Abstract
Mitoferrin 1 (Mfrn1; Slc25a37) and mitoferrin 2 (Mfrn2; Slc25a28) function as essential mitochondrial iron importers for heme and Fe/S cluster biogenesis. A genetic deficiency of Mfrn1 results in a profound hypochromic anemia in vertebrate species. To map the cis-regulatory modules (CRMs) that control expression of the Mfrn genes, we utilized genome-wide chromatin immunoprecipitation (ChIP) datasets for the major erythroid transcription factor GATA-1. We identified the CRMs that faithfully drive the expression of Mfrn1 during blood and heart development and Mfrn2 ubiquitously. Through in vivo analyses of the Mfrn-CRMs in zebrafish and mouse, we demonstrate their functional and evolutionary conservation. Using knockdowns with morpholinos and cell sorting analysis in transgenic zebrafish embryos, we show that GATA-1 directly regulates the expression of Mfrn1. Mutagenesis of individual GATA-1 binding cis elements (GBE) demonstrated that at least two of the three GBE within this CRM are functionally required for GATA-mediated transcription of Mfrn1. Furthermore, ChIP assays demonstrate switching from GATA-2 to GATA-1 at these elements during erythroid maturation. Our results provide new insights into the genetic regulation of mitochondrial function and iron homeostasis and, more generally, illustrate the utility of genome-wide ChIP analysis combined with zebrafish transgenesis for identifying long-range transcriptional enhancers that regulate tissue development.
INTRODUCTION
Iron metabolism and heme synthesis are two key regulators of erythropoiesis. Erythroid differentiation is characterized by the de novo production and activation of numerous enzymes and transporters that are responsible for the synthesis of heme and Fe/S clusters in the mitochondria (2, 42). We previously showed that the anemic zebrafish mutant frascati carries a mutation in the mitochondrial solute carrier, mitoferrin 1 (Mfrn1; Slc25a37), which imports iron into the mitochondria for heme and Fe/S cluster biogenesis in erythroblasts (43). A closely related paralog, mitoferrin 2 (Mfrn2; Slc25a28), functions analogously in nonerythroid tissues and early erythroid progenitors (39, 43). Both Mfrn genes are differentially expressed in a tissue- and developmentally restricted manner. In zebrafish and mouse, Mfrn1 expression is restricted predominantly to hematopoietic tissues, whereas Mfrn2 is ubiquitously expressed (43). Recent studies have demonstrated that Mfrn1 is in part regulated by posttranslational protein stability (39), so that Mfrn1 protein forms an oligomeric complex with Abcb10 to enhance its protein half-life and promote the influx of iron for heme synthesis in erythroblasts (15).
GATA proteins are a family of related transcription factors containing two zinc finger domains: the C-terminal zinc finger is required for DNA binding, while the N-terminal finger stabilizes DNA binding and facilitates physical interaction with their close interacting partners, the FOG (friend-of-GATA) proteins, through direct binding (11, 21). Among the six known members, GATA-1, GATA-2, and GATA-3 are expressed in hematopoietic, reproductive, endocrine, and exocrine cells (8, 12, 31), as well as a specific class of neurons in the central and peripheral nervous system (34, 36). The other GATA factors, GATA-4, -5, and -6, are not expressed exclusively in hematopoietic tissues (30, 35, 40). GATA-1 is highly expressed in mature erythroid cells, where it binds the DNA consensus sequence (T/A)GATA(A/G) in a variety of genes necessary for erythroid maturation (17, 26, 54), including those required for iron acquisition and heme synthesis (44). Therefore, it is likely that GATA-1 participates in regulating mitochondrial iron transport by activating Mfrn1 gene expression.
Critical functions of GATA-1 in erythroid differentiation are achieved through activation and repression of many genes implicated in all steps of erythroid cell maturation (11). Thus, GATA-1 upregulates the erythroid genetic program and suppresses genes involved in the establishment and maintenance of early pluripotential hematopoietic progenitors (10). The precise balance between the transcriptional factors GATA-1 and GATA-2 is part of the erythroid cell fate decision (9). The “GATA switch,” where an increase in the GATA-1 level, facilitated by FOG-1, displaces GATA-2 from its binding site, provides an important mechanism of GATA-1 chromatin occupancy for GATA-related activation, as well as its repression (3, 9, 28, 38).
Genome-wide chromatin immunoprecipitation (ChIP) databases for erythroid chromatin occupancy by GATA-1 provide a powerful resource to identify developmentally important target genes and their associated cis elements or cis-regulatory modules (CRMs) (17, 26, 54). However, these ChIP assays and associated bioinformatics predictions require further in vivo functional validation. Zebrafish offer many practical advantages to facilitate the rapid identification of functional cis elements. These advantages include a large number of progeny that mature rapidly (30, 53), transparent embryos to facilitate identification of embryonic tissues, and highly efficient methods of transgenesis (49).
We used transgenesis in zebrafish and mice to identify the transcriptional regulatory elements that regulate expression of the Mfrn genes during vertebrate development. We first identified a cytosine-phosphate-guanosine (CpG) island, encompassing the promoter region in the Mfrn2 locus, which recapitulates the endogenous expression pattern for the Mfrn2 transcript. We then identified two highly conserved CRMs in the Mfrn1 locus, the −20.4-kb CRM and the −37.5-kb CRM, which specifically control the expression of Mfrn1 during vertebrate hematopoiesis. The −37.5-kb CRM region assayed in this study directed transgene expression to the zebrafish heart, brain, and blood cells and thus recapitulated many of the salient and conserved aspects of endogenous mfrn1 gene expression. A developmental analysis, using fluorescence reporter transgenes and confocal microscopy, revealed that the Mfrn transgenes were restricted in expression to the myocardium of the heart. Finally, using knockdowns, fluorescence-activated cell sorting (FACS), ChIP assays, and mutagenesis of GATA-1 binding cis elements (GBE), we showed that the GATA-1–FOG-1 complex directly regulates the activation of Mfrn1 during erythropoiesis.
MATERIALS AND METHODS
Zebrafish transgenic lines and WISH.
Wild-type (AB* and Tü), cloche (clom39), and transgenic zebrafish (Danio rerio) were maintained according to standard methods (53). The following transgenic zebrafish lines were used: myocardium-specific Tg(cmlc2:DsRed)s879 (18), endothelium-specific Tg(flk1:mCherry-ras)s896, and erythroid-specific Tg(GATA-1:DsRed)sd2. Whole-mount in situ hybridization (WISH) was carried out as described previously (1). All zebrafish studies were conducted under the guidance and approval of the Institutional Animal Care and Use Committee (IACUC) at Children's Hospital Boston.
Zebrafish anti sense MO injections and RT-PCR.
Morpholinos (MOs) against zebrafish GATA-1, FOG-1 (1), and GATA-2 (5′-GACGAGCTAAGAAATATAAGACAGA-3′) were obtained from GeneTools (Philomath, OR); 0.03 to 0.06 pmol MO was injected into embryos at the 1- or 2-cell stage. GATA-2-specific primers were used to perform a semiquantitative reverse transcription (RT)-PCR using total RNA and standard procedures (1).
Identification of promoter-enhancer elements for the Mfrn genes.
Mouse genomic Mfrn1 and Mfrn2 sequences were downloaded from the Ensembl Genome Browser (Sanger Centre, United Kingdom) and displayed using the University of California, Santa Cruz (UCSC) genome database. Candidates for CRMs with GBE associated with high ChIP peaks (17, 26, 54) were identified using the Pennsylvania State University (PSU) Genome Browser (http://main.genome-browser.bx.psu.edu). A candidate CRM was selected in the regions with GBE peaks in both ChIP-on-chip DNA microarray (ChIP-chip) and ChIP sequencing (ChIP-seq) for analysis in transgenic zebrafish.
Gateway subcloning of the mouse CRMs into expression vectors and expression of transgenes in zebrafish.
