor. We compared 6 cell types; for clarity, uTh1 and uTh2 are shown only for the Th1- and Th2-specific genes, respectively (Fig. 7). Several general principles emerged. (i) BRG1 binding regions were more numerous and stronger under conditions where genes were expressed, as with activation-induced genes. (ii) BRG1 binding was found at promoters and distal regions; some genes (e.g., some housekeeping genes and small genes) had only promoter binding or only distal binding (Il17a and Il17f). (iii) BRG1 binding to previously identified elements such as enhancers, LCRs, cohesin binding sites, DHS, and histone modification sites correlated with the regulation of the target genes. (iv) Regulation of BRG1 binding often correlated best with expression of a distal gene. Regulation of BRG1 binding within housekeeping genes (Rad50, Taf3, and Tbkbp1) often correlated better with expression of genes adjacent to tissue-specific genes, suggesting that regulatory regions for tissue-specific genes were often embedded in adjacent housekeeping genes. (v) BRG1 binding was often found at the promoters of active housekeeping genes. (vi) BRG1 binding results determined by ChIP-seq were in good agreement with the results of our previously published assays performed by standard ChIP (63, 88).
Fig. 7.
Lineage-specific BRG1 binding. Signature cytokines (A, C, and E) and transcription factors (B, D, and F) are shown. Th1-specific (A and B), Th2-specific (C and D), and Th17-specific (E and F) genes were compared. ChIP-seq profiles for naïve, Th1S, Th2S, and Th17S cells are shown for each gene; the uTh1 profile is shown for Th1 genes; and the uTh2 profile is shown for Th2 genes. BRG1 occupancy range values (y axis) are identical for all graphs to allow direct comparisons (minimum tag frequency of 0, maximum tag frequency of 1.14 × 10−5). Vertical arrows indicate lineage-specific BRG1 binding identified using CisGenome. Asterisks indicate statistically significant BRG1 binding regions identified using CisGenome. Each panel contains a scale bar for the x axis (genomic location). Genes are indicated as boxes with names below, and arrows inside the boxes indicate the direction of transcription. Note that the Rorc gene produces the ROR-γ mRNA; the ROR-γT (T-cell specific) transcript initiates from an internal, downstream promoter within the Rorc gene. Gata3 promoter A is 10 kb upstream of the indicated Gata3 gene. The genomic coordinates represented (MM9 assembly) are as follows: (A) for Ifnγ, Chr10, nt 117820000 to 117960000; (B) for Tbx21, Chr11, nt 96930000 to 97020000; (C) for Il4, Il5, and Il13, Chr11, nt 53370000 to 53570000; (D) for Gata3, Chr2, nt 8700000 to 10000000; (E) for Il17a and Il17f, Chr1, nt 20680000 to 20820000; (F) for Rorc, Chr3, nt 94140000 to 94230000.
We hypothesize that the BRG1 binding regions we identified may be cis-regulatory elements for transcription. Many of these sites are newly identified in this report. We also note that BRG1 binding overlaps with a subset of the known and putative regulatory regions found in these loci, again suggesting that we are identifying functional regions. For example, BRG1 binding markers CNS4, -5, and -7 are H3 hyperacetylation sites previously identified at the Il17a-Il17f locus (2). At Ifnγ, BRG1 binding at kb +2, 54.5, and 65 was Th1 specific; the positions at kb −21.8, +2, 17.9, and 54.5 appear to overlap previously reported regions of histone modification and cohesin binding (7, 26). At the Il4-Il5-Il13 locus, we detected BRG1 binding at CNS2-HSV, the Il4 promoter, CNS1, Il13 CGRE, RHS7, RHS6b, RHS6a, and the Rad50 promoter, in agreement with our previous standard ChIP results (88). We also detected BRG1 binding at STAT5 binding sites in Kif3a (42). The distal elements RHS7, CNS1, HSV/HSVa, and HSIV regulate the neighboring Il4, Il13, and Il5 genes (5, 40, 49, 71).
We found many BRG1 binding regions at clusters of active genes, e.g., the Il4, Il5, Il13 cluster (chromosome 11 [Chr11], nucleotides [nt] 53350000 to 53600000), the CCR2, CCR3, CCR5 cluster (Chr9, nt 123900000 to 124076172), and the major histocompatibility complex (MHC) cluster (Chr17, nt 34000000 to 36500000) (Fig. 7 and data not shown). We believed that this pattern could be correlated with the activity of these specific genes; alternatively, the correlation could be to gene density or overall gene activity, as previously found in some cases for chromosome structure (23, 45). We examined BRG1 binding at gene-dense clusters of genes that are inactive in Th2S cells (the basic keratin cluster (Chr15, nt 101200000 to 101900000), the acidic keratin cluster (Chr11, nt 99100000 to 100100000), the epidermal differentiation complex (Chr3, nt 91200000 to 93300000), and the olfactory receptor cluster (Chr17, nt 37200000 to 38500000), We found few, if any, BRG1 binding regions (data not shown), suggesting that gene activity correlated with BRG1 binding but not with gene density or gene clustering.
The consistent pattern that emerges from global analysis and from analysis of specific cytokine and transcription factor genes is that more and stronger BRG1 binding regions were found under the same conditions as those in which these genes are expressed. Importantly, many of these binding regions observed in differentiated cells had little or no BRG1 occupancy in the naïve precursor cells. Moreover, BRG1 binding was distinct from the presence of H3K4me3 and H3K27me3 (Fig. 1 [see also Fig. S2 in the supplemental material] and data not shown). Together, these findings suggest that BRG1 might be a marker for regulatory regions of these genes or might directly regulate expression of these genes.
Tissue-specific BRG1 binding.
