Abstract
The chaperone-related AAA ATPase Cdc48 (p97/VCP in higher eukaryotes) segregates ubiquitylated proteins for subsequent degradation by the 26S proteasome or for nonproteolytic fates. The specific outcome of Cdc48 activity is controlled by the evolutionary conserved cofactors Ufd2 and Ufd3, which antagonistically regulate the substrates' ubiquitylation states. In contrast to the interaction of Ufd3 and Cdc48, the interaction between the ubiquitin chain elongating enzyme Ufd2 and Cdc48 has not been precisely mapped. Consequently, it is still unknown whether physiological functions of Ufd2 in fact require Cdc48 binding. Here, we show that Ufd2 binds to the C-terminal tail of Cdc48, unlike the human Ufd2 homologue E4B, which interacts with the N domain of p97. The binding sites for Ufd2 and Ufd3 on Cdc48 overlap and depend critically on the conserved residue Y834 but are not identical. Saccharomyces cerevisiae cdc48 mutants altered in residue Y834 or lacking the C-terminal tail are viable and exhibit normal growth. Importantly, however, loss of Ufd2 and Ufd3 binding in these mutants phenocopies defects of Δufd2 and Δufd3 mutants in the ubiquitin fusion degradation (UFD) and Ole1 fatty acid desaturase activation (OLE) pathways. These results indicate that key cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation require their interaction with Cdc48.
INTRODUCTION
The ubiquitin-proteasome system is the central system for targeted proteolysis in eukaryotic cells. It is not only responsible for bulk protein degradation and protein quality control but also for regulatory proteolysis in numerous cellular processes (25, 26, 53). Furthermore, in addition to proteasomal targeting, covalent modification of proteins with ubiquitin can also elicit nonproteolytic signals (63).
Cdc48 (also known as p97 and VCP in higher eukaryotes) is a central component of many cellular processes involving protein ubiquitylation, both in proteasomal and in nonproteolytic pathways (8, 58). In Saccharomyces cerevisiae, these include the ubiquitin fusion degradation (UFD) (19), endoplasmic reticulum-associated degradation (ERAD) (5, 7, 30, 51, 66), and Ole1 fatty acid desaturase activation (OLE) (52, 60) pathways, transcriptional control (52, 64), the regulation of cell cycle progression at several steps (3, 10, 11, 17, 18, 42), the fusion of endoplasmic reticulum (ER) membranes (36), and ribophagic degradation of the 60S ribosomal subunit (48). Cdc48 is a hexameric member of the AAA+ ATPase superfamily (23). Each monomer consists of an aminoterminal N domain, two ATPase domains, D1 and D2, and an unstructured C-terminal tail. The common mechanism underlying the different cellular functions of Cdc48 is believed to be its “segregase” activity, i.e., the conversion of energy from ATP hydrolysis into conformational changes that mediate the extraction of substrates from their cellular environment (7, 23).
The distinct functions of Cdc48 in diverse cellular processes are tightly controlled by a large number of cofactors (8, 58). Two classes of cofactors can be distinguished. Substrate-recruiting cofactors act as adaptor proteins for substrates and control their binding to Cdc48 in space and time. In contrast, substrate-processing cofactors control the ubiquitylation state of substrates after their recruitment to Cdc48 (9, 56). Yeast Ufd2 and Ufd3 are prototypical substrate-processing cofactors of Cdc48 and possess homologues throughout the eukaryotic kingdom. UFD3 (also known as DOA1) was first identified in a genetic screen for yeast mutants defective in the degradation of the α2 transcriptional repressor (27). Subsequently, UFD3 was identified again, together with UFD2, in a screen for mutants defective in the degradation of the short-lived UFD substrate, ubiquitin-proline-β-galactosidase (Ub-P-β-Gal) (31). Further analysis revealed that the degradation defects of ufd3 mutants are caused by reduced cellular ubiquitin pools and can be suppressed by ubiquitin overexpression (31, 47). The basis underlying ubiquitin depletion in ufd3 mutants is not precisely known but is likely to reflect the inability to recycle ubiquitin in the context of proteasomal protein degradation (47, 56). Δufd3 strains exhibit a number of stress phenotypes and negative genetic interactions, many of which are an indirect consequence of ubiquitin depletion (32, 39, 44, 47, 56). However, defects of Δufd3 in the monoubiquitylation of histone H2B (39), in the sorting of ubiquitylated membrane proteins into multivesicular bodies for vacuolar degradation (54), and in ribophagy (48) were shown to persist upon ubiquitin overexpression, suggesting that Ufd3 is directly involved in these processes.
Ufd2 consists of several Armadillo-like repeats and a C-terminal U box (61), a RING finger-related domain defining a small subfamily of ubiquitin ligases (2). Consistently, Ufd2 was found to catalyze the formation of long ubiquitin chains on Ub-P-β-Gal (35). Intriguingly, this multiubiquitylation activity was strictly dependent on prior mono- or oligoubiquitylation of the substrate by the E3 ubiquitin ligase Ufd4 and was thus termed “E4 activity” (35). In addition to the UFD pathway, the E4 activity of Ufd2 plays important roles in the ERAD and OLE pathways (55, 56). In the latter, the proteolytically processed active p90 form of the Spt23 transcription factor has been proposed to be protected from multiubiquitylation by Ufd3 and the deubiquitylating enzyme Otu1, another cofactor of Cdc48 (56). According to this model, the monoubiquitylated form of p90 is not degraded by the 26S proteasome and can drive the expression of the OLE1 gene, encoding Δ9 fatty acid desaturase. Subsequently, p90 is inactivated by Ufd2-mediated multiubiquitylation and proteasomal degradation. The opposing activities of Ufd2 and Ufd3 with respect to substrate ubiquitylation, together with the finding that both cofactors bind Cdc48 in a mutually exclusive manner, lead to the current model that substrate-processing cofactors can actively regulate the fate of Cdc48 substrates (56).
The physical basis for the interaction between Cdc48 and Ufd3 and between their respective mammalian homologues p97 and phospholipase A2-activating protein (PLAA [also known as PLAP]) has been analyzed in detail (44, 50, 67, 68). Two recent crystallographic studies revealed that the PUL domain of Ufd3/PLAA consists of six Armadillo repeats that bind to the unstructured C terminus of Cdc48/p97 (50, 67). Both groups employed structure-based site-directed mutagenesis to generate ufd3 mutants defective in Cdc48 binding but chose different mutations and arrived at partially opposite conclusions regarding the requirement of Cdc48 binding for cellular functions of Ufd3. Zhao et al. showed that the degradation of Ub-P-β-Gal depends on the interaction between Cdc48 and Ufd3 (67), whereas Qiu et al. concluded from their phenotypic analysis that at least some cellular functions of Ufd3 do not require Cdc48 binding (50).
In contrast to those of Ufd3 and Cdc48, the binding sites of Ufd2 and Cdc48 for each other have not been precisely mapped on either protein. Whereas the human Ufd2 homologue E4B binds to the N domain of p97 (15), yeast Ufd2 has been shown to interact with a Cdc48 fragment lacking the N domain (56). Consequently, it has not been possible to specifically abrogate the interaction between Ufd2 and Cdc48 in vivo in order to address the important open question of whether the E4 activity of Ufd2 requires Cdc48 binding under physiological conditions. In this study, we have mapped the binding sites of Ufd2 and Ufd3 to overlapping residues at the C-terminal tail of Cdc48. Based on these results, we generated and analyzed cdc48 mutant strains defective in Ufd2 and Ufd3 interactions and found that the cellular functions of both cofactors depend on their binding to Cdc48.
MATERIALS AND METHODS
Plasmids.
Plasmids for the bacterial expression of glutathione S-transferase (GST) fusion proteins of yeast Ufd2 and Ufd3 (56), human E4B (15), the PUB domain of human PNGase (1), and the UBX domain of Shp1 (59), for the bacterial expression of hexahistidine fusion proteins of full-length p97 and p97ΔN lacking residues 1 to 199 (33), for the expression of Gal4 activation domain fusions of Ufd2 (35) and Ufd3 (56) in yeast, and for the galactose-inducible expression of Ub-P-β-Gal (4) and GST-His6-Pex29 (40) in yeast have been described previously.
For the bacterial expression of Cdc48 as an N-terminal hexahistidine fusion protein, the coding sequence lacking the start codon was PCR amplified and cloned into the KasI and HindIII sites of pPROEX-HTa (Invitrogen). For the bacterial expression of human PLAA spanning codons 330 to 775 (PLAA330-795), the sequence spanning codon 330 to the stop codon was cloned into the BamHI and EcoRI sites of pGEX-4T2 (GE Healthcare) and mini-pRSETA (49). For yeast two-hybrid experiments, the coding sequences of C-terminal Cdc48 fragments were cloned into the BamHI and PstI sites of pGBD-C1 (29). For the generation of yeast shuffle strains, the CDC48 gene, including the promoter, coding sequence, and terminator, was cloned into the BamHI and PstI sites of YCplac33 and YCplac22 (20). For the expression of N-terminally myc epitope-tagged Cdc48 in yeast, the ADH1 promoter, a cassette encoding a 3× myc epitope tag, and the coding sequence and terminator of wild-type CDC48 and the cdc48-Y834E allele were cloned into the high-copy-number plasmid YEplac112 (20). Cdc48 variants mutated in residue Y834 or lacking the C-terminal six amino acid residues were generated by site-specific mutagenesis using a QuikChange II XL kit (Stratagene) or by appropriate design of PCR primers for cloning. For the generation of the ufd3-R541A,R669A mutant strain, the UFD3 gene, including the promoter, coding sequence, and terminator, was cloned into the KpnI and SphI sites of YIplac128 (20). Codons 541 and 669 were mutated to alanine using the QuikChange II XL kit. The coding sequences of all constructs were confirmed by sequencing.
