Abstract
Fluorescent biosensors for living cells currently require laborious optimization and a unique design for each target. They are limited by the availability of naturally occurring ligands with appropriate target specificity. Here we describe a biosensor based on an engineered fibronectin monobody scaffold that can be tailored to bind different targets via high throughput screening. This Src family kinase (SFK) biosensor was made by derivatizing a monobody specific for activated SFK with a bright dye whose fluorescence increases upon target binding. We identified sites for dye attachment and alterations to eliminate vesiculation in living cells, providing a generalizable scaffold for biosensor production. This approach minimizes cell perturbation because it senses endogenous, unmodified target, and because sensitivity is enhanced by direct dye excitation. Automated correlation of cell velocities and SFK activity revealed that SFK are activated specifically during protrusion. Activity correlates with velocity, and peaks 1–2 microns from the leading edge.
Signaling networks that control cell behavior are tightly regulated in space and time. Fluorescent biosensors for living cells have provided a valuable window on the dynamics of these networks, enabling quantitation of the kinetics and localization of protein activity in vivo. However, their application has been limited because they remain difficult to design, requiring considerable optimization for each target, and identification of ‘affinity reagents’ that bind a specific state of the targeted protein (i.e. phosphorylation or conformation). Biosensors generate a fluorescence readout through attachment of fluorophores at positions unique to each biosensor, and so require extensive optimization. Here we demonstrate that a biosensor can be based on a fixed, engineered scaffold whose binding can be tailored to different target proteins via high throughput screening. By using a uniform scaffold, with only a small variable region to confer specificity, optimization of fluorescence readout mechanisms such as FRET or attachment of environmentally-sensitive dyes can be greatly simplified. Furthermore, high throughput screening can provide biosensors when no naturally occurring affinity reagents are known.
In the application described here, we chose to sense binding of the monobody to its target y attaching a bright, environmentally-sensitive fluorescent dye to the monobody. Binding of the biosensor to endogenous target protein generates a change in fluorescent intensity and/or λmax of the attached dye. Use of a dye-labelled protein domain to sense an endogenous target was previously demonstrated by our lab, using dyes designed for this purpose 1–4. The dyes can be excited at > 580 nm to minimize cell damage and avoid cellular autofluorescence, and they are very bright in hydrophobic environments (quantum yield = 0.17–0.61 and ε > 100,000). This approach minimizes cell perturbation both because it senses endogenous, unaltered target protein and because a bright dye is directly excited, requiring lower concentrations of biosensor. We will use the biosensor to study Src family kinases at the thin leading edge of motile cells, where sensitivity and the ability to use low biosensor concentrations are important.
We selected the fibronectin monobody5 (FN3 monobody) as the scaffold that will be the basis of the new biosensor because it is small (~95 residues), folds stably within living cells, and because the invariable portion need not have cysteines (simplifying dye attachment and folding in the reducing environment of the cell)5. This monobody, derived from the tenth type III domain of human fibronectin (FN3), has an immunoglobulin-like fold composed of seven beta strands which are connected by flexible loops. Several of these flexible loops can be randomized while keeping the core structure intact5–8, thereby creating libraries of 109–1011 variants. FN3 libraries have been screened by phage display8 and other methods to produce binders against a variety of targets5–7. All these features make the FN3 domain an attractive candidate for a generalizable live cell biosensor scaffold (Fig. 1a).
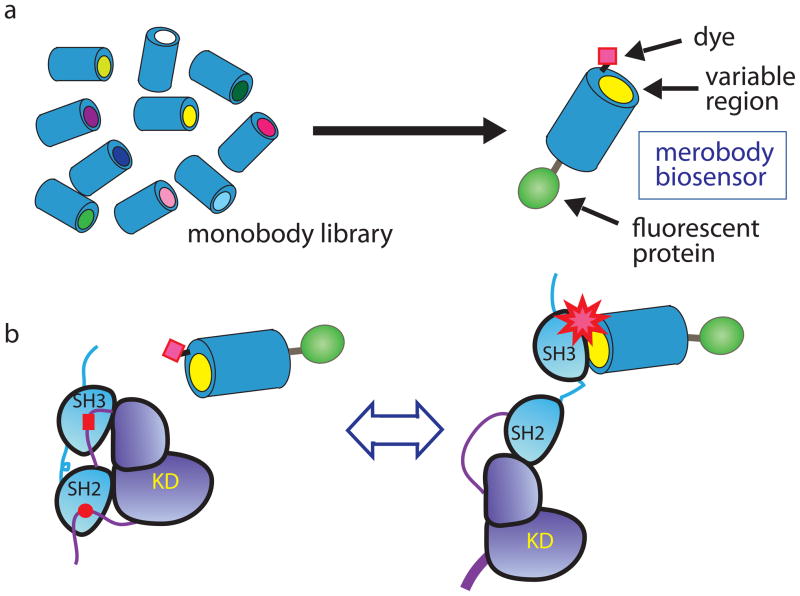
Figure 1. Screening a fibronectin monobody library leads to a biosensor for Src family activity.
a) A library of fibronectin monobodies is screened to find a library member with the appropriate binding selectivity and affinity for the targeted protein state. The library is based on a uniform scaffold stable in living cells and suitable for conversion to biosensors. The appropriate library member is fused to a fluorescent protein (FP) via a flexible linker and further derivatized with an environmentally sensitive dye to report target binding. b) The present biosensor is based on a binder that is specific for the activated conformation of Src family kinases (SFK). Biosensor binding to active SFK leads to increased fluorescence from the merocyanine dye. The ratio of dye fluorescence/protein fluorescence provides a quantitative measure of SFK activation kinetics and localization in living cells.
Src family kinases (SFKs) are regulators of signaling networks impacting cell division, migration, and survival9,10 Because they modulate multiple pathways, their activation must be tightly regulated. SFK stimulation via different cell surface receptors, including integrins11 and receptor tyrosine kinases12, activate SFKs in distinct subcellular locations, with tightly controlled kinetics. A biosensor that enables visualization of SFK activity will be valuable in dissecting the coordinate regulation of these different pathways. Existing biosensors reveal the phosphorylation of Src substrates13–15, but no studies of Src in protrusion/retraction dynamics at the cell’s leading edge have been reported, probably because signal/noise considerations limit quantitative studies in this region of the cell. The new biosensor is shown to provide the sensitivity required for quantitative studies of protrusion/retraction dynamics. Furthermore, it is advantageous that the new biosensor reports SFK conformation, rather than phosphorylation of a substrate liable to diffusion and susceptible to both kinases and phosphatases.
In the following report, we characterize a monobody that binds the Src family kinases, demonstrating that it is specific for the activated ‘open’ conformation of these kinases. We examined dyes at different positions around the monobody binding interface, identifying sites where attachment provides good fluorescence response without interfering with target binding. Structure-activity studies together with modeling provided insights into the interactions of the dyes with the monobody and target that will hopefully be useful in applying the merobody to other targets. The merobody was engineered to eliminate vesiculation in living cells, and to include a fluorescent protein for ratio imaging. Within cells, robust detection of SFK activity at the leading edge, combined with quantitative analysis, reveals changing dynamics correlated with protrusion and retraction velocity.
