Compared with T-ALL diagnosis samples, samples obtained at relapse or after xenograft into immunodeficient mice exhibit additional genomic lesions in oncogenes and/or tumor suppressor genes; these lesions contribute to leukemia-initiating activity.
Abstract
Genomic studies in human acute lymphoblastic leukemia (ALL) have revealed clonal heterogeneity at diagnosis and clonal evolution at relapse. In this study, we used genome-wide profiling to compare human T cell ALL samples at the time of diagnosis and after engraftment (xenograft) into immunodeficient recipient mice. Compared with paired diagnosis samples, the xenograft leukemia often contained additional genomic lesions in established human oncogenes and/or tumor suppressor genes. Mimicking such genomic lesions by short hairpin RNA–mediated knockdown in diagnosis samples conferred a selective advantage in competitive engraftment experiments, demonstrating that additional lesions can be drivers of increased leukemia-initiating activity. In addition, the xenograft leukemias appeared to arise from minor subclones existing in the patient at diagnosis. Comparison of paired diagnosis and relapse samples showed that, with regard to genetic lesions, xenograft leukemias more frequently more closely resembled relapse samples than bulk diagnosis samples. Moreover, a cell cycle– and mitosis-associated gene expression signature was present in xenograft and relapse samples, and xenograft leukemia exhibited diminished sensitivity to drugs. Thus, the establishment of human leukemia in immunodeficient mice selects and expands a more aggressive malignancy, recapitulating the process of relapse in patients. These findings may contribute to the design of novel strategies to prevent or treat relapse.
T cell acute lymphoblastic leukemias (ALLs [T-ALLs]) are aggressive malignancies derived from the transformation of lymphoblastic precursors in the thymus. Despite major therapeutic improvements, a fraction of patients with T-ALL still relapse and experience eventual refractory leukemia (Bailey et al., 2008; Pui et al., 2008). T-ALLs are characterized by the deregulated expression of oncogenic transcription factors combined with the frequent inactivation of the tumor suppressor CDKN2A/P16/ARF and activation of the NOTCH1 pathway (Ferrando et al., 2002; Weng et al., 2004; De Keersmaecker et al., 2005; Soulier et al., 2005; Aifantis et al., 2008). Recently, high-resolution cytogenetic tools have identified frequent additional cryptic deletions and duplications in T-ALL cells (Clappier et al., 2007; Van Vlierberghe et al., 2008; Gutierrez et al., 2009; Tosello et al., 2009; Kleppe et al., 2010). We used our established xenotransplantation assay of human T-ALL in immunodeficient mice (nonobese diabetic [NOD]/scid or NOD/scid/IL-2Rγ null; Armstrong et al., 2009) to test the hypothesis that xenografted leukemia cells could experience clonal evolution.
RESULTS AND DISCUSSION
Establishment of xenograft leukemias frequently selects minor genetic subclones, reminiscent of relapse in patients
We compared by high-density DNA arrays the genome of a series of 18 primary T-ALL cases at the time of diagnosis (diagnosis leukemia) and after primary engraftment in immunodeficient mice (xenograft leukemia; Fig. 1 A). A very good overall stability of the T-ALL genome was observed in all cases, with a pattern of copy number alterations being largely consistent between diagnosis and xenograft leukemia samples (Fig. S1 and Table S2). However, some changes were also observed (Fig. 1 and Table S2). Strikingly, these new genomic abnormalities in xenograft cells mainly targeted genes known as human oncogenes or tumor suppressor genes like PTEN, MYC, MYB, CDKN2A, DLEU7, or WT1. Sequencing T-ALL cancer genes NOTCH1, FBXW7, and PTEN also showed changes in the mutation pattern of xenograft versus diagnosis cells (Fig. 1 D and Table S3). Altogether, genomic changes were detected in 12 out of 18 T-ALL pairs (67%). In some cases, the stepwise sequence of genomic events could be reconstructed and demonstrated that xenograft cells had derived not from the diagnosis cells but from a common prediagnosis ancestor cell (Fig. S2, A and B). To investigate whether the additional genomic abnormalities found in xenograft leukemia cells occurred de novo in the mouse or had preexisted in the patient, we performed highly sensitive lesion-specific genomic PCR backtracking to test for their presence in rare cells at diagnosis. In the four analyzed cases, the genomic lesions were indeed detected at low levels in the diagnosis sample, demonstrating that the engrafted leukemia in the mouse had arisen from a minor genetic subclone present in the patient’s leukemia (Fig. 1 F and Fig. S2 C). These results, along with the fact that the additional lesions target cancer genes, suggest that clonal selection related to the activation of prototypical human oncogenic pathways occurs during leukemia reinitiation and expansion in immunodeficient mice.
Figure 1.
Genomic changes associated with clonal selection in xenografted leukemias. (A) Strategy of sample analysis. (B) Array CGH plots showing focal deletions in a T-ALL case, either in xenografted cells only (WT1 locus at 11p13) or present in both diagnosis and xenografted cells (11q14.2 locus). (C) FISH analysis of TCR-β (red probe) and MYC (green probe) loci in diagnosis and xenografted leukemia cells. Bar, 12 µm. (D) DNA sequence chromatograms showing NOTCH1 and PTEN mutations in xenograft samples. Arrows indicate the mutated nucleotides. (E) List of cancer-associated genes targeted by genomic changes in xenograft leukemias. (F) Representative lesion-specific genomic PCR backtracking in a T-ALL case: focal PTEN deletion as detected by array CGH in the xenograft but not the diagnosis bulk leukemia cells, sequence of the deletion breakpoints junction, and amplification curves from specific quantitative PCR experiments of the undiluted diagnosis sample (indicated by an arrow) compared with 10-fold dilutions of the xenograft sample from 10% to 0.01%. The horizontal green line indicates the threshold for PCR linearity. The horizontal purple line is the cursor of the Genomic Workbench software, centralized on the deletion. Each quantitative PCR point was performed in duplicate. (G) Array CGH plots in a T-ALL triad case showing an 11q deletion in both the xenograft and relapse but not in the diagnosis samples. XLE, unique identification numbers for T-ALL cases.
