Next generation sequencing and copy number analysis provide insights into the complexity of the CLL coding genome, and reveal an association between NOTCH1 mutational activation and poor prognosis.
Abstract
The pathogenesis of chronic lymphocytic leukemia (CLL), the most common leukemia in adults, is still largely unknown. The full spectrum of genetic lesions that are present in the CLL genome, and therefore the number and identity of dysregulated cellular pathways, have not been identified. By combining next-generation sequencing and copy number analysis, we show here that the typical CLL coding genome contains <20 clonally represented gene alterations/case, including predominantly nonsilent mutations, and fewer copy number aberrations. These analyses led to the discovery of several genes not previously known to be altered in CLL. Although most of these genes were affected at low frequency in an expanded CLL screening cohort, mutational activation of NOTCH1, observed in 8.3% of CLL at diagnosis, was detected at significantly higher frequency during disease progression toward Richter transformation (31.0%), as well as in chemorefractory CLL (20.8%). Consistent with the association of NOTCH1 mutations with clinically aggressive forms of the disease, NOTCH1 activation at CLL diagnosis emerged as an independent predictor of poor survival. These results provide initial data on the complexity of the CLL coding genome and identify a dysregulated pathway of diagnostic and therapeutic relevance.
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in the Western world (Chiorazzi et al., 2005; Tam and Keating, 2010; Zenz et al., 2010b). CLL is characterized by a marked degree of clinical heterogeneity, ranging from patients that harbor a highly stable disease with a nearly normal life expectancy, to patients with a rapidly progressive disease that are destined to shortly succumb (Chiorazzi et al., 2005; Tam and Keating, 2010; Zenz et al., 2010b). The variable course of CLL is driven, at least in part, by heterogeneity in the leukemia biology (Tam and Keating, 2010; Zenz et al., 2010b). A fraction of CLL is prone to transformation to diffuse large B cell lymphoma (DLBCL), a condition known as Richter syndrome (RS) that is characterized by a rapidly deteriorating clinical course with an extremely dismal outcome (Tsimberidou and Keating, 2005; Rossi and Gaidano, 2009).
The molecular pathogenesis of CLL is not fully understood. In contrast to other B cell malignancies, CLL is not associated with recurrent balanced chromosomal translocations (Klein and Dalla-Favera, 2010). Deletion of 13q14 affecting the DLEU2/MIR15-16 cluster is conceivably the initiating lesion in nearly 50% of CLL cases, followed by the accumulation of additional alterations during tumor progression, namely del17p13 and del11q22–q23 affecting TP53 and ATM, respectively (Döhner et al., 2000; Calin et al., 2002; Klein et al., 2010; Zenz et al., 2010b). Rare NOTCH1 mutations have been sporadically reported (Sportoletti et al., 2010).
These known genetic lesions, however, do not fully recapitulate the molecular pathogenesis of CLL and do not entirely explain the marked clinical heterogeneity of the disease or the development of severe complications, such as RS transformation and chemorefractoriness, which, despite recent advances, still represent unmet clinical needs (Rossi and Gaidano, 2009; Tam and Keating, 2010; Zenz et al., 2010b; Fangazio et al., 2011). In the instance of CLL transformation, TP53 disruption and MYC alterations are observed in up to 50% of RS cases, but the genetic lesions of the remaining 50% are virtually unknown (Rossi et al., 2011). Also, TP53 disruption is observed in 40% of CLL cases refractory to therapy, but the molecular basis of the remaining 60% CLL is unknown (Zenz et al., 2010a).
Genome-wide methods aimed at the characterization of the entire spectrum of genetic lesions present in the CLL genome may be useful to provide further insights into CLL pathogenesis, and might contribute to elucidate the molecular basis of CLL clinical evolution, including RS transformation and development of chemorefractoriness. On these grounds, we have exploited an integrated approach based on next generation whole-exome sequencing (WES) and genome-wide high-density single-nucleotide polymorphism (SNP) analysis to investigate the CLL coding genome. We report that CLL is characterized by a limited number of structural genetic alterations compared with other B cell malignancies, and carries a restricted number of mutations in genes potentially relevant for CLL pathogenesis.
Among the genes found mutated in CLL, NOTCH1 emerged as a highly recurrent target of genetic lesions in specific phases of the disease. NOTCH1 encodes a class I transmembrane protein functioning as a ligand-activated transcription factor and playing an important role in cell differentiation, proliferation, and apoptosis (Grabher et al., 2006; Aster et al., 2008, 2011; Paganin and Ferrando, 2011). Upon ligand-binding, NOTCH1 undergoes multiple proteolytic cleavages that allow its intracellular portion to translocate to the nucleus, thus leading to transcriptional activation of multiple target genes, including MYC (Palomero et al., 2006). In T cell acute lymphoblastic leukemia (T-ALL), activating mutations of NOTCH1 are the predominant genetic alteration, accounting for up to 60% of the cases (Weng et al., 2004). The present study documents a high frequency of NOTCH1 mutations in aggressive clinical phases of CLL, exemplified by RS transformation and chemorefractoriness, and identifies NOTCH1 alterations as an independent predictor of poor prognosis.
RESULTS
Mutational load by exome sequencing analysis
To identify somatic gene alterations associated with CLL pathogenesis and to gain insights into the overall complexity of the CLL coding genome, we performed exome capture and next generation sequencing analysis of paired tumor and normal DNAs from 5 newly diagnosed, previously untreated CLL patients (Table S1, discovery panel; and Fig. S1). We also analyzed by direct sequencing the coding exons of NOTCH1, as this gene has been previously implicated in the disease (Sportoletti et al., 2010), but was not represented in the capture array.
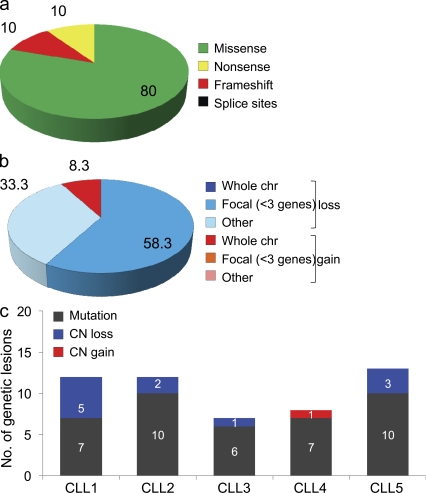
Validation of the candidate variants by Sanger sequencing in the same samples confirmed the presence of 40 somatic, nonsilent mutations involving 39 distinct genes, with NOTCH1 representing the only recurrently mutated one (n = 2/5 cases; Table S2, Table S3, and Supplemental Results). Most of the mutations detected by this approach appeared to be represented in a predominant clone, based on the presence of a clear peak in the chromatogram. The overall mutation load was homogeneous across the five cases (mean = 8, range = 6–10 lesions/case) and included 36 single nucleotide substitutions (n = 32 aa changes and 4 premature stop codons) and 4 frameshift insertions/deletions (Fig. 1 a and Table S3). Although assessed on a limited number of events, the mutational spectrum of CLL was analogous to that reported in other cancer types (e.g., colorectal, pancreatic, and brain tumors; Greenman et al., 2007; Jones et al., 2008; Parsons et al., 2011), with a predominance of transitions over transversions (n = 25:11, ratio 2.2) a preferential targeting of G and C nucleotides (69.4 vs. 30.6%, affecting A/T nucleotides), and a bias for mutations at 5′-CpG-3′ dinucleotides (P < 0.001; Fig. S2).
Figure 1.
Overall load of genetic lesions in CLL. (a) Frequency of Sanger-validated somatic nonsilent mutations identified through WES in the five CLL discovery cases. (b) Overall frequency of somatically acquired CNA according to aberration type in the 5 CLL discovery cases. (c) Combined load of somatically acquired nonsilent mutations and CNA identified in the CLL discovery panel. CN, copy number.
Genome-wide SNP analysis
Analysis of the same 5 CLL samples (and paired normal DNAs) by the Affymetrix SNP 6.0 platform revealed a total of 12 somatic copy number alterations (CNA), with an average of 2.4 lesions per case (range: 1–5; Table S4). 10 of these regions harbored genetic elements, including protein coding genes and/or noncoding RNA genes, and 5 of them encompassed <3 genes that, because of their focal involvement, represent the likely targets of the aberration (Fig. 1 b). Consistent with previous studies (Kujawski et al., 2008; Grubor et al., 2009), the majority of the observed lesions were represented by deletions (11/12, 91.7%), ranging in size from 1.3 kb to loss of a large portion of a chromosomal arm, as in CLL1 that carried a del11q. Deletion of 13q14 was the only recurrent lesion, while trisomy 12 (CLL4) represented the only region of gain (Fig. S3).
Importantly, the SNP array approach successfully identified all four lesions previously detected by FISH analysis and known to associate with CLL, including two del13q14 (CLL2 and CLL5), one trisomy 12 (CLL4), and one del11q (CLL1; Fig. S3). In both instances, del13q14 encompassed the DLEU2/MIR15-16 locus along with 61–68 additional genetic elements, including the RB1 gene. The del11q spanned more than 100 genes, including the tumor suppressor ATM and the negative regulator of the noncanonical NF-κB pathway BIRC3.