Results
The monobody binds to active SFK without kinase inhibition
The monobody biosensor reported here is based upon monobody 1F11, which was shown to bind selectively to the SH3 domains of Src family kinases (SFKs) and contain no cysteines6. 1F11 was generated by phage display screening using the SH3 domain of Src as ‘bait’. It binds to SFKs, but not to closely related kinases6. Because screening was performed against the SH3 domain, we surmised that this monobody might be conformationally sensitive. Crystallography and biochemical data indicate that the Src SH3 may be more exposed upon Src activation (Fig. 1b). Upon activation, two intramolecular linkages are broken to produce a more ‘open’ conformation, enabling auto-phosphorylation that maintains the active conformation16. We first tested whether 1F11 was specific for the open, activated conformation of Src. Figures 2a,b show pull-down assays in which binding of 1F11 to Src was tested in lysates from GN4 rat liver epithelial cells. These cells show robust Src activation when treated with the small molecule ciglitazone17,18. 1F11 or wild-type FN3 monobody (non-binding control) were immobilized on Ni-NTA beads and incubated with cell lysates, then examined for the ability to pull-down endogenous Src. Total levels of Src were unaffected by this treatment17 (Supplementary Results Fig. 1). The 1F11 monobody pulled down substantially more Src from ciglitazone treated cells than from untreated cells, indicating preferential binding to activated kinase (Fig. 2a, supplementary Fig. 2). Wild-type FN3 domain or beads alone showed no Src binding. We showed previously that pre-treatment of GN4 cells with the phosphatase inhibitor Pervanadate prevents ciglitazone-mediated Src activation by blocking dephosphorylation of p-Tyr 529 17,18. Here we found that pre-treatment with pervanadate greatly attenuated 1F11 pulldown of Src (Fig. 2a, supplementary Fig. 2). Therefore, our data indicate that pulldown is indeed sensitive to Src activation state.
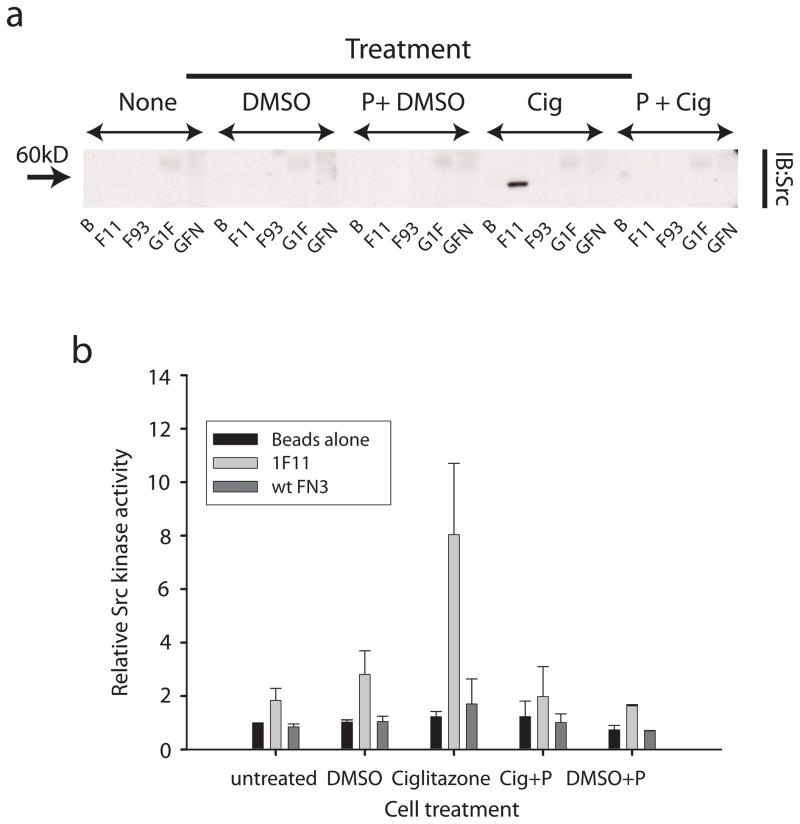
Figure 2. Fibronectin monobody 1F11 preferentially binds active Src.
a) 1F11 monobody binding to Src in lysates from cells +/− the Src activator Ciglitazone. GN4 cells were either untreated, treated with vehicle DMSO, vehicle plus pervanadate pretreatment (P), 50 μM Ciglitazone (Cig), or Ciglitazone with pervanadate pretreatment. Immunoblot was used to assay pulldown of Src by beads alone (B), the 1F11 monobody (1F11), control nonbinding monobody (wt FN3), GFP-1F11 with sub-optimal linker (G1F) or GFP-FN3 sub-optimal linker (GFN). b) Src kinase activity bound to the monobody or controls as in (a). Data shown is an average of three independent experiments.
In-vitro kinase assays showed that the SFKs pulled down by the biosensor were active (Fig. 2b). This was important as it indicated that biosensor binding to the target would not block SFK kinase activity. 1F11 beads that had been incubated with Ciglitazone-treated lysates showed kinase activity several fold higher than wt-FN3 beads, control beads, or 1F11 beads that had been incubated with untreated lysates (Fig. 2b). Together these studies showed that 1F11 binds preferentially to the active form of Src, that 1F11 does not artificially activate the kinase, and that Src remains an active kinase when bound to 1F11.
Optimizing affinity, brightness and fluorescence change
Four solvent-sensitive merocyanine dyes were screened at four positions near the variable loops of the monobody. Single cysteine mutants of the 1F11 monobody fused to m-Cerulean fluorescent protein19 were expressed in E. coli and covalently derivatized with dyes bearing cysteine-reactive iodoacetamide side chains 2,4. These were selected from a set of highly fluorescent fluorophores (Fig. 3a) we published previously, optimized for use as part of biosensors in living cells 2–4. The m-Cerulean was included for ratio imaging in vivo, as explained below. Covalent dye-protein conjugates were separated from free dye using size exclusion chromatography. Dye/protein molar ratios were between 0.8 and 1.1 in all cases, and controls using cysteine-free protein produced dye/protein ratios < 0.05. Polyacrylamide electrophoresis gels, shown to separate protein from unreactive free dye, indicated no detectable free dye in the labeled proteins.
