Abstract
Rationale
Although multiple lines of evidence suggest variable expression of the cardiac sodium channel gene SCN5A plays a role in susceptibility to arrhythmia, little is known about its transcriptional regulation.
Objective
We used in silico and in vitro experiments to identify possible non-coding sequences important for transcriptional regulation of SCN5A. The results were extended to mice in which a putative regulatory region was deleted.
Methods and Results
We identified 92 non-coding regions highly conserved (>70%) between human and mouse SCN5A orthologs. Three conserved non-coding sequences (CNS) showed significant (>5-fold) activity in luciferase assays. Further in vitro studies indicated one, CNS28 in intron 1, as potential regulatory region. Using Recombinase-Mediated Cassette Exchange (RMCE), we generated mice in which a 435 bp region encompassing CNS28 was removed. Animals homozygous for the deletion showed significant increases in SCN5A transcripts, NaV1.5 protein abundance, and sodium current measured in isolated ventricular myocytes. ECGs revealed a significantly shorter QRS (10.7±0.2ms in controls vs. 9.7±0.2ms in knockouts) indicating more rapid ventricular conduction. In vitro analysis of CNS28 identified a short 3′ segment within this region required for regulatory activity and including an E-box motif. Deletion of this segment reduced reporter activity to 3.6±0.3% of baseline in CHO cells and 16±3% in myocytes (both P<0.05), and mutation of individual sites in the E-box restored activity to 62±4% and 57±2% of baseline in CHO cells and myocytes, respectively (both P<0.05).
Conclusions
These findings establish that regulation of cardiac sodium channel expression modulates channel function in vivo, and identify a non-coding region underlying this regulation.
Keywords: Gene Expression Regulation, Sodium Channels, Mice, Transgenic
Introduction
Normal function of the sodium channel encoded by SCN5A is critical to initiation of the action potential and its propagation in atrium and ventricle.1 Mutations that decrease sodium current (INa) by disrupting channel processing or function cause a series of overlapping human arrhythmia syndromes, including Brugada Syndrome and conduction system disease.1,2 In subjects of Asian ancestry, we have described a common variant in the SCN5A core promoter that modulates the duration of the QRS interval, an index of ventricular conduction, in normal subjects.3 In addition, the promoter variant appeared to modulate the extent to which drug challenge prolonged QRS in patients with the Brugada Syndrome. Notably, QRS prolongation is a hallmark of sodium channel block by drugs, and sodium channel blockers are well-recognized to have proarrhythmic potential.4
Taken together, these findings implicate variability in SCN5A expression as a mechanism underlying arrhythmia susceptibility in the whole heart. To date, few studies have addressed mechanisms underlying transcriptional control of SCN5A expression. We have previously identified the core promoter of human SCN5A and common polymorphisms in that region.5,6 Others have reported that transgenic cardiac expression of the putative repressor Snail led to decreased INa and dilated cardiomyopathy; further experiments suggested Scn5a is a Snail target.7 Snail is zinc-finger transcription factor known to target E-box motifs.8 Shang and Dudley reported multiple alternate 5′-splice variants of the murine sodium channel ortholog; these were developmentally regulated and both enhancer and repressor regulatory elements and an alternate promoter were identified.9
In this report, we first identified short sequences highly conserved between mouse and human. Further studies implicated one of these conserved non-coding sequences (CNS), designated CNS28 and located ~1.3 kb upstream of exon 2, as a potential regulator of channel expression. To further test this hypothesis in vivo, we determined the electrophysiologic properties of mice in which the CNS28 region was deleted. We find that the absence of CNS28 results in striking increases in sodium channel expression in the intact heart, with attendant increased sodium current and conduction. Additional experiments in heterologous cells and cardiomyocytes implicate the loss of an E-box binding site as responsible for this increase in sodium channel expression.
Materials and Methods (details for each method are presented in the on-line supplement)
Identification of potential regulatory regions
To identify CNS elements, we compared the human SCN5A locus with its mouse ortholog using the VISTA Genome Browser (http://pipeline.lbl.gov/cgi-bin/gateway2).10,11 Each of 92 CNS elements identified was then PCR amplified and assayed for activity as described below and in the Supplement. Those showing >5-fold increase in reporter activity in luciferase assays were then analyzed for potential muscle-specific transcriptional regulatory modules using the M-SCAN algorithm (http://www.cisreg.ca/cgi-bin/mscan/MSCAN).12,13 For identification of potential repressive transcription factors in CNS28 we used rVista (http://rvista.dcode.org/) to compare human and mouse sequences for conserved transcription factor binding sites.14,15
Reporter constructs
Reporter constructs measuring the activity of all 92 CNS constructs (Online Table I), human CNS28 with the full length human SCN5A promoter (Table 1), and deletion analysis of the alternate mouse Scn5a promoter (Table 1) were generated by cloning PCR fragments into the pGL3-Promoter or pGL3-Basic vectors (Promega). Mutagenesis of the DC3 deletion fragment was performed using the QuickChange XL II kit (Stratagene) using the primers listed in Table 1. Reporter assays were conducted as previously described in CHO cells5,6 and in cardiomyocytes5,6,16 isolated from 1–2 day old mice.
TABLE 1.