Gateway-compatible vectors (Invitrogen) were used to analyze the expression of the candidate CRMs (49). The molecular and transgenic techniques are detailed in our previous studies (1, 49). Embryos were coinjected at the one-cell stage with 25 pg Tol2 transposase cRNA and 50 pg Tol2-flanked destination green fluorescent protein (GFP) reporter plasmid (49). The expression of the transgenic GFP reporter was visualized at 24 h postfertilization (p.f.) under a fluorescence microscope (Leica MZFLIII).
Mutagenesis of the GATA-1 binding sites.
Regions of the −37.5-kb CRM of the mfrn1 gene containing the 3 GATA-1 binding sites are shown in Fig. 1. Mutation of 2 nucleotides at the central core of the GATA sequence (GATA→CTTA) (16, 55) was performed through conventional overlap extension PCR. The resulting PCR products were cloned in the Gateway-compatible vectors (49), and their sequences were confirmed by sequence analysis. The primers used in the overlap extension PCR are listed in Table 1.
Fig. 1.
Genome-wide analysis of ChIP-seq and ChIP-chip data predict CRMs. The Mfrn genes are plotted at the top using the base positions in mouse chromosomes (chr) 14 and 19. The coding regions are represented by the blue boxes. The intronic regions are shown as arrowheads representing the direction of transcription. The TSS is indicated by the bent arrow. The G1R-ER4/G1E inputs are represented by black boxes. The positions of the CpG islands are shown as green boxes. The regulatory-potential scores, based on a seven-species sequence alignment, are shown as blue peaks. The murine amplicons used in zebrafish and mouse transgenesis are shown as red (ChIPs) or green (CpG) rectangles above the line, with the positions of the amplicons relative to the TSS (−20.4 kb, −34.0 kb, and −37.5 kb). Three conserved core sequences for GATA-1 binding sites over the −37.5-kb CRM are shown as pink blocks. The orange numbers indicate the nucleotide positions. The image was generated from the PSU genome bioinformatics site (17).
Table 1.
Sequences of primers used for ChIPs and generating GATA-1 mutations in the GBE
| Primera | Sequence (5′-3′) |
|---|---|
| PCR ChIP | |
| GATA1 HS1-F | TGATGGCTTCTACTAGGCACACG |
| GATA1 HS1-R | GGCTTCACTCCCAGGAATGTAGG |
| Mfrn-1 −35.7 kb-F | CTCTAGGGCCCAGGCTAGAT |
| Mfrn-1 −35.7 kb-R | ACACGGCATGCAGAGGTTAT |
| Mfrn-1 −20.4 kb-F | TCCTTGGTGCTTGATTAGGG |
| Mfrn-1 −20.4 kb-R | TGATGTCAGTGCCACAAGGT |
| Mutagenic GATA-1 PCR | |
| GATA-1 M1-F | ATGTTTCATATCTTAAAGGGCGGCTTGGAGGAGG |
| GATA-1 M1-R | CAAGCCGCCCTTTAAGATATGAAACATATGGATGC |
| GATA-1 M2-F | GAACAGCAGTTTAAGATTCCGTCTCCACCCAACCC |
| GATA-1 M2-R | GGAGACGGAATCTTAAACTGCTGTTCTTCAAAGTC |
| GATA-1 M3-F | CTCCAGAGACTCTTAACCTCTGCATGCCGTGTTTTC |
| GATA-1 M3-R | CATGCAGAGGTTAAGAGTCTCTGGAGAACCTCCTAG |
F, forward primer; R, reverse primer; HS1, upstream GATA-1 gene enhancer; M1, M2, and M3, GATA-1 mutants.
Fluorescence-activated flow cytometry.
The morpholino targeting GATA-1 was injected into fertilized eggs at the 1- or 2-cell stage collected from Tg(Mfrn1:GFP) matings. Single-cell suspensions for flow cytometry from approximately 100 to 200 embryos were prepared from MO-injected and control clutches and sorted as previously described (1).
Mouse transient transgenesis.
Pronuclear injections were carried out at the Transgenic/Gene Targeting Mouse Facility at the University of Utah. Briefly, pronuclei of fertilized eggs were microinjected with linearized DNA, incubated at 37°C overnight, and implanted into a 0.5-day pseudopregnant female. Embryos were dissected in cold 1× phosphate-buffered saline (PBS) at day 8.5 from the day of embryonic implantation (E8.5) and E12.5. GFP emission was immediately acquired on the SZX12 stereomicroscope (Olympus) equipped with a Bh2-RFL-T3 fluorescent lamp (Olympus). The embryos and yolk sac were fixed for 20 min (E8.5 yolk sac) to 1 h (E12.5 embryos) in 4% paraformaldehyde (PFA)/PBS at 4°C with rocking. Sections were coverslipped with Vectashield (Vector Laboratories) and imaged on a BX51 fluorescence microscope (Olympus) or an FV100 confocal microscope (Olympus). All mouse studies were conducted under the guidance and approval of the IACUC at the University of Utah (Salt Lake City, UT) and the Jackson Laboratory (Bar Harbor, ME).
Staining and histology of mouse embryos.
Matings were established between C57BL/6J females (wild-type) and C57BL/6J congenic male mice heterozygous for the Mfrn null allele (43), a gene trap in which the sequence encoding β-galactosidase is inserted downstream of the endogenous Mfrn1 promoter (UCSF BayGenomics mES clone XB454). For whole-mount photography, yolk sacs obtained at E8.5 and embryos obtained at E12.5, E13.5, and E14.5 were fixed briefly and stained for β-galactosidase activity for 48 h (25). All reagents were obtained from major vendors. Abdominal and cardiac cavities were opened to promote contact with the stain. Prior to being imaged (see below), yolk sacs were fixed in 4% paraformaldehyde overnight and transferred through graded ethanol (25). Embryos were cleared by processing them in 25% glycerol and then photographed using a Nikon SMZ1500 dissecting microscope and a Retige 2000R digital camera (12 bit) with QCapture software, version 3.0.
For sectioning, embryos stained as described above were fixed overnight in Bouin's fixative and embedded in paraffin. Digital images of 10-μm sections were collected using a Nikon E600 microscope equipped with Nikon 10×/0.3 and 40×/0.75 objective lenses. Images were captured with a model 2.21 RT-Color Spot digital camera and Spot software version 4.5.
ChIP assay and real-time PCR.
ChIP assays were performed as previously described (1, 54). Friend mouse erythroleukemia (MEL) cells were chemically induced to undergo differentiation by the addition of dimethyl sulfoxide (DMSO) to a final 1.7% (vol/vol) concentration in Dulbecco modified Eagle medium with 10% fetal calf serum, 2% penicillin/streptomycin, and 1% glutamine for 24 h prior to ChIP assay (54). G1ER cells were induced with estradiol as described previously (51). Chromatin from 4 × 106 MEL or G1ER cells was used for each ChIP. For GATA-2 and FOG-1 ChIP, protein A DynaBeads (100.02D; Invitrogen) were incubated with antibodies for 3 h before incubation with chromatin overnight. The GATA-2 antibody (sc-9008) and FOG-1 antibody (sc-9361) were purchased from Santa Cruz Biotechnology. For GATA-1 ChIP, protein A beads were preincubated with rabbit anti-rat IgG (H+L) (Jackson ImmunoResearch) for 1 h before incubation with GATA-1 antibody (sc-265; Santa Cruz Biotechnology). Isotype-matched IgGs were used as controls. The immunoprecipitated DNA was purified and analyzed by quantitative real-time PCR using the QuantiTect SYBR green PCR kit (Qiagen). The primers used for ChIP are listed in Table 1.
Statistical analysis.
Pooled data were calculated as mean ± standard deviation (SD) with the indicated number of repeats. Pairwise comparison was performed by t test and multiple comparisons by analysis of variance (ANOVA) using the SAS 9.1 software.
RESULTS
Locus-wide GATA-1 ChIP analyses identified Mfrn1 and Mfrn2 transcriptional regulatory regions.