Overall complexity of the CLL coding genome
The integration between WES and copy number data provided a total of 52 genetic lesions, ranging from 7 to 13 lesions per case (average = 10.4) and mostly represented by point mutations (Fig. 1 c). Even after considering that some mutations may have been missed because of limited probe density or sequence coverage of the SNP and exome sequencing platforms, this mutation burden is 5–20 times lower than that observed in solid tumor genomes (Jones et al., 2008; Pleasance et al., 2010a; Pleasance et al., 2010b), in multiple myeloma (Chapman et al., 2011), and in DLBCL (unpublished data). Conversely, this load is comparable to that reported in acute myeloid leukemia (Mardis et al., 2009) and in pediatric medulloblastoma (Parsons et al., 2011). This observation, together with the low number of CNAs in general, and whole chromosome alterations in particular, suggests that the CLL genome is relatively stable and homogeneous in terms of mutational load, ploidy, and large chromosomal rearrangements.
Identification of frequently mutated genes
The genes found to be mutated by the aforementioned approach include 32 genes listed in the Catalogue of Somatic Mutations in Cancer database and previously found to be mutated in other cancer genomes (Table S3; Forbes et al., 2011). Although the functional significance of the missense mutations affecting these genes is largely unknown, 22 out of 32 (68.8%) are expected to alter the function of the encoded protein, based on the Polyphen-2 prediction algorithm (Table S3; Adzhubei et al., 2010). The Gene Ontology database (http://www.geneontology.org/) revealed that one third of the identified mutated genes are involved in transcriptional regulation and chromatin remodeling processes (n = 13/39, 33.3%).
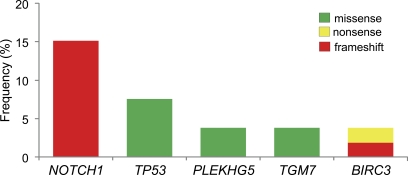
One criterion to assess the pathogenic relevance of the observed genetic lesions is to examine their recurrence in the disease. In the discovery panel, NOTCH1 was the only gene mutated in more than one patient (2/5 cases); however, the lack of recurrence in other mutated genes may be ascribed to the limited number of cases investigated. We therefore expanded the mutational analysis to an independent screening panel of 48 CLL cases with available matched germline DNA (see Table S5 for the detailed characterization of the CLL screening panel). This panel was representative of both the IGHV mutated (n = 24) and unmutated (n = 24) disease categories. The entire coding region and splice sites of the genes found mutated in the discovery panel were studied in the screening phase. This analysis, combined with the results obtained in the 5 cases belonging to the discovery panel, revealed the presence of recurrent mutations (n ≥ 2) in 4 genes, i.e., TGM7, BIRC3, PLEKHG5 (all mutated in ≤5% of cases), and NOTCH1 (mutated in 15.1% of cases; Fig. 2). Although absent in the discovery panel, mutations of TP53 were also tested in the CLL screening panel, as this genetic lesion is known to be recurrent in CLL (Zenz et al., 2010c). This analysis revealed TP53 missense mutations in 4/53 (7.5%) patients.
Figure 2.
Recurrent mutations in CLL. Percentage of CLL primary cases harboring mutations in candidate genes after targeted resequencing of an expanded screening dataset (n = 48 CLL cases). The final number of mutated cases over total analyzed (including the 5 discovery CLL) for each gene is as follows: NOTCH1, 8/53; TP53, 4/53; PLEKHG5, 2/53; TGM7, 2/53; BIRC3, 2/53.
Although the role of the two missense mutations found in the TGM7 gene, which encodes a transglutaminase of unclear specific function, remains to be determined, other mutations provide preliminary evidence of functional significance. BIRC3, a negative regulator of the noncanonical NF-κB pathway that is affected by chromosomal translocations in MALT lymphoma and by focal deletions in multiple myeloma (Zhou et al., 2005; Annunziata et al., 2007; Keats et al., 2007; Rosebeck et al., 2011), was biallelically inactivated in CLL1. In this patient, a copy number loss was observed together with a frameshift deletion located between the third baculovirus IAP repeat (BIR) and the caspase activation and recruitment domain (CARD), leading to removal of the RING domain, which is required for the inhibitory function of BIRC3 (Gyrd-Hansen and Meier, 2010). A second case (CLL45) harbored a nonsense mutation (E424X) truncating the corresponding protein between the third BIR domain and the CARD domain. FISH analysis showed loss of the second BIRC3 allele, indicating biallelic inactivation in this case as well, thus suggesting a tumor suppressor role for BIRC3 in modulating NF-κB activation. Interestingly, another CLL case (CLL5 in the discovery panel) harbored a missense mutation of the MYD88 gene, which encodes an adaptor protein involved in Toll-like receptor signaling and in activation of NF-κB. The same mutation found in case CLL5 (L265P) was recently reported in a large fraction of ABC-DLBCL, where it was shown to be oncogenically active (Ngo et al., 2011). Finally, two cases (CLL4 and CLL13) displayed missense mutations of PLEKHG5, a member of the G protein family that has been suggested to be involved in NF-κB activation (Maystadt et al., 2007). Collectively, these findings suggest that activation of NF-κB may be implicated in at least a fraction of CLL cases.
The recurrence of NOTCH1 mutations (15.1%) appeared potentially relevant, given the well-established involvement of this gene in the pathogenesis of T-ALL and its previously reported association with CLL at low frequency (Weng et al., 2004; Sulis et al., 2008; Sportoletti et al., 2010). This finding, together with the preliminary observation of NOTCH1 mutations in a few cases of RS (see following paragraph), prompted a systematic analysis of NOTCH1 in different clinical phases of CLL.
NOTCH1 mutations at CLL diagnosis occur at low frequency and predict poor outcome
To define the frequency of NOTCH1 mutations at CLL diagnosis, we analyzed a consecutive cohort of CLL patients (n = 120) sampled at diagnosis and followed at the Amedeo Avogadro University of Eastern Piedmont (extension panel). This cohort was representative of the main clinical characteristics of the disease at diagnosis (Table S6). Specifically, IGHV homology ≥98% occurred in 39/120 (32.5%) patients, del13q14 was observed in 61/112 (54.5%) cases, normal FISH in 32/112 (28.6%), trisomy 12 in 17/112 (15.2%), del11q22-q23 in 8/112 (7.1%), and TP53 disruption by mutation and/or deletion in 13/117 (11.1%; Table S6). After a median follow up of 70.3 mo, the 5-yr treatment free survival (TFS) probability of the CLL cohort was 60.0% and the 5-yr overall survival (OS) probability was 83.3%.
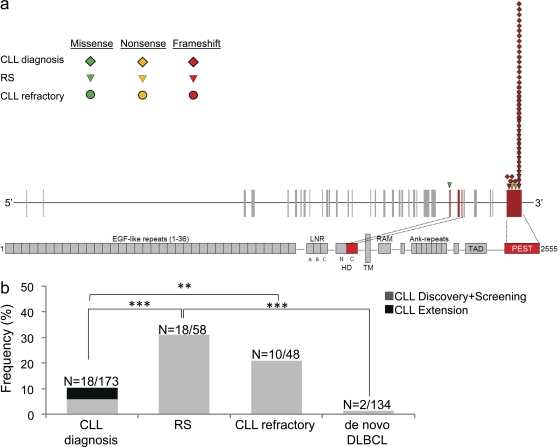
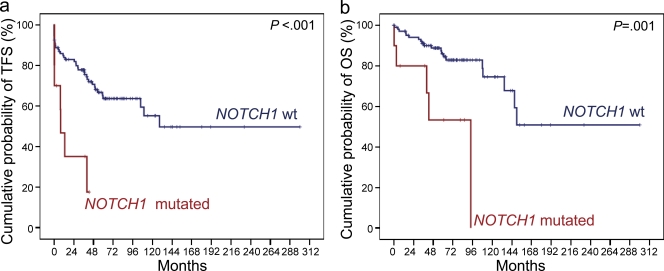
At CLL diagnosis, mutations of NOTCH1 were identified in 10/120 (8.3%) patients, all of which displayed a recurrent 2-bp frameshift deletion (ΔCT7544-7545, P2515fs) that is predicted to cause NOTCH1-impaired degradation through the truncation of the C-terminal PEST domain (Fig. 3 a and Table S7; Weng et al., 2004). NOTCH1 mutations clustered with CLL harboring unfavorable features, including unmutated IGHV genes (7/39, 17.9% vs. 3/81, 3.7% in IGHV mutated CLL; P = 0.013), and TP53 disruption (3/13, 23.1% vs. 7/104, 6.7% in TP53 wild-type CLL; P = 0.047). Consistently, CLL harboring NOTCH1 mutations were characterized by a significantly shorter time to progression to a disease requiring treatment (median TFS, 7.9 mo) compared with CLL cases harboring wild-type NOTCH1 alleles (median TFS, 128.5 mo; P < 0.001; Fig. 4). Also, CLL harboring NOTCH1 mutations were characterized by a significantly shorter OS (median OS, 93.8 mo; 5-yr OS, 53.3%) compared with wild-type CLL (median OS not reached; 5-yr OS, 86.0%; P = 0.001; Fig. 4). By multivariate Cox analysis, NOTCH1 mutations were a predictor of poor OS (HR, 10.1; 95% CI, 2.6–39.0; P = 0.001) independent of confounding covariates, including TP53 disruption (HR, 4.6; 95% CI, 1.6–13.1; P = 0.004) and unmutated IGHV genes (HR, 6.2; 95% CI, 1.4–25.9; P = 0.012).
