Abstract
Dnmt3a1 and Dnmt3a2 are two de novo DNA methyltransferases expressed in mouse embryonic stem cells (mESCs). They differ in that a 219-amino-acid (aa) amino (N)-terminal noncatalytic domain is present only in Dnmt3a1. Here, we examined the unique functions of Dnmt3a1 in mESCs by targeting the coding sequence of the Dnmt3a1 N-terminal domain tagged with enhanced green fluorescent protein (GFP) for insertion into the mouse Rosa26 locus. Using these targeted cells (GFP-3a1Nter), we showed that Dnmt3a1 was efficiently recruited to the silenced Oct3/4 and activated Vtn (vitronectin) gene promoters via its unique N-terminal domain. This recruitment affected the two genes in contrasting ways, compromising Oct3/4 gene promoter DNA methylation to prevent consolidation of the silent state while significantly reducing Vtn transcription. We used this negative effect of the Dnmt3a1 N-terminal domain to investigate the extent of transcriptional regulation by Dnmt3a1 in mESCs by using microarrays. A small group of all-trans retinoic acid (tRA)-inducible genes had lower transcript levels in GFP-3a1Nter cells than in wild-type mESCs. Intriguingly, this group included genes that are important for fetal nutrition, placenta development, and metabolic functions and is enriched for a distinct set of imprinted genes. We also identified a larger group of genes that showed higher transcript levels in the GFP-3a1Nter-expressing cells than in wild-type mESCs, including pluripotency factors and key regulators of primordial germ cell differentiation. Thus, Dnmt3a1 in mESCs functions primarily as a negative and to a lesser extent as a positive regulator of transcription. Our findings suggest that Dnmt3a1 positively affects transcription of specific genes at the promoter level and targets chromosomal domains to epigenetically silence gene clusters in mESCs.
INTRODUCTION
Mammalian cells are characterized by cell type-specific DNA methylation patterns consisting of both intra- and intergenic genomic methylation, forming a molecular fingerprint (5). Ample evidence exists for an inverse correlation between transcription initiation and DNA methylation in promoter regions, and a mandatory DNA demethylation step is normally required before the onset of transcription (4).
The mammalian DNA methylation machinery consists of the maintenance DNA methyltransferase Dnmt1 and “de novo” DNA methyltransferases of the Dnmt3 family. Dnmt1 maintains semiconservative DNA methylation during DNA replication. In contrast, the de novo Dnmts introduce new DNA methylation marks in targeted genomic regions and are key enzymes in the deployment of developmental programs (6, 31, 48).
The two active de novo Dnmts, Dnmt3a and Dnmt3b, are homologous proteins with similar linear arrangements of protein domains. They are encoded from different genomic loci that produce active and inactive proteins through differently or alternatively spliced transcripts. This complex mix of transcripts, combined with the high antigenic similarity of the proteins, makes it difficult to assess the unique and redundant functions of the various Dnmt3 proteins produced in vivo.
The Dnmt3a genomic locus produces two transcripts giving rise to two proteins, the long Dnmt3a1 and the shorter Dnmt3a2, that differ in that a 219-amino-acid (aa) amino (N)-terminal tail is present only in Dnmt3a1 (11, 50). Dnmt3a1 is expressed in both embryonic and adult tissues, and Dnmt3a1 and Dnmt3a2 (here collectively referred to as Dnmt3a) are coexpressed in mouse embryonic stem cells (mESCs). The Dnmt3a locus is responsible for the establishment of genomic imprinting (27) that takes place during primordial germ cell (PGC) formation at a time when the maternal and paternal genomes are physically separated. Genomic imprints include differential DNA methylation marks in specific chromosome regions (differentially methylated regions [DMRs]). These imprints differ between the maternal and paternal chromosomes and survive the wave of DNA demethylation and epigenetic reprogramming that occurs during the first stages of embryonic development (36, 46). Imprinting marks are thought to be preserved by the concerted actions of Dnmt1 and the de novo Dnmts, and they function in part to silence antisense transcription, effectively contributing to monoallelic gene expression (26, 36).
The transcription factors Oct3/4 and Nanog are integral components of the regulatory network sustaining the pluripotency of mESCs and are established Dnmt3 targets during somatic cell differentiation in early embryogenesis (9). The genes encoding these factors undergo differentiation-induced silencing in the epiblast. Treatment of mESCs with all-trans retinoic acid (tRA) also results in stepwise silencing of the Oct3/4 gene promoter, by termination of transcription (18) and by heterochromatinization of the locus involving characteristic histone modifications and de novo DNA methylation (2, 17, 33). Heterochromatinization provides an additional safeguard against reexpression of the Oct3/4 gene and the resulting dedifferentiation of somatic cells. The Oct3/4 gene promoter is methylated by de novo Dnmt3s that are recruited by G9a histone methyltransferase (HMTase) (16, 17). A complex of G9a and glycolipoprotein (GLP) HMTase also recruits Dnmt3s to repetitive DNA sequences that are then methylated and kept in the transcriptionally silent, heterochromatic state, thus securing the integrity and homeostasis of the genome (15, 49).
New roles for de novo Dnmts suggest that these proteins also participate in active transcription. De novo Dnmts are recruited to activated gene promoters along with components of DNA repair machinery (28, 35). For example, in P19 cells, nuclear receptor (COUP-TFI)-dependent transcriptional activation of the vitronectin (Vtn) gene involves the recruitment of Dnmt3a along with base excision repair (BER) components of the promoter (19), resulting in strand-specific DNA demethylation of the promoter.
The specific roles for either Dnmt3a1 or Dnmt3a2 in mESCs are not yet known. In fact, the functions and genomic targets of Dnmt3a can be obscured in knockout cells upon reintroduction of separate genes, because this strategy does not necessarily result in restoration and rescue of the wild-type (wt) situation in the ES cells. For example, such a “rescue” experiment would fail to restore the damaged imprinting marks, because once lost, these marks cannot be regenerated in ESCs (12). Thus, which of the two Dnmt3a forms is responsible for maintaining the Imprints in ESCs is not known.
In an attempt to identify a unique role(s) for Dnmt3a1 in mESCs, we focus on the Dnmt3a1 N-terminal domain, for which no specific function has yet been attributed. Here, we follow an alternative strategy to isolate and study the Dnmt3a1 functional role in mESCs by making use of this unique Dnmt3a1N-terminal domain. We introduced single-copy transgenes encoding the tagged N-terminal domain of Dnmt3a1, resulting in a moderate overexpression of the respective protein that did not overly increase levels of endogenous Dnmt3a1. Our expressed transgenes efficiently competed with endogenous protein for its genomic target sites, without grossly altering the ESC epigenome. Our data confirmed previous suggestions (26, 38) that endogenous Dnmts were not “free” in the nucleus but instead were engaged within the chromatin compartment at all times. Based on our findings, we suggest that Dnmt3a1 upregulates transcription of specific genes at the promoter level, while epigenetically silencing chromosomal gene clusters in mESCs.
MATERIALS AND METHODS
Cell lines and transfections.
Wild-type mouse embryonic stem cells (mESCs-129/3), green fluorescent protein [GFP]-3a2 cells, and GFP-3a1Nter cells were maintained in medium for ES cells (here designated ES medium) containing Dulbecco's modified Eagle medium (DMEM; Gibco) supplemented with 15% fetal bovine serum (FBS; pretested with ES cells; Pan Biotech), 50 U/ml penicillin/streptomycin, 2 mM l-glutamine, 0.1 mM β-mercaptoethanol (β-Me), and 1,000 U/ml mouse leukemia inhibitory factor (LIF; Esgro; Chemicon). The cells were grown on a monolayer of mitotically compromised mouse embryonic fibroblasts (MEFs) in gelatin-coated tissue culture dishes. Growth medium was replaced daily. Transfections were performed (i) by the CaCl2 method, (ii) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and (iii) by electroporation. For electroporation, 1.2 × 107 to 1.5 × 107 cells were resuspended in 800 μl of phosphate-buffered saline (PBS) and mixed with 20 μg of plasmid that had been linearized by restriction enzyme digestion and extracted with phenol, followed by electroporation in a 0.4-mm cuvette (Bio-Rad) at 230 V and 500 μF. Cells were layered over a monolayer of MEFs to recover for 24 h without selection in ES medium. After the recovery period, G418 and ganciclovir was added to the medium at final concentrations of 250 μg/ml and 2 μM, respectively. Resistant ESC colonies were picked after 6 to 7 days, trypsinized, and grown individually in 48-well plates.
For all-trans retinoic acid (tRA) treatment, mESCs-129/3 or clonal cell lines were trypsinized and plated at a density of 105 cells/ml in petri dishes with DMEM–15% FBS–LIF medium for 24 h. Cells were then transferred to new petri dishes in DMEM–15% FBS–1 mM tRA medium that was replaced every second day and kept for 72, 96, or 120 h. Mock-infected cells were also kept in petri dishes in DMEM–15% FBS–LIF medium that was replaced daily.
Short hairpin RNA (shRNA) inhibition of Dnmt3a1 expression in mESCs.
The design of the sh oligonucleotide sequences (scrambled and Dnmt3a1 specific) was performed with the GeneLink online tool. (http://www.genelink.com/sirna/shRNAi.asp). For knockdowns (KDs), two oligonucleotides (64-mers) bearing the coding sequences for the RNA “hairpins” (one scrambled and one targeting specifically the Dnmt3a1 mRNA) were purchased from Invitrogen, and they were phosphorylated, annealed, and cloned into a standard pSUPER vector (Oligoengine).
Twenty micrograms of each of the pSUPER recombinant plasmids bearing the coding sequences for the two RNA hairpins (shRNA) were mixed with 15 μg of a pBL construct bearing the PGK-neo+ selection marker. Both plasmids were linearized with restriction enzyme NotI and were used to electroporate mESCs-129/3. Cells were layered over MEFs to recover for 24 h without selection in ES medium. After recovery, G418 was added to the medium at final concentrations of 250 μg/ml. Resistant ESC colonies were picked after 7 days, trypsinized, and grown individually in 48-well plates.
Plasmids and constructs.
Dnmt3a1 full-length and N-terminal domain cDNAs were obtained by reverse transcription (RT)-PCR from total RNA of L929 cells, and Dnmt3a2 cDNA was similarly obtained from mESCs-129/3. Primer sequences are available upon request. Superscript III reverse transcriptase (Invitrogen) was used for reverse transcription, and Go Taq polymerase (Promega) was used for PCR. For long cDNAs, an Expand Long Template kit (Roche) was used. The cDNAs were cloned into TOPO (Invitrogen) or pGEM T-Easy (Promega) vectors and then subcloned into pGEX (GE), pRSET (Invitrogen), and pEGFPC1 (Clontech) to acquire glutathione S-transferase (GST), His, and enhanced GFP (EGFP) tags, respectively. For knockdowns, the two double-stranded 64-mers bearing the coding sequences for the RNA hairpins were cloned into pSUPER vector (Oligoengine) using BglII and HindIII cloning sites. The G9a expression plasmid and antibody were kind gifts from I. Talianidis. The Rosa26 targeting cassette was provided by the Soriano Laboratory and was engineered to replace the phosphoglycerate kinase (PGK)-Dta negative selection marker with PGK-Tk and to include the bovine growth hormone polyadenylation signal (BGHpA) sequence downstream of the cDNA cloning site.
Immunoprecipitations and Western blots.
