Abstract
Helix-loop-helix (HLH) proteins play a profound role in the process of development and cellular differentiation. Among the HLH proteins expressed in differentiating erythroid cells are the ubiquitous proteins Myc, USF1, USF2, and TFII-I, as well as the hematopoiesis-specific transcription factor Tal1/SCL. All of these HLH proteins exhibit distinct functions during the differentiation of erythroid cells. For example, Myc stimulates the proliferation of erythroid progenitor cells, while the USF proteins and Tal1 regulate genes that specify the differentiated phenotype. This minireview summarizes the known activities of Myc, USF, TFII-I, and Tal11/SCL and discusses how they may function sequentially, cooperatively, or antagonistically in regulating expression programs during the differentiation of erythroid cells.
INTRODUCTION
Adult erythroid cells differentiate from hematopoietic stem cells (HSCs) through a cascade of steps (18, 132). The most primitive HSC is called a long-term HSC (LT-HSC) for its ability to reconstitute HSCs in the bone marrow of irradiated mice over a long period of time. These slowly dividing cells are attached to a niche in the bone marrow and give rise to short-term HSCs, which then differentiate into common lymphoid progenitors (CLPs) or common myeloid progenitors (CMPs). The CMPs go on to differentiate into granulocyte/monocyte precursors (GMPs) or into megakaryocyte/erythroid cell precursors (MEPs). MEPs further differentiate into erythropoietin-responsive BFU-E (blast-forming unit-erythroid) and then CFU-E (CFU-erythroid). The CFU-E cells differentiate to form orthochromatic normoblasts, then reticulocytes, and finally enucleated mature erythrocytes (125). The process of erythropoiesis has been extensively studied in vitro and in vivo and led to the identification of key erythroid cell transcription factors that regulate gene expression programs at the various steps of differentiation. The availability of erythroid cells representing different stages of maturation has rendered this system ideal for studying gene regulatory mechanisms.
Transcription factors are classified based on the presence of specific protein-protein and protein-DNA interaction motifs which allow them to regulate gene expression by binding to DNA in a sequence-specific manner and to recruit coregulator complexes (98). The class of helix-loop-helix (HLH) transcription factors encompasses many proteins that play important roles during development and differentiation (89). The HLH motif is a characteristic dimerization domain which is accompanied by a basic (b) DNA-binding domain. Some HLH proteins contain an additional leucine zipper (ZIP) protein interaction module; these proteins are referred to as bHLHZIP proteins (89). Erythroid cells express many different HLH proteins. Here, we will review the well-characterized proteins USF1, USF2, Myc, TFII-I, and Tal1/SCL but will also discuss how inhibitor of DNA binding (ID) proteins, which only contain the HLH domain, may interfere with the function of HLH transcription factors in erythroid cells.
The HLH proteins discussed here are all capable of interacting with E-box (CANNTG) elements in erythroid cell-specific genes. The sequence of the E box and flanking nucleotides determines the affinity with which individual HLH proteins interact. Therefore, most E boxes will preferentially interact with a specific member of the HLH family of proteins. However, some E-box elements are known to interact with different HLH proteins and the abundance of HLH proteins in the cell, as well as the sequence context of the DNA element they interact with, e.g., the presence of additional transcription factor-binding motifs, will determine which of the HLH proteins occupies a specific site at a given time. In the following sections, we will review the known activities of HLH proteins and then outline mechanisms and factors regulating the abundance and activities of HLH during erythroid cell differentiation.
USF1 AND USF2
USF1 and USF2 are ubiquitously expressed members of the bHLHZIP family of transcription factors (23). These proteins usually interact with DNA as heterodimers but also form homodimers (Fig. 1). Interestingly, the presence and abundance of heterodimers (USF1/USF2) or specific homodimers vary in different cell types and at the various stages of cellular differentiation (119). This suggests that the different dimeric forms of USF could exert unique functions; however, mice deficient in either USF1 or USF2 are viable, whereas the combined deficiency leads to early embryonic death (118). Therefore, homodimers are able to replace many of the vital functions of the USF heterodimer during development and differentiation. Both USF1 and USF2 contain a USF-specific region (USR) at the N terminus, followed by a short basic DNA-binding domain, the HLH domain, and a leucine zipper at the C terminus (Fig. 1) (23). For the rest of this minireview, we will refer to the heterodimer composed of USF1and USF2 as USF.
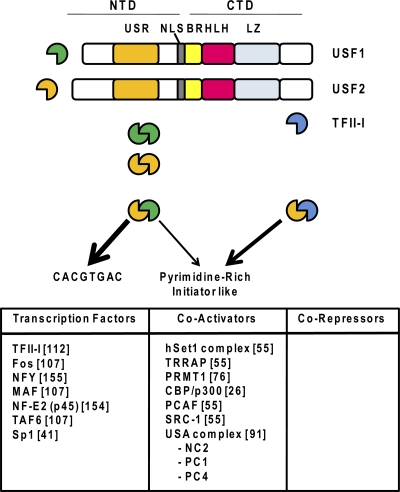
Fig. 1.
Structures and DNA sequence preferences of USF1 and USF2 and proteins that interact with them. USF1 and USF2 contain nuclear localization sequences (NLS), as well as basic region (BR), HLH, and LZ domains in the CTD-specific region and a USR in the N-terminal domain. USF heterodimers preferentially associate with the E-box sequence CACGTGAC but also interact with pyrimidine-rich initiator elements in conjunction with transcription factor TFII-I. Proteins documented to interact with USF proteins are listed at the bottom (references are in brackets).
Early in vitro studies have shown that USF activates transcription by binding to the E-box sequence CACGTG and assisting in the recruitment of the TFIID complex to the promoter (16, 113, 114). Subsequent studies in the Roeder laboratory identified a USF-associated coregulator protein complex, referred to as USA (USF-stimulating activity) (91). Further purification of this coregulator fraction identified both negative cofactors (NCs) and positive cofactors (PCs) (59). One of the NCs, NC2, represses TATA-dependent transcription and mediates transcription complex assembly on genes containing a downstream promoter element (115, 141, 145). The PC fraction included topoisomerase I (PC3) (68) and PC4 (Sub1 in yeast), a 15-kDa protein that activates the transition from initiation to early transcription elongation via interactions with TFIIH (38).
Recent studies have shown that USF interacts with different histone-modifying proteins, including the histone acetyltransferases (HATs) PCAF, SRC-1, and CBP/p300, as well as with the H3K4 methyltransferase-containing Set1 complex and the H4R3-specific methyltransferase Prmt1 (26, 55, 76). Both methylated H3K4 and H4 asymmetrically dimethylated at R3 are associated with permissive or actively transcribed gene loci (44, 67). USF proteins also interact with DNA-binding transcription factors, including the ubiquitously expressed proteins TFII-I (112), nuclear factor Y (NF-Y), Sp1, and the AP1-like transcription factors MafB and NF-E2, an erythroid/myeloid cell-specific heterodimer composed of p45 and a small Maf protein (41, 107, 154, 155).
USF regulates the transcription of many genes during cellular differentiation, mostly through aiding in the recruitment of transcription complexes. FitzGerald et al. (36) recently analyzed the occurrence of 8-mer sequences in 13,010 human promoters that cluster within 100 bp of the transcription start site (TSS). Binding sites for USF were found to be commonly associated with TSSs. Using genome-wide interaction studies, Rada-Iglesias et al. (104) demonstrated that both USF1 and USF2 interact with thousands of genomic loci in a liver cell line. Most of these loci were bound by both USF1 and USF2, emphasizing the fact that the heterodimer is the predominant USF species. However, some genomic loci associate with USF2 but not USF1 (104). Importantly, the USF2-only sites, in contrast to those interacting with both USF1 and USF2, were largely located relatively far away from the genes and were associated with the presence of binding sites for tissue-specific regulatory proteins like HNF4, FOXA2, and FOXA1. It has previously been shown that USF proteins can form bivalent heterotetramers using their LZ domains (35). USF2 homodimers interacting at distal regulatory sites could thus interact with USF heterodimers at gene-proximal sites, which would mediate proximity between distant regulatory DNA elements and gene promoters.
The genome-wide USF interaction map led to two important conclusions with regard to USF's role in transcription complex recruitment (104). First, most of the USF interaction sites are close to the TSSs of genes and binding of USF correlates with active transcription. Second, the interaction of USF positively correlates with increased levels of acetylated histone H3. These data are consistent with the notion that USF exerts mainly positive effects on the transcription of genes and that part of its function is mediated by the recruitment of histone H3-specific HAT activity. In this respect, it is interesting that USF recruits the histone methyltransferase PRMT1 to chromatin and, furthermore, that asymmetric dimethylation of H4R3, mediated by PRMT1, facilitates the acetylation of H3 (76).
Although USF is a ubiquitously expressed transcription factor, it appears to function mostly in the context of differentiated cells. Several reports have documented increased USF protein levels or DNA-binding activity during cellular differentiation. For example, during the differentiation of erythroid cells, there is an increase in USF1 and USF2 protein levels (79). Similarly, Kirito et al. (66) reported that thrombopoietin, the main mediator of platelet production, induces the expression of USF1. Furthermore, increased USF levels have also been observed during the differentiation of osteoclasts, Sertoli cells, and mast cells (85, 144, 152).
In addition to modulating the expression of tissue-specific genes, USF also plays a role in regulating components of the cell cycle, including cyclin-dependent kinases (e.g., Cdk4) and cyclins (e.g., B1) (20, 24, 101, 128). However, although USF promotes transitions during the cell cycle, it has antiproliferative activity as well (83). It antagonizes the function of oncogenes like Myc, E1A, and Ras and activates the expression of the genes for the tumor suppressors p53, BRCA2, and APC (21, 23, 83).