Figure 3.
NOTCH1 is mutated in a large fraction of RS cases and chemorefractory CLL. (a) Schematic diagram of the NOTCH1 gene (top) and protein (bottom), with its conserved functional domains (EGF-like repeats; LNR, LIN-12/NOTCH repeats; HD, heterodimerization; TM, transmembrane; Ank, Ankyrin repeats; TAD, transactivation; PEST). Color-coded shapes indicate the position of the mutations found in CLL at diagnosis, RS, and chemorefractory CLL. (b) Prevalence of NOTCH1 mutations in CLL at diagnosis, RS, chemorefractory CLL, and de novo DLBCL; numbers on top indicate the actual number of mutated samples over total analyzed. ***, P < 0.001; **, P < 0.05.
Figure 4.
Kaplan-Meier estimates of treatment-free survival (TFS) and overall survival (OS), according to the mutational status of NOTCH1.
Collectively, these results confirm previous findings on the low frequency and prognostic relevance of NOTCH1 mutations at CLL diagnosis (Di Ianni et al., 2009; Sportoletti et al., 2010), but identify NOTCH1 mutations as a predictor of unfavorable outcome independent of other biological features of CLL, namely IGHV mutation status and TP53 disruption.
Frequent activating mutations of NOTCH1 in RS
The association between NOTCH1 mutations at CLL diagnosis and unfavorable outcome prompted the investigation of this gene in patients whose disease subsequently acquired a high-risk clinical phenotype. As a model of CLL clinical progression, we initially chose RS transformation because: (a) it represents the most aggressive phase of the clinical spectrum of CLL, as also documented by the fact that RS is rapidly lethal in most patients; (b) the molecular lesions driving CLL transformation to RS are not fully understood (Tsimberidou and Keating, 2005; Rossi and Gaidano, 2009; Fangazio et al., 2011; Rossi et al., 2011).
Based on this rationale, NOTCH1 mutations were analyzed in the diagnostic biopsy of 58 cases of histologically proven RS, all represented by DLBCL transformed from a previous CLL phase. The clinical and biological characteristics of the RS cases are summarized in Table S8. A total of 19 mutations were identified in 18/58 (31.0%) cases (Table S7). The frequency of NOTCH1 mutations in RS was significantly higher compared with the frequency observed at CLL diagnosis in the consecutive series (P < 0.001; Fig. 3b). NOTCH1 mutations in RS were represented by a recurrent 2-bp frameshift deletion (ΔCT7544-7545, P2515fs; n = 15), one frameshift insertion, and two nonsense mutations (Fig. 3 a). On the basis of their distribution along the NOTCH1 protein, all frameshift and nonsense mutations of NOTCH1 are predicted to cause NOTCH1-impaired degradation through the truncation of the C-terminal PEST domain (Table S7 and Fig. 3 a). One single mutation (G5164A, V1722M) affected the heterodimerization domain of NOTCH1 (Fig. 3 a).
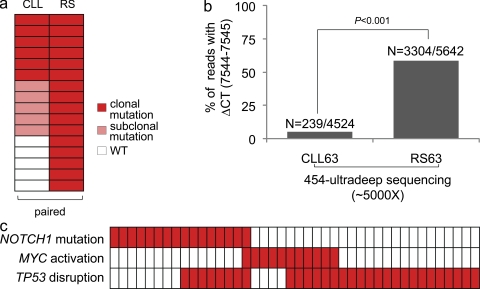
To establish the timing of acquisition of NOTCH1 mutations during the clinical history of RS patients, we investigated paired sequential samples collected at the time of CLL diagnosis. 15 patients harboring 16 NOTCH1 mutations and belonging to the RS series were analyzed by this approach. Of the 16 NOTCH1 mutations observed in the RS phase, 5 (31.3%) were not detectable in the paired CLL phase within the sensitivity threshold of Sanger sequencing (Fig. 5 a). Conversely, 6/16 (37.5%) mutations detected in the RS phase were already present at the time of CLL diagnosis, where their clonal representation appeared to be similar to that observed in RS, as estimated by Sanger sequencing (Fig. 5 a). Interestingly, 5/16 (31.2%) NOTCH1 mutations identified in the RS phase were already present at subclonal levels in the paired CLL phase. To obtain a more quantitative estimate of the clonal representation of NOTCH1 mutated versus germline alleles, the CLL and RS phases from case RS63 were subjected to ultradeep next generation sequencing (sequencing depth ∼5,000 per sample) using the Roche 454 technology. This approach showed that the ΔCT7544-7545 (P2515fs) mutation, occurring in 58.6% (3,304/5,642) of the sequencing reads obtained from the RS phase, was restricted to 5.3% (239/4524) of the reads obtained at the time of CLL diagnosis (Fig. 5 b, Fig. S4, and Fig. S5), indicating an 11-fold increase during the 30-mo interval between CLL diagnosis and RS development. These data suggest that clonally represented mutations of RS might already be present years before RS transformation, and that the clone harboring the mutation is progressively selected during the CLL clinical history ending into RS transformation.
Figure 5.
Timing of NOTCH1 mutations in RS. (a) NOTCH1 mutations in some cases of RS are not identifiable in the preceding CLL phase, whereas in other patients may already be present before transformation, at a clonal/subclonal level. In the heatmap, rows correspond to NOTCH1 mutations and columns represent sequential phases of the disease (paired CLL diagnostic samples and RS samples), color-coded based on the mutational status (for details, see Materials and methods). (b) The NOTCH1 mutation detected in RS case RS63 is present at subclonal levels at the time of CLL diagnosis (sample CLL63). The graph shows a quantitative estimate of the balance between wild-type and mutated (ΔCT7544-7545) NOTCH1 alleles assessed by ultradeep 454 sequencing in the paired CLL-RS samples (CLL63 and RS63). 30 mo elapsed between collection of the CLL sample (diagnosis of CLL) and collection of the RS sample (obtained at diagnosis of RS). Numbers on top indicate the actual number of sequencing reads carrying the mutations over total analyzed (for details, see Materials and methods). (c) Mutations of NOTCH1 are largely mutually exclusive with MYC activating events (translocations and copy number gains) in RS. In the heatmap, rows correspond to the indicated genes, and columns represent individual RS samples, color-coded based on the gene status (white, wild-type; red, mutated or activated or disrupted).
One RS patient (RS86) carrying a NOTCH1 mutation disrupting the PEST domain (ΔCT7544-7545, P2515fs) in the CLL phase and in a first RS biopsy, subsequently acquired a second NOTCH1 mutation (G5164A, V1722M) affecting the heterodimerization domain when RS further progressed from nodal localization to peripheral blood invasion. The detection of PEST and heterodimerization domain mutations occurring in the same patient is of relevance, as mutations in these two distinct domains have been shown to act synergistically in promoting NOTCH1 signaling activation (Weng et al., 2004). The progressive accumulation of multiple NOTCH1 mutations during clinical history further reinforces the notion that NOTCH1 activation might be relevant for CLL progression.
The occurrence of NOTCH1 activation in RS was then compared with the distribution of other genetic alterations that are recurrently associated with this disease. MYC, one of the target genes and major downstream effectors of NOTCH1 (Palomero et al., 2006; Li et al., 2011), was targeted by translocations in 9/51 (17.6%) RS cases and by gene amplification in 2 additional cases. NOTCH1 mutations and MYC abnormalities distributed in a mutually exclusive fashion (Fig. 5 c), although at borderline statistical significance (P = 0.076), conceivably because of the limited number of cases included in the analysis. On these bases, by combining NOTCH1 mutations and MYC abnormalities, 26/51 (51.0%) RS harbor genetic lesions ultimately leading to MYC activation. Among CLL, MYC abnormalities were extremely rare (1/120; 0.8%), in agreement with the notion that they are generally acquired at RS transformation (Huh et al., 2008; Rossi et al., 2011). The sole case of CLL carrying a MYC alteration was devoid of NOTCH1 mutations.
TP53 was disrupted by mutations and/or deletion of the locus in 30/54 (55.5%) RS. TP53 disruption frequently paired with NOTCH1 mutations (8/16, 50%) and MYC abnormalities (7/11, 63.6%) in the same RS sample, thus contributing to a dual-hit genetic mechanism of transformation (Fig. 5 c).
All cases of RS included in this study were histologically classified as DLBCL, a pathological entity characterized by high molecular heterogeneity that may arise either de novo or upon transformation from an indolent B cell malignancy, as in the case of CLL transformation to RS (Swerdlow, 2008). To assess the specificity of NOTCH1 mutations for RS, 134 cases of de novo DLBCL were also investigated. Mutations of NOTCH1 were very rare in this category, being restricted to 2/134 (1.5%) cases (Table S7 and Fig. 3 b).
The frequent recurrence of NOTCH1 mutations in RS prompted the investigation of other NOTCH1-related genes, namely NOTCH2 and FBXW7 (O’Neil et al., 2007; Thompson et al., 2007; Lee et al., 2009). Mutations of NOTCH2 were detected in 2/32 (6.2%) cases of RS, and included one amino acid change (K2121N), that was shown to be acquired at transformation, and a premature stop codon (R2400*; in this case, no material was available from the corresponding diagnostic CLL sample). FBXW7 mutations scores were consistently negative (0/32).