We used a modified protocol to assess the subcellular distribution of endogenous and transgenic proteins. Briefly, ∼108 ESCs were trypsinized and plated on gelatinized plates for 3 to 4 h to minimize MEF contamination. The cells were then harvested and washed twice with phosphate-buffered saline (PBS). The cells were placed in hypotonic buffer A (10 mM HEPES-KOH [pH 7.5], 1.5 mM MgCl2, 10 mM KCl, 1 mM phenylmethylsulfonyl fluoride [PMSF]) to swell for 10 min on ice and were subsequently homogenized with 10 gentle strokes of a Dounce homogenizer with a B pestle. The samples were centrifuged at 2,500 rpm for 5 min at 4°C. The supernatant was the cytoplasmic extract, and the cell nuclei formed the pellet. The pellet was resuspended, washed twice with buffer A, and centrifuged at 2,500 rpm for 5 min at 4°C. The nuclei were resuspended in buffer C (20 mM HEPES-KOH [pH 7.5], 25% glycerol, 0.5 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM PMSF) and left to rotate for 45 min at 4°C. The samples were then centrifuged at 14,000 rpm for 5 min at 4°C. The soluble supernatant was the nuclear extract, and the pellet was the insoluble chromatin. The pellet was washed three times with buffer C and resuspended with 50 harsh strokes of a Dounce homogenizer with pestle B in HSB buffer (20 mM HEPES-KOH [pH 7.5], 0.34 M sucrose, 0.65 M NaCl, 1 mM EDTA, 1 mM β-Μe, 1 mM PMSF). These samples were then dialyzed against 20 mM HEPES-KOH (pH 7.5), 0.1 M NaCl, 1 mM EDTA, 1 mM β-Μe, and 1 mM PMSF, and after the addition of CaCl2 to a final concentration of 3 mM, the samples were treated with 10 U/ml of micrococcal nuclease for 10 min at 37°C and centrifuged at 14,000 rpm at 4°C for 5 min. The salt concentration was subsequently increased to 0.65 M NaCl, and the samples were centrifuged at 14,000 rpm for 5 min at 4°C. The supernatant was the soluble chromatin extract, and the pellet was the insoluble nuclear matrix. All extracts were dialyzed extensively against either PBS or buffer D (20 mM HEPES-KOH [pH 7.5], 20% glycerol, 0.1 M NaCl, 0.2 mM EDTA, 1 mM PMSF), and their protein concentrations were determined by Bradford assays.
For immunoprecipitations, 10 μg of each antibody was incubated with Dynal protein G beads according to the manufacturer's recommendations in 0.1 M sodium citrate buffer, pH 5, for 40 min on ice. The beads were washed with PBS–0.01% Tween and left to interact with 100 μg nuclear extract with continuous rotation at 4°C for 4 h in PBS–0.01% Tween buffer. The supernatant was removed (as flowthrough [FT]), and the beads were washed twice with PBS–0.01% Tween and twice with PBS–0.01% Tween supplemented with 0.3 M NaCl. The beads were boiled for 5 min in SDS electrophoresis buffer and analyzed by Western blotting.
GST pulldowns.
GST fusion proteins were expressed in bacteria and purified from bacterial extracts with Sepharose-GST beads (Sigma) using standard protocols. Sepharose-GST beads loaded with GST fusion proteins and controls were incubated with equal amounts of in vitro-translated [35S]methionine-labeled G9a (prepared with a TNT SP6/T7 high-yield protein kit (Promega) with continuous rotation at 4°C for 4 h in PBS–0.01% Tween buffer supplemented with protease inhibitors. The supernatant was removed, and the beads were washed twice with PBS–0.01% Tween and five times with PBS–0.01% Tween supplemented with 0.3 M NaCl. The beads were boiled for 5 min in SDS electrophoresis buffer and analyzed by SDS-PAGE and autoradiography.
Chromatin immunoprecipitations (ChIPs).
ESCs (∼108) mock or tRA treated for the indicated amounts of time (see the Fig. 3 legend) were harvested and washed twice with PBS. Cells were resuspended in PBS, and formaldehyde was added to a final concentration of 11%. The samples were incubated at 37°C for 12 to 15 min before the addition of quenching buffer (125 mM glycine). The cells were centrifuged at 2,000 rpm, washed with PBS, and resuspended in hypotonic buffer A to swell for 10 min on ice. The samples were then Dounce homogenized by five gentle strokes (pestle B) and centrifuged at 2,500 rpm at 4°C for 5 min. The pellet of nuclei was washed once in buffer C and resuspended in sonication buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 1 mM PMSF). The nuclear preparations were sonicated 15 to 20 times for 20 s each at 80% of the maximum power of a VibraCell sonicator (Sonics) with a microtip. Samples were centrifuged at 4,000 rpm, and the supernatant was adjusted to 0.5% sarcosyl and 0.568 g/ml CsCl2. Ultracentrifugation was carried out at 40,000 rpm for 36 h. The resultant fractions were analyzed on agarose gels. For each sample, fractions containing DNA were pooled and extensively dialyzed against 2 liters of dialysis buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 0.5 mM EGTA, 5% glycerol, 1 mM PMSF). The DNA content of the samples was determined, and it was concentrated if required.
Fig. 3.
Dnmt3a1 is recruited to silent Oct3/4 gene and activated Vtn gene promoters via its N-terminal domain. (A) q-PCR with chromatin-immunoprecipitated (ChIP) samples prepared from untreated and 120-h tRA-treated wt mESCs, GFP-3a2 cells, and GFP-3a1Nter cells. ChIPs were performed with selected antibodies against GFP, 1 to 295 aa of Dnm3a1/3a2, COUP-TF, G9a HMTase, p68/Ddx5 DEAD-box helicase, and tetra-acetylated histone H4 N-terminal tail. Amplification was performed with primers specific for the Oct3/4 gene promoter. (B) q-PCR with ChIP samples prepared from untreated and 120-h tRA-treated wt mESCs, GFP-3a2 cells, and GFP-3a1Nter cells. ChIPs were performed with selected antibodies as in panel A. Amplification was performed with primers specific for Vtn promoter sequences. (C) q-PCR with ChIP samples prepared from untreated and 72-h tRA-treated wt mESCs and GFP-3a1Nter cells. ChIPs were performed with antibodies against GFP and the Dnmt3a C terminus. Amplification was performed with primers specific for promoter sequences of the Nanog and Vtn genes.
For immunoprecipitations, 10 μg of each antibody was incubated with Dynal protein G beads according to the manufacturer's recommendations in 0.1 M sodium citrate buffer (pH 5) for 40 min on ice. The beads were washed with PBS to remove unbound antibody and left to interact with 10- to 20-μg chromatin samples with continuous rotation at 4°C overnight in radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris [pH 8.0], 1% Triton, 140 mM NaCl, 0.1% sodium deoxycholate, 1 mM PMSF). The next day, the supernatant was removed and the beads were washed twice with RIPA buffer, twice with RIPA500 buffer (10 mM Tris [pH 8.0], 1% Triton, 500 mM NaCl, 0.1% sodium deoxycholate, 1 mM PMSF), once with LiCl buffer (10 mM Tris [pH 8.0], 1 mM EDTA, 250 mM LiCl, 0.5% NP-40, 0.5% sodium deoxycholate, 1 mM PMSF), and once with TE buffer (10 mM Tris [pH 8.0], 1 mM EDTA). The beads were eluted twice in 150 μl elution buffer (50 mM Tris [pH 8.0], 0.1 mM EDTA, 50 mM NaHCO3, 1 mM PMSF) at 65°C for 10 min.
The sample volume was increased to 400 μl with water, and NaCl was adjusted to a final concentration of 0.1 M. EDTA and proteinase K were added to final concentrations of 5 mM and 0.4 mg/ml, respectively, and the samples were placed at 42°C for 2 h. The samples were incubated overnight at 65°C to reverse the cross-links. The samples were then phenol-chloroform extracted, and glycogen and 800 μl ethanol were added. The samples were placed at −20°C for more than 5 h and then precipitated by centrifugation at 10,000 × g at 4°C for 30 min. The pellets were washed with 70% ethanol and resuspended in 100 μl TE buffer. Aliquots of 5 μl of each sample were used for quantitative PCR (q-PCR) amplification.
Microarray hybridization and analysis.
Three independent RNA preparations were used for cRNA synthesis and labeling (Enzo) for each condition tested, according to Affymetrix instructions. Labeled samples were used to hybridize mouse Gene 1.0 ST arrays (Affymetrix). The raw data obtained from these experiments were analyzed using the Partek Genomic Suite (Partek). Normalization of the data included gene content robust multiarray average (GCRMA) background correction and probe summarization by the median polish method.
Reverse transcription and q-PCR.
Total RNA was isolated from cell pellets with Trizol reagent (Invitrogen). The RNA samples were DNase treated and further purified with an RNeasy purification system (Qiagen). RNA (5 μg) was used in reverse transcription reactions for 1 h at 37°C using Superscript III reverse transcriptase (Invitrogen). q-PCR was performed for diluted RT samples with the designated primer sets using Evagreen master mix (Bio-Rad). q-PCRs were performed using an miniOpticon real-time PCR detection system (Bio-Rad). All the primers were designed to produce a product 100 to 150 bp long in order to perform all reactions with the following simple two-step program: 95°C for 2 min followed by 40 cycles of 95°C for 30 s, 60°C for 45s, and 70°C for 20 s. The Gapdh mRNA levels were monitored and used as the internal control for quantification in RT-coupled q-PCR experiments. ChIP samples were normalized against input chromatin samples and subjected to proteinase K treatment and reverse cross-linking, as described above.
The results were obtained from duplicates of three independent experiments, and the data were extracted by either MJ Opticon Monitor analysis (v. 3.1) or Bio-Rad CFX manager (v. 1.5.534) and further analyzed in a spreadsheet by the method described in reference 45.
Antibodies.
The following antibodies were purchased: anti-GFP (sc-9996; Santa Cruz), anti-p68 (05-850; Upstate), anti-tetra-acetylated histone Η4 (06-598; Upstate), anti-G9a (ab3939-100 from Abcam and 07-551 from Upstate), anti-Dnmt3a (sc-20703 from Santacruz and ab23565 and ab13888 from Abcam), and anti-FLAG (F3165; Sigma-Aldrich). Anti-COUP-TF antibody was kindly provided by I. Talianidis.
Bisulfite sequencing.
Bisulfite primers were designed with Methyl Primer Express v. 1.0 software, and bisulfite DNA conversion was performed with an EZ methylation DNA Gold kit (Zymo Research) according to the manufacturer's instructions.
Primers and oligonucleotides are available upon request.
Microarray data accession number.
The set of supporting microarray data have been deposited in the ArrayExpress public database, and the assigned accession number is E-MEXP-3012.
RESULTS
Targeting of the Dnmt3a1 N-terminal domain and Dnmt3a2 cDNAs into the Rosa26 locus of mESCs.
Dnmt3a1 and Dnmt3a2 are both expressed in mESCs. To discriminate between them in vivo, we engineered stable mESC clones bearing coding sequences of (i) the enhanced green fluorescent protein (EGFP)-tagged 219-aa Dnmt3a1 N-terminal domain (GFP-3a1Nter cells), (ii) the FLAG and EGFP double-tagged 219-aa Dnmt3a1 N-terminal domain (FLAG-3a1Nter cells), and (iii) the EGFP-tagged Dnmt3a2 (GFP-3a2 cells), with targeted insertion into the mouse Rosa26 locus (Fig. 1 A). Genomic targeting was verified by Southern blotting (not shown), and transgene transcription was driven by the Rosa26 promoter in all cases. To rule out the possibility that expression from single-copy Rosa26 loads the cell with large amounts of transgene mRNA, leading to excessive overexpression, we performed q-PCR for RT total RNA samples from either mock- or 96-h tRA-treated wt mESCs, GFP-3a1Nter cells, and FLAG-3a1Nter cells, using primers specifically amplifying Dnmt3a1 sequences; the primers also amplify the sequences of the tagged 219-aa Dnmt3a1 N-terminal domain in the stable cell lines. Untreated GFP-3a1Nter and FLAG-3a1Nter cells exhibit ∼1.5- to 2-fold-increased Dnmt3a1 levels compared to levels for untreated wt mESCs (see supplementary Fig. 1 at http://www.fleming.gr/files/Agalioti_supplemental_document.pdf, white bars). In contrast, tRA-treated GFP-3a1Nter and FLAG-3a1Nter cells contain levels of Dnmt3a1 similar to those for tRA-treated wt cells, because Dnmt3a1 mRNA is induced by tRA 5- to 6-fold (supplementary Fig. 1 at http://www.fleming.gr/files/Agalioti_supplemental_document.pdf, compare black and white bars). We conclude that the single-copy Rosa-26 EGFP-3a1Nter-inserted transgenes increase merely 2-fold the overall Dnmt3a1 mRNA in uninduced GFP-3a1Nter and FLAG-3a1Nter cells. The 2-fold increase in Dnmt3a1 mRNA levels cannot account for an excessive overexpression of the transgene proteins in the stable cell lines. These findings suggest that the expressed EGFP-3a1Nter and FLAG-3a1Nter proteins are exerting their effects on the mESC epigenome, acting as dominant-negative Dnmt3a1 mutants (see also Discussion).