USF has been shown to regulate genes during the differentiation of erythroid cells, including the gene for HoxB4, an important transcription regulator that stimulates the proliferation and differentiation of erythroid progenitor cells, the gene for glycophorin B, and the gene for adult β-globin (13, 25, 42). Transgenic mice expressing a dominant negative form of USF revealed a defect in erythroid cell differentiation and reduced expression of key erythroid transcription factors, including NF-E2, GATA-1, EKLF, and Tal1, which all contain E-box motifs in DNA regulatory elements (78). Thus, USF regulates the expression of erythroid transcription factors and cooperates with these factors in the activation of erythroid cell-specific genes.
In addition to its role in activating gene expression, USF also contributes to the barrier activity of the HS4 insulator in the chicken β-globin gene locus (55, 140). USF recruits different chromatin-modifying enzymes to cHS4 which are thought to establish active chromatin marks and to prevent the spread of heterochromatin into the active globin gene locus. The involvement of USF in barrier/insulator activity has also been suggested by studies of the human erythroid cell-specific α-spectrin gene locus (39).
TFII-I
Transcription factor TFII-I was originally identified as a protein capable of binding to the initiator (INR) and mediating the transcription of a TATA-less, INR-containing promoter (112). Subsequent cloning and sequencing of its cDNA revealed that it is a relatively large transcription factor of ∼110 kDa. It has six R repeats (R1 to R6), each containing an HLH motif (Fig. 2) (111). Like USF, TFII-I also contains a leucine zipper (LZ), a basic region (b), and a nuclear localization domain. Because of the unusual structure of TFII-I, with its multiple HLH domains, it is not considered a classical HLH protein. Many studies have implicated TFII-I in the positive regulation of gene expression. For example, TFII-I has been shown to interact with USF and to associate with either E-box elements or initiator sequences to activate gene transcription (29, 112). Genes positively regulated by TFII-I include the c-fos gene and genes regulated in response to endoplasmic reticulum stress (63, 100, 108). There are also a number of genes that are transcriptionally suppressed by TFII-I. For example, Roy and colleagues recently demonstrated that TFII-I inhibits the expression of genes that are essential for osteogenesis (71). Also, TFII-I represses the transcription of the vascular endothelial growth factor gene in endothelial cells and of the β-globin gene in erythroid cells by binding to the initiator element of these genes (25, 87). The repression of initiator containing promoters by TFII-I could be brought about by its interaction with Myc (110). Moreover, the inhibitory effect of TFII-I on transcription is mediated by its ability to recruit corepressor complexes, including histone deacetylase 3 (HDAC3) (25, 133), histone H3K4-specific demethylase LSD1 (48), and components of the polycomb repressor complex (26).
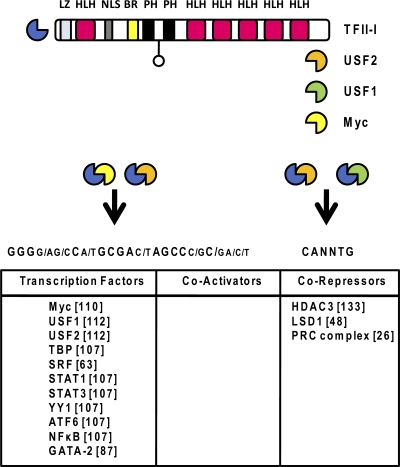
Fig. 2.
Structure and DNA sequence preferences of TFII-I and proteins that interact with it. The gene for TFII-I contains six repeat regions, each encoding an HLH domain. In addition, an LZ motif, a nuclear localization sequence (NLS), a basic region (BR), and two Ph domains are located in the C terminus. TFII-I associates with USF and interacts with pyrimidine-rich initiator elements or E-box motifs. Proteins documented to interact with TFII-I are listed at the bottom (references are in brackets).
The activity of TFII-I is regulated by signal transduction pathways (109), and like USF, it activates the expression of cell cycle regulators, including cyclin D1 and protein kinase C-β, thus mediating cell cycle progression through the G1-to-S and G2-to-M phases of the cell cycle (4). TFII-I is phosphorylated at tyrosine residues, which regulates its abilities to relocate to the nucleus and to interact with specific proteins (19, 96, 149). Phosphorylation by Bruton's tyrosine kinase allows TFII-I to interact with phospholipase C-γ (PLC-γ) in the cytoplasm (14). This interaction prevents the association of PLC-γ with transient receptor potential channel 3, which mediates the PLC-γ-dependent entry of Ca2+ ions into cells. Calcium is an important mediator of signaling pathways and regulates many cellular processes, including the cleavage of transcription factors by Ca2+-dependent proteases (97). Thus, TFII-I's association with PLC-γ inhibits the entry of calcium into the cell and may indirectly change the activity of other transcription factors.
TFII-I is a member of a family of I repeat-containing proteins that are expressed from genes located on human chromosome 7. Haploinsufficiency associated with this genomic region causes Williams-Beuron syndrome (WBS), a disease affecting the development of the neuronal system (108, 109). Recently, the Roy and Bayarsaihan laboratories generated mice with targeted deletions of the gene for TFII-I (Gtf2i) and the gene for the related protein Gtf2ird (34). Both homozygous mutations are embryonic lethal and reveal defects that are consistent with their roles in WBS but also point to important functions of these proteins in nonneuronal tissues.
TFII-I has three different isoforms (alpha, beta, and delta) that are generated by alternative splicing (108, 109). In serum-starved cells, the beta isoform was shown to associate with the c-fos promoter and to keep the promoter accessible while in an inactive state (49). After serum stimulation, the delta isoform is tyrosine phosphorylated, enters the nucleus, and replaces the beta isoform at the c-fos promoter to activate transcription.
TAL1/SCL
Tal1/SCL (referred to here as Tal1) is a hematopoiesis-specific HLH protein that dimerizes with members of the E-protein family, including E2A (E12 and E47), HEB, and E2-2 (Fig. 3) (73, 103). Deficiency of Tal1 causes embryonic lethality at 9.5 days postcoitum (dpc) due to a total lack of yolk sac hematopoiesis (103). Tal1 is among the earliest expressed transcription factors important for the specification of hematopoietic cells. Elegant genetic experiments have shown that, in addition to being required for the early commitment of hematopoietic cells, Tal1 also plays a fundamental role in the regulation of erythroid cell- and megakaryocyte-specific gene expression programs (50, 60, 92). This is consistent with the expression profile of Tal1 during hematopoiesis. It is expressed in HSCs and hematopoietic progenitor cells but downregulated during the differentiation of most hematopoietic cell lineages, except during erythropoiesis and megakaryopoiesis (73, 103). In erythroid cells, at least a fraction of Tal1 interacts with the erythroid cell-specific transcription factor GATA-1 and the coregulators LMO2 and Ldb1 (69, 138, 148). This multiprotein complex associates with composite DNA elements containing an E box and a GATA-binding site separated from each other by 9 or 10 bp (50, 138). E-box/GATA sites have been identified in DNA elements regulating the expression of erythroid cell-specific proteins, including p4.2 and EKLF (3, 138, 146). Interestingly, data from the Brandt laboratory suggest that single-stranded DNA-binding proteins mediate the assembly of the Tal1/GATA-1 multimeric complex at E-box/GATA sites (148).
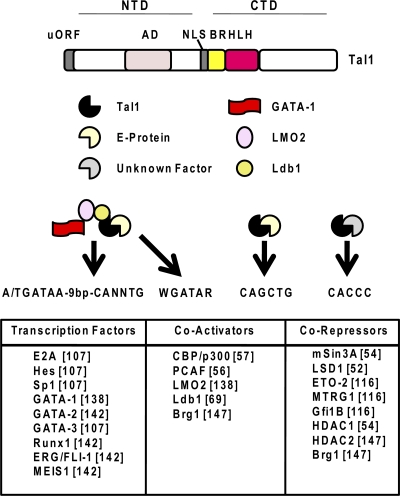
Fig. 3.
Structure and DNA sequence preferences of Tal1 and proteins that interact with it. Tal1 contains an HLH domain and a basic region (BR) in the CTD, as well as an activation domain (AD) and a nuclear localization sequence (NLS) in the N-terminal domain. The Tal1 gene contains a short upstream open reading frame (uORF). Tal1 associates with E proteins and binds to E-box elements, with a preference for the sequence CAGCTG. Tal1 also forms a large complex consisting of GATA-1, LMO2, and Ldb1 and binds to composite elements containing an E box and a GATA site separated by about 9 bp or to a GATA sequence only. The recruitment of Tal1 to CACCC motifs is likely mediated by proteins that have not been identified yet. Proteins documented to interact with Tal1 are listed at the bottom (references are in brackets).
Like other HLH proteins discussed here, Tal1 functions both as an activator and as a repressor of transcription. Early work in the Brandt laboratory characterized Tal1-associated corepressor protein complexes containing HDAC1 and HDAC2, as well as mSin3A and the chromatin remodeling factor Brg1 (54, 147). More recent work identified additional corepressor proteins, especially the histone demethylase LSD1, ETO-2, the ETO-related protein Mtgr1, and Gfi-1b, which associate with Tal1 in erythroid cells (12, 43, 52, 90, 116). ETO-2 is expressed in undifferentiated erythroid progenitor cells and downregulated during erythroid cell differentiation (80). It interacts with HDACs, with the mSin3A corepressor, and with the E proteins of the Tal1 heterodimer (2, 116). Gfi-1b is a DNA-binding oncoprotein containing a SNAG repressor domain which interacts with HLH proteins, including Myc, and represses transcription (70, 106). Previous gain-of-function and loss-of-function experiments have implicated Gfi-1b in erythropoiesis (40).
In addition to interacting with corepressors, Tal1 associates with the coactivators p300 and P/CAF, which both contain HAT activities (56, 57). Tal1 itself is acetylated by P/CAF during the differentiation of erythroid cells and, importantly, acetylation of Tal1 disrupts its interaction with corepressors (56). It is likely that Tal1 exists as part of different protein complexes in erythroid cells. The relative abundance of Tal1-associated repressing and activating protein complexes appears to change during the differentiation of erythroid cells, perhaps with a shift toward association with activating complexes at later stages of erythroid cell differentiation (53).