In vertebrates, the expression of Mfrn1 is largely restricted to the hematopoietic tissues (43), and GATA-1 regulates most of the genes that define the mature erythroid phenotype (11, 30). To map candidate CRMs that direct the expression of the Mfrn genes in teleosts and mammals, we used genome-wide predictions by ChIP analysis to search for candidate GBE (17, 26, 54). The mouse Mfrn1 locus contains three prominent GBE peaks (Fig. 1, red boxes) for both GATA-1 ChIP-seq (Fig. 1, black boxes) and GATA-1 ChIP-chip (Fig. 1, red peaks), designated −20.4 kb, −34.0 kb, and −37.5 kb (for distances in kb relative to the transcriptional start site [TSS]). A high regulatory-potential (RP) score (23, 52) was used to detect similarity to sequence patterns in alignments distinctive of regulatory regions in seven vertebrate species (Fig. 1, blue peaks). The RP score is a computational tool to aid in the identification of putative regulatory sites in vertebrate genomes. Unlike search tools based on known transcription factor binding site motifs, RP scores approach the problem from a comparative-genomics perspective. The scores are computed from a comparison of genome-wide alignments of highly conserved DNA sequences from humans and vertebrate species (23). A fourth putative GBE peak, localized ∼6 kb downstream of the TSS, was not functionally interrogated because of its low RP score. Both Mfrn genes contain CpG-rich regions (Fig. 1, green boxes), which have been associated with promoter activity for expressed genes (4). We examined the −20.4-kb CRM, −34.0-kb CRM, and −37.5-kb CRM sequences and found two or three conserved core GATA-binding motifs (Fig. 1 and data not shown).
Comparative in vivo analysis of promoter regions for Mfrn1 and Mfrn2 genes.
Aside from its hematopoietic expression in the yolk sac blood island equivalent (the intermediate cell mass [ICM]), mfrn1 is also expressed in regions of the central nervous system (CNS) in zebrafish (Fig. 2A and data not shown). In contrast, mfrn2 is more ubiquitously expressed in the CNS and somitic myotomes (Fig. 2B and data not shown). Moreover, the mfrn2 transcript is not detected in the developing ICM (Fig. 2A and B). At 48 h p.f., both mfrn1 and mfrn2 show similar patterns of expression in the CNS (data not shown).
Fig. 2.
Transgenic analysis of the murine CRMs that control the expression of the Mfrn genes in zebrafish. (A and B) Lateral views of embryos at 24 h p.f. processed by WISH showing the expression of the mfrn1 and mfrn2 transcripts. (C and D) GFP expression of the transgenic lines Tg(Mfrn1:GFP) and Tg(Mfrn2:GFP). (A and C) The mfrn1 gene is expressed in the CNS and the ICM. The GFP expression in the ICM is highlighted in the inset. (B and D) The mfrn2 gene is expressed in the CNS and myotomes (M) (magnified in the inset). The GFP expression from both transgenic lines mirrored the endogenous mRNA localization for the corresponding genes. (E to E″) Confocal fluorescence images of a double-transgenic Tg(Mfrn1:GFP)-Tg(GATA-1:DsRed) embryo showing colocalization of GFP and DsRed signals in erythroid progenitors in the ICM.
To investigate the in vivo function of the CpG-rich regions and the GBE in the respective Mfrn1 and Mfrn2 loci, the corresponding mouse genomic sequences were functionally interrogated in transgenic zebrafish. Briefly, each mouse sequence was inserted using the reporter Gateway system into a Tol2 transposon-based vector containing a GFP reporter cassette for coinjection with Tol2 mRNA as previously described (1). The resulting constructs were injected into one-cell zebrafish embryos and evaluated for GFP reporter expression at 24 h p.f. The CpG island surrounding the promoter region of Mfrn2 gave rise to transgenic fish expressing GFP in the retina, myotome, and CNS that recapitulate the endogenous mfrn2 expression pattern (Fig. 2B and D). A majority of the injected embryos that survived showed the same consistent expression pattern between 24 and 96 h p.f. (data not shown). The surviving embryos were screened for germ line integration to generate the stable Tg(Mfrn2:GFP) transgenic fish (Fig. 2D and data not shown). No GFP expression was observed in embryos injected with the corresponding CpG island from Mfrn1, indicating that other transcriptional regulatory elements might be necessary for its expression (data not shown).
Occupancy by GATA-1 accurately predicts Mfrn1 CRM in zebrafish and mouse.
To test the in vivo functions of the three candidate CRMs in the Mfrn1 locus, we proceeded as described above. Injections of the −37.5-kb CRM and −20.4-kb CRM produce transient transgenic zebrafish expressing GFP in the ICM (Fig. 3 and Table 2). Importantly, the −37.5-kb CRM directed the reporter gene expression in the same tissues in which mfrn1 mRNA is normally expressed (Fig. 2A and C). In contrast, the −20.4-kb CRM resulted in a lower level of GFP expression (Table 2). Injection of the −37.5-kb CRM also produced a high frequency of promiscuous expression in the hatching gland (HG), where the mfrn1 mRNA is not normally expressed (Fig. 3). The GFP expression using a construct that contains the −37.5-kb CRM linked with its native CpG island more effectively recapitulated the endogenous mfrn1 expression pattern, excluding the leaky expression in the HG (Fig. 3). Therefore, we generated the stable Tg(Mfrn1:GFP) using the −37.5-kb CRM combined with the endogenous Mfrn1 minimal promoter (CPG island). Henceforth, we refer to the Tg(Mfrn1:GFP) transgenic construct as containing the −37.5-kb CRM and the endogenous Mfrn1 minimal promoter. No consistent or significant expression of the GFP reporter was seen in any of the zebrafish transgenic embryos injected with the −34.0-kb CRM (Table 2). To confirm that the Tg(Mfrn1:GFP) drives the expression in the hematopoietic compartment, double Tg (Mfrn1:GFP)-Tg(GATA-1:DsRed) transgenic fish were generated (Fig. 2E to E″). At 24 h p.f., erythroid progenitor cells in the posterior ICM expressing the red fluorescent protein DsRed also coexpressed GFP (Fig. 2E″).
Fig. 3.
The mouse −37.5-kb CRM plus its CpG island confer specificity of GFP reporter expression in zebrafish embryos comparable to that of endogenous mfrn1 mRNA. Lateral views of zebrafish embryos at 22 (A) and 26 (B and C) h p.f. are displayed. Shown are fluorescence (A, B, and C) and bright-field merged (A′, B′, and C′) images. (A) The −20.4-kb CRM Mfrn1 transgene directs GFP reporter gene expression in the ICM (arrow). (B) The −37.5-kb CRM Mfrn1 transgene directs GFP expression in the ICM (white arrow) and CNS (yellow arrow) and promiscuously in the HG (red arrow). (C) The −37.5-kb CRM, combined with its CpG island (+CpG), restricts the GFP reporter to the ICM and CNS, thereby more faithfully reflecting its endogenous expression.
Table 2.
Functional analysis of Mfrn1 candidate CRMs by GFP transgenesis
| CRM | Stage | No. of injected embryos | No. of implanted embryos | No. of surviving embryos | No. of GFP+ embryos | % (GFP+/ implanted embryos) |
|---|---|---|---|---|---|---|
| Mouse | ||||||
| −20.4 kb | E8.5 | 95 | 50 | 1 | 2 | |
| E12.5 | 103 | 72 | 0 | 0 | ||
| −37.5 kb | E8.5 | 68 | 23 | 3 | 13 | |
| E12.5 | 115 | 70 | 10 | 14 | ||
| Zebrafish | ||||||
| −20.4 kb | 24 h p.f. | 274 | 156 | 36 | 23 | |
| −34.0 kb | 24 h p.f. | 118 | 92 | 0 | 0 | |
| −37.5 kb | 24 h p.f. | 578 | 392 | 344 | 88 |
To determine the fidelity and general application of our observations to mammalian transcriptional regulation, we assayed the activities of Mfrn1 CRMs in mouse embryos. We focused our analysis on the −37.5-kb CRM construct, which accounts for the most robust filial expression of endogenous Mfrn1 in teleosts. Briefly, the Mfrn1 CRM construct containing GFP as a reporter was injected into mouse pronuclei. The GFP reporter was analyzed at E8.5 and E12.5 developmental stages and compared to the endogenous mouse Mfrn1 expression. Consistent with our previous observations in the zebrafish, the Mfrn1 −37.5-kb CRM directs GFP expression in the mouse tissues where Mfrn1 is normally expressed (Fig. 4). At E8.5, during primitive hematopoiesis, the Mfrn1 CRM drives GFP expression in the yolk blood islands, the site of primitive erythropoiesis (Fig. 4A and B). At E12.5, the Mfrn1 CRM activates GFP expression in the fetal liver, the site of definitive erythropoiesis (Fig. 4C to F).
