Prostaglandin E2 inhibits the expression of retinal dehydrogenase, thus inhibiting retinoic acid production and the priming of gut-tropic T cells by dendritic cells.
Abstract
The production of retinoic acid (RA) by dendritic cells (DCs) is critical for the induction of gut-tropic immune responses by driving the expression of intestinal-specific homing receptors, such as α4β7 and CCR9, upon T and B cell activation. However, how RA production is regulated during DC development remains unclear. We describe an unexpected role for prostaglandin E2 (PGE2) as a negative regulator of retinal dehydrogenases (RALDH), the enzymes responsible for RA synthesis. The presence of PGE2 during DC differentiation inhibited RALDH expression in mouse and human DCs, abrogating their ability to induce CCR9 expression upon T cell priming. Furthermore, blocking PGE2 signaling increased the frequency of RALDH+ DCs in vitro, and reducing PGE2 synthesis in vivo promoted the systemic emergence of RA-producing DCs and the priming of CCR9+ T cells in nonintestinal sites such as the spleen. Finally, we found that PGE2 stimulated the expression of the inducible cyclic AMP early repressor, which appears to directly inhibit RALDH expression in DCs, thus providing mechanistic insight into how PGE2 signaling down-modulates RALDH. Given the role of PGE2 in regulating the development of RA-producing DCs, modulating this pathway may prove a novel means to control the development of gut-tropic immune responses.
It has been >40 yr since the observation that lymphoblasts isolated from intestinal LNs preferentially home to the intestine when compared with their counterparts derived from skin-draining peripheral LNs (pLNs; Griscelli et al., 1969; Hall et al., 1977). It has subsequently been shown that such gut-specific migration is a result of T and B cells activated within the gut-associated-lymphoid tissue (GALT) selectively up-regulating the expression of homing receptors, such as α4β7 and CCR9, whose ligands (MadCAM and CCL25, respectively) are largely restricted to the intestinal tract (Campbell and Butcher, 2002; Johansson-Lindbom et al., 2003). In addition to inducing the expression of gut-specific homing receptors, intestinal LNs (i.e., mesenteric LN [mLN] and Peyer’s patch) have been reported to support the development of peripherally generated FoxP3+ regulatory T cells (Annacker et al., 2005; Coombes et al., 2007) and isotype class switching to Ig-A (Shikina et al., 2004; Mora et al., 2006). Consequently, the concept that the GALT is specialized in driving immune responses tailored to the intestinal mucosa has emerged.
It is generally accepted that the DCs that localize to the GALT are the architects of this gut-tropism. Reductionist studies have demonstrated that DCs taken from the intestinal LNs, but not splenic or skin-draining pLNs, induce α4β7 and CCR9 expression upon T and B cell priming (Johansson-Lindbom et al., 2003; Mora et al., 2003, 2006) and also promote FoxP3 expression in a TGF-β–dependent manner (Annacker et al., 2005; Coombes et al., 2007; Mucida et al., 2007). This activity has been further restricted to a subset of CD103-expressing DCs that originate from the lamina propria and migrate to the draining LNs in a CCR7-dependent manner (Annacker et al., 2005; Johansson-Lindbom et al., 2005; Coombes et al., 2007). CD103+ DCs appear to be the major DC subset involved in trafficking antigen from intestine to LN and are potent activators of T cells within the LN (Jaensson et al., 2008; Schulz et al., 2009).
The mechanisms by which intestinal DCs drive the expression of gut-homing receptors was first described by Iwata et al. (2004), with the seminal observation that retinoic acid (RA) triggered the expression of the gut-tropic homing receptors during T cell priming. Subsequent studies have shown that RA similarly drives α4β7/CCR9 expression on B and T cells in mice and humans (Jaensson et al., 2008) and is at least partly involved in the generation of FOX-P3+ regulatory T cells and Ig-A switching (Coombes et al., 2007; Mucida et al., 2007). RA is the active metabolite of vitamin A, being synthesized via a multi-step pathway whereby retinol is converted into the intermediate retinal by the ubiquitously expressed alcohol dehydrogenase enzymes, and then into RA by a family of retinal dehydrogenase enzymes (RALDH) that are expressed in a more restricted manner (Napoli, 1999; Duester, 2000). Critically, the RALDH enzymes that control RA synthesis are highly expressed by CD11c+ DCs from intestinal LNs but are absent from splenic DCs, correlating with the former being able to produce RA (Iwata et al., 2004). A more detailed analysis has demonstrated that RALDH expression is restricted to the CD103+ subset of mLN DCs (Coombes et al., 2007). Collectively, these findings suggest that CD103+ intestinal DCs, by virtue of RALDH expression, are uniquely endowed with the capacity to produce RA, which in turn drives the expression of genes appropriate for intestinal immunity. Recently however, a population of RALDH-expressing CD103−CD11b+ DCs that originate in the dermis and lung has also been previously described (Guilliams et al., 2010). These DCs were capable of priming FoxP3+ regulatory T cells in vitro, suggesting that a similar mechanism is in place for the generation of regulatory T cells against lung- and skin-derived self-antigens.
What controls the expression of the RALDH enzymes, and thus the ability to synthesize RA, remains unclear. One possibility is that RALDH expression is restricted to lineage-specific DCs. Alternatively, RALDH expression may be regulated by the local microenvironment. Because RALDH-expressing intestinal CD103+ DCs and RALDH-negative conventional lymphoid CD8α DCs appear to be developmentally related—both are derived from common DC precursors and are dependant upon the transcription factors Irf8 and Batf3 for development (Bogunovic et al., 2009; Varol et al., 2009; Edelson et al., 2010)—it is unlikely that RALDH expression is restricted to DCs of a unique lineage. Instead, the observations that feeding mice a vitamin A–deficient diet dramatically reduces DC RALDH expression in vivo (Yokota et al., 2009) and that multiple factors, including GM-CSF, IL-4, IL-13, and the TLR-2 ligand zymosan, can induce in vitro RALDH expression (Manicassamy et al., 2009; Yokota et al., 2009) suggest that the gut microenvironment may be capable of inducing RALDH expression through the provision of soluble factors. Indeed, it has been reported that intestinal CD103+ DC development is dependent on GM-CSF (Bogunovic et al., 2009), a potent inducer of RALDH expression in vitro. However, how the expression of the RALDH enzymes by DCs is regulated in vivo remains to be clarified, as the intestine does not switch on RALDH expression ubiquitously. Indeed, not all intestinal DCs express the RALDH enzyme during the steady state (Jaensson et al., 2008; Schulz et al., 2009), whereas monocyte-derived DCs (mo-DCs) infiltrating the intestine during periods of inflammation do not acquire RALDH activity (Siddiqui et al., 2010). Furthermore, dermal-derived CD103+CD11b− DCs are also dependent on GM-CSF for development (King et al., 2010) but do not express the RALDH enzyme in vivo (Guilliams et al., 2010).
To address the mechanisms regulating the development of RA-producing DCs in vivo, we sought to identify potential factors that regulate RALDH expression during DC differentiation. In this paper, we describe an unexpected role for prostaglandin E2 (PGE2) as a negative regulator of RALDH expression in both mice and human DCs. Consequently, modulating PGE2 signaling during DC development dramatically altered the ability of DCs to prime gut-homing T cells. Given the central role for RA-producing DCs in promoting gut-tropic immunity, our findings suggest that modulating PGE signaling may be a powerful approach by which to control the tissue specificity of developing immune responses.
RESULTS
Multiple factors can induce RALDH expression in a wide range of DCs in vitro
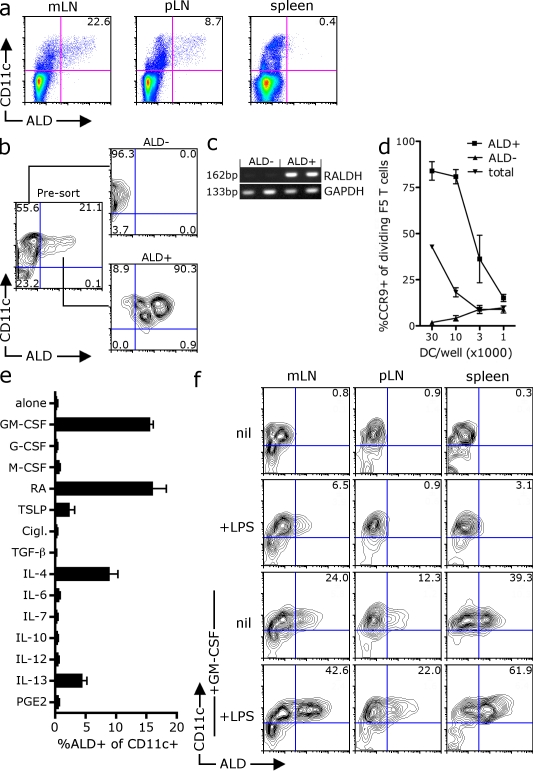
To investigate the development of RA-producing DCs, we initially screened a panel of factors for their ability to induce the expression of the RALDH enzymes that convert retinal to RA. To measure RALDH activity, we used the commercially available substrate aldefluor (ALD), which is converted into a fluorescent product by RALDH enzymes, allowing the identification of RALDH-expressing cells by flow cytometry. Consistent with previous studies (Yokota et al., 2009; Guilliams et al., 2010), we found that although the spleen was devoid of ALD fluorescent cells, a subset of CD11c+ cells in both the mLNs (∼23%) and skin-draining pLNs (∼9%) converted the ALD substrate and fluoresced, indicating functional RALDH enzymes (Fig. 1 a). To confirm that ALD fluorescence correlated with RALDH activity and RA production, we sorted mesenteric CD11c+ DCs into ALD-positive and -negative subsets (Fig. 1 b) and found that only the ALD+ DCs expressed the RALDH2 transcript (Fig. 1 c). Second, although both subsets stimulated robust peptide-specific T cell proliferation in vitro (Fig. S1 a), the ability to drive CCR9 expression upon T cell priming was enriched and restricted to ALD+ DCs (Fig. 1 d). Thus, ALD fluorescence appears to mark the subset of DCs that express RALDH2 and have a unique ability to prime CCR9 expression, which is consistent with the synthesis of RA (for convenience herein we shall refer to the ALD+/− cells as RALDH+ and RALDH−). Consistent with a previous study (Guilliams et al., 2010), the RALDH+ CD11c+ cells from the mLN were positive for CD103 and heterogenous for CD11b, whereas the RALDH+ DCs isolated from the skin-draining pLNs were CD103−CD11b+, suggesting that RALDH activity was common to differing DC subsets in vivo (Fig. S1 b).