PCR and Mutagenesis Primers
| Primer | Sequence | |
|---|---|---|
| CNS28−/− mouse genotyping | F | ATGGAGGCCAAAGGTCAGCTTGCAG |
| R | TGAGCATGTTGAAGAGCGAGTGAACCAG | |
| Human CNS28 for pGL3-SCN5A Promoter | F_ SalI | TGAGGTACCGTTCTGAATCTTTTGAGGCC |
| R_ BamHI | TCAAGATCTGATTCTAAAGACGGGAAATG | |
| Alternate mouse promoter deletion constructs | DC0-F | TGAGGTACCCTTATAGGGGTCACTAATGACATGCC |
| DC1-F | TGAGGTACCCCCTTGGCTGACAGGAAGAGAGTGTG | |
| DC2-F | TGAGGTACCGTGGTAATTAGCGGTGCAGCCTCCT | |
| DC3-F | TGAGGTACCGTTCTGAATCTTTTGAGGCCACCAGG | |
| DC4-F | TGAGGTACCAGTCTAGCTAGGGACGGTGCTGC | |
| DC-R | TCAAGATCTCACAGGCTCTCCTCAGGCTGCCT | |
| E-Box Mutagenesis | F | ACGTCACACACTTAAGCCTGTTGGAAGTCC |
| R | GGACTTCCAACAGGCTTAAGTGTGTGACGT | |
Generation of CNS28−/− mice
We used mouse embryonic stem (ES) cells in which a region of the Scn5a gene was modified to allow Recombinase-Mediated Cassette Exchange (RMCE)17,18 to be used to easily generate animals containing allelic variants of human sodium channels under control of Scn5a regulatory sequences. Using this approach, we previously generated H/H mice in which the targeted region was replaced by the human SCN5A full-length cDNA.19 For the present experiments, we modified the original H exchange construct to delete bases 1720 to 2154 (that encompass the CNS28 region), and then performed RMCE as previously described to generate CNS28+/− animals.19 After removal of the hygromycin resistance cassette by breeding to a FlpE-expressing line, mice were then interbred to generate CNS28−/− animals. Experiments began after three backcrosses using littermate H/H mice as controls. All experiments performed on mice were approved by the institutional animal care and use committee.
Quantitative Real Time RT-PCR
Total mRNA from adult mice atria or ventricles was isolated using the TRIzol method (Invitrogen) and cDNA was prepared using the Transcriptor First Strand cDNA Synthesis kit (Roche). Quantitative Real Time RT-PCR (qPCR) on SCN5A was performed with TaqMan probes targeting human SCN5A (Hs00165693_m1) using beta actin (Mm00607939_s1) as a reference gene. qPCR targeting mouse Scn1b was performed using TaqMan probe (Mm00441210_m1) with Hprt1 (Mm00446968_m1) as the reference gene. Analysis was performed using SDS 2.2.2 software (Applied Biosystems).
Western blotting
Western blotting protocol and semi-quantitative protein analysis are described in the Online Supplement.
Electrocardiogram (ECG) recordings and lidocaine challenge
ECGs and drug challenges were recorded as previously described.20
Isolation of mouse ventricular cardiomyocytes and sodium current recordings
Adult H/H and CNS28−/− mouse cardiac ventricular myocytes were isolated by a modified collagenase/protease method,21 and sodium current studied as previously described.19 Protocols and data analysis are presented in the Online Supplement.
Echocardiogram
Transthoracic echocardiograms were performed on resting conscious mice at the Murine Cardiovascular Core, Vanderbilt University as previously described.22 Signals were acquired using a 15-MHz transducer (Sonos 5500 system, Agilent) and analyzed by a sonographer who was blind to the genotype.
Data analysis
Results are presented as mean±SE, and statistical comparisons were made using the unpaired Student’s t test. A value of P<0.05 was considered statistically significant.
Results
Conserved nucleotide sequences
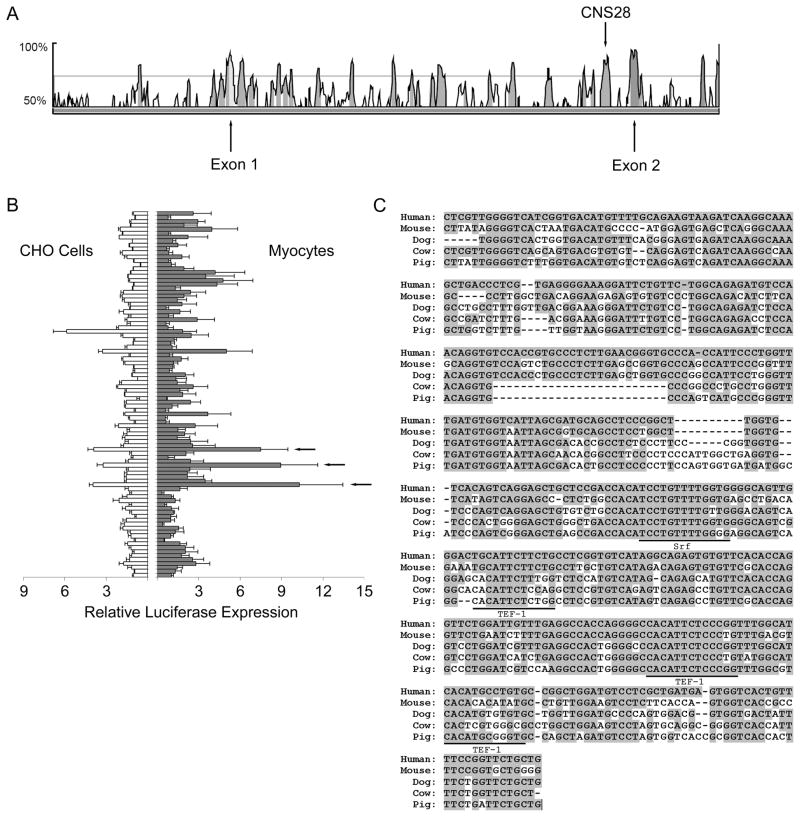
In order to identify genomic elements that may play a role in the transcriptional regulation of SCN5A, we compared human and mouse SCN5A sequences to identify conserved regions. Using the genomic sequence comparison program VISTA, we identified 92 CNS elements 77–446 bp long with >70% nucleotide identity between the human locus and its mouse ortholog. 12/92 CNS elements were identified in the 47.7 kb 5′ upstream region between SCN5A exon 1 and the terminal exon of SCN10A gene, which is just upstream, and 15/92 CNS elements were identified in the 26.4kb 3′ downstream region between the terminal exon (exon 28) of SCN5A and the exon 1 of the downstream gene ENDOGL1. The highest density was in intron 1 where 23 CNS elements were contained in 16kb (Figure 1A). 10 introns contained no CNS elements.
Figure 1.
Identification of CNS28. A. A portion of the VISTA human to mouse sequence comparison for SCN5A with the locations of Exon 1, Exon 2, and CNS28 marked. The peaks show regions of high conservation across species. Peaks that are shaded pink satisfy the CNS selection criteria of 70% identity between sequences. B. Initial luciferase experiments of all 92 CNS constructs in CHO cells and myocytes with CNS23, CNS28, and CNS32 marked by arrows. C. Human CNS28 aligned with mouse, dog, cow, and pig orthologs with the TEF-1 and SRF sites suggested by MSCAN marked.