Fig. 4.
Analysis of the murine CRMs in transgenic mice. (A) The mouse Mfrn1 gene is highly expressed in the yolk sac (YS) blood islands. (B, D, and F) Transgenic-mouse embryos carrying the murine −37.5-kb CRM for the Mfrn1 gene show strong expression in the YS, as well as the hematopoietic fetal liver (FL), comparable to the endogenous Mfrn1 mRNA expression in the embryonic YS (A) and FL (C). (E and F) Higher magnifications of the transversely sectioned fetal liver from panels C and D are shown. The mouse embryonic stages are indicated in the bottom left corners.
We evaluated various CRM combinations to determine their levels of GFP reporter expression. The tandem combination of −34.0-kb CRM plus −37.5-kb CRM or −20.4-kb CRM plus −37.5-kb CRM did not result in any difference in the expression of the GFP reporter in zebrafish embryos compared to −37.5-kb CRM alone in terms of tissue fidelity or intensity (data not shown).
Occupancy by GATA factors at the distal Mfrn1 CRMs.
To correlate the transgenic in vivo enhancer activity with the in vivo transcriptional-factor occupancy during erythroid maturation, ChIP experiments were performed with characterized antibodies (1, 54) by using β-estradiol-inducible G1ER and MEL cell lines. The G1ER (G1R-ER4) cells are derived from GATA-1-null embryonic cells, which stably express a conditional form of GATA-1 in which the full-length protein is fused to the ligand-binding domain of the estrogen receptor (51). The addition of estradiol to G1ER cells activates GATA-1 and triggers erythroid maturation by nuclear translocation of the fusion protein (51, 52). In undifferentiated erythroid cells, the −37.5-kb CRM is occupied by GATA-2, which functions in hematopoietic stem cell (HSC) and early erythroid precursors (22) (Fig. 5A). In β-estradiol-induced G1ER and DMSO-treated MEL cells, GATA-1 was significantly enriched at both −20.4-kb and −37.5-kb CRMs in these more mature erythroid cells (Fig. 5A and B). Similarly, FOG-1 was strongly enriched at both CRMs in G1ER and MEL cells induced to undergo erythroid differentiation (Fig. 5A and B). The level of GATA-2 occupancy at both CRMs was very low in MEL cells (data not shown). Similar results for low GATA-2 expression in uninduced and induced MEL cells have been reported (54).
Fig. 5.
GATA-1 and FOG-1 bind Mfrn1 CRM sites in vivo. (A) The relative occupancy levels of GATA-2, GATA-1, and FOG-1 from ChIP analysis with G1ER cells are indicated by the fold enrichment at each site, normalized to levels in the negative-control region (a 2-kb fragment upstream of the GATA-1 gene enhancer hypersensitive site 1 [HS1]). The bar graphs represent quantification of the ChIP analyses (means + SD from 3 independent experiments). The addition of estradiol to G1ER cells activates GATA-1 and triggers erythroid maturation (uninduced [U] and induced [I]). The asterisks indicate the changes of significance in GATA-2, GATA-1, and FOG-1 occupancy at the −20.4-kb and −37.5-kb CRMs in the Mfrn1 locus compared to the serum control (*, P < 0.05). (B) The relative occupancy levels of GATA-1 and FOG-1 from ChIP analysis of differentiated (DMSO-induced) MEL cells are indicated by the fold enrichment at the −20.4-kb and −37.5-kb CRM sites (*, P < 0.05).
The GATA-1 binding sites are involved in −37.5-kb CRM enhancer-like activity.
As shown in Fig. 1, the −37.5-kb CRM contains three GBE, designated GBE1, GBE2, and GBE3 (Fig. 6A). To determine whether these binding sites are functionally responsible for the hematopoietic-cell-specific enhancer activity, the GATA core binding sites were mutagenized (GATA→CTTA) (16, 55). The mutant constructs were interrogated in vivo using microinjections in wild-type embryos at the one-cell developmental stage. The mutations in the three GBE were labeled M1, M2, and M3 (Fig. 6A). The effects of the GBE mutations on the −37.5-kb CRM transcriptional activity were evaluated by expression of the GFP reporter. The embryos injected with either control or mutated constructions were placed in 3 different categories (normal, reduced, and absent), depending on the intensity levels of GFP expression in the ICM (Fig. 6B to B″). The single GBE mutant M2 construct resulted in a marked reduction of GFP expression, indicating GBE2 is a critical regulatory cis element (Fig. 6A, G, and H compared with C and D). The M1 and M3 single mutations resulted in normal GFP reporter expression compared with the wild-type construct (Fig. 6A, E, F, I, and J). The M2 mutation resulted in ∼65% reduction in transcriptional activity compared with the wild-type construct, whereas the double M1-M2 and M2-M3 mutations resulted in complete absence of GFP reporter expression (Fig. 6A, K, L, O, and P). The M1-M3 double mutation resulted in 90% absence of GFP expression (Fig. 6A, M, and N). These results suggest that GBE2 is necessary but not sufficient for full transcriptional activity. GBE2 requires cooperative interaction with one of its adjacent GBE (either GBE1 or GBE3) for full GATA-mediated transactivation at the −37.5-kb CRM.
Fig. 6.
GBE sites are functionally necessary for GATA-1-mediated transactivation of the −37.5-kb CRM enhancer. (A) A −37.5-kb CRM containing three putative GBE sites (GBE1 to -3) was constructed to drive expression of a GFP reporter. The GATA-binding core sequence in each of the three GBE was mutagenized (GATA→CTTA) singly or in combination, as indicated by “X,” and labeled M1, M2, or M3 for the respective GBE. Reporter plasmids with two mutated GBE are labeled accordingly. The GFP expression in transgenic zebrafish embryos injected with either wild-type or mutant constructs at the one-cell stage is quantified at the right of the corresponding construct. (B to B″) Embryos injected with the various GBE constructs were placed in three different categories, depending on the level of GFP expression: normal, reduced, and absent. (C to P) Lateral views of representative embryos injected with GBE control (wt) or mutant constructs are shown at 24 h p.f. The images on the left are merged bright-field and fluorescence images. The images on the right are fluorescence images. Higher-magnification views of the ICM are shown in the insets.
GATA-1 and FOG-1 transcriptionally regulate mfrn1 during early erythropoiesis.
In zebrafish, the cells destined for hematopoiesis express GATA-1, FOG-1, and GATA-2 transcription factors in the lateral-plate mesoderm (LPM), which subsequently gives rise to the ICM (1, 27). We used WISH to compare the expression patterns of hematopoietic markers and mfrn1 at early stages of embryogenesis. The zebrafish GATA-2 and FOG-1 are expressed as maternal transcripts (1); in contrast, GATA-1 and mfrn1 are expressed later in development when zygotic gene transcription is activated. Between the 15- and 20-somite stages, mfrn1 is coexpressed with GATA-1, GATA-2, FOG-1, lmo2, and scl (Fig. 7). These results, combined with our bioinformatics ChIP-seq and GBE mutagenesis studies shown in Fig. 1, 2, 5, and 6, suggest that mfrn1 expression is dependent on GATA and FOG-1 factors during erythropoiesis.