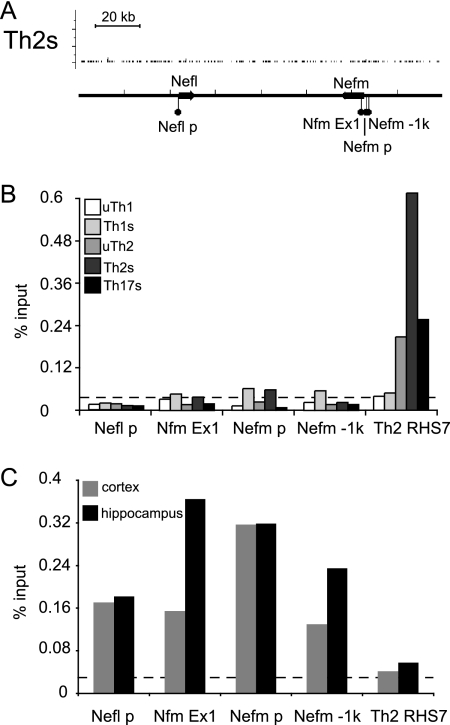
The analyses presented above were performed using 6 T helper subsets that are developmentally similar. We asked whether BRG1 binding might be regulated in a tissue-specific manner. First, we examined more distantly related tissue by performing ChIP using samples from mouse brain tissue. Analysis performed using ChIP-seq and standard ChIP T cells revealed the absence of BRG1 binding to loci in the brain-specific neurofilament genes Nefl and Nefm (Fig. 8A and B). In contrast, strong BRG1 binding was detected in mouse brain tissue within the neuron-specific neurofilament genes, including binding at Nefm exon 1, one of our standard negative controls for BRG1 binding in T cell studies (Fig. 8C). Next, we compared our BRG1 binding results to the findings of a recent study performed using ES cells (30). There was little if any BRG1 binding to the Il4, -5, and -13 genes in ES cells, in contrast to the results seen in experiments investigating the same region in Th2S cells (see Fig. S3 in the supplemental material). Examination of the Nanog pluripotency gene revealed BRG1 binding regions in ES cells, as reported, but not in Th2S cells (see Fig. S3 in the supplemental material). Global correlation of BRG1 binding in Th2S cells with binding in ES cells was much weaker than correlation with binding in other T cell fates (Table 4) or with the presence of H3K4me3 in Th2S cells (Table 2). Together, these results suggest that the BRG1 binding pattern is specific to the tissue type, as well as to the differentiation and activation states.
Fig. 8.
Tissue-specific BRG1 binding. (A) ChIP-seq profiles for Th2S cells are shown. BRG1 occupancy (y axis) is again plotted from the minimum tag frequency of 0 to a maximum tag frequency of 1.14 × 10−5. A scale bar indicates 20 kb along the x axis. No statistically significant BRG1 binding regions were identified using CisGenome in this interval for any T helper subset. Genes are indicated as arrows on the profiles, indicating the direction of transcription; Chr18, nt 35014000 to 35025000 (MM9) is shown. (B) BRG1 binding in mouse T cells was measured by standard ChIP and normalized to the input samples. The value representing 2 times the average control IgG IP result is indicated by the dashed line; Nfm is a negative-control locus in T cells (Nefm exon 1). The locus data represent details of features shown in panel A. This experiment was a biological replicate of the ChIP-seq whose results are shown in panel A (but with different mice, with cells harvested on different days, etc.). (C) BRG1 binding in mouse brain tissue was measured by standard ChIP and normalized to the input samples. The value representing 2 times the average control IgG IP result is indicated by the dashed line. RHS7 is a positive-control locus in Th2 cells. The locus data represent details of features shown in panel A.
BRG1 binds to miRNA genes in T helper subsets.
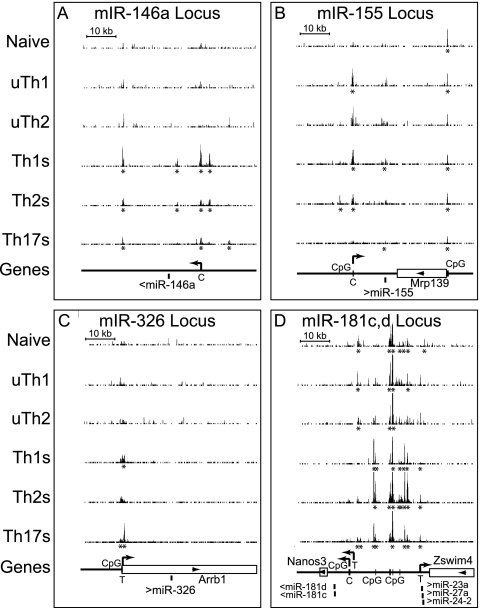
We hypothesized that BRG1 might regulate expression of noncoding RNA genes. We asked whether BRG1 binding was regulated in the vicinity of the miRNA transcription units shown to help determine T helper function. A number of studies have found an important role for miRNAs in T helper cells. Differentiated T helper cells from miR-155-null mice produced more IL-4 and IL-5, whereas IFN-γ levels were reduced, suggesting that miR-155 plays a role in T helper polarization (10, 66, 78). Th17 differentiation and multiple sclerosis (MS) were found to be regulated by miR-326 expression; MS was also linked to the overlapping beta-arrestin gene (18, 56, 68). miR-146a was found to be misregulated in cases of systemic lupus erythematosus (76). Expression of miR146 is upregulated in Th1 cells but not Th2 cells, and miR-155 is upregulated in Th1 and Th2 cells (10, 50). Recently, many miRNA promoters were mapped using chromatin structure markers (11, 59).
We found that BRG1 binding in the vicinity of several miRNA transcription units correlated with the reported expression pattern. BRG1 binding at the miR-146am, miR-155, and miR326 loci was consistent with the reported expression pattern (Fig. 9). We also find BRG1 at a limited number of miRNA processing sites, such as miR-155. As miRNA processing is initially cotranscriptional (51), it is possible that the chromatin structure is regulating RNA processing events, as recently suggested (73). BRG1 was found at the miR-181c,d cluster preferentially in activated cells and in the Let-7a-1, -f-1, and -d cluster (Fig. 9 and data not shown). These findings are in agreement with a recent report that BRG1 regulates myoblast miRNA transcription (46). Together, these findings suggest that BRG1 may regulate RNA-coding genes.
Fig. 9.
BRG1 binding to miRNA loci is specific with respect to lineage and activation. BRG1 binding to miRNA loci was measured by ChIP-seq, and the results obtained for T helper subsets were compared. The BRG1 occupancy range values (y axis) are identical for all graphs to allow direct comparison (minimum tag frequency of 0, maximum tag frequency of 1.14 × 10−5). Asterisks indicate statistically significant BRG1 binding regions identified using CisGenome. Each panel contains a scale bar for the x axis (genomic location). Genes are indicated as boxes with names below, and arrows inside the boxes indicate the direction of transcription. Mature miRNA locations are marked as bars; the direction of transcription is indicated with arrows. Predicted transcriptional start sites are indicated with arrows (“T” represents T cell predictions [11], “C” represents cancer cell predictions [59]). CpG islands are marked as vertical bars. (A) For miR-146a, Chr11, nt 43157899 to 43217963. (B) For miR-155, Chr16, nt 84684385 to 84744449. (C) For miR-326, Chr7, nt 106670000 to 106730000. (D) For miRNA 181c, 181d, 23a, 27a, and 24-2 clusters, Chr8, nt 86690000 to 86750000.