We tested the effect of each CNS on the SV40 promoter driving luciferase both in Chinese hamster ovary (CHO) cells and cardiomyocytes. There were three elements, CNS23, CNS28 and CNS32, that demonstrated a >5-fold increase in reporter activity (Figure 1B). The MSCAN analysis, which we used to identify potential functionally important clusters of muscle-specific transcription factors, designated 3 tandem TEF-1 (Transcriptional Enhancer Factor 1) sites and a SRF (Serum Response Factor) recognition site in CNS28 (Figure 1C), and no elements in the other two. There was a high degree of conservation across species at the SRF and one of the TEF-1 sites. Accordingly, our further studies focused on the activity of CNS28, a 435bp region located in the 5′ portion of intron 1.
Generating CNS28−/− mice
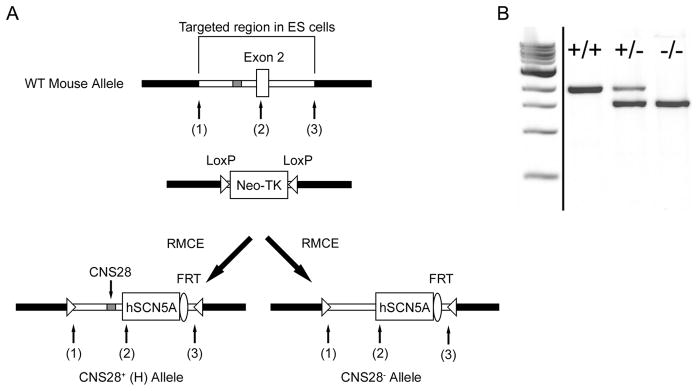
In our previous studies, we replaced a region flanking the endogenous exon 2 transcription site with a construct that contained approximately 2kb of mouse intron 1 along with the human SCN5A cDNA. In homozygous mice, designated H/H, no murine Scn5a expression was detected; ventricular myocyte sodium current and electrocardiograms (ECGs) were identical to those in wild-type animals, indicating that the human channel generated physiologic channel function.19 In the present experiments, we used site-directed mutagenesis to remove the CNS28 element from our original exchange construct (Figure 2A) and then used Recombinase Mediated Cassette Exchange (RMCE) to generate CNS28+/− mice. Loss of CNS28 was confirmed by genotyping (Figure 2B), and matings of CNS28+/− x CNS28+/− mice yielded pup distributions in Hardy Weinberg equilibrium with 32 H/H, 64 CNS28+/−, and 41 CNS28−/−.
Figure 2.
Generation of CNS28−/− mice. A. General overview showing generation of the CNS28−/− mouse using Recombinase Mediated Cassette Exchange (RMCE). Briefly, the region of the wild-type mouse Scn5a locus between sites (1) and (3), including exon 2 and portions of introns 1 and 2, was previously modified by gene targeting to contain two inversely-oriented LoxP sites flanking a cassette that contains a neomysin resistant gene and the thymidine kinase (TK) gene.19. An exchange vector that includes the human SCN5A cDNA flanked by the portions of introns 1 and 2 in the targeted regions was previously used to generate H/H mice. In the present experiment, the exchange vector was modified to remove a 435 bp fragment in intron 1 to generate the CNS28−/− mice. The 5′ end of CNS28 is located 1361 bp upstream from the start of exon 2. A hygromycin-resistance cassette in the exchanged DNA was removed by breeding to FlpE-expressing animals leaving a single residual FRT site. B. Genotyping reaction across CNS28 showing H/H, CNS28+/−, and CNS28−/−. These lanes were run on the same gel but were noncontiguous.
Sodium channel expression in CNS28−/− mice
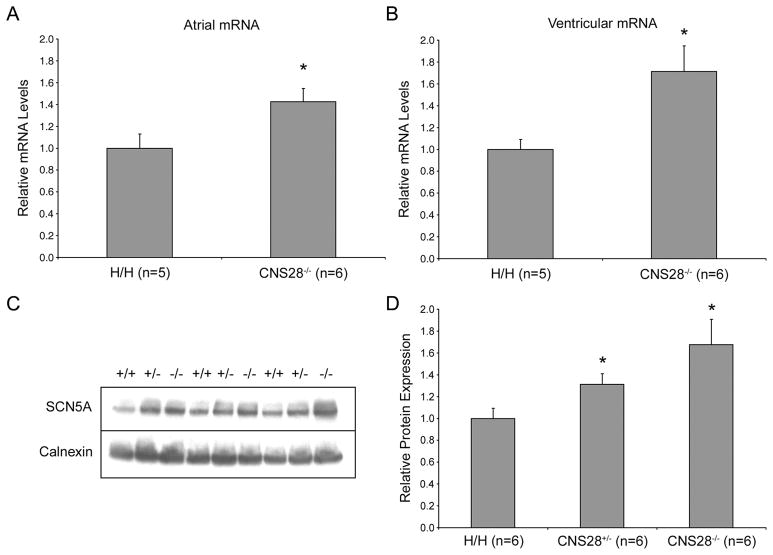
Our initial results indicated that the CNS28 fragment increased SV40-mediated reporter activity. However, in CNS28−/− mouse hearts, quantitative Real-Time PCR (qPCR), showed that SCN5A transcripts levels were 42±12% more abundant in atria and 71±23% more abundant in ventricles than in H/H mice (P<0.05 in both tissues, Figures 3A and 3B). Western blotting showed concordant results: calnexin-normalized Nav1.5 abundance in heart was 31±10% higher in CNS28+/− and 67±23% higher in CNS28−/− mice than in H/H mice (P<0.05 for both CNS28+/− and CNS28−/− when compared to H/H, Figures 3C and 3D).
Figure 3.
Relative transcript and protein amounts in H/H and CNS28−/−. SCN5A atrial mRNA (A) and ventricular mRNA (B) levels analyzed by quantitative Real-Time PCR. CNS28−/− mice had 42±12% more expression in atria and 71±23% more in ventricles compared to H/H. C. Whole heart protein expression levels analyzed by Western Blot. Bands were analyzed on the same gel. D. Band densitometry analysis of the western blots showing relative NaV1.5 amounts normalized to Calnexin. NaV1.5 abundance in heart was 31±10% higher in CNS28+/− mice and 67±23% higher in CNS28−/− mice. *P<0.05 compared to H/H.