Yeast strains.
The strains used in this study are listed in Table 1. Δufd2 and Δufd3 mutants were generated by disruption of the respective coding sequences in the parental DF5a strain using standard techniques (62). CDC48 shuffle strains were constructed by generating a DF5a/α heterozygote with an exact deletion of one of the two chromosomal CDC48 coding sequences (62), followed by transformation with the centromeric plasmid YCplac33-CDC48 carrying a URA3 marker. Following sporulation and tetrad dissection, haploid MATa progeny carrying the chromosomal deletion of CDC48 was transformed with the centromeric plasmid YCplac22 carrying wild-type CDC48 or cdc48 mutant alleles and a TRP1 marker. Counterselection against YCplac33 on 5-fluoroorotic acid plates resulted in strains carrying the respective YCplac22-derived plasmids as the sole source of Cdc48. Alternatively, for some experiments, strains carrying mutations in the chromosomal CDC48 locus were constructed as follows (21, 22). A loxP-LEU2-loxP knockout cassette was PCR amplified from pUG73 using primers with flanking sequences corresponding to the 3′ region of the CDC48 coding sequence, with codon 834 encoding tyrosine, phenylalanine, or glutamic acid. The PCR products were transformed into the DF5a/α strain and transformants were selected for integration of the loxP-LEU-loxP cassette at the 3′ end of the coding sequence of one chromosomal CDC48 locus. LEU-positive transformants were transformed with pSH47 for the galactose-inducible expression of Cre recombinase, grown in the presence of galactose, and selected for loss of the LEU marker. Subsequently, pSH47 was removed by counterselection on 5-fluoroorotic acid plates, and haploid progeny was obtained by sporulation and tetrad dissection. The identities of the CDC48, cdc48-Y834F, and cdc48-Y834E coding sequences followed by a single loxP site were verified by PCR amplification of the corresponding chromosomal region, followed by sequencing of the PCR products. The ufd3-R541A,R669A mutant strain was generated by linearization of YIplac128-ufd3-R541A,R669A at the ClaI site and integration into the leu2 locus of the Δufd3 strain YAB1173.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| PJ69-4a | MATatrp1-901 leu2-3,112 ura3-52 his3Δ200 gal4Δ gal80Δ GAL2-ADE2 LYS2::GAL1-HIS3 met2::GAL7-lacZ | 29 |
| DF5a | MATaura3-52 leu2-3,112 lys2-801 trp1-1 his3Δ200 | 16 |
| YAB1173 | DF5a Δufd3::HIS3MX6 | This work |
| YAB1170 | DF5a Δufd3::HIS3MX6 YIplac128-ufd3-R541A,R669A::LEU2 | This work |
| YAB1157 | DF5a Δufd2::HIS3MX6 | This work |
| YAB1164 | DF5a Δufd2::hph-NT1 | This work |
| YAB1158 | DF5a CDC48::loxP | This work |
| YAB1159 | DF5a cdc48-Y834E::loxP | This work |
| YAB1160 | DF5a cdc48-Y834F::loxP | This work |
| YAB1020 | DF5a Δcdc48::HIS3MX6 YCplac22-CDC48 | This work |
| YAB1021 | DF5a Δcdc48::HIS3MX6 YCplac22-cdc48-Y834E | This work |
| YAB1022 | DF5a Δcdc48::HIS3MX6 YCplac22-cdc48-Y834F | This work |
| YAB1023 | DF5a Δcdc48::HIS3MX6 YCplac22-cdc48-Y834A | This work |
| YAB1024 | DF5a Δcdc48::HIS3MX6 YCplac22-cdc48ΔC6 | This work |
| YAB1165 | DF5a Δcdc48::HIS3MX6Δufd2::hph-NT1 YCplac22-CDC48 | This work |
| YAB1166 | DF5a Δcdc48::HIS3MX6Δufd2::hph-NT1 YCplac22-cdc48-Y834E | This work |
| YAB1167 | DF5a Δcdc48::HIS3MX6Δufd2::hph-NT1 YCplac22-cdc48-Y834F | This work |
| YAB1168 | DF5a Δcdc48::HIS3MX6Δufd2::hph-NT1 YCplac22-cdc48-Y834A | This work |
| YAB1169 | DF5a Δcdc48::HIS3MX6Δufd2::hph-NT1 YCplac22-cdc48ΔC6 | This work |
| Δptp1 strain | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0Δptp1::kanMX4 | EUROSCARF |
| PTP1 | MATahis3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PTP1 | EUROSCARF |
Proteins and peptides.
Recombinant GST and hexahistidine fusion proteins were expressed in Escherichia coli BL21(DE3) pRIL and purified on glutathione and Ni-nitrilotriacetic acid (Ni-NTA) affinity matrices, respectively, using standard protocols. p97 and E4B were purified exactly as described previously (15). The hexahistidine tag of Cdc48 fusion proteins was removed by incubation with TEV protease, resulting in the following N terminus for the untagged Cdc48 proteins: Gly-Ala-(derived from KasI restriction site)-Gly2, etc. Purified proteins were dialyzed against Tris-buffered saline (TBS) and 5 mM dithiothreitol (DTT) (Ufd2 and Ufd3) or 50 mM HEPES-NaOH [pH 7.8], 100 mM NaCl, 5 mM MgCl2, 1 mM 2-mercaptoethanol, and 10% glycerol (Cdc48, p97, and E4B), flash frozen in liquid nitrogen, and stored at −80°C. Cdc48-derived peptides C11 (biotin-SNAEEDDDLYS-COOH), C11-pY (biotin-SNAEEDDDLpYS-COOH), C11-A (biotin-SNAEEDDDLAS-COOH), and C17 (biotin-AGAAFGSNAEEDDDLYS-COOH), the p97-derived peptide C16 (biotin-TGGSVYTEDNDDDLYG-COOH), and the E4B-derived VCP binding motive (VBM) peptide (acetyl-SADEIRRRRLARLAG-NH2) were synthesized in-house and purified to >90% purity by high-pressure liquid chromatography (HPLC). Peptides were dissolved in and dialyzed against TBS, flash frozen in liquid nitrogen, and stored at −80°C. Peptide concentrations were determined photometrically and by quantitative amino acid analysis.
In vitro binding assays.
Glutathione-Sepharose pulldown experiments were performed exactly as described previously (33), using 0.76 nmol GST fusion proteins, 20 μl glutathione-Sepharose, and 0.1 nmol Cdc48 or p97. In competition experiments, Cdc48 or p97 was preincubated for 10 min at room temperature with a 50-fold or 75-fold (see Fig. 2b) molar excess of competitor peptide or hexahistidine-tagged PLAA330-795 (see Fig. 2b) relative to the GST fusion proteins. Streptavidin-Sepharose pulldown experiments were performed in analogy to the glutathione-Sepharose pulldown experiments, using 10 nmol biotinylated peptides, 20 μl streptavidin-Sepharose, and 0.76 nmol GST fusion proteins.
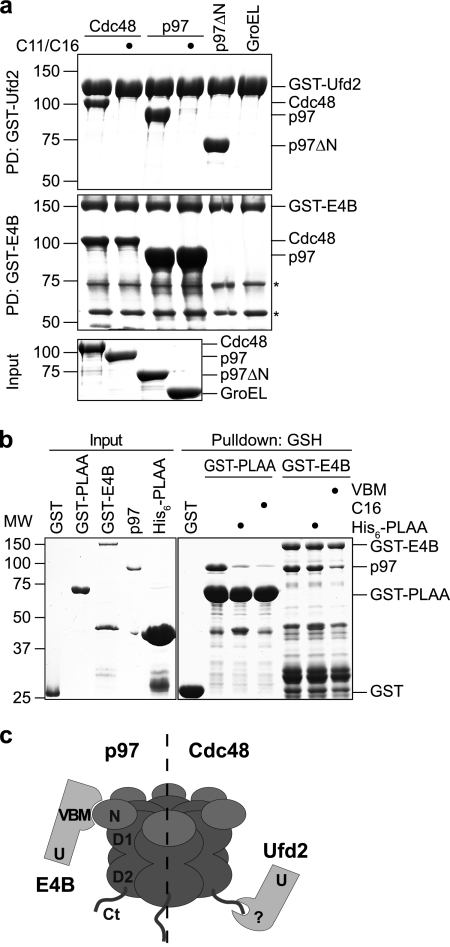
Fig. 2.