Intraclonal heterogeneity at diagnosis and clonal evolution at relapse have been reported in human ALL (Zuna et al., 2004; Choi et al., 2007; Mullighan et al., 2008; Yang et al., 2008; Tosello et al., 2009; Anderson et al., 2011). Genomic analysis of relapsing ALLs has suggested that the relapse frequently does not simply result from mutations of specific drug-resistance genes but rather from additional oncogenic abnormalities that contribute to the emergence of a leukemic subclone (Mullighan et al., 2008). Reminiscent of this process, our results suggest that development of human leukemia in the immunodeficient mouse does not simply result from specific adaptation to the mouse environment but rather from the oncogenic selection and expansion of a preexisting leukemic subclone. Therefore, we hypothesized that leukemia reinitiation in the mouse could mirror the leukemia relapse process in patients. We investigated a series of T-ALL patients who had further relapsed; mice were injected with the patient diagnosis sample, and the resulting xenograft, diagnosis, and relapse DNA triads were compared (n = 8 triads). In all triads but one, relapse and xenograft samples harbored a range of new genomic lesions compared with diagnosis cells (Figs. 1 G and 2 and Tables S1 and S3). Oncogenetic trees showed that the xenograft and the relapse leukemia cells had derived not from the bulk diagnosis cells but from a common prediagnosis ancestor cell in seven of eight T-ALL triads, as did the diagnosis cells, which is consistent with a branched rather than linear clonal evolution (Fig. 2). In five out of eight cases (XLE_61, XLE_92, XLE_95, XLE_96, and XLE_98), the xenograft and relapse clones were located in the same branch, whereas in two cases, the xenograft clone was closer to the bulk diagnosis clone (XLE_93 and XLE_97), and in one case the three clones were equally distinct (XLE_94). It is noteworthy that distinct genetic lesions targeting the same gene could be found in several samples from the same patient (like CDKN2A/P16/ARF deletions, MYB duplications, and NOTCH1 mutations), demonstrating that they had independently arisen more than once (Fig. 2). Altogether, these results are consistent with a neo-Darwinian genetic diversification of leukemia, in accordance with the model of Anderson et al. (2011), that is especially uncovered by relapse or xenograft.
Figure 2.
Oncogenetic trees for the diagnosis, xenograft, and relapse T-ALL triads. The stepwise sequence of genomic events could be reconstructed from the DNA copy number and sequence data in the samples. XLE, unique identification numbers for the T-ALL cases; D, diagnosis; X, xenograft; R, relapse samples; R*, in the case of the triad XLE_61, limited genomic DNA at relapse allowed only to backtrack the PTEN lesions seen in the xenograft. Superscript letters indicate genomic alterations that involve different breakpoints or sequence mutation of the same locus or gene in the diagnosis, xenograft, and/or relapse samples from the same T-ALL case.
Additional genomic lesions of xenograft leukemias can be drivers of increased leukemia-initiating activity
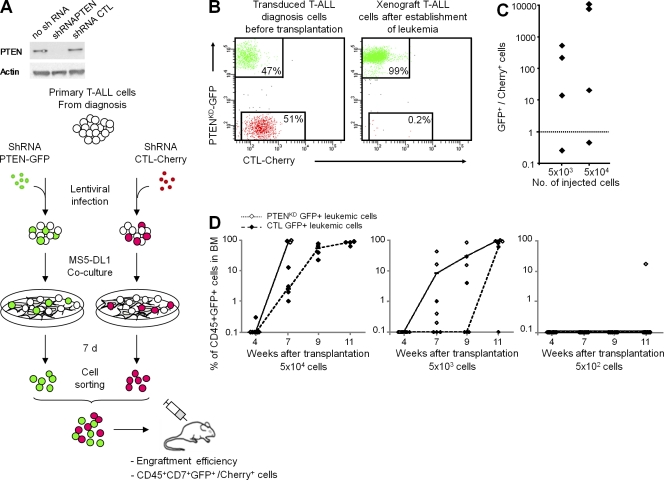
At this point, we reasoned that if some additional genomic abnormalities observed in the engrafted subclones were responsible for their preferential engraftment, this advantage should be conserved when primary xenograft cells are reinjected into secondary recipients. Indeed, xenografted cells reestablished leukemia more rapidly than diagnosis cells in secondary versus primary xenografts experiments (Fig. S3). Although increased proliferation could be involved in such accelerated secondary engraftment, clonal selection may also have enriched the xenograft leukemia in leukemic stem cells, as suggested by limiting-dilution analyses (Fig. S3 C). Interestingly, Boiko et al. (2010) have suggested that the more cancers are advanced, the more efficiently they engraft, which is consistent with a progressive enrichment in cancer stem cells and the loss of hierarchical structure as tumors progress.
