Abstract
The Aryl hydrocarbon receptor (AhR) has been best known for its role in mediating the toxicity of dioxin. Here we show that AhR overexpression is found among estrogen receptor (ER)α-negative human breast tumors and that its overexpression is positively correlated to that of the NF-κB subunit RelB and Interleukin (IL)-8. Increased DNA binding activity of the AhR and RelB is coupled to IL-8 overexpression in primary breast cancer tissue, which was also supported by in situ hybridization. Activation of AhR in vitro by 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) induced IL-8 expression in MDA-MB 436 and MCF-7 cells in an AhR and RelB dependent manner. Consistently, downregulation of RelB or AhR by small interfering RNAs (siRNA) decreased the level of IL-8 but increased expression of ERα in vitro in MCF-7 cells. Our results strongly suggest that RelB and AhR have a critical role in the regulation of IL-8 and reveal a supportive role of RelB and AhR in the anti-apoptotic response in human breast cancer cells. AhR and RelB may present a novel therapeutic target for inflammatory driven breast carcinogenesis and tumor progression. Overexpression of pro-survival factors AhR and RelB may explain the process of the development of environmentally-induced type of breast cancers.
Introduction
Recent reports have shown that the development of certain types of breast tumor is associated with inflammation and an inappropriate production of chemokines especially IL-8, which appears to be a critical phase in breast tumor metastasis [1–3]. Previous reports from our own group have demonstrated the critical role of the AhR in the transcriptional induction of IL-8 mediated by an activated AhR [4]. Interestingly, it was reported initially by David H. Sherr’s group that high levels of AhR expression is detected in chemical-induced mammary tumors in vivo in rats [5] as well as in several mouse and human cell lines exhibiting malignant phenotypes [6]. It was pointed out by the same group of scientists [7] that there is likelihood of the AhR contributing greatly to ongoing mammary tumor cell growth and apoptosis resistance while promoting transition to an invasive, metastatic phenotype in these cases. The above observations at the same time have raised some important questions: how does the overexpressed AhR contributes to those cellular changes, even in the absence of its ligands, how it coordinates its actions with signaling of growth factors and hormones, how it causes inflammatory responses in those cells, and how it contributes to malignant progression of those cells? Meanwhile there have been a number of recent discoveries regarding the physiological roles of the AhR itself (i.e. its natural biological activities that is not induced by exogenous ligands) mostly based on studies on AhR knockout mice [8–10] as well as the roles of AhR as one of the major coordinators of cellular stress responses, particularly inflammatory responses [11]. The role of AhR as a pro-survival factor conferring cells to acquire apoptosis resistance, even in the absence of any exogenously ligand of this receptor, has been reported in an ERα-negative MCF10AT1 breast cancer cells from our group [12]. These recent developments point to the possibility that the overexpression and activation of the AhR could play an important role in aiding the process of malignant transformation of those transformed mammary epithelial cells.
Despite the clear-cut demonstration of overexpression of AhR in DMBA-induced breast tumors in rats and mice, and a number of established human breast cancer cell lines [5–6], data about a possible role of AhR in human primary breast tumor samples are scarce. Accordingly the major objective of this study has been to identify the type of breast tumors that overexpresses AhR as well as RelB and to gain insight into the mechanism of the AhR-dependent progression of those mammary epithelial cells leading to overexpression of IL-8 and the malignant phenotype.
Materials and methods
Reagents and Antibodies
Dimethylsulfoxide (Me2SO) and Phorbol-12-myristate-13-acetate (TPA) were obtained from SIGMA (St. Louis, MO). [y-32P] ATP (6000 Ci/mmol) was purchased from ICN (Costa Mesa, CA). TCDD (>99% purity) was originally obtained from Dow Chemicals Co (Midland, MI). Other molecular biological reagents were purchased from Qiagen (Valencia, CA) and Roche (Indianapolis, IN). Polyclonal RelA, CEBPβ (Santa Cruz Biotechnology, Santa Cruz, CA), NF-κB member RelB, p50 (Active Motif, Carlsbad, CA) and polyclonal AhR (Novus Biologicals, Littleton, CO) antibodies were used for supershift in EMSA. Actin AC-15 antibody was purchased from Sigma.
Breast tissue samples
Frozen human breast tissues with pathology reports were obtained from the National Cancer Institute Cooperative Human Tissue Network and the University of California at Davis Cancer Center Specimen Repository. All of the samples were approved for laboratory use by the institutional review board at the University of California at Davis School of Medicine. Samples were homogenized and analyzed via Western blot analysis as described [13].
Cell Culture, chemical treatment and transient transfection experiments
Human breast cancer cell lines were obtained from A.T.C.C. (Manassas, VA) and maintained in DMEM medium containing 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Two ERα-negative cell lines have been developed through long-term exposure to 1 nM of 17β-estradiol (E2) (for 20 passages for P20E) from MCF10AT1 [12]. In all cases cells were treated with TCDD to make the final concentration of 10 nM in the medium, with 1 μl of dimethylsulfoxide (DMSO) as the vehicle. The standard methods of treating those cells with specific chemical inhibitors have already been described [12]. The MCF10AT1 cell line was originally developed by Dr. Fred Miller’s group at Wayne State University by stably transfecting MCF10A cells with a mutant c-Ha-ras (codon 12 valine) [14–15]. While the transfection of c-Ha-ras alone caused a modest level of ERα (about 1/10 of MCF-7 cells) expression in these cells, it did not give them the ability to spontaneously form pre-neoplastic lesions in nude mice. After xenografting the MCF10A cells into athymic mice, hosts treated with E2 produced pre-neoplastic colonies after one year, which sporadically progressed to a proliferative state; from these colonies the MCF10AT1 line was isolated [15–16]. Upon exposure to 1 nM E2 for multiple passages MCF10AT1 cells gradually lost ERα functions, and gained overexpression of AhR after 17 to 20 passages [12]. It must be noted that no other artificial alteration than the E2 exposure for 20 passages was necessary to produce this unique genotype. For transient transfection MCF-7 cells were plated in DMEM with 10% FBS.Transfection of short interfering RNA (siRNA) or plasmid DNA was performed using jetPEI™ (PolyTransfection, Qbiogene, Irvine, CA), according to the manufacturer’s instructions. The transfection was allowed to proceed for 24 h and cells were treated with 10 nM TCDD for another 24 h where indicated. Cells transfected with the IL-8 luciferase reporter construct were then washed twice with PBS and lysed with 100 μl passive lyses buffer. Luciferase activities were measured with the Luciferase Reporter Assay System (Promega, Madison, WI) using a luminometer (Berthold Lumat LB 9501/16, Pittsburgh, PA). In case of siRNA transfection, the reduction of the target RNA and protein was detected by quantitative real-time RT-PCR and Western blot. siRNA to target human AhR (catalog no. M-004990) was designed and synthesized by Dharmacon (Lafayette, CO). siRNA to target human RelB (5′-GGAUUUGCCGAAUUAACAA-3′) and a negative control siRNA (catalog no. 10272280) were synthesized by Qiagen (Valencia, CA) as described earlier [4].