The association of coregulator complexes with known histone-modifying activities suggests that one function of Tal1 is to establish chromatin states that are compatible or not compatible with the assembly of elongation-competent transcription complexes. Moreover, the Tal1/GATA-1-associated protein Ldb1 is required for the establishment of proximity between the locus control region (LCR) and the actively transcribed adult β-globin gene and mediates the efficient recruitment of pTEFb (transcription elongation factor B) to the β-globin gene promoter (121, 122). pTEFB stimulates transcription elongation by phosphorylating the C-terminal domain (CTD) of RNA polymerase II (Pol II), as well as the elongation factor DSIF (DRB-sensitive inducing factor) (9).
Not all of the functions associated with Tal1 during hematopoiesis and erythropoiesis require Tal1's ability to bind DNA directly. In fact, mutant mice expressing a DNA-binding-deficient Tal1 protein survive beyond 9.5 dpc, the time when Tal1-null embryos die (102). Analysis of these mice revealed that direct DNA binding of Tal1 is not required for its function during the earlier specification of hematopoietic cells but may be more important for activating gene expression at later stages of erythroid cell differentiation. The Porcher and Vyas laboratories recently conducted an elegant and important study to identify target genes of Tal1 during the differentiation of erythroid cells (61). Ter119-negative erythroid progenitor cells were isolated from the livers of fetal wild-type mice and from mice expressing the DNA-binding mutant form of Tal1 and subjected to chromatin immunoprecipitation-sequencing analysis using antibodies specific for Tal1 and to expression analysis by microarrays. The data demonstrate that Tal1 associates with thousands of genomic loci in erythroid progenitor cells. However, only a small fraction of genes proximal to Tal1-binding sites change expression in cells expressing the DNA-binding mutant form of Tal1. Among the genes that did reveal expression changes between wild-type cells and cells expressing the DNA-binding mutant form of Tal1 were those that express proteins whose functions include transcription regulation, signaling, cell cycle regulation, nucleosome assembly, ubiquitination, apoptosis, and cell proliferation. Interestingly, Tal1 associates with many genes encoding transcription regulators, including GATA-1, LMO2, Ldb1, E2A, Bcl11a, NF-E2, EKLF1, BACH1, Myb, and Myc (61). All of these proteins have been implicated in the regulation of erythropoiesis. However, the transcription of these genes is not perturbed by the DNA-binding-defective mutant Tal1 protein, despite the fact that some of these genes, such as those for EKLF and Tal1, contain functional E boxes in regulatory DNA elements.
The study by Porcher and colleagues also identified novel DNA sequences that constitute preferred genomic Tal1-binding sites (61). The preferred E box for Tal1 appears to be CAGCTG; however, genomic Tal1-binding sites were also enriched for CACCC and CTGCCA/TGNNG motifs, the latter of which has previously been associated with GATA-1 occupancy in erythroid cells (153). This further demonstrates that many of the functions of Tal1 are not dependent on direct DNA binding but may instead be mediated by the interaction of Tal1 with other DNA-binding activities, e.g., GATA-1. Interestingly, most of the sites that associate with the DNA-binding-defective mutant form of Tal1 are located in distal regulatory elements, suggesting that enhancer elements recruit Tal1 mainly by protein-protein interactions rather than by direct DNA binding (61).
Adult hematopoietic stem cells express both Tal1 and the related HLH protein Lyl1, which is important for B-cell development (123). Tal1 and Lyl1 function redundantly in maintaining HSC function. A recent report by Wilson et al. demonstrated that Tal1 and Lyl1 function within the context of five other transcription factors that play important roles in hematopoiesis, LMO2, GATA2, Runx1, ERG, and FLI-1 (142). Direct interactions between Tal1 and Runx1, GATA2, and ERG have been documented, suggesting that these proteins function together in regulating gene expression patterns in stem and hematopoietic progenitor cells.
MYC
Myc proteins are a family of highly related oncoproteins that play important roles in cellular proliferation and cell cycle control (27, 31). The most prominent member of this family is c-Myc, here referred to as Myc. Like USF, Myc interacts with classical E-box elements. Flanking nucleotides have predictive value for the interaction of USF and are thus able to favor or disfavor interactions with USF or Myc (28). Myc has been shown to regulate transcription mediated by all of the three major RNA Pols (I, II, and III) (27, 31).
The structure of Myc is similar to that of USF (Fig. 4). It contains a basic region, an HLH domain, an LZ domain at the C terminus, and regulatory domains in the N terminus, including two specific regulatory domains that are called Myc box 1 (MB1) and MB2. MB1 is subject to phosphorylation, which stabilizes Myc and modulates its ability to interact with other proteins (17, 84, 135). Myc dimerizes with Max, another bHLHZIP transcription factor. Heterodimerization with Max is required for DNA binding (5, 120). Although the Myc/Max heterodimer can function as an activator or a repressor of transcription, in embryonic stem cells, this complex predominantly stimulates transcription elongation at genes that harbor paused RNA Pol II (45, 105). Myc-mediated stimulation of transcription elongation is brought about by its recruitment of transcription elongation factor pTEFB (30, 105). Many proteins have been shown to directly interact with Myc, including transcription factors, coregulators like CBP/p300 and DNMT3A, and components of the basal transcription machinery (Fig. 4) (8, 107, 136).
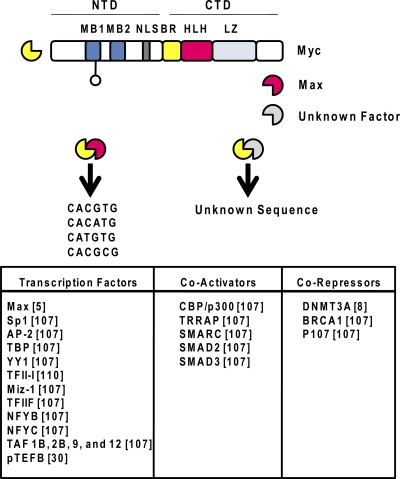
Fig. 4.
Structure and DNA sequence preferences of Myc and proteins that interact with it. Myc contains a basic region (BR), an HLH domain, and an LZ in the CTD. Two Myc-binding elements (MB1 and MB2) and a nuclear localization sequence (NLS) are located in the N-terminal domain (NTD). Myc interacts with Max and binds to E-box motifs preferentially with CG, CA, TG, and CG residues in the center. It also interacts with genomic regions that lack E-box sequences; these associations are likely mediated by the interaction of Myc with other DNA-binding proteins. Proteins documented to interact with Myc are listed at the bottom (references are in brackets).
The genome-wide analysis of Myc binding revealed that it binds to regions containing E-box elements and to regions not containing E-box elements (88, 150). This suggests that Myc is recruited to genomic DNA either directly or via protein-protein interactions, similar to Tal1. Current estimates suggest that Myc directly or indirectly regulates more than 20% of the genes in stem cells (27, 31).
Although Myc stimulates the proliferation of cells and in general represses cellular differentiation, Myc activity is required for the expansion and differentiation of erythroid progenitor cells (37, 46). Expression of Myc is low in slow-cycling LT-HSCs but increases during the differentiation of ST-HSCs and erythroid progenitor cells until the erythroblast stage, after which Myc expression declines (37, 46). This coincides with the proliferation status of stem/progenitor cells. Overexpression of Myc in bone marrow leads to an increase in LT-HSC release from the bone marrow niche and differentiation to ST-HSCs and hematopoietic progenitor cells (37, 46). In addition to promoting proliferation of erythroid progenitor cells, Myc also prevents the premature differentiation of these cells (1). This is mediated by the inhibition of p27, a negative regulator of cyclin-dependent kinases, and by blocking of the expression of erythroid cell-specific transcription factors and coregulators, including NF-E2, GATA-1, and LMO2. Interestingly, inhibition of p27-mediated induction of erythroid cell differentiation by Myc occurred without reversal of the p27-induced arrest of the cell cycle (1).
ID PROTEINS
ID proteins contain the HLH dimerization motif but lack a basic DNA-binding domain (62). These proteins interact with and antagonize the function of tissue-specific E proteins in a dominant negative fashion. As discussed previously, E proteins are heterodimeric partners of Tal1. There are four members in the ID family of proteins, referred to as ID1 to ID4. ID2 has been shown to promote erythroid cell development (58). This protein is expressed in CMPs, at reduced levels in GMPs, and at increased levels in MEPs, suggesting a role for ID2 in regulating early myeloid lineage decisions. ID2 has been shown to interact with Pu.1, a hematopoiesis-specific transcription factor that interacts with and inhibits GATA-1. The interaction between ID2 and Pu.1 prevents inhibition of GATA-1, which then promotes erythroid cell differentiation (126). Another study demonstrated that overexpression of ID1 promotes the survival and expansion of erythroid progenitor cells (143).
EXPRESSION AND REGULATION OF HLH PROTEINS DURING ERYTHROID CELL DIFFERENTIATION
During the early stages of erythroid cell differentiation, all of the HLH proteins discussed here are expressed. The dynamics of TFII-I expression in the course of erythroid cell differentiation are unknown. Previous studies have shown that the expression of USF and Tal1 increases while the expression of Myc decreases during the differentiation of erythroid cells (Fig. 5) (37, 46, 54, 79). Phosphorylation of Tal1 stimulates its ubiquitination, which may allow rapid turnover of Tal1 in early erythroid progenitor cells (95, 129, 130). The turnover of Tal1 may convert its function from that of a repressor to that of an activator. Several studies have shown that Tal1, Myc, and USF are subject to proteolytic cleavage by calcium-dependent proteases during early stages of cellular differentiation (22, 79, 151). USF is subject to cleavage by calpain, a calcium-dependent protease, in undifferentiated erythroid cells (79). Likewise, Eisenman and colleagues demonstrated that Myc is subject to calpain-dependent proteolytic cleavage during early stages of muscle cell differentiation (22). The Myc cleavage product Nick, which lacks a nuclear localization sequence, was shown to stimulate cellular differentiation. If a similar mechanism operates in erythroid cells, it could explain Myc's role during early erythroid cell differentiation. Furthermore, caspase 3 has been shown to cleave Tal1 (151) and, importantly, caspases are known to be regulated by calpains (15). This suggests that calcium-dependent proteases regulate the activity of three important HLH proteins (Tal1, USF, and Myc) during early erythroid cell differentiation.