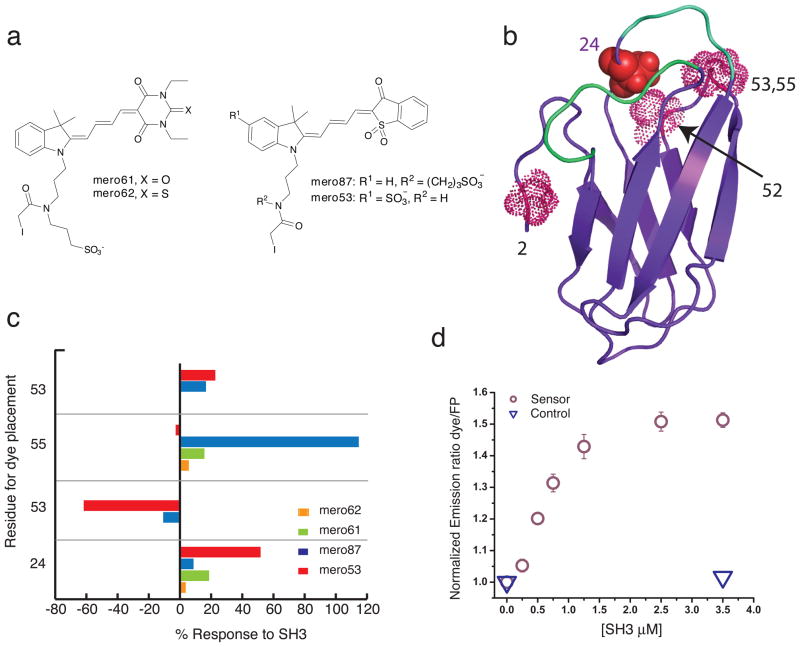
Figure 3. Screening for responsive sensor variants – selecting dye and site for dye labeling.
a) Structures of the environmentally sensitive merocyanine dyes tested on the monobody. b) Ribbon representation of the active state binder 1F11 (based on published FN3 domain crystal structure PDB: 1NFNA). Residues 2, 24, 52, 53, and 55 where cysteine was incorporated for dye attachment and testing are shown as space-filling side chains. The alanine shown in bold marks the position of dye attachment in the final merobody biosensor. The putative target binding loops are shown in cyan. c) Ratiometric fluorescence response (dye emission/m-Cerulean emission) of the various combinations of mero dyes and residues labeled. d) Titration showing the change in normalized emission ratio Mero-53/m-Cerulean for the biosensor or control (0.5 μM) with increasing c-Src SH3. The control sensor has a P80A mutation in the FG binding loop of 1F11.
The four sites selected for dye attachment were on or near the putative binding interface. This interface is comprised of residues on the BC and FG loops of 1F11, including those that were randomized for SH3 binding 6. Three of the dye attachment sites (52, 53 and 55) were on the DE loop adjacent to the BC and FP loops, proximal to the binding interface (Fig. 3b). The fourth site, alanine 24, is part of the BC binding loop but is not among the residues that were randomized for phage-displayed screening. This latter site was selected because it would be proximal to the putative binding interface but less likely to disrupt SH3 binding. Each dye conjugate was evaluated for fluorescence change in response to recombinant c-Src SH3, as well as brightness in the bound and unbound state (Fig. 3).
Of all the variations tested, three showed substantial response: dye mero87 at position 53 (C53-m87), dye mero53 at position 52 (C52-m53), and dye mero53 at position 24 (C53-m24) (Fig. 3c). At least for this target, the two closely related dyes mero87 and mero53 showed far stronger response than other structures, albeit at different positions. These two dyes differ only in the positioning of the sulfate group used to confer water solubility on the dye. Modeling of dye-protein interactions and more detailed analysis is in the Discussion section. Control biosensors lacking a critical proline residue in the FG binding loop of 1F11 (P78A) showed no response to SH3, for all three of the well responding variants (Fig. 3d).
The three biosensors that showed substantial response to Src SH3 were titrated with the SH3 to examine whether sequence modification or dye attachment had affected the monobody’s affinity for the SH3 domain (Fig. 3d, supplementary Fig. 3). The apparent binding constants for C53-m87, C52-m53 and C24-m53 were 0.97, 0.26 and 0.69 micromolar respectively, compared to 0.25 micromolar for the native 1F11 monobody (as measured by isothermal titration calorimetry6). The dye-labeled P78A binding-incompetent mutant of the monobody showed no affinity for the SH3 in these assays. These data showed that labeling 1F11 with merocyanine dyes and fusing it to mCerulean minimally perturbed target binding.
Biosensor C24-mero53 was selected for live cell studies, and is referred to simply as the SFK merobody biosensor henceforth. Although this biosensor did not undergo the greatest change in response to SH3 binding, its 50% increase was nonetheless substantial and compares favorably with many biosensors that have proven valuable in vivo14,20,21 This biosensor was selected because it remained extremely bright when the dye was attached to the monobody. Even in aqueous environments prior to target binding, the dye emission was greater than that of the m-Cerulean on the biosensor (supplementary Fig. 4, supplementary table 1), and upon target binding, dye emission increased to ~ 2.4-fold that of cerulean. Usefulness in living cells is a function of both dynamic range and brightness. A dye with impressive fluorescence change can be too dim to discern at non-perturbing biosensor concentrations in vivo.
Optimizing the fibronectin scaffold for living cells
Initial studies in cells showed that the dye-conjugated native monobody suffered from problems making it unsuitable for live cell imaging. Although the 1F11 monobody conjugated to mero87 showed a response to SH3 domain in vitro (supplementary Fig. 5), it showed extensive vesiculation in living cells (supplementary Fig. 6). This has been observed previously with dye-labeled proteins (data not shown), and is likely due to autophagy22 or proteolytic processing23. Use of the fibronectin monobody as a broadly applicable scaffold for biosensors depends critically on eliminating this vesiculation, as fluorescent puncta hinder quantitation in vivo, and could obscure real interactions with punctate cell structures. To examine whether this problem was caused by the dye or the merobody structure, 1F11 was conjugated at the same position to commercially available Alexa 488, a water soluble dye used frequently to generate fluorescent protein conjugates for live cell imaging 24. This conjugate also showed vesiculated and nonuniform distribution in cells (supplementary Fig. 6). We concluded that further modification of the fibronectin monobody would be required before it could be used as a biosensor.
The monobody was fused to GFP and GFP variants in an attempt to enhance solubility or alter the balance of charges that might contribute to autophagy. This also provided a second fluorophore for ratiometric imaging of dyes that responded to SFK by changing their fluorescence intensity; the ratio of dye fluorescence to fluorescent protein fluorescence could be used to normalize out effects of cell thickness, uneven illumination etc. on dye brightness1,25,26. GFP variants without surface-exposed cysteines were prepared so that cysteine labeling could still be used for site-specific dye incorporation. The fluorescent protein was attached to the N terminus, as our earlier studies had shown that attachment at the C terminus interfered with SH3 interactions (data not shown). Attachment of GFP did eliminate the vesiculation, but surprisingly also completely abrogated the fluorescence response of the dye (supplementary Fig. 7). Increasing the length of the linker between the monobody and the fluorescent protein (supplementary Fig. 8) restored fluorescence response without losing uniform monobody distribution in cells (supplementary Fig. 6). The distribution of fluorescence appeared the same at both the dye wavelengths and the wavelengths of the fluorescent protein, indicating that the puncta were not simply a concentration of free dye. In the final, optimized monobody, m-Cerulean fluorescent protein with a C48S mutation (to prevent dye labeling) was attached to the N terminus of 1F11.
Spatio-temporal dynamics of SFK activity in motile cells
Although Src family members have been clearly implicated in motility, analysis of their dynamics within protrusions and retractions has not been reported, perhaps due to the difficulty of obtaining sufficient signal within the thin cell edge. We first applied the SFK monobody biosensor in NIH 3T3 mouse embryo fibroblasts (MEF). Ratio imaging and cell handling were as previously described1,25, with SFK activity indicated by an increase in the dye/cerulean emission ratio.