Overall, these data document that mutations activating the Notch pathway are a frequent event in the pathogenesis of RS. In the context of the clinicopathological spectrum of DLBCL subtypes, NOTCH1 mutations appear to preferentially associate with transformed cases.
Frequent activating mutations of NOTCH1 in chemorefractory CLL
Because RS is a highly chemorefractory condition (Rossi and Gaidano, 2009; Tsimberidou and Keating, 2005), we hypothesized that NOTCH1 mutations might also provide a marker of chemorefractoriness in patients who clinically progressed because of treatment failure without transformation to RS. To address this issue, NOTCH1 mutations were investigated in 48 cases of progressive and chemorefractory CLL in which RS had been ruled out based on clinical picture, lymph node biopsy, and/or 18FDG-PET studies (Table S9). Samples from these cases were obtained at the time of progression immediately before starting the treatment to which the patient eventually failed to respond. Mutations were identified in 10/48 (20.8%) cases of chemorefractory CLL (Table S7), including 8/35 (22.8%) fludarabine-refractory and 2/13 (15.3%) alkylator-refractory cases. The frequency of NOTCH1 mutations in chemorefractory CLL was significantly higher compared with the frequency observed at CLL diagnosis in the consecutive series (P = 0.033; Fig. 3 b). All mutations led to a shift in the reading frame and, as observed in RS, were mostly (8/10) represented by the 2-bp frameshift deletion ΔCT7544-7545 (P2515fs). On the basis of their distribution along the NOTCH1 protein, mutations observed in chemorefractory CLL are all predicted to increase NOTCH1 stability via truncation of the C-terminal PEST domain (Fig. 3 a; Weng et al., 2004).
In CLL, the molecular mechanisms of chemorefractoriness are represented by TP53 disruption in up to 40% of cases, indicating that other currently unknown mechanisms might be operative in the remaining 60% of cases. In the current series of chemorefractory CLL, TP53 was disrupted by mutations and/or deletions in 19/48 (39.6%) cases. NOTCH1 mutations occurred in 6/29 (20.7%) TP53 wild-type chemorefractory CLL, whereas they overlapped with TP53 disruption in 4/19 cases (21.1%).
Based on these data, NOTCH1 mutations represent a frequent genetic lesion of CLL associated with progressive disease failing treatment.
DISCUSSION
The goal of this study was to characterize the nature and frequency of somatic genetic alterations affecting the CLL coding genome by integrating the analysis of structural and sequence mutations of DNA. This combined approach has allowed the definition of the degree of complexity characterizing the CLL coding genome, with implications for the mechanisms leading to genetic alterations in this disease. This analysis has also revealed the involvement of the NOTCH1 pathway in disease progression and transformation, with diagnostic and therapeutic implications.
Although analysis of larger cohorts of patients will be required to conclusively assess the precise number of genetic lesions that are present in the CLL genome, the combined set of gene alterations (i.e., copy number changes and mutations) identified in the CLL discovery panel provides initial information about the order of magnitude of the lesions associated with this disease. Given the presence of ∼8 nonsilent mutations and ∼2 CNAs per CLL case, our results suggest that the coding genome of CLL harbors at least 10 potentially relevant genetic aberrations. As an additional ∼40% of nonsilent mutations may have escaped detection by the WES approach used, because of relatively low depth of coverage, the total number of gene alterations present in a CLL genome may be <20. This estimate refers to those lesions that are clonally represented in the CLL genome at diagnosis, and which may have thus contributed to the malignant transformation process. As such, this preliminary CLL genome picture may serve as an initial database for the determination of frequency in additional panel of cases.
The order of magnitude of lesions detected in the coding genome of CLL appears considerably lower than that reported for common epithelial cancers. Among hematologic malignancies, the complexity of the CLL genome is also on the lower side, markedly smaller than that of DLBCL (<100) and multiple myeloma (∼50), and comparable to that of some acute leukemias (∼10; Ley et al., 2008; Mardis et al., 2009; Chapman et al., 2011; unpublished data). The predominance of point mutations over CNAs is notable, as it is unusual compared with most cancer types (Mullighan et al., 2007; Lenz et al., 2008; Parsons et al., 2011). This observation, together with the well-documented rarity of reciprocal balanced chromosomal translocations in CLL (Klein and Dalla-Favera, 2010), is consistent with previous models suggesting a derivation from a B cell counterpart in which immunoglobulin gene remodeling mechanisms are not active and those controlling ploidy remain intact.
On the other hand, similar to most cancer types are: (a) the observed predominance of transitions over transversions; (b) the preferential targeting of C:G and G:C base pairs; and (c) the significant bias toward alterations at CpG dinucleotides. The distribution of the mutations along the entire span of transcribed regions does not seem to reflect the abnormal and/or ectopic activity of members belonging to the APOBEC family of deaminases, including activation induced deaminase (Albesiano et al., 2003; Delker et al., 2009), which targets the 5′ portion of genes within 2 kb from the transcription initiation site. Instead, the observed pattern of alterations is more consistent with a derivation from endogenous biochemical processes, such as the spontaneous deamination of 5-methylcytosine residues (Parsons et al., 2011).
The screening of the mutations observed in the five cases of the discovery panel into an extended panel of cases has shown that only very few of these mutations display some minimal degree of recurrence, even when the entire gene is analyzed. This outcome appears different from that obtained in other malignancies, e.g., in DLBCL, where an analogous approach has led to the identification of a few highly recurrent genetic lesions (Pasqualucci et al., 2011). This result may reflect the fact that the most significant alterations involved in CLL pathogenesis have already been identified, including del13q14 involving the DLEU2/MIR15-16 cluster, trisomy 12, and del11q. Alternatively, the rarity in recurrence may reflect the relationship of the observed lesions in functional pathways that are not presently recognized. One example in support of this notion is the observation of three lesions (BIRC3, MYD88, and PLEKHG5) each detected at very low frequency, yet all linked to perturbation of the NF-κB activity, a well-documented oncogenic pathway (Staudt, 2010). Each of the genetic alterations observed in this initial study represents the basis for extensive analysis of additional lesions in their respective cellular pathways.
A novel finding emerging form this study is the association between somatic mutations of NOTCH1 and different aspects of disease progression. The mutations observed in CLL have consequences on NOTCH1 function that have been documented in T-ALL, where their oncogenic activity has been extensively demonstrated in vitro and in vivo (Weng et al., 2004; O’Neil et al., 2006). In fact, the presence of NOTCH1 mutations in rare cases of CLL at diagnosis has been previously reported and is confirmed in our study (Di Ianni et al., 2009; Sportoletti et al., 2010). Conversely, the association of these mutations at a significantly higher frequency with both RS and CLL investigated at the time of chemorefractoriness strongly suggests that NOTCH1 mutations are selected during disease progression. At CLL diagnosis, NOTCH1 mutations identify a disease subgroup enriched for poor risk genetic features, and characterized by an aggressive clinical phenotype. Future investigations involving large and prospective CLL cohorts are required to show how NOTCH1 mutations interact with other high-risk genetic features.
In the case of RS, our data cannot conclusively distinguish whether NOTCH1 mutations are already present at subclonal levels early during CLL development and are subsequently selected (as demonstrated for a few cases), or whether they are acquired de novo during disease transformation to RS. A conclusive demonstration of the precise timing of NOTCH1 mutations in RS awaits studies aimed at tracking NOTCH1 mutations with high sensitivity techniques in disease phases preceding RS development (Campbell et al., 2008). Also, the recurrent presence of NOTCH1 mutations in chemorefractory CLL might be caused either by selection of more malignant clones during disease progression or by a role of these mutations in determining drug resistance. This issue may be addressed by dedicated studies of NOTCH1 mutations in prospective cohorts of CLL, ideally in the context of controlled clinical trials.
Concerning the pathogenesis of RS, our results show that NOTCH1 mutations are largely mutually exclusive with MYC oncogenic activation. This finding is consistent with the observation that NOTCH1 directly stimulates MYC transcription and suggests that activation of oncogenic MYC may be one common final pathway selected for tumorigenesis (Palomero et al., 2006). Conversely, both NOTCH1 mutational activation and MYC deregulation often coexist with inactivation of the TP53 tumor suppressor gene, an event that is commonly observed in association with MYC activation in tumors, which may be selected to prevent the apoptotic effects and the response to genomic instability both induced by MYC overexpression (Eischen et al., 1999; Schmitt et al., 2002; Dominguez-Sola et al., 2007; Rossi et al., 2011).