Fig. 7.
Mfrn1 expression pattern during early erythropoiesis. Dorsal and lateral views of embryos at the 15-, 17-, and 20-somite stages (the vertical arrow indicates the anterior-posterior axis; the horizontal arrows indicate the rostral-caudal axis). The zygotic expression pattern of mfrn1 is coincident with the expression of GATA-1, FOG-1, GATA-2, scl, and lmo2 in the LPM and ICM (arrowheads).
To investigate whether GATA-1, FOG-1, or GATA-2 regulates the expression of mfrn1, we depleted zebrafish embryos of these transcription factors by injecting antisense MOs. The MO-injected embryos (morphants) were processed by WISH to reveal the expression of mfrn1 in erythroid progenitor cells between the 15- and 20-somite stages. Control embryos showed normal expression of mfrn1 (Fig. 7), whereas the embryos injected with either GATA-1 or FOG-1 MO showed markedly reduced mfrn1 expression in the LPM and ICM (Fig. 8A). The absence of mfrn1 expression noted in GATA-1 and FOG-1 morphants was not observed in GATA-2 morphants (Fig. 8A), indicating that the loss of mfrn1 expression is specific to the loss of either GATA-1 or FOG-1 function. These results suggest that the occupancy of GATA-2 at the mfrn1 enhancer elements (Fig. 5A) does not activate its transcription in blood. As a control for full knockdown of GATA-2 in morphant embryos, we used RT-PCR to show that the GATA-2 MO resulted in the production of an aberrantly spliced mRNA, leading to subsequent nonsense-mediated mRNA decay (data not shown). To exclude the possibility that the loss of mfrn1 expression was caused by the absence of hematopoietic progenitors in the GATA-1 morphants, we examined the expression of early hematopoietic markers, scl and lmo2, between the 15- and 20-somite stages. There were no changes in scl and lmo2 expression in the GATA-1 morphants compared with control embryos (Fig. 8A), indicating that GATA-1 specifically regulates mfrn1 expression during erythroid maturation. These results are consistent with our previous analysis, where loss of the FOG-1 cofactor did not perturb early erythroid formation in the LMP and ICM (1). Our data suggest that GATA-1 and FOG-1 transcription factors activate the expression of Mfrn1 in the developing erythron. Our combined ChiP (Fig. 5) and morpholino (Fig. 8) data approximate normal erythroid differentiation, where GATA-2 is expressed at relatively high levels in early erythroid progenitors and is displaced by GATA-1 and FOG-1 during erythroid maturation, as schematically shown in Fig. 8D.
Fig. 8.
GATA-1 and FOG-1 regulate the expression of Mfrn1 during erythropoiesis. (A) The GATA-1 morphants show normal scl and lmo2 expression in hematopoietic progenitors; in contrast, the loss of either GATA-1 or FOG-1 results in the loss of mfrn1 expression in the LPM and ICM. Loss of GATA-2 function does not affect mfrn1 expression. The numbers of morphants with expression of the hematopoietic marker and total numbers of injected embryos are displayed in the right upper corners. (B) Lateral views at 22 h p.f. of Tg(Mfrn1:GFP) embryos uninjected or injected with GATA-1 MO. The arrowheads denote GFP expression in the ICM (highlighted in the insets). (C) The GFP+ cells from Tg(Mfrn1:GFP) embryos were purified by flow cytometry. Quantitative analysis of sorted cells from GATA-1 morphants (GATA-1 MO) shows a reduction in the number of GFP+ cells compared with control Tg(Mfrn1:GFP) embryos. (D) Model for Mfrn1 regulation by GATA factors. Initially, the Mfrn1-CRM is occupied by GATA-2, which does not activate Mfrn1 transcription. After the level of GATA-1 reaches a certain threshold level, GATA-1 binds the Mfrn1-CRM, together with FOG-1, displacing GATA-2 and activating the expression of Mfrn1 during erythroid maturation.
GATA-1 regulates the Mfrn1 −37.5-kb CRM enhancer in vivo.
By 22 h p.f., the expression of GFP in the Tg(Mfrn1:GFP) embryos is localized to the CNS and erythroid progenitors in the ICM (Fig. 2C and E″). Although GATA-1 MO-injected Tg(Mfrn1:GFP) transgenic embryos show no difference in the GFP reporter signal in the CNS, there is complete absence of GFP expression in the ICM (Fig. 8B). GATA-1 is therefore exclusively required for transcriptional activation of the Mfrn1 −37.5-kb CRM in erythroid progenitors (Fig. 8A and B). We used FACS to more precisely quantify the number of GFP-expressing (GFP+) cells in the GATA-1 morphant using the Tg(Mfrn1:GFP) transgenic line. FACS analysis of uninjected Tg(Mfrn1:GFP) embryos represents the total number of GFP+ cells, including those in the CNS and ICM (Fig. 8C). In contrast, GATA-1 morphants show a drastic reduction in the number GFP+ cells in the ICM (Fig. 8B and C). The FACS results, along with our earlier WISH and GBE mutagenesis data, indicate that the Mfrn1 enhancer is a direct transcriptional target of GATA-1 during erythropoiesis.
Mfrn CRMs regulate expression of the GFP reporter in the developing heart.
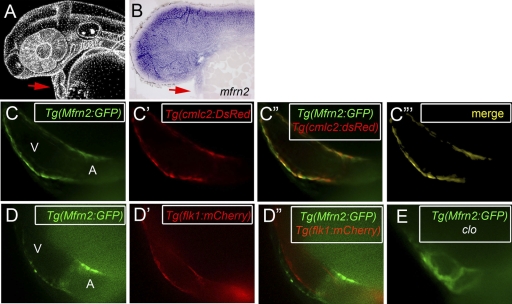
Both transgenic lines, Tg(Mfrn1:GFP) and Tg(Mfrn2:GFP), drive the expression of GFP in the developing cardiac atrium and ventricle at 48 h p.f. (Fig. 9 and 10). Transverse sections of a 48-h p.f. zebrafish embryo revealed that mfrn1 is expressed in the heart and kidney and at the midbrain-hindbrain boundary (Fig. 9B). During early development, the vertebrate heart is composed of two layers, the outer myocardium and the inner endocardium (Fig. 9C and D). We used a transgenic line that expresses the red fluorescent protein DsRed in the myocardial cell layer, Tg(cmlc2:DsRed) (Fig. 9E′), to generate a double-transgenic line, Tg(Mfrn1:GFP)-Tg(cmlc2:DsRed) (Fig. 9E to E″). Whole-mount confocal fluorescence images show that the GFP signal is colocalized in the myocardial cell layer (Fig. 9E‴). A similar expression pattern was observed for Tg(Mfrn2:GFP) (Fig. 10). We next examined the Tg(Mfrn1:GFP) expression (Fig. 9F) in a transgenic line that delineates the endothelial cell layer, Tg(flk1:mCherry-ras) (Fig. 9F′) by generating a double-transgenic fish, Tg(Mfrn1:GFP)-Tg(flk1:mCherry-ras) (Fig. 9F″). Neither Mfrn transgenic line directed GFP expression in the endocardial cell layer (Fig. 9F″ and 10D″), shown by the lack of colocalization signals. Because the cloche mutant specifically lacks an endocardial layer in its developing heart (45), we examined the expression of our Mfrn transgenes in this genetic background. Both GFP reporters regulated by the Mfrn CRMs were observed in clo mutants (Fig. 9G and 10E). Together, these results indicate that the GFP expression regulated by both Mfrn transgenic lines is in the myocardial layer (Fig. 9 and 10).
Fig. 9.