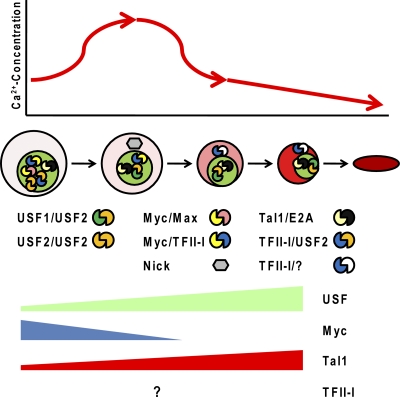
Fig. 5.
Model outlining a correlation of erythroid cell differentiation, calcium concentration, and HLH transcription factor activity. During the differentiation of erythroid cells, there is a transient increase in the intracellular calcium concentration. This activates m-calpain, causing proteolytic cleavage of USF and Myc. The Myc cleavage product Nick, localized in the cytoplasm, may regulate processes involved in the differentiation of erythroid cells. At later erythroid cell differentiation stages, the expression of Myc is repressed. The decrease in the calcium concentration, which may or may not be regulated by the relocation of TFII-I to the cytoplasm, inhibits calpain activity, allowing increased expression of full-length USF. Tal1 expression increases during the differentiation of erythroid cells, but proteolytic degradation in erythroid progenitor cells likely facilitates the conversion of Tal1 from a repressor to an activator. If and how the expression of TFII-I changes during erythroid cell maturation is unknown. It is postulated that during the early stages of erythropoiesis, TFII-I, perhaps in conjunction with USF or Myc, functions as a repressor of erythroid cell-specific genes.
Previous studies have shown that the calcium concentration within erythroid cells changes during differentiation (Fig. 5) (139). The concentration of calcium is low in early erythroid progenitor cells, increases for a short period of time, and then decreases thereafter, reaching its lowest concentration in mature red blood cells. Other reports suggest that there may be several short periods during the differentiation of red blood cells that are characterized by a transient increase in the intracellular calcium concentration (93, 131). Nevertheless, changes in the calcium concentration during erythroid cell differentiation may be important for the regulation of Myc, Tal1, and USF. The low concentration of calcium in erythroid progenitor cells would allow Myc to function in the nucleus to activate genes involved in cell proliferation and to repress genes involved in differentiation. As the cells differentiate, the increase in the calcium concentration activates calpains, leading to the cleavage of USF, Tal1, and Myc. Cleavage of Myc would prevent this protein from acting as a repressor of differentiation in the nucleus and would allow the Myc cleavage product Nick to participate in mechanisms leading to the differentiation of erythroid cells. Cleavage of USF and Tal1 leads to rapid turnover of these proteins, which facilitates their conversion from repressors to activators. The decrease in the calcium concentration at later erythroid cell differentiation stages increases the activity of USF and, perhaps, Tal1. The increased expression of USF may favor the formation of active USF1/USF2 heterodimers and would decrease the formation of repressive USF/TFII-I protein complexes.
The regulation of USF, Tal1, and Myc by calcium-dependent proteases is interesting in light of the fact that TFII-I is located in the cytoplasm during the differentiation of lymphocytes and prevents the influx of calcium by interacting with PLC-γ (14). A similar mechanism may operate during the differentiation of erythroid cells, and the relocation of TFII-I to the cytoplasm and inhibition of calcium influx would increase the levels of full-length USF and Tal1, allowing these proteins to participate in the activation of erythroid cell-specific genes. In this context, it is interesting that Myc has previously been shown to stimulate the proliferation and differentiation of B lymphocytes by increasing the intracellular concentration of calcium in B-cell progenitors (47). The data suggest common mechanisms involved in the Myc/TFII-I-regulated proliferation and differentiation of erythroid and lymphoid progenitor cells.
REGULATION OF GENE EXPRESSION BY HLH PROTEINS DURING ERYTHROID CELL DIFFERENTIATION
Changes in gene expression programs during cellular differentiation are regulated by a complex network of transcription factors (81, 82). The abundance and functionality of transcription factors are critical for the finely tuned balance of activating and repressing activities that mediate gene expression patterns at specific differentiation stages. The available evidence suggests that the HLH proteins discussed here play specific and nonredundant roles in regulating gene expression programs during erythroid cell differentiation.
The transcription factor Myc functions early during the proliferation and differentiation of HSCs (37, 46) and promotes the release of LT-HSCs within their niche in bone marrow. Myc mediates the activation of cell cycle and antiapoptotic genes, thereby allowing rapid proliferation of progenitor cells and inhibiting the premature differentiation of erythroid progenitor cells. Because Myc is capable of interacting with TFII-I and inhibits the transcription of initiator-containing promoters, these two proteins may prevent the premature transcription of erythroid cell-specific genes in erythroid progenitor cells (110). This repressive complex could also include USF2, which interacts with TFII-I and is detectable at low levels at the β-globin gene promoter in undifferentiated murine erythroleukemia (MEL) cells (154).
The functions of Tal1 in early hematopoietic progenitor cells likely include the regulation of genes required for the maintenance of hematopoietic progenitor cells (73, 103, 132). It appears that Tal1-associated repressor activity is more prominent in early progenitor cells, suggesting that one of its functions in these cells is to prevent the expression of lineage-specific genes. We must note that the heterodimerization partner of Tal1, the E2A gene product, is subject to inhibition by the ID proteins (62). Because many functions of Tal1 in erythroid progenitor cells are independent of the DNA-binding capability of Tal1, we speculate that ID proteins may inhibit or reduce the sequence-specific binding of Tal1 in these cells. Instead, Tal1 and associated coregulators, including ETO-2, may be recruited to their sites of action via other DNA-binding transcription factors. E2A proteins are expressed in HSCs and in subsets of hematopoietic progenitor cells. HSC repopulation experiments showed that E2A-deficient bone marrow significantly reduced LT-HSC, GM progenitors, and erythroid/megakaryocytic progenitor cells, as well as pre CFU-E colonies (117). The data reveal that E2A proteins play important roles in maintaining the HSC pool and promoting the differentiation of both myelolymphoid and myeloerythroid progenitor cells. Thus, Tal1 and its heterodimeric partner contribute important functions during HSC pool formation and ID proteins may transiently interfere with this function during the earliest stages of erythroid cell lineage specification.
It has been shown that USF interacts with regulatory E-box elements in the Tal1 gene locus in undifferentiated MEL cells (78). This interaction decreases upon the differentiation of these cells, suggesting that USF regulates the expression of Tal1 in undifferentiated progenitor cells. At later differentiation stages, when the ID proteins no longer inhibit Tal1 heterodimers, Tal1 may autoactivate its own gene (53).
Tal1 and USF both interact with E-box elements and have been shown to regulate the expression of erythroid cell genes. Because these proteins are not known to interact with each other, they are unlikely to interact with the same E box at the same time. Recent genome-wide analysis of DNA interactions of these proteins suggests that they have different preferred target sequences (61, 104). Tal1 can be recruited to DNA directly via a specific E box that exhibits a consensus motif slightly different from the one identified for USF. In addition, Tal1 is often recruited to chromatin in a DNA-binding-independent manner (61), a situation that has so far not been observed for USF. The interactions of USF and Tal1 with NF-E2 and GATA-1, respectively, could further determine which DNA sequences they interact with in vivo. Therefore, it appears that USF and Tal1 are recruited to different genomic locations due to differences in target site preferences and interactions with different partner proteins that may stabilize binding to regulatory DNA elements. If USF and Tal1 bind to the same E box, as has been demonstrated in in vitro studies for an E box located in the β-globin gene locus (10, 32, 72, 137), the abundance of the proteins, the location in the nucleus, and the interaction with other proteins will likely determine if an E box is occupied by USF or Tal1.
The relationship between USF and TFII-I is incompletely understood. These proteins interact with each other but appear to exhibit antagonistic activities in erythroid cells (25, 112). For example, USF activates the transcription of the β-globin gene, while TFII-I inhibits the expression of this gene (25). A low concentration of USF in erythroid progenitor cells may favor the formation of heterodimers with TFII-I. USF proteins may thus contribute to the repressive activity of TFII-I. Upon differentiation, the increase in USF proteins would lead to increased formation of USF heterodimers, which exert positive effects on erythroid cell-specific gene expression patterns. Therefore, similar to the situation described for Tal1, a switch of protein-binding partners may increase the activating potential of USF during the differentiation of erythroid cells. The available evidence suggests that the main function of USF in differentiated erythroid cells is to cooperate with erythroid cell-specific transcription factors in the recruitment of transcription complexes (78, 154).
REGULATION OF GLOBIN GENE EXPRESSION BY HLH PROTEINS
The main component of a differentiated red blood cell is hemoglobin, which is a heterotetramer composed of two α- and two β-globin chains. Each globin chain associates with a heme group, which binds oxygen with the help of iron atoms. The α- and β-globin gene loci contain multiple genes that are expressed in a developmental stage-specific manner (124). High-level expression of the globin genes is regulated by cis-acting DNA elements that are located distal or proximal to the genes (51, 99). The β-globin LCR is a powerful DNA regulatory element that is composed of several 200- to 400-bp DNase I-hypersensitive sites that function together to achieve high-level expression of the β-type globin genes in an adult erythroid cell (11, 33). Many transcription factors have been identified over the last 20 years that participate in the transcriptional regulation of the globin genes (64, 77, 132). Many of these same transcription factors are also involved in the regulation of other erythroid cell-specific genes, including those expressing red cell-specific membrane proteins (94, 132).