Stat binding and BRG1 binding.
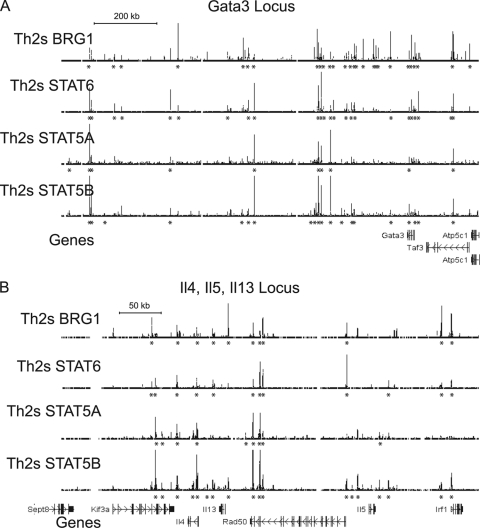
Previously we found that STAT6 could bind BRG1 and recruit BRG1 to the LCR in the Il4-Il5Il13 cluster (88). We compared BRG1 binding to recently published results for STAT4, -5a, -5b, and -6 (42, 86). Many BRG1 binding regions in the Gata3 locus and the Th2 cluster were occupied by STAT6 and/or STAT5 in Th2S cells, including the LCR in the Il4-Il5Il13 cluster (Fig. 10). Globally, Stat6 binding in stimulated Th2 cells correlated best with BRG1 binding in stimulated Th2 cells; correlation was weaker in resting Th2 cells, and poor in naïve cells (Table 5). This was consistent with results showing Stat6 recruitment of BRG1 to a subset of binding regions. This did not agree well with the model that BRG1 might bind to sites in naïve cells to facilitate later binding of Stat transcription factors. At the Tbx21 and miR-146a loci, many BRG1 binding regions were also occupied by STAT4 in Th1S cells (see Fig. S4A and B in the supplemental material). Tbx21 and miR-146a are strongly expressed in Th1 cells, which is consistent with the known role of Stat4 in expression of Th1-specific genes. Stat6 and Stat4 were also found at BRG1 binding regions in the activation-specific gene Egr1 (see Fig. S4C in the supplemental material), suggesting that the function of these transcription factors extended beyond regulation of lineage-specific genes. We also identified regions that appeared to bind only BRG1 or STATs.
Fig. 10.
BRG1 binding occurs at STAT binding sites. ChIP-seq profiles from Th2S cells for BRG1, STAT6, STAT5A, and STAT5B are shown. BRG1 data are from this study, while Stat6 data are from reference 86 and Stat5 data are from reference 42. Occupancy range values (y axis) are identical for all graphs to allow direct comparison (minimum tag frequency of 0, maximum tag frequency of 1.14 × 10−5). Asterisks indicate statistically significant binding regions identified using CisGenome. Each panel contains a scale bar for the x axis (genomic location). Gene exons are indicated as vertical bars with names on the side, and arrows indicate the direction of transcription. Gata3 promoter A is 10 kb upstream (rightward) of the indicated Gata3 gene. The BRG1 data from Fig. 6 are repeated here to facilitate direct comparison. The genomic coordinates represented (MM9 assembly) are as follows: (A) for Gata3, Chr2, nt 8700000 to 10000000; (B) for Il4, Il5, and Il13, Chr11, nt 53370000 to 53570000.
Table 5.
Correlation of Stat6 binding to BRG1 binding in different cell fatesa
| Comparison | Correlation (R2) |
|---|---|
| Th2S BRG1 vs Th2S Stat6 | 0.27 |
| uTh2 BRG1 vs Th2S Stat6 | 0.18 |
| Naïve BRG1 vs Th2S Stat6 | 0.05 |
For each gene, the amount of BRG1 binding and the amount of Stat6 binding were compared. The number of sequence reads in statistically significant regions in Th2S, uTh2, naïve, and Stat6 in Th2S cells was determined using CisGenome and summed using Excel for BRG1. Th2S, uTh2, and naïve data were from this study; Stat6 data were previously published (86). Linear curve fit was determined by least-squares regression.
Novel BRG1 binding sites distal to the Gata3 promoters possess enhancer-like activity.
We frequently noted the pattern of distal BRG1 binding regions that positively correlated with gene activity, suggesting a functional role. Often, those distal regions were located in neighboring housekeeping genes or gene-free regions. We used Gata3 as a test case to investigate whether these distal BRG1 binding regions might be functional.
Gata3 is required for T cell development (27, 81) and Th2 fate (90) as well as development of heart, brain, kidney, and mammary epithelia. A 625-kb fragment that rescues Gata3 expression at many regions in the body appears to lack the sequences required to direct expression in the thymus and at other locations (39), suggesting that distal unidentified elements are critical for T cell expression of Gata3 and that the known proximal sequences are insufficient for that expression.
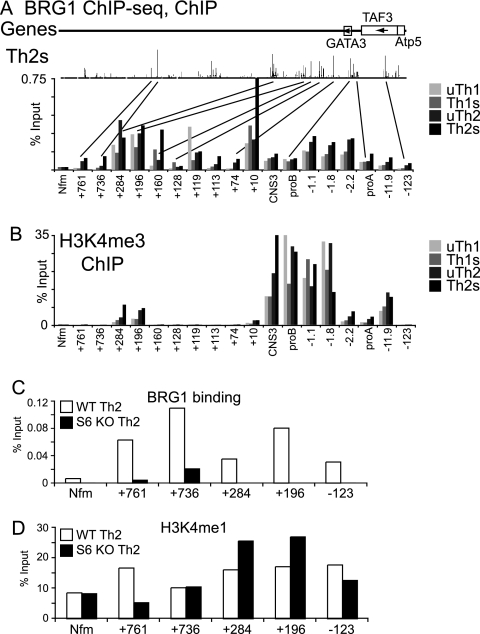
First, we used standard ChIP to examine BRG1 binding to several sites we had identified by ChIP-seq. As is consistent with the ChIP-seq data, we found all of the tested sites bound BRG1 and that binding in Th2S cells was stronger at most sites than in Th1S cells (Fig. 11A), validating some of the ChIP-seq data using a biologically independent sample. BRG1 occupancy at distal sites was similar to or greater than BRG1 binding to the GATA3 promoters.