The binding mode of Ufd2 is not conserved in mammals. (a) Glutathione-Sepharose pulldown (PD) assay. Yeast Cdc48 and human p97 in the absence or presence of a 100-fold molar excess of the respective C-terminal peptide (C11 for Cdc48 and C16 for p97) as indicated, a human p97 fragment lacking the N domain (residues 1 to 199; p97ΔN), and the bacterial chaperone GroEL were incubated with immobilized GST fusion proteins of yeast Ufd2 (PD: GST-Ufd2 [upper panel]) and human E4B (PD: GST-E4B [middle panel]). Binding was analyzed by SDS-PAGE, followed by Coomassie brilliant blue staining. The bottom panel shows the input levels. Asterisks in the middle panel indicate degradation products of GST-E4B. (b) Human PLAA does not compete with E4B for binding to p97. Glutathione-Sepharose pulldown assays with GST-PLAA (residues 330 to 795) and GST-E4B were performed in the absence or presence of excess hexahistidine-tagged PLAA (residues 330 to 795; His6-PLAA), p97 C-terminal peptide (C16), and an E4B-derived VBM peptide as indicated and analyzed by SDS-PAGE, followed by Coomassie brilliant blue staining. (c) Model illustrating different binding modes between E4B-p97 and Ufd2-Cdc48. In yeast, Ufd2 binds via an unknown site (?) to the C-terminal tail (Ct) of Cdc48 (right). In higher eukaryotes, E4B binds via a VBM to the N domain of p97 (left).
Yeast two-hybrid assays.
Yeast two-hybrid interactions were analyzed exactly as described previously (33).
Antibodies.
Rabbit polyclonal antibodies were raised against recombinant, full-length yeast Cdc48, Ufd2, Ufd3, and Shp1 proteins and affinity purified on antigen-coupled Sepharose matrices using standard protocols. Antibodies against yeast Ufd1 and Ubx2 have been described previously (57). The following commercially available antibodies were used: anti-Gal4BD (RK5C1; sc-510; Santa Cruz), 4G10 (Millipore), PY20 (Abcam), anti-β-galactosidase (Promega), unspecific rabbit IgGs (Bethyl Laboratories), anti-GST (GE Healthcare), anti-RGS-His (Qiagen), and anti-myc (9E10; M5546; Sigma).
Fluorescence-activated cell sorter analysis.
Yeast strains were grown in yeast extract-peptone-dextrose (YPD) at 25°C to an optical density at 600 nm (OD600) between 0.6 and 0.8. Cells corresponding to an OD600 of 0.6 were harvested, fixed overnight in 70% ethanol, sonicated, and incubated for at least 8 h with 0.5 mg/ml RNase A (Sigma) at 37°C. The DNA was stained overnight with 50 μg/ml propidium iodide in phosphate-buffered saline (PBS). The propidium iodide concentration was decreased to 5 μg/ml, and the samples were sonicated, diluted 4-fold with PBS, and analyzed on a Beckman Coulter FC500 flow cytometer, with 10,000 total ungated events counted.
Immunoprecipitation.
Yeast cultures were grown in YPD to an OD600 of 1. Cells were harvested and lysed using glass beads in buffer B (50 mM Tris-HCl [pH 7.5], 100 mM KCl, 5 mM MgCl2, 0.1% NP-40, 10% glycerol, 10 mM NaF, 1 mM sodium orthovanadate, 2 mM phenylmethylsulfonyl fluoride [PMSF], 2 mM benzamidine, complete protease inhibitor cocktail [Roche]). The NP-40 concentration was increased to 1%, and the extracts were centrifuged at 2,600 × g for 5 min and then at 20,000 × g for 30 min. The supernatants were incubated with 1 μg anti-Ufd2, anti-Ufd3, anti-myc, or anti-Cdc48 antibody or with 2 μg unspecific rabbit IgGs overnight. Immunocomplexes were immobilized on protein A-Sepharose beads (GE Healthcare), and bound proteins were analyzed by immunoblotting.
For the immunoprecipitation experiment whose results are shown in Fig. 4d, buffer B was replaced with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, 1 mM EDTA) containing 60 mM β-glycerophosphate, 2 mM benzamidine, 1 mM sodium orthovanadate, 10 mM NaF, 2 mM PMSF, 20 mM N-ethylmaleimide (NEM), and complete protease inhibitor cocktail. No additional detergent was added after glass bead lysis.
Fig. 4.
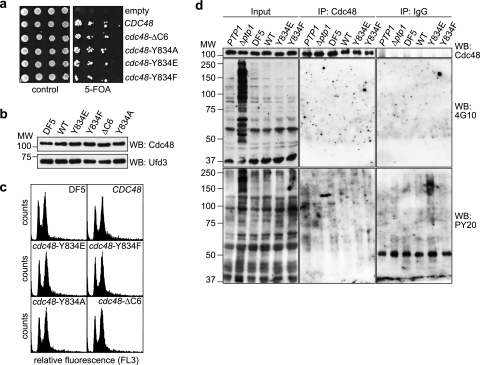
Residue Y834 is not required for normal growth and is not significantly phosphorylated. (a) Generation of cdc48 shuffle strains. Δcdc48 cells carrying a URA3-based CDC48 expression plasmid together with a TRP1-based plasmid for the expression of the indicated wild-type and mutant CDC48 alleles were spotted in serial dilutions onto control plates or plates containing 5-fluoroorotic acid (5-FOA) to counterselect against the URA3-based CDC48 plasmid. (b) Western blot (WB) analysis of Cdc48 expression levels in the parental DF5 strain and the shuffle strains. The Ufd3 blot serves as a loading control. (c) Fluorescence-activated cell sorter analysis. The cell cycle distribution of logarithmically growing cells of the parental DF5 strain and the indicated shuffle strains was analyzed by fluorescence-activated cell sorting. (d) Analysis of Y834 phosphorylation. Cdc48 was immunoprecipitated under stringent conditions from lysates of the parental DF5 strain and the indicated shuffle strains. Immunoprecipitates were analyzed by Western blot analysis using the phosphotyrosine-specific antibodies 4G10 and PY20 and an anti-Cdc48 antibody as indicated. Lysates of the phosphotyrosine phosphatase-deficient Δptp1 strain accumulating phosphotyrosine-modified proteins and an isogenic PTP1 strain served as positive controls for the specificity of 4G10 and PY20. Immunoprecipitations (IPs) using unspecific IgGs served as negative controls. Note that very long exposure times resulting in significant background staining were deliberately chosen for the 4G10 and PY20 blots of the IP samples in order to check for even very weak signals.
Cycloheximide shutoff experiments.
For Ub-P-β-Gal degradation assays, yeast strains harboring plasmid pUb-P-β-Gal were grown in synthetic complete galactose medium lacking uracil to an OD600 of 0.8. The expression of Ub-P-β-Gal was shut off by changing the medium to YPD, and translation was simultaneously stopped by adding cycloheximide to a final concentration of 0.5 mg/ml. For each time point, yeast cells corresponding to an OD600 of 1 were harvested, and protein extracts were prepared as described previously (34). Ub-P-β-Gal levels were detected by Western blot analysis and quantified using ImageJ (W. S. Rasband, National Institutes of Health, Bethesda, MD; http://rsb.info.nih.gov/ij/).
For GST-His6-Pex29 degradation assays, yeast strains harboring pEGH-Pex29 (40) were grown in synthetic complete raffinose medium lacking uracil to an OD600 of 0.8, and the expression of GST-His6-Pex29 was induced for 3 h by addition of galactose. Expression was shut off as described above, and for each time point, yeast cells corresponding to an OD600 of 30 were harvested. Cells were lysed in buffer C (50 mM HEPES [pH 7.5], 5 mM EDTA, 150 mM NaCl, 1% Triton X-100, 2 mM PMSF, complete protease inhibitor cocktail) using glass beads, and extracts were incubated with 20 μl glutathione-Sepharose 4B (GE Healthcare). Glutathione-Sepharose pulldown experiments were performed as described previously (33), and bound material was eluted by heating in HU buffer (34). GST-His6-Pex29 was detected by Western blot analysis and quantified as described above.
RESULTS
Ufd2 and Ufd3 bind to the C-terminal tail of Cdc48.
In an effort to identify binding partners of the carboxyl-terminal region of Cdc48, we performed two-hybrid screens using a bait comprising the 50 C-terminal residues. The only robust interactors identified were the known substrate-processing cofactors Ufd2 and Ufd3 (data not shown). To narrow down their binding site on Cdc48, we performed direct two-hybrid interaction tests using a series of C-terminal fragments of Cdc48 (Fig. 1a). Both Ufd2 and Ufd3 interacted with Cdc48 fragments spanning at least the 15 C-terminal residues but not with fragments lacking the last 10 or 20 residues. Remarkably, Ufd2 also interacted with Cdc48 fragments spanning the last 10 and, very weakly, the last 5 residues, whereas Ufd3 failed to interact with these fragments, suggesting that Ufd2 and Ufd3 bind to overlapping but nonidentical sites at the C terminus of Cdc48. All Cdc48 two-hybrid constructs were expressed to similar levels (Fig. 1b), excluding the possibility of false-negative results.
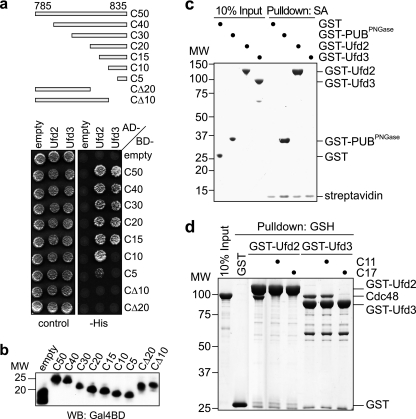
Fig. 1.