Figure 1.
Identification of factors that induce RALDH expression in vitro. (a) mLN, skin-draining pLN, or spleens from C57BL/6 mice were digested, treated with ALD, and stained with anti-CD11c antibodies for analysis by flow cytometry. A representative dot plot of ALD and CD11c expression from at least seven experiments is shown with inset values indicating the percentage of live (PI−) CD11c+ DCs that are ALD+. The mean was taken from between 11 and 23 mice. (b–d) CD11c+ mLN cells sorted into ALD+ and ALD− subsets. (c) The expression of RALDH2 mRNA by ALD+ and ALD− DC subsets was examined by RT-PCR (values indicate molecular mass of PCR product). (d) NP366–374 peptide–pulsed total CD11c+ mLN DCs or CD11c+ DCs sorted into ALD+ or ALD− subsets were used to stimulate CFSE-labeled CD8+ F5 T cells in vitro. After 4 d, the expression of CCR9 on dividing T cells was analyzed by flow cytometry. The graph shows the mean percentage of dividing T cells that express CCR9 with SEM pooled from two independent experiments. (e) BM cells were cultured with FLT3-L in the presence of the indicated factors. After 3 d, LPS was added and, 18 h later, cells were treated with ALD and stained with CD11c. Shown is the percentage of CD11c+ DCs that are ALD positive. The mean was taken from two experiments with SEM. (f) ALD− CD11c+ DCs isolated from the mLN (mes), pLN, or spleen were cultured with GM-CSF and/or LPS for 48 h before analysis of RALDH activity by ALD staining. A representative contour plot of ALD and CD11c expression is shown. Inset values show the mean percentage of CD11c cells that are ALD+ pooled from three experiments.
To examine what factors can induce RALDH expression during DC development, we differentiated DCs from BM precursors with FLT3-L in the presence of a panel of immune factors. Although FLT3-L can drive the differentiation of CD11c+ DCs from BM precursors, FLT3-L alone does not induce RALDH expression (Fig. 1 e). Consistent with an earlier study (Yokota et al., 2009), the addition of anyone of GM-CSF, RA, IL-4, IL-13, and, to a lesser degree, thymic stromal lymphopoietin (TSLP) resulted in the emergence of RALDH+ DCs, suggesting that each of these factors can induce the expression of RALDH during DC development (Fig. 1 e). To examine whether GM-CSF would also induce RALDH activity in differentiated lymphoid DCs, we sorted RALDH− CD11c+ cells from the mLN, skin draining pLN, and spleen and cultured them with GM-CSF for 48 h before measuring RALDH activity. As seen in Fig. 1 f, a large fraction of sorted RALDH− DCs from all organs up-regulated the RALDH enzyme in response to GM-CSF, an activity which appears to be enhanced by LPS. Together, this data shows that multiple factors can induce RALDH expression in vitro and that DCs that do not express RALDH in vivo have the potential to do so in response to these positive signals.
Stromal cell–derived factors inhibit RALDH expression during DC differentiation
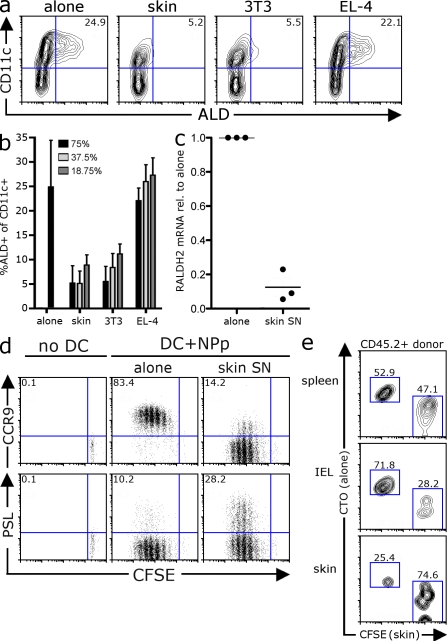
Given that RALDH activity is restricted to a minor subset of DCs in vivo, it was surprising that such a large proportion of DCs could be induced to express RALDH in vitro by a wide panel of factors including GM-CSF. We therefore postulated that RALDH expression may be actively suppressed in vivo by negative regulators. The stroma is emerging as major source of molecules that modulate DC activity, and we wondered whether stromal-derived factors might also be involved in regulating DC RALDH expression. To this end, we differentiated DCs from BM precursors with GM-CSF in the presence of supernatants (SNs) derived from a panel of stromal or lymphocyte cell lines. Again, culturing BM with GM-CSF alone drives the differentiation of RALDH-expressing DCs, which account for ∼25% of CD11c+ cells (Fig. 2 a). However, when BM-derived DCs (BM-DCs) were differentiated with GM-CSF in the presence of SN taken from a primary skin stromal line or the 3T3 fibroblast line, RALDH+ DCs failed to develop (Fig. 2 a). This suppressive activity was restricted to SN purified from stromal cell lines, as SN from the thymoma EL-4 did not impair the development of RALDH-expressing DCs at any concentration (Fig. 2, a and b). Consistent with the loss of RALDH+ activity, CD11c+ DCs differentiated in the presence of skin SN expressed substantially less RALDH2 mRNA, suggesting that RALDH inhibition occurs at a transcriptional level (Fig. 2 c). Importantly, although CD11c+ DCs cultured with skin SN do not develop RALDH expression, the expression of MHC class II and CD86 by CD11c+ DCs was equivalent to that of control DCs (Fig. S2 a). Furthermore, skin SN–conditioned DCs produced equivalent or enhanced levels of IL-12 upon TLR stimulation, suggesting that the loss of RALDH activity was not a consequence of general dysfunction (Fig. S2 b). Consistent with this, when pulsed with the influenza NP366–374 peptide, both control and skin SN–conditioned DCs could drive the proliferation of the CD8+ NP366–374–specific F5 T cells in vitro, as measured by the sequential loss of CFSE fluorescence (Fig. 2 d). However, only control BM-DCs induced CCR9 expression on dividing T cells, which is consistent with their intact RALDH expression. Indeed, although skin SN–conditioned DCs had lost the ability to induce CCR9 expression, they had an enhanced ability to drive P-selectin ligand (PSL) expression, a lectin involved in tethering to nonintestinal endothelial (Fig. 2 d). These phenotypic differences correlated with distinct homing potential in vivo. When transferred into congenic recipients at equivalent frequencies, although both T cell populations were found at equal numbers in the spleen, T cells primed by control DCs homed more effectively to the small intestine epithelial, whereas T cells primed by skin SN–conditioned DCs had an enhanced capacity to traffic into inflamed skin (Fig. 2 e). In summary, this data shows that the stroma can produce soluble factors that suppress RALDH expression during DC differentiation, limiting the development DCs capable of priming gut-homing T cells.
Figure 2.
Stromal-derived factors inhibit DC RALDH expression. (a and b) BM from C57BL/6 mice was cultured with GM-CSF alone or in the presence of SNs derived from primary mouse skin lines (skin), 3T3 fibroblasts, or EL-4 cells. After 3 d, LPS was added and, 18 h later, cells were stained with ALD and CD11c to measure DC RALDH activity. (a) A representative contour plot depicting the expression of CD11c and ALD is shown with inset values indicating the percentage of CD11c+ DCs that are ALD+. (b) BM was differentiated, as in a, with SNs at various concentrations. Bar graphs show the mean percentages of CD11c+ DCs that are ALD+ with SD. Data were pooled from between 2 and 18 experiments. (c) DCs were differentiated from BM with GM-CSF alone or in the presence of skin SN. Cells were treated with LPS at day 3. 16–18 h later, DCs were purified with anti-CD11c or MHC class II microbeads and analyzed for RALDH2 expression by real-time qPCR. RALDH2 expression was normalized to GAPDH. Shown is relative expression of RALDH2 by skin SN versus control DCs, with each dot representing an individual experiment and horizontal bars the pooled mean. (d) DCs were differentiated from BM with GM-CSF alone, or in the presence of skin SN. After 3 d, LPS was added and, 18 h later, MHC class II+ DCs were purified, pulsed with NP366–374-peptide, and used to stimulate CFSE-labeled CD8+ F5 T cells. 4 d later, the expression of CCR9 and PSL was analyzed by flow cytometry. Dot plots show the expression of homing receptors versus CFSE on live (PI−) TCR+ cells. Inset values are the mean percentage of dividing T cells positive for each receptor pooled from between 7 and 17 experiments. (e) CD8+ F5 T cells were activated as in d. After 4 d, T cells were labeled with CFSE or CTO, pooled at a 1:1 ratio, and transferred into congenic B6.Ly5.1 recipients (∼2 × 106/recipient) that had been primed with oxazolone and challenged on the ear. After 16–18 h, the relative frequency of donor (CD45.2+) T cells primed by control versus skin SN-conditioned DCs at the various sites was determined by flow cytometry. A representative contour plot gated on live (PI−) CD45.2+ donor T cells is shown, where control T cells are labeled with CTO and skin SN DC-primed T cells with CFSE. Inset is the mean frequency of each population pooled from three independent experiments.