From a clinical standpoint, NOTCH1 mutations cluster with CLL subgroups that are currently scored at extremely high risk because of TP53 disruption, refractoriness to fludarabine, or transformation to RS. In the context of these highly unfavorable clinical settings, the appearance or selection of NOTCH1 mutations during disease progression has diagnostic and therapeutic implications. The detection of these mutations at the subclonal level using high sensitivity methods, such as the ultradeep next generation sequencing shown in Fig. 5 b, Fig. S4, and Fig. S5, may provide an objective and measurable biomarker for the early detection of the risk of progression or chemorefractoriness and therefore influence the clinical management of CLL. The validity of this attractive hypothesis needs to be explored by dedicated studies aimed at assessing the role of NOTCH1 as a predictor of RS and/or chemorefractoriness at the time of CLL diagnosis. Finally, NOTCH1 represents a well-established therapeutic target with some drugs already available, such as those inhibiting its enzymatic conversion to active transcription factor, and others under active development (Real et al., 2009). Our results add CLL to the cadre of common diseases in which these drugs should be tested for their efficacy alone or in combination with available therapeutic regimens.
MATERIALS AND METHODS
Patients.
Samples from 5 newly diagnosed and previously untreated CLL patients (discovery panel) were obtained from the Division of Hematology, Department of Clinical and Experimental Medicine, Amedeo Avogadro University of Eastern Piedmont, as frozen peripheral blood mononuclear cells isolated by Ficoll-Paque gradient centrifugation. Diagnosis of CLL was based on IWCLL-NCI Working Group criteria and confirmed by a flow cytometry score >3 (Hallek et al., 2008). In all cases, the fraction of tumor cells corresponded to >80%, as assessed by FACS analysis of CD19/CD5 expression. Matched normal DNA was obtained from peripheral blood granulocytes that had been shown to be devoid of tumor cells by PCR analysis of patient-specific IGHV-D-J rearrangements. The clinical and biological characteristics of cases belonging to the discovery panel are summarized in Table S1. Patients were representative of the two major immunogenetic subgroups of CLL (IGHV mutated, two cases; IGHV unmutated, three cases), and harbored common CLL-associated genetic abnormalities.
The screening panel used to assess the recurrence of mutations affecting genes that were identified through WES was composed of 48 newly diagnosed and previously untreated CLL patients provided by Amedeo Avogadro University of Eastern Piedmont. In all cases, the fraction of tumor cells corresponded to >80%, as assessed by FACS analysis of CD19/CD5 expression. Matched normal DNA was obtained for all cases from purified granulocytes or saliva and was confirmed to be devoid of tumor-derived material by PCR analysis of tumor-specific IGHV-D-J rearrangements. The clinical and biological characteristics of cases belonging to the screening panel are summarized in Table S5. Patients were representative of the two major immunogenetic subgroups of CLL (IGHV mutated, 24 cases; IGHV unmutated, 24 cases).
The prevalence of NOTCH1 mutations was further assessed in 3 clinical CLL panels representative of different disease phases, and including a consecutive series of newly diagnosed and previously untreated CLL (n = 120, extension panel), a cohort of chemorefractory CLL (n = 48), and a cohort of CLL transformed to RS (n = 58). Diagnosis of chemorefractoriness was according to guidelines (Hallek et al., 2008). Diagnosis of RS was based on the histology of lymph node or extranodal tissue excisional biopsies. After institutional pathological review, all RS cases were classified as DLBCL according to the World Health Organization Classification of Tumours of the Hematopoietic and Lymphoid Tissues (Swerdlow, 2008). In all RS cases, molecular studies were performed on the biopsy used for RS diagnosis. Biological samples from the paired CLL phase were available in 45 (77.5%) cases.
The clinical and biological characteristics of the consecutive series of newly diagnosed and previously untreated CLL (n = 120) are summarized in Table S6. After a median follow up from diagnosis for living patients of 70.3 mo, the 5 yr-treatment free survival probability of the CLL cohort was 60.0% and the 5-yr overall survival probability was 83.3%. The clinical and biological characteristics of the chemorefractory CLL cohort (n = 48) are summarized in Table S9. The median overall survival from chemorefractoriness was 1.6 yr. The clinical and biological characteristics of the RS cohort (n = 58) are summarized in Table S8. The median overall survival from RS transformation was 1.1 yr.
The study was approved by the Institutional Review Board of Columbia University and by the Ethical Committee of the Azienda Ospedaliera Maggiore della Carità di Novara, Amedeo Avogadro University of Eastern Piedmont.
IGHV mutational status.
IGHV mutational status was performed as previously reported (Rossi et al., 2009). PCR products were directly sequenced with the ABI PRISM BigDye Terminator v1.1 Ready Reaction Cycle Sequencing kit (Applied Biosystems) using the ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). Sequences were aligned to the ImMunoGeneTics sequence directory (Chiorazzi et al., 2005) and considered mutated if homology to the corresponding germline gene was <98% (Damle et al., 1999; Hamblin et al., 1999).
FISH analysis.
The following probes were used for FISH analysis of CLL and RS cases: LSID13S319, CEP12, LSIp53, LSIATM, LSI IGH/BCL2, LSI IGH/CCND1, LSI BCL6, LSI, IGH/MYC/CEP8, MYC break-apart, LSI N-MYC, CEP2, CEP3, CEP11, CEP18, CEP19 (Abbott); a BCL-3 split signal probe (Dako); 6q21/α-satellite (Kreatech Biotechnology); BAC clones 373L24-REL, 440P05-BCL11A, RP11-177O8-BIRC3. The detailed protocol used for FISH studies is described elsewhere (Rossi et al., 2009).
Whole-exome capture and massively parallel sequencing.
Whole-exome capture and next generation sequencing of the five CLL tumor/normal DNA samples were performed using the NimbleGen Sequence Capture 2.1M Human Exome Array and the 454 Genome Sequencer FLX instrument, according to the manufacturer’s protocol. The array features oligonucleotides for ∼180,000 coding exons and 551 miRNA exons, corresponding to ∼85% of nonrepetitive sequences in the CCDS database (total size of target region, 34 Mb). The resulting target-enriched pool was amplified and subjected to high-throughput sequencing on the Genome Sequencer FLX Instrument of 454 Life Sciences using 2 PicoTiterPlate runs per sample (∼1 million reads/plate, at 400-bp reads). All procedures were performed at Roche and 454 Life Sciences.
454 sequencing data analysis.
The computational pipeline used for the analysis of the deep sequencing data is reported in Fig. S1.
The sequencing reads obtained from the Roche FLX454 Sequencer were aligned to the human reference genome (National Center for Biotechnology Information build 36; hg18 assembly) with the Genome Sequencer Reference Mapper, version 2.3 (Roche). This application discards sequencing reads aligning to multiple positions of the reference sequence. Furthermore, because duplicate reads of a single DNA origin are a known potential artifact of the emulsion PCR step, groups of reads mapping to the same basepair of the reference sequence are counted as a single read in the subsequent computation of variants.
The alignments of the uniquely mapped reads to the reference genome were used to define the genetic variants in the CLL tumor and normal DNAs as single base-pair substitutions, small insertions, and small deletions (indels). The Mapper application declares the presence of a variant allele with high-confidence, if there are at least three nonduplicate reads harboring the variant (two in one sense and one in the opposite). Variant candidates in tumor samples were identified with the GSMapper software, using high confidence variant detection method. Germline variants were filtered using a less conservative approach (presence in ≥1 read) and by comparison with common SNPs reported in the National Center for Biotechnology Information database. Using this observation, one can predict the sensitivity of the high-confidence criteria to detect heterozygous variant alleles in a sample containing 100% cancer genetic material as the fraction of sites covered with a depth of at least 6. Assuming that the two alleles of a heterozygous variant are covered with equal probability, a more precise prediction for the sensitivity to detect such variants is given by the high-confidence coverage (HCC) computed by the formula:
where Nk corresponds to the number of basepairs covered by k sequencing reads and NT corresponds to the total number of base-airs covered.
Variants obtained by the high-confidence criterion of the Mapper application were filtered further for nonsilent exonic mutations and mutations affecting a nucleotide within 4 nt of a consensus splice site. Furthermore, germline variants reported in publicly available databases (dbSNP, Ensembl Database, and UCSC Genome Browser) and/or identified in the paired normal DNAs were removed. Finally, variants caused by known systematic errors of the 454 sequencing technology (small indels affecting homopolymer repeats) and variants below the sensitivity of direct Sanger sequencing (i.e., observed in <20% of the reads), were also discarded.
Validation of candidate somatic mutations by Sanger sequencing.
Candidate nonsilent somatic mutations were subjected to validation by conventional Sanger-based resequencing analysis of PCR products obtained from both normal and tumor high molecular weight genomic DNA using primers specific for the exon encompassing the variant. The sequences surrounding the genomic locations of the candidate tumor-specific nonsilent mutations identified through WES were obtained from the UCSC Human Genome database (validation phase). The validation screen allowed us to eliminate additional private polymorphisms that had been missed by the next generation sequencing approach, presumably because of low coverage of the normal sample, as well as false positives caused by errors introduced during the sequencing procedure and/or by mapping errors, as it may occur in the case of repetitive regions or highly homologous regions in the genome. Confirmed nonsilent mutations were tested for their functional consequences in silico by using the PolyPhen-2 (polymorphism phenotyping) algorithm, which is based on structure/sequence conservation.
Mutation screening of identified genes.
The complete coding sequences and exon/intron junctions of genes identified through the WES approach, including NOTCH1 (under GenBank/EMBL/DDBJ accession no. NM_017617.2), were analyzed in a larger dataset (screening panel, n = 48 CLL cases) by PCR amplification and direct sequencing of whole genome amplified DNA obtained from two independent aliquots of high molecular weight genomic DNA (12 ng each) using the REPLI-g Mini kit (QIAGEN).