Ufd2 and Ufd3 bind to the carboxyl-terminal tail of Cdc48. (a) Yeast two-hybrid analysis. Yeast PJ69-4A cells transformed with the indicated combinations of bait (AD) and prey (BD) plasmids were spotted onto synthetic complete medium agar plates lacking uracil and leucine (control) or lacking uracil, leucine, and histidine (−His). Growth was monitored after 3 days. The analyzed C-terminal fragments of Cdc48 between residues 785 and 835 are schematically shown at the top. (b) Bait protein levels in panel a were analyzed by Western blot (WB) analysis using an antibody against the Gal4 DNA binding domain (WB: Gal4BD) (c) Streptavidin (SA)-Sepharose pulldown assay using a biotinylated peptide spanning the 11 C-terminal residues of Cdc48 and the indicated recombinant GST fusion proteins. Binding to the immobilized peptide was analyzed by SDS-PAGE, followed by Coomassie brilliant blue staining of the gel. PUBPNGase, PUB domain of human peptide-N-glycanase. (d) Glutathione (GSH)-Sepharose pulldown assay using recombinant Cdc48 and GST fusion proteins of Ufd2 and Ufd3. Incubation of the immobilized GST fusion proteins with Cdc48 was performed in the absence or presence of a 50-fold molar excess of the indicated Cdc48 C-terminal peptides (C11 and C17). Binding of Cdc48 was analyzed by SDS-PAGE, followed by Coomassie brilliant blue staining of the gel. MW, molecular weight (in thousands).
In order to verify that Ufd2 and Ufd3 bind directly to the C terminus of Cdc48, we performed in vitro pulldown experiments (Fig. 1c) using a synthetic peptide spanning the 11 C-terminal residues of Cdc48 (C11). The peptide was immobilized on streptavidin-Sepharose beads via an N-terminal biotin group and incubated with recombinant, purified GST-Ufd2 and GST-Ufd3 fusion proteins. As a positive control, we used a GST fusion of the PUB domain from human peptide-N-glycanase (PNGase), which binds with high affinity to a highly homologous C-terminal peptide from human p97 (68; our unpublished data). GST-Ufd2 and the positive control bound to the C11 peptide with similar efficiencies, demonstrating direct, robust binding of Ufd2 to the C terminus of Cdc48. In contrast, no binding of GST-Ufd3 could be observed in this assay, despite the well-documented binding of the human Ufd3 homologue PLAA to the corresponding p97-derived peptide (50, 67, 68). In order to avoid potential steric hindrance due to the streptavidin-Sepharose matrix, we analyzed binding of GST-Ufd2 and GST-Ufd3 to full-length, recombinant Cdc48 in a glutathione-Sepharose pulldown assay in the presence of excess Cdc48 C-terminal peptides as competitors (Fig. 1d). Binding of Cdc48 to GST-Ufd2 was completely abrogated by peptides spanning the 11 or 17 C-terminal residues of Cdc48, in line with the robust binding of GST-Ufd2 to the immobilized C11 peptide. Of note, an excess of peptide C17, but not peptide C11, abolished binding of Cdc48 to GST-Ufd3 in this assay. This result is consistent with our two-hybrid interaction data for Ufd3 (Fig. 1a) and with a previous report demonstrating (i) efficient binding of the Ufd3 PUL domain to a Cdc48-derived C14 peptide and (ii) a 10-fold-higher affinity of a p97-derived C13 peptide than that of a C10 peptide for the PUL domain of mouse PLAA (67). In summary, our data show that the C-terminal region of Cdc48 is necessary and sufficient for direct binding to both Ufd2 and Ufd3. They furthermore demonstrate that the interactions of Ufd2 and Ufd3 with Cdc48 differ in detail, inasmuch as additional critical determinants for Ufd3 binding are located between residues 820 and 824 of Cdc48. Finally, the competition experiments with excess peptide indicate that the C-terminal tail of Cdc48 is the only binding site for Ufd2 and Ufd3.
The binding mode of Ufd2 is not conserved in mammals.
Whereas overlapping binding sites for Ufd2 and Ufd3 had previously been postulated based on their competitive binding to Cdc48 (56), the finding that Ufd2 binds to the C-terminal tail of Cdc48 was still puzzling in light of the well-characterized binding of its mammalian homologue E4B/Ufd2a to the N domain of p97 via a VCP binding motive (VBM) (6, 15, 43). In order to elucidate this apparent discrepancy, we directly compared the binding modes of the yeast and human binding partners in parallel glutathione-Sepharose pulldown experiments using purified, recombinant proteins (Fig. 2 a). Yeast GST-Ufd2 bound robustly to Cdc48 as well as to human full-length p97 and a p97 variant lacking its N domain but not to the unrelated bacterial chaperone GroEL. These interactions were efficiently competed by an excess of the C11 and C16 peptides derived from the C termini of Cdc48 and p97, respectively, indicating that the heterologous binding between yeast Ufd2 and human p97 is exclusively mediated by the highly conserved C terminus of p97 and not by its N domain. Conversely, human GST-E4B bound robustly to p97 and somewhat weaker to yeast Cdc48 but not to p97 lacking its N domain. Importantly, these interactions could not be suppressed by competition with the corresponding C-terminal peptides, indicating that they are not mediated by the C-terminal tails of Cdc48 and p97, respectively. If E4B and PLAA interact with different sites on p97, then their binding to p97 should, unlike that of the yeast homologs (56), not be mutually exclusive. Indeed, in a competition pulldown experiment using GST-E4B and GST-PLAA330-795, an excess of a hexahistidine fusion of PLAA330-795 competed for binding of p97 to GST-PLAA330-795 but not to GST-E4B, while an excess of an E4B-derived VBM peptide did (Fig. 2b). In summary, these results show for the first time directly that the evolutionarily highly conserved binding partners Cdc48/p97 and Ufd2/E4B interact via nonconserved binding sites; in yeast, the C terminus of Cdc48 binds to a yet-unknown site in Ufd2, while in mammals, the N domain of p97 binds to the VBM of E4B (Fig. 2c).
Cdc48 residue Y834 is essential for Ufd2 and Ufd3 binding.
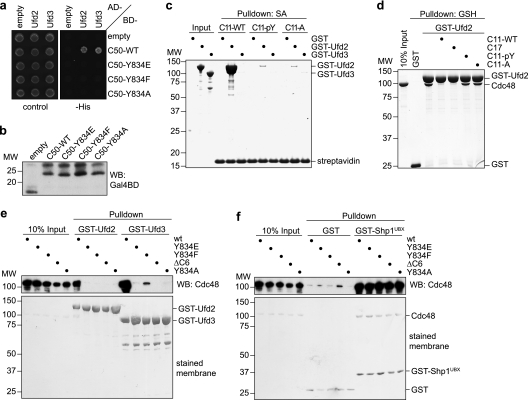
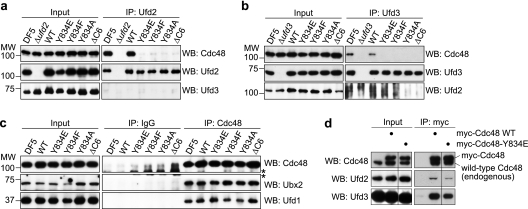
Binding of human PNGase and PLAA to the C terminus of p97 has been shown to be sensitive to phosphorylation or mutation of the penultimate tyrosine residue Y805 of p97 (68). We therefore tested the importance of the corresponding residue Y834 of Cdc48 for binding of Ufd2 and Ufd3. In a two-hybrid assay using the C-terminal 50-residue fragment of Cdc48, replacement of the tyrosine residue with glutamate, phenylalanine, or alanine completely abolished the interaction with Ufd2 and Ufd3 (Fig. 3a and b), indicating that residue Y834 is indeed critical for the binding of both cofactors. In a streptavidin-Sepharose pulldown assay with biotinylated C11 peptides, the interaction with GST-Ufd2 was almost completely lost upon replacement of Y834 with phosphotyrosine or alanine (Fig. 3c). Consistently, the C11 peptides altered in Y834 were unable to compete with full-length Cdc48 for binding to GST-Ufd2 in a glutathione pulldown assay (Fig. 3d). As GST-Ufd3 does not bind to wild-type C11 under these conditions (see above), the importance of Y834 for the interaction with GST-Ufd3 could not be analyzed by this approach. To further confirm the central role of Y834 in cofactor binding, we performed glutathione-Sepharose pulldown assays using mutant Cdc48 proteins carrying the Y834E, Y834F, or Y834A amino acid changes or lacking the six C-terminal residues (ΔC6) (Fig. 3e). Whereas wild-type Cdc48 bound efficiently to GST-Ufd2, the Cdc48 mutant proteins completely failed to interact with GST-Ufd2. Similarly, the Cdc48 mutant proteins were unable to bind GST-Ufd3, with the exception of the Cdc48-Y834F protein, which exhibited a strongly reduced binding (Fig. 3e). All wild-type and mutant Cdc48 proteins were proficient in binding to a GST fusion of the UBX domain from the N domain binding cofactor Shp1 (59) but not to GST alone, excluding the possibility that the mutations cause gross structural defects in Cdc48 (Fig. 3f). Together, these results show that residue Y834 plays a pivotal role in the binding of Cdc48 to Ufd2 and Ufd3 and that mutation or phosphorylation of Y834 leads to a complete or very substantial loss of cofactor binding.