Fig. 11.
Distal BRG1 binding sites in Gata3 are programmed by Stat6. (A) Standard ChIP was used to measure BRG1 binding in resting Th1, stimulated Th1, resting Th2, and stimulated Th2 cells. The samples were biological replicates of the ChIP-seq samples (but with different mice, with cells harvested on different days, etc.). x axis labels indicate distances in kilobases from promoter B: promoter A is at kb −10.2; CNS3 is at kb +1.7. Nfm is a negative-control locus for BRG1 in T cells (Nefm exon 1 in Fig. 8). The results of ChIP-seq for BRG1 from Fig. 7D are shown again to facilitate direct comparisons. (B) Standard ChIP was used to measure H3K4 trimethylation in the Gata3 locus in resting Th1, stimulated Th1, resting Th2, and stimulated Th2 cells. Chromatin from the same cells whose results are shown in panel A was analyzed at the same time, to facilitate direct comparisons. (C and D) Standard ChIP was used to measure BRG 1 binding (C) and H3K4 monomethylation (D) in the Gata3 locus in stimulated cells cultured under stimulated Th2 conditions and made from cells of wild-type (WT) mice (white bars) and STAT6-null mice (black bars). The same chromatin was analyzed in both panels to facilitate direct comparisons.
Next, we examined targeting of BRG1 to the Gata3 locus. BRG1 binding to GATA3 at kb 761, 731, 284, 196, and −123 was reduced or absent when naïve cells from STAT6 knockout (KO) mice were differentiated under Th2 conditions (Fig. 11C). All of these sites appear to have STAT6 binding (Fig. 10A). This indicated that Stat6 was necessary for BRG1 binding to these sites, probably through direct recruitment. H3K4 monomethylation (an enhancer marker) was not different from the neuronal control locus at kb +736, suggesting that H3K4me1 was not necessary for BRG1 binding; moreover, H3K4me1 was not lost at kb +284 or +196 in Stat6 KO cells, suggesting that H3K4me1 was not sufficient for BRG1 recruitment (Fig. 11D). BRG1 binding was also distinct from that of H3K4 trimethylation (a promoter marker) and H3K9,14ac (an active chromatin marker), suggesting that these features were not sufficient for BRG1 binding (Fig. 11B and data not shown). H3K4me3 was strongest around promoter B; in our system, most of the GATA3 mRNA was produced from promoter B in Th2S cells (data not shown). H3K4 trimethylation results were in good agreement with published ChIP-seq data (85) (see also Fig. S2D in the supplemental material).
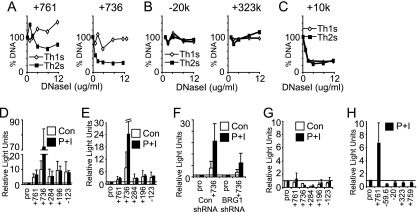
Next, we asked whether the BRG1 binding regions were in open or closed chromatin conformation; open chromatin, detected as DNase I hypersensitivity (DHS), frequently correlates with active cis elements (4, 16, 25, 67). The five tested distal BRG1 elements at kb +761, +736, +284, +196 and −123 were all DNase I hypersensitive (Fig. 12A and data not shown). In particular, kb +761 and +736 were Th2 specific, which is consistent with their BRG1 binding specificity. Four regions that did not bind BRG1 were not DHS in Th1 or Th2 cells (Fig. 12B and data not shown). DHS was found at kb +10 in Th1 and Th2 cells (Fig. 12C), which is consistent with BRG1 binding to this element in Th1 and Th2 cells. These findings expand on our previous observations that both GATA3 promoters and CNS3 bound BRG1 that and promoter A exhibited BRG1-dependent DHS (88).
Fig. 12.
Novel BRG1 binding sites in the Gata3 locus are in open chromatin and possess enhancer-like activity. (A, B, and C) Distal BRG1 binding sites have an open chromatin conformation. Nuclei from stimulated Th1 and Th2 cells were digested with DNase I; DNA was quantified using Q-PCR primers detecting the indicated regions. For each region, a representative dose response is shown; similar results were obtained by averaging three biological replicates digested with 8 μg/ml DNase I (data not shown). (A) Distal BRG1 binding regions are DHS in Th2 cells but not in Th1 cells. (B) Control regions lacking BRG1 binding are not DHS. (C) A control region with BRG1 binding in Th1 and Th2 cells is DHS in both. (D and E) Distal BRG1 binding sites possess enhancer-like activity. The indicated elements were placed immediately upstream (D) or 2.2 kb downstream (E) of the SV40 promoter in the pRep4-luc episomal vector and transfected into CEM cells (human lymphoblastoid T cell line), and the reporter was assayed with (black bars) and without (white bars) stimulation. Averages and standard deviations of the results from two biological replicate experiments are shown; all values are shown relative to the promoter values (also indicated as a horizontal line). (F) Enhancer-like activity is BRG1 dependent. The indicated elements were placed upstream of the SV40 promoter in the pRep4-luc episomal vector and transfected into CEM cells (human lymphoblastoid T cell line), and the reporter was assayed with (black bars) and without (white bars) stimulation. Cells were treated with BRG1 shRNA or a control, as indicated. Averages and standard deviations of the results from two biological replicate experiments are shown; all values are shown relative to the promoter values (also indicated as a horizontal line). (G) Distal BRG1 binding sites lack enhancer-like activity in a nonepisomal vector. The indicated elements were placed upstream of the SV40 promoter in the pGL3 promoter vector and transfected into CEM cells (human lymphoblastoid T cell line), and the reporter was assayed with (black bars) and without (white bars) stimulation. The horizontal line at 1 on the y axis indicates the value obtained with the promoter alone. Averages and standard deviations of the results from two biological replicate experiments are shown; all values are shown relative to the promoter values (also indicated as a horizontal line). These same elements exhibited increased expression using episomal reporters (D and E). None of the five tested regions were positive for enhancer-like activity in this assay. (H) Distal regions without BRG1 binding in Th2 cells lack enhancer-like activity. The indicated elements were placed upstream of the SV40 promoter in the pRep4-luc episomal vector and transfected into CEM cells (human lymphoblastoid T cell line), and the reporter was assayed with stimulation. The horizontal line at 1 on the y axis indicates the value obtained with the promoter alone. Averages and standard deviations of the results from two biological replicate experiments are shown; all values are shown relative to the promoter values. All four tested regions lacking BRG1 binding were negative for enhancer-like activity in this assay.