PGE2 inhibits RALDH expression during DC differentiation
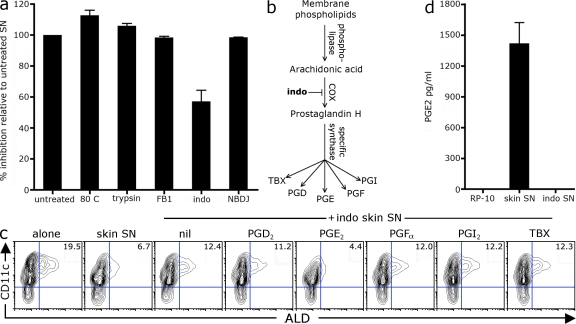
We next sought to identify the soluble factor produced by skin stroma that inhibits DC RALDH expression. The ability to suppress RALDH expression was maintained in skin SN that had been heated or digested with trypsin, indicating that the active factor was unlikely to be a protein or a peptide (Fig. 3 a). We next tested whether a lipid may be responsible for RALDH inhibition. To this end, we grew skin stroma lines in the presence of lipid inhibitors to block the production of ceramides (fumonisin B1; He et al., 2006), prostaglandins (indomethacin; Nakata et al., 1981), or glycosphingolipids (N-butyldeoxynojirimycin [NBDJ]; Andersson et al., 2000), thus generating skin SN depleted of these various lipid species. As seen in Fig. 3 a, although skin SN depleted of ceramides or sphingolipid retained maximal suppressive activity, SN from indomethacin-treated skin stroma had a substantially reduced ability to suppress the development of RALDH-expressing DCs. Indomethacin inhibits the cyclooxygenase (COX) enzymes, which convert arachidonic acid to prostaglandin H, which is then converted into one of five active metabolites: PGD2, PGE2, PGFα, PGI, or thromboxane (Katler and Weissmann, 1977; Fig. 3 b). We asked whether these prostaglandins could rescue the suppressive activity of indomethacin-treated skin SN. As seen in Fig. 3 c, the addition of exogenous PGE2, but not of the other prostaglandins, restored the suppressive ability of indomethacin-treated skin stroma SN. Consistent with PGE2 as the active factor, endogenous PGE2 was detected in the skin SN (at ∼2 ng/ml or 5–10 nM), but not in SN, from indomethacin-treated skin stroma (Fig. 3 d). Intriguingly, exogenous PGE2 alone, in the absence of skin SN, had a reduced suppressive activity (Fig. S3), suggesting that the skin SN enhances the delivery or responsiveness to PGE2. Nonetheless, this data suggests that stromal-derived PGE2 inhibits the differentiation of RALDH-expressing DCs.
Figure 3.
Stroma-derived PGE2 inhibits RALDH expression during DC differentiation. (a) BM-DCs were differentiated with GM-CSF in the presence of skin stroma SN that had been heated at 80°C or digested with trypsin, or SN derived from skin stroma grown in the presence of fumonisin B1 (FB1), indomethacin (indo), or NBDJ. After 3 d, LPS was added and, 18 h later, cells were stained with ALD and CD11c to analyze DC RALDH activity. Bar graphs show the mean relative inhibition of ALD+ CD11c+ DCs between treated SN and untreated parent SN with SD. Relative inhibition was calculated as described in Materials and methods. Values were pooled from 2–10 experiments. (b) Pathway of prostaglandin synthesis. (c) BM cells were cultured with GM-CSF alone or in the presence of SN from WT or indomethacin-treated skin stroma (SN at 25% culture volume). 0.1 µM of exogenous individual prostaglandins subtypes was added at the beginning of culture. After 3 d, LPS was added and, 18 h later, cells were stained with ALD and CD11c to analyze DC RALDH activity. A representative contour plot of CD11c versus ALD staining is shown with inset values of the mean percentage of CD11c+ DCs that are ALD+ pooled from three experiments. (d) The concentration of PGE2 in skin stromal SN was determined by ELISA. Shown is the mean with SD from seven individual lines.
Deficiency in PGE2 receptor subtype 4 (EP-4) enhances DC RALDH expression
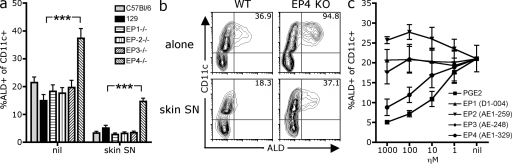
To further examine whether PGE2 signaling negatively regulated the expression of the RALDH enzymes, we asked whether mice deficient in PGE2 receptors had aberrant RALDH activity. PGE2 signals through four receptors: EP-1, -2, -3, or -4. These are G-protein–coupled receptors that have distinct downstream signaling events (Sugimoto et al., 2000; Sugimoto and Narumiya, 2007). We used genetically engineered mice that lack each individual EP receptor to examine whether a PGE2 receptor deficiency enhanced RALDH expression (Segi et al., 1998; Ushikubi et al., 1998; Hizaki et al., 1999). To this end, we differentiated DCs with GM-CSF from BM isolated from either WT or EP-KO mice in the presence or absence of skin SN before measuring RALDH activity. As seen in Fig. 4, compared with their WT 129 controls, BM from EP-4–deficient mice gave rise to a significantly enhanced frequency of RALDH+ DCs when cultured with GM-CSF alone (Fig. 4, a and b). Second, although the skin SN completely abolished the development of RALDH+ DCs from WT or EP-1, -2, or -3 KO BM, a population of RALDH+ DCs did emerge from EP-4 KO BM cultured with skin SN (Fig. 4, a and b). EP-4 deficiency did not alter other aspects of BM-DC development, in terms of DC frequency or phenotype, with equivalent levels of CD86, CD11b, or CD103 expression by EP-4 KO and WT BM-DCs (Fig. S4 a). Thus, the absence of EP-4 signaling appeared to enhance DC RALDH expression selectively. Consistent with PGE2 signaling via EP-4 to inhibit RALDH expression, the EP-4–specific agonist ONO-AE1-329 (Sugimoto and Narumiya, 2007; Jones et al., 2009), but not EP-1, -2, or -3 agonists, inhibited DC RALDH expression similar to PGE2 (Fig. 4 c). However, it is important to note that in vivo, EP-4 KO mice only showed a statistically significant increase in the frequency of RALDH+ DCs in the pLNs but not the mLN or spleen (Fig. S4). Given this modest phenotype in vivo, it is probable that some redundancy between other PGE2 receptors, notably EP-2, which, like EP-4, enhances cAMP (Sugimoto et al., 2000), or alternate factors may exist in vivo.
Figure 4.
EP-4 deficiency enhances BM-DC RALDH expression. (a and b) BM cells from EP-KO or WT C57BL/6 and 129 mice were cultured with GM-CSF alone or in the presence of skin SN. After 3 d, LPS was added and, 18 h later, cells were stained with CD11c and ALD to measure DC RALDH activity. (a) Bar graph shows the mean percentage of CD11c+ DCs that are ALD+ for each mouse genotype with SEM pooled from four experiments (***, P < 0.001). (b) A representative contour plot of CD11c and ALD expression of EP-4 KO and WT 129 BM-DCs is shown. Inset values indicate the MFI of ALD expression by CD11c+ DCs. (c) BM cells were cultured with GM-CSF in the presence of exogenous PGE2 or EP-specific agonists (cultures comprised 25% SN from indomethacin-treated skin stroma to enhance PGE2 responsiveness). Indomethacin was added at the beginning of culture to block endogenous PGE2 synthesis. After 3 d, LPS was added and, 18 h later, cells were stained with CD11c and ALD to measure DC RALDH activity. Graph shows the mean ± SEM of the percentage of CD11c+ DCs that are ALD+ pooled from three experiments.
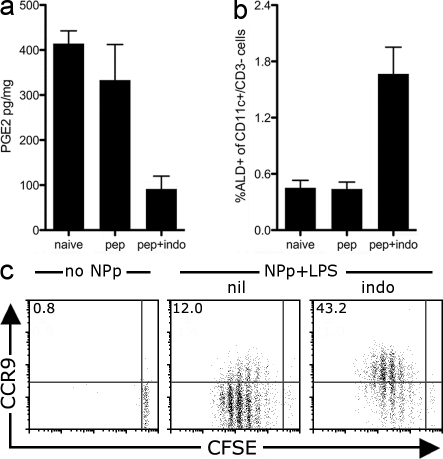
Limiting PGE2 levels in vivo promotes the emergence of RALDH-expressing DCs in the spleen and the priming of gut-homing T cells systemically
Given our findings in vitro, we wished to examine whether reducing PGE2 synthesis would increase the frequency of RA-producing DCs in vivo. Splenic DCs do not express RALDH and consequently fail to induce the expression of the gut-homing receptor CCR9 during T cell priming (Johansson-Lindbom et al., 2003; Iwata et al., 2004). Given that PGE2 is present within the spleen in the steady state (Fig. 5 a), we asked whether limiting PGE2 systemically—through the i.v. injection of indomethacin—would result in the emergence of RALDH-expressing DCs in the spleen that would in turn drive CCR9 expression during T cell activation. To this end, we transferred CFSE-labeled CD8+ F5 T cells into C57BL/6 recipients and, 1 d later, elicited priming within the spleen through the i.v. injection of the NP366–374 peptide with LPS in combination with indomethacin treatment. 60 h later, we analyzed RALDH activity by splenic CD11c+ DCs and the expression of CCR9 on dividing F5 T cells. As seen in Fig. 5, indomethacin administration reduces splenic PGE2 levels (Fig. 5 a) and results in a four to fivefold increase in RALDH-expressing splenic DCs (Fig. 5 b). Critically, in comparison with untreated mice where dividing F5 T cells do not express CCR9, a large fraction (∼43%) of dividing T cells up-regulate CCR9 in indomethacin-treated mice, which is consistent with the enhanced RALDH expression by local splenic DCs (Fig. 5 c). Thus, inhibiting prostaglandin synthesis allows the emergence of RALDH-expressing DCs in nonintestinal sites such as the spleen, which in turn prime gut-homing T cells at this site.
Figure 5.
Indomethacin administration promotes the emergence of splenic RALDH+ DCs in vivo and the priming of CCR9+ T cells systemically. (a–c) 5 × 105 CFSE-labeled CD8+ F5 T cells were transferred into C57BL/6 mice and, 1 d later, mice received 50 µg NP366–374-peptide plus 10 µg LPS via i.v. tail vein injection (NPp+LPS). Indomethacin-treated mice received 100 µg indomethacin i.v. 1 h before peptide and then once daily. 60 h after NP366–374 peptide injection, mice were sacrificed and the spleens analyzed. (a) The concentration of PGE2 was determined in spleen homogenate by ELISA. Shown is the mean with SEM from three to five mice pooled from two experiments. (b) Splenocytes were treated with ALD and stained with CD11c and CD3 (to gate out to CFSE+ T cells). Shown is the mean percentage of CD3-CD11c+ DCs that are ALD+ in the spleen. Mean with SEM from 7–11 mice per group is shown, pooled from four experiments. (c) Splenocytes were stained with antibodies against CD8α and CCR9. Dot plots were gated on live (PI−) CD8+ CFSE+ T cells and show the expression of CCR9 and CFSE on donor F5 T cells. Inset value is the mean percentage of dividing T cells that are CCR9+ pooled from between 6 and 13 mice per group, acquired over four experiments.