RNA isolation, cDNA synthesis and real time PCR
Total RNA from primary breast tissue and breast cancer cell lines was isolated and reverse transcribed into cDNA as described before (4) by using Trizol and the RNeasy Mini kit (Qiagen, Valencia, CA). Quantitative detection of mRNA level was performed with a LightCycler Instrument (Roche Diagnostics, Mannheim, Germany) using the QuantiTect SYBR Green PCR Kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The primers for each gene were designed using OLIGO primer analysis software, provided by Steve Rosen and Whitehead Institute/MIT Center for Genome Research. All PCR assays were performed in triplicate. The intra-assay variability was <7%.
Nuclear protein extraction and EMSA
Briefly, 300 mg of frozen tissue were pulverized in liquid nitrogen. The tissue powders were resuspended in homogenization buffer (1 g/ml) in 10 mM HEPES (pH 7.9), 10 mM KCl, 0.1 mM EDTA, 0.1mM EGTA, 50 mM sucrose, 1 mM dithiothreitol (DTT), 0.5 mM phenylmethylsulfonylfluoride (PMSF), leupeptin (5 μg/ml), and aprotinin (5μg/ml). Samples were Dounce homogenized for 30 strokes with a loose pestle and then 30 strokes with a tight pestle. The KCl concentration was then adjusted to 100 mM and the nuclei were washed twice with the homogenization buffer with 100 mM KCl. Nuclear proteins were extracted on ice for 30 min in 2 packed nuclear volumes containing 10 mM HEPES (pH 7.9), 400 mM NaCl, 0.1 mM EDTA, 0.1 mM EGTA, 20% glycerol, 1 mM DTT, 0.5 mM PMSF, leupeptin (0.5 μg/ml), and aprotinin (5 μg/ml). Nuclear proteins from cell lines were extracted as described earlier [4]. DNA-protein binding reactions were carried out in a total volume of 15 μl containing 10 μg nuclear protein, 80,000 cpm of DNA oligonucleotide, 25 mM Tris buffer (pH 7.5), 50 mM NaCl, 1 mM EDTA, 0.5 mM dithiothreitol, 5% glycerol, and 1 μg poly (dI-dC). The samples were incubated at room temperature for 20 min. Supershift analysis were performed by adding 2 μg of the corresponding antibodies to the reaction mixtures. Competition experiments were performed in the presence of a 100-fold molar excess of unlabeled DNA fragments. Protein-DNA complexes were resolved on a 4% nondenaturating polyacrylamide gel and visualized by exposure of the dehydrated gels to X-ray films. For quantitative analysis, respective bands were quantified using a ChemiImager™4400 (Alpha Innotech Corporation, San Leandro, CA).
Immunohistochemistry
Immunohistochemistry with paraffin sections of breast tissue was performed with Vectastain Kits (Burlingame, CA) according to the manufacturer’s instructions. In brief, sections were deparaffinized and incubated with primary antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA), Novus Biologicals (Littleton, CO), and R&D Sytems (Minneapolis, MN) for immuno-histochemical staining of RelB (clone C-19), AhR (NB100-39027), and IL-8 (clone 6217), respectively. After 18h at 4 °C, sections were incubated with a biotinylated Anti-Ig and avidin-biotin-HRP for 30 min each. Tissue sections from tumors and non-tumors were stained with DAB used for visualization of the antibody binding. In each tissue section, hotspots with nuclear staining were searched. Digital images were acquired with an Olympus microscope (model BH-2, Olympus, New York) and digital image transfer software (Leica Application suite 2.7).
Antibodies and Western blotting
A polyclonal anti-human RelB and AhR, a polyclonal antibody against human ACTIN (SC-1616), a horseradish peroxidase conjugated secondary antibody, and pre-stained standard markers (SC-2361) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Whole cell lysates (30 μg) were separated on a 10% SDS–polyacrylamide gel and blotted onto a PVDF membrane (Bio-Rad, Herkules, CA). The antigen–antibody complexes were visualized using the chemiluminescence substrate SuperSignal®, West Pico (Pierce, Rockford, IL) as recommended by the manufacturer. For quantitative analysis, respective bands were quantified using a ChemiImager™4400 (Alpha Innotech Corporation, San Leandro, CA).
Apoptosis Detection by Annexin V Staining
Cells were seeded at 1×105 cells in 60mm culture dishes in 2 ml medium after 24 hrs media was refreshed. After 21 hrs, cells were treated with various inhibitors and incubated for and additional 3 hrs. Apoptosis was induced by UV light (100 mJ/cm2 for 100 seconds) that was generated by a mercury lamp, but filtered through the petri dish cover before reaching the cells in culture and incubated at 37°C for 4 hrs. This treatment effectively gives cell enough exposure to UVB to cause apoptosis. Afterwards, 2 μl CaCl2 (20 mM), 2 μl annexin V-fluorescein isothiocyanate (50 μg/ml, Sigma) and 2 μl of propidium iodide (PI) solution (50 μg/ml) were added directly to each plate. Apoptotic cells were counted directly by using the fluorescence microscope (Leitz, Wetzlar, Germany). Both annexin V-positive and annexin V-PI-double-positive cells were considered to be apoptotic.
Statistical analysis
All quantitative experiments were repeated a minimum of three times and results were expressed as means ± standard deviations. Data were evaluated statically by Student’s t-test at the significant level of at least P < 0.05.