Both Tal1 and USF interact with the LCR element HS2 and with the adult β-globin gene promoter (32, 61, 72, 137, 154). There is a conserved E box in the downstream promoter region of the adult β-globin gene which fits the USF but not the Tal1 preferred E-box sequence (72). USF has the most prominent, if not the only, binding activity in erythroid cell extracts that interacts with this site in vitro (72). Thus, Tal1 could be recruited to the LCR by direct DNA binding, but its association with the adult β-globin gene promoter could be mediated by other DNA-binding proteins, such as GATA-1. The interaction profile of USF proteins during erythroid cell differentiation is also of interest (154). It appears that USF2 interacts with LCR HS2 and with the adult β-globin promoter in undifferentiated erythroid cells. The coactivator CBP, which interacts with USF2 in erythroid cells, as well as TFIIB and Pol II, is already recruited to LCR HS2 in undifferentiated cells. Because USF has been shown to be required for the efficient recruitment of transcription complexes to the β-globin gene locus, the data suggest that USF2 is involved in the priming of the globin gene locus for activation at subsequent differentiation stages. The increase in USF1 expression during erythroid cell differentiation could increase the number of USF heterodimers interacting with the globin gene locus. The USF heterodimers recruit Prmt1 to LCR HS2 and to the adult β-globin gene promoter (76). Prmt1-mediated H4R3 methylation facilitates the subsequent acetylation of H3 (76), and these histone modifications could facilitate the transfer of activities from the LCR to the β-globin gene promoter, including elongation-competent transcription complexes (154). Furthermore, increased USF1 expression and interaction with the β-globin gene locus correlate with the binding of Pol II to the adult β-globin gene promoter, suggesting that the USF heterodimer is required for the efficient recruitment of the transcription complex to the globin gene promoter (154). Tal1 and associated cofactors mediate proximity between the LCR and the adult β-globin gene promoter and regulate transcription elongation via recruitment of the Pol II CTD kinase pTEFB (121, 122).
It appears that gene loci that are expressed in differentiated cells are primed for activation in progenitor cells (6, 127). This is best illustrated by the α- and β-globin gene loci, in which distal regulatory elements recruit transcription factors and coregulators early during the differentiation of erythroid cells (7, 74, 86, 134, 137, 154). The available data suggest that HLH proteins, specifically, USF and Tal1 but perhaps also TFII-I, play a profound role in priming gene loci for activation. As mentioned before, USF2 interacts with the β-globin LCR before the β-globin gene becomes activated in undifferentiated MEL cells (154). Furthermore, Tal1 interacts with the β-globin LCR, as well as with HS12 of the α-globin gene locus, in erythroid progenitor cells (61, 132). Expression of a DNA-binding mutant form of Tal1 severely reduces its interaction with these sites and leads to increased α- and β-globin gene expression (132, 61). This suggests that Tal1 and associated corepressors occupy erythroid cell-specific cis-regulatory DNA elements and keep genes in a primed but inactive configuration. After differentiation, Tal1 is converted to an activator and contributes to the high-level expression of the globin genes. A recent study by Higgs and colleagues found that TFII-I associates with the α-globin gene in differentiated erythroid cells (134). Interestingly, TFII-I localizes to a broad region within the transcribed α-globin gene, suggesting that TFII-I may play a role in the process of transcription elongation. In this respect, like Tal1 and USF, TFII-I may be converted from a repressor to an activator during the differentiation of erythroid cells.
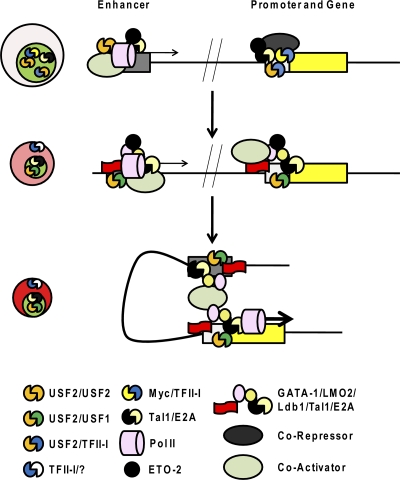
Figure 6 outlines a hypothesis of how the HLH proteins regulate the differentiation-dependent expression of a hypothetical erythroid cell-specific gene that is under the control of a promoter and an enhancer. It is proposed that USF2 binds to the enhancer region and recruits coactivators and transcription complexes in erythroid progenitor cells. Transcription of the enhancer maintains its accessibility through subsequent cell divisions during erythroid cell differentiation (65, 75). The Tal1/E-protein/ETO2 complex also interacts with the enhancer at early differentiation stages and contributes to keeping the enhancer in a poised but inactive configuration. At this stage, TFII-I and Myc may be involved in preventing expression by binding to the promoter and recruiting corepressors. At subsequent differentiation stages, USF heterodimers and the heteromeric Tal1 complex containing LMO2 and Ldb1 are recruited to the enhancer and the promoter, replacing the repressive activities. The USF and Tal1 complexes mediate proximity between the enhancer and promoter, facilitate the transfer of the transcription complex to the promoter, and stimulate transcription elongation.
Fig. 6.
Hypothetical model of the regulation of an erythroid cell-specific gene by HLH proteins during the differentiation of erythroid cells. It is proposed that USF2 homodimers and Tal-1/E-protein heterodimers occupy enhancer elements of erythroid cell-specific genes in erythroid progenitor cells. At this stage, Pol II is recruited at low levels and enhancer transcription may assist in maintaining an open configuration (65, 75). The Tal1/E-protein/ETO complex and TFII-I/Myc heterodimers bind to the promoter regions and, through corepressors, keep the promoter in an inactive configuration. During the differentiation of erythroid cells, both enhancer and promoter regions are sequentially activated, which is facilitated by the following processes: (i) relocation of TFII-I to the cytoplasm, (ii) repression of Myc expression, (iii) an increase in the activity of USF, and (iv) conversion of Tal1 from a repressor to an activator. This leads to increased binding of activating HLH proteins, including USF heterodimers and Tal1/GATA-1/LMO2/Lbd1, as well as coactivators, to the enhancer and the promoter. Ldb1 and other activities then mediate proximity between the enhancer and promoter, and activities are transferred from the enhancer to the promoter to position transcription complexes, to enhance transcription elongation, and/or to facilitate reinitiation of transcription. This is a simplified scenario that, in reality, involves many other erythroid cell-specific and ubiquitous activities.
CONCLUSIONS
The evidence discussed here points to an important role for HLH proteins in priming erythroid cell-specific gene loci during early differentiation stages. Subsequently, protease-mediated turnover of HLH proteins and increased expression of interacting erythroid cell-specific transcription factors convert HLH proteins from repressors to activators. The recent identification of Tal1- and USF-associated proteins, as well as genome-wide interaction studies of these proteins, led to great advances in our understanding of the roles these proteins have in gene regulation. It is important to continue this line of investigation and to determine the protein- and DNA-binding profiles of HLH proteins at different stages of erythroid cell differentiation. Further mechanistic studies are needed to uncover how the HLH proteins repress and activate transcription. It appears that USF is directly involved in the recruitment of transcription complexes to DNA, while the Tal1/Gata-1/Ldb1 complex mediates efficient transcription elongation by recruiting the CTD kinase pTEFB and the chromatin remodeling complex facilitator of chromatin transcription (122). The analysis of TFII-I-interacting proteins and its genome-wide interaction profile in erythroid cells at specific differentiation stages will shed light on its various functions throughout this process. Also, the ID proteins should be further studied to evaluate if and how they affect the function of Tal1 in erythroid progenitor cells. Finally, HLH proteins function in the context of a protein network that involves many other transcription factors within erythroid cells. Further studies must be performed to elucidate how HLH proteins coordinate erythroid cell-specific gene expression programs in conjunction with other erythroid cell-specific or ubiquitous transcription factors.
ACKNOWLEDGMENTS
We thank our colleagues in the Huang and Bungert laboratories for stimulating discussions.
This work was supported by grants from the NIH (J.B., RO1DK052356; J.B. and J.S., RO1DK083389; S.H., RO1HL090589 and RO1HL091929) and the American Heart Association (J.B.).
Footnotes
Published ahead of print on 31 January 2011.