Fig. 3.
Tyrosine residue Y834 of Cdc48 is essential for binding of Ufd2 and Ufd3. (a) Yeast two-hybrid analysis. Yeast PJ69-4A cells transformed with the indicated combinations of bait (AD) and prey (BD) plasmids were analyzed as for Fig. 1a. (b) Western blot analysis of bait expression levels. (c) Streptavidin-Sepharose pulldown assay. Binding of GST fusion proteins of Ufd2 and Ufd3 to the indicated C-terminal Cdc48 peptides immobilized to streptavidin-Sepharose beads was analyzed as for Fig. 1c. C11-WT, wild-type C11 peptide; C11-pY, C11 with phosphotyrosine at the position corresponding to Y834; C11-A, C11 with alanine at the position corresponding to Y834. (d) Glutathione-Sepharose competition pulldown assay. Binding of Cdc48 to GST-Ufd2 in the absence or presence of a 50-fold molar excess of the indicated C-terminal Cdc48 peptides was analyzed as for Fig. 1d. (e and f) Glutathione-Sepharose pulldown assays. Binding of the indicated wild-type and mutant Cdc48 proteins to immobilized GST fusion proteins of Ufd2 or Ufd3 (e) and the UBX domain of Shp1 or GST only (f) was analyzed as described for Fig. 1d.
The C-terminal tail and residue Y834 of Cdc48 are dispensable for viability.
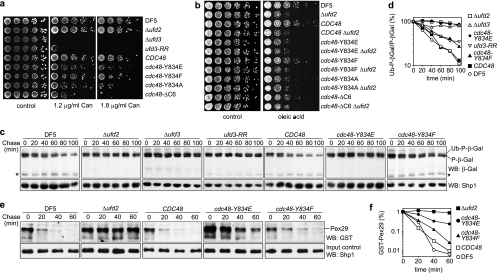
We next wished to investigate the importance of the C-terminal tail of Cdc48 in vivo. To that end, we generated yeast shuffle strains expressing wild-type CDC48 or the mutant cdc48-Y834E, cdc48-Y834F, cdc48-Y834A, and cdc48ΔC6 alleles under the control of the authentic CDC48 promoter and terminator sequences. All shuffle strains are viable (Fig. 4a), express Cdc48 at levels similar to that of the DF5 parental strain (Fig. 4b), and grow with identical generation times in rich medium (data not shown). Furthermore, the cell cycle distributions of the shuffle strains are indistinguishable from each other (Fig. 4c). Together, these data rule out the possibility that the mutant cdc48 alleles are deficient in any essential cellular function of Cdc48.
Residue Y805 of mammalian p97 is a well-documented phosphorylation target (14, 37, 38), and yeast Cdc48 was reported to be phosphorylated on residue Y834 as well (41). To test whether our shuffle strains are affected in Cdc48 phosphorylation, we prepared total cell lysates under denaturing conditions and performed Western blot analyses with the phosphotyrosine-specific antibodies 4G10 and PY20. 4G10 and PY20 have been successfully used for the detection of p97 phosphorylation on Y805 (28, 37; E. Chevet, personal communication), and 4G10 was used by Madeo et al. to detect phosphotyrosine-modified Cdc48 (41; K.-U. Fröhlich, personal communication). Surprisingly, however, in our hands neither antibody detected a signal in the 100-kDa region that differed between strains expressing wild-type and mutant Cdc48 (data not shown). In addition, we were unable to detect any systematic differences in the migration pattern of wild-type and Y834-mutated Cdc48 in Western blot analyses using purified Cdc48 antibody (Fig. 4d and data not shown). To further support these results, we immunoprecipitated endogenous wild-type and mutant Cdc48 under stringent conditions in the presence of phosphatase inhibitors and performed Western blot analyses using 4G10 and PY20 (Fig. 4d). As a control, we included a strain lacking the broad-specificity phosphotyrosine phosphatase Ptp1 (65). In the Δptp1 lysate, an accumulation of phosphotyrosine-modified proteins in addition to presumably unspecific background bands was detected by 4G10 and, less prominently, PY20, proving that the experimental conditions were principally compatible with the detection of phosphotyrosine-modified proteins (Fig. 4d, Input). However, despite the efficient immunoprecipitation of Cdc48, neither antibody produced a signal in the 100-kDa region that was specifically missing in strains expressing mutant Cdc48 (Fig. 4d). We conclude that phosphorylation of Y834 can at best affect a very minor subpopulation of Cdc48 that evaded our detection methods.
Next, we analyzed the interaction of the untagged wild-type and mutant Cdc48 proteins expressed in the shuffle strains with endogenous, untagged Ufd2 and Ufd3 in immunoprecipitation experiments using purified antibodies directed against Ufd2 and Ufd3 (Fig. 5 a and b). Whereas wild-type Cdc48 expressed in the CDC48 shuffle strain bound to Ufd2 and Ufd3 as efficiently as in the DF5 parental strain, none of the mutant Cdc48 proteins coimmunoprecipitated with Ufd2 or Ufd3. In contrast, Cdc48 immunoprecipitation experiments showed that the mutant Cdc48 proteins were unaffected in their ability to coimmunoprecipitate the N domain binding cofactors Ufd1 and Ubx2 (Fig. 5c). Together, these results are highly consistent with the binding experiments performed in vitro. They show for the first time that the C-terminal tail and specifically residue Y834 of Cdc48 are required for a stable interaction with Ufd2 and Ufd3 under physiological conditions, and they indicate that the shuffle strains are valid tools for studying the consequences of loss of Ufd2 and Ufd3 binding in vivo.
Fig. 5.
cdc48 C-terminal mutants are specifically deficient in Ufd2 and Ufd3 binding. (a and b) Immunoprecipitation of endogenous Ufd2 (a) and Ufd3 (b). Lysates (Input) of the DF5 parental strain and the indicated shuffle strains were subjected to immunoprecipitation (IP) with Ufd2 and Ufd3 antibodies, respectively. Coimmunoprecipitation of Cdc48 with Ufd2 and Ufd3 was analyzed by Western blot (WB) analysis. Lysates from Δufd2 (a) and Δufd3 (b) strains served as negative controls. Additionally, Western blots against Ufd3 (a) and Ufd2 (b) demonstrate the specificity of the IPs. (c) Immunoprecipitation of endogenous Cdc48. The binding of the N domain binding cofactors Ufd1 and Ubx2 to wild-type and mutant Cdc48 was analyzed as described above. IPs with unspecific IgGs served as negative controls. The asterisks indicate unspecific background signals from the IgG chains. (d) Immunoprecipitation of myc epitope-tagged wild-type and Y834E mutant Cdc48 expressed in the DF5 parental strain expressing endogenous wild-type Cdc48. Binding of Ufd2 and Ufd3 to the myc immunoprecipitates was analyzed as described above. The separating vertical line indicates that an irrelevant lane on the original blot was cut out. Note that all lanes of a panel are from the same blot and were processed identically, allowing us to directly compare the amounts of precipitated proteins.
To obtain further insight into the requirements for Ufd2 and Ufd3 binding to Cdc48, we performed anti-myc immunoprecipitations with lysates of the DF5 parental yeast strain expressing N-terminally myc epitope-tagged wild-type Cdc48 or the Cdc48-Y834E mutant protein in addition to endogenous, untagged wild-type Cdc48 (Fig. 5d). The presence of untagged Cdc48 in the immunoprecipitates indicates that mixed hexamers consisting of myc-tagged and untagged protomers were readily formed in vivo. Thus, every single hexamer in the myc-Cdc48-Y834E immunoprecipitate must contain at least one mutated protomer. The coprecipitation of reduced but significant amounts of Ufd2 and Ufd3 with myc-Cdc48-Y834E proves that a single mutant protomer is insufficient to abolish Ufd2/Ufd3 binding. In fact, the overall reduction of Ufd2/Ufd3 binding would be consistent with the possibility that cofactor binding is proportional to the content of wild-type protomers. However, because the actual distribution of differently composed mixed hexamers in the immunoprecipitate cannot be determined, one can merely conclude that between one and five wild-type protomers per Cdc48 hexamer are required for Ufd2/Ufd3 binding, even though the latter extreme is highly unlikely.
Key cellular functions of Ufd2 and Ufd3 require binding to Cdc48.
