Abstract
We report a genome-wide association study to iron status. We identify an association of SNPs in TPMRSS6 to serum iron (rs855791, combined P = 1.5×10−20), transferrin saturation (combined P = 2.2×10−23), and erythrocyte mean cell volume (MCV, combined P = 1.1×10−10). We also find suggestive evidence of association with blood haemoglobin levels (combined P = 5.3×10−7). These findings demonstrate the involvement of TMPRSS6 in control of iron homeostasis and in normal erythropoiesis.
Mutations in genes which code for components of iron homeostasis mechanisms can cause overload in hereditary haemochromatosis (commonly associated with HFE, but also with HJV, HAMP, TFR2, SLC40A1) or deficiency in iron-refractory iron deficiency anaemia (associated with TMPRSS6). Although most attention has been paid to variants with major effects leading to inherited disease, variation in iron status within the general population 1,2 is important in relation to risk of iron deficiency anaemia, oxidative stress, liver disease and metabolic syndrome.
Investigation of iron status in humans can be based on quantitative tests including serum iron, serum transferrin, transferrin saturation with iron, and serum ferritin. We aimed to identify polymorphisms causing variation in iron status in the general population using genome-wide association methods in 2,516 adolescent and 2,302 adult individuals from 2,277 Australian twin families (Supplementary Table 1). The genotyping was performed using Human610-Quadv1 chips (~582K SNPs) and HumanCNV370-Quadv3 chips (~351K SNPs) (Supplementary Methods). We replicated previously reported SNPs in TF (rs3811647) with serum transferrin and in HFE (rs1800562) with iron, transferrin and transferrin saturation 2 in both the adolescent and adult samples (Table 1, Supplementary Fig. 1 & 2).
Table 1.
Additive effects (in SD) of the three SNPs in TMPRSS6, HFE and TF on serum iron, transferrin, transferrin saturation, ferritin, and blood haemoglobin (Hb) and erythrocyte mean cell volume (MCV)
| Adolescent |
Adult |
Combined |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | N | Beta | SE | R2 (%) | P | N | Beta | SE | R2 (%) | P | Beta | SE | P |
| rs855791(TMPRSS6) | |||||||||||||
| Iron | 2505 | −0.183 | 0.027 | 2.2 | 1.6×10−11 | 2298 | −0.191 | 0.030 | 1.9 | 9.9×10−11 | −0.187 | 0.020 | 1.5×10−20 |
| Transferrin | 2513 | 0.078 | 0.029 | 0.4 | 7.1×10−3 | 2299 | 0.069 | 0.031 | 0.2 | 0.023 | 0.074 | 0.021 | 4.9×10−4 |
| Saturation | 2503 | −0.196 | 0.027 | 2.5 | 9.7×10−13 | 2294 | −0.198 | 0.029 | 2.0 | 1.8×10−11 | −0.197 | 0.020 | 2.2×10−23 |
| Log10(ferritin) | 2512 | −0.092 | 0.028 | 0.5 | 8.8×10−4 | 2301 | −0.040 | 0.030 | 0.1 | 0.17 | −0.068 | 0.021 | 9.3×10−4 |
| Hb | 2468 | −0.151 | 0.032 | 1.1 | 2.3×10−6 | 3188 | −0.067 | 0.025 | 0.2 | 7.7×10−3 | −0.099 | 0.020 | 5.3×10−7 |
| MCV | 2467 | −0.139 | 0.032 | 0.9 | 1.3×10−5 | 3181 | −0.120 | 0.025 | 0.7 | 1.8×10−6 | −0.127 | 0.020 | 1.1×10−10 |