We then sought to model the selective advantage and gain of malignancy likely conferred by the acquisition of an additional genomic lesion in leukemia cells. The tumor suppressor gene PTEN was a good candidate here as (i) it is involved in both proliferation and leukemia stem cell properties (Yilmaz et al., 2006), (ii) it has been associated with cancer progression and relapse including in T-ALL (Palomero et al., 2007; Gutierrez et al., 2009), and (iii) we found it newly mutated or deleted in xenograft leukemia cells from several patients (Fig. 1 F and Tables S2 and S3). Using lentiviral transduction, we genetically modified the diagnosis primary leukemia cells (Gerby et al., 2010) of a patient who had further relapsed and harbored a PTEN deletion in both his relapse and xenografted cells but not in the bulk diagnosis cells. Competitive engraftment experiments clearly demonstrated the selective advantage of PTEN knockdown leukemia cells over control diagnosis leukemia cells (Fig. 3, A–C). Moreover, kinetics and limiting-dilution analyses in noncompetitive engraftment experiments showed an increased leukemia initiating activity caused by PTEN knockdown in primary leukemia cells (Fig. 3 D). These results demonstrate that the additional genomic lesions in xenograft leukemia can be drivers of the increased malignancy and related clonal selection of the human leukemia cells in mice. Importantly, this original experimental strategy faithfully reproduces the process of tumor progression from diagnosis toward relapse.
Figure 3.
PTEN knockdown confers a selective advantage to diagnosis leukemia cells. (A) Schematic of the protocol for competitive engraftment experiments shown in B and C. Case XLE_61 T-ALL diagnosis cells transduced with control shRNA (shRNA CTL and CTL-Cherry+ cells) and shRNA PTEN (PTENKD-GFP+ cells) lentivectors were sorted and co-transplanted in equal number into NSG mice. Two cell doses (5 × 104 and 5 × 103 cells per mouse) were tested; three to five mice were injected per cell dose. Immunoblots show PTEN silencing efficiency. (B) Flow cytometry analyses showing the proportion of PTENKD-GFP+ and CTL-Cherry+ cells in leukemia cells immediately before transplantation (left) and in BM cells of a representative mouse after establishment of leukemia (right). (C) PTENKD-GFP+ versus CTL-Cherry+ CD45+ cells ratios are indicated, and each dot represents data of one mouse. The dotted line indicates ratio 1. (D) Kinetics of engraftment of cells transduced with shRNA CTL (CTL-GFP+ cells) and shRNA PTEN (PTENKD-GFP+ cells) lentivectors, injected in parallel (noncompetitive) experiments with decreasing cell doses, as measured by the percentage of CD45+GFP+ cells in BM cells. Each dot represents data of one mouse, dashes represent medians, and lines connect the medians.
Xenograft samples strongly express cell cycle and mitosis genes and display in vitro diminished sensitivity to drugs, reminiscent of leukemias at relapse
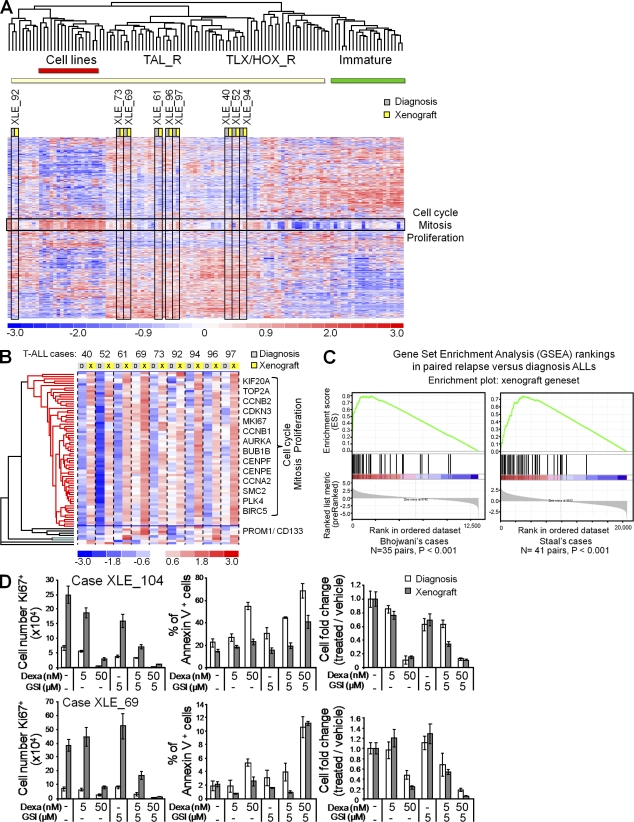
To gain insight into the functional changes of the xenograft cells relative to diagnosis cells, we profiled by large-scale expression analysis 9 paired xenograft and diagnosis T-ALL samples along with 93 other human T-ALL samples. First, unsupervised analysis showed that the diagnosis and xenograft samples from the same patient clustered next to one another for all pairs, demonstrating the excellent overall relevance of the xenograft model (Fig. 4 A). We then performed a paired supervised analysis that sorted out a list of up-regulated genes that individually differentiated the xenografts from the corresponding diagnosis leukemias (Fig. 4 B; the list of genes is in the supplemental material). Strikingly, this list was dramatically enriched in a cluster of genes clearly linked to cell cycle and mitosis (see Gene Ontology annotations in Table S1), suggesting that an increased proliferation contributes to the selective advantage of the engrafted cells. The xenograft signature also included a few other unrelated genes, including CD133 (PROMD1/Prominin-1), which is a marker of hematopoietic stem cells and of cancer stem cells (Singh et al., 2004). Interestingly, the xenograft signature was strongly expressed in all of the nine xenograft samples relative to the paired diagnosis samples, including in three cases without identified additional genomic changes in the xenograft sample (cases XLE_40, XLE_52, and XLE_69; Fig. 4 B). Cryptic genomic lesions or epigenetic mechanisms could be involved in these cases, also converging on cell cycle and proliferation.
Figure 4.