During initial studies, SFK activity was observed at the edges of extending protrusions and in dorsal ruffles (large circular actin-based protrusions). We stimulated MEFs with platelet derived growth factor (PDGF)27,28 to induce dorsal ruffling and cell protrusions and to verify previous finding of Src activation there. Immuno-staining has shown active Src to be specifically localized within dorsal ruffles29 and at the cell edge28–31. Also, Src activity is thought to be necessary for ruffle formation and cell protrusion28,29. The merobody showed elevated SFK activity within the ruffles from the time of their appearance until they closed to become macropinosomes (Fig. 4a, supplementary video 1, supplementary Fig. 9). Closure was accompanied by a sudden drop in activation. These experiments were able to directly demonstrate that Src is activated within ruffles specifically during periods of ruffle formation and movement, as would be expected given Src’s role in regulating actin polymerization 27.
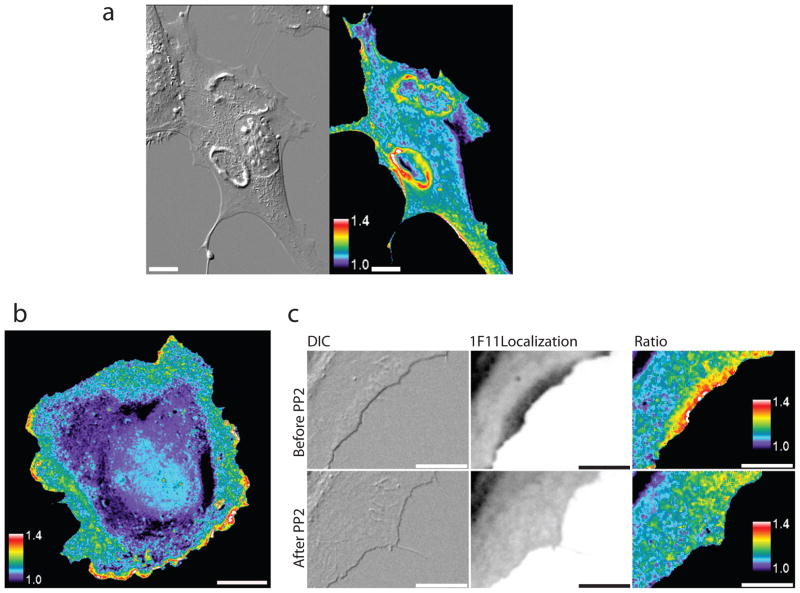
Figure 4. Src activation dynamics in living cells.
a) DIC (left panel) and ratio images (right panel) of a PDGF-stimulated NIH 3T3 MEF microinjected with the SFK merobody biosensor. Scale baris 20 μm. Note prominent circular dorsal ruffles b) Ratio image of a PTK1 cell microinjected with the biosensor. Scale bar is 20μm. c) DIC (left panels), m-Cerulean (merobody localization, middle panels), and ratio images (right panels) from representative frames of a movie in which a PTK1 cell microinjected with the biosensor was treated with the Src inhibitor PP2. Scale bar is 10 μm.
We next focused our attention on protrusion and retraction at the cell edge, where velocity could be more readily quantified, enabling detailed correlation of SFK activity with specific aspects of cell movement. SFK are likely involved in controlling both actin and adhesion dynamics9,10,28,32–34. A more detailed understanding of SFK dynamics during extension and retraction could help to differentiate potential roles and mechanisms. By titrating down the amount of biosensor in cells (roughly determined as brightness per unit area, appropriate for the flat cells used) we were able to define brightness ranges with good signal/noise where no apparent perturbation of extension/protrusion occurred. Controls indicated that, at the biosensor concentrations we used, the velocities of protrusion and retraction were not correlated with the amount of biosensor present in the cell (supplementary Fig. 10). There was no visible difference in cell edge dynamics between cells injected with the SFK monobdy, non-injected cells, or cells injected with P78A control biosensor incapable of binding Src.
For quantitation of protrusion/retraction dynamics we switched to PTK1 kangaroo rat kidney epithelial cells because of their large, flat lamellipodia. During constitutive protrusion, we discovered a band of high SFK activity immediately adjacent to the cell edge (Fig. 4b, c; also see supplementary videos 3 and 4 and Supplementary Fig.11). We also observed that the biosensor itself localized to the cell edge in protrusions, as detected by monitoring m-Cerulean fluorescence (Fig. 4c and supplementary video 4). This indicated significant enrichment of active SFK along the edge. The nonbinding control biosensor P78Ashowed no change in fluorescence ratio (Supplementary Fig. 12) and no cell edge enrichment. The specificity of 1F11 for different SH3 domains was extensively examined in the original paper describing the 1F11 monobody6, but we confirmed specificity for SFK in living cells by examining the effect of the Src kinase inhibitor PP2. As shown in Figure 4c and supplementary videos 4, 5 and 6, PP2 abolished both high fluorescence ratios and localization of the probe to the cell edge within 3 minutes after PP2 addition.
To quantify the relationship between SFK dynamics and protrusion or retraction, we correlated the distribution of SFK activity with edge velocity. Briefly, our automated approach first involved tracking the cell edge and deriving the velocity at each pixel along the cell boundary. This was based on methods described elsewhere35,36. From a given pixel at the edge, ‘line scans’ were generated by bi-linearly interpolating pixel values along vectors oriented into the cell and normal to the edge (supplementary Fig. 13). For every pixel along the edge of the cell and for each time point there was a line scan and an edge velocity. Line scan values were calculated for 1 μm from the edge to 20 μm into the cell. The first portion of the line scan (0- 1 μm) was found to be subject to a lower signal-to-noise ratio in both fluorescence channels, potentially leading to artifacts in the ratio image. We therefore excluded from our analysis regions within 1 micron of the edge. The portion of the line scan 1μm-20μm from the cell edge was sufficient to clearly delineate regions of SFK activation, and differences between protrusion and retraction. The line scans were sorted by velocity into bins of width = 0.2 pixels/frame. Then, for each cell and each velocity bin, a mean line scan was calculated.