| rs1800562 (HFE) | |||||||||||||
| Iron | 2502 | 0.343 | 0.052 | 2.2 | 4.7×10−11 | 2299 | 0.315 | 0.053 | 1.5 | 2.5×10−9 | 0.329 | 0.037 | 8.2×10−19 |
| Transferrin | 2510 | −0.602 | 0.056 | 6.0 | 4.3×10−27 | 2300 | −0.605 | 0.055 | 5.4 | 2.9×10−28 | −0.604 | 0.039 | 2.2×10−53 |
| Saturation | 2500 | 0.598 | 0.053 | 6.6 | 2.3×10−29 | 2295 | 0.523 | 0.054 | 4.2 | 3.0×10−22 | 0.561 | 0.038 | 8.5×10−50 |
| Log10(ferritin) | 2509 | 0.168 | 0.053 | 0.5 | 1.6×10−3 | 2302 | 0.103 | 0.054 | 0.2 | 0.06 | 0.136 | 0.038 | 3.4×10−4 |
| Hb | 2465 | 0.236 | 0.061 | 0.8 | 1.2×10−4 | 3451 | 0.169 | 0.051 | 0.3 | 9.0×10−4 | 0.197 | 0.039 | 5.1×10−7 |
| MCV | 2464 | 0.148 | 0.061 | 0.3 | 0.015 | 3442 | 0.273 | 0.051 | 0.8 | 7.7×10−8 | 0.222 | 0.039 | 1.5×10−8 |
| rs3811647 (TF) | |||||||||||||
| Iron | 2505 | 0.073 | 0.029 | 0.3 | 0.012 | 2299 | 0.042 | 0.032 | 0.1 | 0.18 | 0.059 | 0.022 | 0.006 |
| Transferrin | 2513 | 0.460 | 0.031 | 11.2 | 4.4×10−50 | 2300 | 0.371 | 0.032 | 6.1 | 2.1×10−30 | 0.417 | 0.022 | 2.1×10−78 |
| Saturation | 2503 | −0.089 | 0.029 | 0.5 | 2.4×10−3 | 2295 | −0.093 | 0.031 | 0.4 | 3.1×10−3 | −0.091 | 0.021 | 1.8×10−5 |
| Log10(ferritin) | 2512 | −0.053 | 0.029 | 0.2 | 0.07 | 2302 | −0.030 | 0.032 | 0.1 | 0.35 | −0.044 | 0.022 | 0.04 |
| Hb | 2468 | −0.019 | 0.034 | 0.02 | 0.58 | 3470 | 0.032 | 0.026 | 0.04 | 0.21 | 0.013 | 0.021 | 0.52 |
| MCV | 2467 | −0.041 | 0.034 | 0.1 | 0.23 | 3461 | −0.042 | 0.026 | 0.1 | 0.10 | −0.042 | 0.021 | 0.04 |
Note: The results are from the Australian cohorts, except for Hb and MCV in adults (which are from the Dutch cohort). The results in the adolescent cohort are from the mean of measurements from up to 4 visits. The rs855791 variant was not genotyped in the Dutch panel and so was imputed using PLINK 11 as described previously 12. Genotypes were imputed with high confidence (information score of 0.97) in 92% of individuals. Minor allele frequencies (MAFs) for rs855791 (T), rs1800562 (A) and rs381164 (A) in the adolescent cohort are 0.42, 0.08 and 0.33, respectively. R2 is the proportion of the phenotypic variance explained by each SNP. After correction for multiple testing, there is no significant evidence for heterogeneity between the effect sizes in adolescent and adult data.
We identified significant associations between SNPs in TMPRSS6 and serum iron (adolescents P = 1.6×10−11, adults P = 9.9×10−11) and transferrin saturation (adolescents P = 9.7×10−13, adults P = 1.8×10−11) (Table 1). The strongest TMPRSS6 association was with rs855791, a non-synonymous coding SNP (A736V) in exon 17 (Fig. 1). The effects of rs855791 on serum iron and transferrin saturation were additive with each T allele decreasing serum iron and transferrin saturation by 0.18 and 0.20 SD of the mean phenotypes, respectively. The SNP explains 2.2% and 2.5% of the variation in the mean of serum iron and transferrin saturation in the adolescent cohort and 1.9 % and 2.0 % in the adult cohort.