Sequences for all annotated exons and flanking introns were retrieved from the UCSC Human Genome database, using the corresponding mRNA accession no. as a reference. PCR primers, located ≥50 bp upstream or downstream to target exon boundaries, were either derived from previously published studies or designed in the Primer 3 program, and filtered using UCSC In Silico PCR to exclude pairs yielding more than a single product. All PCR primers and conditions are available upon request. The oligonucleotides used for the validation phase produced PCR products and chromatograms evaluable in ∼99.3% of the target regions in the 5 CLL cases belonging to the discovery panel. The vast majority of the oligonucleotides used for the screening phase (∼98%) yielded PCR products and high quality sequencing in 40 or more of the 48 screening samples. The NOTCH1 gene was also screened in an independent panel of 120 CLL diagnostic samples, 58 RS cases, and 48 chemorefractory CLL. NOTCH2 (accession no. NM_024408.3) and FBXW7 (accession no. NM_033632.2) were screened in 32 RS cases. All PCR primers and conditions are available upon request.
Purified amplicons were sequenced by conventional Sanger method (Genewiz, Inc.) and compared with the corresponding germline sequences using the Mutation Surveyor Version 2.41 software package (SoftGenetics) after automated and/or manual curation. All variants were sequenced from both strands on independent PCR products obtained from high molecular weight genomic DNA. Synonymous mutations, previously reported polymorphisms, and changes present in the matched normal DNA, which was available for all samples, were removed from the analysis. To rule out the possibility of contamination across samples, DNA of selected RS cases harboring the highly recurrent ΔCT(7544–7545) NOTCH1 mutation was re-extracted from the original tissue biopsy and retested in an independent laboratory, which confirmed the presence of the mutation.
Copy number analysis by high-density SNP array analysis.
Genome-wide DNA profiles were obtained from high molecular weight tumor and normal genomic DNA of the five CLL cases of the discovery panel using the Genome-Wide Human SNP Array 6.0 (Affymetrix). In brief, DNA was restriction enzyme digested, ligated, PCR-amplified, purified, labeled, fragmented, and hybridized to the arrays according to the manufacturer’s instructions. Data analysis was performed as previously described (Mullighan et al., 2007; Pounds et al., 2009).
RNA extraction and gene expression profile analysis.
Adequate material for extraction of RNA was available for three of the five CLL discovery cases, which were characterized by gene expression profile analysis. Total RNA extraction was performed by TRIzol (Invitrogen) following the manufacturer’s instruction. Gene expression profile analysis of normal mature B cells (naive, memory, centroblasts, and centrocytes) and primary CLL cases was performed using Affymetrix HG-U133Plus 2.0 arrays. The cRNA was labeled and hybridized to the array according to the manufacturer’s protocol. The data were analyzed with Microarray Suite version 5.0 (MAS 5.0) using Affymetrix default analysis settings and global scaling as normalization method. Probe sets that had a mean (mu) <50, or a coefficient of variation (CV) <0.3, as assessed in a panel of 259 independent B cell–related gene expression profiles analyzed on the same array, were filtered out and considered not informative. Genes found mutated in the WES analysis were considered expressed if the corresponding probes were called “Present” in the CLL case carrying the mutation and/or in >90% of CLL cases from an independent panel hybridized to the same platform (n = 16).
Backtracking of RS-associated NOTCH1 mutation in the paired CLL diagnostic sample through 454-ultradeep sequencing.
Oligonucleotides containing the NOTCH1 specific sequences, along with the 10-bp MID tag and amplicon library A and B sequencing adapters, were used to amplify the 349-bp target region of NOTCH1 exon 34 carrying the recurrent NOTCH1 mutation ΔCT(7544–7545), P2515fs (see Results) in the CLL and RS phases from case 63. PCR primers and conditions are available upon request. Purified PCR products were quantitated, diluted, pooled, and amplified by emulsion PCR. The obtained amplicon library was subjected to ultradeep sequencing on the Genome Sequencer Junior instrument (454 Life Sciences), according to the manufacturer’s instructions. The obtained sequencing reads were mapped to the reference sequence by Amplicon Variant Analyzer (Roche).
To identify subclonal variants in the CLL phase, we used the BackTracker algorithm, developed in the Rabadan laboratory at Columbia University. We used the long-read component of BWA aligner (Li and Durbin, 2010) to map the reads to the NOTCH1 reference genome (hg18). Next, using in-house developed software for variant detection, we compiled a list of substitutions, insertions, and deletions. Positions with Phred score below 30 were removed. For deletions, we assigned the mean Phred score of the neighboring mapped positions. Excluding variants not seen in reads mapped to both forward and reverse strands, we fitted a negative binomial or Luria-Delbruck distribution to variant depths, for which we applied a nonlinear least square fit to values larger than one. Unlike the approach presented by Campbell et al. (Campbell et al., 2008), homo-polymers were not eliminated (NOTCH1 mutation was located in a homo-polymer region), no artificial depth cut-offs were imposed, and higher Phred score values were considered. Probabilities for candidate variants were corrected to take into account multiple positions (Bonferroni or Sidac). Three threshold values were established for substitutions, deletions, and insertions (Fig. 5 b, Fig. S4, and Fig. S5). In the CLL phase, we found the C deletion at position 7544 at 5.3% frequency and the T deletion at 7545 at 6.6% frequency, both passing the cutoffs.
Functional categories and pathway analysis of the mutated genes.
The genes found to be mutated in CLL were assigned to functional categories or to annotated pathways using the gene ontology Gene Ontology database (http://www.geneontology.org/), the publicly available bioinformatic tool DAVID 2008 (Database for Annotation, Visualization and Integrated Discovery; http://david.abcc.ncifcrf.gov/), and the Molecular Signatures Database from the Broad Institute (MSigDB; http://www.broadinstitute.org/gsea/msigdb/index.jsp).
Analysis of clinical correlations.
OS of the consecutive series of newly diagnosed and previously untreated CLL was measured from date of diagnosis to date of death or last follow up (censoring). Treatment free survival (TFS) was measured from date of CLL diagnosis to date of progressive and symptomatic disease requiring treatment according to IWCLL-NCI Working Group guidelines, death or last follow up (censoring; Hallek et al., 2008). Refractory disease was defined as treatment failure (stable disease or progressive disease during treatment) or disease progression within 6 mo from antileukemic therapy (Hallek et al., 2008). OS of the chemorefractory CLL cohort was measured from date of chemorefractoriness to date of death or last follow up (censoring). RS survival was measured from date of RS diagnosis to date of death or last follow up (censoring). The date of RS diagnosis was defined as the date of the diagnostic biopsy. Survival analysis was performed by the Kaplan-Meier method. Multivariate analysis was performed by the Cox regression model. All statistical tests were two-sided. Statistical significance was defined as P value <0.05. The analysis was performed with the Statistical Package for the Social Sciences (SPSS) software v.18.0 (Chicago, IL).
Accession nos.
The WES data and the SNP array data from the five discovery CLL cases will be deposited in dbGaP under accession no. phs000364.v1.p1.
Online supplemental material.
Fig. S1 shows the approach used for WES analysis. Fig. S2 shows the mutation spectrum in the five CLL discovery cases. Fig. S3 shows the known CLL-associated copy number aberrations identified by SNP array analysis in the 5 CLL discovery cases. Figs. S4 and S5 show the results of the ultradeep 454 DNA sequencing analysis of NOTCH1 in one CLL/RS pair. Table S1 summarizes the features of the 5 CLL discovery cases. Table S2 shows the results of the 454 sequencing and mapping after whole-exome capture in the five CLL discovery cases. Table S3 lists the validated somatic mutations identified by WES in the CLL discovery panel. Table S4 lists the tumor-acquired copy number changes identified in the five CLL discovery cases. Tables S5, S6, S8, and S9 summarize the clinical and biological characteristics of the CLL screening panel, CLL extension panel, RS cohort, and chemorefractory CLL cohort, respectively. Table S7 lists the NOTCH1 mutations found in the study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20110921/DC1.
Acknowledgments
We would like to thank T. Palomero, E. Tzilianos, V. Miljkovic, and the Genomics Technologies Shared Resource of the Herbert Irving Comprehensive Cancer Center at Columbia University and D. Burgess at Roche NimbleGen and 454 Life Sciences for assistance with the whole-exome capture and sequencing procedure. We would also like to thank M. Iacono at Roche Italy for assistance with the Roche 454 Junior backtrack experiments.
This study was supported by National Institutes of Health grant CA-37295 (to R. Della-Favera); the Italian Association for Cancer Research, Special Program Molecular Clinical Oncology, 5 x 1000, no. 10007, Milan, Italy (to G. Gaidano and to R. Foa); Progetto FIRB-Programma “Futuro in Ricerca” 2008 (to D. Rossi) and PRIN 2008 (to G. Gaidano), MIUR, Rome, Italy; Progetto Giovani Ricercatori 2008, Ministero della Salute, Rome, Italy (to D. Rossi); Novara-AIL Onlus, Novara (to G. Gaidano); Helmut Horten Foundation and San Salvatore Foundation (to F. Bertoni). The work of V. Trifonov, H. Khiabanian, and R. Rabadan is supported by the Northeast Biodefence Center (U54-AI057158), the National Institutes of Health (U54 CA121852-05), and the National Library of Medicine (1R01LM010140-01). S. Monti and S. Cresta are being supported by fellowships from Novara-AIL Onlus. L. Pasqualucci is on leave from the Institute of Hematology, University of Perugia Medical School.