REFERENCES
- 1. Acosta J. C., et al. 2008. Myc inhibits p27-induced erythroid differentiation of leukemia cells by repressing erythroid master genes without reversing p27-mediated cell cycle arrest. Mol. Cell. Biol. 28:7286–7295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Amann J. M., et al. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 21:6470–6483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anderson K. P., Crable S. C., Lingrel J. B. 2000. The GATA-E box-GATA motif in the EKLF promoter is required for in vivo expression. Blood 95:1652–1655 [PubMed] [Google Scholar]
- 4. Ashworth T., Roy A. L. 2009. Phase specific functions of the transcription factor TFII-I during cell cycle. Cell Cycle 8:596–605 [DOI] [PubMed] [Google Scholar]
- 5. Blackwood E. M., Eisenman R. N. 1991. Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251:1211–1217 [DOI] [PubMed] [Google Scholar]
- 6. Bonifer C., Hoogenkamp M., Krysinska H., Tagoh H. 2008. How transcription factors program chromatin—lessons from studies of the regulation of myeloid-specific genes. Semin. Immunol. 20:257–263 [DOI] [PubMed] [Google Scholar]
- 7. Bottardi S., Ross J., Pierre-Charles N., Blank V., Milot E. 2006. Lineage-specific activators affect beta-globin locus chromatin in multipotent hematopoietic progenitors. EMBO J. 25:3586–3595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brenner C., et al. 2005. Myc represses transcription through recruitment of DNA methyltransferase corepressor. EMBO J. 24:336–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Brès V., Yoh S. M., Jones K. A. 2008. The multi-tasking P-TEFb complex. Curr. Opin. Cell Biol. 20:334–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bresnick E. H., Felsenfeld G. 1993. Evidence that the transcription factor USF is a component of the human beta-globin locus control region heteromeric protein complex. J. Biol. Chem. 268:18824–18834 [PubMed] [Google Scholar]
- 11. Bulger M., Groudine M. 1999. Looping versus linking: toward a model for long-distance gene activation. Genes Dev. 13:2465–2477 [DOI] [PubMed] [Google Scholar]
- 12. Cai Y., et al. 2009. Eto2/MTG16 and MTGR1 are heteromeric corepressors of the TAL1/SCL transcription factor in murine erythroid progenitors. Biochem. Biophys. Res. Commun. 390:295–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Camara-Clayette V., Rahuel C., Bertrand O., Cartron J. P. 1999. The E-box of the human glycophorin B promoter is involved in the erythroid-specific expression of the GPB gene. Biochem. Biophys. Res. Commun. 265:170–176 [DOI] [PubMed] [Google Scholar]
- 14. Caraveo G., van Rossum D. B., Patterson R. L., Snyder S. H., Desiderio S. 2006. Action of TFII-I outside the nucleus as an inhibitor of agonist-induced calcium entry. Science 314:122–125 [DOI] [PubMed] [Google Scholar]
- 15. Carlile G. W., Smith D. H., Wiedmann M. 2004. Caspase-3 has a nonapoptotic function in erythroid maturation. Blood 103:4310–4316 [DOI] [PubMed] [Google Scholar]
- 16. Carthew R. W., Chodosh L. A., Sharp P. A. 1985. An RNA polymerase II transcription factor binds to an upstream element in the adenovirus major late promoter. Cell 43:439–448 [DOI] [PubMed] [Google Scholar]
- 17. Channavajhala P., Seldin D. C. 2002. Functional interaction of protein kinase CK2 and c-Myc in lymphomagenesis. Oncogene 21:5280–5288 [DOI] [PubMed] [Google Scholar]
- 18. Chasis J. A., Mohandas N. 2008. Erythroblastic islands: niches for erythropoiesis. Blood 112:470–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cheriyath V., Desgranges Z. P., Roy A. L. 2002. c-Src-dependent transcriptional activation of TFII-I. J. Biol. Chem. 277:22798–22805 [DOI] [PubMed] [Google Scholar]
- 20. Cheung E., Mayr P., Coda-Zabetta F., Woodman P. G., Boam D. S. 1999. DNA-binding activity of the transcription factor upstream stimulatory factor 1 (USF-1) is regulated by cyclin-dependent phosphorylation. Biochem. J. 344(Pt. 1):145–152 [PMC free article] [PubMed] [Google Scholar]
- 21. Choe C., Chen N., Sawadogo M. 2005. Decreased tumorigenicity of c-Myc-transformed fibroblasts expressing active USF2. Exp. Cell Res. 302:1–10 [DOI] [PubMed] [Google Scholar]
- 22. Conacci-Sorrell M., Ngouenet C., Eisenman R. N. 2010. Myc-nick: a cytoplasmic cleavage product of Myc that promotes alpha-tubulin acetylation and cell differentiation. Cell 142:480–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Corre S., Galibert M. D. 2005. Upstream stimulating factors: highly versatile stress-responsive transcription factors. Pigment Cell Res. 18:337–348 [DOI] [PubMed] [Google Scholar]
- 24. Corre S., et al. 2009. Target gene specificity of USF-1 is directed via p38-mediated phosphorylation-dependent acetylation. J. Biol. Chem. 284:18851–18862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Crusselle-Davis V. J., Vieira K. F., Zhou Z., Anantharaman A., Bungert J. 2006. Antagonistic regulation of beta-globin gene expression by helix-loop-helix proteins USF and TFII-I. Mol. Cell. Biol. 26:6832–6843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Crusselle-Davis V. J., et al. 2007. Recruitment of coregulator complexes to the beta-globin gene locus by TFII-I and upstream stimulatory factor. FEBS J. 274:6065–6073 [DOI] [PubMed] [Google Scholar]
- 27. Dang C. V., et al. 2006. The c-Myc target gene network. Semin. Cancer Biol. 16:253–264 [DOI] [PubMed] [Google Scholar]
- 28. Desbarats L., Gaubatz S., Eilers M. 1996. Discrimination between different E-box-binding proteins at an endogenous target gene of c-myc. Genes Dev. 10:447–460 [DOI] [PubMed] [Google Scholar]
- 29. Du H., Roy A. L., Roeder R. G. 1993. Human transcription factor USF stimulates transcription through the initiator elements of the HIV-1 and the Ad-ML promoters. EMBO J. 12:501–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Eberhardy S. R., Farnham P. J. 2002. Myc recruits P-TEFb to mediate the final step in the transcriptional activation of the cad promoter. J. Biol. Chem. 277:40156–40162 [DOI] [PubMed] [Google Scholar]
- 31. Eilers M., Eisenman R. N. 2008. Myc's broad reach. Genes Dev. 22:2755–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Elnitski L., Miller W., Hardison R. 1997. Conserved E boxes function as part of the enhancer in hypersensitive site 2 of the beta-globin locus control region. Role of basic helix-loop-helix proteins. J. Biol. Chem. 272:369–378 [DOI] [PubMed] [Google Scholar]
- 33. Engel J. D., Tanimoto K. 2000. Looping, linking, and chromatin activity: new insights into beta-globin locus regulation. Cell 100:499–502 [DOI] [PubMed] [Google Scholar]
- 34. Enkhmandakh B., et al. 2009. Essential functions of the Williams-Beuren syndrome-associated TFII-I genes in embryonic development. Proc. Natl. Acad. Sci. U. S. A. 106:181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ferré-D'Amaré A. R., Pognonec P., Roeder R. G., Burley S. K. 1994. Structure and function of the b/HLH/Z domain of USF. EMBO J. 13:180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. FitzGerald P. C., Shlyakhtenko A., Mir A. A., Vinson C. 2004. Clustering of DNA sequences in human promoters. Genome Res. 14:1562–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fröjmark A. S., et al. 2010. Cooperative effect of ribosomal protein s19 and Pim-1 kinase on murine c-Myc expression and myeloid/erythroid cellularity. J. Mol. Med. 88:39–46 [DOI] [PubMed] [Google Scholar]
- 38. Fukuda A., et al. 2004. Transcriptional coactivator PC4 stimulates promoter escape and facilitates transcriptional synergy by GAL4-VP16. Mol. Cell. Biol. 24:6525–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gallagher P. G., et al. 2009. An insulator with barrier-element activity promotes alpha-spectrin gene expression in erythroid cells. Blood 113:1547–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Garçon L., et al. 2005. Gfi-1B plays a critical role in terminal differentiation of normal and transformed erythroid progenitor cells. Blood 105:1448–1455 [DOI] [PubMed] [Google Scholar]
- 41. Ge Y., Jensen T. L., Matherly L. H., Taub J. W. 2003. Physical and functional interactions between USF and Sp1 proteins regulate human deoxycytidine kinase promoter activity. J. Biol. Chem. 278:49901–49910 [DOI] [PubMed] [Google Scholar]
- 42. Giannola D. M., et al. 2000. Hematopoietic expression of HOXB4 is regulated in normal and leukemic stem cells through transcriptional activation of the HOXB4 promoter by upstream stimulating factor (USF)-1 and USF-2. J. Exp. Med. 192:1479–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goardon N., et al. 2006. ETO2 coordinates cellular proliferation and differentiation during erythropoiesis. EMBO J. 25:357–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Goldberg A. D., Allis C. D., Bernstein E. 2007. Epigenetics: a landscape takes shape. Cell 128:635–638 [DOI] [PubMed] [Google Scholar]
- 45. Guenther M. G., Levine S. S., Boyer L. A., Jaenisch R., Young R. A. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 130:77–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guo Y., et al. 2009. c-Myc-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Blood 114:2097–2106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Habib T., et al. 2007. Myc stimulates B lymphocyte differentiation and amplifies calcium signaling. J. Cell Biol. 179:717–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hakimi M. A., Dong Y., Lane W. S., Speicher D. W., Shiekhattar R. 2003. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 278:7234–7239 [DOI] [PubMed] [Google Scholar]
- 49. Hakre S., et al. 2006. Opposing functions of TFII-I spliced isoforms in growth factor-induced gene expression. Mol. Cell 24:301–308 [DOI] [PubMed] [Google Scholar]
- 50. Hall M. A., et al. 2003. The critical regulator of embryonic hematopoiesis, SCL, is vital in the adult for megakaryopoiesis, erythropoiesis, and lineage choice in CFU-S12. Proc. Natl. Acad. Sci. U. S. A. 100:992–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Higgs D. R., Wood W. G. 2008. Long-range regulation of alpha globin gene expression during erythropoiesis. Curr. Opin. Hematol. 15:176–183 [DOI] [PubMed] [Google Scholar]
- 52. Hu X., et al. 2009. LSD1-mediated epigenetic modification is required for TAL1 function and hematopoiesis. Proc. Natl. Acad. Sci. U. S. A. 106:10141–10146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hu X., Ybarra R., Qiu Y., Bungert J., Huang S. 2009. Transcriptional regulation by TAL1: a link between epigenetic modifications and erythropoiesis. Epigenetics 4:357–361 [DOI] [PubMed] [Google Scholar]