The absence of any general growth defects in the cdc48 shuffle strains enabled us to specifically analyze the effects of mutating the C-terminal tail of Cdc48 on known activities of Ufd2 and Ufd3 in vivo. ufd3 mutants are sensitive to a number of stress conditions, mostly as an indirect consequence of reduced ubiquitin pools in these strains (32, 39, 44, 47, 56). We compared the sensitivities of our shuffle strains to the arginine analogue canavanine with that of a Δufd3 strain (Fig. 6a). Whereas the CDC48 wild-type shuffle strain showed growth on canavanine plates similar to that of the DF5 parental strain, the cdc48 mutant shuffle strains were hypersensitive to canavanine, with the cdc48-Y834F strain being less sensitive than the cdc48-Y834E, cdc48-Y834A, and cdc48ΔC6 strains. The ufd3-R541A,R669A mutant strain expressing the Ufd3-R541A,R669A protein impaired in Cdc48 binding (67) was slightly more sensitive to canavanine than the cdc48 mutant strains (Fig. 6a). This difference in canavanine sensitivity could be due to differential ubiquitin depletion, which in Δufd3 has been shown to be attenuated upon additional deletion of UFD2 (56). Thus, the cdc48 mutants are likely to exhibit less severe ubiquitin depletion than ufd3-R541A,R669A because of their inability to bind both Ufd2 and Ufd3. In contrast to the hypersensitive ufd3 strains, Δufd2 cells grew normally on canavanine plates, indicating that the hypersensitivity of the cdc48 shuffle strains can be fully attributed to the impaired Ufd3 binding of the mutant Cdc48 proteins.
Fig. 6.
cdc48 mutants deficient in Ufd2 and Ufd3 binding phenocopy Δufd2 and Δufd3 mutants. (a and b) Sensitivity to canavanine and oleic acid. Cultures of the indicated wild-type and mutant strains were serially diluted and spotted onto agar plates containing the indicated canavanine (Can) concentrations (a) or 0.2% oleic acid (b) or the respective control plates. Plates were incubated at 30°C (a) or 33°C (b) for 4 days. ufd3-RR, ufd3 mutant strain expressing Ufd3-R541A,R669A impaired in Cdc48 binding. (c) Degradation of a UFD substrate. Protein expression in logarithmically growing cultures of the indicated wild-type and mutant strains expressing ubiquitin-proline-β-galactosidase (Ub-P-β-Gal) under the control of a galactose-induced promoter was stopped by transfer to glucose-containing medium and addition of cycloheximide. The degradation of Ub-P-β-Gal over time was analyzed by Western blot analysis. Note the existence of a very long-lived P-β-Gal and of a metastable β-Gal degradation product (asterisk). A Western blot against Shp1 served as a loading control. (d) Quantification of the results in panel c. The signal of Ub-P-β-Gal was normalized to the signal of the stable P-β-Gal species and plotted against time. (e) Degradation of a bona fide ERAD substrate. Degradation of GST-His6-Pex29 expressed under the control of a galactose-induced promoter was analyzed as described above. At the indicated time points, GST-His6-Pex29 was pulled down with glutathione-Sepharose beads and detected in Western blots against GST. A Western blot against Shp1 served as a control for equal input of cell lysates into the pulldown. Note that GST-His6-Pex29 could be detected only after enrichment by pulldown and not directly in the lysates. (f) Quantification of results in panel e.
Ufd2 is involved in the degradation of proteins via the UFD, OLE, and ERAD pathways (9, 31, 45, 55, 56). In the OLE pathway, Ufd2 is required for the efficient degradation of the activated Spt23 transcription factor, which drives the expression of the OLE1 gene encoding Δ9 fatty acid desaturase. Consequently, Δufd2 cells are hypersensitive to elevated oleic acid concentrations in the medium (55). Growth of the CDC48 shuffle strain on plates containing oleic acid was comparable to that of the DF5 parental strain, whereas the cdc48 mutant shuffle strains exhibited different sensitivities (Fig. 6b). While the cdc48-Y834E, cdc48-Y834A, and cdc48-ΔC6 strains were as hypersensitive as the Δufd2 strain, the cdc48-Y834F strain was less sensitive than the other mutant strains. As Δufd3 cells are not hypersensitive to oleic acid (56; data not shown), the observed hypersensitivity of the cdc48 shuffle strains is specifically attributable to the impaired Ufd2 binding in these mutants. Importantly, deletion of UFD2 in the cdc48 mutant strains did not aggravate their oleic acid hypersensitivity, thus formally proving that the function of Ufd2 in the OLE pathway is strictly dependent on Cdc48 binding.
In order to directly analyze the consequences of impaired cofactor binding in the cdc48 mutant strains on protein degradation, we determined the in vivo half-life of the UFD model substrate Ub-P-β-Gal by performing cycloheximide shutoff experiments (Fig. 6c and d). The CDC48 wild-type shuffle strain was proficient in Ub-P-β-Gal degradation, as judged by degradation kinetics that were indistinguishable from those of the DF5 parental strain (Fig. 6c and d) and by the appearance of a β-galactosidase fragment diagnostic of its degradation (Fig. 6c, asterisk). In contrast, Ub-P-β-Gal was stable in the Δufd2 and Δufd3 strains, as expected (19, 31). Importantly, the cdc48-Y834E strain showed a strong stabilization of Ub-P-β-Gal that was comparable to that of the Δufd2 and Δufd3 strains, indicating that binding of Ufd2 and/or Ufd3 to Cdc48 is required for their function(s) in the UFD pathway. The cdc48-Y834F strain again showed an intermediate phenotype in that Ub-P-β-Gal was degraded over time, albeit with slower kinetics (Fig. 6d). Interestingly, the ufd3-R541A,R669A mutant strain also exhibited intermediate degradation kinetics, suggesting either that the UFD phenotype of Δufd3 is partially Cdc48 independent or, more likely, that some residual Cdc48 binding to Ufd3-R541A,R669A exists, perhaps bridged by the overexpressed UFD substrate.
Finally, we analyzed the degradation kinetics of the recently characterized Ufd2-dependent bona fide ERAD substrate Pex29 (40) in the cdc48 mutant strains. In line with the results noted above, Pex29 was strongly stabilized in the cdc48-Y834E strain and moderately but significantly stabilized in the cdc48-Y834F strain (Fig. 6e and f).
In summary, the results of the sensitivity assays and degradation assays show that known cellular functions of the Cdc48 substrate-processing cofactors Ufd2 and Ufd3 depend critically on their interaction with Cdc48.
DISCUSSION
In this study, we mapped the binding of the prototypical substrate-processing cofactors Ufd2 and Ufd3 to overlapping but nonidentical sites at the C terminus of Cdc48. We showed for the first time that the C-terminal tail of Cdc48 is both necessary and sufficient for Ufd2 binding and that it is the only binding site for Ufd2. Importantly, the mapping of the binding sites allowed us to generate cdc48 mutant yeast strains defective in Ufd2 and Ufd3 binding and to demonstrate that key cellular functions of Ufd2 and Ufd3 depend on their interaction with Cdc48.
Within the six C-terminal residues of Cdc48 mediating binding to Ufd2 and Ufd3, residue Y834 was found to be of particular importance. The homologous residue Y805 of p97 has previously been shown to be critical for binding of the structurally unrelated PUB and PUL domains to p97 (68). We show here that Y834 is crucial for Ufd2 binding as well, identifying this residue as a key determinant for binding of all cofactors known to interact with the C terminus of Cdc48/p97. Importantly, however, our results indicate that Ufd3, in contrast to Ufd2, possesses additional binding determinants, in agreement with a recent study reporting that a p97-C10 peptide has a lower affinity for the PUL domain of PLAA than either p97-C13 or full-length p97 (67). The higher affinity of the longer peptide suggests that critical contacts between the PUL domain and p97 may be missing in a recent crystal structure of the PUL domain from PLAA (50). This structure was obtained with the p97-C10 peptide and revealed contacts between the PUL domain and residues L804 and Y805 of p97, whereas the five highly conserved acidic residues preceding L804 were disordered. It is tempting to speculate that residues upstream of the sequence covered by p97-C10 make additional contacts with the PUL domain, thus permitting electrostatic interactions between the highly conserved acidic p97 residues and a conserved positively charged groove of the PUL domain adjacent to the binding site for L804 and Y805.
Cell cycle-dependent phosphorylation of Cdc48 residue Y834 has been reported to control subcellular localization of Cdc48 and cell growth (41). We failed to detect significant phosphorylation of Y834 using Cdc48 and phosphotyrosine antibodies that sensitively detected their respective antigens (Fig. 4d), suggesting that, at best, only a very minor subpopulation of Cdc48 is phosphorylated on Y834. Using strains that express mutated Cdc48 under the control of the CDC48 promoter at wild-type levels, we also failed to detect a slow-growth phenotype for the cdc48-Y834F mutant, in contrast to Madeo et al. (41), and demonstrated normal cell cycle progression for all mutants tested (Fig. 4c). These results clearly indicate that either phosphorylation of Y834 is not required for the cell cycle-dependent localization of Cdc48 or cell cycle-dependent localization of Cdc48 is not essential for normal cell cycle progression. However, a potential physiological role of Y834 phosphorylation (if existent) in controlling cofactor binding cannot be formally excluded, in part because the binding stoichiometry between Cdc48 and Ufd2/Ufd3 in vivo is unknown. We found that wild-type and Y834E protomers form mixed Cdc48 hexamers that still bind Ufd2 and Ufd3, proving that Ufd2/Ufd3 binding is not abolished by a single mutant protomer and suggesting that one or a few wild-type protomers per hexamer may be sufficient for Ufd2/Ufd3 binding (Fig. 5d). If a single unphosphorylated C-terminal binding site within a Cdc48 hexamer were indeed sufficient for a normal interaction with Ufd2 and Ufd3, then an efficient binding control would require the concerted phosphorylation of Y834 on all six subunits of the hexamer. This scenario is highly unlikely to represent a major mechanism for the control of Ufd2/Ufd3 binding, given our failure to detect significant levels of Y834 phosphorylation in unstressed, logarithmically growing yeast cells even upon enrichment of Cdc48 by immunoprecipitation (Fig. 4d). It should be noted though that our analysis of tyrosine phosphorylation in whole-cell extracts does not exclude the formal possibility that very small subpopulations of Cdc48 are phosphorylated on all six Y834 residues to control cofactor interactions in a spatially and/or temporally restricted manner. Such a regulatory mechanism would presumably rely on a pulse of tyrosine kinase activity, and phosphorylation of p97 residue Y805 is indeed induced in response to T cell activation (13). The future identification of the kinase(s) catalyzing the phosphorylation of Cdc48 on residue Y834 (if existent) will be a prerequisite to further analyzing the significance of Y834 phosphorylation in yeast.