Finally, we tested whether these newly identified distal elements could increase expression of a reporter construct. Five novel, distal BRG1 binding elements were tested in an episomal reporter assay, as episomes have been reported to more closely recapitulate native chromatin structure than standard transient-transfection templates (70). All five elements activated expression of a reporter when located immediately upstream Fig. 12D) or 2.2 kb downstream (Fig. 12E) of a reporter. These BRG1 binding regions also exhibit DHS (Fig. 12A and data not shown). The kb +736 element was strongly induced by stimulation; consistent with this finding, kb +736 DNase I HS was inducible by activation in Th2 cells, and this was the only element in this group with activation-induced BRG1 binding in Th2 cells (Fig. 11A). Cotransfection of short hairpin RNA (shRNA) for BRG1 with the kb +736 element reduced enhancer function, revealing that the activity of the kb +736 element was dependent on the presence of BRG1 (Fig. 12F). None of these elements were active in nonepisomal vectors, suggesting that their function might require chromatin with physiologic properties (Fig. 12G). Four regions lacking BRG1 binding that were also insensitive to DNase I (Fig. 12B and data not shown) were tested in the episomal reporter assay; none were able to increase reporter activity (Fig. 12H). Together, these results indicate that distal BRG1 binding regions have BRG1-dependent enhancer-like activity.
DISCUSSION
Previously, we found that BRG1 was a regulator of Th2 differentiation and directly regulated cytokine transcription and chromatin structure (88). Here, we expanded our analysis of BRG1 to include other T helper subsets, and we expanded our studies from two loci to the entire genome. We used primary cells because the chromatin markers are stronger and more dynamic than in cell lines. We used global analysis to determine the general principles of BRG1 binding at all T helper genes, as well as analysis of specific genes of known importance to determine whether these general rules were typical of key T cell genes. Our comparison of our data with recently published data for histone modifications and transcription factors in these cells reveals both important similarities and differences among these factors. Activation signals and lineage signals both play important roles in programming BRG1 binding. BRG1 binding was also tissue specific, as T helper cell BRG1 binding was different from that seen with brain and ES cells. BRG1 binding was dynamic, as activation of T helper cells resulted in redistribution of BRG1 after only 1.5 h. In contrast, binding of the remodeling enzyme SNF2H in T cells is not strongly regulated by activation (63). We found that BRG1 binding positively correlated with gene activity at proximal and distal regions, suggesting that both of these types of regions are involved in gene activation. BRG1 binding correlated with tissue-specific and activation-specific expression of protein-coding and miRNA genes. Newly identified distal BRG1 binding regions in the GATA3 locus were specific with respect to fate and hypersensitive to DNase I and possessed BRG1-dependent enhancer-like activity. BRG1 binding to these regions was dependent on the presence of STAT6. Because we found that BRG1 bound to several known enhancers when they were active and that the tested BRG1 binding regions were able to activate transcription from reporters, we suggest that BRG1 binding appears to be a novel marker for cis-regulatory regions.
BRG1 binding was found at discrete sites averaging 170 bp in length. This is approximately the size of a nucleosome, enhancer, or DNase I HS. We did not observe gene clusters, entire genes, or transcribed gene bodies that were coated with BRG1. BRG1 binding regions often corresponded to a subset of sites of enhancers, histone modifications, DNase I HS, and cohesin binding sites in well-annotated regions of the T helper cell genome. Transcription factor recruitment was also found at the tested BRG1 binding sites. While both BRG1 binding and H3K4 trimethylation were enriched at active genes, many BRG1 binding regions were not enriched for H3K4me3, suggesting independent targeting and function. This was especially true of distal BRG1 sites, consistent with the promoter-proximal location of most H3K4me3 sites.
Globally, we observed BRG1 binding associated with gene expression, suggesting that BRG1 served as an activator in most cases. However, we do not exclude the possibility that BRG1 may play a more limited role in transcriptional repression as well. For example, we observed BRG1 binding to HSIV of the Th2 cluster in Th1 cells; HSIV is an IL-4 silencer in Th1 cells (5). BRG1 has been found to be a repressor of CD4 expression during T cell development (14). We hypothesize that BRG1 binding is a marker for active chromatin rather than for active genes. BRG1 binding to a particular locus likely exerts combinatorial gene regulation by the activity of multiple positive and negative cis-regulatory elements. This model is also consistent with our detection of lineage-specific and nonrestricted BRG1 binding sites in the signature cytokine loci and transcription factors. BRG1 binding was enriched in a lineage-specific manner, though it was not absolutely restricted. This is in contrast to the results seen with H3K4me3, which appears to be a marker of recent promoter activity.
We hypothesize that we have been measuring active BRG1 complexes at many of the binding regions we have identified. Our previous analysis of BRG1- and SNF2H-remodeling enzymes at several loci found that partial loss of function induced using knockdown reagents altered chromatin structure and gene expression at a subset of the loci bound by the remodeling enzymes, suggesting that direct binding was important for chromatin remodeling (63, 88). However, we note that others have identified BRG1 binding to chromatin by analysis using mutants lacking ATPase activity (17, 32), suggesting that BRG1 ChIP-seq would also detect inactive forms of SWI/SNF and ATPase-independent SWI/SNF activity, if present. Little is known about regulation of SWI/SNF activity, though calcium signaling can activate SWI/SNF and phosphorylation can inhibit SWI/SNF (37, 69), raising the possibility that BRG1 is bound and poised for later activation at some sites. To our knowledge, no reagent is available to enable researchers to distinguish these forms of BRG1 by ChIP. At lineage-specific genes, there was little BRG1 binding in naïve cells, and for activation-induced genes, there was more BRG1 binding in activated cells than resting cells. Analysis of different cell fates and states revealed that BRG1 recruitment frequently parallels gene induction, which is consistent with the idea that transcription factor-mediated recruitment of SWI/SNF is a key mode of remodeling regulation.