Fig. 1.
Targeting of the tagged Dnmt3a1-N-terminal domain and Dnmt3a2 coding sequences into the mouse Rosa26 locus. (A) Top, schematic representation of Dnmt3a1 and Dnmt3a2 proteins. Bottom, schematic representation of the targeting strategy to introduce the EGFP- or FLAG/EGFP-tagged Dnmt3a1-N-terminal domain or EGFP-tagged Dnmt3a2 cDNAs into the nonessential Rosa26 mouse genomic locus. (B) Western blot of whole-cell protein extracts from wild-type mESCs (lanes 1 and 3) and GFP-3a2 cells (lanes 2 and 4). Lanes 1 and 2, a monoclonal anti-GFP (α-GFP) antibody was used for detection; lanes 3 and 4, an antibody against the C-terminal part of Dnmt3a (epitope between aa 750 and 908) was used. A double band marked by asterisks corresponds to two different forms of Dnmt3a1. The blue and the red arrowheads indicate EGFP-Dnmt3a2 and endogenous Dnmt3a2, respectively. (C) Western blot of fractionated protein extracts from wild-type mESCs and GFP-3a2 cells. Lanes 1 and 2, nuclear (Nucl.) extracts. Lanes 3 and 4, chromatin (Chrom.) extracts. Upper panel, a monoclonal anti-GFP antibody was used to detect EGFP-Dnmt3a2. Efficient subcellular fractionation was verified by immunoblotting with Pab 204 antibody (Upstate) (28a) recognizing the nuclear Ddx5/p68 DEAD-box RNA helicase (middle panel), and with an antibody against the chromatin-associated tetra-acetylated histone H4 N-terminal tail (lower panel) (Upstate). (D) Western blot of nuclear (lanes 1 to 3) and chromatin (lanes 4 to 6) extracts from HEK-293T cells transiently transfected with constructs encoding EGFP-Dnmt3a1 (lanes 1 and 4) or EGFP-Dnmt3a2 (lanes 2 and 5) or with empty EGFP-encoding vector (lanes 3 and 6). Antibody detection was as in panel C. (E) Western blot of cytoplasmic (lanes 1 and 2), nuclear (lanes 3 and 4), and chromatin (lanes 5 and 6) extracts from wild-type mESCs (lanes 1, 3, and 5) and GFP-3a1Nter cells (lanes 2, 4, and 6). Upper panel, the H-295 antibody (Santa Cruz) against the N-terminal Dnmt3a1/3a2 domain (aa 1 to 295) was used for detection. Lower panel, detection was performed with a monoclonal anti-GFP antibody. The black arrowhead indicates endogenous Dnmt3a1 protein. Gray arrowheads indicate EGFP-3a1Nter protein.
Endogenous Dnmt3a1 and Dnmt3a2 are chromatin-associated proteins. Their solubilization requires chromatin degradation and high salt extraction. We used Western blotting to detect endogenous Dnmt3a1 and Dnmt3a2 as well as transgene protein products in protein extracts (Fig. 1) before and after subcellular fractionation (see supplementary Fig. 2 at http://www.fleming.gr/files/Agalioti_supplemental_document.pdf). EGFP-3a1Nter was soluble and present in the nucleoplasm and chromatin and to a lesser extent in the cytoplasm (Fig. 1E, lanes 2, 4, and 6). This protein, however, appeared at a higher molecular mass than anticipated (∼82 kDa instead of an estimated 51 kDa) (see supplementary Fig. 3A at http://www.fleming.gr/files/Agalioti_supplemental_document.pdf). This apparent discrepancy in mass can be caused either by posttranslational modifications imposed specifically by the eukaryotic cell to the transgene protein or by factors inherent to the particular protein sequence, causing the anomalous protein migration in SDS-PAGE. To discriminate between the two possibilities, we compared the migration of the GST-3a1Nter protein produced in bacteria and the EGFP-3a1Nter protein from the GFP-3a1Nter cell extract. Both GST-3a1Nter and EGFP-3a1Nter proteins have anticipated molecular masses of ∼51 kDa, and both migrate anomalously above 64 kDa in the Western blot shown in supplementary Fig. 3A (http://www.fleming.gr/files/Agalioti_supplemental_document.pdf). Additionally, we compared the elution profile of the double-tagged FLAG/EGFP-3a1Nter protein from FLAG-3a1Nter cell nuclear extracts with the elution profiles of protein standards which were analyzed with a Superose 12 gel filtration column (GE Healthcare). The FLAG/EGFP-3a1Nter protein is eluted at 12.5 to 13 ml (corresponding to fractions 15 to 16), and the 44-kDa peak for the protein standards is detected at 13.72 ml of the Superose 12 column elution volume (see supplementary Fig. 3B at http://www.fleming.gr/files/Agalioti_supplemental_document.pdf). Despite the anomalous migration, the EGFP-3a1Nter protein is eluted from gel filtration according to its anticipated molecular weight. From the above findings, we conclude that the EGFP-3a1Nter protein migration pattern is probably due to its anomalous migration in SDS-PAGE and not attributed to eukaryotic cell-specific posttranslational modifications.
The 97-kDa EGFP-Dnmt3a2 protein was detected by anti-GFP antibody in GFP-3a2 cells (Fig. 1B, lane 2) and was also chromatin bound (Fig. 1C, compare lane 2 with lane 4). Surprisingly, EGFP-Dnmt3a1 and -Dnmt3a2 proteins were detected in both nuclear extracts and chromatin of transiently transfected HEK-293T cells (Fig. 1D, lanes 1 and 2 and lanes 4 and 5), presumably because they were being expressed at amounts exceeding the saturation levels of their genomic target sites. In contrast, GFP-3a2 cells only moderately overexpressed EGFP-Dnmt3a2 protein, at net Dnmt3a2 levels below saturation of genomic target sites.
EGFP-3a1Nter expression compromised silencing of the Oct3/4 and Nanog genes and transcriptional activation of the Vtn gene.
tRA treatment of mESCs results in methylation of the Oct3/4 and Nanog promoters through G9a-dependent recruitment of de novo Dnmts (Dnmt3a and Dnmt3b) (16, 17). Promoter DNA methylation consolidates the transcriptionally silent state of the Oct3/4 and Nanog genes. However, Dnmt3a is also recruited to the promoter of the inducible Vtn gene in P19 cells (19).
To test the effect of EGFP-3a1Nter protein expression on silencing of the Oct3/4 and Nanog genes and transcriptional activation of the Vtn gene, we performed q-PCR using specific primers and RT total RNA samples from either mock- or 120-h tRA-treated wt mESCs, GFP-3a1Nter cells, or GFP-3a2 cells. EGFP-Dnmt3a2 expression affected neither Oct3/4 or Nanog gene mRNA levels in untreated GFP-3a2 cells (Fig. 2 A) nor transcriptional downregulation after 96 h of tRA treatment. In contrast, higher levels of both Oct3/4 and Nanog mRNAs persisted in GFP-3a1Nter cells compared to wt cells after tRA treatment (Fig. 2A, gray bars; comparisons are indicated by single and double asterisks), suggesting that expression of the EGFP-3a1Nter protein compromised permanent silencing of the Oct3/4 and Nanog genes in GFP-3a1Nter cells.
Fig. 2.
EGFP-Dnmt3a1Nter expression compromised Oct3/4 and Nanog gene silencing and Vtn gene transcription. (A) q-PCR using reverse-transcribed total RNA samples from untreated or 120-h tRA-treated wt mESCs, GFP-3a1Nter cells, or GFP-3a2 cells. Amplification was performed with primers specific for Oct3/4, Nanog, or Vtn gene mRNAs. Rel., relative. (B) q-PCR using reverse-transcribed total RNA samples from wild-type mESCs, GFP-3a1Nter cells, and GFP-3a2 cells. Cells were untreated or tRA treated for 120 h. After 120 h of tRA treatment, the cells were incubated with LIF for 8 more days. Amplification was carried out with primers specific for Oct3/4, Nanog, and Vtn gene mRNAs. (C) Bisulfite sequencing of the 375-bp region around the transcription start site (TSS; +1) of the Oct3/4 gene promoter. The region contains 13 CpGs (arbitrarily numbered from 1 to 13). PCR was performed with specific primers using bisulfite-converted DNA from 96-h tRA-treated wt mESCs and GFP-3a1Nter cells. Six and seven individual PCR products derived from wild-type mESCs and GFP-3a1Nter cells, respectively, were cloned into the TOPO II PCR vector (Invitrogen) and sequenced with Sp6 and T7 primers. Methylation was calculated to be 30% for this genomic area in wt mESCs and 14% in GFP-3a1Nter-derived clones. (D) Bisulfite sequencing of the 279-bp region (−370 to −90) upstream of the transcription start site (noncoding strand) of the Vtn gene promoter. The region contains 11 CpGs arbitrarily numbered 6 to 16 (top). PCR was performed with specific primers using bisulfite-converted genomic DNA from untreated and 96-h tRA-treated wt mESCs and GFP-3a1Nter cells. Individual PCR products were cloned into the TOPO II PCR vector (Invitrogen) and sequenced with Sp6 and T7 primers. Methylation was calculated to be 75% in this genomic area for untreated wt mESCs and GFP-3a1Nter cells, 44% for tRA-treated wt mESCs, and 77% for tRA-treated GFP-3a1Nter cells.
To verify that silencing was compromised, we assessed all three cell lines for their ability to reactivate Oct3/4 and Nanog gene transcription upon readdition of LIF to the cell culture medium after 120 h of tRA treatment. LIF treatment resulted in partial transcriptional reactivation of the Oct3/4 gene but not the Nanog gene in GFP-3a1Nter cells, but not in wt mESCs (Fig. 2B, single and double asterisks). We performed bisulfite sequencing of 375 bp around the transcription start site (TSS; +1) of the Oct3/4 gene promoter in 96-h tRA-treated GFP-3a1Nter cells and wt mESCs to monitor the methylation status of the 13 CpGs of this region (Fig. 2C). This region was found to be 14% methylated in GFP-3a1Nter cells, compared to 30% methylation in wt cells. We concluded that methylation of the Oct3/4 gene promoter was inefficient in GFP-3a1Nter cells, presumably due to competition of EGFP-3a1Nter with endogenous Dnmts (Fig. 2C). The tRA-inducible Vtn gene was silent in both wt mESCs and GFP-3a2 cells. Strikingly, GFP-3a1Nter cells exhibited suboptimal tRA-induced Vtn transcription compared to that for wt mESCs (Fig. 2A, triple asterisks).
In P19 cells, induction of Vtn gene transcription is associated with strand-specific promoter demethylation (19). We therefore examined the methylation status of a 279-bp region containing 11 CpGs (arbitrarily numbered 6 to 16) corresponding to the noncoding strand (from −370 to −90 relative to the TSS) of the Vtn promoter (Fig. 2D) by bisulfite sequencing. This region is heavily methylated in untreated wt mESCs, and the methylation percentage calculated with bisulfite sequencing was 75%. This methylation percentage dropped to 44% in tRA-treated wt mESCs (Fig. 2D). Strikingly, the methylation percentage is calculated to be 75% in untreated GFP-3a1Nter cells, as is the case in wt mESCs, and remains high (77%) after 96 h of tRA treatment of these cells. These data show that the Vtn promoter is not demethylated upon tRA treatment of GFP-3a1Nter cells and that the suboptimal Vtn tRA-dependent transcriptional induction in GFP-3a1Nter cells correlated with high levels of strand-specific methylation of the Vtn promoter. This result is in agreement with data provided by Gallais et al. (19), suggesting that Dnmt3a plays a role in the demethylation of Vtn promoter in P19 cells, presumably acting as a deaminase instead of a DNA methyltransferase.