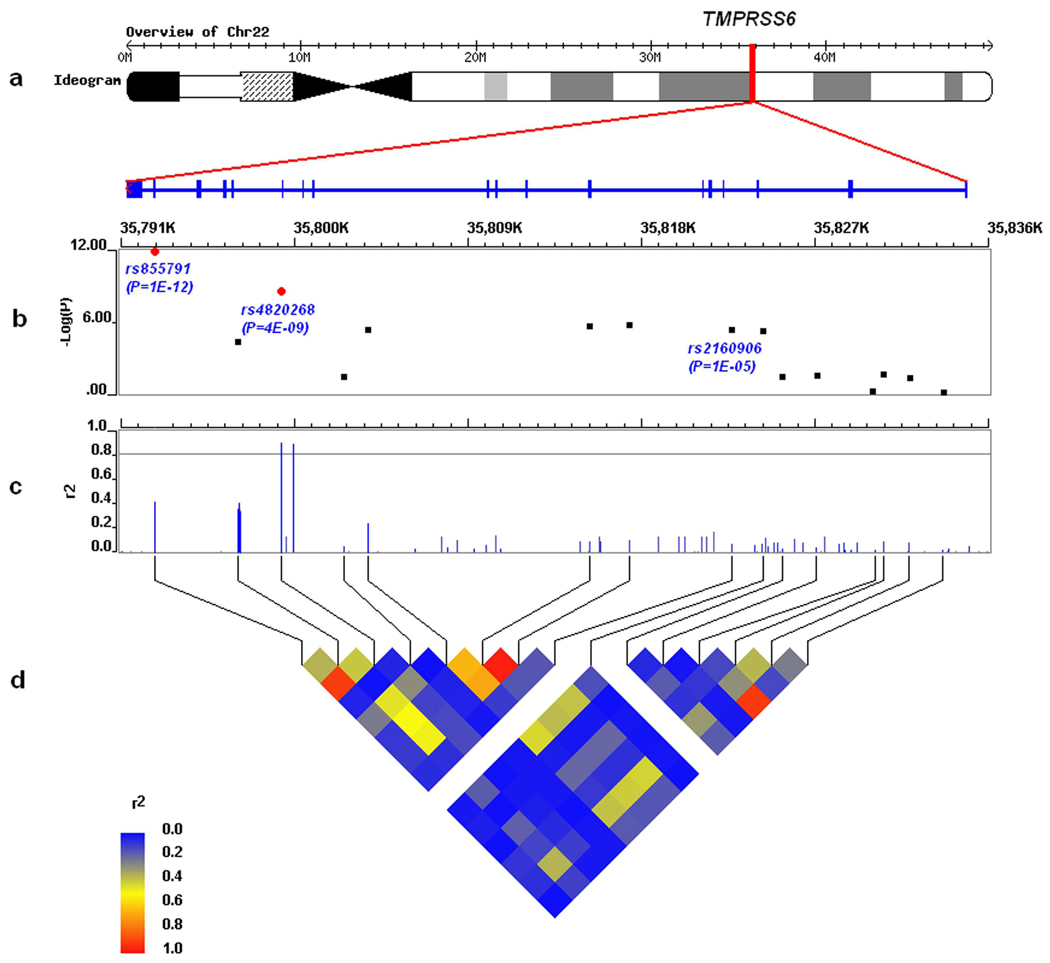
Figure 1.
Detailed results of the associations between SNPs within TMPRSS6 and transferring saturation in the adolescent cohort: (a) The genomic location of TMPRSS6 at 22q12-q13 on Chromosome 22; (b) −log10(P) of the association tests between SNPs and transferring saturation, where red dots indicate genome-wide significant SNPs. rs2160906 has a significant cis-acting effect on TMPRSS6 expression (P = 8.4×10−8) 10 ; (c) Linkage disequilibrium (LD) (r2) between rs855791 and all other HapMap SNPs within TMPRSS6; (d) HapMap-LD plot (r2) between genotyped SNPs within TMPRSS6.
Because of the involvement of TMPRSS6 mutations in iron-refractory iron deficiency anaemia 3 and because low values for blood haemoglobin (Hb) and erythrocyte mean cell volume (MCV) occur in iron-deficiency states, we next checked for associations between rs855791 and Hb or MCV in the adolescents. Strong associations were found (P = 2.3×10−6 and 1.3×10−5, respectively). The T allele of rs855791 decreased mean Hb and MCV by 0.15 and 0.14 SD, respectively. These correspond to 1.1 and 0.9% of the total variance in mean Hb and MCV. We replicated these associations in an independent Adult Dutch cohort 4 consisting of 3,470 unrelated individuals (P = 7.7×10−3 and 1.8×10−6 for Hb and MCV, respectively) (Table 1).
Genotypic means for each of the marker phenotypes for the most significant SNPs in TF, HFE and TMPRSS6, by sex and study cohort, are presented in Supplementary Fig. 3A & 3B. The direction of the allelic effects was consistent with expectation, in that the alleles or genotypes causing low iron or transferrin saturation were the same in the adults and adolescents and were also those associated with lower Hb and MCV. The estimated effect sizes stratified by gender after correcting for the effect of age are presented in Supplementary Table 2. Except for the effect of rs1800562 on Hb in the adult cohort, there appear to be no significant differences between the estimates for males and females.