Results
Western Blot analysis of AhR and RelB in primary breast tissues
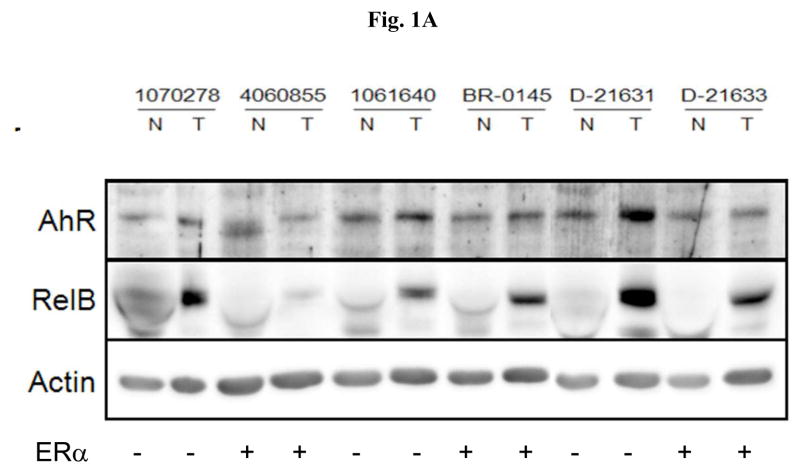
To analyze the expression level of AhR and RelB in primary breast tissue we performed Western blot analysis with whole cell protein from 6 pairs of breast tumor samples from the genotype of ERα-negative and ERα-positive samples. For this purpose we compared their protein expression of AhR and RelB in both the tumor sample (designated by T) and the matched normal breast tissue samples (designated as N) from each patient (Fig. 1A). A relatively high level of RelB and AhR protein was found in ERα-negative tumor tissue (lanes 2, 6 and 10). In five of the six samples we found a significantly higher level of RelB protein in the tumor tissue compared to normal tissue (Fig. 1A). We observed a less pronounced but significant increase of AhR protein in 4 out of 6 patients compared to the corresponding normal tissue.
Fig. 1.
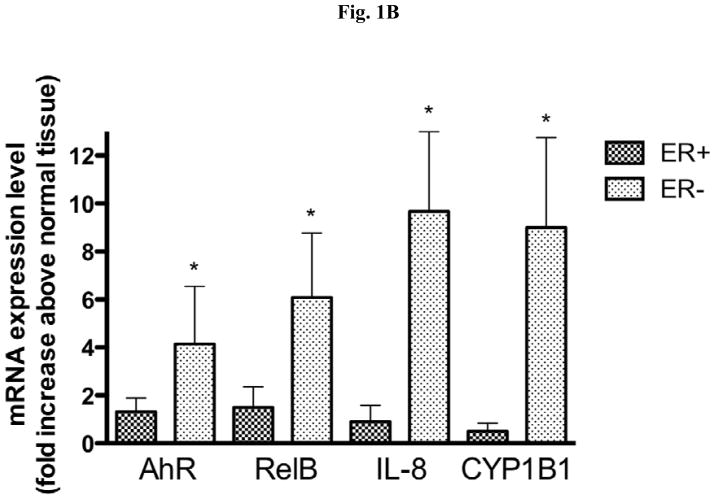
Expression of AhR and RelB in ER positive and ER negative breast tumors. (A) Protein level of AhR and RelB in primary breast tissue samples. Whole tissue protein was used to analyze expression of AhR and RelB in both the tumor sample (designated by T) and the matched normal breast tissue samples (designated as N) from 6 different patients. The expression level of β-actin was used as a loading control. (B) Total RNA was extracted from a total of 20 (10 ER-negative and 10 ER-positive) breast tumor samples and their corresponding normal tissue as described under Material and Methods. The mRNA expression of AhR, RelB, IL-8, and CYP1B1 in ERα-negative (dotted bars) and ERα-positive (shaded bars) samples was analyzed by real time PCR. *, significantly different from mRNA expression in corresponding normal tissue (p<0.01) (C) Nuclear and cytoplasmic staining of breast tumor tissue sections using antibodies against AhR and NF-κB member RelB. For each slide, a breast tumor with corresponding normal tissue was selected which was identified as ERα-negative (ID#D21631). Images of random fields were taken at a magnification of 100× for RelB and AhR.
Relative gene expression differences of NF-κB and AhR target genes in breast tumors versus normal breast tissue
Here we investigated the mRNA expression level of the transcription factors RelB and AhR, as well as their downstream regulated genes IL-8 and cytochrome P450 (CYP)1B1 in tumor tissues of breast cancer patients, by analyzing the mRNA level of normal breast tissue and tumor tissue from a total of 20 patients. The samples were analyzed according to their ERα expression status and it became apparent that the ERα-negative tumors show a consistently higher expression levels of the markers studied compared tumor tissue identified as ERα-positive (Fig. 1B). The elevated levels of the expressions of RelB, AhR, IL-8 and CYP1B1 were statistically significant in ERα-negative tissue samples as compared to the corresponding normal tissues, each collected from the same patient. Interestingly, the expression of CYP1A1, which is well known to be regulated by the classical AhR pathway together with ARNT, was not increased in ERα-positive or ERα-negative tumor samples (data not shown).
In situ hybridization of AhR, RelB and IL-8 proteins
To visualize the localization of AhR, RelB, and IL-8 proteins within breast tissues, we have conducted in situ hybridization experiments on a representative pair of normal versus tumor tissue from the same patient. This tissue sample (ID#D21631) is a representative ERα-negative tissue sample and was identified to express high mRNA levels of RelB and IL-8 in tumor tissue compared to the corresponding normal tissue. The result of AhR hybridization (Fig. 1C) showed that AhR protein is expressed abundantly in the tumor tissue examined. AhR overexpressing cells in tumor samples were spread all over the tumor tissue, and not confined to the organized ductal tubular structures. A similar result was obtained after hybridization with an IL-8 antibody, showing an abundant IL-8 expression in the tumor tissue compared to the normal tissue. In contrast to the tumor tissue, the corresponding normal tissue showed organized ductal structures along with stroma cells. While there were sporadic dark spots in both celltypes, the overall expression of AhR and IL-8 appears to be low as compare to the corresponding tumor tissue. Regarding RelB hybridization (Fig. 1C), it appears that in the normal tissue sample the RelB protein is found only around the cells lining the ductal structures. In contrast, we found a much higher level of expression of RelB in the tumor sample as a whole compared to the normal tissue. Upon close examination the main characteristic of the tumor sample was found to be the tremendous number of cancer cells overexpressing RelB protein packed tightly.