PGE2 inhibits RALDH expression in human mo-DCs
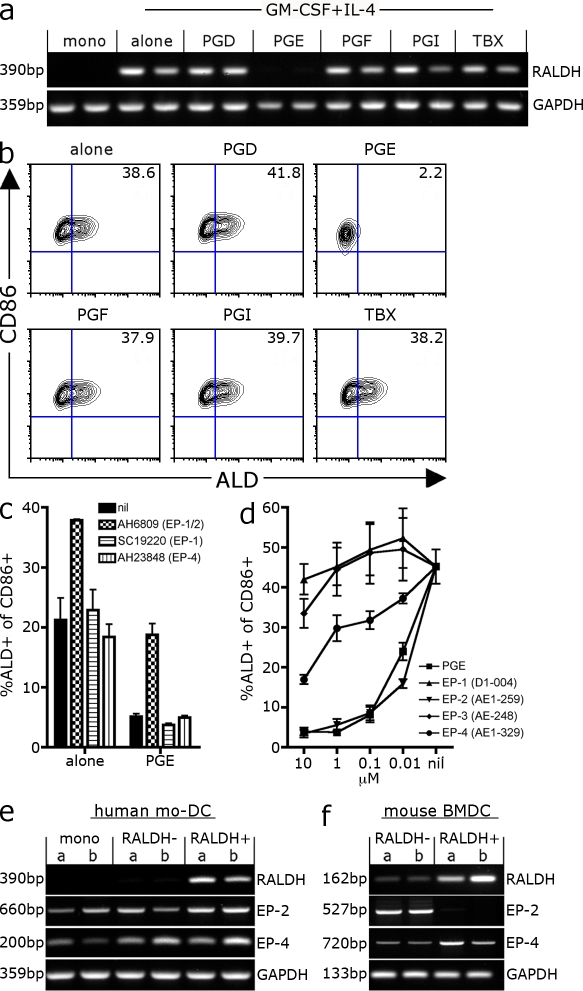
We next wished to examine whether PGE2 similarly regulated RALDH expression in human DCs. To this end, we cultured CD14+ monocytes with GM-CSF and IL-4 in the presence or absence of prostaglandins and asked: (a) does GM-CSF/IL-4 induce RALDH expression in human mo-DCs; and (b) does PGE2 inhibit this induction? As seen in Fig. 6, although monocytes do not express RALDH2 mRNA, differentiation with GM-CSF and IL-4–induced RALDH expression by mo-DCs as measured by both ALD staining and RT-PCR (Fig. 6, a and b). Consistent with our previous experiments, PGE2, but none of the other prostaglandins, inhibited RALDH expression at a protein and mRNA level, suggesting that PGE2 suppressed RALDH transcription in human mo-DCs (Fig. 6, a and b). We next asked which PGE2 receptor was signaling RALDH inhibition in human mo-DCs. Using a panel of EP-specific antagonists, we found that AH6809, an antagonist for EP-1 and EP-2 (Jones et al., 2009), enhanced the frequency of RALDH-positive DCs in the absence of exogenous PGE2, suggesting that some level of endogenous PGE2 was limiting RALDH expression in culture (Fig. 6 c). Second, AH6809 blocked the complete suppression of RALDH+ DCs after the addition of exogenous PGE2. Given that the EP-1–specific antagonist SC1990 (Jones et al., 2009) had no effect, AH6809 is presumably acting by blocking EP-2 signaling. Surprisingly, the EP-4 antagonist AH23848 also had no effect on RALDH expression. We extended this analysis by using EP-specific agonists to ask which agonists could recapitulate PGE2 activity. It is of note that these experiments were performed in the presence of indomethacin to limit endogenous PGE2 production. As seen in Fig. 6 d, agonists for EP-1 or EP-3 showed negligible activity, whereas EP-4–specific agonists (AE1-329) partially limited the development of RALDH+ DCs (Sugimoto and Narumiya, 2007). Consistent with the antagonist data, the EP-2–specific agonist (AE1-259) was substantially more potent, completely inhibiting RALDH expression with an activity equivalent to PGE2 (Fig. 6 d). Thus, similar to what we observe in mice, PGE2 inhibits RALDH transcription during DC differentiation. However, in contrast to mouse BM-DCs, this activity appears to be mediated predominantly through PGE2–EP-2 signaling. Given this distinction, we compared the PGE2 receptor expression by mouse BM-DCs and human mo-DCs sorted into RALDH+ and RALDH− subsets (Fig. 6, e and f). Consistent with their sensitivity to EP-2 signaling, human monocytes and both RALDH-negative and -positive mo-DCs express EP-2 in addition to EP-4 (Fig. 6 e). However, in the case of mouse BM-DCs, although both DC subsets expressed EP-4, EP-2 expression was restricted to RALDH-negative DCs (Fig. 6 f). Therefore, although human mo-DCs express both EP-2 and EP-4 and are highly sensitive to EP-2-PGE2 signaling, mouse RALDH+ BM-DCs have diminished EP-2 receptor expression and are consequently refractory to EP-2–specific agonists.
Figure 6.
PGE2 inhibits RALDH expression in human mo-DCs. (a and b) DCs were differentiated from CD14+ monocytes purified from buffy coats with GM-CSF and IL-4 alone or in the presence of 0.1 µM of exogenous prostaglandin subtypes. Indomethacin was included to stop endogenous prostaglandin synthesis. After 3 d, LPS was added and, 18 h later, cells were harvested and analyzed for RALDH expression by RT-PCR and ALD staining. (a) RALDH2 expression analyzed by RT-PCR. Duplicate samples were derived from two independent experiments using different donors. (b) A representative contour plot of ALD and CD86 expression is shown with the mean percentage of ALD+ of CD86+ DCs inset. Values were pooled from two independent experiments with different donors. (c) CD14+ monocytes were differentiated with GM-CSF/IL-4 alone or with 25 µM PGE2 in the presence of antagonists against EP-1/2 (AH6809), EP-1 (SC19220), or EP-4 (AH23848). After 3 d, LPS was added and, 18 h later, cells were treated with ALD and stained with CD86 to measure DC RALDH activity. Bar graphs show mean percentage of CD86+ DCs that are ALD+ with SEM. Data were pooled from three independent experiments. (d) CD14+ monocytes were differentiated with GM-CSF/IL-4 with indomethacin in the presence of PGE2 or EP-specific agonists at varying concentrations. After 3 d, LPS was added and, 18 h later, cells were treated with ALD and stained with CD86 to measure DC RALDH activity. Line graph shows the mean percentage of CD86+ DCs that are ALD+ with SEM, pooled from three independent experiments. (e and f) Human mo-DCs or mouse BM-DCs generated as described were treated with ALD and stained with CD86 or CD11c, respectively, and sorted into CD86+ RALDH+/− or CD11c+ RALDH+ and RALDH− subsets. The expression of EP-2 and EP-4 was examined by RT-PCR. Duplicates were from different cDNA obtained from two independent experiments and donors (values indicate molecular mass of PCR product).
PGE2 elicits inducible cAMP early repressor (ICER), which inhibits RALDH expression and is expressed by RALDH− but not RALDH+ DCs in vivo
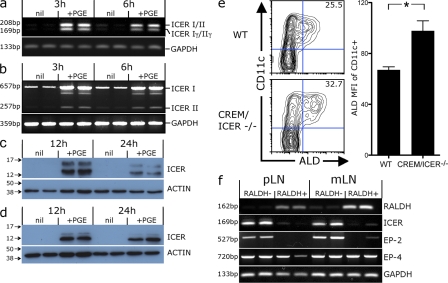
Our findings demonstrate that PGE2 inhibits RALDH transcription signaling via either EP-2 (in human mo-DCs) or EP-4 (for mouse BM-DCs). Both EP-2 and EP-4 are Gαs protein–coupled receptors that increase intracellular cAMP (Sugimoto et al., 2000). This suggests that the suppression of RALDH expression is likely to be downstream of an increase in intracellular cAMP. Consistent with this, the addition of the dibutyryl cAMP analogue (dB-cAMP) during mo-DC differentiation similarly suppressed the development of RALDH+ DCs in vitro (Fig. S5). How would increased cAMP inhibit RALDH expression? Elevated cAMP regulates gene expression via the phosphorylation of downstream nuclear transcription factors of the cAMP response element (CRE) binding protein (CREB)/CRE modulator (CREM)/activating transcription factor 1 (ATF-1) family (Borrelli et al., 1992; Mayr and Montminy, 2001). These proteins bind at conserved CRE binding sites (TGACGTCA), initiating the transcription of CRE-responsive genes (Borrelli et al., 1992; Mayr and Montminy, 2001). However, cAMP can also give rise to proteins that repress the transcription of CRE-responsive genes. Notably, cAMP can drive the expression of the ICER, a truncated CREM protein that lacks a trans-activation domain and thereby represses the transcription of genes whose promoters contain CRE-binding sites (Foulkes et al., 1991; Foulkes and Sassone-Corsi, 1992; Molina et al., 1993). Given that RALDH2 transcription is repressed by PGE2, and that the RALDH2 promoter contains multiple CRE-binding sites (Wang et al., 2001), we speculated that PGE2 might repress RALDH2 transcription through the induction of ICER. Consistent with this possibility, PGE2 induced ICER mRNA and protein expression in both CD11c+ mouse BM-DCs and human mo-DCs in vitro (Fig. 7, a–d). To test whether ICER regulates RALDH expression, we differentiated DCs with GM-CSF from BM of either WT or CREM/ICER-deficient mice and compared their RALDH activity. Consistent with ICER inhibiting RALDH transcription, CREM/ICER KO BM-DCs had significantly higher levels of RALDH activity compared with their WT counterparts (Fig. 7 e). Given that ICER inhibits DC RALDH expression in vitro, we next asked whether the subset of DCs that express RALDH in vivo do so in part by an ability to avoid ICER expression. To this end, we measured ICER expression directly ex vivo comparing RALDH+ and RALDH− DC subsets sorted from mLNs and skin-draining pLNs (Fig. 7 f). Intriguingly, we found a reciprocal expression between ICER and RALDH2, with little or no ICER mRNA being detected in RALDH+ DCs, whereas ICER was expressed by RALDH− DCs isolated from both mLN and skin-draining pLNs (Fig. 7 f). Furthermore, ICER expression coincided with EP-2, with both being expressed exclusively by RALDH− DCs. This is consistent with our observation for BM-DCs, again indicating that RALDH+ DCs do not express EP-2. These patterns of expression are consistent with the possibility that those DCs that express the RALDH enzymes in vivo do so, in part, by limiting ICER expression, possibly by virtue of reduced PGE–EP-2 signaling.