To assess whether the effects of rs855791 on Hb and MCV are explained by effects on iron status, or whether there may be direct effects of HFE or TMPRSS6 variants on erythropoesis, we repeated the SNP association analysis for Hb and MCV in the Australian adolescents using the residuals after adjustment for the effect of transferrin saturation (chosen because this measure shows the strongest association with rs855791). This first required assessment of the relationships between transferrin saturation, Hb and MCV (Supplementary Fig. 4); there were essentially linear relationships across the range found in this population sample. Results showed that linear adjustment for transferrin saturation reduced but did not eliminate the effects of rs855791 in TMPRSS6 on Hb (from R2 = 1.2%, P = 1.4×10−6 to R2 = 0.5%, P = 0.003), and MCV (from R2 = 0.8%, P = 6.9×10−5 to R2 = 0.4%, P = 0.008). Similar adjustment essentially eliminated the effects of rs1800562 in HFE (Hb from R2 = 0.8%, P = 1.1×10−4 to R2 = 0.2%, P = 0.09, MCV from R2 = 0.4%, P = 0.007 to R2 < 0.1%), P = 0.40). The residual allelic association with rs855791, together with the strong overall association between transferrin saturation and Hb or MCV (Supplementary Table 3 and Supplementary Fig. 4), suggest that the effects of this TMPRSS6 polymorphism on Hb and MCV may not be solely due to the availability of transferrin-bound iron being rate-limiting for erythropoesis.
We have shown genome-wide significant association of a common SNP in TMPRSS6 with serum iron, transferrin saturation and MCV, and suggestive association with Hb, extending previous data 2 implicating TMPRSS6 in variation in serum iron and transferring saturation in the general population. Data from other sources 3,5–7 shows that knockout (in mice) or mutations with major effect (in humans) in TMPRSS6 greatly affect iron status. Loss of function in this gene produces iron deficiency and anaemia, probably through a combination of protease action on hemojuvelin 6 and regulation of hepcidin expression 8. We have now found significant and consistent TMPRSS6 effects on both iron status (serum iron and transferrin saturation) and erythropoesis (Hb and MCV) in adolescents and adults from the general population. The effects of this non-synonymous coding variant on both Hb and MCV raise important questions about the relationship between iron status and normal haemoglobin synthesis. One implication is that transferrin saturation may play a role in the control of erythropoesis, and alleles which increase it allow an increase in haemoglobin concentration and MCV even in people who show no evidence of iron deficiency.
There are now confirmed effects on iron markers in three genes, each previously known from human case studies or mouse experiments to affect iron homeostasis. TF mainly affects transferrin concentration, and this may in turn affect the concentration of diferric transferrin, which regulates hepcidin expression in the liver by interacting with HFE and TFR2 gene products 9. HFE likewise regulates hepcidin, and clinical or experimentally induced mutations lead to iron overload; the effects in these population samples are significant for serum iron, serum transferrin and transferrin saturation but not (genome-wide) on serum ferritin. The effects of the three SNPs on serum ferritin were weak compared to other traits. With the current sample size we do not yet have power to detect any significant ferritin-associated SNPs at genome-wide significance levels. Note that rs855791 and rs1800562 were associated with serum ferritin with combined P values around 10−4 (Table 1).
Loss-of-function mutations in the protease domain of TMPRSS6 gene product, matripase-2, lead to increased hepcidin expression and iron deficiency 5,6,7. The variant which has the largest effect in this study produces an amino acid change, from alanine to valine at amino acid 736. This too is in the protease domain and we speculate that it may lead to changes in protease activity. Other variants of TMPRSS6 may also affect iron status. A report on gene expression in human liver 10 found a cis-acting effect of rs2160906 (P = 8.4×10−8, HapMap CEU r2 with rs855791 is 0.06) at 35.82 MB on TMPRSS6 expression and we found strong supportive evidence (combined P = 9.8×10−8 with transferrin saturation) for an effect of this polymorphism on iron status (Fig. 1).