DNA-binding activity of nuclear transcription factors in tumor versus normal breast tissue
To assess the nuclear protein binding activities to specific response elements, we used electromobility shift assay (EMSA) with nuclear protein preparations from an ERα-negative tumor sample in comparison to the normal tissue counter-part (ID#D21631). For this purpose we selected several oligonucleotide probes which could be involved in the regulation of IL-8 as follows: a) a RelB and AhR binding sequence (designated as “RelBAhRE”) in the −50 to +1 base pair region of the human IL-8 promoter [17–18] a consensus element for NF-κB, c) AP-1, d) DRE (dioxin response element), and e) CEBP. The most outstanding feature of the data shown in Fig. 2A is that nuclear protein binding to the RelBAhRE oligonucleotide is clearly increased in the tumor (T) tissue compared to normal (N) tissue (lanes 1 and 2). This band contains the heterodimer of RelB and AhR [17], which was confirmed in the super-shift assay (Fig. 2B). It is worthy to point out that a similar complex also exists in EMSA of nuclear protein binding to the consensus NF-κB-response element (lanes 3 and 4, Fig. 2A). The increased binding in the tumor sample appears to be accompanied by a simultaneous decrease in the intensity of the upper complex compared to the normal sample. The upper complex is expected to include RelA (p65), and the above results may suggest that the increase in the titer of RelB and AhR represents the negative cellular feedback response to the hyper-inflammatory status of those transformed cells [19]. Interestingly the patterns of nuclear protein binging to AP-1 (lanes 7 and 8) and C/EBP consensus oligonucleotides (lanes 10 and 11) were opposite; the former showing less, whereas the latter showed increased DNA-binding activity in the tumor sample compared to the normal counter-part. The DRE binding activity was comparable for the nuclear protein preparation from both, normal or tumor tissues. In order to confirm that the protein band showing a conspicuous increase in binding to the labeled RelBAhRE probe (Fig. 2A, lanes 1 and 2) contains AhR and RelB proteins, we have run an EMSA supershift assay. The result shown in Fig. 2B demonstrates that the intensity of this band decreases significantly when the nuclear protein preparation has been pre-treated with either AhR (lanes 3 and 4) antibody or RelB (lanes 7 and 8) antibody. As for the meaning of the increased protein binding of nuclear proteins from tumor tissue to the consensus C/EBP response element (Fig. 2A lanes 10 and 11), we performed supershift assay with a specific antibody against the C/EBPβ protein (Fig. 2C, lanes 3 and 4). From the results we concluded that the increased protein binding to the C/EBP response element in tumor tissue is due mostly to that of C/EBPβ protein.
Fig. 2.
Increased DNA binding activity of RelB and AhR in breast tumor tissue. (A) DNA-binding activity of a RelB/AhR response element and other consensus elements relevant for the activity of IL-8 in tumor versus normal breast tissue. Nuclear protein extracts of normal (N) or tumor− (T) breast tissue from a cancer patient (ID#D21631) was incubated with oligonucleotides containing the AhR/RelB-binding site identified on the promoter of IL-8 as well as consensus elements of NF-κB, AP-1, C/EBP or DRE. (B) Supershift analysis of nuclear protein extracts from normal and tumor tissue with AhR/RelB probe of IL-8. A possible binding of AhR, and NF-κB subunits RelA, and RelB was tested by supershift analyses using specific antibodies for the corresponding proteins. To confirm specificity a 100-fold excess of the unlabeled oligonucleotides was added. (C) Supershift analysis of nuclear protein extracts from normal and tumor tissue with a C/EBP consensus probe. Binding of C/EBPβ was identified by supershift analyses using specific antibody for C/EBPβ protein.
Regulation of IL-8 by AhR and RelB in human breast cancer cells
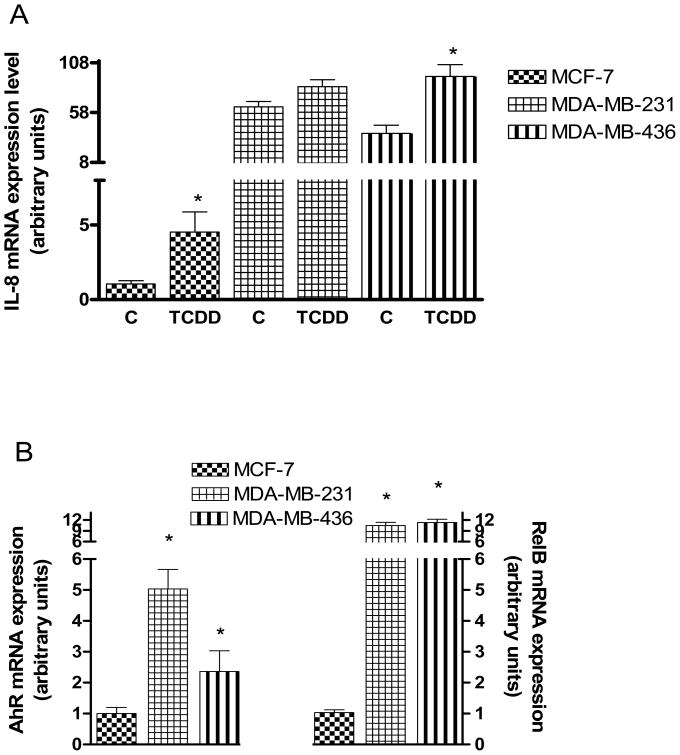
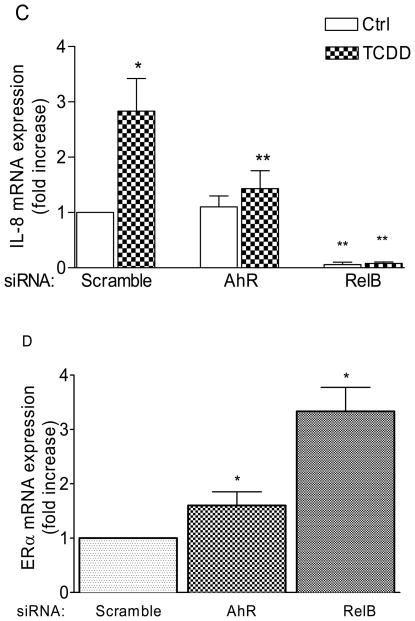
An important item, still requiring confirmation is the functional correlation between the expression of IL-8 and that of RelB in association with AhR in mammary epithelial cells. For this purpose we surveyed the existing, well established lines preferably one of which should exhibit a high titer of ERα and two showing very low levels of ERα that is coupled to elevated expression of AhR and RelB. After a search we identified ERα-positive MCF-7 and ERα-negative MDA-MB-231 and MDA-MB-436 cells fit reasonably to the above requirements. Both ERα-negative cells showed much higher levels of constitutive expression of IL-8 mRNA than MCF-7 cells (Fig. 3A). When cells were treated with TCDD to activate AhR, IL-8 expression increased significantly in MCF-7 and MDA-MB-436 cell lines. We also confirmed the elevated levels of the mRNA expressions of AhR and RelB (Fig. 3B) in these two ER-negative breast cancer cell lines, which showed that AhR and especially RelB are significantly higher expressed in ER-negative MDA-MB-231 and MDA-MB-436 as compared to ER-positive MCF-7 cells. Transfection experiments with siRNA to target AhR and RelB suggest that IL-8 expression in MCF-7 largely depends on AhR and RelB, particularly the latter, suppressing TCDD-induced expression of IL-8 (Fig. 3C). Transfection of MCF-7 with siRelB also suppressed the constitutive expression of IL-8. On the other hand, the mRNA level of ERα was significantly increased in MCF-7 cells transfected with siRNA specific for AhR and RelB (Fig. 3D), confirming the inverse relationship between RelB and ERα. Overexpression of AhR and RelB in MCF-7 cells caused a further increase of IL-8 mRNA expression supporting the role of AhR and RelB in the regulation of IL-8 (Fig. 3E). As measured by ELISA, in MDA-MB-231 cells, only LPS was able to increase the secretion of IL-8, whereas TCDD but not LPS stimulated the secretion of IL-8 significantly only in MCF-7 cells (Fig. 3F), demonstrating that the IL-8 gene is fully functional in MCF-7 and MDA-MB-231 cells.