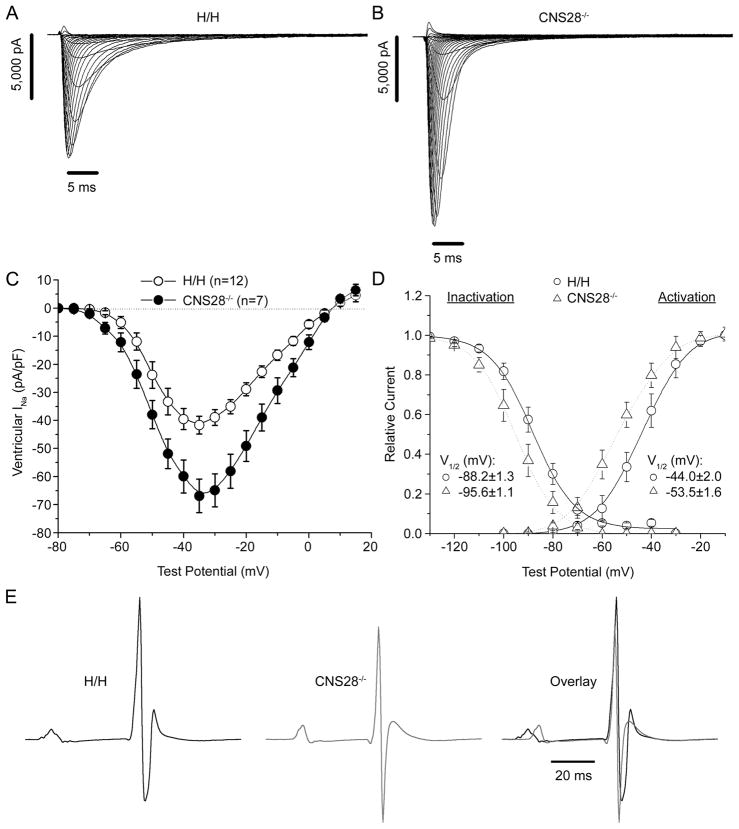
These changes translated into increases in functional sodium channels (Figures 4A and 4B). Peak INa amplitude measured at −30 mV was 59±14% greater in CNS28−/− ventricular myocytes compared to that in H/H cells (P<0.05, Figure 4C). Interestingly, we also observed small but significant negative shifts in the voltage-dependence of sodium channel activation and inactivation in CNS28−/− myocytes (Figure 4D).
Figure 4.
Increased amounts of NaV1.5 in CNS28−/− are functional. Representative traces of INa from a ventricular myocyte isolated from either an H/H (A) or CNS28−/− (B) mouse. C. Current-Voltage relationship was evaluated in H/H and CNS28−/− mice. INa amplitude measured at −30 mV was 59±14% greater in CNS28−/− ventricular myocytes compared to that in H/H cells. D. Voltage dependence of activation and inactivation measured in isolated ventricular myocytes where we observed small but significant negative shifts in the voltage-dependence of sodium channel activation and inactivation in CNS28−/−. E. Representative ECG traces averaged over 10 second intervals for H/H and CNS28−/− mice. CNS28−/− mice display 12.3% shorter PR intervals, 9.3% shorter QRS intervals, and 7.0% shorter QT intervals compared to H/H. *P<0.05 compared to H/H.
CNS28−/− mice also showed ECG changes consistent with these findings. Compared with WT, homozygotes displayed 12.3% shorter PR intervals (38.8±0.6ms vs. 34.1±0.7ms), 9.3% shorter QRS intervals (10.7±0.2ms vs. 9.7±0.2ms), and 7.0% shorter QT intervals (52.9±0.8ms vs 49.2±1.2ms) (all P<0.05, Figure 4E). After challenge with the sodium channel blocker lidocaine, these intervals increased to a similar absolute extent in both genotypes, prolonging PR by 14.1 and 13.9 ms and QRS by 4.1 and 3.2 ms in H/H and CNS28−/− respectively. Flecainide challenge showed similar results (data not shown).
In vitro transcriptional regulation by CNS28
The finding that deleting CNS28 increased channel transcripts and protein, with functional consequences, indicates that this region includes sequences that suppress sodium channel expression in vivo. This result is at odds with the initial screening experiment that identified CNS28 as a potential positive regulatory sequence.
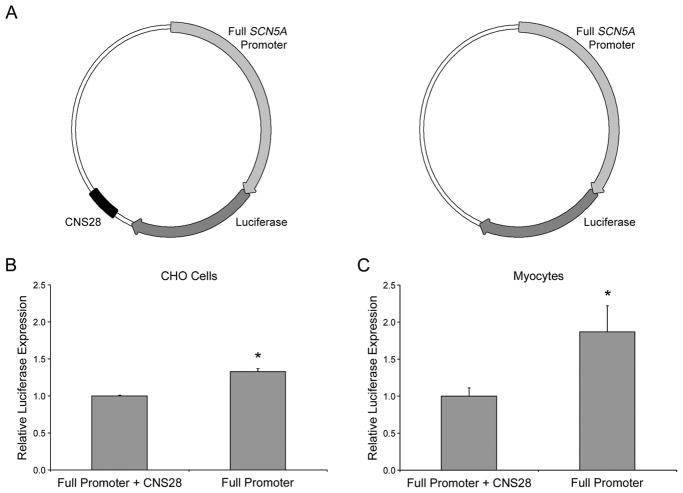
Based on these results we assessed reporter activity of constructs in which human CNS28 was included with the full length SCN5A promoter6 rather than the SV40 promoter used in the initial experiments. We compared activity of two constructs, each containing the SCN5A promoter and luciferase, one with and one without CNS28 (Figure 5A). The construct without CNS28 increased luciferase expression 33±4% in CHO cells and 87±35% in myocytes (Figures 5B and 5C, both P<0.05), consistent with the repressor function indicated by the in vivo findings.
Figure 5.
Activity of human CNS28 on the full length human SCN5A promoter. A. Graphical representation of the vectors used. Luciferase activity driven by the full length human SCN5A promoter with and without CNS28 in CHO cells (B) and neonatal cardiomyocytes (C). SCN5A promoter with CNS28 is designated 100% activity. When CNS28 is not present luciferase activity increases by 33±4% in CHO cells and 87±35% in myocytes. *P<0.05.