Estimates of heritability 1 indicate other loci or gene interactions must exist, but their identification will require larger studies or meta-analysis of multiple studies. Polygenic effects on iron status may have clinical importance for iron overload states; both primary (as risk modifiers in HFE C282Y homozygotes) and secondary to liver disease or metabolic syndrome, and case-control studies on the effects of TMPRSS6 variation in these conditions are needed.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to the twins and their families for their generous participation in these studies. We would like to thank staff at the Queensland Institute of Medical Research: Dixie Statham, Ann Eldridge and Marlene Grace for the Australian sample collection, Lisa Bowdler, Steven Crooks and staff of the Molecular Epidemiology Laboratory for sample processing and preparation, David Smyth and Harry Beeby for IT support, Margie Wright for supervision, Allan McRae and Sarah Medland for discussion. For the Australian study we acknowledge funding from the Australian National Health and Medical Research Council (NHMRC grants 241944, 389875, 389891, 389892, 389938, 442915, 442981, 496739 and 552485), U.S. National Institutes of Health (NIH grants AA07535, AA10248 and AA014041) and the Australian Research Council (ARC grant DP0770096). For the Netherlands Twin Registry (NTR) and the Netherlands Study of Depression and Anxiety (NESDA) samples, we acknowledge support from The Netherlands Society for Scientific Research (NWO 904-61-090; 904-61-193; 480-04-004; 400-05-717; SPI 56-464-14192), Center for Medical Systems Biology (NWO Genomics); Geestkracht program of ZonMW (10-000-1002); matching funds from universities and mental health care institutes involved in NESDA (GGZ Buitenamstel-Geestgronden, Rivierduinen, University Medical Center Groningen, GGZ Lentis, GGZ Friesland, GGZ Drenthe); Centre for Neurogenomics and Cognitive Research VU University (CNCR-VU); European Science Foundation (EU/QLRT-2001-01254); NIMH (R01 MH059160) and matching funds from participating institutes in NTR and NESDA. Genotyping of NTR and NESDA samples was funded by the Genetic Association Information Network (GAIN) of the Foundation for the US National Institutes of Health, and analysis was supported by grants from GAIN and the NIMH (MH081802). B.B. is the recipient of an NHMRC Biomedical Postdoctoral Fellowship (552498). D.R.N., G.W.M and P.M.V. are supported by the NHMRC Fellowship Scheme.
Footnotes
The authors declare that they have no competing financial interests.
AUTHOR CONTRIBUTIONS
Study design and coordination: A.C.H., A.K.H., B.W.P., D.I.B., D.R.N., E.J.C. de G., G.W.M., I.H.F., J.B.W., N.G.M. and P.M.V. Obtaining study funding: A.C.H., B.W.P., D.I.B., D.R.N., E.J.C.de G., G.W.M., I.H.F., J.B.W., N.G.M. and P.M.V. Sample collection and phenotype data collection: A.C.H., A.K.H., D.R.N., G.W.M., I.H.F., J.B.W., L.W., M.J.C. and N.G.M. (Australian): B.W.P., D.I.B., E.J.C.de G., G.W. and J-J.H. (Dutch). Data preparation: A.K.H., B.B., B.P.M., D.R.N., J.B.W., J-J.H., L.W., M.J.C., R.P.S.M. and S.G. Statistical analyses: B.B., B.P.M., D.R.N., J.B.W., M.A.R.F., P.M.V. and S.G. Results interpretation: B.B., D.R.N., G.W.M., J.B.W., M.A.R.F., N.G.M. and P.M.V. Manuscript writing: B.B., M.A.R.F. and J.B.W. Review and revision of the manuscript: A.C.H., B.B., B.W.P., D.I.B., D.R.N., E.J.C.de G., G.W., G.W.M., J.B.W., M.A.R.F., N.G.M. and P.M.V. All authors contributed to the final version of the paper.
REFERENCES
- 1.Whitfield JB, et al. Am J Hum Genet. 2000;66:1246–1258. doi: 10.1086/302862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benyamin B, et al. Am J Hum Genet. 2009;84:60–65. doi: 10.1016/j.ajhg.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Finberg KE, et al. Nat Genet. 2008;40:569–571. doi: 10.1038/ng.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boomsma DI, et al. Eur J Hum Genet. 2008;16:335–342. doi: 10.1038/sj.ejhg.5201979. [DOI] [PubMed] [Google Scholar]
- 5.Melis MA, et al. Haematologica. 2008;93:1473–1479. doi: 10.3324/haematol.13342. [DOI] [PubMed] [Google Scholar]
- 6.Silvestri L, et al. Cell Metab. 2008;8:502–511. doi: 10.1016/j.cmet.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramsay AJ, Hooper JD, Folgueras AR, Velasco G, Lopez-Otin C. Haematologica. 2009;94:840–849. doi: 10.3324/haematol.2008.001867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Muckenthaler MU. Cell Metab. 2008;8:1–3. doi: 10.1016/j.cmet.2008.06.009. [DOI] [PubMed] [Google Scholar]
- 9.Gao J, et al. Cell Metab. 2009;9:217–227. doi: 10.1016/j.cmet.2009.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schadt EE, et al. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Purcell S, et al. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ferreira MA, et al. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.