Figure 7.
PGE2 induces ICER expression, which limits RALDH activity in vitro and is selectively expressed by RALDH- DCs in vivo. (a–d) Mouse CD11c+ BM-DCs or human mo-DCs were treated with 1 µM PGE2 and analyzed for ICER expression by RT-PCR and Western blot analysis at the indicated times after treatment. A representative from at least two independent experiments is shown. (a) RT-PCR for ICER expression by mouse BM-DCs. Isoforms I & II run as a 208-bp product and isoforms Iγ/IIγ as a 169-bp product. (b) RT-PCR for ICER expression in human mo-DCs. Isoforms I & II run as 657- and 257-bp product, respectively. (c and d) Western blot for ICER protein expression by mouse BM-DCs (c) and human mo-DCs (d). Values indicate the position of protein ladder markers (kD). (e) BM cells from CREM/ICER null mutant mice or WT controls were cultured with GM-CSF. After 3 d, LPS was added and, 18 to 24 h later, cells were stained with CD11c and ALD to measure DC RALDH activity. A representative contour plot of CD11c and ALD expression of CREM/ICER KO and WT (WT) BM-DCs is shown. Inset values indicate the mean percentage of CD11c+ DCs that are ALD+. Bar graph shows the ALD MFI of CD11c+ DCs with SEM. Data were pooled from two experiments (*, P < 0.05). (f) CD11c+ cells were purified from the mLN or pLN of C57BL/6 mice, treated with ALD, and sorted into CD11c+RALDH+ and RALDH− subsets by flow cytometric sorting. The expression of RALDH2, ICER, and PGE2 receptors was analyzed by RT-PCR. Duplicates are with cDNA derived from different mice, and a representative of at least four independent experiments is shown (values indicate MW of PCR product).
DISCUSSION
The production of RA by DCs is critical for driving the development of gut-tropic immune responses (Iwata et al., 2004; Coombes et al., 2007; Jaensson et al., 2008). However, the factors that regulate RA synthesis by DCs remain poorly defined. Consistent with others (Yokota et al., 2009), we show that in reductionist in vitro settings, multiple factors, most notably GM-CSF, can induce RALDH expression in both mouse and human DCs. Indeed GM-CSF was able to induce RALDH activity even by RALDH-negative lymphoid DCs stimulated ex vivo. Despite this observation that multiple factors—many of which are widely available in vivo—can elicit RALDH expression in vitro, the capacity to produce RA is highly restricted to defined subsets of DCs in vivo. We therefore speculated that in vivo, negative regulators of RALDH might act to limit the systemic emergence of RA-producing DCs. Consistent with this possibility we discovered that PGE2 blocked RALDH expression induced by GM-CSF, repressing the transcription of the RALDH enzymes in both mouse and human DCs. Consistent with an inhibitory role, blocking PGE2 signaling via receptor antagonists or using BM precursors that were deficient in the PGE2 receptor subtype 4 (EP-4) gave rise to an enhanced frequency of RALDH+ DCs in vitro, whereas reducing PGE2 levels in vivo resulted in the emergence of RA-producing DCs in the spleen that were capable of priming CCR9+ gut-homing T cells at this site. These findings are consistent with an earlier observation in madine-darby canine kidney cells (Napoli, 1993) and suggest that PGE2 is a negative regulator of RALDH expression in DCs that effectively limits the emergence of RA-producing DCs.
PGE2 appears to inhibit RALDH expression at the transcriptional level. Of the four PGE2 receptors (EP-1–4), PGE2 signaled predominantly through EP-2 to inhibit RALDH transcription in human mo-DCs and EP-4 in mouse BM-DCs. Both EP-2 and EP-4 are Gαs protein–coupled receptors whose engagement results in adenyl cyclase release and an increase in intracellular cAMP (Sugimoto et al., 2000). Given this, in addition to the fact that the cAMP analogue similarly suppressed RALDH, it is likely that the inhibition of RALDH expression is downstream of cAMP. Elevated cAMP leads to the phosphorylation of nuclear transcription factors of the CREB/CREM/ATF-1 family (Mayr and Montminy, 2001). When phosphorylated, these can recruit cofactors that in complex bind at conserved CRE binding sites (TGACGTCA) and thereby initiate the transcription of CRE-responsive genes (Borrelli et al., 1992; Mayr and Montminy, 2001). However, CREB/CREM/ATF-1 will also drive the expression of the ICER that is transcribed from the intronic CRE-P2 promoter of CREM. ICER is a truncated protein that lacks the trans-activation domain of CREM but retains the DNA-binding motif and thereby acts as a repressor of CRE-responsive genes (Foulkes et al., 1991; Foulkes and Sassone-Corsi, 1992; Molina et al., 1993). Given that the RALDH2 promoter contains multiple CRE binding sites (Wang et al., 2001), the induction of ICER proteins has the potential to repress RALDH2 transcription. Consistent with this, our findings that BM-DCs from mice lacking ICER had significantly enhanced RALDH activity suggest that ICER actively limits RALDH expression during DC development. Given that PGE2 stimulates ICER expression in mouse and human DCs, we propose that PGE2 most likely represses the transcription of RALDH via the induction of ICER. Second, our finding that ICER was absent or expressed at substantially reduced levels by RALDH+ DCs in vivo suggests that RA-producing DCs express the RALDH enzyme, in part as a result of their unique ability to limit or avoid ICER expression. Collectively, these results provide a mechanistic insight into how PGE2 signaling would drive the transcriptional repression of RALDH.
Why those RA-producing DCs that emerge in vivo do not express ICER and how they escape PGE-mediated RALDH inhibition remains an outstanding question. One possibility is that these DCs may develop in microenvironments devoid of PGE2. However, PGE2 is present constitutively in the small intestine at levels similar to that found in the skin and spleen (Fig. S6). Furthermore, it is well established that DCs, including those found in the mLNs, produce PGE2 (Broere et al., 2009). For these reasons, we feel it likely that RALDH+ DCs are exposed to PGE2 in vivo. Alternatively, PGE2 signaling may fail to induce ICER expression in this DC subset, thus leaving the transcription of RALDH unhindered. A third possibility is that RALDH+ DCs are themselves refractory to PGE2. Although this issue warrants further studies, it is intriguing that mouse RALDH+ DCs that emerge both in vitro and in vivo do not express the EP-2 receptor, whereas their RALDH-negative counterparts do. Although EP-4 signaling does drive RALDH inhibition in mouse BM-DCs, in the case of human monocytes (which express both EP-2 and -4), PGE2–EP-2 signaling was clearly the dominant pathway for RALDH inhibition. Blocking signaling through EP-2, but not EP-4, abrogated PGE2-mediated RALDH inhibition, whereas EP-2 agonists were substantially more potent in comparison with their EP-4 counterparts on human mo-DCs. The enhanced activity of EP-2 may relate to PGE2–EP-2 signaling, providing a more sustained signal compared with EP-4, as the latter is internalized after stimulation, whereas EP-2 does not undergo agonist induced desensitization (Nishigaki et al., 1996; Desai et al., 2000). Given that EP-2 is the dominant receptor for signaling RALDH inhibition in human mo-DCs, coupled with the finding that RALDH+ DCs are devoid of EP-2, it is tempting to speculate that RALDH-expressing DCs arise from a unique precursor that does not express EP-2 and are thus refractory to PGE2. Although this is possible, it is important to note that mice lacking the EP-2 receptor did not show an increase in RALDH+ DCs in vivo. Thus, the absence of EP-2 does not alone explain the development of RALDH+ DCs, potentially as the result of a level of redundancy between EP-2 and -4 or additional inhibitory factors that also drive the phosphorylation of CREB proteins. Consequently, the exact mechanism that enables RA-producing DCs to escape PGE2-mediated RALDH inhibition remains unclear and is an area of ongoing research.
Early inflammatory molecules, such as IL-1 and TNF, induce the expression of COX-2 and thereby increase local PGE2 production (Alaaeddine et al., 1999; Dinarello, 2002). As such, DCs differentiating at the site of inflammation, including inflammatory monocytes which infiltrate inflamed tissues, are likely to be exposed to high concentrations of PGE2. With this in mind, it is interesting that inflammatory mo-DCs infiltrating the GALT during periods of colitis do not acquire RALDH activity (Siddiqui et al., 2010). Instead, these inflammatory mo-DCs produce high levels of IL-23, a cytokine which is enhanced by PGE2 (Khayrullina et al., 2008), and appear to augment Th-17 responses that in turn exacerbate intestinal pathology (Siddiqui et al., 2010). Our findings, which establish a link between PGE2 and RALDH expression, may help to explain why inflammatory mo-DCs fail to acquire the capacity to produce RA. Given that RA-producing DCs potently inhibit Th-17 differentiation and instead favor the differentiation of FOXP-3 regulatory T cells (Mucida et al., 2007), defining the processes which underpin the loss of RA-producing DCs during periods of chronic inflammation may have important implications in understanding the onset immunopathology.
Our results clearly demonstrate that PGE2 treatment modulates how DCs program homing receptor expression during T cell priming. Reducing PGE2 levels in vivo enhanced the ability of DCs to drive CCR9 expression upon T cell activation, whereas DCs differentiated in the presence of exogenous PGE2 lost the capacity to induce CCR9, instead showing an enhanced ability to elicit the expression of nonintestinal homing receptors, such as PSL, upon activated T cells. Given that PGE2 is commonly used in clinical trials to mature mo-DCs used for immunotherapy (Jonuleit et al., 1997; Schadendorf et al., 2006), our findings have immediate implications within this setting. PGE2 treatment would reduce RALDH activity and thereby enhance the capacity of DCs to activate T cells capable of migrating to nonintestinal sites, such as the skin, and thus would be highly appropriate when eliciting T cells response to cancers such melanoma. In contrast, blocking PGE2 signaling would enhance RALDH expression and, thus, the ability of DCs to prime CCR9+ gut-homing T cells that would be most effective against intestinal cancers. Second, our findings that indomethacin treatment promoted the systemic priming of CCR9+ T cells raises the possibility that the administration of COX inhibitors in combination with vaccines may be a means to elicit gut-tropic immune responses after parenteral inoculations. Given that most vaccines are delivered via subcutaneous or intramuscular injections, routes which do not typically elicit gut-homing T cells, this approach maybe beneficial when vaccinating against pathogens, such as HIV, that replicate within the intestinal tract. Thus, we suggest that manipulating DCs’ exposure to PGE2 may be a means to enhance the efficacy of T cell immunity via controlling the tissue specificity of such responses.