Fig. 3.
Analysis of IL-8, AhR and RelB mRNA expression in ERα-negative MDA-MB-231 and MDA-MB-436 cells in comparison to ERα-positive MCF-7 cells. (A) Quantitative IL-8 mRNA expression analyses in MCF-7, MDA-MB-231 and MDA-MB-436 cells after treatment with TCDD for 24 h. (B) Relative expression of AhR and RelB mRNA in MCF-7, MDA-MB-231 and MDA-MB-436 cells. Expression of mRNA was analyzed by real-time PCR as described above.
Induction of mRNA expression of IL-8 by TCDD is RelB- and AhR-dependent. (C) Total RNA was prepared 48 h post-transfection with either a scrambled siRNA or a specific siRNA targeted against AhR or RelB and IL-8 mRNA expression was analyzed after treatment with 10 nM TCDD for 24 h (shaded bars). (D) ERα mRNA was analyzed 48 h post-transfection with either a scrambled siRNA or a specific siRNA targeted against AhR or RelB in MCF-7 cells. (E) IL-8 expression was analyzed in AhR− and RelB transfected MCF-7 cells after treatment with 10 nM TCDD for 24 h (shaded bars). (F) Secretion of IL-8 protein stimulated in MCF-7 and MDA-MB-231 cells. Cells were treated with TCDD (10 nM) or LPS (100 ng/ml) as a positive control. After 72h the cell culture supernatant was collected to evaluate IL-8 protein levels by ELISA. Results are expressed as pg IL-8/106 cells.
*, significantly different from control (p<0.01); **, significantly lower than TCDD-treated cells transfected with scrambled siRNA
Based on the report from Wang et al [20] that suppression of ERα leads to up-regulation of RelB, we addressed this question by modulating the expression of AhR and RelB in MCF-7 cells. The result confirmed the importance of these two genes in controlling the expression of IL-8 expressions in MCF-7 cells (Fig. 3C and E). Furthermore, we could show that suppression of AhR and RelB results in up-regulation of ERα (Fig. 3D) confirming the inverse relationship between RelB (and to a lesser extent AhR) and ERα, as reported earlier [20].
Establishment of an in vitro model that express a similar genotype as ERα-negative primary breast tumors
Although the above studies on those three existing breast cancer cell lines have helped us to ascertain the importance of AhR and ERα in controlling the expression of RelB and IL-8, which were achieved by the use of artificial means such as transfecting them with siRNA, overexpression plasmids and TCDD, there are limitations in the above approach, since besides those genes we have identified there could be a host of differences in their basic genotypes that are not known to us. A better approach would be to have a pair of genetically matched cell lines, one ER-positive line with normal expression of AhR-RelB, and the other ER-negative line showing this AhR-RelB overexpressing genotype. Fortunately we have such a MCF breast cancer line (i.e. P20C/P20E) that was already available in our laboratory as described in our previous publications [12]. The ERα-negative cell line, P20E from MCF10AT1 [12] turned out to be a suitable in vitro model for this sub-type of breast tumor. To ascertain the differences in the protein expression levels of AhR and RelB between ER+ and ER− sub-lines within P20C and P20E, Western blotting experiments were conducted with specific antibodies directed AhR and RelB. It was found that both AhR and RelB protein levels were increased in the E2-selected cells (P20E) as compared to the matched control P20C (Fig. 4A).
Fig. 4.
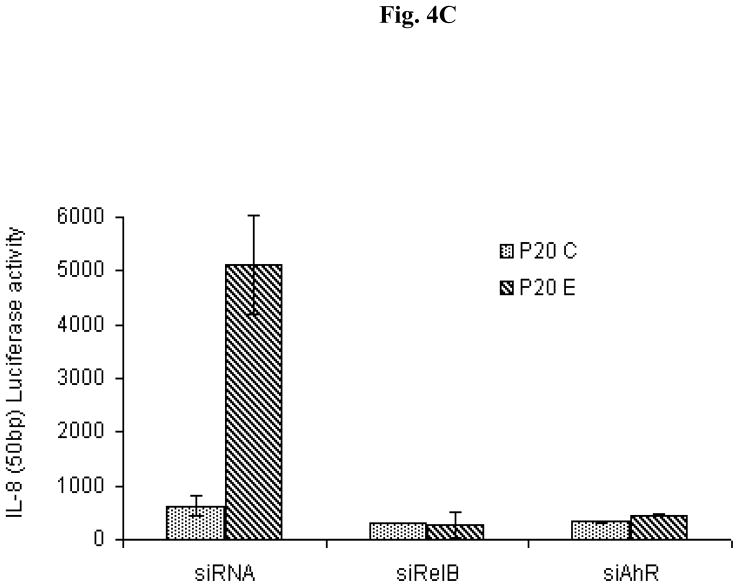
Increased expression of AhR and RelB in MCF10AT1 after long-term treatment with E2 (P20E). (A) Western blot of total cell protein was prepared from untreated P20C and P20E cells derived from MCF10AT1 to analyze protein level of AhR and RelB. (B) EMSA with nuclear proteins from P20C and P20E cells using a NF-κB oligonucleotide. Super-shift assay was performed with specific antibodies against AhR and RelB proteins to identify the major constituent nuclear transcription factor proteins. (C) Suppressive effect of siRNA targeting AhR and RelB on P20E expression of IL-8 gene activity in comparison to P20C as assessed by luciferase activity of the reporter construct containing the RelBAhRE site of the IL-8 promoter.