Our current work suggests a widespread role for BRG1 in differentiated cells, extending our previous work in Th2 cells. This is perhaps unexpected, as BRG1 function could have been limited to the critical period of cell fate choice, followed by a BRG1-independent epigenetic memory of that choice. Cell-free chromatin studies have found that ATP-dependent remodeling is not required to maintain nucleosome positioning or hypersensitivity (31, 61). Consistent with these findings, analysis of DHS in cells is frequently performed using isolated nuclei in the absence of ATP. Here we found that BRG1 was rapidly lost from activation-repressed genes, and, consistent with our earlier work, we found that BRG1 was recruited to activation-induced genes. We suggest that BRG1 is required to establish new chromatin structures or to reestablish transient chromatin structures, leading to gene activation. These changes could be increases or decreases in accessibility, depending on the locus. We found that the remodeling ATPase SNF2H could increase and decrease accessibility at different loci in the same cells (63). The results of cell-free chromatin studies also support the idea that remodeling enzymes can decrease accessibility under certain circumstances (31, 61). A corollary of these ideas is that the epigenetic memory resides at least in part with the transcription factors that recruit BRG1 to sites of action rather than to the chromatin structure per se.
Early long-term studies of Th cell clones suggested that these cells are terminally differentiated and that the corresponding forms of lineage-specific cytokine expression are mutually exclusive (52). However, this long-held view has been questioned recently by studies that demonstrate that more flexibility is built into Th subset specification than had been appreciated before (58). We suggest that dynamic but lineage-restricted BRG1 recruitment to sites in the Th transcription factor and cytokine loci facilitates plasticity. As BRG1 responds to both activation and cytokine signals, BRG1 can integrate extracellular signals through rapid changes in chromatin structure.
The transcriptional regulation of Gata3 is complex. Gata3 is required for CD4+ T cell development (27, 81) and determination of Th2 fate (90) as well as for heart, brain, kidney, and mammary epithelia. Within differentiated Th cells, Gata3 expression is elevated in the Th2 subset. Loss of Gata3 results in embryonic lethality (60). A 120-kb Gata3 genomic fragment delays lethality by merely a single day, while a 625-kb fragment that rescues Gata3 expression at many regions in the body plan lacks the sequences required to direct expression in the thymus and other locations, suggesting that the boundaries of the Gata3 gene and the regulatory elements required to direct Gata3 expression in T cells are not yet known (39). We found Th2-specific BRG1 binding to numerous sites in the Gata3 locus as far as 760 kb downstream of the promoter. BRG1 binding did not obviously correlate with the presence of H3K4me3 or H3K4me1 markers, and recent investigations of the monomethyl marker suggest that, unlike the results seen with BRG1 binding, it does not respond to the activation state of cells (22, 35). However, the distal BRG1 binding sites have been shown to correlate with nuclease hypersensitivity in Th2 cells as well as with enhancer-like activity in a chromatin-based reporter assay in T cells. Additionally, BRG1 binding was reduced or absent at many of these distal sites in the absence of Stat6 (data not shown). These findings suggest that these elements may represent previously unidentified cis-regulatory regions required for Gata3 expression in Th2 cells, many of which reside hundreds of kilobases from the Gata3 promoters. Given that in the Gata3 region there are 760 CNSs but only 27 BRG1 binding regions, 16 of which are Th2 specific, identification of BRG1 binding regions might be an efficient way to identify positive cis-acting regions.
We detected many BRG1 binding regions that were distal from transcription start sites (TSS). Frequently, these correlated with expression of a nearby gene. In some cases (e.g., Il17a and Il17f), there were only distal binding regions and no binding regions at the promoter. This is consistent with the recent suggestion that enhancer chromatin is more cell specific than are promoters (28). In other cases, we found the distal sites were in housekeeping genes and that BRG1 binding correlated with expression of the neighboring lineage-specific or activation-specific gene rather than the housekeeping gene. We hypothesize that cis-regulatory regions are often found in neighboring genes. We found BRG1 binding to known distal regulatory regions in the Th2 cluster, some located in the Rad50 DNA repair gene. We also found that binding to distal regions positively correlated with gene activity in a genome-wide manner. Finally, the distal regions we tested all possessed enhancer activity. Taking these findings together, this suggests that at least some of the distal sites are functional. In this way, BRG1 is a key component in the molecular logic of T cell signal response.
Supplementary Material
ACKNOWLEDGMENTS
We thank Sebastian Fugmann, Nan-ping Weng, and Rebecca Potts for helpful discussions, Weidong Wang for providing J1 antisera, and Keji Zhao for pRep vectors. We thank the reviewers for very helpful suggestions.
This research was supported by the Intramural Research Program of the NIH, National Institute on Aging.
We declare no competing financial interests.
Footnotes
Supplemental material for this article may be found at http://mcb.asm.org/.
Published ahead of print on 24 January 2011.