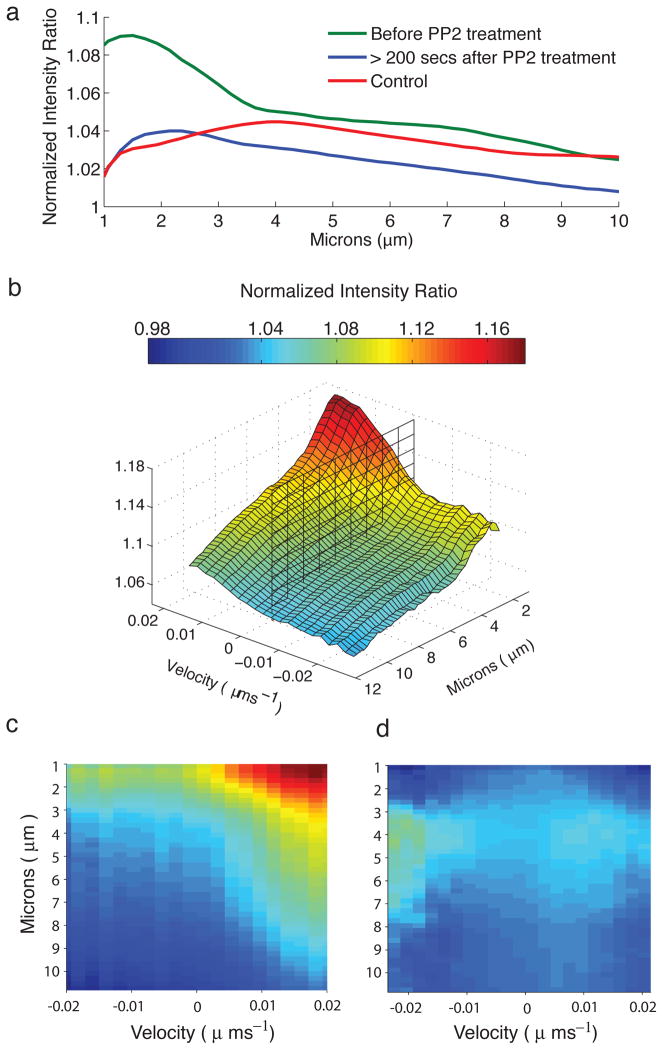
Figure 5 shows the average SFK activity as a function of distance from the cell edge during protrusion. SFK activity peaked between 1 and 2 microns from the cell edge, with a gradual diminution of activity at points further within the cell. This activation was greatly diminished when using the control monobody incapable of target binding (P78A -see above), or when cells were treated with PP2. The distribution of SFK activity was dependent on the velocity and direction of edge movement. Figure 5b shows the correlation of cell edge velocity and the SFK activity profile. Using a total of 270,460 line scans for the merobody biosensor, the data indicated that SFK activation is greater during protrusion than retraction, that activation level is proportional to the velocity of the protrusion, and that the distribution of activity relative to the cell edge is consistently highest at a single peak 1–2microns from the edge (for both retraction and protrusion). Again here the control biosensor (266,623 line scans) showed greatly reduced activity (Fig. 5d) and the Src inhibitor PP2 (145,240 lines scans pretreatment and 138,320 scans post-treatment) flattened the activity profile (supplementary Fig. 14). Further, edge velocity gradually decreased after addition of PP2 (supplementary Fig. 15).
Figure 5. Automated edge analysis and line scans reveals distinct zone of SFK activity that is correlated with protrusion velocity.
a) SFK activity as a function of microns from the cell edge. SFK activity is localized towards the edge of the cell and is inhibited by PP2 Treatment. For all data points the standard error is less than 0.1%. At 1μm, the difference between the mean normalized intensity ratio, before and after PP2 treatment, is ~0.08 and is statistically significant (p<0.001). b) SFK activity as a function of distance from the cell edge and velocity. SFK activity is approximately proportional to protrusion speed. The vertical plane is at velocity = 0. The standard error, for a given velocity and distance, is less than 0.3%. c, d) Response of the merobody biosensor (c) compared to the non-binding control (d). The merobody biosensor reports both higher activity than the non-binding control, and a stronger dependence on velocity.The standarderror for for a given velocity and distance is less than 0.3% (c) and less than 1% (d).
Discussion
The goal of these studies was a biosensor based on an engineered scaffold designed for high throughput screening. This proof of principle study can pave the way for generating other biosensors via screening, i.e. for targets where no suitable naturally occurring binders are known, and with greatly simplified biosensor engineering. To study Src family signaling at the cell’s leading edge, we based this prototype biosensor on a known fibronectin monobody that binds specifically to the SH3 domains of Src family kinases 6. The fibronectin monobody was a good choice for a generally applicable biosensor scaffold because it contains no native cysteines (facilitating site-specific dye attachment) and folds well in living cells. This contrasts with scFv and other antibody fragments 37. The FN3 monobody has flexible loops that accommodate insertion and randomization of amino acid residues, and has proven capacity to generate binders against diverse protein targets 5,6,38. We used a solvent-sensitive merocyanine dye to report target binding, which proved to provide enhanced sensitivity, with brightness > 2 times higher than direct excitation of cerulean fluorescent protein, and therefore substantially higher than fluorescent proteins indirectly excited for FRET. This proved valuable at the thin edge of cells where signal/noise is an important limitation. This biosensor design could report activation of endogenous target protein, thereby reducing cell perturbation.
We showed that the monobody had the desired specificity for active SFKs, and then examined where dyes could be attached around the binding site to report target binding without greatly diminishing affinity. Three different sites were suitable, indicating that dyes will likely be suitable as readouts for monobodies binding to different proteins. The four fluorophores tested were designed for use in living cells --bright (ε > 100,000, QY = 0.17–0.61 in hydrophobic environments), with excitation at ≥ 550 nm to avoid auto-fluorescence and minimize cell damage, and with solvent-sensitive fluorescence suitable for reporting protein binding in vivo. 2,4. Remarkably, for the two dyes based on the coupled indolenine and benzothiophen-3-one-1,1-dioxide rings (mero87 and mero53), shifting the position of attachment determined whether the dye showed an increase or decrease of fluorescence upon target binding. The attachment site producing a decrease in fluorescence (position 52) is positioned at the interface between the beta sheet and the flexible DE loop. Dye attachment may have caused partial unfolding of the monobody, leading the dye to find a hydrophobic pocket in the monobody before interaction with target protein. Binding to the target could force the dye out of the pocket, thereby decreasing fluorescence. This would be partially driven by restoration of binding interactions that stabilize the monobody. For positions and dyes showing an increase in emission intensity upon binding (C53-m87 and C24-m53), the dye likely experienced a more hydrophobic environment on target binding. There were marked differences in the response and brightness exhibited by two dyes that differed only in placement of a sulphonate moiety(mero53 and 87). Repositioning of the sulphonate could have altered the dyes’ photophysics4 or interaction with the monobody interface (see below). The environment of the dye on the monobody also influenced its brightness. Although dye mero87 at position 53 gave the largest response (Fig. 3), its low overall brightness on the monobody led us to select dye mero53 at position 24 for use in living cells. This dye showed a 50% fluorescence increase upon target binding and was 2.4x as bright as Cerulean fluorescent protein. The labeled monobody had 500–600 nM affinity for Src SH3, a range proven valuable for biosensor reversibility and specificity in previous studies 1,21,39.