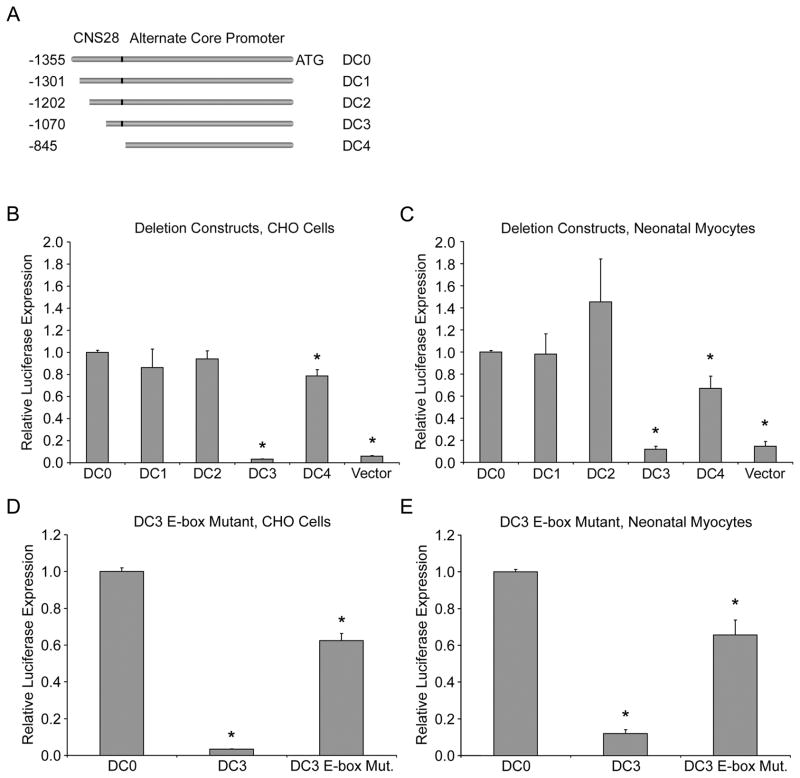
We have previously identified a region upstream of exon 1 as the core SCN5A promoter.5,6 Experiments conducted by Shang and Dudley revealed a second, 1363 bp Scn5a promoter in the mouse.9 The 5′ 435bp of this promoter are what we identified in our screen as CNS28. Their analysis suggested that the 5′ 480bp contained a repressive element.9 We hypothesized that CNS28 contained this repressive activity and generated a series of smaller deletions of the alternate promoter they described (Figure 6A). The rVista analysis identified 30 possible transcription factor binding sites conserved between the human and mouse and therefore break points were chosen between clusters of these sites (Online Figure I). The full alternate promoter-luciferase construct (1,361 bp) was designated deletion construct 0 (DC0), and contains all the features of the Shang and Dudley promoter stated above. Deletion construct 1 (DC1) removes the initial 50 bp and deletion construct 2 (DC2) an additional 100bp. Deletion construct 3 (DC3) eliminates 285 bp of the alternate promoter. Deletion construct 4 (DC4) takes away the entirety of CNS28 from the alternate promoter by deleting the first 435 bp.
Figure 6.
Progressive deletion construct analysis the alternate promoter. A. A schematic showing the progressive truncations of the CNS28 portion of the mouse alternate promoter in the luciferase constructs used. The translation start site is designated as +1. Analysis of the deletion constructs in CHO cells (B) or neonatal myocytes (C). DC0 is designated 100% activity. DC3, which deletes the majority of CNS28 from DC0, reduced reporter activity in both cell types to 3.3±0.1% of DC0 in CHO cells, and 12±3% in myocytes. Removal of the last portion of CNS28, DC4, restored activity close to baseline. Mutating the E-box consensus sequence in DC3 restores reporter activity to 62±4% of the activity of DC0 in CHO cells (D) and 66±10% in myocytes (E). *P<0.05 compared to DC0.
In our luciferase reporter assay, the activity of DC0 was used as the baseline reference in both CHO cells and myocytes. Constructs DC1 and DC2 showed no difference in activity versus DC0 in either cell type. The DC3 construct virtually abolished reporter activity in both cell types reducing activity to 3.3±0.1% of DC0 in CHO cells, and 12±3% in myocytes (both P<0.05). The last construct, DC4, restored activity to 79±6% and 67±11% of DC0 in CHO cells and myocytes respectively (Figures 6B and 6C, both P<0.05). There are two points illustrated by this data. First, between DC2 and DC3 there is likely an enhancer element that can counter the negative transcriptional activity we observe with CNS28; therefore, DC3 shows the highest repressive activity. Second, removal of 150 base pairs between DC3 and DC4 results in loss of that repressive activity. Sequence analysis of this region shows that it contains an E-box motif (consensus sequence CANNTG).23 The repressor Snail has previously been shown to bind E-boxes in the core promoter of SCN5A.7 Therefore we tested the hypothesis that this E-box may be important for repressor binding. When the motif was mutated in DC3 (CATATG to CTTAAG), activity was restored to 62±4% of the activity of DC0 in CHO cells and 66±10% in myocytes (Figures 6D and 6E, both P<0.05). This increase in activity is similar to the increase we see between DC3 and DC4 where the final segment containing the E-box is removed further supporting our hypothesis.
Discussion
Sodium channels are required for normal cardiac function
Nav1.5 expression is absolutely required for normal cardiac function; knockouts in mice and in fish are embryolethal and cause cardiac developmental abnormalities.24,25 More modest reduction of sodium current slows cardiac conduction and creates an arrhythmia-prone heart in the setting of monogenic disease26,27 or during therapy with sodium channel blocking drugs.4 We have previously found an association between a variant haplotype in a regulatory region of the cardiac sodium channel promoter commonly observed in Asian subjects and variable QRS duration, and recent genome-wide association studies have identified an association between the SCN5A-10A locus and variable PR and QRS durations.28–30 These data support the general hypothesis31,32 that variable cardiac ion channel transcription modulates the electrophysiologic properties of the heart. A contrary view is that feedback mechanisms in the mammalian heart regulate transcription to achieve a tight range of normal electrophysiologic behaviors,32 although monogenic diseases producing striking electrocardiographic changes argue against such tight control. Previous work in this area has focused on in vitro approaches and on establishing the functional consequences of coding region variants in genetically-modified mice. However, no study to date has directly evaluated the functional consequences of deleting non-coding potential regulatory regions of a cardiac ion channel gene.