REFERENCES
- 1. Agarwal S., Rao A. 1998. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity 9:765–775 [DOI] [PubMed] [Google Scholar]
- 2. Akimzhanov A. M., Yang X. O., Dong C. 2007. Chromatin remodeling of interleukin-17 (IL-17)-IL-17F cytokine gene locus during inflammatory helper T cell differentiation. J. Biol. Chem. 282:5969–5972 [DOI] [PubMed] [Google Scholar]
- 3. Amsen D., Spilianakis C. G., Flavell R. A. 2009. How are T(H)1 and T(H)2 effector cells made? Curr. Opin. Immunol. 21:153–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ansel K. M., Djuretic I., Tanasa B., Rao A. 2006. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 24:607–656 [DOI] [PubMed] [Google Scholar]
- 5. Ansel K. M., et al. 2004. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 5:1251–1259 [DOI] [PubMed] [Google Scholar]
- 6. Araki Y., et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity 30:912–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Aune T. M., Collins P. L., Chang S. 2009. Epigenetics and T helper 1 differentiation. Immunology 126:299–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avni O., et al. 2002. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 3:643–651 [DOI] [PubMed] [Google Scholar]
- 9. Baguet A., Bix M. 2004. Chromatin landscape dynamics of the Il4-Il13 locus during T helper 1 and 2 development. Proc. Natl. Acad. Sci. U. S. A. 101:11410–11415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Banerjee A., Schambach F., DeJong C. S., Hammond S. M., Reiner S. L. 2010. Micro-RNA-155 inhibits IFN-gamma signaling in CD4+ T cells. Eur. J. Immunol. 40:225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barski A., et al. 2009. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 19:1742–1751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Biddie S. C., John S., Hager G. L. 2010. Genome-wide mechanisms of nuclear receptor action. Trends Endocrinol. Metab. 21:3–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chi T. H., et al. 2003. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity 19:169–182 [DOI] [PubMed] [Google Scholar]
- 14. Chi T. H., et al. 2002. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418:195–199 [DOI] [PubMed] [Google Scholar]
- 15. Clapier C. R., Cairns B. R. 2009. The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78:273–304 [DOI] [PubMed] [Google Scholar]
- 16. Crawford G. E., et al. 2004. Identifying gene regulatory elements by genome-wide recovery of DNase hypersensitive sites. Proc. Natl. Acad. Sci. U. S. A. 101:992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. de la Serna I. L., et al. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Du C., et al. 2009. MicroRNA miR-326 regulates T(H)-17 differentiation and is associated with the pathogenesis of multiple sclerosis. Nat. Immunol. 10:1252–1259 [DOI] [PubMed] [Google Scholar]
- 19. Fields P. E., Kim S. T., Flavell R. A. 2002. Cutting edge: changes in histone acetylation at the IL-4 and IFN-gamma loci accompany Th1/Th2 differentiation. J. Immunol. 169:647–650 [DOI] [PubMed] [Google Scholar]
- 20. Fields P. E., Lee G. R., Kim S. T., Bartsevich V. V., Flavell R. A. 2004. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity 21:865–876 [DOI] [PubMed] [Google Scholar]
- 21. Flaus A., Martin D. M., Barton G. J., Owen-Hughes T. 2006. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34:2887–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghisletti S., et al. 2010. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32:317–328 [DOI] [PubMed] [Google Scholar]
- 23. Gilbert N., et al. 2004. Chromatin architecture of the human genome: gene-rich domains are enriched in open chromatin fibers. Cell 118:555–566 [DOI] [PubMed] [Google Scholar]
- 24. Grogan J. L., Locksley R. M. 2002. T helper cell differentiation: on again, off again. Curr. Opin. Immunol. 14:366–372 [DOI] [PubMed] [Google Scholar]
- 25. Gross D. S., Garrard W. T. 1988. Nuclease hypersensitive sites in chromatin. Annu. Rev. Biochem. 57:159–197 [DOI] [PubMed] [Google Scholar]
- 26. Hadjur S., et al. 2009. Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460:410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hattori N., Kawamoto H., Fujimoto S., Kuno K., Katsura Y. 1996. Involvement of transcription factors TCF-1 and GATA-3 in the initiation of the earliest step of T cell development in the thymus. J. Exp. Med. 184:1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Heintzman N. D., et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ho L., Crabtree G. R. 2010. Chromatin remodelling during development. Nature 463:474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ho L., et al. 2009. An embryonic stem cell chromatin remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U. S. A. 106:5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ishii H., et al. 2009. Mi2beta shows chromatin enzyme specificity by erasing a DNase I-hypersensitive site established by ACF. J. Biol. Chem. 284:7533–7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jani A., et al. 2008. A novel genetic strategy reveals unexpected roles of the Swi-Snf-like chromatin-remodeling BAF complex in thymocyte development. J. Exp. Med. 205:2813–2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ji H., et al. 2008. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat. Biotechnol. 26:1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kidder B. L., Palmer S., Knott J. G. 2009. SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem Cells 27:317–328 [DOI] [PubMed] [Google Scholar]
- 35. Kim T. K., et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Koyanagi M., et al. 2005. EZH2 and histone 3 trimethyl lysine 27 associated with Il4 and Il13 gene silencing in Th1 cells. J. Biol. Chem. 280:31470–31477 [DOI] [PubMed] [Google Scholar]
- 37. Lai D., et al. 2009. Induction of TLR4-target genes entails calcium/calmodulin-dependent regulation of chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 106:1169–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lake R. J., Geyko A., Hemashettar G., Zhao Y., Fan H. Y. 2010. UV-induced association of the CSB remodeling protein with chromatin requires ATP-dependent relief of N-terminal autorepression. Mol. Cell 37:235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lakshmanan G., et al. 1999. Localization of distant urogenital system-, central nervous system-, and endocardium-specific transcriptional regulatory elements in the GATA-3 locus. Mol. Cell. Biol. 19:1558–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Lee G. R., Spilianakis C. G., Flavell R. A. 2005. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 6:42–48 [DOI] [PubMed] [Google Scholar]
- 41. Letimier F. A., Passini N., Gasparian S., Bianchi E., Rogge L. 2007. Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rbeta2 expression during human Th1 cell differentiation. EMBO J. 26:1292–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Liao W., et al. 2008. Priming for T helper type 2 differentiation by interleukin 2-mediated induction of interleukin 4 receptor alpha-chain expression. Nat. Immunol. 9:1288–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lim P. S., et al. 2009. Defining the chromatin signature of inducible genes in T cells. Genome Biol. 10:R107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Liu R., et al. 2001. Regulation of CSF1 promoter by the SWI/SNF-like BAF complex. Cell 106:309–318 [DOI] [PubMed] [Google Scholar]