Collectively, our data showed that Dnmt3a1 in mESCs positively affects Vtn gene transcription and simultaneously contributes to the silencing of the Oct3/4 and Nanog genes.
Dnmt3a1 is recruited to Oct3/4 and Vtn gene promoters via its N-terminal domain.
To test whether EGFP-tagged proteins were recruited to Oct3/4 and Vtn promoters upon tRA treatment, we performed chromatin immunoprecipitation (ChIP) analysis of these elements using wild-type mESC, GFP-3a1Nter cell, and GFP-3a2 cell chromatin preparations and specific antibodies. We used anti-GFP antibody in addition to anti-G9a and anti-COUP-TF antibodies, since G9a and COUP-TF are thought to tether Dnmts to the Oct3/4 and Vtn gene promoters, respectively, upon tRA stimulation (1, 16, 17).
As expected, Oct3/4 but not Vtn promoter sequences were enriched in all ChIP samples from 120-h tRA-treated cells using the anti-G9a antibody (Fig. 3 A). In addition, the Vtn promoter was enriched in anti-COUP-TF immunoprecipitated samples from all chromatin preparations from tRA-treated cells (Fig. 3A, wt, GFP-3a2, and GFP-3a1Nter). Interestingly, Oct3/4 gene promoter sequences were also enriched in COUP-TF ChIP samples (Fig. 3A), suggesting that COUP-TFI was recruited to both the silenced Oct3/4 promoter and the activated Vtn promoter. This is consistent with the previously suggested role of COUP-TFI in Oct3/4 gene transcriptional repression observed in embryonal carcinoma (EC) cells (3). Oct3/4 and Vtn promoter sequences from tRA-treated wt mESC, GFP-3a2 cell, and GFP-3a1Nter cell chromatin preparations were enriched in ChIP reactions performed with a polyclonal antibody recognizing both Dnmt3a1 and Dnmt3a2 (H-295; Santa Cruz) (Fig. 3A).
Anti-GFP antibody immunoprecipitated Oct3/4 and Vtn promoter sequences from GFP-3a1Nter but not from GFP-3a2 tRA-treated cells (Fig. 3A). The same antibody failed to precipitate either Oct3/4 or Vtn promoter sequences from wild-type mESC chromatin preparations (Fig. 3A and B, respectively), confirming the specificity of the ChIP reactions.
It is known that Dnmt3a is targeted to the mouse major pericentromeric repeats via its PWWP domain (13). We tested our ChIP reactions by performing ChIPs monitoring the mouse major repeats.
In line with the above evidence, we show that PCR from ChIP samples with a monoclonal antibody against the C-terminal (Cter) part of Dnmt3a protein (ab13888; Abcam) (Fig. 1B, lanes 3 and 4) recognizing both Dnmt3a isoforms, results in detecting major repeat sequences from untreated and 120-h tRA-treated wt ESCs (see supplementary Fig. 4 at http://www.fleming.gr/files/Agalioti_supplemental_document.pdf, lanes 15 and 16). Major repeats from GFP-3a2 chromatin preparations from untreated and tRA-treated samples were also enriched in ChIP reactions performed with anti-GFP antibody (see supplementary Fig. 4, lanes 11 and 12). In contrast, anti-GFP antibody failed to immunoprecipitate the mouse major repeat sequences in wt mESCs and GFP-3a1Nter chromatin preparations from untreated and tRA-treated samples (see supplementary Fig. 4, lanes 3 and 4 and lanes 7 and 8). Our results are in line with evidence indicating that the mouse major repeats are targeted by Dnmt3a via its PWWP domain and that the EGFP-3a1 Nter protein is not recruited to this genomic region. Moreover, our experiments indicate that our anti-GFP chromatin immunoprecipitations are highly specific.
To show that competition between EGFP-3a1Nter and endogenous Dnmt3a1 protein took place specifically on the promoters of the affected genes, we employed ChIP using antibodies recognizing either EGFP-3a1Nter protein or endogenous Dnmt3a1. Anti-GFP antibody immunoprecipitated both Nanog and Vtn promoter sequences specifically from 72-h tRA-treated GFP-3a1Nter cells (Fig. 3C, α-GFP antibody), indicating that EGFP-3a1Nter protein was efficiently recruited to both these sites. We subjected the same chromatin samples to ChIP using the antibody against the C-terminal (Cter) part of Dnmt3a protein (Fig. 1B, lanes 3 and 4). This anti-Dnmt3a-Cter antibody precipitated Nanog and Vtn gene sequences from tRA-treated wild-type mESCs with a 5-fold higher efficiency than that for uninduced cells (Fig. 3C, α-Dnmt3a-Cter antibody). In contrast, anti-Dnmt3a-Cter antibody failed to specifically precipitate Nanog and Vtn gene promoter sequences in tRA-treated GFP-3a1Nter chromatin samples (Fig. 3C, comparisons indicated by single and double asterisks), suggesting that tRA-induced recruitment of EGFP-3a1Nter to the Nanog and Vtn promoters efficiently outcompeted the recruitment of endogenous Dnmt3a1, thus depriving the anti-Dnmt3a-Cter antibody from encountering its epitope at these genomic sites.
Taken together, these results showed that EGFP-3a1Nter but not EGFP-Dnmt3a2 protein was efficiently recruited to Oct3/4, Nanog, and Vtn gene promoters, where it outcompeted endogenous Dnmt3a1. DNA methylation of the Oct3/4 and Nanog gene promoters was compromised, and these genes were not permanently silenced.
EGFP-3a1Nter protein interacts in vivo and in vitro with G9a HMTase.
G9a HMTase tethers Dnmt3a to the Oct3/4 gene promoter upon tRA-induced gene silencing in mESCs (17). G9a interacts with the carboxy-terminal domain of Dnmt3a1 in vitro (16). We reasoned that EGFP-3a1Nter protein must contain an interaction site, since it is efficiently recruited to the Oct3/4 gene promoter in GFP-3a1Nter cells in vivo. We therefore performed protein coimmunoprecipitation assays (co-IPs) using anti-G9a and anti-FLAG antibodies in GFP-3a1Nter and FLAG-3a1Nter cell nuclear extracts. FLAG-3a1Nter cells express a fusion protein of the 219-aa Dnmt3a1 N terminus with both an N-terminal FLAG tag and a C-terminal EGFP tag from a single-copy insertion at the Rosa26 locus (Fig. 1A).
EGFP-3a1Nter protein was detected with anti-GFP antibody in co-IPs using anti-G9a and GFP-3a1Nter nuclear extracts (Fig. 4 A, lane 6). In the complementary reaction, G9a was detected with anti-G9a antibody in co-IPs using anti-FLAG and nuclear and chromatin FLAG-3a1Nter cell extracts (Fig. 4B, lanes 5 and 8). We therefore concluded that Dnmt3a1 N-terminal domain coprecipitated with G9a in vivo in mESCs.
Fig. 4.
Dnmt3a1 N-terminal domain coimmunoprecipitates with G9a in mESCs. (A) Western blot of input (lanes 1 and 2), flowthrough (lanes 3 and 4), and immunoprecipitation (lanes 5 and 6; beads) samples from wt mESC (lanes 1, 3, and 5) and GFP-3a1Nter cell (lanes 2, 4, and 6) nuclear extracts. Immunoprecipitation reactions were performed with anti-G9a antibody (07-551; Upstate). Upper panel, detection was performed with a monoclonal anti-GFP antibody (Santa Cruz). Lower panel, detection was performed with anti-G9a antibody (Abcam). (B) Western blot of input (lanes 4 and 6), flowthrough (lanes 1, 3, and 7), and immunoprecipitation (lanes 2, 5, and 8) samples from FLAG-3a1Nter cell cytoplasmic, nuclear, and chromatin extracts. FLAG-3a1Nter cells express a double-tagged FLAG-3a1Nter-EGFP protein which can be detected with anti-FLAG and anti-GFP antibodies. Immunoprecipitation reactions were performed with anti-FLAG polyclonal antibody (Sigma). Upper panel, detection was performed with a monoclonal anti-GFP antibody. Lower panel, detection was performed with anti-G9a antibody (Abcam). (C) Schematic representation showing Dnmt3a1 domains and deletion proteins fused to GST. Numbers indicate amino acid positions. The GST pull-down experiment was performed using bacterially produced and purified corresponding GST-Dnmt3a1 domains and in vitro-translated G9a protein labeled with [35S]methionine. The results of this experiment are summarized in the table on the left. Frg, fragment number.
Next, we used a series of deletion constructs of Dnmt3a1 fused to glutathione S-transferase (GST) to investigate direct interactions with G9a (Fig. 3C). High-stringency GST pull-down experiments with these deletion proteins and in vitro-translated radioactively labeled G9a revealed multiple contacts between Dnmt3a and G9a. We observed strong interactions between G9a and either the Dnmt3a1 N-terminal domain (aa 1 to 219) or sequences spanning the PWWP domain of Dnmt3a (aa 242 to 467) (Fig. 4C, fragments 4 to 6). An interaction between the PWWP domain of Dnmt3a and G9a is suggested by Dong et al. (15). Neither the extreme C-terminal sequences (aa 567 to 903) nor the middle part of the C-rich region of Dnmt3a (aa 487 to 542), showed detectable interactions with G9a. However, the region between aa 487 and 903 did interact strongly (Fig. 4C, fragment 7). We propose that an additional G9a interaction region may require aa 542 to 567 of Dnmt3a1/3a2, contributing either to direct contacts to G9a or to the correct folding of the corresponding deletion protein.
Quantitative differences in the positive and negative transcriptional effects of Dnmt3a1.
Dnmt3a positively affects transcription when participating in demethylation cycles at active promoters (28, 35) and Dnmt3a-mediated demethylation has been shown to exert a positive effect on tRA-activated transcription of the Vtn gene (19). Moreover, Dnmt3a has a well-established role in permanent transcriptional silencing, for example, in the case of the Oct3/4 and Nanog genes. An interesting question is to what extent ESCs make use of Dnmt3a's positive versus negative activities and what the target genes are in each case. To gain insight into this question, we compared the transcription profiles of wt mESCs and GFP-3a1Nter cells, under either untreated or 120-h tRA-treated conditions (Fig. 5 A). Expression of the N-terminal domain of Dnmt3a1 is expected to compromise wt Dnmt3a1 function, thus revealing positive and negative transcriptional targets in the differential comparison. Labeled cRNAs corresponding to each condition were used to hybridize mouse Gene 1.0 ST Arrays (Affymetrix). The raw data were subjected to normalization and background correction.
Fig. 5.
mESCs utilize Dnmt3a1 positive and negative effects in transcription to different extents. (A) Schematic representation of the pairwise transcription profile (microarray) comparisons between untreated wt mESC and GFP-3a1Nter cell samples (green) and between tRA-treated wt mESC and GFP-3a1Nter cell samples (blue). The numbers correspond to one-way ANOVA-selected probe sets (p.s) that are differentially expressed between conditions and fulfill the selection criteria (P values, ≤0.05; expression change, ±1.5-fold across conditions). (B) Venn diagram comparison between differentially expressed probe sets (p.s) for untreated (green) and tRA-treated (blue) wt mESC versus GFP-3a1Nter cells. There were 12 probe sets that changed between the two cell lines under both untreated and tRA-treated conditions, 143 differentially expressed probe sets fulfilling the selection criteria (A) exclusively in untreated cells, and 364 probe sets differentially expressed in tRA-treated cells. In sum, 519 probe sets changed across conditions. (C) The 519 probe sets that changed between mESCs and GFP-3a1Nter cells were divided according to their upregulation (green) or downregulation (pink) in GFP-3a1Nter cells. A comparison of these categories under undifferentiated (LIF) or tRA-treated conditions revealed differences in both the ratio and the extent of up- and downregulation after tRA treatment differentiation.
To identify specific genes that were affected by Dnmt3a1, one-way analysis of variance (ANOVA) comparisons were performed between wt mESC and GFP-3a1Nter samples that were either untreated or tRA treated (P values, ≤0.05; expression change, ±1.5-fold across conditions). We identified 155 probe sets (PS) that were differentially expressed between untreated wt mESCs and GFP-3a1Nter cells, and 376 PS that were differentially expressed between tRA-treated wt mESCs and GFP-3a1Nter cells (Fig. 5A). The Venn diagram in Fig. 5B illustrates that there were 12 PS in common between these two categories of PS, in addition to 143 PS unique to untreated cells and 364 PS unique to tRA-treated cells.