The transgenic Tg(Mfrn1:GFP) expresses the GFP reporter in the myocardium. (A and B) Lateral views of zebrafish embryos at 48 h p.f. (A) Schematic representation of the zebrafish embryo (http://www.zfin.org). (B) WISH of mfrn1 in embryonic sections showing its expression in the heart (H; red arrow), kidney, a hematopoietic tissue in teleosts (K; red arrowhead), and the midbrain-hindbrain boundary (asterisk). (C and D) By 48 h p.f., cardiac chambers (atrium [A] and ventricle [V]) and two layers (myocardium [green] and endocardium [red]) form. (E to E‴) Merged confocal images of a double-transgenic Tg(Mfrn1:GFP)-Tg(cmlc2:DsRed) embryo reveal colocalized expression of mfrn1 in the myocardial layer (yellow). (F to F″) Merged confocal images of a double-transgenic Tg(Mfrn1:GFP)-Tg(flk1:mCherry) embryo suggest that Mfrn1 is not expressed in the endothelial cell layer. (G) GFP expression for transgenic Tg(Mfrn1:GFP) in cloche indicates that mfrn1 is expressed in the myocardium. (H and I) Bright-field and fluorescence images of the transgenic Tg(Mfrn1:GFP) mouse embryo at E12.5 showing expression of the GFP reporter in the heart (H; arrows) and liver (L; arrowheads). (J to L) LacZ reporter expression from an Mfrn1 gene trap transgenic mouse at embryonic stages E12.5 to E14, confirming endogenous Mfrn1 expression (blue) in the atria (A) and ventricles (V).
Fig. 10.
The transgenic Tg(Mfrn2:GFP) expresses the GFP reporter in the myocardium. (A and B) Lateral views of zebrafish embryos at 48 h p.f. (A) Schematic representation of the zebrafish embryo (http://www.zfin.org). (B) WISH of mfrn2 in sectioned embryos showing expression in the heart (red arrow). (C and C′) Confocal fluorescence images showing the transgenic Tg(Mfrn2:GFP) expression in the heart (green) (C) and the transgenic Tg(cmlc2:DsRed) marker in the myocardium (red) (C′). (C″ and C‴) Merged confocal images of a double-transgenic Tg(Mfrn2:GFP)-Tg(cmlc2:DsRed) embryo reveal colocalized expression of Mfrn2 in the myocardial layer (yellow). (D and D′) Confocal images of the transgenic Tg(Mfrn2:GFP) (green) (D) and the endocardium-specific transgenic Tg(flk1:mCherry) (red) (D′). (D″) Merged confocal images in a double-transgenic Tg(Mfrn2:GFP)-Tg(flk1:mCherry) embryo indicate Mfrn2 is not expressed in the endothelial-cell layer. (E) Confocal microscopy of transgenic Tg(Mfrn2:GFP) in the endocardium-deficient mutant (clo) show GFP expression, supporting the idea that Mfrn2 is specifically expressed in the myocardium.
In E12.5 transgenic mice, the −37.5-kb CRM GFP reporter is expressed in the fetal heart and liver at levels comparable to those in the endogenous mouse Mfrn1 (Fig. 9H and I). Examination of mouse strains carrying a lacZ gene trap marker in the Mfrn1 gene (43) showed β-galactosidase staining in embryos at E12.5 to E14.5 confined to the heart (Fig. 9J to L). Both atrial and ventricular chambers express the lacZ surrogate marker for Mfrn1 initially, although its expression is subsequently restricted to the atria by E14.5 (Fig. 9L).
DISCUSSION
The closely related Mfrn genes encode inner mitochondrial solute transporters that are critical for iron assimilation in the genesis of heme and Fe/S clusters. Their fundamental function is highly conserved in the eukaryotic phyla, since the ΔMRS3/4 yeast deficiency strain is complemented by either vertebrate Mfrn (33, 43). In contrast, Mfrn1 plays a crucial nonredundant role in vertebrate erythropoiesis (43), when tremendous quantities of iron are required for heme synthesis and hemoglobin production. During erythroid maturation, differential protein turnover accounts for part of the regulation of Mfrn1 in undifferentiated erythroid cells (39). An increase in Mfrn1 protein stability is greatly enhanced by its oligomerization with another inner mitochondrial protein, Abcb10 (15), which is a GATA-1-inducible ATP-binding cassette transporter (44). The identification of ferrochelatase, the terminal enzyme that catalyzes the insertion of iron into protoprophyrin IX to form heme, in the same protein complex with Mfrn1 and Abcb10 directly integrates the import of iron with heme synthesis (14).
The regulation and function of the Mfrn transporters in coordinating the production of heme molecules with terminal stages of erythroid cell differentiation is reminiscent of 5-aminolevulinate synthase (ALAS), the first and rate-limiting enzyme of the heme biosynthetic pathway. Like the Mfrn genes, there are two separate ALAS genes encoding distinct ALAS isoenzymes. The housekeeping gene, ALAS1, is ubiquitously expressed and regulated by heme via the peroxisome proliferator-activated receptor γ coactivator 1α (PGC-1α) (24, 29). The erythroid-specific gene, ALAS2, is expressed in erythroid tissues to supply heme for hemoglobin production, and its transcription is not regulated by heme. Similar to Mfrn1 distal enhancers, the promoter and enhancer in intron 8 of ALAS2 have several erythroid-specific cis elements for GATA factors to confer its tissue-restricted expression (19, 46, 47). The posttranslational regulation of ALAS2 is mediated by the intracellular levels of iron and iron-responsive protein 1 (IRP1) binding to its cognate iron-responsive element (IRE) in the 5′ untranslated region (UTR) of the ALAS2 mRNA (19). No IRE motifs have been identified in the UTR of either Mfrn transporter cDNA to which their posttranslational regulation by iron can be attributed.
In vertebrate genomes, CpG islands typically occur near the TSSs of genes, surrounding their respective promoter regions (4). Histones bound to CpG islands are the sites of specific modifications (6), and they frequently bind ubiquitous transcription factors (13). The CpG regions exhibit particularly accessible chromatin structures (20). For this reason, they play a fundamental role in predicting promoters (5). Both the Mfrn1 and Mfrn2 genes contain CpG islands surrounding their TSSs; however, only the Mfrn2 CpG island directed expression of the GFP reporter in an manner identical to its endogenous transcript in transgenic zebrafish embryos. This observation is consistent with the notion of CpG islands functioning as promoter elements for housekeeping genes (4). In contrast, the Mfrn1 CpG island failed to direct expression of the GFP reporter to hematopoietic tissues. Similarly, most minimal GATA-regulated promoters (or larger regions) are themselves not sufficient to direct expression in transgenic assays, and they often require enhancers to augment their transcriptional activities (37).
We used genome-wide predictions by chromatin occupancy (17, 26, 54) to identify distal candidate enhancer CRMs with preserved GBE in the mouse Mfrn1 locus. Using this strategy, we found two genomic segments, the −20.4-kb CRM and −37.5-kb CRM, that directed GFP expression in developing erythroid cells during zebrafish and mouse embryogenesis. Whereas the −37.5-kb CRM was sufficient to account for the erythroid expression of Mfrn1, the CpG island surrounding the TSS was required to confer specificity and repress promiscuous expression in the HG. The −37.5-kb CRM murine genomic DNA fragment also faithfully directed expression of the GFP reporter in the cardiac myocardial layer. Our analysis represents the first demonstration that noncoding sequences with preservation of GBE in the Mfrn1 locus can direct expression in erythroid and myocardial populations.
In order to evaluate the fidelity of our findings in zebrafish, we tested both Mfrn1 CRMs using transgenesis in mouse embryos. In zebrafish and mice, the −37.5-kb CRM directed the reporter GFP in all tissues where the endogenous Mfrn1 transcripts are normally expressed, whereas the −20.4-kb CRM displayed less consistent GFP expression. The data generated in both species were highly concordant and suggest that transgenesis in zebrafish serves as a rapid, robust system to validate regulatory elements from mammalian species. The subset of GATA-1-occupied sites active as enhancers shows strong association with the evolutionary preservation for the binding-site motif across mammalian lineages. While the genome-wide ChIP data permit the identification of candidate DNA segments occupied by GATA-1 (17, 26, 54), the use of zebrafish transgenesis allows faster functional validation. We initially evaluated our CRM constructions for enhancer activity in K562 human erythroleukemia cells, and the addition of the −37.5-kb CRM caused only a modest increase in reporter luciferase activity above that of a control vector containing only the γ-globin promoter (data not shown). This result indicated that the context-dependent functions of the CRM could be more reliably evaluated in vivo using transgenic models, validating the premise of our studies. Similar cautions for using cell lines to functionally interrogate CRMs have been noted by other investigators (7, 17, 48, 50).