This study shows that cdc48 mutants altered in the Ufd2/Ufd3 binding site are impaired in the OLE, UFD, and ERAD pathways and in cellular stress responses, indicating that Cdc48 binding is required for well-known cellular functions of Ufd2 and Ufd3. Ufd2 is a prototypical E4 ubiquitin chain elongating enzyme and Cdc48 substrate-processing cofactor (35, 56). Nevertheless, its ability to directly interact with E2 ubiquitin-conjugating enzymes (46) as well as with the ubiquitin binding protein Rad23 (24, 55) raised the possibility that Ufd2 may bind and multiubiquitylate preubiquitylated substrates independently of Cdc48. However, the phenotypes of the cdc48 mutants clearly indicate that Ufd2 cannot function independently of Cdc48 in the OLE and UFD protein degradation pathways (Fig. 6b to d) and suggest that all cellular functions of Ufd2 depend on Cdc48 binding. This would imply that all Ufd2 substrates are also Cdc48 substrates, a prediction supported by a recent study characterizing novel Ufd2 substrates (40).
Our phenotypic analysis of canavanine sensitivities indicated that cdc48 C-terminal mutants are nearly as sensitive as the ufd3-R541A,R669A mutant expressing Ufd3 mutated in residues R541 and R669 but less sensitive than the Δufd3 mutant (Fig. 6a). While these data might suggest that the defects leading to the hypersensitivity of the Δufd3 mutant are partially Cdc48 independent, the interpretation is complicated by the fact that phenotypes of Δufd3 are attenuated by deletion of UFD2 (56). Because the cdc48 C-terminal mutants are deficient in Ufd2 and Ufd3 binding, it is not possible to differentiate between the possibilities of additional Cdc48-independent defects in the Δufd3 mutant on one hand and attenuation caused by impaired Ufd2 binding in the cdc48 mutants on the other hand. A recent study employing a ufd3 strain mutated in Ufd3 residues I483, D538, and L571 came to the conclusion that Cdc48-independent functions of Ufd3 exist (50). While more detailed structural insights into the interaction between Cdc48 and Ufd3 have to await a cocrystal structure analysis of both full-length proteins, the phenotypic analysis by Qiu et al. is difficult to reconcile, as the well-characterized canavanine hypersensitivity of ufd3 mutants (Fig. 6a) (39, 44) was not observed in their experimental system (50). The authors also failed to demonstrate mutual loss of binding for the ufd3 and cdc48 mutants used, leaving the possibility that residual interactions between Cdc48 and Ufd3 caused the lack of some phenotypes. Thus, while we cannot formally exclude the existence of Cdc48-independent functions of Ufd3, unequivocal evidence supporting such a possibility is lacking.
Interestingly, the Y834F mutation caused only a partial loss of function in vivo, whereas the phenotypes of all other cdc48 C-terminal mutants resembled those of Δufd2 and Δufd3 mutants. A likely explanation is that the Y834F mutation does not completely abolish Cdc48 binding to Ufd2 and Ufd3. While atomic details of the interaction between Cdc48 and Ufd2 are not available, the interaction between the phenolic ring of residue Y834/Y805 with the PUB and PUL domains is mediated by both hydrogen bonds of the hydroxyl group and van der Waals interactions of the aromatic ring (50, 68). The latter may well be preserved in the Y834F mutant but not the other mutants, raising the possibility of a weak residual binding of Y834F to Ufd2 and Ufd3 that would be consistent with its milder phenotypes. Indeed, weak binding of Y834F suggestive of residual, transient interactions could be observed for Ufd3 in vitro (Fig. 3e) and, albeit variably, for Ufd2 in some but not all immunoprecipitation experiments (data not shown).
Surprisingly, and in contrast to that of Ufd3, the Cdc48 binding site of Ufd2 is not conserved throughout evolution. In higher eukaryotes, binding relies on the interaction of a VBM in Ufd2/E4B with the N domain of Cdc48/p97 (Fig. 2) (6, 15, 43). Remarkably, E4B does not bind to the C-terminal tail of p97 (15), even though the sequence of the tail is extremely conserved from yeast to humans. This suggests that an active counterselection against the (elusive) Cdc48 binding site of yeast Ufd2 must have existed during evolution. One can only speculate about potential evolutionary driving forces behind this unusual finding. However, it seems plausible that the transformation of fungal Ufd2 proteins into N domain interactors in higher eukaryotes was coupled to the emergence of the PUB domain, which binds to the C-terminal tail of Cdc48/p97 and is found in all eukaryotes but fungi (12). Perhaps the benefits of a diversification of Cdc48/p97-cofactor complexes and their cellular functions gained from PUB domain proteins outweighed the costs of having to adapt to a new Ufd2 binding mechanism.
ACKNOWLEDGMENTS
We thank Stefan Jentsch for continued support, Yvonne Hufnagel and Sebastian Höpfl for excellent technical assistance, Hermann Schindelin for critical reading of the manuscript, Gang Zhao for providing reagents, and Chang Liu and Hai Rao for providing reagents and for experimental advice.
Financial support for this work was provided by the Deutsche Forschungsgemeinschaft (grant BU951/3-1 to A.B.), the European Union (Erasmus fellowship to G.L.), and the Basque Government (postdoctoral fellowship to V.F.-S.).
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1. Allen M. D., Buchberger A., Bycroft M. 2006. The PUB domain functions as a p97 binding module in human peptide N-glycanase. J. Biol. Chem. 281:25502–25508 [DOI] [PubMed] [Google Scholar]
- 2. Aravind L., Koonin E. V. 2000. The U box is a modified RING finger—a common domain in ubiquitination. Curr. Biol. 10:R132–134 [DOI] [PubMed] [Google Scholar]
- 3. Archambault V., et al. 2004. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell 14:699–711 [DOI] [PubMed] [Google Scholar]
- 4. Bachmair A., Finley D., Varshavsky A. 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234:179–186 [DOI] [PubMed] [Google Scholar]
- 5. Bays N. W., Wilhovsky S. K., Goradia A., Hodgkiss-Harlow K., Hampton R. Y. 2001. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Mol. Biol. Cell 12:4114–4128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Boeddrich A., et al. 2006. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. EMBO J. 25:1547–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Braun S., Matuschewski K., Rape M., Thoms S., Jentsch S. 2002. Role of the ubiquitin-selective CDC48(UFD1/NPL4) chaperone (segregase) in ERAD of OLE1 and other substrates. EMBO J. 21:615–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Buchberger A. 2006. Cdc48 (p97) and its cofactors, p. 194–211In Mayer R. J., Ciechanover A. J., Rechsteiner M. (ed.), Protein degradation, vol. 3: cell biology of the ubiquitin-proteasome system. Wiley-VCH, Weinheim, Germany [Google Scholar]
- 9. Buchberger A. 2010. Control of ubiquitin conjugation by Cdc48 and its cofactors. Subcell. Biochem. 54:17–30 [DOI] [PubMed] [Google Scholar]
- 10. Cao K., Nakajima R., Meyer H. H., Zheng Y. 2003. The AAA-ATPase Cdc48/p97 regulates spindle disassembly at the end of mitosis. Cell 115:355–367 [DOI] [PubMed] [Google Scholar]
- 11. Cheng Y. L., Chen R. H. 2010. The AAA-ATPase Cdc48 and cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J. Cell Sci. 123:2025–2034 [DOI] [PubMed] [Google Scholar]
- 12. Doerks T., Copley R. R., Schultz J., Ponting C. P., Bork P. 2002. Systematic identification of novel protein domain families associated with nuclear functions. Genome Res. 12:47–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Egerton M., et al. 1992. VCP, the mammalian homolog of cdc48, is tyrosine phosphorylated in response to T cell antigen receptor activation. EMBO J. 11:3533–3540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Egerton M., Samelson L. E. 1994. Biochemical characterization of valosin-containing protein, a protein tyrosine kinase substrate in hematopoietic cells. J. Biol. Chem. 269:11435–11441 [PubMed] [Google Scholar]
- 15. Fernandez-Saiz V., Buchberger A. 2010. Imbalances in p97 co-factor interactions in human proteinopathy. EMBO Rep. 11:479–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Finley D., Ozkaynak E., Varshavsky A. 1987. The yeast polyubiquitin gene is essential for resistance to high temperatures, starvation, and other stresses. Cell 48:1035–1046 [DOI] [PubMed] [Google Scholar]
- 17. Frohlich K. U., et al. 1991. Yeast cell cycle protein CDC48p shows full-length homology to the mammalian protein VCP and is a member of a protein family involved in secretion, peroxisome formation, and gene expression. J. Cell Biol. 114:443–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fu X., Ng C., Feng D., Liang C. 2003. Cdc48p is required for the cell cycle commitment point at Start via degradation of the G1-CDK inhibitor Far1p. J. Cell Biol. 163:21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghislain M., Dohmen R. J., Levy F., Varshavsky A. 1996. Cdc48p interacts with Ufd3p, a WD repeat protein required for ubiquitin-mediated proteolysis in Saccharomyces cerevisiae. EMBO J. 15:4884–4899 [PMC free article] [PubMed] [Google Scholar]