The authors have no competing financial interests.
Footnotes
Abbreviations used:
- BIR
- baculovirus IAP repeat
- CLL
- chronic lymphocytic leukemia
- CNA
- copy number alteration
- DLBCL
- diffuse large B cell lymphoma
- RS
- Richter syndrome
- SNP
- single-nucleotide polymorphism
- T-ALL
- T cell acute lymphoblastic leukemia
- WES
- whole-exome sequencing
References
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. 2010. A method and server for predicting damaging missense mutations. Nat. Methods. 7:248–249 10.1038/nmeth0410-248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albesiano E., Messmer B.T., Damle R.N., Allen S.L., Rai K.R., Chiorazzi N. 2003. Activation-induced cytidine deaminase in chronic lymphocytic leukemia B cells: expression as multiple forms in a dynamic, variably sized fraction of the clone. Blood. 102:3333–3339 10.1182/blood-2003-05-1585 [DOI] [PubMed] [Google Scholar]
- Annunziata C.M., Davis R.E., Demchenko Y., Bellamy W., Gabrea A., Zhan F., Lenz G., Hanamura I., Wright G., Xiao W., et al. 2007. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 12:115–130 10.1016/j.ccr.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster J.C., Pear W.S., Blacklow S.C. 2008. Notch signaling in leukemia. Annu. Rev. Pathol. 3:587–613 10.1146/annurev.pathmechdis.3.121806.154300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aster J.C., Blacklow S.C., Pear W.S. 2011. Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223:262–273 10.1002/path.2789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calin G.A., Dumitru C.D., Shimizu M., Bichi R., Zupo S., Noch E., Aldler H., Rattan S., Keating M., Rai K., et al. 2002. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA. 99:15524–15529 10.1073/pnas.242606799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P.J., Pleasance E.D., Stephens P.J., Dicks E., Rance R., Goodhead I., Follows G.A., Green A.R., Futreal P.A., Stratton M.R. 2008. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl. Acad. Sci. USA. 105:13081–13086 10.1073/pnas.0801523105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.A., Lawrence M.S., Keats J.J., Cibulskis K., Sougnez C., Schinzel A.C., Harview C.L., Brunet J.P., Ahmann G.J., Adli M., et al. 2011. Initial genome sequencing and analysis of multiple myeloma. Nature. 471:467–472 10.1038/nature09837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiorazzi N., Rai K.R., Ferrarini M. 2005. Chronic lymphocytic leukemia. N. Engl. J. Med. 352:804–815 10.1056/NEJMra041720 [DOI] [PubMed] [Google Scholar]
- Damle R.N., Wasil T., Fais F., Ghiotto F., Valetto A., Allen S.L., Buchbinder A., Budman D., Dittmar K., Kolitz J., et al. 1999. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 94:1840–1847 [PubMed] [Google Scholar]
- Delker R.K., Fugmann S.D., Papavasiliou F.N. 2009. A coming-of-age story: activation-induced cytidine deaminase turns 10. Nat. Immunol. 10:1147–1153 10.1038/ni.1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Ianni M., Baldoni S., Rosati E., Ciurnelli R., Cavalli L., Martelli M.F., Marconi P., Screpanti I., Falzetti F. 2009. A new genetic lesion in B-CLL: a NOTCH1 PEST domain mutation. Br. J. Haematol. 146:689–691 10.1111/j.1365-2141.2009.07816.x [DOI] [PubMed] [Google Scholar]
- Döhner H., Stilgenbauer S., Benner A., Leupolt E., Kröber A., Bullinger L., Döhner K., Bentz M., Lichter P. 2000. Genomic aberrations and survival in chronic lymphocytic leukemia. N. Engl. J. Med. 343:1910–1916 10.1056/NEJM200012283432602 [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D., Ying C.Y., Grandori C., Ruggiero L., Chen B., Li M., Galloway D.A., Gu W., Gautier J., Dalla-Favera R. 2007. Non-transcriptional control of DNA replication by c-Myc. Nature. 448:445–451 10.1038/nature05953 [DOI] [PubMed] [Google Scholar]
- Eischen C.M., Weber J.D., Roussel M.F., Sherr C.J., Cleveland J.L. 1999. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in Myc-induced lymphomagenesis. Genes Dev. 13:2658–2669 10.1101/gad.13.20.2658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fangazio M., De Paoli L., Rossi D., Gaidano G. 2011. Predictive markers and driving factors behind Richter syndrome development. Expert Rev. Anticancer Ther. 11:433–442 10.1586/era.10.237 [DOI] [PubMed] [Google Scholar]
- Forbes S.A., Bindal N., Bamford S., Cole C., Kok C.Y., Beare D., Jia M., Shepherd R., Leung K., Menzies A., et al. 2011. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 39:D945–D950 10.1093/nar/gkq929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabher C., von Boehmer H., Look A.T. 2006. Notch 1 activation in the molecular pathogenesis of T-cell acute lymphoblastic leukaemia. Nat. Rev. Cancer. 6:347–359 10.1038/nrc1880 [DOI] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., Davies H., Teague J., Butler A., Stevens C., et al. 2007. Patterns of somatic mutation in human cancer genomes. Nature. 446:153–158 10.1038/nature05610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubor V., Krasnitz A., Troge J.E., Meth J.L., Lakshmi B., Kendall J.T., Yamrom B., Alex G., Pai D., Navin N., et al. 2009. Novel genomic alterations and clonal evolution in chronic lymphocytic leukemia revealed by representational oligonucleotide microarray analysis (ROMA). Blood. 113:1294–1303 10.1182/blood-2008-05-158865 [DOI] [PubMed] [Google Scholar]
- Gyrd-Hansen M., Meier P. 2010. IAPs: from caspase inhibitors to modulators of NF-kappaB, inflammation and cancer. Nat. Rev. Cancer. 10:561–574 10.1038/nrc2889 [DOI] [PubMed] [Google Scholar]
- Hallek M., Cheson B.D., Catovsky D., Caligaris-Cappio F., Dighiero G., Döhner H., Hillmen P., Keating M.J., Montserrat E., Rai K.R., Kipps T.J.; International Workshop on Chronic Lymphocytic Leukemia 2008. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 111:5446–5456 10.1182/blood-2007-06-093906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamblin T.J., Davis Z., Gardiner A., Oscier D.G., Stevenson F.K. 1999. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 94:1848–1854 [PubMed] [Google Scholar]
- Huh Y.O., Lin K.I., Vega F., Schlette E., Yin C.C., Keating M.J., Luthra R., Medeiros L.J., Abruzzo L.V. 2008. MYC translocation in chronic lymphocytic leukaemia is associated with increased prolymphocytes and a poor prognosis. Br. J. Haematol. 142:36–44 10.1111/j.1365-2141.2008.07152.x [DOI] [PubMed] [Google Scholar]
- Jones S., Zhang X., Parsons D.W., Lin J.C., Leary R.J., Angenendt P., Mankoo P., Carter H., Kamiyama H., Jimeno A., et al. 2008. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 321:1801–1806 10.1126/science.1164368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keats J.J., Fonseca R., Chesi M., Schop R., Baker A., Chng W.J., Van Wier S., Tiedemann R., Shi C.X., Sebag M., et al. 2007. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell. 12:131–144 10.1016/j.ccr.2007.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Dalla-Favera R. 2010. New insights into the pathogenesis of chronic lymphocytic leukemia. Semin. Cancer Biol. 20:377–383 10.1016/j.semcancer.2010.10.012 [DOI] [PubMed] [Google Scholar]
- Klein U., Lia M., Crespo M., Siegel R., Shen Q., Mo T., Ambesi-Impiombato A., Califano A., Migliazza A., Bhagat G., Dalla-Favera R. 2010. The DLEU2/miR-15a/16-1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 17:28–40 10.1016/j.ccr.2009.11.019 [DOI] [PubMed] [Google Scholar]