The mutational analyses of three GBE in the −37.5-kb CRM demonstrated that mutation of the GBE2 site is necessary but not sufficient for full enhancer activity. We also showed that erythroid-specific transcriptional regulation was mediated by at least two GATA-1 binding sites, namely, GBE2 with either one of its adjacent GBE, in the central core of the −37.5-kb CRM. The observation that the three GBE are conserved in mammals underscores the importance of this regulation (Fig. 1). The consensus GBE3 is fully conserved in rat and mouse, while GBE1 and GBE2 are fully conserved in human, dog, rat, and mouse. These functional studies in transgenic zebrafish suggest that the GBE within the −37.5-kb CRM regulate Mfrn1 expression during erythroid maturation.
Our data with morphant embryos indicate that activation of mfrn1 expression is directly dependent on GATA-1 and FOG-1 activity (Fig. 8A). Previous experiments have shown that GATA-1 occupancy of chromatin sites bound by GATA-2 is facilitated by FOG-1 (32, 38). Our ChIP analysis demonstrated that FOG-1 occupies the −37.5-kb CRM in the same regions bound by GATA factors, consistent with FOG-1 facilitating the displacement of GATA-2 by GATA-1 during erythroid maturation (9, 28, 38). Based on these data, we propose a model for erythroid Mfrn1 expression in which at early developmental stages the maternal GATA-2 factor binds to the −20.4-kb and −37.5-kb CRMs without activating Mfrn1 transcription. Later, during zygotic transition, the activation of GATA-1 expression, facilitated by FOG-1, induces an exchange of nuclear factors at both CRMs. GATA-2 is displaced by the GATA-1-FOG-1 complex, resulting in the activation of Mfrn1 transcription (Fig. 8D). This sequence of events approximates previous in vitro observations in which GATA-2 is expressed at relatively high levels in early erythropoiesis and is gradually replaced by GATA-1 during late erythropoiesis (52). This is consistent with models in which GATA factors nucleate the binding of various protein complexes and changes in protein/DNA binding ultimately lead to a rapid increase in transcription at the α-globin promoter (3).
The expression of the GFP reporter by the Mfrn1 distal CRM in the heart and CNS is not surprising, since other GATA factors, in combination with their FOG cofactors, could regulate expression in those tissues. In vertebrates, the GATA family members GATA-4, GATA-5, and GATA-6 are predominantly expressed during heart development to regulate cardiac morphogenesis, and they share ∼85% similarity in their DNA-binding domains at the protein level (30, 35, 40). In addition, GATA-2 and GATA-3 are expressed during early stages of CNS development, indicating that these factors might collaborate during neural-tissue differentiation (34, 36). Using ChIP assays, we demonstrated that the Mfrn1 CRM is bound by GATA-2. Thus, GATA-2 could bind to the Mfrn1 enhancer to potentially activate its expression in the CNS. Similar to the “cardiac” and “neuronal” GATA factors, their FOG-1-FOG-2 partners are also expressed in both tissues, and their loss of function contributes to several cardiac and neurogenic defects (11, 12). Finally, in the Mfrn1 CRM sequence, we also identified binding motifs for the helix-loop-helix transcriptional factors, MyoD, myogenin, and Myf-5, which are critical for specification and differentiation of muscle cells (41). On the basis of these observations, we propose that those motifs are key elements for driving Mfrn1 CRM myotomal expression. Together, our results demonstrate that analysis of mouse genomic element in zebrafish is suitable for rapid identification of long-range cis-regulatory elements that regulate tissue-specific gene expression.
ACKNOWLEDGMENTS
We thank Christian Lawrence, Jason Best, Adam Cardullo, and Paul McBride for help with zebrafish animal husbandry; Jessica Wagner for help with confocal microscopy; Kathleen Soltis for screening cloche and Tg(Mfrn:GFP) zebrafish; Shuo Lin for the Tg(GATA-1:GFP) transgenic line; David Traver for the Tg(GATA-1:DsRed) transgenic line; and Susan Tamowski for the pronuclear injections. We thank Iman Schultz, Heidi Anderson, Adrianne Kolpak, and John Kanki for reviewing the manuscript. Confocal fluorescence microscopy was performed at the Harvard Digestive Disease Center Imaging Facility (Children's Hospital Boston).
This work was supported by grants from the American Heart Association (J.D.C.) and the March of Dimes Foundation (B.H.P.) and by National Institutes of Health grants K08 HL074891 (N.C.C.), U01 HL066611 (L.L.P.), R01 DK052380 (J.K.), R01 DK070838 (B.H.P.), P01 HL032262 (A.B.C. and B.H.P.), and P30 DK072437 (J.K. and B.H.P.).
We declare no financial conflict of interest.
Footnotes
Published ahead of print on 19 January 2011.
REFERENCES
- 1. Amigo J. D., et al. 2009. The role and regulation of friend of GATA-1 (FOG-1) during blood development in the zebrafish. Blood 114:4654–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrews N. C. 2008. Forging a field: the golden age of iron biology. Blood 112:219–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Anguita E., et al. 2004. Globin gene activation during haemopoiesis is driven by protein complexes nucleated by GATA-1 and GATA-2. EMBO J. 23:2841–2852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Antequera F. 2003. Structure, function and evolution of CpG island promoters. Cell. Mol. Life Sci. 60:1647–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bajic V. B., Tan S. L., Suzuki Y., Sugano S. 2004. Promoter prediction analysis on the whole human genome. Nat. Biotechnol. 22:1467–1473 [DOI] [PubMed] [Google Scholar]
- 6. Bernstein B. E., et al. 2005. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell 120:169–181 [DOI] [PubMed] [Google Scholar]
- 7. Blow M. J., et al. 2010. ChIP-Seq identification of weakly conserved heart enhancers. Nat. Genet. 42:806–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bresnick E. H., Lee H.-Y., Fujiwara T., Johnson K. D., Keles S. 2010. GATA switches as developmental drivers. J. Biol. Chem. 285:31087–31093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bresnick E. H., Martowicz M. L., Pal S., Johnson K. D. 2005. Developmental control via GATA factor interplay at chromatin domains. J. Cell. Physiol. 205:1–9 [DOI] [PubMed] [Google Scholar]
- 10. Cantor A. B., et al. 2008. Antagonism of FOG-1 and GATA factors in fate choice for the mast cell lineage. J. Exp. Med. 205:611–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cantor A. B., Orkin S. H. 2005. Coregulation of GATA factors by the Friend of GATA (FOG) family of multitype zinc finger proteins. Semin. Cell Dev. Biol. 16:117–128 [DOI] [PubMed] [Google Scholar]
- 12. Cantor A. B., Orkin S. H. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene 21:3368–3376 [DOI] [PubMed] [Google Scholar]
- 13. Cawley S., et al. 2004. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell 116:499–509 [DOI] [PubMed] [Google Scholar]
- 14. Chen W., Dailey H. A., Paw B. H. 2010. Ferrochelatase forms an oligomeric complex with mitoferrin-1 and Abcb10 for erythroid heme biosynthesis. Blood 116:628–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen W., et al. 2009. Abcb10 physically interacts with mitoferrin-1 (Slc25a37) to enhance its stability and function in the erythroid mitochondria. Proc. Natl. Acad. Sci. U. S. A. 106:16263–16268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheng G., Hagen T. P., Dawson M. L., Barnes K. V., Menick D. R. 1999. The role of GATA, CArG, E-box, and a novel element in the regulation of cardiac expression of the Na+-Ca2+ exchanger gene. J. Biol. Chem. 274:12819–12826 [DOI] [PubMed] [Google Scholar]