Increased proliferation and overall diminished drug sensitivity in xenografted leukemias. (A) Paired diagnosis and xenograft leukemias were analyzed by unsupervised large-scale gene expression profiling. 9 pairs of diagnosis and xenograft T-ALL samples along with a series of 73 primary T-ALL cases and 20 T cell lines were analyzed; the 9 pairs are boxed vertically in black. TAL_R, TLX/HOX_R, and immature T-ALL refer to the classification of T-ALL subtypes based on gene expression and genomic annotations as reported for another T-ALL series (Soulier et al., 2005). The cell cycle and mitosis cluster is boxed horizontally in black. (B) Gene clustering of the 59 unique genes of the xenograft signature, as defined by reference to the paired primary samples (“overexpressed in T-ALL xenografts gene set”). Representative genes are shown on the right of the heat map. (C) Gene set enrichment analysis (GSEA) of the xenograft gene set in the relapse versus diagnosis samples of two independent ALL series (Bhojwani et al., 2006; Staal et al., 2010). (D) Proliferation, apoptosis, and dose-dependent inhibition of paired diagnosis and xenograft leukemia samples treated with dexamethasone (dexa), γ-secretase inhibitor (GSI), or both, compared with vehicle only (DMSO; -). Proliferation was assessed as the number of KI67+ cells, apoptosis as the percentage of annexin V+ cells, and dose-dependent inhibition as the cell number fold change (treated/vehicle). Error bars represent means ± SD of triplicate experiments.
Several studies have found cell cycle, mitosis, and proliferation signatures in relapsing ALL patients (Bhojwani et al., 2006; Kirschner-Schwabe et al., 2006; Staal et al., 2010). We analyzed the relevance of the xenograft signature in relapse samples from patients. We performed gene set enrichment analyses using the expression data from two independent series of paired diagnosis and relapse ALLs (Bhojwani et al., 2006; Staal et al., 2010). Strikingly, the xenograft signature was significantly enriched in the relapse cells relative to the diagnosis cells in both series (Fig. 4 C). These data indicate that xenograft and relapse leukemias share a unique profile of increased proliferation and highlight the relevance of the xenograft leukemia system to model patient relapse.
Although the majority of ALL patients at diagnosis respond well to treatment and reach complete remission, patients who relapse frequently experience drug resistance and eventual refractory leukemia (Klumper et al., 1995; Choi et al., 2007; Bailey et al., 2008). Additional oncogenic abnormalities, like PTEN inactivation, could be involved in acquired resistance to treatment (Palomero et al., 2007). Because we found evidence of clonal selection and gain of malignancy in xenografted leukemia cells, we assessed whether this oncogenic progression was also associated with an acquired diminished sensitivity to drugs. We therefore assayed the sensitivity of xenografted leukemia samples to glucocorticoids, which are essential components of ALL treatment, and to γ-secretase inhibitors, which synergistically target T-ALL cells as the result of oncogenic activation of the NOTCH1 pathway (Weng et al., 2004; Real et al., 2009). In vitro cultures of leukemia samples on the Notch ligand–expressing stromal cells MS5-DL1 (Armstrong et al., 2009) first confirmed that xenografted cells were much more proliferative than diagnosis cells (Fig. 4 D). Moreover, although xenograft cells were still inhibited in a dose-dependent manner, high doses of glucocorticoids were necessary to efficiently reduce the cell number, and the addition of γ-secretase inhibitors was required to increase apoptosis (Fig. 4 D). These data suggest that increased proliferation and inhibition of apoptosis result in an overall lesser sensitivity of xenograft samples to treatment, as observed in patients at relapse. This is strengthened by our finding that xenograft cells, like relapsing cells, often have additional genomic lesions targeting cancer genes that are related to cell cycle and proliferation rather than specific drug-resistance genes. Such mechanisms could contribute to the poor response to treatment of relapsing leukemia. It is noteworthy that additional mutations in PTEN, MYC, or NOTCH1, as found in this study in xenograft leukemias, may also modulate the NOTCH pathway and thus the sensitivity of the leukemia cells to γ-secretase inhibitors (Weng et al., 2004; Palomero et al., 2006, 2007).
Despite major therapeutic improvement in ALL, relapsing patients still have a dismal prognosis, highlighting the need for more efficient strategies to treat these aggressive tumors. Our data indicate that the development of human T-ALL in immunodeficient mice favors the emergence of leukemic subclones with enhanced leukemia-initiating activity, increased proliferation, and diminished overall sensitivity to drugs and therefore recapitulates key biological features of the posttherapeutic tumor progression process in patients (Fig. S4). Importantly, the additional genetic lesions found in xenografted cells target human cancer–associated genes like in relapsing cells and they can be drivers of tumor progression. Future antileukemic strategies will have to consider the subclonal origin of leukemia relapse. In particular, it would be critical to identify and eradicate minor leukemia subclones during the initial phase of treatment (Bailey et al., 2008; Freyer et al., 2011). The biological mechanisms underlying relapse will have to be extensively analyzed at the genomic (including whole genome sequencing), epigenetic, and functional levels. Interestingly, our gene expression profiling analysis suggests that despite the complexity of the genomic lesions, the pathways of tumor progression in xenograft and in patients at relapse share unique features. The xenograft leukemia model is thus relevant and should be helpful to design new strategies to prevent or treat relapse in patients (Fig. S4).
Cancer cells are functionally heterogeneous, and in several cancers, a cellular hierarchy has been described with cancer stem cells at the apex (Dick, 2008). Xenotransplantation in immunodeficient mice is currently the key assay to evaluate human cancer stem cells activity. By demonstrating that genetic subclones are frequently selected in xenograft of T-ALL cells and that genetic lesions in these subclones can modulate leukemia-initiating activity and drive a gain of malignancy, our results add a new level of complexity that should be considered when characterizing human ALL leukemic stem cells (Dick, 2008; Shackleton et al., 2009; Anderson et al., 2011).
MATERIALS AND METHODS
T-ALL patients.