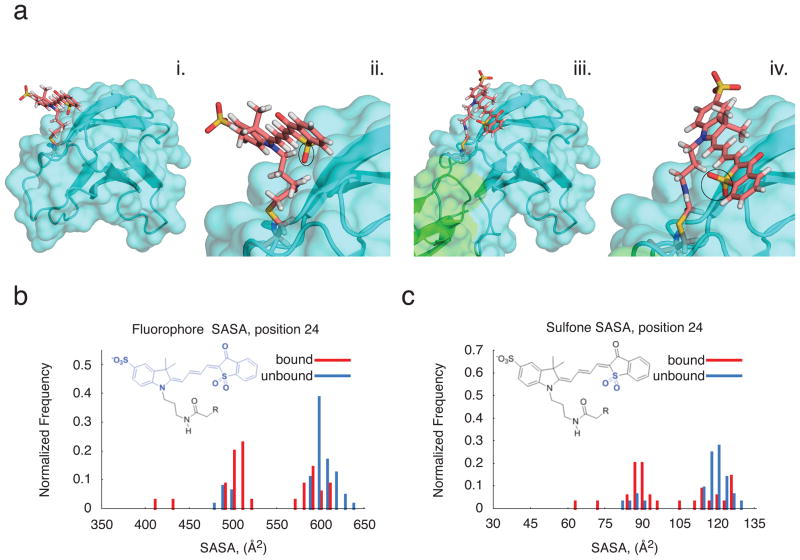
We generated computer models of 1F11-mero53 conjugates and docked these models to Src SH3, thereby examining the merobody-target interface (Fig. 6, supplementary Figs. 16, 17). In the biosensor used for live cell studies, our modeling suggests that the dye does not directly interact with the SH3 domain. Rather, it experiences a change in local environment due to differing interactions with the FN3 monobody itself (Fig. 6a). Modeling suggests that the water-exposed surface area of the dye decreases as the merobody binds the target SH3 (Fig.6b,c), consistent with the observed increase in dye emission upon binding. Merocyanine dyes, including mero53, generally show enhancement in emission intensity when shifting from polar to a polar environments. The decrease in solvent-accessible surface area (see Fig. 6c) is in fact more pronounced for the specific moiety on the merocyanine (the sulphone) that is believed to confer sensitivity to solvent polarity2. The models do not show changes in solvation for mero53 attached at positions that produced poor fluorescence response (Supplementary Fig. 16, 17).
Figure 6. Modeling of the 1F11-dye/SH3 interface.
a) Computermodels of either unbound 1F11-mero53 conjugate alone (i and ii) or in complex with cSrc-SH3 (iii and iv). Dye is attached to residue 24 as in the final ‘merobody’ biosensor. c-Src SH3 is green, 1F11 is blue, and dye is salmon. In ii,iv (magnified versions), the sulphone group of the dye is highlighted (closed circles). The model of unbound biosensor is part of the sub-population in which the dye has higher solvent accessible surface area (SASA). The model of bound biosensor shown here is the highest-scoring model and member of the low-SASA cluster. b,c) SASA distribution for the top 0.5% of models for the bound and unbound states, either for whole fluorophore (b) or the sulphone group (c).
Finally, we attached a fluorescent protein via an optimized linker to eliminate the formation of fluorescent puncta, potentially due to autophagy, that would have severely hindered live cell imaging. We are hopeful that these changes, and the identification of optimum dye attachment sites, have generated a scaffold that can now be targeted to other intracellular proteins, providing a generalizable tool to study endogenous protein conformation. This work has demonstrated the feasibility of generating practical biosensors from engineered scaffolds. It is important to note that essentially all biosensors perturb cell physiology, as they must interact with the molecules whose behaviors they report. Different designs either inhibit or mimic normal protein action. The SFK monobody biosensor described here may compete with endogenous ligands that normally bind to the SH3 domain of Src family proteins. This could generate either ‘false negative’ data, in which native ligands outcompete biosensor, or the biosensor could inhibit normal interactions. The enhanced sensitivity of the SFK merobody will enable us to use less biosensor, more closely approaching the equilibria in unperturbed cells.
The SFK monobody revealed localized and transient activation of SFK at the cell edge and in PDGF-induced dorsal ruffles. Immunostaining has shown that phosphorylated, active Src localizes to dorsal ruffles and at the cell edge28–31, where it phosphorylates cortactin or N-WASP, leading to Arp2/3 activation and consequent actin polymerization.27,40–42 Src is known to be necessary for the formation of dorsal ruffles, and SFKs are known to regulate signaling molecules involved in actin assembly and organization within these ruffles27 (Abl tyrosine kinase43, Rac GTPase 44). The merobody biosensor provided direct evidence, consistent with these previous studies, that endogenous SFK are in the active conformation within ruffles specifically during actin-based protrusion.
The biosensor was used to study SFK activation in the lamellipodia of migrating cells. Through development of a quantitative line scanning approach, statistically valid correlations of protrusion velocity and Src activity distribution could be based on thousands of line scans. Though it was elevated in both protrusion and retraction, SFK activity was significantly higher during cell protrusion. Most strikingly, during protrusion the activity was proportional to the rate of lamellipodial extension. SFK activity may regulate protrusion speed by controlling the rate and extent of actin polymerization, potentially through phosphorylation of actin-regulatory proteins (potentially the WAVE complex40,41, cortactin40,45, gelsolin, pCAS46,47, Abl tyrosine kinase29,43, or regulators of Rho family GTPases12,34,44,48–50). The biosensor showed that activation occurred with a defined profile, peaking within 2 microns of the edge in the lamellipodium of the PTK1 cells. Src kinases play a critical role in regulating both actin dynamics and the assembly and disassembly of adhesions 33,34,47, so SFK at this position may regulate actin, focal adhesions, or their coordination. Further work will be required to define the interactions of SFK with specific molecules at the leading edge and their positions relative to actin and adhesion dynamics. SFK activation in retraction is much less pronounced and may be part of constitutive signaling responsible for edge retraction 48.
In conclusion, we have demonstrated the feasibility of producing a biosensor based on modification of the fibronectin monobody, a scaffold suitable for high throughput screening and for use in living cells. This exemplifies a generalizable approach capable of producing biosensors when no suitable affinity reagents are known, to increase the throughput of biosensor production, and to greatly simplify biosensor design. These biosensors report the activation of endogenous, unmodified proteins, thereby reducing perturbation of cell physiology. Dyes here provided exceptional sensitivity, but made it more difficult to introduce the biosensor into living cells. Ultimately it may be possible to use fluorescent proteins for genetically encoded readouts of endogenous target binding. Automated image analysis revealed that SFK are more strongly activated during protrusion than retraction, and that the level of activity is proportional to the velocity of the extending edge. Automated analysis of multiple points along the cell edge revealed an activity profile with a single peak of maximal activation at the edge of constitutively migrating PTK cells.
Methods
Additional methods are described in the Supplementary Methods section.
Dye labeling
DMSO solutions of cysteine-reactive merocyanine dyes (10–20 mM) were added to 1F11-mCerulean fusion proteins (200–300 μM) in 50 mM HEPES, 100 mM NaCl buffer pH 7.4 such that the dye was present in 5–10 fold molar excess and the DMSO in the reaction mixture was less than 10%. After reaction for 4–5 hours, excess dye was separated from labeled protein using size exclusion G-25 (GE healthcare) columns. During G-25 size exclusion, a clear separation was seen between the labeled protein band and the relatively immobile free dye. Labeled proteins were subjected to SDS-PAGE electrophoresis and a single fluorescent band was observed (controls in which labeled monobody and dye were mixed showed that free dye could be observed as a separate band of lower molecular weight). Coomassie labeling was also used to verify homogeneity of the biosensor preparations. Labeling efficiency was calculated by measuring the dye and protein concentrations of the labeled conjugate. Dye concentration was estimated using dye absorbance at absorption maxima after dissolving the conjugate in DMSO. Protein concentration was estimated by using absorbance due to the mCerulean FP (molar extinction coefficient 43,000). Labeling efficiency was estimated to be in the range of 0.9–1.2 dye/protein molar ratio, for the various preparations tested. Binding to SH3 was compared for individual batches and similar results were obtained. Also, independent batches gave consistent results in live cell imaging experiments.