- 45. Mahy N. L., Perry P. E., Bickmore W. A. 2002. Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J. Cell Biol. 159:753–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mallappa C., et al. 2010. Myogenic microRNA expression requires ATP-dependent chromatin remodeling enzyme function. Mol. Cell. Biol. 30:3176–3186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mikkelsen T. S., et al. 2007. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mohrmann L., Verrijzer C. P. 2005. Composition and functional specificity of SWI2/SNF2 class chromatin remodeling complexes. Biochim. Biophys. Acta 1681:59–73 [DOI] [PubMed] [Google Scholar]
- 49. Mohrs M., et al. 2001. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2:842–847 [DOI] [PubMed] [Google Scholar]
- 50. Monticelli S., et al. 2005. MicroRNA profiling of the murine hematopoietic system. Genome Biol. 6:R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morlando M., et al. 2008. Primary microRNA transcripts are processed co-transcriptionally. Nat. Struct. Mol. Biol. 15:902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Mosmann T. R., Cherwinski H., Bond M. W., Giedlin M. A., Coffman R. L. 1986. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136:2348–2357 [PubMed] [Google Scholar]
- 53. Mowen K. A., Glimcher L. H. 2004. Signaling pathways in Th2 development. Immunol. Rev. 202:203–222 [DOI] [PubMed] [Google Scholar]
- 54. Murphy K. M., Reiner S. L. 2002. The lineage decisions of helper T cells. Nat. Rev. Immunol. 2:933–944 [DOI] [PubMed] [Google Scholar]
- 55. Ochs H. D., Oukka M., Torgerson T. R. 2009. TH17 cells and regulatory T cells in primary immunodeficiency diseases. J. Allergy Clin. Immunol. 123:977–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ohguro H., et al. 1993. Beta-arrestin and arrestin are recognized by autoantibodies in sera from multiple sclerosis patients. Proc. Natl. Acad. Sci. U. S. A. 90:3241–3245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Reference deleted. [Google Scholar]
- 58. O'Shea J. J., Paul W. E. 2010. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science 327:1098–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ozsolak F., et al. 2008. Chromatin structure analyses identify miRNA promoters. Genes Dev. 22:3172–3183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Pandolfi P. P., et al. 1995. Targeted disruption of the GATA3 gene causes severe abnormalities in the nervous system and in fetal liver haematopoiesis. Nat. Genet. 11:40–44 [DOI] [PubMed] [Google Scholar]
- 61. Pazin M. J., Bhargava P., Geiduschek E. P., Kadonaga J. T. 1997. Nucleosome mobility and the maintenance of nucleosome positioning. Science 276:809–812 [DOI] [PubMed] [Google Scholar]
- 62. Placek K., et al. 2009. Integration of distinct intracellular signaling pathways at distal regulatory elements directs T-bet expression in human CD4+ T cells. J. Immunol. 183:7743–7751 [DOI] [PubMed] [Google Scholar]
- 63. Precht P., Wurster A. L., Pazin M. J. 2010. The SNF2H chromatin remodeling enzyme has opposing effects on cytokine gene expression. Mol. Immunol. 47:2038–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ramirez-Carrozzi V. R., et al. 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138:114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Reiner S. L. 2007. Development in motion: helper T cells at work. Cell 129:33–36 [DOI] [PubMed] [Google Scholar]
- 66. Rodriguez A., et al. 2007. Requirement of bic/microRNA-155 for normal immune function. Science 316:608–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sabo P. J., et al. 2004. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl. Acad. Sci. U. S. A. 101:16837–16842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Shi Y., et al. 2007. Critical regulation of CD4+ T cell survival and autoimmunity by beta-arrestin 1. Nat. Immunol. 8:817–824 [DOI] [PubMed] [Google Scholar]
- 69. Sif S., Stukenberg P. T., Kirschner M. W., Kingston R. E. 1998. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 12:2842–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Smith C. L., Hager G. L. 1997. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J. Biol. Chem. 272:27493–27496 [DOI] [PubMed] [Google Scholar]
- 71. Solymar D. C., Agarwal S., Bassing C. H., Alt F. W., Rao A. 2002. A 3′ enhancer in the IL-4 gene regulates cytokine production by Th2 cells and mast cells. Immunity 17:41–50 [DOI] [PubMed] [Google Scholar]
- 72. Sotelo J., et al. 2010. Long-range enhancers on 8q24 regulate c-Myc. Proc. Natl. Acad. Sci. U. S. A. 107:3001–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Spies N., Nielsen C. B., Padgett R. A., Burge C. B. 2009. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 36:245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stritesky G. L., Yeh N., Kaplan M. H. 2008. IL-23 promotes maintenance but not commitment to the Th17 lineage. J. Immunol. 181:5948–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Takemoto N., et al. 2000. Cutting edge: chromatin remodeling at the IL-4/IL-13 intergenic regulatory region for Th2-specific cytokine gene cluster. J. Immunol. 165:6687–6691 [DOI] [PubMed] [Google Scholar]
- 76. Tang Y., et al. 2009. MicroRNA-146A contributes to abnormal activation of the type I interferon pathway in human lupus by targeting the key signaling proteins. Arthritis Rheum. 60:1065–1075 [DOI] [PubMed] [Google Scholar]
- 77. Tarakhovsky A. 2010. Tools and landscapes of epigenetics. Nat. Immunol. 11:565–568 [DOI] [PubMed] [Google Scholar]
- 78. Thai T. H., et al. 2007. Regulation of the germinal center response by microRNA-155. Science 316:604–608 [DOI] [PubMed] [Google Scholar]
- 79. Thompson M. 2009. Polybromo-1: the chromatin targeting subunit of the PBAF complex. Biochimie 91:309–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Tillo D., et al. 2010. High nucleosome occupancy is encoded at human regulatory sequences. PLoS One 5:e9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ting C. N., Olson M. C., Barton K. P., Leiden J. M. 1996. Transcription factor GATA-3 is required for development of the T-cell lineage. Nature 384:474–478 [DOI] [PubMed] [Google Scholar]
- 82. Wan M., et al. 2009. Molecular basis of CD4 repression by the Swi/Snf-like BAF chromatin remodeling complex. Eur. J. Immunol. 39:580–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Wan Y. Y., Flavell R. A. 2009. How diverse—CD4 effector T cells and their functions. J. Mol. Cell. Biol. 1:20–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Weaver C. T., Murphy K. M. 2007. The central role of the Th17 lineage in regulating the inflammatory/autoimmune axis. Semin. Immunol. 19:351–352 [DOI] [PubMed] [Google Scholar]
- 85. Wei G., et al. 2009. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 30:155–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wei L., et al. 2010. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32:840–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Witte J. S. 2007. Multiple prostate cancer risk variants on 8q24. Nat. Genet. 39:579–580 [DOI] [PubMed] [Google Scholar]
- 88. Wurster A. L., Pazin M. J. 2008. BRG1-mediated chromatin remodeling regulates differentiation and gene expression of T helper cells. Mol. Cell. Biol. 28:7274–7285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhang F., Boothby M. 2006. T helper type 1-specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-gamma promoter are Stat4 dependent. J. Exp. Med. 203:1493–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Zheng W., Flavell R. A. 1997. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell 89:587–596 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.