- 20. Gietz R. D., Sugino A. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527–534 [DOI] [PubMed] [Google Scholar]
- 21. Gueldener U., Heinisch J., Koehler G. J., Voss D., Hegemann J. H. 2002. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 30:e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guldener U., Heck S., Fielder T., Beinhauer J., Hegemann J. H. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24:2519–2524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hanson P. I., Whiteheart S. W. 2005. AAA+ proteins: have engine, will work. Nat. Rev. Mol. Cell Biol. 6:519–529 [DOI] [PubMed] [Google Scholar]
- 24. Hanzelmann P., Stingele J., Hofmann K., Schindelin H., Raasi S. 2010. The yeast E4 ubiquitin ligase Ufd2 interacts with the ubiquitin-like domains of Rad23 and Dsk2 via a novel and distinct ubiquitin-like binding domain. J. Biol. Chem. 285:20390–20398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hershko A., Ciechanover A. 1998. The ubiquitin system. Annu. Rev. Biochem. 67:425–479 [DOI] [PubMed] [Google Scholar]
- 26. Hochstrasser M. 1996. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30:405–439 [DOI] [PubMed] [Google Scholar]
- 27. Hochstrasser M., Varshavsky A. 1990. In vivo degradation of a transcriptional regulator: the yeast alpha 2 repressor. Cell 61:697–708 [DOI] [PubMed] [Google Scholar]
- 28. Humphreys D., Hume P. J., Koronakis V. 2009. The Salmonella effector SptP dephosphorylates host AAA+ ATPase VCP to promote development of its intracellular replicative niche. Cell Host Microbe 5:225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. James P., Halladay J., Craig E. A. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jarosch E., et al. 2002. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nat. Cell Biol. 4:134–139 [DOI] [PubMed] [Google Scholar]
- 31. Johnson E. S., Ma P. C., Ota I. M., Varshavsky A. 1995. A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem. 270:17442–17456 [DOI] [PubMed] [Google Scholar]
- 32. Keil R. L., Wolfe D., Reiner T., Peterson C. J., Riley J. L. 1996. Molecular genetic analysis of volatile-anesthetic action. Mol. Cell. Biol. 16:3446–3453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kern M., Fernandez-Saiz V., Schafer Z., Buchberger A. 2009. UBXD1 binds p97 through two independent binding sites. Biochem. Biophys. Res. Commun. 380:303–307 [DOI] [PubMed] [Google Scholar]
- 34. Knop M., et al. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15:963–972 [DOI] [PubMed] [Google Scholar]
- 35. Koegl M., et al. 1999. A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96:635–644 [DOI] [PubMed] [Google Scholar]
- 36. Latterich M., Frohlich K. U., Schekman R. 1995. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell 82:885–893 [DOI] [PubMed] [Google Scholar]
- 37. Lavoie C., et al. 2000. Tyrosine phosphorylation of p97 regulates transitional endoplasmic reticulum assembly in vitro. Proc. Natl. Acad. Sci. U. S. A. 97:13637–13642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Li G., Zhao G., Schindelin H., Lennarz W. J. 2008. Tyrosine phosphorylation of ATPase p97 regulates its activity during ERAD. Biochem. Biophys. Res. Commun. 375:247–251 [DOI] [PubMed] [Google Scholar]
- 39. Lis E. T., Romesberg F. E. 2006. Role of Doa1 in the Saccharomyces cerevisiae DNA damage response. Mol. Cell. Biol. 26:4122–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liu C., et al. 2010. Ubiquitin chain elongation enzyme Ufd2 regulates a subset of Doa10 substrates. J. Biol. Chem. 285:10265–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Madeo F., Schlauer J., Zischka H., Mecke D., Frohlich K. U. 1998. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Mol. Biol. Cell 9:131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Moir D., Stewart S. E., Osmond B. C., Botstein D. 1982. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics 100:547–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Morreale G., Conforti L., Coadwell J., Wilbrey A. L., Coleman M. P. 2009. Evolutionary divergence of valosin-containing protein/cell division cycle protein 48 binding interactions among endoplasmic reticulum-associated degradation proteins. FEBS J. 276:1208–1220 [DOI] [PubMed] [Google Scholar]
- 44. Mullally J. E., Chernova T., Wilkinson K. D. 2006. Doa1 is a Cdc48 adapter that possesses a novel ubiquitin binding domain. Mol. Cell. Biol. 26:822–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nakatsukasa K., Huyer G., Michaelis S., Brodsky J. L. 2008. Dissecting the ER-associated degradation of a misfolded polytopic membrane protein. Cell 132:101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nordquist K. A., et al. 2010. Structural and functional characterization of the monomeric U-box domain from E4B. Biochemistry 49:347–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ogiso Y., et al. 2004. Lub1 participates in ubiquitin homeostasis and stress response via maintenance of cellular ubiquitin contents in fission yeast. Mol. Cell. Biol. 24:2324–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ossareh-Nazari B., et al. 2010. Cdc48 and Ufd3, new partners of the ubiquitin protease Ubp3, are required for ribophagy. EMBO Rep. 11:548–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Perrett S., Freeman S. J., Butler P. J. G., Fersht A. R. 1999. Equilibrium folding properties of the yeast prion protein determinant Ure2. J. Mol. Biol. 290:331–345 [DOI] [PubMed] [Google Scholar]
- 50. Qiu L., et al. 2010. Structure and function of the PLAA/Ufd3-p97/Cdc48 complex. J. Biol. Chem. 285:365–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rabinovich E., Kerem A., Frohlich K. U., Diamant N., Bar-Nun S. 2002. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Mol. Cell. Biol. 22:626–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Rape M., et al. 2001. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell 107:667–677 [DOI] [PubMed] [Google Scholar]
- 53. Ravid T., Hochstrasser M. 2008. Diversity of degradation signals in the ubiquitin-proteasome system. Nat. Rev. Mol. Cell Biol. 9:679–690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ren J., Pashkova N., Winistorfer S., Piper R. C. 2008. DOA1/UFD3 plays a role in sorting ubiquitinated membrane proteins into multivesicular bodies. J. Biol. Chem. 283:21599–21611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Richly H., et al. 2005. A series of ubiquitin binding factors connects CDC48/p97 to substrate multiubiquitylation and proteasomal targeting. Cell 120:73–84 [DOI] [PubMed] [Google Scholar]
- 56. Rumpf S., Jentsch S. 2006. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Mol. Cell 21:261–269 [DOI] [PubMed] [Google Scholar]
- 57. Schuberth C., Buchberger A. 2005. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nat. Cell Biol. 7:999–1006 [DOI] [PubMed] [Google Scholar]
- 58. Schuberth C., Buchberger A. 2008. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell. Mol. Life Sci. 65:2360–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schuberth C., Richly H., Rumpf S., Buchberger A. 2004. Shp1 and Ubx2 are adaptors of Cdc48 involved in ubiquitin-dependent protein degradation. EMBO Rep. 5:818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shcherbik N., Haines D. S. 2007. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Mol. Cell 25:385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Tu D., Li W., Ye Y., Brunger A. T. 2007. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. U. S. A. 104:15599–15606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wach A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259–265 [DOI] [PubMed] [Google Scholar]
- 63. Welchman R. L., Gordon C., Mayer R. J. 2005. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat. Rev. Mol. Cell Biol. 6:599–609 [DOI] [PubMed] [Google Scholar]
- 64. Wilcox A. J., Laney J. D. 2009. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nat. Cell Biol. 11:1481–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wilson L. K., Benton B. M., Zhou S., Thorner J., Martin G. S. 1995. The yeast immunophilin Fpr3 is a physiological substrate of the tyrosine-specific phosphoprotein phosphatase Ptp1. J. Biol. Chem. 270:25185–25193 [DOI] [PubMed] [Google Scholar]
- 66. Ye Y., Meyer H. H., Rapoport T. A. 2001. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414:652–656 [DOI] [PubMed] [Google Scholar]
- 67. Zhao G., Li G., Schindelin H., Lennarz W. J. 2009. An Armadillo motif in Ufd3 interacts with Cdc48 and is involved in ubiquitin homeostasis and protein degradation. Proc. Natl. Acad. Sci. U. S. A. 106:16197–16202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Zhao G., et al. 2007. Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation. Proc. Natl. Acad. Sci. U. S. A. 104:8785–8790 [DOI] [PMC free article] [PubMed] [Google Scholar]