Microscopy
For imaging experiments MEFs and PTK1 cells were plated onto coverslips coated with 5μg/mL Fibronectin (Sigma) overnight. Culture media was exchanged for imaging media for one hour prior to microinjection. Cells were microinjected using a biosensor concentration of 40μM in the microinjection needle, and were allowed to recover for 30–60min prior to imaging experiments. MEFs were stimulated using 30ng/mL PDGF (Sigma). For inhibiting SFKs PTK1 cells were treated with 10μM PP2 (Sigma).
Automated linescanning and analysis of Src activity
The response of the biosensor was analyzed using custom automated line scanning software. Line scans of length 100 pixels (~ 20 um) were calculated at every pixel around the edge of the cell and orientated into the cell in a direction normal to the edge. For each line scan we assigned a velocity by using the previous frame to calculate the velocity of cell edge at that location (see supplementary methods). Line scans were then grouped, and averaged, by velocity (Figure 6).
Supplementary Material
Acknowledgments
We gratefully acknowledge Christopher MacNevin for assistance with dye synthesis, Feimo Shen for help with imaging studies, Doug Renfrew for assistance with computational modeling, Alan Nguyen for technical assistance, Betsy Clarke for expert administrative assistance, and funding from the American Heart Association(AG) and the National Institutes of Health (GM GM082288 and GM057464 to KMH).
Footnotes
Competing financial interests. The authors declare no competing financial interests.
Author contributions. Akash Gulyani developed the biosensors, with help from Brian Dewar and Lee Graves with phosphorylation assays. Eric Vitriol carried out live cell imaging studies, assisted by Akash Gulyani. Richard Allen and Tim Elston developed the image analysis algorithms and carried out automated analysis of SFK activity, with help from Jianrong Wu. Steven Lewis and Brian Kuhlman modeled dye-protein interactions. Dmitry Gremiachinskiy synthesized the dyes. Brian Kay provided protein constructs and input regarding fibronectin screening and structure. Klaus Hahn conceived the study, directed the research and wrote the final manuscript based on contributions from all authors.
References
- 1.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–9. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 2.Toutchkine A, Kraynov V, Hahn K. Solvent-sensitive dyes to report protein conformational changes in living cells. J Am Chem Soc. 2003;125:4132–45. doi: 10.1021/ja0290882. [DOI] [PubMed] [Google Scholar]
- 3.Toutchkine A, Nguyen DV, Hahn KM. Merocyanine dyes with improved photostability. Org Lett. 2007;9:2775–7. doi: 10.1021/ol070780h. [DOI] [PubMed] [Google Scholar]
- 4.Toutchkine A, Nguyen DV, Hahn KM. Simple one-pot preparation of water-soluble, cysteine-reactive cyanine and merocyanine dyes for biological imaging. Bioconjug Chem. 2007;18:1344–8. doi: 10.1021/bc060376n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koide A, Bailey CW, Huang X, Koide S. The fibronectin type III domain as a scaffold for novel binding proteins. J Mol Biol. 1998;284:1141–51. doi: 10.1006/jmbi.1998.2238. [DOI] [PubMed] [Google Scholar]
- 6.Karatan E, et al. Molecular recognition properties of FN3 monobodies that bind the Src SH3 domain. Chem Biol. 2004;11:835–44. doi: 10.1016/j.chembiol.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 7.Koide A, Abbatiello S, Rothgery L, Koide S. Probing protein conformational changes in living cells by using designer binding proteins: application to the estrogen receptor. Proc Natl Acad Sci U S A. 2002;99:1253–8. doi: 10.1073/pnas.032665299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sidhu SS, Koide S. Phage display for engineering and analyzing protein interaction interfaces. Curr Opin Struct Biol. 2007;17:481–7. doi: 10.1016/j.sbi.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 9.Parsons SJ, Parsons JT. Src family kinases, key regulators of signal transduction. Oncogene. 2004;23:7906–9. doi: 10.1038/sj.onc.1208160. [DOI] [PubMed] [Google Scholar]
- 10.Thomas SM, Brugge JS. Cellular functions regulated by Src family kinases. Annu Rev Cell Dev Biol. 1997;13:513–609. doi: 10.1146/annurev.cellbio.13.1.513. [DOI] [PubMed] [Google Scholar]
- 11.Playford MP, Schaller MD. The interplay between Src and integrins in normal and tumor biology. Oncogene. 2004;23:7928–46. doi: 10.1038/sj.onc.1208080. [DOI] [PubMed] [Google Scholar]
- 12.Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957–68. doi: 10.1038/sj.onc.1208079. [DOI] [PubMed] [Google Scholar]
- 13.Ouyang M, Sun J, Chien S, Wang Y. Determination of hierarchical relationship of Src and Racat subcellular locations with FRET biosensors. Proc Natl Acad Sci U S A. 2008;105:14353–8. doi: 10.1073/pnas.0807537105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, et al. Visualizing the mechanical activation of Src. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- 15.Ting AY, Kain KH, Klemke RL, Tsien RY. Genetically encoded fluorescent reporters of protein tyrosine kinase activities in living cells. Proc Natl Acad Sci U S A. 2001;98:15003–8. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cowan-Jacob SW, et al. The crystal structure of a c-Src complex in an active conformation suggests possible steps in c-Srcactivation. Structure. 2005;13:861–71. doi: 10.1016/j.str.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 17.Gardner OS, Dewar BJ, Earp HS, Samet JM, Graves LM. Dependence of peroxisome proliferator-activated receptor ligand-induced mitogen-activated protein kinase signaling on epidermal growth factor receptor transactivation. J Biol Chem. 2003;278:46261–9. doi: 10.1074/jbc.M307827200. [DOI] [PubMed] [Google Scholar]
- 18.Dewar BJ, et al. Capacitative calcium entry contributes to the differential transactivation of the epidermal growth factor receptor in response to thiazolidinediones. Mol Pharmacol. 2007;72:1146–56. doi: 10.1124/mol.107.037549. [DOI] [PubMed] [Google Scholar]
- 19.Rizzo MA, Springer GH, Granada B, Piston DW. An improved cyan fluorescent protein variant useful for FRET. Nat Biotechnol. 2004;22:445–9. doi: 10.1038/nbt945. [DOI] [PubMed] [Google Scholar]