Further analysis of these 519 PS revealed a balance of Dnmt3a1-positive and -negative functions in transcription in undifferentiated mESCs (Fig. 5C, LIF), because the 155 affected PS in the GFP-3a1Nter samples were divided nearly equally into two groups of 78 and 77 PS, exhibiting higher and lower transcript levels, respectively, than those for the wt mESC samples. In contrast, upon tRA induction, the amount of affected PS in the GFP-3a1Nter samples doubled (Fig. 5C, tRA). In addition, a much larger group of PS (279) exhibited higher levels of transcription than those for the wt samples, with only 97 PS representing Dnmt3a1-positive targets (PS with lower levels than the wt ESC samples) (Fig. 5C, tRA, green versus pink).
In summary, our data present a quantitative picture of the positive and negative effects of Dnmt3a1 in untreated and tRA-stimulated mESC transcription. We find that across the 519 PS identified as differentially expressed as a result of Dnmt3a1-compromised function in GFP-3a1Nter cells, the majority, 347 PS, representing 1% of the total number of the inquired PS, are upregulated, whereas 172 PS, representing 0.47% of the inquired PS, are repressed (Fig. 5C). Based on our data, we suggest that Dnmt3a1 plays a major role in induced differentiation of mESCs by exerting its negative functions to consolidate transcriptional silencing. Dnmt3a1 also assumes a minor role in gene expression in the undifferentiated mESC transcriptome.
A small but distinct set of genes requires Dnmt3a1 for optimal tRA-dependent transcription induction.
The up- and downregulated probe sets identified as differentially expressed between wt mESCs and GFP-3a1Nter cells were further analyzed as to the cell functions affected by the expression of the tagged Dnmt3a1 N-terminal domain in mESCs. Among the downregulated probe sets in untreated and tRA-treated GFP-3a1Nter cells, there were 46 annotated genes that shared some remarkable features (Table 1). These genes are all related to trophectodermal lineage functions influencing the overall fitness of the embryo during gestation. These genes are scattered throughout the genome in terms of chromosomal position and are sometimes associated with various types of imprinting (Table 1). Five of these genes are known to be imprinted (Grb10, Igf2r, Cdkn1c, Zim1, H13), and others have well-documented functions in the placenta (Htra1, Ppap2b, Ihh, Gpr50, Cited, Dusp9). The placenta-specific expression of a subgroup of genes in this category (Gpr50, Cited, and Dusp9) is also imprinted due to X inactivation. Several other genes function in insulin signaling, adipose tissue, and liver development (Dkk1, Klb, Dgat2).
Table 1.
Annotated genes among the downregulated probe setsa
| Name | Function | CHR |
|---|---|---|
| Grb10 | Maternally expressed in all tissues, paternally expressed in the brain; insulin receptor signaling component | 11A1 |
| Gpr50 | Orphan receptor resembling melatonin receptors; involved in energy metabolism | XA7.2 |
| Cited1 | Encodes a transcriptional cofactor interacting with Smad4, ER, TFAP2, and CBP/p300; functions in the placenta, where due to X inactivation its expression is imprinted | XD |
| Htra1 | Encodes serine protease; inhibitor of TGF-β signaling with a function in ovulation and placenta development | 7F3 |
| Klb | Encodes single-pass transmembrane protein expressed in adipocytes, interacting with the FGF21 receptor; controls FGF-stimulated glucose uptake in adipocytes | 5C3.1 |
| Myo7a | Deafness gene having to do with inner-ear development; in humans, causes Usher syndrome; mice bearing its mutation are called waltzer mice | 7T2 |
| Runx1 | Transcription factor; null mice lack hemopoietic cells | 16C3.3 |
| Dkk1 | Encodes a secreted protein which binds to Kremen receptors and antagonizes canonical Wnt signaling via swift sweeping out of Lpr6 receptors; functions in bone marrow and adipocyte differentiation | 19C1 |
| Folr1 | Receptor; controls folate metabolism; important for hemopoiesis | 7T3 |
| Foxq1 | Encodes “satin hair” mouse mutant protein with a role in regulating gastric epithelium acid secretion and hair shaft development | 13A3.2 |
| 1110032E23Rik | Unknown gene downregulated by FGF signaling with embryonic expression in the gastrointestinal tract | |
| Gas6 | Negative regulator of chondrogenesis through ERK signaling; also functions in endothelial cells in enhancing the interactions between endothelial cells, platelets, and leukocytes during inflammation | 8A1.1 |
| BC026682 | Predicted to encode a membrane protein | 4C7 |
| Amn | Encodes a single-pass transmembrane protein indispensable for gastrulation in mice; nullizygous mice lack the mesoderm that forms the “trunk”; its N terminus is required for absorption of vitamin B12 | 12F1 |
| Wfikkn1 | Encodes a serine protease activity inhibitor | 17A3 |
| Metrn | Neurotropic factor involved in gliogenesis | 17A3.3 |
| Rbmx | Hepatic transcriptional regulator of the SREBP-1c gene in response to a high-fructose diet, which induces what resembles metabolic syndrome in mice | XA5 |
| Wnt11 | Antagonist of the canonical Wnt pathway and a known positive factor in heart development; indirect evidence shows that it causes abortions when ectopically expressed (by dicer knockout) in the glandular epithelium of the uterus | |
| Zim1 | Transcription factor; maternally expressed imprinted gene | 7A1 |
| Hoxd8 | Expression is increased in midgestational wounds compared with that for healthy control groups and late gestational wounds, suggesting that it may play a role in scarless wound repair | 2C3 |
| Dgat2 | Encodes an enzyme catalyzing the final step in mammalian triglyceride synthesis; has something to do with obesity and adipocyte hypertrophy | 7T2 |
| 4930444G20Rik | No information; gene product has homology to a peptidase | 10A3 |
| Punc | Encodes a novel NCAM; dispensable for normal embryogenesis but expressed in the inner ear and in brain regions controlling balance | 9B |
| Vstm2b | Encodes a membrane protein; no other information | 7B3 |
| Ascl1 | Encodes a very famous protein; proneural gene; bHLH transcription factor functioning in the context of the Notch pathway | 10C1 |
| Igf2r | Paternally imprinted gene, maternally expressed | 17A1 |
| Cpm | Encodes a GPI-anchored protein; angiotensin-converting enzyme may have something to do with its release by attacking GPI | 10D2 |
| Dusp9 | Encodes a phosphatase with a protective effect against insulin resistance; during embryonic development, functions in the placenta where due to X inactivation its expression is imprinted | XA7.3 |
| Gbgt1 | Encodes a Forssman antigen of the ABO blood group-determining antigens; membranic in topology and a transferase with unknown substrate; the murine gene might confer protection against Shiga toxin and participate in carbohydrate metabolism | 2A3 |
| F630110N24Rik | No information; encodes a transmembrane protein and possibly functions in transport | 10C1 |
| Serpinf2 | Also known as the alpha 2-antiplasmin gene; TGF-β-signaling agonist in inducing fibrosis | 11B5 |
| H13 | Imprinted gene; gene product is an aspartyl peptidase, also called Spp; member of the presenilin family; confers histocompatibility-based rejection of allografts | 2T5 |
| Mex3d | Based on its homology, this gene is involved in destabilization of RNA and RNA binding | 10C1 |
| Ihh | Functions in ossification along with the Wnt pathway; also has roles in epithelial-mesenchymal transition, in the intestine crypt-epithelium developmental axis in an Rb-regulated manner, and in implantation due to its critical expression in the uterine epithelium | 1C3 |
| Ppap2b | Gene product is also called Lpp3; mice lacking its lipid phosphate phosphatase 3 fail to form chorioallantoic placenta and yolk sac vasculature; also shows some characteristic canonical Wnt signaling impairment | 4C6 |
| Ctsz | Gene product is also called cathepsin Z or cathepsin S; encodes a cysteine-type endopeptidase that functions in lysosomes; may have a role in antigen recognition | |
| AA986860 | Encodes an androgen-regulated protein | 1T4 |
| XM_001479553 | No information; homology with HMG14 and HMG17 molecules | |
| Plxnc1 | Encodes plexins, which, comprising plexin A, B, C, and D subfamilies, are receptors for semaphorins governing cell adhesion, migration, and axon guidance | 10C2 |
| Rnf182 | No information; targeted knockout exists | 13A4 |
| Frem1 | Fraser syndrome mutants; important for epithelial adhesion | 4C3 |
| Cdkn1c | Also called p57Kip; imprinted; the mutant mouse is the model of preeclampsia leading to miscarriages, symptoms of which appear not only when mothers bear the p57 deletion but also when the fetuses are nullizygous; gene also has a role in ossification | 7F2 |
| Serpinb9f | Serine peptidase activity inhibitor | 13A2 |
| Atoh8 | bHLH transcription factor which is embryonic lethal; may function in kidney and pancreas; also called Math6; also expressed in the brain | 6C1 |
| Otfr566 | Encodes a G-protein olfactory receptor; no other information | 7T3 |
| Plekhh1 | No information | 12C3 |
These genes are all related to trophectodermal lineage functions influencing the overall fitness of the embryo during gestation and are scattered throughout the genome. CHR, chromosome; ER, estrogen receptor; TGF-β, transforming growth factor β; FGF, fibroblast growth factor; ERK, extracellular signal-regulated kinase; NCAM, neural cell adhesion molecule; bHLH, basic helix-loop-helix protein; GPI, glycosylphosphatidylinositol.
We verified our microarray results by performing reverse transcription (RT)-coupled q-PCR to compare the mRNA levels for selected genes of Table 1 between wt mESCs and GFP-3a1Nter cells under untreated or tRA-treated conditions (Fig. 6 A). The tested genes, the Grb10, Gpr50, Ihh, Dkk1, Igf2r, and Cdkn1c genes, are tRA inducible in mESCs, and their mRNA levels were significantly reduced in tRA-treated GFP-3a1Nter cells compared to levels for tRA-treated wt mESCs (Fig. 6A).
Fig. 6.
Genes with compromised tRA-dependent transcriptional activation in GFP-3a1Nter cells require Dnmt3a1 at the promoter level. (A) q-PCR from reverse-transcribed total RNA samples from untreated and 120-h tRA-treated wt mESC and GFP-3a1Nter cells. Amplification was performed with primers specific for Dkk1, Grb10, Igf2r, Cdkn1c, Ihh, and Gpr50 gene mRNAs. (B) Bisulfite sequencing of the Grb10 imprinting genomic domain residing between the first and second exons. Sequenced clones were PCR derived using specific primers from bisulfite-treated wild-type mESCs and GFP-3a1Nter cells. Methylated clones presumably correspond to the active (maternal) allele of the Grb10 gene, while unmethylated sequences correspond to the inactive (paternal) allele. (C) Dnmt3a1 positively affects the transcriptional activation of Gpr50, Vtn, and Grb10 genes at the promoter level. PCR with “up” and “down” sets of primers mapping near the promoter and the last exon of the genes, respectively (position shown on the upper scheme) indicated that the same amount of mRNA was produced irrespective of the primer position.
Imprinted genes are regulated by differentially methylated CG-rich regions (DMRs) existing within their loci. These CpG islands, when unmethylated, serve as antisense transcripts (Air) and as neural specific gene promoters in the cases of the Igf2r and Grb10 genes, respectively.
We tested the integrity of the methylated imprint present in the first intron of the Grb10 gene by bisulfite sequencing. Bisulfite-converted genomic DNA was subjected to PCR using specific primers which amplify a region of 282 bp within the Grb10 gene DMR, containing 25 CpGs.