T-ALL samples were obtained from patients who were treated at Hôpital Saint-Louis or Hôpital Trousseau (Paris, France) according to the national protocols. Informed consent was obtained from the patients or their relatives. The Institutional Review Board of the Institut Universitaire d’Hématologie, Université Paris Diderot approved the study. BM or peripheral blood samples from 18 T-ALL patients were used for the first set of xenograft experiments. For a second set of experiments, seven more cases with available cryopreserved paired samples both at diagnosis and at relapse were studied. There were 22 children and 3 adults. For the large-scale gene expression analysis, 9 pairs of T-ALL diagnosis and xenograft samples were examined in addition to 73 cryopreserved T-ALL samples of patients from Hôpital Saint-Louis, 20 human T-ALL cell lines, and 1 normal BM sample. Only samples with at least 80% leukemia cells were used. The T-ALL cell lines were obtained from DSMZ.
Mice.
Immunodeficient NOD.CB17-Prkdc(scid) (abbreviated NOD/scid [NS]) and NOD.Cg-Prkdc(scid)Il2rg(tm1Wjll)/SzJ (abbreviated NOD/scid/IL-2Rγ null [NSG]) were both obtained from the Jackson Laboratory. Mice were housed in pathogen-free animal facilities at the Institut de Radiobiologie Cellulaire et Moléculaire (Commissariat à l’Energie Atomique, Fontenay-aux-Roses, France). All experimental procedures were performed in compliance with the French Ministry of Agriculture regulations (animal facility registration number A920322) for animal experimentation.
Xenotransplantation.
Mice (6–8 wk old) were sublethally irradiated at 3 Gy (IBL 637 CisBio International; dose rate 0.61 Gy/min) and anesthetized with isoflurane before injection. Human leukemia cells were transplanted into the retro-orbital sinus. Development of human T-ALL in mice was followed up using intrafemoral BM sampling. Conjugated mouse anti–human-specific monoclonal antibodies PE-CD45 and PE.Cy7-CD7 (Beckman Coulter) and FACS analysis (FACSCalibur; BD) were used to identify human leukemia cells. Mice were euthanized when they reached endpoints set to meet accepted animal care guidelines. For the initial series of xenotransplantation, 5 × 104 to 5 × 106 leukemia cells were transplanted into either NS or NSG mice; for all further experiments, NSG mice were used.
Sample preparation and DNA and RNA extraction.
White blood cells from patient samples were isolated by Ficoll centrifugation and cryopreserved using standard procedures. After thawing, cells were spun down, and DNA was extracted with the DNA blood mini kit (QIAGEN). RNA extraction for large-scale analysis was performed with the miRNeasy (QIAGEN). Nucleic acid quantity and quality was assessed using a spectrophotometer (ND-1000; NanoDrop).
Genome-wide DNA array analysis.
Genomic DNAs from paired diagnosis and xenograft samples (n = 25 patients) and from corresponding relapse leukemia samples (in 7 of these patients) were analyzed by high-density array comparative genomic hybridization (CGH) technologies using the 1 × 1M Microarray SurePrint G3 Catalog (Agilent Technologies) or by single nucleotide polymorphism (SNP) array technologies using the Genome-Wide Human SNP Array 6.0 (Affymetrix) according to the manufacturers’ recommendations (total n = 42 and n = 14 DNA samples, respectively; each sample originating from the same patient was analyzed by the same technology). All Microarray SNP/CGH array data files are available from ArrayExpress under accession no. E-MTAB-593. Analyses were performed using the Genomic Workbench software (Agilent Technologies) with the help of the ADM-2 algorithm for array CGH data, Genomic Suite 6.5 software (Partek), and the Hidden Markov Model and Segmentation algorithms for SNP-based copy number and genotyping analyses. The final retained abnormalities were validated by two investigators (E. Clappier and J. Soulier) by visual analysis considering the size and Log2 ratios of the abnormalities with respect to the individual background noise of each array at each particular chromosomal location, as reported previously (Clappier et al., 2007; Kleppe et al., 2010). Final validated data are provided in Tables S2 and S3. The University of California, Santa Cruz Genome Browser was used for locating genes on the human genome. Polymorphic copy number variations were excluded using the Database of the Genomic Variants tracks in the University of California, Santa Cruz Genome Browser or in the Genomic Workbench software.
Fluorescent in situ hybridization (FISH) analysis.
The unbalanced translocation TCRB-MYC suspected on the array CGH profile in the xenograft sample of case XLE_22 was confirmed by FISH analysis using the bacterial artificial chromosome probes RP11-460A11 (green; at the centromeric part of the MYC locus on 8q24) and RP11-78G15 (red; at the telomeric part of the TCRB locus on 7q34) by demonstrating a fusion in the xenograft but not diagnosis nuclei. Probes were obtained from AmpliTech.
NOTCH1, FBXW7, PTEN, and WT1 mutations screening.
Mutational screening of the NOTCH1, FBXW7, and WT1 genes was performed. NOTCH1 mutations were searched for in the N- and C-terminal regions of the heterodimerization domain (exons 26 and 27, respectively), the transcriptional activation domain, and the PEST domains (distal part of exon 34); extracellular juxtamembrane expansion mutations (exon 28) were also investigated. For FBXW7, mutations in the WD40 domain were examined by sequencing exons 9 and 10. PTEN mutations were assessed in exon 7, and WT1 mutations were assessed in exon 7.
Quantitative genomic PCR for specific backtracking of abnormalities.
Lesion-specific quantitative genomic PCR assays were designed based on the breakpoint or mutation sequences of four genomic abnormalities identified in xenografted T-ALL cells. Genomic deletion breakpoints were mapped by tiling primers outward from the genomic locations of CGH array probes defining the minimal regions of deletion at ∼500-bp intervals to beyond the next nondeleted probes telomeric and centromeric to the deletion. A combination of forward and reverse PCR primers was used to amplify the region of genomic deletion, and the genomic deletion breakpoints were determined by direct sequencing of PCR products. A specific real-time PCR assay using TaqMan probes or SYBR Green reagents (Applied Biosystems) was designed for each lesion analyzed. Quantification of the genomic lesion in diagnosis sample cells was performed using 10-fold dilutions of the xenograft DNA harboring the lesion. Experiments and interpretation of results were performed according to guidelines for minimal residual disease assessment.