- 20.Fosbrink M, Aye-Han NN, Cheong R, Levchenko A, Zhang J. Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. Proc Natl Acad Sci U S A. 2010;107:5459–64. doi: 10.1073/pnas.0909671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–72. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 22.Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol. 221:3–12. doi: 10.1002/path.2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Reis RC, Sorgine MH, Coelho-Sampaio T. A novel methodology for the investigation of intracellular proteolytic processing in intact cells. Eur J Cell Biol. 1998;75:192–7. doi: 10.1016/S0171-9335(98)80061-7. [DOI] [PubMed] [Google Scholar]
- 24.Panchuk-Voloshina N, et al. Alexa dyes, a series of new fluorescent dyes that yield exceptionally bright, photostable conjugates. J Histochem Cytochem. 1999;47:1179–88. doi: 10.1177/002215549904700910. [DOI] [PubMed] [Google Scholar]
- 25.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr Protoc Cell Biol. 2010;Chapter 14(Unit 14):11, 1–26. doi: 10.1002/0471143030.cb1411s46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bright GR, Fisher GW, Rogowska J, Taylor DL. Fluorescence ratio imaging microscopy. Methods Cell Biol. 1989;30:157–92. doi: 10.1016/s0091-679x(08)60979-6. [DOI] [PubMed] [Google Scholar]
- 27.Buccione R, Orth JD, McNiven MA. Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol. 2004;5:647–57. doi: 10.1038/nrm1436. [DOI] [PubMed] [Google Scholar]
- 28.Veracini L, et al. Two functionally distinct pools of Src kinases for PDGF receptor signalling. Biochem Soc Trans. 2005;33:1313–5. doi: 10.1042/BST0331313. [DOI] [PubMed] [Google Scholar]
- 29.Veracini L, et al. Two distinct pools of Src family tyrosine kinases regulate PDGF-induced DNA synthesis and actin dorsal ruffles. J Cell Sci. 2006;119:2921–34. doi: 10.1242/jcs.03015. [DOI] [PubMed] [Google Scholar]
- 30.Sandilands E, Brunton VG, Frame MC. The membrane targeting and spatial activation of Src, Yes and Fyn is influenced by palmitoylation and distinct RhoB/RhoD endosome requirements. J Cell Sci. 2007;120:2555–64. doi: 10.1242/jcs.003657. [DOI] [PubMed] [Google Scholar]
- 31.Sandilands E, et al. RhoB and actin polymerization coordinate Src activation with endosome-mediated delivery to the membrane. Dev Cell. 2004;7:855–69. doi: 10.1016/j.devcel.2004.09.019. [DOI] [PubMed] [Google Scholar]
- 32.Cary LA, Klinghoffer RA, Sachsenmaier C, Cooper JA. SRC catalytic but not scaffolding function is needed for integrin-regulated tyrosine phosphorylation, cell migration, and cell spreading. Mol Cell Biol. 2002;22:2427–40. doi: 10.1128/MCB.22.8.2427-2440.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fincham VJ, Frame MC. The catalytic activity of Src is dispensable for translocation to focal adhesions but controls the turnover of these structures during cell motility. EMBO J. 1998;17:81–92. doi: 10.1093/emboj/17.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frame MC, Fincham VJ, Carragher NO, Wyke JA. v-Src’s hold over actin and cell adhesions. Nat Rev Mol Cell Biol. 2002;3:233–45. doi: 10.1038/nrm779. [DOI] [PubMed] [Google Scholar]
- 35.Machacek M, Danuser G. Morphodynamic profiling of protrusion phenotypes. Biophys J. 2006;90:1439–52. doi: 10.1529/biophysj.105.070383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Machacek M, et al. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Renard M, et al. Knowledge-based design of reagentless fluorescent biosensors from recombinant antibodies. J Mol Biol. 2002;318:429–42. doi: 10.1016/S0022-2836(02)00023-2. [DOI] [PubMed] [Google Scholar]
- 38.Koide A, Jordan MR, Horner SR, Batori V, Koide S. Stabilization of a fibronectin type III domain by the removal of unfavorable electrostatic interactions on the protein surface. Biochemistry. 2001;40:10326–33. doi: 10.1021/bi010916y. [DOI] [PubMed] [Google Scholar]
- 39.Kraynov VS, et al. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–7. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 40.Martinez-Quiles N, Ho HY, Kirschner MW, Ramesh N, Geha RS. Erk/Src phosphorylation of cortactin acts as a switch on-switch off mechanism that controls its ability to activate N-WASP. Mol Cell Biol. 2004;24:5269–80. doi: 10.1128/MCB.24.12.5269-5280.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cory GO, Garg R, Cramer R, Ridley AJ. Phosphorylation of tyrosine 291 enhances the ability of WASp to stimulate actin polymerization and filopodium formation. Wiskott-Aldrich Syndrome protein. J Biol Chem. 2002;277:45115–21. doi: 10.1074/jbc.M203346200. [DOI] [PubMed] [Google Scholar]
- 42.Suetsugu S, et al. Sustained activation of N-WASP through phosphorylation is essential for neurite extension. Dev Cell. 2002;3:645–58. doi: 10.1016/s1534-5807(02)00324-6. [DOI] [PubMed] [Google Scholar]
- 43.Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM. c-Abl is activated by growth factors and Src family kinases and has a role in the cellular response to PDGF. Genes Dev. 1999;13:2400–11. doi: 10.1101/gad.13.18.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sini P, Cannas A, Koleske AJ, Di Fiore PP, Scita G. Abl-dependent tyrosine phosphorylation of Sos-1 mediates growth-factor-induced Rac activation. Nat Cell Biol. 2004;6:268–74. doi: 10.1038/ncb1096. [DOI] [PubMed] [Google Scholar]
- 45.Yang L, Kowalski JR, Zhan X, Thomas SM, Luscinskas FW. Endothelial cell cortactin phosphorylation by Src contributes to polymorphonuclear leukocyte transmigration in vitro. Circ Res. 2006;98:394–402. doi: 10.1161/01.RES.0000201958.59020.1a. [DOI] [PubMed] [Google Scholar]
- 46.Bouton AH, Riggins RB, Bruce-Staskal PJ. Functions of the adapter protein Cas: signal convergence and the determination of cellular responses. Oncogene. 2001;20:6448–58. doi: 10.1038/sj.onc.1204785. [DOI] [PubMed] [Google Scholar]
- 47.Polte TR, Hanks SK. Complexes of focal adhesion kinase (FAK) and Crk-associated substrate (p130(Cas)) are elevated in cytoskeleton-associated fractions following adhesion and Src transformation. Requirements for Src kinase activity and FAK proline-rich motifs. J Biol Chem. 1997;272:5501–9. doi: 10.1074/jbc.272.9.5501. [DOI] [PubMed] [Google Scholar]
- 48.DerMardirossian C, Rocklin G, Seo JY, Bokoch GM. Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling. Mol Biol Cell. 2006;17:4760–8. doi: 10.1091/mbc.E06-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Timpson P, Jones GE, Frame MC, Brunton VG. Coordination of cell polarization and migration by the Rho family GTPases requires Src tyrosine kinase activity. Curr Biol. 2001;11:1836–46. doi: 10.1016/s0960-9822(01)00583-8. [DOI] [PubMed] [Google Scholar]
- 50.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.