In wt mESCs, this genomic region appeared either totally unmethylated (0% methylation, with the unmethylated sequences presumably corresponding to the silent paternal allele) or fully methylated (95.2% methylation, with the methylated sequences presumably corresponding to the active maternal allele) (Fig. 6B). For GFP-3a1Nter cells, we found either fully unmethylated clones (0% methylation) or severely hypomethylated clones (16.8% methylation). Thus, the methylation of this DMR is compromised in GFP-3a1Nter cells compared to that in wt mESCs. These results suggest that endogenous Dnmt3a1 may be retaining the methylation pattern of the Grb10 locus imprint by tethering to this sequence via its N-terminal domain.
We tested whether transcription of the Dnmt3a1 positively regulated genes was compromised at the initiation step or at elongation through methylated areas, such as DMRs. We reasoned that if preservation of the Grb10 gene imprint depended on transcriptional read-through of the area, then it could affect transcriptional elongation in GFP-3a1Nter cells. In the case of compromised mRNA elongation, the RT-coupled q-PCR of GFP-3a1Nter cells should reveal different transcript levels, depending on whether the mRNA region monitored is near the promoter or further downstream near the last exon of the gene.
The genes depicted in Fig. 6C were selected for this test. The active (maternal) copy of the Grb10 gene contains the methylated DMR within its first intron (52). The Gpr50 gene also contains a CpG island within its first intron, and the Vtn gene has a methylated promoter, although no CpG islands are found within its coding sequences (reference 19 and data presented here). To test for mRNA elongation efficiency, we designed two sets of primers for each of these genes, an “up” set mapped near the TSS (5′ untranslated region [UTR]) and a “down” set at the 3′ UTR (Fig. 6C).
The relative mRNA levels obtained by the “up” and “down” primer sets of the Gpr50, Vtn, and Grb10 genes were within the same range in wild-type mESCs and GFP-3a1Nter cells and failed to detect different mRNA levels with respect to relative mRNA positions in either wt or GFP-3a1Nter cells. In all three cases, the mRNA levels monitored with the “up” set of primers mapping near the gene promoter were significantly lower in tRA-treated GFP-3a1Nter cells than in wild-type mESCs, as was also found for all of the other primer sets used in the experiments depicted in Fig. 6A and C. We conclude that the transcriptional defect measured for Gpr50, Vtn, and Grb10 genes cannot be attributed to compromised elongation but may be due to suboptimal transcriptional initiation of these three genes in tRA-treated GFP-3a1Nter cells.
Dnmt3a1 targets whole chromosomal regions, including gene clusters for epigenetic silencing in mESCs.
Among the upregulated probe sets in GFP-3a1Nter cells compared to wild-type mESCs, there are 179 annotated genes that are presented in Table 2. Remarkably, upon examining the chromosomal locations of these genes, we noticed that in the vast majority of cases, two or more of them cluster together. The most interesting gene cluster is on chromosome 6 and involves the important ES cell pluripotency-determining Nanog, Dppa3 (also known as Stella), and Gdf3 genes (Fig. 7 A, schematic representation). In addition, the Kif3a and Rad50 genes, which show transcriptional upregulation in GFP-3a1Nter cells, map to chromosome 11 and constitute the boundaries of a genomic region containing interleukin-4 (IL-4) and IL-13 cytokine genes, which are important for T-cell differentiation and maturation. Importantly, the Kif3a, IL-4, IL-13, and Rad50 genes are distributed over an ∼100-kb region also containing a locus control region (LCR) located at the 3′ end of the Rad50 gene (29). The region (schematically depicted in Fig. 7A) exhibits different chromatin structures in different T-cell lineages, and our results may implicate Dnmt3a as having an as-yet-unidentified role in the proper “archiving” or “bookmarking” of this region at a very early developmental stage, such as that of mESCs.
Table 2.
Annotated genes among the upregulated probe setsa
| Name | CHR |
|---|---|
| Plagl1 | 10A2 |
| Ctgf | 10A4 |
| Enpp3 | 10A4 |
| Prdm1 | 10B2 |
| Grik2 | 10B3 |
| Fabp7 | 10B4 |
| Spic | 10C1 |
| Erbb3 | 10D3 |
| Upp1 | 11A1 |
| 2410008K03Rik | 11A1 |
| B3gnt2 | 11A3.2 |
| Zrsr1 | 11A3.2 |
| Papolg | 11A3.2 |
| Ccdc88a | 11A3.3 |
| Ccdc99 | 11A4 |
| Cpeb4 | 11A4 |
| Ranbp17 | 11A4 |
| Ublcp1 | 11B1.1 |
| Glra1 | 11B1.3 |
| Kif3a | 11B1.3 |
| Rad50 | 11B1.3 |
| Zcchc10 | 11B1.3 |
| Ulk2 | 11B2 |
| Rich2 | 11B3 |
| Hs3st3a1 | 11B3 |
| Pipox | 11B5 |
| Ssh2 | 11B5 |
| Rad51c | 11C |
| Calcoco2 | 11D |
| Cntd1 | 11D |
| Fbxo47 | 11D |
| Gm1564 | 11D |
| Ccdc46 | 11T1 |
| Itgb3 | 11T1 |
| Pecam1 | 11T1 |
| Sstr2 | 11T2 |
| Tex19.1 | 11T2 |
| ENSMUSG0000005 | 12A1.1 |
| Six1 | 12C3 |
| Pfkp | 13A1 |
| Zfp640 | 13B3 |
| Vmn2r-ps105 | 13B3 |
| 2410141K09Rik | 13B3 |
| C330022B21Rik | 13B3 |
| ENSMUSG0000007 | 13B3 |
| Duxbl | 14A4 |
| Otx2 | 14C1 |
| Ccnb1ip1 | 14C1 |
| Clu | 14D1 |
| ENSMUSG0000 | 15A1 |
| Aard | 15C |
| Ly6a | 15D3 |
| Smc1b | 15T2 |
| Etv5 | 16B1 |
| Fetub | 16B1 |
| Liph | 16B1 |
| 676915 | 16B1 |
| Morc1 | 16B5 |
| Fndc1 | 17A1 |
| Hsf2bp | 17A3.3 |
| Dazl | 17C |
| Psma8 | 18A1 |
| Spry4 | 18B3 |
| Dmrt1 | 19B |
| 6530404N21Ril | 19C1 |
| Mybl1 | 1A2 |
| Aim2 | 1A3 |
| Prdm14 | 1A3 |
| Rims1 | 1A5 |
| Mgat4a | 1B |
| Nab1 | 1C1.1 |
| Stat4 | 1C1.1 |
| Inpp5d | 1D |
| Rgs4 | 1H2.2 |
| Lefty1 | 1H5 |
| Lefty2 | 1H5 |
| Nr5a2 | 1T4 |
| Pax8 | 2A3 |
| Rif1 | 2C1.1 |
| Gca | 2C1.3 |
| Nnat | 2H2 |
| Tcfap2c | 2H3 |
| Thbd | 2T5 |
| Chmp4c | 3A1 |
| Lrrc34 | 3A3 |
| Mnd1 | 3F1/3E3 |
| Adh1 | 3G3 |
| Bche | 3T3 |
| Aqp3 | 4A5 |
| Ttc39b | 4C4 |
| Dnajc6 | 4C6 |
| L1td1 | 4C6 |
| C130073F10Rik | 4C6 |
| Zyg11a | 4C7 |
| Grhl3 | 4D3 |
| Slc30a2 | 4D3 |
| Rex2 | 4T1 |
| 2610305D13Rik | 4T1 |
| OTTMUSG00000010694 | 4T1 |
| OTTMUSG00000011061 | 4T1 |
| OTTMUSG00000011070 | 4T1 |
| OTTMUSG00000010681 | 4T1 |
| OTTMUSG00000011097 | 4T1 |
| Kit | 5C3.2 |
| Spp1 | 5T5 |
| BC020002 | 6A1 |
| Gkn2 | 6D1 |
| Trh | 6D1 |
| Apobec1 | 6F2 |
| Gdf3 | 6F2 |
| Nanog | 6F2 |
| Dppa3 | 6F2 |
| Hsn2 | 6T1 |
| AU018091 | 7A1 |
| EG627821 | 7A1 |
| Nlrp4c | 7A1 |
| Tdrd12 | 7B1 |
| Hsd17b14 | 7B4 |
| C330019L16Rik | 7B4 |
| ENSMUSG00000071952 | 7B4 |
| BC066028 | 7F5 |
| Ifitm1 | 7F5 |
| Nlrp14 | 7T3 |
| C330021F23Rik | 8A1.1 |
| Tacc1 | 8A2 |
| Wrn | 8A3 |
| Acsl1 | 8B1.1 |
| Zfp42 | 8B1.1 |
| Gpm6a | 8B1.3 |
| Neil3 | 8B1.3 |
| Gbp6 | 8B2 |
| Galnt7 | 8B2 |
| Cbr4 | 8B3.1 |
| 2700029M09Rik | 8B3.1 |
| Klf2 | 8B3.3 |
| Lpl | 8B3.3 |
| Psd3 | 8B3.3 |
| ENSMUSG000 | 8B3.3 |
| Isx | 8C1 |
| Slc10a7 | 8C1 |
| Zfp330 | 8C2 |
| Chd9 | 8C5 |
| Mt2 | 8C5 |
| Polr2c | 8C5 |
| Slc6a2 | 8C5 |
| Nqo1 | 8D3 |
| Cdyl2 | 8T1 |
| Plcg2 | 8T1 |
| Slc35f2 | 9A5.3 |
| 2410076l21Rik | 9B |
| Rab27a | 9D |
| Cdv3 | 9F1 |
| Lrrc2 | 9F2 |
| Tdgf1 | 9F3 |
| Pcolce2 | 9T3.3 |
| Efhc2 | XA1.2 |
| Maob | XA1.2 |
| Gm9 | XA3.3 |
| Rhox4b | XA3.3 |
| 1700013H16Ri | XA5 |
| Xlr5a | XA7.3 |
| Slc7a3 | XC3 |
| Dmrtc1c | XD |
| Taf9b | XD |
| AV320801 | XE3 |
| EG331529 | XF1 |
| Nxf3 | XF1 |
| Pramel3 | XF1 |
| Acot9 | XF3 |
| Ott | XF3 |
| Scml2 | XF4 |
| Zfy1 | YA1 |
| Ube1y1 | YA1 |
In the vast majority of cases, two or more of these genes are clustered together. See the text for discussions of entries shown in boldface type. CHR, chromosome.
Fig. 7.
Genes with higher mRNA levels in GFP-3a1Nter cells compared to wild-type mESCs cluster together in large chromosomal domains. (A) Schematic representation (not to scale) of two chromosome domains containing Nanog, Dppa3, Gdf3, and Apobec genes on chromosome 6 and Rad50, IL-13, IL-4, and Kif3a genes on chromosome 11. Black vertical lines indicate the presence of CpG islands within these domains. (B) q-PCR from reverse-transcribed total RNA samples from untreated and 120-h tRA-treated wt mESCs and GFP-3a1Nter cells. Amplification was performed with primers specific for mRNAs of Dppa3, Rad50, Gdf3, Kif3a, Apobec, Dazl, and Plagl1 genes.
We verified our microarray results for a selected list of the upregulated genes by performing RT-coupled q-PCR (Fig. 7B). Although not all the genes tested were affected by tRA, all mRNA levels were elevated in the GFP-3a1Nter cells compared to levels for wild-type mESCs. Our data suggest that Dnmt3a1 targets specific chromosomal regions that contain ESC pluripotency genes and primordial germ cell (PGC) determinants for methylation-dependent permanent silencing during somatic ESC differentiation. Future high-definition mapping of these areas should reveal the specific sequences which are targeted by Dnmt3a1 in these cells.
Dnmt3a1 protein KD affects silencing and tRA transcription induction of selected microarray-identified Dnmt3a1 gene targets.