Large-scale gene expression analyses.
9 pairs of xenograft and diagnosis T-ALL samples along with a series of 93 T-ALL samples (73 primary T-ALL and 20 T cell lines) were profiled using large-scale gene expression analysis on the GeneChip Human Genome U133 Plus 2.0 Array (Affymetrix). RNA extraction, labeling, and microarray hybridization were processed in the same manner for all samples in the same set of experiments. Microarray data files are available from ArrayExpress under accession no. E-MTAB-604. After robust multichip average normalization, unsupervised hierarchical clustering was performed using Dchip tools as previously reported on another set of T-ALLs (Soulier et al., 2005). Probe sets for the unsupervised clustering shown in Fig. 4 A were selected on expression variability and reliability among samples after robust multichip average normalization on the following criteria: SD > 1.5 and gene expression >6 in at least 10% of cases and nonredundant genes, resulting in 880 probe sets. To avoid spurious clustering caused by contaminating BM cells, probe sets with high expression in normal BM samples were identified using unsupervised hierarchical classification and excluded, as well as genes from chromosome Y, resulting in a final list of 749 probe sets. The Cell cycle and Mitosis cluster identified from this analysis largely overlaps with the Cluster 6 previously defined (Soulier et al., 2005). Comparison between the paired xenografts and diagnosis samples was performed using the SAM procedure (target proportion of false discoveries, 0.005; 500 permutations; percentile for determining falsely called genes, 90), resulting in 100 probe sets “overexpressed T-ALL xenograft” (58 unique genes; see list in supplemental material). Biological pathways which were statistically linked with this gene set were tested using Gene Ontology and the Broad institute tools. Gene set enrichment analysis of this gene set in relapse versus diagnosis leukemias was performed using two series of paired ALL. These series include n = 35 ALL pairs (32 B cell progenitor ALLs and 3 T-ALLs; Bhojwani et al., 2006) and n = 41 pairs (27 B cell progenitor ALLs and 14 T-ALLs; Staal et al., 2010). Means of probe set expression were ranked using paired Student’s t tests, and 1,000 permutations were performed to calculate nominal probabilities as indicated.
PTEN gene silencing in primary T-ALL cells.
The pSuper construct containing PTEN short hairpin RNA (shRNA) was generated by cloning the following human PTEN-specific RNA interference target sequence into the pSuper plasmid: 5′-AGACAAAGCCAACCGATAC-3′. The H1-shRNAPTEN fragment was then subcloned into the pTRIP/ΔU3-MND-GFP where the green fluorescent protein coding sequence is under the control of the MND promoter as previously done for a control shCTL directed against the human hepatitis B virus. A lentiviral vector encoding the red fluorescent protein Cherry was subsequently derived from this plasmid. Human T-ALL samples were transduced with unconcentrated G/IL-7SUx envelope pseudotyped vectors as previously reported (Gerby et al., 2010). In brief, human T-ALL cells were co-cultured during 48 h with the mouse stromal cells expressing the Notch ligand DL1 MS5-DL1 in reconstituted α-MEM supplemented with 10% FCS and 10% human AB serum, in the presence of stem cell factor, rhFlt3-L, insulin, and rhIL-7 as reported previously (Gerby et al., 2010). Leukemia cells were then spinoculated for 2 h in the presence of plastic-bound human DL1, 2.5 µg/ml protamine sulfate, and G/IL-7SUx pseudotyped lentiviral vectors, and transduction was continued for a further 48 h. Cells were again co-cultured with MS5-DL1 cells plus cytokines during 1 wk for maximal reporter fluorescent protein expression. Transduced leukemia cells were then sorted (Flow cytometer Influx; BD) based on the CD45+CD7+ and GFP+ or Cherry+ phenotype before transplantation into immunodeficient mice.
In vitro drug sensitivity assay.
Diagnosis and xenograft leukemia cells were co-cultured for 1 wk with MS5-DL1 cells, as described previously (Gerby et al., 2010), in the presence of 5 and 50 nM dexamethasone (Dexa; D-4902; Sigma-Aldrich) and/or 5 µM γ-secretase inhibitor (N-[N-(3,5-difluorophenacetyl)-l-alanyl]-S-phenylglycine t-butyl ester [DAPT]; EMD) or with the DMSO/vehicle alone. Cell cycle activation was quantified using KI67 labeling, and apoptosis was measured by annexin V–FITC staining (BD) on harvested leukemic cells and analyzed by FACS.
Online supplemental material.
Fig. S1 shows examples of identical genomic lesions in diagnosis and xenograft leukemia samples. Fig. S2 shows that diagnosis and xenograft leukemias can derive from a common prediagnosis ancestor clone and also shows specific backtracking data in diagnosis samples of genomic abnormalities identified in xenograft samples. Fig. S3 shows that xenografted leukemias have increased leukemia-initiating activity relative to diagnosis leukemias. Fig. S4 shows a model for clonal selection and gain of malignancy in patient and xenograft. Table S1 lists the enriched Gene Ontology terms in the overexpressed in T-ALL xenografts gene set. Table S2, included as an Excel file, is the list of copy number aberrations in T-ALL samples. Table S3, included as an Excel file, is the list of NOTCH1, FBXW7, PTEN, and WT1 mutations in T-ALL samples. The overexpressed in T-ALL xenografts gene set is also listed in the supplemental material. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110105/DC1.