Accordingly, EMSA analyses were performed with nuclear proteins from P20C and P20E cells. The results revealed that a nuclear protein complex containing RelB and AhR exist only in the nuclear protein preparation from AhR overexpressing P20E cells, but not in control P20C cells, as judged by nuclear protein binding to the NF-κB probe (Fig. 4B), confirming the similar observation made in breast tumors (Fig. 2AB). The functional significance of the AhR-RelB interaction in P20E cells in comparison to P20C cells was studied next by using the IL-8 reporter plasmid containing the RelBAhRE binding site. The results showed that the increased IL-8 promoter activity in P20E compared to P20C cells is clearly susceptible to inhibition either by siAhR or by siRelB (Fig. 4C) as in the case of MCF-7 cells (Fig. 3C), except that in the case of P20C/P20E there was no need to rely on the use of TCDD as the elevated AhR in P20E has already been shown to be constitutively activated without exogenous ligands [12] The significant role of the RelB:AhR complex in expressing the IL-8 in P20E cells verified from this series of experiments indeed support our previous observation that IL-8 expression in severl types of cells is mediated through nuclear protein binding of AhR and RelB [4].
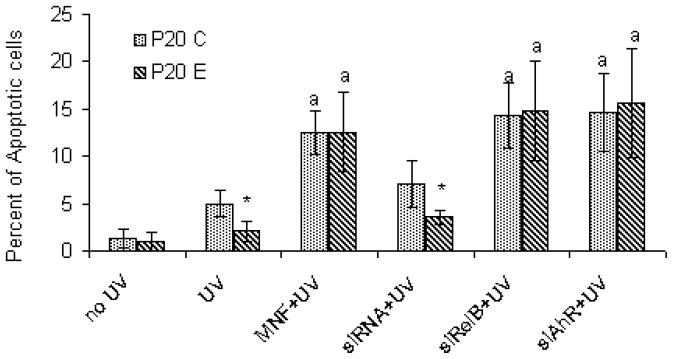
To assess the phenotypic significance of the AhR/RelB interaction, we further examined the sensitivity of these two lines derived from MCF10AT1 cells to UVB-induced apoptosis. The results showed that P20E cells are rather resistant to UVB-induced apoptosis compared to P20C cells (Fig. 5). Treatment with either an AhR antagonist 3′-methoxy-4′-nitroflavone (MNF), siRNA targeting AhR (siAhR) or RelB (siRNA), restores the apoptotic response and even further increased the number of apoptotic cells toward UV in P20E as well as P20C cells, which indicates the critical role of AhR and RelB for the anti-apoptotic effect in P20E cells.
Fig. 5.
Study on the contribution of AhR and RelB on the apoptotic response in P20E cells P20C cells. For this purpose cells were exposed to UV-irradiation after treatment with AhR antagonist (10 μM MNF), or siRNA transfection targeting AhR (siAhR) or RelB (siRelB), as described in Method and materials. Apoptotic cells were counted 4 hrs after UVB-irradiation. *, significantly different between P20 C and P20 E (p<0.01); a, significantly different between UVB treated alone and UVB treated with MNF or siRNA.
Discussion
The initial observation that the AhR and RelB overexpressing breast tumors are found mainly among the ERα-negative type of tumors has already provided an important clue. A literature search for a similar sub-type of breast cancer overexpressing RelB showed that the case of inflammatory breast cancers [21] appears to fit best to this sub-type. In that work it has been demonstrated that overexpression of RelB and translocation of RelB into the nucleus of inflammatory breast cancer cases are found only among ERα-negative tumors. However, it is premature to conclude that AhR overexpression belongs to this subtype of breast tumor and more work will be needed to further characterize this sub-type of breast tumors.
Regarding the question on the validity of comparing directly the marker expressions of the tumor tissue versus the equivalent tissues from the same patient as a pair, we acknowledge that the proportion of the mammary epithelial cells in relation to other types of cells in tumors is expected to be significantly higher that that found in the normal tissue counterpart. This difference in the cellular composition could bring in a potential bias in our interpretation of the results. However, the fact that we could not find a significant difference between tumor versus normal tissue, in terms of expressions of other markers such as CYP1A1 or RelA (data not shown) compared to markers reported in this work, argues against the contribution of a uniform bias affecting our interpretation. Additionally the finding that the same genotype of cultured breast cancer cells helps greatly to conclude that those markers are indeed expressed by this sub-type of transformed mammary epithelial cells.
There is additional evidence for the importance of the loss of ERα function leading to the development of this sub-type in vitro. First, AhR and RelB overexpressing P20E cells of transformed mammary epithelial cells were obtained through their long-term exposure to E2 to down-regulate ERα [12]. Wang et al. [20] obtained a similar genotype by artificially overexpressing RelB in MCF-7 breast cancer cells, which showed the repressed state of ERα expression. The observation of an inverse relationship of ERα and RelB was also confirmed in the current study by silencing RelB in MCF-7 cells, which lead to increased expression of ERα. ERα-negative type of breast tumors has been known to be associated with high production of inflammatory cytokines that include IL-8 [22]. IL-8 has been known to be also associated with Cox-2 overexpressing type of breast tumors [23], which is involved in metastasis. Results from the current study demonstrate for the first time a tight association of the AhR with RelB and the overexpression of IL-8 with this inflammatory type of breast tumors.