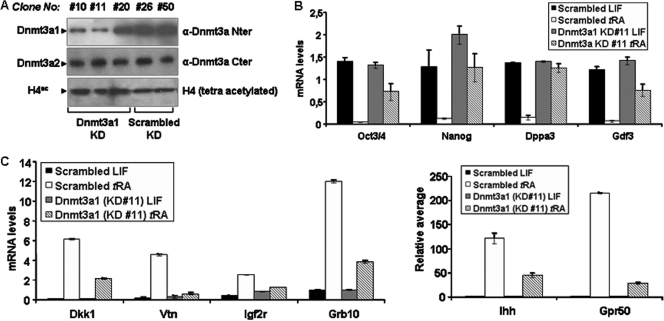
The above-described experiments indicate that the expression of the catalytically inactive Dnmt3a1 N-terminal domain in mESCs interferes with the functions of the endogenous Dnmt3a1 protein. By using the GFP-3a1Nter cells, we were able to identify developmentally significant genes and genes important for mESC differentiation, respectively, for which Dnmt3a1 exerts positive and negative effects to their transcription. To corroborate these data, we proceeded to reduce the protein levels of the endogenous Dnmt3a1 in mESCs and to test the effect of this reduction in selected identified Dnmt3a1 gene targets. To this end, stable mESC cell lines which express either a scrambled or a Dnmt3a1-specific mRNA targeting hairpin (shRNA) were obtained. These stable clones were tested for the expression of Dnmt3a1 and Dnmt3a2 by Western blotting. Dnmt3a1 knockdown (KD) clones 10 and 11 show 80%-reduced Dnm3a1 mRNA levels upon tRA treatment (data not shown) and minimized expression of endogenous Dnmt3a1 (Fig. 8 A, upper panel), while Dnmt3a2 levels remain intact (Fig. 8A, middle panel). RNA samples from untreated or 96-h tRA-treated KD clone 11, along with those from control clone 50 expressing the scrambled shRNA, were analyzed by RT-coupled q-PCR with primers specific for the Oct3/4, Nanog, Dppa3, and Gdf3 genes. We found that the mRNA levels for all four genes were elevated in tRA-treated KD clone 11 compared to levels for tRA-treated control clone 50 cells (Fig. 8B). We conclude that in the cells with reduced Dnmt3a1 protein levels, none of the four genes tested are optimally silenced upon tRA treatment. Furthermore, we tested the tRA-dependent transcription induction of the Dkk1, Vtn, Igf2r, Grb10, Ihh, and Gpr50 genes for KD clone 11 and control clone 50. We found that none of the tested genes was optimally induced in 96-h tRA-treated KD clone 11 cells (Fig. 8C). We therefore conclude that Dnmt3a1 protein level reduction compromises the transcriptional induction of the microarray-identified genes representing Dnmt3a1-specific targets.
Fig. 8.
Dnmt3a1 knockdown compromises the tRA-mediated activation and silencing of specific genes. (A) Western blot of chromatin protein samples from stable clones expressing Scrambled (Scrambled KD; clones 26 and 50) or Dnmt3a1-specific shRNA (Dnmt3a1 KD; clones 10, 11, and 20). Upper panel, detection was performed with the H-295 antibody (Santa Cruz) against the N-terminal Dnmt3a1/3a2 domain (aa 1 to 295). Lower panel, detection was performed with an antibody against the chromatin-associated tetra-acetylated histone H4 N-terminal tail (Upstate). (B) q-PCR from reverse-transcribed total RNA samples from untreated and 96-h tRA-treated stable mESC clones expressing Scrambled (clone 50) and Dnmt3a1-specific shRNA (clone11). Amplification was performed with primers specific for mRNAs of the Oct3/4, Nanog, Dppa3, and Gdf3 genes. (C) q-PCR from reverse-transcribed total RNA samples from untreated and 96-h tRA-treated stable mESC clones expressing Scrambled (clone 50) and Dnmt3a1-specific shRNA (clone 11). Amplification was performed with primers specific for Dkk1, Vtn, Igf2r, Grb10, Ihh, and Gpr50 gene mRNAs.
DISCUSSION
How is the epigenome created during cell differentiation? One unresolved question involves understanding the interrelationship among and the order of the various epigenetic modifications relative to transcriptional silencing. Another is how the de novo Dnmts, enzymes binding DNA with no apparent sequence specificity, are recruited to their target genomic sites to create the DNA methylation patterns that form a cell type-specific molecular fingerprint within the organism.
De novo Dnmts are recruited to specific genomic sites via different protein regions. For example, the de novo Dnmt PWWP domain directs Dnmt3a and Dnmt3b to the pericentromeric major repeats (13). On the other hand, the Dnmt3L ATRX domain-plant homology domain (PHD) binds specifically to the extreme N terminus of the histone H3 tail when lysine 4 of this tail is unmethylated (H3K40me) (24, 40, 42). Crystallographic and biochemical data suggest that Dnmt3a enzymatic activity is stimulated by Dnmt3L and that Dnmt3a contacts the H3K40me tail associated with the Dnmt3L PHD (21, 25, 40). In addition, the PHD of Dnmt3a binds preferentially to arginine 3 on the histone H4 tail when it has been dimethylated (H4R3me2) by PRMT5 (53).
The N-terminal domain of Dnmt3a is called a “regulatory” domain because it does not possess enzymatic DNA methyltransferase activity. This domain does not share significant homology with any other known protein. Here, we explore some of the unique functions of Dnmt3a1 in ESCs by using this N-terminal domain to demarcate some of its specific functions in vivo. Specific target genes for Dnmt3a1 have not been identified for mESCs, and although the Dnmt3a genomic locus is responsible for the establishment of genomic imprinting in PGCs, it is not clear which one of the two proteins (Dnmt3a1 or Dnmt3a2) mediates this function (27, 30, 47).
EGFP-3a1Nter is a soluble nuclear protein, unlike endogenous Dnmts. Dnmt3a1 is known to be SUMOylated at its N-terminal domain (32); however, the appearance of EGFP-3a1Nter at a higher-than-anticipated molecular weight in this study is probably due to the protein's anomalous migration in SDS-PAGE rather than being the result of eukaryotic cell-specific posttranslational modifications.
We showed that expression of the catalytically inactive EGFP-3a1Nter protein from a single-copy allele of the Rosa26 locus efficiently compromised Dnmt3a1 functions in mESCs by exerting a dominant-negative effect. An alternative explanation for our observations could be the case of a neomorphic effect (a newly acquired property) of the EGFP-3a1Nter protein. If there were a neomorphic effect of this protein, the results obtained with the GFP-3a1Nter cells should significantly deviate from those obtained from Dnmt3a1 knockdown cells. For the Oct3/4, Nanog, Dppa3, Gdf3, Dkk1, Vtn, Igf2r, Grb10, Ihh, and Gpr50 genes tested in this study, this is not the case. Specifically, we find that the transcription of these genes is affected similarly by EGFP-3a1Nter transgene expression and by the Dnmt3a1-specific knockdown in the corresponding cells.
EGFP-3a1Nter is recruited to the Oct3/4 gene promoter where it outcompetes endogenous Dnmt3a1, resulting in hypomethylation of this genomic region. The remaining DNA methylation we observe at this promoter for tRA-treated GFP-3a1Nter cells could be due to DNA methylation attributed to Dnmt3b (2). Dnmt3a and Dnmt3b are both recruited to the Oct3/4 and Nanog gene promoters (33); however, in the absence of Dnmt3a, the Oct3/4 gene promoter methylation is unstable (2). Since GFP-Dnmt3a2 on the Oct3/4 gene promoter is not detected, we suggest that the recruitment of Dnmt3a on this promoter is mediated by tethering the N-terminal part of Dnmt3a1 to these sites.
G9a HMTase recruits Dnmt3a to the Oct3/4 promoter (16, 17). The Dnmt3a C-terminal domain is known to bind to G9a in vitro (16). We discovered an additional in vitro interaction between G9a and the unique N-terminal domain of Dnmt3a1. Our data suggest that this newly identified interaction is tethering Dnmt3a1 protein to the Oct3/4 gene promoter.
Dnmt-mediated DNA methylation is associated largely with transcriptional silencing and heterochromatinization, but experimental evidence suggests that de novo Dnmt3 enzymatic activity can be modified in vivo. More specifically, Dnmts can exploit the ability of nuclear receptor-dependent promoters to bind 5-methylcytosine (5meC) and deaminate it toward thymine. This triggers the base excision repair (BER) pathway, which ultimately results in cytosine demethylation (28, 35, 54). Although the details of when and how deamination is favored over the methyltransferase reaction are not understood, this mechanism is associated with transcriptional activation rather than silencing (6, 20). It is unclear if either Dnmt3a1 or Dnmt3a2 or both of them function as deaminases (20, 41), and the positive effect of Dnmt on transcription is limited to a number of nuclear receptor target genes, such as the vitronectin (Vtn) gene. Upon transcription induction, the demethylation of the Vtn promoter is impaired in GFP-3a1Nter cells, as is the case in P19 cells (19). Our data suggest that Dnmt3a1 functions as a deaminase in the case of the Vtn gene.
Dnmt3a1 is a central player exerting both negative and positive functions in mESC transcription. An interesting question is to what extent Dnmt3a1 affects transcription in mESCs, either positively or negatively. Here, we compared the transcription profiles of wt mESCs and the GFP-3a1Nter cells to identify gene targets requiring Dnmt3a1 for their efficient transcriptional activation or silencing upon tRA induction. We find that the expression of the EGFP-3a1Nter transgene affects the transcription of 1.4% of the probe sets investigated in this study. More specifically, 1% of these PS requires Dnmt3a1 for silencing, whereas 0.4% of these PS depends on Dnmt3a1 for efficient transcription. Thus, Dnmt3a1 functions principally as a negative and to a lesser extent as a positive regulator of transcription in mESCs, as is the case in postnatal neural stem cells (NSCs) (51).
Our results suggest that the transcription of a large group of genes is negatively regulated by Dnmt3a1 in mESCs. Importantly, we find many examples of clustering of these genes on specific chromosomes. This is an interesting finding, since Dnmt3a1 is suggested to target whole chromosomal domains by an as-yet-unidentified mechanism. It is possible that these chromosomal territories contain Dnmt3a1 target genes which are responsive to signals other than tRA.
Interestingly, among the Dnmt3a1 negatively regulated genes, we identified the Apobec1 (AID family) and Dppa3 (also called Stella) genes. These two genes have enzymatic functions associated with the undifferentiated phenotype of mESCs and play important roles in genomic DNA demethylation (7, 37, 43). This result suggests that Dnmt3a1, besides being a dual methyltransferase/deaminase enzyme, could also affect global DNA demethylation by regulating the expression of these genes.
Dnmt3a1 positively affects the transcription of an interesting set of genes. This set includes some well-studied imprinted genes, important to the organism for acquiring resources in the womb for early postnatal development and for adult life (14). Dnmt3a and Dnmt3L are known to be responsible for establishing the imprints at specific chromosomal regions (DMRs) during PGC differentiation (10, 22, 27). For the Igf2r and Grb10 imprinted genes, one allele is transcribed. The transcribed alleles of the Igf2r and Grb10 genes contain methylated CpG islands (DMRs) within their coding sequences (8, 23). The Igf2r DMR is one of the few known Dnmt3a genomic targets (12, 39). Here, we find that Grb10 gene transcription and Grb10 gene DMR methylation are severely compromised in GFP-3a1Nter cells.
In Dnmt3L−/− ES cells, the Grb10 imprint is demethylated, and this demethylation abolishes the Grb10 gene expression (10, 22, 23, 44, 51), as is the case for GFP-3a1Nter cells. Additionally, our data suggest that Dnmt3a1 targets Grb10 DMR via its N-terminal domain. The current notion is that Dnmt3a proteins, by interacting with Dnmt3L via their C-terminal domain, are tethered to their target sequences (25). On the other hand, Nimura et al. (38) show that Dnmt3L is not responsible for tethering Dnmt3a2 in the mESC genome. The data presented in the present study suggest essentially the same for Dnmt3a1. The targeting of Dnmt3a1 to the Grb10 gene DMR may be mediated not by the C-terminal domain interaction with Dnmt3L but rather through a novel interaction of Dnmt3a1 N-terminal domain with an unidentified molecule. We propose that Dnmt3a1 targets and maintains the Grb10 gene DMR methylation either directly or indirectly by recruiting another Dnmt.
We note that in addition to the Grb10 gene, the Cdkn1c imprinted gene could fall in the same category. Besides our finding that Dnmt3a1 positively regulates the transcription of the Grb10 and Cdkn1c genes, both these genes also share common regulation by the Eed Polycomb group protein (34).