CNS28 regulates cardiac sodium channel expression
Our initial in silico screen identified 92 highly conserved non-coding sequences and preliminary reporter experiments, using the SV40 promoter, suggested that one, CNS28, included positive regulatory elements for SCN5A. Driven by this initial result, we generated CNS28−/− mice and unexpectedly these animals demonstrate dramatic increases in sodium channel transcripts and in channel protein expression. This increased Nav1.5 is functional: the CNS28−/− mice have significantly larger peak INa in isolated ventricular myocytes and shorter PR and QRS, indicating rapid ventricular conduction. The increased sodium current also displayed unexpected shifts in the voltage-dependence of channel activation and inactivation; the mechanism requires further study but one possibility is a change in the extent of interaction with ancillary proteins, such as beta-subunits, that are known to modulate channel gating.33 For example, using real-time PCR we found no alterations in expression of Scn1b in CNS28−/− mouse hearts (Online Figure II). This supports the notion that while more sodium channels are present concomitant increases in function modifying protein partners may not occur. Changes in the NaV1.5 macromolecular complex could lead to alteration in channel gating. Further experiments examining changes in expression and localization of other NaV1.5 partners34 may assist in elucidating the underlying mechanism. Another possibility is raised by the clustering of Scn5a with Scn10a and 11a in the mouse and human genomes; thus, it is conceivable that removal of CNS28 impacts the level of expression of these channels in heart. Such a change in the channel profile in the heart could result in shifts in the inactivation and activation of cardiac sodium current. However, in preliminary experiments we observe no change in Scn11a expression in the two lines, and have been unable to amplify Scn10a, suggesting this channel is not abundantly expressed in heart. Importantly, heart wall measurements, examination of contractile function (Online Table II), and survival rates were no different in CNS28−/− mice compared to H/H animals.
Molecular basis for CNS28 actiivity
Our study identified a 435bp segment of DNA whose deletion increases channel expression, and reporter experiments using the more physiologically relevant SCN5A promoter are consistent with this result. Our analysis of deletion constructs demonstrates that a repressor element lies within a 226bp fragment at the 3′ end of CNS28. The five binding sites conserved between mouse and human in this region identified by rVista include: E47, E12, E2A, MYOD, and T3R. None of these factors are good candidates to act as a repressor. However, in the 226bp segment, we also identified a single E-box motif in the mouse and two in the human. Mice overexpressing Snail, which binds E-box sites in the SCN5A core promoter, exhibit a reduction in Scn5A transcript and protein.7 We observed an increase in luciferase expression following mutation of the E-box, arguing this motif is critical for repression of SCN5A. E-boxes are typically bound by basic helix-loop-helix factors, such as HAND proteins.23 However, given the previous studies linking Snail to SCN5A, our work raises the possibility that the CNS28 E-box is a Snail binding region and this interaction is needed in vivo to regulate SCN5A transcript levels.
Limitations
One limitation of our study is that in order to identify elements relevant to SCN5A transcription we choose to examine highly conserved elements between the mouse and human and replace the mouse Scn5a allele with a human SCN5A. However, in the animals we generated, potential mouse regulatory sequences, including the core promoter, are intact and therefore the effects we see following deletion of CNS28 are in the murine context and may or may not apply to humans. However, E-boxes are evolutionarily conserved in both mouse and human promoter regions so likely have similar functions in both species.
Summary
We hypothesize that increased sodium channel expression should translate to protection against arrhythmias that are mediated by decreased sodium channel function; settings in which this mechanism is thought to be operative include acute ischemia especially with sodium channel block4 and the Brugada Syndrome.3,25,35 As a first test of this hypothesis, we examined the extent to which challenge with a sodium channel blocker slowed conduction in H/H and CNS28−/− mice. In this experiment, both lidocaine and flecainide prolonged PR and QRS to a similar extent; thus, with drug challenge, absolute conduction times remained shorter in the CNS28−/− animals. The further elucidation of mechanisms affecting SCN5A transcription may point to entirely novel ways in which to intervene to stabilize cardiac electrophysiologic activity.
Supplementary Material
Novelty and Significance.
What is known?
Decreased cardiac sodium current, through mutations or drug block, increases susceptibility to brady- and tachyarrhythmias.
While in vitro experiments have identified putative regulatory elements in the cardiac sodium channel genes SCN5A, the extent to which these alter channel function in vivo is unknown
What new information does this article contribute?
We identify multiple non-coding regions in SCN5A displaying cross-species sequence conservation.
Informatic and promoter-reporter studies implicate one of these, conserved non-coding sequence 28 (CNS28), as a potential regulator of channel expression.
We have compared SCN5A expression and function in mice with humanized cardiac sodium channels to mice that are identical except they lack a 435 base pair (bp) fragment, which includes CNS28, in intron 1.
Removal of these 435 bp increases SCN5A expression and sodium current amplitude, and speeds cardiac conduction.
Deletion of a putative Snail binding region within the 435 bp element partially relieves its repressive activity.
Multiple lines of evidence suggest that reduced expression of the human cardiac sodium channel gene SCN5A can lead to arrhythmias. However, no study to date has explored the in vivo consequences of deleting potential regulatory non-coding regions of a cardiac ion gene. In this study we first identified conserved non-coding sequences (CNS) when the mouse and human genes are compared. Initial experiments identified one, CNS28, as a potential regulator of channel expression. We have previously generated mice in which the murine ortholog Scn5a is ablated and the human SCN5A cDNA expressed in its place. In the present experiments, we generated mice in which a 435 bp region encompassing CNS28 was removed. Removing CNS28 increased SCN5A mRNA, protein, and sodium current, and was associated with enhanced conduction (decreased PR and QRS intervals). Promoter-reporter experiments showed that removal of an E-box in CNS28 increased reporter activity. These findings support the hypothesis that variable expression of cardiac ion channels can alter the electrophysiologic properties of the intact heart. Understanding the mechanisms underlying ion channel regulation could lead to potential new therapies for arrhythmias.