AhR has been known mostly for its role in mediating the toxicity of dioxin. However, in recent years enough evidence has been accumulated to shed light on its multiple biological roles. One of the major functions of AhR being proposed is its role as a mediator of “stress response” and regulator of inflammatory responses to promote cell survival [11]. For instance, data from the literature show the potential role of the AhR to prevent apoptosis, which is associated with mammary tumor development [7]. A number of reports, including from our own laboratory, have shown that activation of AhR by dioxin leads to a significant inhibition of apoptosis in breast epithelial cells [24] as well as in lymphoma cells [14]. According to our recent findings the AhR may interact with RelB in the nucleus in U937 macrophages, which is stimulated through activation of AhR and regulates gene expression of certain chemokines including IL-8 [4, 18]. Independently others have shown that overexpression of the NF-κB subunit RelB causes prevention of apoptosis, which is associated with tumorigenesis including lymphoma, mammary, and prostate tumor development [25–27], although the possible involvement of AhR has not been considered in these studies. For instance, activation of the alternative NF-κB signaling pathway via the NF-kB subunits RelB and p52 has been associated with up-regulation of anti-apoptotic proteins in Non-Hodgkin lymphoma cells [25]. Ectopic expression of relB, or RelB knockdown using small interfering RNA, demonstrated the important role of this subunit in control of tumor cell survival and implicated activation of the anti-apoptotic factors survivin and manganese superoxide dismutase [28]. An aberrant constitutive expression of NF-κB subunits including RelB has been reported in over 90% of breast cancers. RelB complexes were observed in mouse mammary tumors induced by either ectopic c-Rel expression or carcinogen exposure [26] and a constitutive de novo RelB synthesis is selectively active in invasive ERα-negative breast cancer cells. Induction of Bcl-2 by RelB promoted the more invasive phenotype of ERα-negative cancer cells. Indeed the inhibition of de novo RelB synthesis may represents a new mechanism whereby ERα prevents epithelial to mesenchymal transition [20].
It must be pointed out that despite these recent findings only little had been known about the role of RelB interacting with the AhR in human breast tumorigenesis. Together, data of the current study indicate that the anti-apoptotic effect mediated through an activated AhR and the promotion of tumor development involves the interaction of AhR and RelB associated with overexpression of IL-8. In this regard, it is interesting to note that, in addition to AhR, both IL-8 [29] and RelB [30] are regarded as pro-survival factors contributing to carcinogenesis. When viewed in this way, the participation of AhR, a known environmental sensor and an effective pro-survival factor, in the formation of this sub-type of ER-negative breast tumors may provide a valuable hint to the selective advantages of this sub-type of breast cancer cells in surviving in the hostile cellular environment.
It is well known that breast cancer is a disease that can be affected by the environment. While genetic factors are known to contribute, the contemporary view emerging is that carcinogenesis is the result of accumulation of many cellular changes -- even when each contributing a small change --occurring during long time periods. This appears to be particularly applicable to the cases of hormonal carcinogenesis [31]. While it should be considered that those cellular changes can be caused by mutations including those occurring during DNA repair [32], the findings of current study point to the possibility that long periods of accumulation of a number of pro-survival factors and their functional integration, without involving any known mutations, may contribute to the process of malignant transformation of mammary epithelial cells as well. Particularly intriguing is the role of AhR interacting with RelB to express this inflammatory type of breast cancers.
Acknowledgments
Funding
This study has been supported by Research Grant FAS0703859 from the Susan G. Komen Foundation for the Cure, American Heart Association Western Affiliate 0765056Y and NIEHS grant R21ES15846.
We like to thank Laura Van Winkle (University of California Davis, CA) for technical assistance. We thank Ulrich Siebenlist (National Institute of Health, Bethesda, MD), David Sherr and Gail Sonenshein (Boston University, MA) for providing us with critical plasmids.
Footnotes
Conflict of Interest Statement: None declared.
References
- 1.Ali S, Lazennec G. Chemokines: novel targets for breast cancer metastasis. Cancer Metastasis Rev. 2007;26:401–420. doi: 10.1007/s10555-007-9073-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freund A, Jolivel V, Durand S, Kersual N, Chalbos D, Chavey C, Vignon F, Lazennec G. Mechanisms underlying differential expression of interleukin-8 in breast cancer cells. Oncogene. 2004;23:6105–6114. doi: 10.1038/sj.onc.1207815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bièche I, Chavey C, Andrieu C, Busson M, Vacher S, Le Corre L, Guinebretière JM, Burlinchon S, Lidereau R, Lazennec GCXC. Chemokines located in the 4q21 region are up-regulated in breast cancer. Endocr Relat Cancer. 2007;14:1039–1052. doi: 10.1677/erc.1.01301. [DOI] [PubMed] [Google Scholar]
- 4.Vogel CF, Sciullo E, Li W, Wong P, Lazennec G, Matsumura F. RelB, a new partner of aryl hydrocarbon receptor-mediated transcription. Mol Endocrinol. 2007;21:2941–2955. doi: 10.1210/me.2007-0211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Trombino AF, Near RI, Matulka RA, Yang S, Hafer LJ, Toselli PA, Kim DW, Rogers AE, Sonenshein GE, Sherr DH. Expression of the aryl hydrocarbon receptor/transcription factor (AhR) and AhR-regulated CYP1 gene transcripts in a rat model of mammary tumorigenesis. Breast Cancer Res Treat. 2000;63:117–131. doi: 10.1023/a:1006443104670. [DOI] [PubMed] [Google Scholar]
- 6.Shin SR, Sánchez-Velar N, Sherr DH, Sonenshein GE. 7,12-dimethylbenz(a)anthracene treatment of a c-rel mouse mammary tumor cell line induces epithelial to mesenchymal transition via activation of nuclear factor-kappaB. Cancer Res. 2006;66:2570–2575. doi: 10.1158/0008-5472.CAN-05-3056. [DOI] [PubMed] [Google Scholar]
- 7.Schlezinger JJ, Liu D, Farago M, Seldin DC, Belguise K, Sonenshein GE, Sherr DH. A role for the aryl hydrocarbon receptor in mammary gland tumorigenesis. Biol Chem. 2006;387:1175–1187. doi: 10.1515/BC.2006.145. [DOI] [PubMed] [Google Scholar]