The transcription of the Gpr50 gene and that of the Ihh gene share some common mechanistic details (data not shown), and we show here that Dnmt3a1 likely exerts a positive effect at the promoter level. The mechanistic details for the Gpr50 and Ihh genes will be revealed in future experiments.
Finally, we note that the qualitative analysis of our data does not discriminate between Dnmt3a1 direct or indirect effects in the mESC transcriptome. However, this analysis suggests that the gene targets and the functions of Dnmt3a1 in mESCs are markedly different from Dnmt3a1 target genes in postnatal NSCs (51).
We conclude that by generating a cell line bearing the unique Dnmt3a1-specific functional mutation, we identified some of the unique functions and the genes targeted by Dnmt3a1 in mESCs. Strikingly, our experiments reveal that Dnmt3a1 targets whole chromosomal regions for epigenetic silencing at a very early developmental stage. Moreover, Dnmt3a1 is responsible for the optimal transcriptional activation of a distinct set of unique genes, many of which are imprinted. Our data implicate Dnmt3a1 in imprint maintenance in mESCs, a finding which is expected to shape our view on how the mouse embryonic stem cell targets and uses the de novo DNA methylation apparatus.
ACKNOWLEDGMENTS
We thank Vaggellis Harokopos for expression profiling, Dimitris Papageorgiou for help with gel filtration experiment, and Iannis Talianidis, John Srouboulis, and E. M. C. Skoulakis for helpful discussions and critical readings of the manuscript.
This work was supported by Greek General Secretariat of Research and Technology (GSRT) grants from the 3rd Community Support Program 2000–2006 (PENED-03ED804, PENED-03ED542 to T.A.), by excellence grants for the GSRT Research Institutes, and in part by the Marie Curie “INTEGER” Initial Training Network (PITN-GA-2008-214902).
Footnotes
Published ahead of print on 24 January 2011.
REFERENCES
- 1. Adam F., et al. 2000. COUP-TFI regulates cell migration and axogenesis in differentiating P19 embryonal carcinoma cells. Mol. Endocrinol. 14:1918–1933 [DOI] [PubMed] [Google Scholar]
- 2. Athanasiadou R., et al. 2010. Targeting of de novo DNA methylation throughout Oct4 gene regulatory region in differentiating embryonic stem cells. PLoS One 5:e9937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ben-Shushan E., Sharir H., Pikarski E., Berghman Y. 1995. A dynamic balance between ARP-1/COUP-TFII, EAR-3/COUP-TFI, and retinoic acid receptor:retinoid X receptor heterodimers regulates Oct3/4 expression in embryonal carcinoma cells. Mol. Cell. Biol. 15:1034–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bergman Y., Mostoslavsky R. 1998. DNA demethylation: turning genes on. Biol. Chem. 379:401–407 [DOI] [PubMed] [Google Scholar]
- 5. Bernstein B. E., Meissner A., Lander E. 2007. The mammalian epigenome. Cell 128:669–681 [DOI] [PubMed] [Google Scholar]
- 6. Bestor T. H. 2000. The DNA methyltransferase of mammals. Hum. Mol. Gen. 9:2395–2402 [DOI] [PubMed] [Google Scholar]
- 7. Bhutani N., et al. 2010. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463:1042–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Birger Y., Shemer R., Perk J., Razin A. 1999. The imprinting box of the mouse Igf2r gene. Nature 397:84–88 [DOI] [PubMed] [Google Scholar]
- 9. Boiani M., Schöler H. R. 2005. Regulatory networks in embryo-derived pluripotent stem cells. Nat. Rev. Mol. Cell Biol. 6:872–884 [DOI] [PubMed] [Google Scholar]
- 10. Bourc'his D., Xu G. L., Lin C. S., Bollman B., Bestor T. H. 2001. Dnmt3L and the establishment of maternal genomic imprints. Science 294:2536–2539 [DOI] [PubMed] [Google Scholar]
- 11. Chen T., Ueda Y., Xie S., Li E. 2002. A novel Dnmt3a isoform produced from an alternative promoter localizes to euchromatin and its expression correlates with active de novo methylation. J. Biol. Chem. 277:38746–38754 [DOI] [PubMed] [Google Scholar]
- 12. Chen T., Ueda, Dodge Y. J. E., Wang Z., Li E. 2003. Establishment and maintenance of DNA methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol. Cell. Biol. 23:5594–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chen T., Tsujimoto N., Li E. 2004. The PWWP domain of Dnmt3a and Dnmt3b is required for directing DNA methylation to the major satellite repeats at pericentric heterochromatin. Mol. Cell. Biol. 24:9048–9058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Constância M., Kelsey G., Reik W. 2004. Resourceful imprinting. Nature 432:53–57 [DOI] [PubMed] [Google Scholar]
- 15. Dong K. B., et al. 2008. DNA methylation in ES cells requires the lysine methyltransferase G9a but not its catalytic activity. EMBO J. 22:2691–2701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Epsztejn-Litman S., et al. 2008. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat. Struct. Mol. Biol. 15:1176–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feldman N., et al. 2006. G9a-mediated irreversible epigenetic inactivation of Oct3/4 during early embryogenesis. Nat. Cell Biol. 8:188–194 [DOI] [PubMed] [Google Scholar]
- 18. Fuhrmann G., et al. 2001. Mouse germline restriction of Oct4 expression by germ cell nuclear factor. Dev. Cell 1:377–384 [DOI] [PubMed] [Google Scholar]
- 19. Gallais R., et al. 2007. Dnmt3a and 3b associate with the nuclear orphan receptor COUP-TFI during gene activation. Mol. Endocrinol. 21:2085–2098 [DOI] [PubMed] [Google Scholar]
- 20. Gehring M., Reik W., Henikoff S. 2009. DNA demethylation by DNA repair. Trends Genet. 25:82–90 [DOI] [PubMed] [Google Scholar]
- 21. Gowher H., Liebert K., Hermann A., Xu G., Jeltsch A. 2005. Mechanism of stimulation of catalytic activity of Dnmt3a and Dnmt3b DNA (cytosine C5) methyltransferases by Dnmt3L. J. Biol. Chem. 280:13341–13348 [DOI] [PubMed] [Google Scholar]
- 22. Hata K., Okano M., Lei H., Li E. 2002. Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice. Development 129:1983–1993 [DOI] [PubMed] [Google Scholar]
- 23. Hikichi T., Konda T., Kaneko-Ishino T., Ishino F. 2003. Imprinting regulation of the Meg1/Grb10 and human GRB10 genes; roles of brain-specific promoters and mouse specific CTCF binding sites. Nucleic Acid Res. 31:1398–1406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hu J. L., Zhou B. O., Zhang R. R., Zhou J. Q., Xu G. L. 2009. The N-terminus of histone H3 is required for de novo DNA methylation in chromatin. Proc. Natl. Acad. Sci. U. S. A. 106:22187–22192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jia D., Jurkowska R. Z., Zhang X., Jelsch A., Cheng X. 2007. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature 449:248–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Jones P. A., Liang G. 2009. Rethinking how DNA methylation patterns are maintained. Nat. Rev. Genet. 10:805–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kaneda M., et al. 2004. Essential role for de novo DNA methyltransferase Dnm3a in genomic imprinting. Nature 429:900–903 [DOI] [PubMed] [Google Scholar]
- 28. Kangaspeska S., et al. 2008. Transient cyclical methylation of promoter DNA. Nature 452:112–116 [DOI] [PubMed] [Google Scholar]
- 28a. Lane D. P., Hoeffler W. K. 1980. SV40 large T shares an antigenic determinant with a cellullar protein of molecular weight 68,000. Nature 288:167–170 [DOI] [PubMed] [Google Scholar]
- 29. Lee D. U., Rao A. 2004. Molecular analysis of a locus control region in the T-helper 2 cytokine gene cluster: a target for STAT6 but not GATA3. Proc. Nat. Acad. Sci. 101:16010–16015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lees-Murdock D. J., Shovlin T. C., Gardiner T., de Felici M., Walsh C. P. 2005. DNA methyltransferase expression in the mouse germ line during periods of de novo methylation. Dev. Dyn. 232:992–1002 [DOI] [PubMed] [Google Scholar]
- 31. Li E. 2002. Chromatin modification and epigenetic reprogramming in mammalian development. Nat. Rev. Genet. 3:662–673 [DOI] [PubMed] [Google Scholar]
- 32. Li B., et al. 2007. Polycomb protein Cbx4 promotes SUMO modification of de novo methyltransferase Dnmt3a. Biochem. J. 405:369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li J.-Y., et al. 2007. Synergistic function of DNA methyltransferases Dnmt3a and Dnmt3b in the methylation of Oct4 and Nanog. Mol. Cell. Biol. 27:8748–8759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mager J. N. D., Montgomery F., de Villena P., Magnuson T. 2003. Genome imprinting regulated by the mouse Polycomb group protein Eed. Nat. Genet. 33:502–507 [DOI] [PubMed] [Google Scholar]
- 35. Metivier R., et al. 2008. Cyclical DNA methylation of a transcriptionally active promoter. Nature 452:45–52 [DOI] [PubMed] [Google Scholar]
- 36. Morgan D. H., Santos F., Green K., Dean W., Reik W. 2005. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 14(Spec. no. 1):R47–R58 [DOI] [PubMed] [Google Scholar]
- 37. Nakamura T., et al. 2007. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat. Cell Biol. 9:64–71 [DOI] [PubMed] [Google Scholar]
- 38. Nimura K., et al. 2006. Dnmt3a2 targets endogenous Dnmt3L to ES cell chromatin and induces regional DNA methylation. Genes Cells 11:1225–1237 [DOI] [PubMed] [Google Scholar]
- 39. Okano M., Bell D. W., Haber D. A., Li E. 1999. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell 99:247–257 [DOI] [PubMed] [Google Scholar]
- 40. Ooi S. K., et al. 2007. Dnmt3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448:714–717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ooi S. K., Bestor T. H. 2008. The colorful history of active DNA demethylation. Cell 133:1145–1148 [DOI] [PubMed] [Google Scholar]
- 42. Otani J., et al. 2009. Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain. EMBO Rep. 10:1235–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Popp C., et al. 2010. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature 463:1101–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Phillips J. E., Corces V. G. 2009. CTCF: master weaver of the genome. Cell. 137:1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ramakers C., Ruijters J. M., Lekanne-Deprez R. H., Moorman A. F. M. 2003. Assumption-free analysis of quantitative real time polymerase reaction (PCR) data. Neurosci. Lett. 339:62–66 [DOI] [PubMed] [Google Scholar]
- 46. Reik W., Dean W., Walter J. 2001. Epigenetic reprogramming in mammalian development. Science 293:1089–1093 [DOI] [PubMed] [Google Scholar]
- 47. Sakai Y., Suetake I., Shinozaki F., Yamashina S., Tajima S. 2004. Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos. Gene Expr. Patterns 5:231–237 [DOI] [PubMed] [Google Scholar]
- 48. Siedlecki P., Zielenkiewicz P. 2006. Mammalian DNA methyltransferases. Acta Biochim. Pol. 53:245–256 [PubMed] [Google Scholar]
- 49. Tachibana M., Matsumura, Fukuda Y. M., Kimura H., Shinkai Y. 2008. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. EMBO J. 27:2681–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weisenberger D. J., et al. 2002. Identification and characterization of alternatively spliced variants of DNA methyltransferase 3a in mammalian cells. Gene 298:91–99 [DOI] [PubMed] [Google Scholar]
- 51. Wu H., et al. 2010. Dnmt3a-dependent nonpromoter DNA methylation facilitates transcription of neurogenic genes. Science 329:444–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yamasaki-Ishizaki Y., et al. 2007. Role of DNA methylation and histone H3 Lysine 27 methylation in tissue-specific imprinting of mouse Grb10. Mol. Cell. Biol. 27:732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhao Q., et al. 2009. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat. Struct. Mol. Biol. 16:304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhu J.-K. 2009. Active DNA demethylation mediated by DNA glycosylases. Annu. Rev. Genet. 43:143–166 [DOI] [PMC free article] [PubMed] [Google Scholar]