Acknowledgments
We acknowledge the patients who agreed to use of their cells for research and all the physicians who sent biological samples. We thank Thierry Leblanc (Hôpital Robert Debré, Paris, France), Daniela Geromin (Hôpital Saint-Louis, Assistance Publique–Hôpitaux de Paris, Paris, France), Julien Calvo, Julien Tillet, and Christophe Joubert (all from Institut de Radiobiologie Cellulaire et Moléculaire, Commissariat à l’Energie Atomique, Fontenay-aux-Roses, France), Samuel Quentin (Institut Universitaire d’Hématologie/Hôpital Saint-Louis, Assistance Publique–Hôpitaux de Paris), and Lydia Suarez and Kelly Massmalo (both from the Hôpital Robert Debré). Cell sorting was performed at the flow cytometry platform of Institut de Radiobiologie Cellulaire et Moléculaire supervised by Jan Baijer. We are grateful to Paul-Henri Romeo and Claude Gazin for helpful discussions and to Hugues de Thé, Jean-Claude Gluckman, and Michelle Goodhardt for critical reading of the manuscript.
This work was supported by Institut National de la Santé et de la Recherche Médicale, Commissariat à l’Energie Atomique, Universités Paris Diderot and Paris Sud, Institut National du Cancer (INCA), Cancéropôle d’Ile de France, the program Carte d’Identité des Tumeurs from the Ligue Nationale contre le Cancer, and the Association Laurette Fugain. E. Clappier was supported by INCA, and B. Gerby was supported by a PhD fellowship from Ligue Nationale contre le Cancer and Société Francaise d’Hématologie.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- ALL
- acute lymphoblastic leukemia
- CGH
- comparative genomic hybridization
- FISH
- fluorescent in situ hybridization
- NOD
- nonobese diabetic
- shRNA
- short hairpin RNA
- SNP
- single nucleotide polymorphism
- T-ALL
- T cell ALL
References
- Aifantis I., Raetz E., Buonamici S. 2008. Molecular pathogenesis of T-cell leukaemia and lymphoma. Nat. Rev. Immunol. 8:380–390 10.1038/nri2304 [DOI] [PubMed] [Google Scholar]
- Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., et al. 2011. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 469:356–361 10.1038/nature09650 [DOI] [PubMed] [Google Scholar]
- Armstrong F., Brunet de la Grange P., Gerby B., Rouyez M.C., Calvo J., Fontenay M., Boissel N., Dombret H., Baruchel A., Landman-Parker J., et al. 2009. NOTCH is a key regulator of human T-cell acute leukemia initiating cell activity. Blood. 113:1730–1740 10.1182/blood-2008-02-138172 [DOI] [PubMed] [Google Scholar]
- Bailey L.C., Lange B.J., Rheingold S.R., Bunin N.J. 2008. Bone-marrow relapse in paediatric acute lymphoblastic leukaemia. Lancet Oncol. 9:873–883 10.1016/S1470-2045(08)70229-8 [DOI] [PubMed] [Google Scholar]
- Bhojwani D., Kang H., Moskowitz N.P., Min D.J., Lee H., Potter J.W., Davidson G., Willman C.L., Borowitz M.J., Belitskaya-Levy I., et al. 2006. Biologic pathways associated with relapse in childhood acute lymphoblastic leukemia: a Children’s Oncology Group study. Blood. 108:711–717 10.1182/blood-2006-02-002824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko A.D., Razorenova O.V., van de Rijn M., Swetter S.M., Johnson D.L., Ly D.P., Butler P.D., Yang G.P., Joshua B., Kaplan M.J., et al. 2010. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 466:133–137 10.1038/nature09161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S., Henderson M.J., Kwan E., Beesley A.H., Sutton R., Bahar A.Y., Giles J., Venn N.C., Pozza L.D., Baker D.L., et al. 2007. Relapse in children with acute lymphoblastic leukemia involving selection of a preexisting drug-resistant subclone. Blood. 110:632–639 10.1182/blood-2007-01-067785 [DOI] [PubMed] [Google Scholar]
- Clappier E., Cuccuini W., Kalota A., Crinquette A., Cayuela J.M., Dik W.A., Langerak A.W., Montpellier B., Nadel B., Walrafen P., et al. 2007. The C-MYB locus is involved in chromosomal translocation and genomic duplications in human T-cell acute leukemia (T-ALL), the translocation defining a new T-ALL subtype in very young children. Blood. 110:1251–1261 10.1182/blood-2006-12-064683 [DOI] [PubMed] [Google Scholar]
- De Keersmaecker K., Marynen P., Cools J. 2005. Genetic insights in the pathogenesis of T-cell acute lymphoblastic leukemia. Haematologica. 90:1116–1127 [PubMed] [Google Scholar]
- Dick J.E. 2008. Stem cell concepts renew cancer research. Blood. 112:4793–4807 10.1182/blood-2008-08-077941 [DOI] [PubMed] [Google Scholar]
- Ferrando A.A., Neuberg D.S., Staunton J., Loh M.L., Huard C., Raimondi S.C., Behm F.G., Pui C.H., Downing J.R., Gilliland D.G., et al. 2002. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 1:75–87 10.1016/S1535-6108(02)00018-1 [DOI] [PubMed] [Google Scholar]
- Freyer D.R., Devidas M., La M., Carroll W.L., Gaynon P.S., Hunger S.P., Seibel N.L. 2011. Postrelapse survival in childhood acute lymphoblastic leukemia is independent of initial treatment intensity: a report from the Children’s Oncology Group. Blood. 117:3010–3015 10.1182/blood-2010-07-294678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerby B., Armstrong F., de la Grange P.B., Medyouf H., Calvo J., Verhoeyen E., Cosset F.L., Bernstein I., Amselem S., Boissel N., et al. 2010. Optimized gene transfer into human primary leukemic T cell with NOD-SCID/leukemia-initiating cell activity. Leukemia. 24:646–649 10.1038/leu.2009.235 [DOI] [PubMed] [Google Scholar]