In summary, we show in this paper that although multiple factors can elicit the expression of RALDH during DC differentiation, PGE2 provides a dominant inhibitory signal that represses the transcription of RALDH. The development of RA-producing DCs may therefore reflect a balance between the ability to respond to positive stimuli, such as GM-CSF which drives RALDH expression, while ignoring or subverting the inhibitory stimuli such as PGE2 that elicit transcriptional repression. Given the central role that RA-producing DCs play in the induction of gut-specific immunity, we believe that modulating PGE2 signaling during DC differentiation may therefore be a powerful means to manipulate the tissue tropism of immune responses.
MATERIALS AND METHODS
Mice.
C57BL/6, F5, and B6.SJL-PtprcaPep3b/BoyJ (B6Ly5.1) mice were maintained at the Biological Services Unit at John Radcliffe Hospital (University of Oxford). F5 TCR transgenic mice express a TCR specific for the H2-Db-restricted A/NT/60/68 influenza nucleoprotein peptide NP68366–374-ASNENMDAM (Mamalaki et al., 1993). Mice with deletions in one of Ptger1 (EP-1), Ptger2 (EP-2), Ptger3 (EP-3), or Ptger4 (EP-4) were generated as previously described (Segi et al., 1998; Ushikubi et al., 1998; Hizaki et al., 1999). With the exception of EP-4−/−, these mice were backcrossed at least eight times onto the C57BL/6 background. Ptger4−/− mice do not survive on the C57BL/6 background as a result of patent ductus arteriosus (Segi et al., 1998) and were backcrossed on a mixed background of 129/Ola × C57BL/6. Mice were provided by S. Narumiya (Kyoto University, Kyoto, Japan), and breeding colonies were maintained by M. Belvisi at Imperial College London (London, England, UK). CREM/ICER-deficient mice were generated as previously described (Blendy et al., 1996; Conti et al., 2004). Mutant and WTs are F1 hybrids (129SVEV:C57BL/6) obtained by crossing heterozygous CREM/ICER +/− 129SVEV N12 × CREM/ICER+/− C57BL/6 N15. Mice were provided by J. Blendy (University of Pennsylvania School of Medicine, Philadelphia, PA) and colonies were maintained at the Department of Pharmacology (University of Pennsylvania School of Medicine). All mice were used under institutional guidelines with the authority of a UK Home Office project license.
Reagents and media.
Recombinant mouse G-CSF, M-CSF, TGF-β, IL-4, IL-6, IL-7, IL-10, IL-12, and IL-13 and human FLT3-L and GM-CSF were purchased from PeproTech. Mouse TSLP and the PGE2 ELISA kit were obtained from R&D Systems and CPG1826 from InvivoGen. Ciglitazone, all-trans RA, Escherichia coli LPS, trypsin-coated agarose beads, SC19220, and AH23848 were obtained from Sigma-Aldrich. PGD, PGF, PGI, thromboxane, and AH6809 were obtained from Cayman Chemical and PGE2 was from the John Radcliffe Hospital Pharmacy (Oxford, London, UK). The EP-specific agonists ONO-D1-004 (EP-1), ONO-AE1-259 (EP-2), ONO-AE-248 (EP-3), and ONO-AE1-329 (EP-4) were a gift from the ONO Pharmaceutical Company. Mouse GM-CSF and human IL-4 were produced in house. IL-12p40 ELISA was performed with capture (C15.6) and detection (C17.8) antibodies purchased from eBioscience.
Cell lines.
Primary skin stromal lines were generated from adult C57BL/6 mice by digesting full-thickness skin in 1 mg/ml of type II collagenase (Worthington Biochemical Corporation) and 10 µg/ml DNase (Sigma-Aldrich) and grown in complete DME (DME supplemented with 1 mM sodium pyruvate, 25 mM Hepes, MEM-NEA, 50 µM 2-mercaptoethanol, l-glutamine, and antibiotics) with 10% FCS (DME-10) until confluent adherent monolayers developed. 3T3 fibroblasts were maintained in DME-10 and the EL-4 thymoma in complete RPMI with 10% FCS (RP-10). To generate cell line SNs, cells were grown until 50–70% confluent and media was replaced with fresh RP-10. Cells were cultured for a further 2 d before 0.22 µM SN was harvested and filtered for use. In some instances, cells were grown for the final 2 d in the presence of 1 µM fumonisin B1 (Sigma-Aldrich), 1 µM NBDJ (Sigma-Aldrich), or 1 µM indomethacin (Sigma-Aldrich), or SN was subsequently heated (80°C for 20 min) and digested with 5–10 U/ml trypsin-agarose beads for 2 h at 37°C (Sigma-Aldrich). The following formulae was used to compare the relative inhibitory activity of treated versus untreated SN: [(without SNa – treated SNa)/(without SNa – untreated SNa)] × 100 = % relative inhibition by treated SN, where a = % ALD+ of CD11c+ cells.
Flow cytometry.
Antibodies for flow cytometry were as follows: anti-CD11c APC (N418), anti-TCR APC (H57-597), anti-CD3 eFluor 450 (17A2), anti-CD45.2 APC (104), anti-CD103 biotin (M290), anti–CD11b-APC (M1/70), anti-CD8α APC (53.67), anti-Ia/Ie FITC (M5/114), anti-CD86 PE (GL-1), rat-IgG2a PE isotype control, and anti–human Fc PE (eBioscience). Anti-CCR9 PE (242503) and the P-selectin/human Fc chimeric protein were obtained from R&D Systems. Anti–human CD86 APC (2331) was obtained from BD. ALD staining (STEMCELL Technologies) was used as per the manufacturer’s instructions. 10 µg/ml propidium iodide (Sigma-Aldrich) was added immediately before analysis on FACSCalibur or Cyan (BD) flow cytometers. Data were analyzed with FlowJo software (Tree Star).
Isolation of CD11c+ DCs from tissues.
CD11c+ DCs were isolated from tissues essentially as previously described (Vremec and Shortman, 1997). In brief, organs were digested in 1 mg/ml of type II collagenase (Worthington Biochemical Corporation) with 10 µg/ml DNase (Sigma-Aldrich) for 20 min at room temperature before the addition of 10 µM EDTA for a final 5 min. In some instances, DCs were subsequently enriched with anti-CD11c microbeads (Miltenyi Biotec) as per the manufacturer’s instructions.
Generation of mouse BM-DCs.
BM was isolated from the femur and tibia of mice and cultured in 6-well plates (106 cells/well). For FLT3-L DCs, cells were cultured with 100 ng/ml of human recombinant FLT3-L in RP-10 with GM-CSF, G-CSF, M-CSF, TSLP, TGF-β, IL-4, IL-6, IL-7, IL-10, IL-12, IL-13, or PGE2 at 20 ng/ml, or RA and 100 µM Ciglitizone. After 3 d, 0.25 ng/ml LPS was added and cells were harvested the next day. For GM-CSF–cultured BM-DCs, cells were cultured with 20 ng/ml GM-CSF in RP10 alone or with SN at 75% of culture volume (unless otherwise stated). After 3 d, 0.25 ng/ml LPS was added and cells were harvested the next day. For in vitro proliferation assays, DCs were subsequently enriched with anti-CD11c or anti–MHC class II microbeads (Miltenyi Biotec). Human mo-DCs were generated essentially as previously described (Sallusto and Lanzavecchia, 1994). In brief, CD14+ monocytes were purified from buffy coats with CD14+ microbeads (Miltenyi Biotec) and cultured in 6-well plates (106 cells/well) in RP-10 with 50 ng/ml of human recombinant GM-CSF (PeproTech) and IL-4 (5%). After 3 d, 1 ng/ml LPS was added and cells were harvested the next day. In some instances, 1 µM indomethacin was added at the beginning of culture to block the endogenous prostaglandin.
PCR.
RNA was extracted with RNA easy kit (QIAGEN) as per manufacturer’s instructions. cDNA was synthesized from 20 ng RNA with oligo-dT primers (Invitrogen) and Superscript Reverse transcription (Invitrogen). Primers and conditions are listed in Table S1. ICER sense primer corresponds to the 5′ untranslated region of ICER and the anti-sense to bp 496–516 of CREM (Tetradis et al., 1998).
For quantitative (q) PCR, GAPDH and RALDH2 mRNA levels were determined with primers/probe (Roche; Table S1) using TaqMan Gene Expression assays (Applied Biosystems) with TaqMan Gene Expression Master Mix (Applied Biosystems), according to the manufacturer’s instructions. Changes in gene expression relative to the endogenous control GAPDH were calculated using the formula 2-ΔCT. Fold changes in expression relative to control DCs were calculated according to the 2-ΔΔCT method.
In vitro T cell proliferation assays.
CD8+ T cells were enriched from splenocytes of F5 mice using a CD8+ negative isolation kit (Dynal; Invitrogen) and labeled with 1 µM CFSE for 10 min at 37°C. DCs were purified with anti-CD11c or MHC class II microbeads beads (Miltenyi Biotec) according to the manufacturer’s instructions, and pulsed with 0.1 µg/ml NP366–374-peptide for 45 min at 37°C. 25 × 103 DCs were plated into 96-well flat-bottom plates with 75 × 103 CFSE-labeled CD8+ F5 T cells. After 4 d, cells were stained with anti-TCR and anti-CCR9 or -PSL antibodies with appropriate secondary antibodies and analyzed by flow cytometry.
In vivo homing experiments.
F5 T cells activated in vitro by NP366–374 peptide-pulsed control or SN-conditioned DCs were labeled with 0.1 µM CFSE or 20 µM CTO, pooled at a 1:1 ratio, and ∼2 × 106 total donor cells were transferred i.v. into B6.Ly5.1 mice that had been sensitized with oxazolone and challenged on the ear as previously described (Johnson et al., 2006). After 16 h, recipient mice were sacrificed, perfused, and single cell suspension made from the spleen, ear, and intestine as previously described (Weigmann et al., 2007). Cells were stained with anti–mouse CD45.2 for analysis by flow cytometry.
Adoptive transfer experiments.