Acknowledgments
We would like to acknowledge Wei Zhang for animal care and technical assistance.
Sources of Funding
This work was supported by grants from the United States Public Health Service (HL49989, HL65962).
Non-standard Abbreviations and Acronyms
- CHO
Chinese Hamster Ovary
- CNS
Conserved Non-coding Sequence
- ECG
Electrocardiogram
- H
Humanized SCN5A Allele
- Nav1.5
Voltage-gated Cardiac Sodium Channel
- RMCE
Recombinase Mediated Cassette Exchange
Footnotes
Disclosures
The authors have no conflicts of interest to declare.
References
- 1.George AL. Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balser JR. Inherited sodium channelopathies. novel therapeutic and proarrhythmic molecular mechanisms. Trends Cardiovasc Med. 2001;11:229–237. doi: 10.1016/s1050-1738(01)00116-5. [DOI] [PubMed] [Google Scholar]
- 3.Bezzina CR, Shimizu W, Yang P, Koopmann TT, Tanck MWT, Miyamoto Y, Kamakura S, Roden DM, Wilde AAM. Common Sodium Channel Promoter Haplotype in Asian Subjects Underlies Variability in Cardiac Conduction. Circulation. 2006;113:338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- 4.Cardiac Arrhythmia Suppression Trial (CAST) Investigators. Preliminary report: effect of encainide and flecainide on mortality in a randomized trial of arrhythmia suppression after myocardial infarction. The Cardiac Arrhythmia Suppression Trial (CAST) Investigators. N Engl J Med. 1989;321:406–412. doi: 10.1056/NEJM198908103210629. [DOI] [PubMed] [Google Scholar]
- 5.Yang P, Koopmann TT, Pfeufer A, Jalilzadeh S, Schulze-Bahr E, Kaab S, Wilde AA, Roden DM, Bezzina CR. Polymorphisms in the cardiac sodium channel promoter displaying variant in vitro expression activity. Eur J Hum Genet. 2007;16:350–357. doi: 10.1038/sj.ejhg.5201952. [DOI] [PubMed] [Google Scholar]
- 6.Yang P, Kupershmidt S, Roden DM. Cloning and initial characterization of the human cardiac sodium channel (SCN5A) promoter. Cardiovascular Research. 2004;61:56–65. doi: 10.1016/j.cardiores.2003.09.030. [DOI] [PubMed] [Google Scholar]
- 7.Hesse M, Kondo CS, Clark RB, Su L, Allen FL, Geary-Joo CTM, Kunnathu S, Severson DL, Nygren A, Giles WR, Cross JC. Dilated cardiomyopathy is associated with reduced expression of the cardiac sodium channel Scn5a. Cardiovascular Research. 2007;75:498–509. doi: 10.1016/j.cardiores.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 8.Nieto MA. The snail superfamily of zinc-finger transcription factors. Nat Rev Mol Cell Biol. 2002;3:155–166. doi: 10.1038/nrm757. [DOI] [PubMed] [Google Scholar]
- 9.Shang LL, Dudley SC. Tandem Promoters and Developmentally Regulated 5′ and 3′ mRNA Untranslated Regions of the Mouse Scn5a Cardiac Sodium Channel. Journal of Biological Chemistry. 2005;280:933–940. doi: 10.1074/jbc.M409977200. [DOI] [PubMed] [Google Scholar]
- 10.Dubchak I, Brudno M, Loots GG, Pachter L, Mayor C, Rubin EM, Frazer KA. Active Conservation of Noncoding Sequences Revealed by Three-Way Species Comparisons. Genome Research. 2000;10:1304–1306. doi: 10.1101/gr.142200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mayor C, Brudno M, Schwartz JR, Poliakov A, Rubin EM, Frazer KA, Pachter LS, Dubchak I. VISTA: visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 12.Alkema WBL, Johansson O, Lagergren J, Wasserman WW. MSCAN: identification of functional clusters of transcription factor binding sites. Nucl Acids Res. 2004;32:W195–W198. doi: 10.1093/nar/gkh387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson O, Alkema W, Wasserman WW, Lagergren J. Identification of functional clusters of transcription factor binding motifs in genome sequences: the MSCAN algorithm. Bioinformatics. 2003;19:i169–i176. doi: 10.1093/bioinformatics/btg1021. [DOI] [PubMed] [Google Scholar]
- 14.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucl Acids Res. 2004;32:W217–W221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ovcharenko I, Loots GG, Hardison RC, Miller W, Stubbs L. zPicture: Dynamic Alignment and Visualization Tool for Analyzing Conservation Profiles. Genome Research. 2004;14:472–477. doi: 10.1101/gr.2129504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lowe JS, Palygin O, Bhasin N, Hund TJ, Boyden PA, Shibata E, Anderson ME, Mohler PJ. Voltage-gated Nav channel targeting in the heart requires an ankyrin-G dependent cellular pathway. J Cell Biol. 2008;180:173–186. doi: 10.1083/jcb.200710107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones JR, Shelton KD, Magnuson MA. Strategies for the Use of Site-Specific Recombinases in Genome Engineering. In: Su GH, editor. Pancreatic Cancer. Humana Press; 2005. [DOI] [PubMed] [Google Scholar]
- 18.Seibler J, Schubeler D, Fiering S, Groudine M, Bode J. DNA Cassette Exchange in ES Cells Mediated by FLP Recombinase: An Efficient Strategy for Repeated Modification of Tagged Loci by Marker-Free ConstructsΓÇá. Biochemistry. 1998;37:6229–6234. doi: 10.1021/bi980288t. [DOI] [PubMed] [Google Scholar]
- 19.Liu K, Hipkens S, Yang T, Abraham R, Zhang W, Chopra N, Knollmann B, Magnuson MA, Roden DM. Recombinase-mediated cassette exchange to rapidly and efficiently generate mice with human cardiac sodium channels. Genesis. 2006;44:556–564. doi: 10.1002/dvg.20247. [DOI] [PubMed] [Google Scholar]