- Gutierrez A., Sanda T., Grebliunaite R., Carracedo A., Salmena L., Ahn Y., Dahlberg S., Neuberg D., Moreau L.A., Winter S.S., et al. 2009. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 114:647–650 10.1182/blood-2009-02-206722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner-Schwabe R., Lottaz C., Tödling J., Rhein P., Karawajew L., Eckert C., von Stackelberg A., Ungethüm U., Kostka D., Kulozik A.E., et al. 2006. Expression of late cell cycle genes and an increased proliferative capacity characterize very early relapse of childhood acute lymphoblastic leukemia. Clin. Cancer Res. 12:4553–4561 10.1158/1078-0432.CCR-06-0235 [DOI] [PubMed] [Google Scholar]
- Kleppe M., Lahortiga I., El Chaar T., De Keersmaecker K., Mentens N., Graux C., Van Roosbroeck K., Ferrando A.A., Langerak A.W., Meijerink J.P., et al. 2010. Deletion of the protein tyrosine phosphatase gene PTPN2 in T-cell acute lymphoblastic leukemia. Nat. Genet. 42:530–535 10.1038/ng.587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumper E., Pieters R., Veerman A.J., Huismans D.R., Loonen A.H., Hählen K., Kaspers G.J., van Wering E.R., Hartmann R., Henze G. 1995. In vitro cellular drug resistance in children with relapsed/refractory acute lymphoblastic leukemia. Blood. 86:3861–3868 [PubMed] [Google Scholar]
- Mullighan C.G., Phillips L.A., Su X., Ma J., Miller C.B., Shurtleff S.A., Downing J.R. 2008. Genomic analysis of the clonal origins of relapsed acute lymphoblastic leukemia. Science. 322:1377–1380 10.1126/science.1164266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., Lim W.K., Odom D.T., Sulis M.L., Real P.J., Margolin A., Barnes K.C., O’Neil J., Neuberg D., Weng A.P., et al. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA. 103:18261–18266 10.1073/pnas.0606108103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., Sulis M.L., Cortina M., Real P.J., Barnes K., Ciofani M., Caparros E., Buteau J., Brown K., Perkins S.L., et al. 2007. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat. Med. 13:1203–1210 10.1038/nm1636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pui C.H., Robison L.L., Look A.T. 2008. Acute lymphoblastic leukaemia. Lancet. 371:1030–1043 10.1016/S0140-6736(08)60457-2 [DOI] [PubMed] [Google Scholar]
- Real P.J., Tosello V., Palomero T., Castillo M., Hernando E., de Stanchina E., Sulis M.L., Barnes K., Sawai C., Homminga I., et al. 2009. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat. Med. 15:50–58 10.1038/nm.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shackleton M., Quintana E., Fearon E.R., Morrison S.J. 2009. Heterogeneity in cancer: cancer stem cells versus clonal evolution. Cell. 138:822–829 10.1016/j.cell.2009.08.017 [DOI] [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. 2004. Identification of human brain tumour initiating cells. Nature. 432:396–401 10.1038/nature03128 [DOI] [PubMed] [Google Scholar]
- Soulier J., Clappier E., Cayuela J.M., Regnault A., García-Peydró M., Dombret H., Baruchel A., Toribio M.L., Sigaux F. 2005. HOXA genes are included in genetic and biologic networks defining human acute T-cell leukemia (T-ALL). Blood. 106:274–286 10.1182/blood-2004-10-3900 [DOI] [PubMed] [Google Scholar]
- Staal F.J., de Ridder D., Szczepanski T., Schonewille T., van der Linden E.C., van Wering E.R., van der Velden V.H., van Dongen J.J. 2010. Genome-wide expression analysis of paired diagnosis-relapse samples in ALL indicates involvement of pathways related to DNA replication, cell cycle and DNA repair, independent of immune phenotype. Leukemia. 24:491–499 10.1038/leu.2009.286 [DOI] [PubMed] [Google Scholar]
- Tosello V., Mansour M.R., Barnes K., Paganin M., Sulis M.L., Jenkinson S., Allen C.G., Gale R.E., Linch D.C., Palomero T., et al. 2009. WT1 mutations in T-ALL. Blood. 114:1038–1045 10.1182/blood-2008-12-192039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Vlierberghe P., Pieters R., Beverloo H.B., Meijerink J.P. 2008. Molecular-genetic insights in paediatric T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 143:153–168 10.1111/j.1365-2141.2008.07314.x [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Yang J.J., Bhojwani D., Yang W., Cai X., Stocco G., Crews K., Wang J., Morrison D., Devidas M., Hunger S.P., et al. 2008. Genome-wide copy number profiling reveals molecular evolution from diagnosis to relapse in childhood acute lymphoblastic leukemia. Blood. 112:4178–4183 10.1182/blood-2008-06-165027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O.H., Valdez R., Theisen B.K., Guo W., Ferguson D.O., Wu H., Morrison S.J. 2006. Pten dependence distinguishes haematopoietic stem cells from leukaemia-initiating cells. Nature. 441:475–482 10.1038/nature04703 [DOI] [PubMed] [Google Scholar]
- Zuna J., Ford A.M., Peham M., Patel N., Saha V., Eckert C., Köchling J., Panzer-Grümayer R., Trka J., Greaves M. 2004. TEL deletion analysis supports a novel view of relapse in childhood acute lymphoblastic leukemia. Clin. Cancer Res. 10:5355–5360 10.1158/1078-0432.CCR-04-0584 [DOI] [PubMed] [Google Scholar]