CD8+ T cells were enriched from splenocytes of F5 mice, labeled with 1 µM CFSE, and 5 × 105 cells were transferred into C57BL/6 recipients by i.v. tail vein injections. 1 d later, mice were injected i.v. with 50 µg NP366–374–peptide with 10 µg LPS. After 60 h, spleens were harvested, digested with collagenase, and stained with ALD and CD11c, as previously described, or stained with anti-CD8 and anti–CCR9 antibodies for analysis by flow cytometry.
Western blots.
Cell pellets were lysed in 1% Igepal, 0.5% sodium deoxycholate, and 0.1% SDS with inhibitors (1 mM phenylmethylsulfonyl fluoride, 50 mM sodium fluoride, 2 mM sodium orthovanadate, and complete protease inhibitor [Roche]). Lysate was separated on 15% polyacrylamide gel via SDS-PAGE, transferred to a PVDC membrane (Hybond; GE Healthcare), and blocked with PBS, 5% BSA, and 0.1% Tween-20. Anti–CREM-1 antibody (X-12; 1:500; Santa Cruz Biotechnology, Inc.) was added in blocking buffer overnight at 4°C and detected with horseradish peroxidase–conjugated secondary antibody before developing using Super Signal chemiluminescent reagent (Thermo Fisher Scientific).
Statistical analysis.
Data were analyzed using a two-tailed Student’s t test.
Online supplemental material.
Fig. S1 shows that both ALD-positive and -negative subsets stimulated robust peptide-specific T cell proliferation in vitro and that RALDH activity is common to differing DC subsets in vivo. Fig. S2 shows that CD11c+ DCs have equivalent expression of MHC class II and CD86 to control DCs and that skin SN-conditioned DCs produced equivalent or enhanced levels of IL-12 upon TLR stimulation. Fig. S3 shows that exogenous PGE2 alone, in the absence of skin SN, has reduced suppressive activity. Fig. S4 shows that EP-4 deficiency did not change the frequency or phenotype of DCs during BM-DC development and appeared to enhance DC RALDH expression selectively. In vivo, EP-4 KO mice showed a statistically significant increase in the frequency of RALDH+ DCs in the pLNs, but not in mLNs or the spleen. Fig. S5 shows that, when added during mo-DC differentiation, the cAMP analogue dibutyryl-cAMP suppressed the development of RALDH+ DCs in vivo. Fig. S6 shows that PGE2 is present constitutively in the small intestine at levels similar to those found in the skin and the spleen. Table S1 lists the primers and conditions used in PCR. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20101967/DC1.
Acknowledgments
We would like to thank ONO Pharmaceuticals Co. (Osaka Japan) for the provision of the EP-specific agonists ONO-D1-004, ONO-AE1-259, ONO-AE-248, and ONO-AE1-329. We are also very grateful to Mark Birrell and Maria Belvisi (Imperial College, London, UK) for generously providing EP-KO mice and Julie Blendy (University of Pennsylvania School of Medicine) for generously providing the CREM/ICER-null mice. We would also like to thank Ellen Pure and Irene Crichton (Wistar Institute, Pennsylvania) for their support, Craig Waugh for flow cytometric sorting, and Giorgio Napolitani and Carmen De Santo for critical reading and discussion.
A. Stock was supported by a CJ Martin Fellowship from the NHMRC (Australia). This work was supported by Cancer Research UK (C399/A2291) and the UK Medical Research Council.
The authors have no competing financial interests to declare.
Footnotes
Abbreviations used:
- ALD
- aldefluor
- ATF-1
- activating transcription factor 1
- BM-DC
- BM-derived DC
- COX
- cyclooxygenase
- CRE
- cAMP response element
- CREB
- CRE binding protein
- CREM
- CRE modulator
- GALT
- gut-associated-lymphoid tissue
- ICER
- inducible cAMP early repressor
- mLN
- mesenteric LN
- mo-DC
- monocyte-derived DC
- NBDJ
- N-butyldeoxynojirimycin
- PGE2
- prostaglandin E2
- pLN
- peripheral LN
- PSL
- P-selectin ligand
- RA
- retinoic acid
- RALDH
- retinal dehydrogenase
- SN
- supernatant
- TSLP
- thymic stromal lymphopoietin
References
- Alaaeddine N., Di Battista J.A., Pelletier J.P., Kiansa K., Cloutier J.M., Martel-Pelletier J. 1999. Inhibition of tumor necrosis factor alpha-induced prostaglandin E2 production by the antiinflammatory cytokines interleukin-4, interleukin-10, and interleukin-13 in osteoarthritic synovial fibroblasts: distinct targeting in the signaling pathways. Arthritis Rheum. 42:710–718 [DOI] [PubMed] [Google Scholar]
- Andersson U., Butters T.D., Dwek R.A., Platt F.M. 2000. N-butyldeoxygalactonojirimycin: a more selective inhibitor of glycosphingolipid biosynthesis than N-butyldeoxynojirimycin, in vitro and in vivo. Biochem. Pharmacol. 59:821–829 10.1016/S0006-2952(99)00384-6 [DOI] [PubMed] [Google Scholar]
- Annacker O., Coombes J.L., Malmstrom V., Uhlig H.H., Bourne T., Johansson-Lindbom B., Agace W.W., Parker C.M., Powrie F. 2005. Essential role for CD103 in the T cell–mediated regulation of experimental colitis. J. Exp. Med. 202:1051–1061 10.1084/jem.20040662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blendy J.A., Kaestner K.H., Weinbauer G.F., Nieschlag E., Schütz G. 1996. Severe impairment of spermatogenesis in mice lacking the CREM gene. Nature. 380:162–165 10.1038/380162a0 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrelli E., Montmayeur J.P., Foulkes N.S., Sassone-Corsi P. 1992. Signal transduction and gene control: the cAMP pathway. Crit. Rev. Oncog. 3:321–338 [PubMed] [Google Scholar]
- Broere F., du Pré M.F., van Berkel L.A., Garssen J., Schmidt-Weber C.B., Lambrecht B.N., Hendriks R.W., Nieuwenhuis E.E., Kraal G., Samsom J.N. 2009. Cyclooxygenase-2 in mucosal DC mediates induction of regulatory T cells in the intestine through suppression of IL-4. Mucosal Immunol. 2:254–264 10.1038/mi.2009.2 [DOI] [PubMed] [Google Scholar]
- Campbell D.J., Butcher E.C. 2002. Rapid acquisition of tissue-specific homing phenotypes by CD4+ T cells activated in cutaneous or mucosal lymphoid tissues. J. Exp. Med. 195:135–141 10.1084/jem.20011502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti A.C., Kuo Y.C., Valentino R.J., Blendy J.A. 2004. Inducible cAMP early repressor regulates corticosterone suppression after tricyclic antidepressant treatment. J. Neurosci. 24:1967–1975 10.1523/JNEUROSCI.4804-03.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coombes J.L., Siddiqui K.R., Arancibia-Cárcamo C.V., Hall J., Sun C.M., Belkaid Y., Powrie F. 2007. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-β and retinoic acid–dependent mechanism. J. Exp. Med. 204:1757–1764 10.1084/jem.20070590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai S., April H., Nwaneshiudu C., Ashby B. 2000. Comparison of agonist-induced internalization of the human EP2 and EP4 prostaglandin receptors: role of the carboxyl terminus in EP4 receptor sequestration. Mol. Pharmacol. 58:1279–1286 [DOI] [PubMed] [Google Scholar]
- Dinarello C.A. 2002. The IL-1 family and inflammatory diseases. Clin. Exp. Rheumatol. 20:S1–S13 [PubMed] [Google Scholar]
- Duester G. 2000. Families of retinoid dehydrogenases regulating vitamin A function: production of visual pigment and retinoic acid. Eur. J. Biochem. 267:4315–4324 10.1046/j.1432-1327.2000.01497.x [DOI] [PubMed] [Google Scholar]
- Edelson B.T., Kc W., Juang R., Kohyama M., Benoit L.A., Klekotka P.A., Moon C., Albring J.C., Ise W., Michael D.G., et al. 2010. Peripheral CD103+ dendritic cells form a unified subset developmentally related to CD8α+ conventional dendritic cells. J. Exp. Med. 207:823–836 10.1084/jem.20091627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes N.S., Sassone-Corsi P. 1992. More is better: activators and repressors from the same gene. Cell. 68:411–414 10.1016/0092-8674(92)90178-F [DOI] [PubMed] [Google Scholar]
- Foulkes N.S., Borrelli E., Sassone-Corsi P. 1991. CREM gene: use of alternative DNA-binding domains generates multiple antagonists of cAMP-induced transcription. Cell. 64:739–749 10.1016/0092-8674(91)90503-Q [DOI] [PubMed] [Google Scholar]
- Griscelli C., Vassalli P., McCluskey R.T. 1969. The distribution of large dividing lymph node cells in syngeneic recipient rats after intravenous injection. J. Exp. Med. 130:1427–1451 10.1084/jem.130.6.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M., Crozat K., Henri S., Tamoutounour S., Grenot P., Devilard E., de Bovis B., Alexopoulou L., Dalod M., Malissen B. 2010. Skin-draining lymph nodes contain dermis-derived CD103(-) dendritic cells that constitutively produce retinoic acid and induce Foxp3(+) regulatory T cells. Blood. 115:1958–1968 10.1182/blood-2009-09-245274 [DOI] [PubMed] [Google Scholar]
- Hall J.G., Hopkins J., Orlans E. 1977. Studies on the lymphocytes of sheep. III. Destination of lymph-borne immunoblasts in relation to their tissue of origin. Eur. J. Immunol. 7:30–37 10.1002/eji.1830070108 [DOI] [PubMed] [Google Scholar]
- He Q., Suzuki H., Sharma N., Sharma R.P. 2006. Ceramide synthase inhibition by fumonisin B1 treatment activates sphingolipid-metabolizing systems in mouse liver. Toxicol. Sci. 94:388–397 10.1093/toxsci/kfl102 [DOI] [PubMed] [Google Scholar]