- 54. Huang S., Brandt S. J. 2000. mSin3A regulates murine erythroleukemia cell differentiation through association with the TAL1 (or SCL) transcription factor. Mol. Cell. Biol. 20:2248–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang S., Li X., Yusufzai T. M., Qiu Y., Felsenfeld G. 2007. USF1 recruits histone modification complexes and is critical for maintenance of a chromatin barrier. Mol. Cell. Biol. 27:7991–8002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Huang S., Qiu Y., Shi Y., Xu Z., Brandt S. J. 2000. P/CAF-mediated acetylation regulates the function of the basic helix-loop-helix transcription factor TAL1/SCL. EMBO J. 19:6792–6803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Huang S., Qiu Y., Stein R. W., Brandt S. J. 1999. p300 functions as a transcriptional coactivator for the TAL1/SCL oncoprotein. Oncogene 18:4958–4967 [DOI] [PubMed] [Google Scholar]
- 58. Ji M., et al. 2008. Id2 intrinsically regulates lymphoid and erythroid development via interaction with different target proteins. Blood 112:1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kaiser K., Meisterernst M. 1996. The human general co-factors. Trends Biochem. Sci. 21:342–345 [PubMed] [Google Scholar]
- 60. Kassouf M. T., Chagraoui H., Vyas P., Porcher C. 2008. Differential use of SCL/TAL-1 DNA-binding domain in developmental hematopoiesis. Blood 112:1056–1067 [DOI] [PubMed] [Google Scholar]
- 61. Kassouf M. T., et al. 2010. Genome-wide identification of TAL1's functional targets: insights into its mechanisms of action in primary erythroid cells. Genome Res. 20:1064–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kee B. L. 2009. E and ID proteins branch out. Nat. Rev. Immunol. 9:175–184 [DOI] [PubMed] [Google Scholar]
- 63. Kim D. W., Cochran B. H. 2000. Extracellular signal-regulated kinase binds to TFII-I and regulates its activation of the c-fos promoter. Mol. Cell. Biol. 20:1140–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kim S. I., Bresnick E. H. 2007. Transcriptional control of erythropoiesis: emerging mechanisms and principles. Oncogene 26:6777–6794 [DOI] [PubMed] [Google Scholar]
- 65. Kim T. K., et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Kirito K., Fox N., Kaushansky K. 2003. Thrombopoietin stimulates Hoxb4 expression: an explanation for the favorable effects of TPO on hematopoietic stem cells. Blood 102:3172–3178 [DOI] [PubMed] [Google Scholar]
- 67. Kouzarides T. 2007. Chromatin modifications and their function. Cell 128:693–705 [DOI] [PubMed] [Google Scholar]
- 68. Kretzschmar M., Meisterernst M., Roeder R. G. 1993. Identification of human DNA topoisomerase I as a cofactor for activator-dependent transcription by RNA polymerase II. Proc. Natl. Acad. Sci. U. S. A. 90:11508–11512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lahlil R., Lécuyer E., Herblot S., Hoang T. 2004. SCL assembles a multifactorial complex that determines glycophorin A expression. Mol. Cell. Biol. 24:1439–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Laurent B., et al. 2009. Gfi-1B promoter remains associated with active chromatin marks throughout erythroid differentiation of human primary progenitor cells. Stem Cells 27:2153–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lazebnik M. B., Tussié-Luna M. I., Hinds P. W., Roy A. L. 2009. Williams-Beuren syndrome-associated transcription factor TFII-I regulates osteogenic marker genes. J. Biol. Chem. 284:36234–36239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Leach K. M., et al. 2003. Characterization of the human beta-globin downstream promoter region. Nucleic Acids Res. 31:1292–1301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lécuyer E., Hoang T. 2004. SCL: from the origin of hematopoiesis to stem cells and leukemia. Exp. Hematol. 32:11–24 [DOI] [PubMed] [Google Scholar]
- 74. Levings P. P., Bungert J. 2002. The human beta-globin locus control region. Eur. J. Biochem. 269:1589–1599 [DOI] [PubMed] [Google Scholar]
- 75. Levings P. P., Zhou Z., Vieira K. F., Crusselle-Davis V. J., Bungert J. 2006. Recruitment of transcription complexes to the beta-globin locus control region and transcription of hypersensitive site 3 prior to erythroid differentiation of murine embryonic stem cells. FEBS J. 273:746–755 [DOI] [PubMed] [Google Scholar]
- 76. Li X., et al. 2010. H4R3 methylation facilitates beta-globin transcription by regulating histone acetyltransferase binding and H3 acetylation. Blood 115:2028–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Liang S., Moghimi B., Yang T. P., Strouboulis J., Bungert J. 2008. Locus control region mediated regulation of adult beta-globin gene expression. J. Cell. Biochem. 105:9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Liang S. Y., et al. 2009. Defective erythropoiesis in transgenic mice expressing dominant-negative upstream stimulatory factor. Mol. Cell. Biol. 29:5900–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Lin I. J., et al. 2009. Calpeptin increases the activity of upstream stimulatory factor and induces high level globin gene expression in erythroid cells. J. Biol. Chem. 284:20130–20135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lindberg S. R., Olsson A., Persson A. M., Olsson I. 2005. The leukemia-associated ETO homologues are differently expressed during hematopoietic differentiation. Exp. Hematol. 33:189–198 [DOI] [PubMed] [Google Scholar]
- 81. Loose M., Patient R. 2006. Global genetic regulatory networks controlling hematopoietic cell fates. Curr. Opin. Hematol. 13:229–236 [DOI] [PubMed] [Google Scholar]
- 82. Loose M., Swiers G., Patient R. 2007. Transcriptional networks regulating hematopoietic cell fate decisions. Curr. Opin. Hematol. 14:307–314 [DOI] [PubMed] [Google Scholar]
- 83. Luo X., Sawadogo M. 1996. Antiproliferative properties of the USF family of helix-loop-helix transcription factors. Proc. Natl. Acad. Sci. U. S. A. 93:1308–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lutterbach B., Hann S. R. 1994. Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol. Cell. Biol. 14:5510–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ma Z., Jhun B., Jung S. Y., Oh C. K. 2008. Binding of upstream stimulatory factor 1 to the E-box regulates the 4G/5G polymorphism-dependent plasminogen activator inhibitor 1 expression in mast cells. J. Allergy Clin. Immunol. 121:1006–1012 e2 [DOI] [PubMed] [Google Scholar]
- 86. Mahajan M. C., Karmakar S., Newburger P. E., Krause D. S., Weissman S. M. 2009. Dynamics of alpha-globin locus chromatin structure and gene expression during erythroid differentiation of human CD34(+) cells in culture. Exp. Hematol. 37:1143–1156 e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Mammoto A., et al. 2009. A mechanosensitive transcriptional mechanism that controls angiogenesis. Nature 457:1103–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Margolin A. A., et al. 2009. ChIP-on-chip significance analysis reveals large-scale binding and regulation by human transcription factor oncogenes. Proc. Natl. Acad. Sci. U. S. A. 106:244–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Massari M. E., Murre C. 2000. Helix-loop-helix proteins: regulators of transcription in eucaryotic organisms. Mol. Cell. Biol. 20:429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Meier N., et al. 2006. Novel binding partners of Ldb1 are required for haematopoietic development. Development 133:4913–4923 [DOI] [PubMed] [Google Scholar]
- 91. Meisterernst M., Roy A. L., Lieu H. M., Roeder R. G. 1991. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell 66:981–993 [DOI] [PubMed] [Google Scholar]
- 92. Mikkola H. K., et al. 2003. Haematopoietic stem cells retain long-term repopulating activity and multipotency in the absence of stem-cell leukaemia SCL/tal-1 gene. Nature 421:547–551 [DOI] [PubMed] [Google Scholar]
- 93. Miller B. A., Cheung J. Y., Tillotson D. L., Hope S. M., Scaduto R. C., Jr 1989. Erythropoietin stimulates a rise in intracellular-free calcium concentration in single BFU-E derived erythroblasts at specific stages of differentiation. Blood 73:1188–1194 [PubMed] [Google Scholar]
- 94. Mohandas N., Gallagher P. G. 2008. Red cell membrane: past, present, and future. Blood 112:3939–3948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Nie L., Wu H., Sun X. H. 2008. Ubiquitination and degradation of Tal1/SCL are induced by notch signaling and depend on Skp2 and CHIP. J. Biol. Chem. 283:684–692 [DOI] [PubMed] [Google Scholar]
- 96. Novina C. D., Cheriyath V., Roy A. L. 1998. Regulation of TFII-I activity by phosphorylation. J. Biol. Chem. 273:33443–33448 [DOI] [PubMed] [Google Scholar]
- 97. Oh-hora M., Rao A. 2008. Calcium signaling in lymphocytes. Curr. Opin. Immunol. 20:250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Orphanides G., Reinberg D. 2002. A unified theory of gene expression. Cell 108:439–451 [DOI] [PubMed] [Google Scholar]
- 99. Palstra R. J., de Laat W., Grosveld F. 2008. Beta-globin regulation and long-range interactions. Adv. Genet. 61:107–142 [DOI] [PubMed] [Google Scholar]
- 100. Parker R., et al. 2001. Identification of TFII-I as the endoplasmic reticulum stress response element binding factor ERSF: its autoregulation by stress and interaction with ATF6. Mol. Cell. Biol. 21:3220–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Pawar S. A., Szentirmay M. N., Hermeking H., Sawadogo M. 2004. Evidence for a cancer-specific switch at the CDK4 promoter with loss of control by both USF and c-Myc. Oncogene 23:6125–6135 [DOI] [PubMed] [Google Scholar]
- 102. Porcher C., Liao E. C., Fujiwara Y., Zon L. I., Orkin S. H. 1999. Specification of hematopoietic and vascular development by the bHLH transcription factor SCL without direct DNA binding. Development 126:4603–4615 [DOI] [PubMed] [Google Scholar]
- 103. Porcher C., et al. 1996. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 86:47–57 [DOI] [PubMed] [Google Scholar]
- 104. Rada-Iglesias A., et al. 2008. Whole-genome maps of USF1 and USF2 binding and histone H3 acetylation reveal new aspects of promoter structure and candidate genes for common human disorders. Genome Res. 18:380–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Rahl P. B., et al. 2010. c-Myc regulates transcriptional pause release. Cell 141:432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Randrianarison-Huetz V., et al. 2010. Gfi-1B controls human erythroid and megakaryocytic differentiation by regulating TGF-beta signaling at the bipotent erythro-megakaryocytic progenitor stage. Blood 115:2784–2795 [DOI] [PubMed] [Google Scholar]