- 17. Cheng Y., et al. 2009. Erythroid GATA1 function revealed by genome-wide analysis of transcription factor occupancy, histone modifications, and mRNA expression. Genome Res. 19:2172–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chi N. C., et al. 2008. Genetic and physiologic dissection of the vertebrate cardiac conduction system. PLoS Biol. 6:e109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Cox T. C., Bawden M. J., Martin A., May B. K. 1991. Human erythroid 5-aminolevulinate synthase: promoter analysis and identification of an iron-responsive element in the mRNA. EMBO J. 10:1891–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Crawford G. E., et al. 2006. Genome-wide mapping of DNase hypersensitive sites using massively parallel signature sequencing (MPSS). Genome Res. 16:123–131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Crispino J. D., Lodish M. B., MacKay J. P., Orkin S. H. 1999. Use of altered specificity mutants to probe a specific protein-protein interaction in differentiation: the GATA-1:FOG complex. Mol. Cell 3:219–228 [DOI] [PubMed] [Google Scholar]
- 22. Dore L. C., et al. 2008. A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. U. S. A. 105:3333–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Elnitski L., et al. 2003. Distinguishing regulatory DNA from neutral sites. Genome Res. 13:64–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Estall J. L., et al. 2009. PGC-1alpha negatively regulates hepatic FGF21 expression by modulating the heme/Rev-Erb(alpha) axis. Proc. Natl. Acad. Sci. U. S. A. 106:22510–22515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Frasch M., Chen X., Lufkin T. 1995. Evolutionary-conserved enhancers direct region-specific expression of the murine Hoxa-1 and Hoxa-2 loci in both mice and Drosophila. Development 121:957–974 [DOI] [PubMed] [Google Scholar]
- 26. Fujiwara T., et al. 2009. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36:667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gering M., Yamada Y., Rabbitts T. H., Patient R. K. 2003. Lmo2 and Scl/Tal1 convert non-axial mesoderm into haemangioblasts which differentiate into endothelial cells in the absence of Gata1. Development 130:6187–6199 [DOI] [PubMed] [Google Scholar]
- 28. Grass J. A., et al. 2003. GATA-1-dependent transcriptional repression of GATA-2 via disruption of positive autoregulation and domain-wide chromatin remodeling. Proc. Natl. Acad. Sci. U. S. A. 100:8811–8816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Handschin C., et al. 2005. Nutritional regulation of hepatic heme biosynthesis and porphyria through PGC-1alpha. Cell 122:505–515 [DOI] [PubMed] [Google Scholar]
- 30. Heicklen-Klein A., McReynolds L. J., Evans T. 2005. Using the zebrafish model to study GATA transcription factors. Semin. Cell Dev. Biol. 16:95–106 [DOI] [PubMed] [Google Scholar]
- 31. LaVoie H. A. 2003. The role of GATA in mammalian reproduction. Exp. Biol. Med. (Maywood) 228:1282–1290 [DOI] [PubMed] [Google Scholar]
- 32. Letting D. L., Chen Y.-Y., Rakowski C., Reedy S., Blobel G. A. 2004. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl. Acad. Sci. U. S. A. 101:476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li F. Y., et al. 2001. Characterization of a novel human putative mitochondrial transporter homologous to the yeast mitochondrial RNA splicing proteins 3 and 4. FEBS Lett. 494:79–84 [DOI] [PubMed] [Google Scholar]
- 34. Meng A., Tang H., Ong B. A., Farrell M. J., Lin S. 1997. Promoter analysis in living zebrafish embryos identifies a cis-acting motif required for neuronal expression of GATA-2. Proc. Natl. Acad. Sci. U. S. A. 94:6267–6272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Molkentin J. D. 2000. The zinc finger-containing transcription factors GATA-4, -5, and -6. Ubiquitously expressed regulators of tissue-specific gene expression. J. Biol. Chem. 275:38949–38952 [DOI] [PubMed] [Google Scholar]
- 36. Nardelli J., Thiesson D., Fujiwara Y., Tsai F. Y., Orkin S. H. 1999. Expression and genetic interaction of transcription factors GATA-2 and GATA-3 during development of the mouse central nervous system. Dev. Biol. 210:305–321 [DOI] [PubMed] [Google Scholar]
- 37. Nishimura S., et al. 2000. A GATA box in the GATA-1 gene hematopoietic enhancer is a critical element in the network of GATA factors and sites that regulate this gene. Mol. Cell. Biol. 20:713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pal S., et al. 2004. Coregulator-dependent facilitation of chromatin occupancy by GATA-1. Proc. Natl. Acad. Sci. U. S. A. 101:980–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paradkar P. N., Zumbrennen K. B., Paw B. H., Ward D. M., Kaplan J. 2009. Regulation of mitochondrial iron import through differential turnover of mitoferrin 1 and mitoferrin 2. Mol. Cell. Biol. 29:1007–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Patient R. K., McGhee J. D. 2002. The GATA family (vertebrates and invertebrates). Curr. Opin. Genet. Dev. 12:416–422 [DOI] [PubMed] [Google Scholar]
- 41. Pownall M. E., Gustafsson M. K., Emerson C. P. 2002. Myogenic regulatory factors and the specification of muscle progenitors in vertebrate embryos. Annu. Rev. Cell Dev. Biol. 18:747–783 [DOI] [PubMed] [Google Scholar]
- 42. Schultz I. J., Chen C., Paw B. H., Hamza I. 2010. Iron and porphyrin trafficking in heme biogenesis. J. Biol. Chem. 285:26753–26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shaw G. C., et al. 2006. Mitoferrin is essential for erythroid iron assimilation. Nature 440:96–100 [DOI] [PubMed] [Google Scholar]
- 44. Shirihai O. S., Gregory T., Yu C., Orkin S. H., Weiss M. J. 2000. ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 19:2492–2502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stainier D. Y., Weinstein B. M., Detrich H. W., Zon L. I., Fishman M. C. 1995. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development 121:3141–3150 [DOI] [PubMed] [Google Scholar]
- 46. Surinya K. H., Cox T. C., May B. K. 1998. Identification and characterization of a conserved erythroid-specific enhancer located in intron 8 of the human 5-aminolevulinate synthase 2 gene. J. Biol. Chem. 273:16798–16809 [DOI] [PubMed] [Google Scholar]
- 47. Surinya K. H., Cox T. C., May B. K. 1997. Transcriptional regulation of the human erythroid 5-aminolevulinate synthase gene. Identification of promoter elements and role of regulatory proteins. J. Biol. Chem. 272:26585–26594 [DOI] [PubMed] [Google Scholar]
- 48. Suzuki M., Moriguchi T., Ohneda K., Yamamoto M. 2009. Differential contribution of the Gata1 gene hematopoietic enhancer to erythroid differentiation. Mol. Cell. Biol. 29:1163–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Villefranc J. A., Amigo J., Lawson N. D. 2007. Gateway compatible vectors for analysis of gene function in the zebrafish. Dev. Dyn. 236:3077–3087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Visel A., et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Weiss M. J., Yu C., Orkin S. H. 1997. Erythroid-cell-specific properties of transcription factor GATA-1 revealed by phenotypic rescue of a gene-targeted cell line. Mol. Cell. Biol. 17:1642–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Welch J. J., et al. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood 104:3136–3147 [DOI] [PubMed] [Google Scholar]
- 53. Westerfield M. 1995. The zebrafish book, 3rd ed. University of Oregon Press, Eugene, OR [Google Scholar]
- 54. Yu M., et al. 2009. Insights into GATA-1-mediated gene activation versus repression via genome-wide chromatin occupancy analysis. Mol. Cell 36:682–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zon L. I., et al. 1991. GATA-binding transcription factors in mast cells regulate the promoter of the mast cell carboxypeptidase A gene. J. Biol. Chem. 266:22948–22953 [PubMed] [Google Scholar]