- Hizaki H., Segi E., Sugimoto Y., Hirose M., Saji T., Ushikubi F., Matsuoka T., Noda Y., Tanaka T., Yoshida N., et al. 1999. Abortive expansion of the cumulus and impaired fertility in mice lacking the prostaglandin E receptor subtype EP(2). Proc. Natl. Acad. Sci. USA. 96:10501–10506 10.1073/pnas.96.18.10501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata M., Hirakiyama A., Eshima Y., Kagechika H., Kato C., Song S.Y. 2004. Retinoic acid imprints gut-homing specificity on T cells. Immunity. 21:527–538 10.1016/j.immuni.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Jaensson E., Uronen-Hansson H., Pabst O., Eksteen B., Tian J., Coombes J.L., Berg P.L., Davidsson T., Powrie F., Johansson-Lindbom B., Agace W.W. 2008. Small intestinal CD103+ dendritic cells display unique functional properties that are conserved between mice and humans. J. Exp. Med. 205:2139–2149 10.1084/jem.20080414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson-Lindbom B., Svensson M., Wurbel M.A., Malissen B., Márquez G., Agace W. 2003. Selective generation of gut tropic T cells in gut-associated lymphoid tissue (GALT): requirement for GALT dendritic cells and adjuvant. J. Exp. Med. 198:963–969 10.1084/jem.20031244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson-Lindbom B., Svensson M., Pabst O., Palmqvist C., Marquez G., Förster R., Agace W.W. 2005. Functional specialization of gut CD103+ dendritic cells in the regulation of tissue-selective T cell homing. J. Exp. Med. 202:1063–1073 10.1084/jem.20051100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L.A., Clasper S., Holt A.P., Lalor P.F., Baban D., Jackson D.G. 2006. An inflammation-induced mechanism for leukocyte transmigration across lymphatic vessel endothelium. J. Exp. Med. 203:2763–2777 10.1084/jem.20051759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones R.L., Giembycz M.A., Woodward D.F. 2009. Prostanoid receptor antagonists: development strategies and therapeutic applications. Br. J. Pharmacol. 158:104–145 10.1111/j.1476-5381.2009.00317.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonuleit H., Kühn U., Müller G., Steinbrink K., Paragnik L., Schmitt E., Knop J., Enk A.H. 1997. Pro-inflammatory cytokines and prostaglandins induce maturation of potent immunostimulatory dendritic cells under fetal calf serum-free conditions. Eur. J. Immunol. 27:3135–3142 10.1002/eji.1830271209 [DOI] [PubMed] [Google Scholar]
- Katler E., Weissmann G. 1977. Steroids, aspirin, and inflammation. Inflammation. 2:295–307 10.1007/BF00921009 [DOI] [PubMed] [Google Scholar]
- Khayrullina T., Yen J.H., Jing H., Ganea D. 2008. In vitro differentiation of dendritic cells in the presence of prostaglandin E2 alters the IL-12/IL-23 balance and promotes differentiation of Th17 cells. J. Immunol. 181:721–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King I.L., Kroenke M.A., Segal B.M. 2010. GM-CSF-dependent, CD103+ dermal dendritic cells play a critical role in Th effector cell differentiation after subcutaneous immunization. J. Exp. Med. 207:953–961 10.1084/jem.20091844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamalaki C., Elliott J., Norton T., Yannoutsos N., Townsend A.R., Chandler P., Simpson E., Kioussis D. 1993. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev. Immunol. 3:159–174 10.1155/1993/98015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy S., Ravindran R., Deng J., Oluoch H., Denning T.L., Kasturi S.P., Rosenthal K.M., Evavold B.D., Pulendran B. 2009. Toll-like receptor 2-dependent induction of vitamin A-metabolizing enzymes in dendritic cells promotes T regulatory responses and inhibits autoimmunity. Nat. Med. 15:401–409 10.1038/nm.1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr B., Montminy M. 2001. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2:599–609 10.1038/35085068 [DOI] [PubMed] [Google Scholar]
- Molina C.A., Foulkes N.S., Lalli E., Sassone-Corsi P. 1993. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 75:875–886 10.1016/0092-8674(93)90532-U [DOI] [PubMed] [Google Scholar]
- Mora J.R., Bono M.R., Manjunath N., Weninger W., Cavanagh L.L., Rosemblatt M., Von Andrian U.H. 2003. Selective imprinting of gut-homing T cells by Peyer’s patch dendritic cells. Nature. 424:88–93 10.1038/nature01726 [DOI] [PubMed] [Google Scholar]
- Mora J.R., Iwata M., Eksteen B., Song S.Y., Junt T., Senman B., Otipoby K.L., Yokota A., Takeuchi H., Ricciardi-Castagnoli P., et al. 2006. Generation of gut-homing IgA-secreting B cells by intestinal dendritic cells. Science. 314:1157–1160 10.1126/science.1132742 [DOI] [PubMed] [Google Scholar]
- Mucida D., Park Y., Kim G., Turovskaya O., Scott I., Kronenberg M., Cheroutre H. 2007. Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science. 317:256–260 10.1126/science.1145697 [DOI] [PubMed] [Google Scholar]
- Nakata K., Ashida K., Nakazawa K., Fujiwara M. 1981. Effects of indomethacin on prostaglandin synthesis and on contractile response of the guinea pig gallbladder. Pharmacology. 23:95–101 10.1159/000137535 [DOI] [PubMed] [Google Scholar]
- Napoli J.L. 1993. Prostaglandin E and 12-O-tetradecanoylphorbol-13-acetate are negative modulators of retinoic acid synthesis. Arch. Biochem. Biophys. 300:577–581 10.1006/abbi.1993.1080 [DOI] [PubMed] [Google Scholar]
- Napoli J.L. 1999. Interactions of retinoid binding proteins and enzymes in retinoid metabolism. Biochim. Biophys. Acta. 1440:139–162 [DOI] [PubMed] [Google Scholar]
- Nishigaki N., Negishi M., Ichikawa A. 1996. Two Gs-coupled prostaglandin E receptor subtypes, EP2 and EP4, differ in desensitization and sensitivity to the metabolic inactivation of the agonist. Mol. Pharmacol. 50:1031–1037 [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. 1994. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor α. J. Exp. Med. 179:1109–1118 10.1084/jem.179.4.1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadendorf D., Ugurel S., Schuler-Thurner B., Nestle F.O., Enk A., Bröcker E.B., Grabbe S., Rittgen W., Edler L., Sucker A., et al. ; DC study group of the DeCOG 2006. Dacarbazine (DTIC) versus vaccination with autologous peptide-pulsed dendritic cells (DC) in first-line treatment of patients with metastatic melanoma: a randomized phase III trial of the DC study group of the DeCOG. Ann. Oncol. 17:563–570 10.1093/annonc/mdj138 [DOI] [PubMed] [Google Scholar]
- Schulz O., Jaensson E., Persson E.K., Liu X., Worbs T., Agace W.W., Pabst O. 2009. Intestinal CD103+, but not CX3CR1+, antigen sampling cells migrate in lymph and serve classical dendritic cell functions. J. Exp. Med. 206:3101–3114 10.1084/jem.20091925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segi E., Sugimoto Y., Yamasaki A., Aze Y., Oida H., Nishimura T., Murata T., Matsuoka T., Ushikubi F., Hirose M., et al. 1998. Patent ductus arteriosus and neonatal death in prostaglandin receptor EP4-deficient mice. Biochem. Biophys. Res. Commun. 246:7–12 10.1006/bbrc.1998.8461 [DOI] [PubMed] [Google Scholar]
- Shikina T., Hiroi T., Iwatani K., Jang M.H., Fukuyama S., Tamura M., Kubo T., Ishikawa H., Kiyono H. 2004. IgA class switch occurs in the organized nasopharynx- and gut-associated lymphoid tissue, but not in the diffuse lamina propria of airways and gut. J. Immunol. 172:6259–6264 [DOI] [PubMed] [Google Scholar]
- Siddiqui K.R., Laffont S., Powrie F. 2010. E-cadherin marks a subset of inflammatory dendritic cells that promote T cell-mediated colitis. Immunity. 32:557–567 10.1016/j.immuni.2010.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y., Narumiya S. 2007. Prostaglandin E receptors. J. Biol. Chem. 282:11613–11617 10.1074/jbc.R600038200 [DOI] [PubMed] [Google Scholar]
- Sugimoto Y., Narumiya S., Ichikawa A. 2000. Distribution and function of prostanoid receptors: studies from knockout mice. Prog. Lipid Res. 39:289–314 10.1016/S0163-7827(00)00008-4 [DOI] [PubMed] [Google Scholar]
- Tetradis S., Nervina J.M., Nemoto K., Kream B.E. 1998. Parathyroid hormone induces expression of the inducible cAMP early repressor in osteoblastic MC3T3-E1 cells and mouse calvariae. J. Bone Miner. Res. 13:1846–1851 10.1359/jbmr.1998.13.12.1846 [DOI] [PubMed] [Google Scholar]
- Ushikubi F., Segi E., Sugimoto Y., Murata T., Matsuoka T., Kobayashi T., Hizaki H., Tuboi K., Katsuyama M., Ichikawa A., et al. 1998. Impaired febrile response in mice lacking the prostaglandin E receptor subtype EP3. Nature. 395:281–284 10.1038/26233 [DOI] [PubMed] [Google Scholar]
- Varol C., Vallon-Eberhard A., Elinav E., Aychek T., Shapira Y., Luche H., Fehling H.J., Hardt W.D., Shakhar G., Jung S. 2009. Intestinal lamina propria dendritic cell subsets have different origin and functions. Immunity. 31:502–512 10.1016/j.immuni.2009.06.025 [DOI] [PubMed] [Google Scholar]
- Vremec D., Shortman K. 1997. Dendritic cell subtypes in mouse lymphoid organs: cross-correlation of surface markers, changes with incubation, and differences among thymus, spleen, and lymph nodes. J. Immunol. 159:565–573 [PubMed] [Google Scholar]
- Wang X., Sperkova Z., Napoli J.L. 2001. Analysis of mouse retinal dehydrogenase type 2 promoter and expression. Genomics. 74:245–250 10.1006/geno.2001.6546 [DOI] [PubMed] [Google Scholar]
- Weigmann B., Tubbe I., Seidel D., Nicolaev A., Becker C., Neurath M.F. 2007. Isolation and subsequent analysis of murine lamina propria mononuclear cells from colonic tissue. Nat. Protoc. 2:2307–2311 10.1038/nprot.2007.315 [DOI] [PubMed] [Google Scholar]
- Yokota A., Takeuchi H., Maeda N., Ohoka Y., Kato C., Song S.Y., Iwata M. 2009. GM-CSF and IL-4 synergistically trigger dendritic cells to acquire retinoic acid-producing capacity. Int. Immunol. 21:361–377 10.1093/intimm/dxp003 [DOI] [PMC free article] [PubMed] [Google Scholar]