- 107. Ravasi T., et al. 2010. An atlas of combinatorial transcriptional regulation in mouse and man. Cell 140:744–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Roy A. L. 2001. Biochemistry and biology of the inducible multifunctional transcription factor TFII-I. Gene 274:1–13 [DOI] [PubMed] [Google Scholar]
- 109. Roy A. L. 2007. Signal-induced functions of the transcription factor TFII-I. Biochim. Biophys. Acta 1769:613–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Roy A. L., Carruthers C., Gutjahr T., Roeder R. G. 1993. Direct role for Myc in transcription initiation mediated by interactions with TFII-I. Nature 365:359–361 [DOI] [PubMed] [Google Scholar]
- 111. Roy A. L., et al. 1997. Cloning of an inr- and E-box-binding protein, TFII-I, that interacts physically and functionally with USF1. EMBO J. 16:7091–7104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Roy A. L., Meisterernst M., Pognonec P., Roeder R. G. 1991. Cooperative interaction of an initiator-binding transcription initiation factor and the helix-loop-helix activator USF. Nature 354:245–248 [DOI] [PubMed] [Google Scholar]
- 113. Sawadogo M. 1988. Multiple forms of the human gene-specific transcription factor USF. II. DNA binding properties and transcriptional activity of the purified HeLa USF. J. Biol. Chem. 263:11994–12001 [PubMed] [Google Scholar]
- 114. Sawadogo M., Van Dyke M. W., Gregor P. D., Roeder R. G. 1988. Multiple forms of the human gene-specific transcription factor USF. I. Complete purification and identification of USF from HeLa cell nuclei. J. Biol. Chem. 263:11985–11993 [PubMed] [Google Scholar]
- 115. Schluesche P., Stelzer G., Piaia E., Lamb D. C., Meisterernst M. 2007. NC2 mobilizes TBP on core promoter TATA boxes. Nat. Struct. Mol. Biol. 14:1196–1201 [DOI] [PubMed] [Google Scholar]
- 116. Schuh A. H., et al. 2005. ETO-2 associates with SCL in erythroid cells and megakaryocytes and provides repressor functions in erythropoiesis. Mol. Cell. Biol. 25:10235–10250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Semerad C. L., Mercer E. M., Inlay M. A., Weissman I. L., Murre C. 2009. E2A proteins maintain the hematopoietic stem cell pool and promote the maturation of myelolymphoid and myeloerythroid progenitors. Proc. Natl. Acad. Sci. U. S. A. 106:1930–1935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Sirito M., Lin Q., Deng J. M., Behringer R. R., Sawadogo M. 1998. Overlapping roles and asymmetrical cross-regulation of the USF proteins in mice. Proc. Natl. Acad. Sci. U. S. A. 95:3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Sirito M., Lin Q., Maity T., Sawadogo M. 1994. Ubiquitous expression of the 43- and 44-kDa forms of transcription factor USF in mammalian cells. Nucleic Acids Res. 22:427–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Sommer A., Bousset K., Kremmer E., Austen M., Lüscher B. 1998. Identification and characterization of specific DNA-binding complexes containing members of the Myc/Max/Mad network of transcriptional regulators. J. Biol. Chem. 273:6632–6642 [DOI] [PubMed] [Google Scholar]
- 121. Song S. H., Hou C., Dean A. 2007. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell 28:810–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Song S. H., et al. 2010. Multiple functions of Ldb1 required for beta-globin activation during erythroid differentiation. Blood 116:2356–2364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Souroullas G. P., Salmon J. M., Sablitzky F., Curtis D. J., Goodell M. A. 2009. Adult hematopoietic stem and progenitor cells require either Lyl1 or Scl for survival. Cell Stem Cell 4:180–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Stamatoyannopoulos G. 2005. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 33:259–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Stamatoyannopoulos G., Nienhuis A. W. 1994. Hemoglobin switching, p. 107–155In Stamatoyannopoulos G., Neinhuis A. W., Majerus P., Varmus H. (ed.), The molecular basis of blood diseases, 2nd ed. The W. B. Saunders Co., Philadelphia, PA [Google Scholar]
- 126. Stopka T., Amanatullah D. F., Papetti M., Skoultchi A. I. 2005. PU. 1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 24:3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Szutorisz H., Dillon N., Tora L. 2005. The role of enhancers as centres for general transcription factor recruitment. Trends Biochem. Sci. 30:593–599 [DOI] [PubMed] [Google Scholar]
- 128. Takeda D. Y., Dutta A. 2005. DNA replication and progression through S phase. Oncogene 24:2827–2843 [DOI] [PubMed] [Google Scholar]
- 129. Tang T., Arbiser J. L., Brandt S. J. 2002. Phosphorylation by mitogen-activated protein kinase mediates the hypoxia-induced turnover of the TAL1/SCL transcription factor in endothelial cells. J. Biol. Chem. 277:18365–18372 [DOI] [PubMed] [Google Scholar]
- 130. Terme J. M., Lhermitte L., Asnafi V., Jalinot P. 2009. TGF-beta induces degradation of TAL1/SCL by the ubiquitin-proteasome pathway through AKT-mediated phosphorylation. Blood 113:6695–6698 [DOI] [PubMed] [Google Scholar]
- 131. Tong Q., et al. 2008. TRPC3 is the erythropoietin-regulated calcium channel in human erythroid cells. J. Biol. Chem. 283:10385–10395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132. Tsiftsoglou A. S., Vizirianakis I. S., Strouboulis J. 2009. Erythropoiesis: model systems, molecular regulators, and developmental programs. IUBMB Life 61:800–830 [DOI] [PubMed] [Google Scholar]
- 133. Tussié-Luna M. I., Bayarsaihan D., Seto E., Ruddle F. H., Roy A. L. 2002. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASxbeta. Proc. Natl. Acad. Sci. U. S. A. 99:12807–12812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Vernimmen D., De Gobbi M., Sloane-Stanley J. A., Wood W. G., Higgs D. R. 2007. Long-range chromosomal interactions regulate the timing of the transition between poised and active gene expression. EMBO J. 26:2041–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Vervoorts J., Lüscher-Firzlaff J., Lüscher B. 2006. The ins and outs of MYC regulation by posttranslational mechanisms. J. Biol. Chem. 281:34725–34729 [DOI] [PubMed] [Google Scholar]
- 136. Vervoorts J., et al. 2003. Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep. 4:484–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Vieira K. F., et al. 2004. Recruitment of transcription complexes to the beta-globin gene locus in vivo and in vitro. J. Biol. Chem. 279:50350–50357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Wadman I. A., et al. 1997. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 16:3145–3157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Wang J., et al. 2006. Biochemical and biophysical studies on the precursor cells of mouse erythrocytes at different stages. Cell Biochem. Biophys. 45:147–156 [DOI] [PubMed] [Google Scholar]
- 140. West A. G., Huang S., Gaszner M., Litt M. D., Felsenfeld G. 2004. Recruitment of histone modifications by USF proteins at a vertebrate barrier element. Mol. Cell 16:453–463 [DOI] [PubMed] [Google Scholar]
- 141. Willy P. J., Kobayashi R., Kadonaga J. T. 2000. A basal transcription factor that activates or represses transcription. Science 290:982–985 [DOI] [PubMed] [Google Scholar]
- 142. Wilson N. K., et al. 2010. Combinatorial transcriptional control in blood stem/progenitor cells: genome-wide analysis of ten major transcriptional regulators. Cell Stem Cell 7:532–544 [DOI] [PubMed] [Google Scholar]
- 143. Wood A. D., et al. 2009. ID1 promotes expansion and survival of primary erythroid cells and is a target of JAK2V617F-STAT5 signaling. Blood 114:1820–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Wood M. A., Walker W. H. 2009. USF1/2 transcription factor DNA-binding activity is induced during rat Sertoli cell differentiation. Biol. Reprod. 80:24–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Xie J., Collart M., Lemaire M., Stelzer G., Meisterernst M. 2000. A single point mutation in TFIIA suppresses NC2 requirement in vivo. EMBO J. 19:672–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Xu Z., Huang S., Chang L. S., Agulnick A. D., Brandt S. J. 2003. Identification of a TAL1 target gene reveals a positive role for the LIM domain-binding protein Ldb1 in erythroid gene expression and differentiation. Mol. Cell. Biol. 23:7585–7599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Xu Z., Meng X., Cai Y., Koury M. J., Brandt S. J. 2006. Recruitment of the SWI/SNF protein Brg1 by a multiprotein complex effects transcriptional repression in murine erythroid progenitors. Biochem. J. 399:297–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Xu Z., et al. 2007. Single-stranded DNA-binding proteins regulate the abundance of LIM domain and LIM domain-binding proteins. Genes Dev. 21:942–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Yang W., Desiderio S. 1997. BAP-135, a target for Bruton's tyrosine kinase in response to B cell receptor engagement. Proc. Natl. Acad. Sci. U. S. A. 94:604–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Zeller K. I., et al. 2006. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl. Acad. Sci. U. S. A. 103:17834–17839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Zeuner A., et al. 2003. Control of erythroid cell production via caspase-mediated cleavage of transcription factor SCL/Tal-1. Cell Death Differ. 10:905–913 [DOI] [PubMed] [Google Scholar]
- 152. Zhang Y., et al. 2008. Intricate gene regulatory networks of helix-loop-helix (HLH) proteins support regulation of bone-tissue related genes during osteoblast differentiation. J. Cell. Biochem. 105:487–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Zhang Y., et al. 2009. Primary sequence and epigenetic determinants of in vivo occupancy of genomic DNA by GATA1. Nucleic Acids Res. 37:7024–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Zhou Z., et al. 2010. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the beta-globin gene locus. J. Biol. Chem. 285:15894–15905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Zhu J., Giannola D. M., Zhang Y., Rivera A. J., Emerson S. G. 2003. NF-Y cooperates with USF1/2 to induce the hematopoietic expression of HOXB4. Blood 102:2420–2427 [DOI] [PubMed] [Google Scholar]