- 20.Knollmann BC, Chopra N, Hlaing T, Akin B, Yang T, Ettensohn K, Knollmann BEC, Horton KD, Weissman NJ, Holinstat I, Zhang W, Roden DM, Jones LR, Franzini-Armstrong C, Pfeifer K. Casq2 deletion causes sarcoplasmic reticulum volume increase, premature Ca2+ release, and catecholaminergic polymorphic ventricular tachycardia. J Clin Invest. 2006;116:2510–2520. doi: 10.1172/JCI29128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mitra R, Morad M. A uniform enzymatic method for dissociation of myocytes from hearts and stomachs of vertebrates. Am J Physiol. 1985;249:H1056–60. doi: 10.1152/ajpheart.1985.249.5.H1056. [DOI] [PubMed] [Google Scholar]
- 22.Rottman JN, Ni G, Brown M. Echocardiographic Evaluation of Ventricular Function in Mice. Echocardiography. 2007;24:83–89. doi: 10.1111/j.1540-8175.2006.00356.x. [DOI] [PubMed] [Google Scholar]
- 23.Massari ME, Murre C. Helix-Loop-Helix Proteins: Regulators of Transcription in Eucaryotic Organisms. Mol Cell Biol. 2000;20:429–440. doi: 10.1128/mcb.20.2.429-440.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chopra SS, Stroud DM, Watanabe H, Bennett JS, Burns CG, Wells KS, Yang T, Zhong TP, Roden DM. Voltage-Gated Sodium Channels Are Required for Heart Development in Zebrafish. Circ Res. 2010;106:1342–1350. doi: 10.1161/CIRCRESAHA.109.213132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papadatos GA, Wallerstein PMR, Head CEG, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AEO, Huang CLH, Vandenberg JI, Colledge WH, Grace AA. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Antzelevitch C, Brugada P, Borggrefe M, Brugada J, Brugada R, Corrado D, Gussak I, LeMarec H, Nademanee K, Perez Riera AR, Shimizu W, Schulze-Bahr E, Tan H, Wilde A. Brugada Syndrome: Report of the Second Consensus Conference: Endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:659–670. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 27.Kapplinger JD, Tester DJ, Alders M, Benito Ba, Berthet M, Brugada J, Brugada P, Fressart V, Guerchicoff A, Harris-Kerr C, Kamakura S, Kyndt F, Koopmann TT, Miyamoto Y, Pfeiffer R, Pollevick GD, Probst V, Zumhagen S, Vatta M, Towbin JA, Shimizu W, Schulze-Bahr E, Antzelevitch C, Salisbury BA, Guicheney P, Wilde AAM, Brugada R, Schott JJ, Ackerman MJ. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7:33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chambers JC, Zhao J, Terracciano CMN, Bezzina CR, Zhang W, Kaba R, Navaratnarajah M, Lotlikar A, Sehmi JS, Kooner MK, Deng G, Siedlecka U, Parasramka S, El-Hamamsy I, Wass MN, Dekker LRC, de Jong JSSG, Sternberg MJE, McKenna W, Severs NJ, de Silva R, Wilde AAM, Anand P, Yacoub M, Scott J, Elliott P, Wood JN, Kooner JS. Genetic variation in SCN10A influences cardiac conduction. Nat Genet. 2010;42:149–152. doi: 10.1038/ng.516. [DOI] [PubMed] [Google Scholar]
- 29.Holm H, Gudbjartsson DF, Arnar DO, Thorleifsson G, Thorgeirsson G, Stefansdottir H, Gudjonsson SA, Jonasdottir A, Mathiesen EB, Njolstad I, Nyrnes A, Wilsgaard T, Hald EM, Hveem K, Stoltenberg C, Lochen ML, Kong A, Thorsteinsdottir U, Stefansson K. Several common variants modulate heart rate, PR interval and QRS duration. Nat Genet. 2010;42:117–122. doi: 10.1038/ng.511. [DOI] [PubMed] [Google Scholar]
- 30.Pfeufer A, van Noord C, Marciante KD, Arking DE, Larson MG, Smith AV, Tarasov KV, Muller M, Sotoodehnia N, Sinner MF, Verwoert GC, Li M, Kao WHL, Kottgen A, Coresh J, Bis JC, Psaty BM, Rice K, Rotter JI, Rivadeneira F, Hofman A, Kors JA, Stricker BHC, Uitterlinden AG, van Duijn CM, Beckmann BM, Sauter W, Gieger C, Lubitz SA, Newton-Cheh C, Wang TJ, Magnani JW, Schnabel RB, Chung MK, Barnard J, Smith JD, Van Wagoner DR, Vasan RS, Aspelund T, Eiriksdottir G, Harris TB, Launer LJ, Najjar SS, Lakatta E, Schlessinger D, Uda M, Abecasis GR, Muller-Myhsok B, Ehret GB, Boerwinkle E, Chakravarti A, Soliman EZ, Lunetta KL, Perz S, Wichmann HE, Meitinger T, Levy D, Gudnason V, Ellinor PT, Sanna S, Kaab S, Witteman JCM, Alonso A, Benjamin EJ, Heckbert SR. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–159. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arking DE, Chugh SS, Chakravarti A, Spooner PM. Genomics in Sudden Cardiac Death. Circ Res. 2004;94:712–723. doi: 10.1161/01.RES.0000123861.16082.95. [DOI] [PubMed] [Google Scholar]
- 32.Rosati B, McKinnon D. Regulation of Ion Channel Expression. Circ Res. 2004;94:874–883. doi: 10.1161/01.RES.0000124921.81025.1F. [DOI] [PubMed] [Google Scholar]
- 33.Aman TK, Grieco-Calub TM, Chen C, Rusconi R, Slat EA, Isom LL, Raman IM. Regulation of Persistent Na Current by Interactions between {beta} Subunits of Voltage-Gated Na Channels. J Neurosci. 2009;29:2027–2042. doi: 10.1523/JNEUROSCI.4531-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abriel H. Cardiac sodium channel Nav1. 5 and interacting proteins: Physiology and pathophysiology. Journal of Molecular and Cellular Cardiology. 2010;48:2–11. doi: 10.1016/j.yjmcc.2009.08.025. [DOI] [PubMed] [Google Scholar]
- 35.Ruan Y, Liu N, Priori SG. Sodium channel mutations and arrhythmias. Nat Rev Cardiol. 2009;6:337–348. doi: 10.1038/nrcardio.2009.44. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.