- 8.Gonzalez FJ, Fernandez-Salguero P. The aryl hydrocarbon receptor: studies using the AHR-null mice. Drug Metab Dispos. 1998;26:1194–1198. [PubMed] [Google Scholar]
- 9.McMillan BJ, Bradfield CA. The aryl hydrocarbon receptor sans xenobiotics: endogenous function in genetic model systems. Mol Pharmacol. 2007;72:487–498. doi: 10.1124/mol.107.037259. [DOI] [PubMed] [Google Scholar]
- 10.Kawajiri K, Fujii-Kuriyama Y. Cytochrome P450 gene regulation and physiological functions mediated by the aryl hydrocarbon receptor. Arch Biochem Biophys. 2007;464:207–212. doi: 10.1016/j.abb.2007.03.038. [DOI] [PubMed] [Google Scholar]
- 11.Matsumura F. The significance of the nongenomic pathway in mediating inflammatory signaling of the dioxin-activated Ah receptor to cause toxic effects. Biochem Pharmacol. 2009;77:608–626. doi: 10.1016/j.bcp.2008.10.013. [DOI] [PubMed] [Google Scholar]
- 12.Wong PS, Li W, Vogel CF, Matsumura F. Characterization of MCF mammary epithelial cells overexpressing the Arylhydrocarbon receptor (AhR) BMC Cancer. 2009;9:234. doi: 10.1186/1471-2407-9-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller JK, Shattuck DL, Ingalla EQ, Yen L, Borowsky AD, Young LJ, Cardiff RD, Carraway KL, Sweeney C. Suppression of the negative regulator LRIG1 contributes to ErbB2 overexpression in breast cancer. Cancer Res. 2008;68:8286–8294. doi: 10.1158/0008-5472.CAN-07-6316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shekhar MP, Nangia-Makker P, Wolman SR, Tait L, Heppner GH, Visscher DW. Direct action of estrogen on sequence of progression of human preneoplastic breast disease. Am J Pathol. 1998;152:1129–1132. [PMC free article] [PubMed] [Google Scholar]
- 15.Miller FR. Xenograft models of premalignant breast disease. J Mammary Gland Biol Neoplasia. 2000;5:379–391. doi: 10.1023/a:1009577811584. [DOI] [PubMed] [Google Scholar]
- 16.Dawson PJ, Wolman SR, Tait L, Heppner GH, Miller FR. MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am J Pathol. 1996;148:313–319. [PMC free article] [PubMed] [Google Scholar]
- 17.Vogel CF, Li W, Sciullo E, Newman J, Hammock B, Reader JR, Tuscano J, Matsumura F. Pathogenesis of aryl hydrocarbon receptor-mediated development of lymphoma is associated with increased cyclooxygenase-2 expression. Am J Pathol. 2007;171:1538–1548. doi: 10.2353/ajpath.2007.070406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vogel CF, Sciullo E, Matsumura F. Involvement of RelB in aryl hydrocarbon receptor-mediated induction of chemokines. Biochem Biophys Res Commun. 2007;363:722–726. doi: 10.1016/j.bbrc.2007.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vogel CF, Matsumura F. A new cross-talk between the aryl hydrocarbon receptor and RelB, a member of the NF-kappaB family. Biochem Pharmacol. 2009;77:734–745. doi: 10.1016/j.bcp.2008.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang X, Belguise K, Kersual N, Kirsch KH, Mineva ND, Galtier F, Chalbos D, Sonenshein GE. Oestrogen signalling inhibits invasive phenotype by repressing RelB and its target BCL2. Nat Cell Biol. 2007;9:470–478. doi: 10.1038/ncb1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Van Laere SJ, Van der Auwera I, Van den Eynden GG, Elst HJ, Weyler J, Harris AL, van Dam P, Van Marck EA, Vermeulen PB, Dirix YY. Nuclear factor-kappaB signature of inflammatory breast cancer by cDNA microarray validated by quantitative real-time reverse transcription-PCR, immunohistochemistry, and nuclear factor-kappaB DNA-binding. Clin Cancer Res. 2006;12:3249–3256. doi: 10.1158/1078-0432.CCR-05-2800. [DOI] [PubMed] [Google Scholar]
- 22.Chavey C, Bibeau F, Gourgou-Bourgade S, Burlinchon S, Boissière F, Laune D, Roques S, Lazennec G. Oestrogen receptor negative breast cancers exhibit high cytokine content. Breast Cancer Res. 2007;9:R15. doi: 10.1186/bcr1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh B, Berry JA, Vincent LE, Lucci A. Involvement of IL-8 in COX-2-mediated bone metastases from breast cancer. J Surg Res. 2006;134:44–51. doi: 10.1016/j.jss.2006.03.018. [DOI] [PubMed] [Google Scholar]
- 24.Park S, Matsumura F. Characterization of anti-apoptotic action of TCDD as a defensive cellular stress response reaction against the cell damaging action of ultra-violet irradiation in an immortalized normal human mammary epithelial cell line, MCF10A. Toxicology. 2006;217:139–146. doi: 10.1016/j.tox.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Lwin T, Hazlehurst LA, Li Z, Dessureault S, Sotomayor E, Moscinski LC, Dalton WS, Tao J. Bone marrow stromal cells prevent apoptosis of lymphoma cells by upregulation of anti-apoptotic proteins associated with activation of NF-kappaB (RelB/p52) in non-Hodgkin’s lymphoma cells. Leukemia. 2007;21:1521–1531. doi: 10.1038/sj.leu.2404723. [DOI] [PubMed] [Google Scholar]
- 26.Demicco EG, Kavanagh KT, Romieu-Mourez R, Wang X, Shin SR, Landesman-Bollag E, Seldin DC, Sonenshein GE. RelB/p52 NF-kappaB complexes rescue an early delay in mammary gland development in transgenic mice with targeted superrepressor IkappaB-alpha expression and promote carcinogenesis of the mammary gland. Mol Cell Biol. 2005;25:10136–1047. doi: 10.1128/MCB.25.22.10136-10147.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Y, Josson S, Fang F, Oberley TD, St Clair DK, Wan XS, Sun Y, Bakthavatchalu V, Muthuswamy A, St Clair WH. RelB enhances prostate cancer growth: implications for the role of the nuclear factor-kappaB alternative pathway in tumorigenicity. Cancer Res. 2009;69:3267–3271. doi: 10.1158/0008-5472.CAN-08-4635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mineva ND, Rothstein TL, Meyers JA, Lerner A, Sonenshein GE. CD40 ligand-mediated activation of the de novo RelB NF-kappaB synthesis pathway in transformed B cells promotes rescue from apoptosis. J Biol Chem. 2007;282:17475–1785. doi: 10.1074/jbc.M607313200. [DOI] [PubMed] [Google Scholar]
- 29.Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, Scharpé S, Vermeulen PB, Dirix LY. Increased serum interleukin-8 in patients with early and metastatic breast cancer correlates with early dissemination and survival. Clin Cancer Res. 2004;10:7157–7162. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 30.Baud V, Jacque E. The alternative NF-kB activation pathway and cancer: friend or foe? Med Sci. 2008;24:1083–1088. doi: 10.1051/medsci/200824121083. [DOI] [PubMed] [Google Scholar]
- 31.Henderson BE, Feigelson HS. Hormonal carcinogenesis. Carcinogenesis. 2000;21:427–433. doi: 10.1093/carcin/21.3.427. [DOI] [PubMed] [Google Scholar]
- 32.Alberts B. Redefining Cancer Research. Science. 2009;325:1319. doi: 10.1126/science.1181224. [DOI] [PubMed] [Google Scholar]