- Kujawski L., Ouillette P., Erba H., Saddler C., Jakubowiak A., Kaminski M., Shedden K., Malek S.N. 2008. Genomic complexity identifies patients with aggressive chronic lymphocytic leukemia. Blood. 112:1993–2003 10.1182/blood-2007-07-099432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.Y., Kumano K., Nakazaki K., Sanada M., Matsumoto A., Yamamoto G., Nannya Y., Suzuki R., Ota S., Ota Y., et al. 2009. Gain-of-function mutations and copy number increases of Notch2 in diffuse large B-cell lymphoma. Cancer Sci. 100:920–926 10.1111/j.1349-7006.2009.01130.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G., Wright G.W., Emre N.C., Kohlhammer H., Dave S.S., Davis R.E., Carty S., Lam L.T., Shaffer A.L., Xiao W., et al. 2008. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc. Natl. Acad. Sci. USA. 105:13520–13525 10.1073/pnas.0804295105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley T.J., Mardis E.R., Ding L., Fulton B., McLellan M.D., Chen K., Dooling D., Dunford-Shore B.H., McGrath S., Hickenbotham M., et al. 2008. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 456:66–72 10.1038/nature07485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 26:589–595 10.1093/bioinformatics/btp698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Sanda T., Look A.T., Novina C.D., von Boehmer H. 2011. Repression of tumor suppressor miR-451 is essential for NOTCH1-induced oncogenesis in T-ALL. J. Exp. Med. 208:663–675 10.1084/jem.20102384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis E.R., Ding L., Dooling D.J., Larson D.E., McLellan M.D., Chen K., Koboldt D.C., Fulton R.S., Delehaunty K.D., McGrath S.D., et al. 2009. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 361:1058–1066 10.1056/NEJMoa0903840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maystadt I., Rezsöhazy R., Barkats M., Duque S., Vannuffel P., Remacle S., Lambert B., Najimi M., Sokal E., Munnich A., et al. 2007. The nuclear factor kappaB-activator gene PLEKHG5 is mutated in a form of autosomal recessive lower motor neuron disease with childhood onset. Am. J. Hum. Genet. 81:67–76 10.1086/518900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullighan C.G., Goorha S., Radtke I., Miller C.B., Coustan-Smith E., Dalton J.D., Girtman K., Mathew S., Ma J., Pounds S.B., et al. 2007. Genome-wide analysis of genetic alterations in acute lymphoblastic leukaemia. Nature. 446:758–764 10.1038/nature05690 [DOI] [PubMed] [Google Scholar]
- Ngo V.N., Young R.M., Schmitz R., Jhavar S., Xiao W., Lim K.-H., Kohlhammer H., Xu W., Yang Y., Zhao H., et al. 2011. Oncogenically active MYD88 mutations in human lymphoma. Nature. 470:115–119 10.1038/nature09671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J., Calvo J., McKenna K., Krishnamoorthy V., Aster J.C., Bassing C.H., Alt F.W., Kelliher M., Look A.T. 2006. Activating Notch1 mutations in mouse models of T-ALL. Blood. 107:781–785 10.1182/blood-2005-06-2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., Hardwick J., Welcker M., Meijerink J.P., Pieters R., et al. 2007. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J. Exp. Med. 204:1813–1824 10.1084/jem.20070876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paganin M., Ferrando A. 2011. Molecular pathogenesis and targeted therapies for NOTCH1-induced T-cell acute lymphoblastic leukemia. Blood Rev. 25:83–90 10.1016/j.blre.2010.09.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palomero T., Lim W.K., Odom D.T., Sulis M.L., Real P.J., Margolin A., Barnes K.C., O’Neil J., Neuberg D., Weng A.P., et al. 2006. NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional network promoting leukemic cell growth. Proc. Natl. Acad. Sci. USA. 103:18261–18266 10.1073/pnas.0606108103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons D.W., Li M., Zhang X., Jones S., Leary R.J., Lin J.C., Boca S.M., Carter H., Samayoa J., Bettegowda C., et al. 2011. The genetic landscape of the childhood cancer medulloblastoma. Science. 331:435–439 10.1126/science.1198056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L., Dominguez-Sola D., Chiarenza A., Fabbri G., Grunn A., Trifonov V., Kasper L.H., Lerach S., Tang H., Ma J., et al. 2011. Inactivating mutations of acetyltransferase genes in B-cell lymphoma. Nature. 471:189–195 10.1038/nature09730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance E.D., Cheetham R.K., Stephens P.J., McBride D.J., Humphray S.J., Greenman C.D., Varela I., Lin M.L., Ordóñez G.R., Bignell G.R., et al. 2010a. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 463:191–196 10.1038/nature08658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleasance E.D., Stephens P.J., O’Meara S., McBride D.J., Meynert A., Jones D., Lin M.-L., Beare D., Lau K.W., Greenman C., et al. 2010b. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 463:184–190 10.1038/nature08629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pounds S., Cheng C., Mullighan C., Raimondi S.C., Shurtleff S., Downing J.R. 2009. Reference alignment of SNP microarray signals for copy number analysis of tumors. Bioinformatics. 25:315–321 10.1093/bioinformatics/btn624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real P.J., Tosello V., Palomero T., Castillo M., Hernando E., de Stanchina E., Sulis M.L., Barnes K., Sawai C., Homminga I., et al. 2009. Gamma-secretase inhibitors reverse glucocorticoid resistance in T cell acute lymphoblastic leukemia. Nat. Med. 15:50–58 10.1038/nm.1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosebeck S., Madden L., Jin X., Gu S., Apel I.J., Appert A., Hamoudi R.A., Noels H., Sagaert X., Van Loo P., et al. 2011. Cleavage of NIK by the API2-MALT1 fusion oncoprotein leads to noncanonical NF-kappaB activation. Science. 331:468–472 10.1126/science.1198946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi D., Gaidano G. 2009. Richter syndrome: molecular insights and clinical perspectives. Hematol. Oncol. 27:1–10 10.1002/hon.880 [DOI] [PubMed] [Google Scholar]
- Rossi D., Spina V., Cerri M., Rasi S., Deambrogi C., De Paoli L., Laurenti L., Maffei R., Forconi F., Bertoni F., et al. 2009. Stereotyped B-cell receptor is an independent risk factor of chronic lymphocytic leukemia transformation to Richter syndrome. Clin. Cancer Res. 15:4415–4422 10.1158/1078-0432.CCR-08-3266 [DOI] [PubMed] [Google Scholar]
- Rossi D., Spina V., Deambrogi C., Rasi S., Laurenti L., Stamatopoulos K., Arcaini L., Lucioni M., Rocque G.B., Xu-Monette Z.Y., et al. 2011. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 117:3391–3401 10.1182/blood-2010-09-302174 [DOI] [PubMed] [Google Scholar]
- Schmitt C.A., Fridman J.S., Yang M., Baranov E., Hoffman R.M., Lowe S.W. 2002. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 1:289–298 10.1016/S1535-6108(02)00047-8 [DOI] [PubMed] [Google Scholar]
- Sportoletti P., Baldoni S., Cavalli L., Del Papa B., Bonifacio E., Ciurnelli R., Bell A.S., Di Tommaso A., Rosati E., Crescenzi B., et al. 2010. NOTCH1 PEST domain mutation is an adverse prognostic factor in B-CLL. Br. J. Haematol. 151:404–406 10.1111/j.1365-2141.2010.08368.x [DOI] [PubMed] [Google Scholar]
- Staudt L.M. 2010. Oncogenic activation of NF-kappaB. Cold Spring Harb. Perspect. Biol. 2:a000109 10.1101/cshperspect.a000109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulis M.L., Williams O., Palomero T., Tosello V., Pallikuppam S., Real P.J., Barnes K., Zuurbier L., Meijerink J.P., Ferrando A.A. 2008. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 112:733–740 10.1182/blood-2007-12-130096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swerdlow S.H., Campo E., Harris N.L., Jaffe E.S., Pileri S.A., Stein H., Thiele J., Vardiman J.W., 2008. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. International Agency for Research on Cancer (IARC), Lyon: 439 pp [Google Scholar]
- Tam C.S., Keating M.J. 2010. Chemoimmunotherapy of chronic lymphocytic leukemia. Nat Rev Clin Oncol. 7:521–532 10.1038/nrclinonc.2010.101 [DOI] [PubMed] [Google Scholar]
- Thompson B.J., Buonamici S., Sulis M.L., Palomero T., Vilimas T., Basso G., Ferrando A., Aifantis I. 2007. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J. Exp. Med. 204:1825–1835 10.1084/jem.20070872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsimberidou A.M., Keating M.J. 2005. Richter syndrome: biology, incidence, and therapeutic strategies. Cancer. 103:216–228 10.1002/cncr.20773 [DOI] [PubMed] [Google Scholar]
- Weng A.P., Ferrando A.A., Lee W., Morris J.P., IV, Silverman L.B., Sanchez-Irizarry C., Blacklow S.C., Look A.T., Aster J.C. 2004. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 306:269–271 10.1126/science.1102160 [DOI] [PubMed] [Google Scholar]
- Zenz T., Eichhorst B., Busch R., Denzel T., Häbe S., Winkler D., Bühler A., Edelmann J., Bergmann M., Hopfinger G., et al. 2010a. TP53 mutation and survival in chronic lymphocytic leukemia. J. Clin. Oncol. 28:4473–4479 10.1200/JCO.2009.27.8762 [DOI] [PubMed] [Google Scholar]
- Zenz T., Mertens D., Küppers R., Döhner H., Stilgenbauer S. 2010b. From pathogenesis to treatment of chronic lymphocytic leukaemia. Nat. Rev. Cancer. 10:37–50 [DOI] [PubMed] [Google Scholar]
- Zenz T., Vollmer D, Trbusek M, Smardova J, Benner A, Soussi T, Helfrich H, Heuberger M, Hoth P, Fuge M, et al. 2010c. TP53 mutation profile in chronic lymphocytic leukemia: evidence for a disease specific profile from a comprehensive analysis of 268 mutations. Leukemia. 24:2071–2079 [DOI] [PubMed] [Google Scholar]
- Zhou H., Du M.Q., Dixit V.M. 2005. Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell. 7:425–431 10.1016/j.ccr.2005.04.012 [DOI] [PubMed] [Google Scholar]