A study of the consequences of removing one copy of Mad2 in diploid budding yeast shows that MAD2 haploinsufficiency is due to subunit imbalance in the Mad1–Mad2 complex and that a normal Mad2:Mad1 ratio is essential for cells to respond to a loss of tension on mitotic chromosomes but is dispensable for a response to unattached chromosomes.
Abstract
Chromosome segregation depends on the spindle checkpoint, which delays anaphase until all chromosomes have bound microtubules and have been placed under tension. The Mad1–Mad2 complex is an essential component of the checkpoint. We studied the consequences of removing one copy of MAD2 in diploid cells of the budding yeast, Saccharomyces cerevisiae. Compared to MAD2/MAD2 cells, MAD2/mad2Δ heterozygotes show increased chromosome loss and have different responses to two insults that activate the spindle checkpoint: MAD2/mad2Δ cells respond normally to antimicrotubule drugs but cannot respond to chromosomes that lack tension between sister chromatids. In MAD2/mad2Δ cells with normal sister chromatid cohesion, removing one copy of MAD1 restores the checkpoint and returns chromosome loss to wild-type levels. We conclude that cells need the normal Mad2:Mad1 ratio to respond to chromosomes that are not under tension.
INTRODUCTION
Errors in chromosome segregation produce aneuploidy, which causes Down syndrome and contributes to cancer progression. Accurate chromosome segregation depends on the spindle checkpoint, which delays anaphase until all sister chromatid pairs have attached to opposite poles of the spindle (biorientation), and was first identified in budding yeast, Saccharomyces cerevisiae (Li and Murray, 1991; Hoyt et al., 1991). Inactivating the checkpoint causes chromosome loss in yeast, and chromosome loss, or embryonic lethality, in Drosophila, Caenorhabditis elegans, and mice (for review see Musacchio and Salmon, 2007).
Chromosomes are attached to the spindle by their kinetochores—multiprotein complexes that assemble on centromeric DNA and capture microtubules. When sister chromatids attach to microtubules emanating from opposite spindle poles, the spindle forces that pull the kinetochores toward opposite poles are balanced by cohesion between the chromatids, resulting in tension across the kinetochores. If both sister kinetochores attach to the same pole, they cannot generate tension. This relaxation induces kinetochores to release their microtubules (Li and Nicklas, 1995; Pinsky and Biggins, 2005), a process that depends on the activity of the protein kinase Ipl1, the yeast homologue of Aurora B (Biggins et al., 1999; Tanaka et al., 2002; Hauf et al., 2003; Dewar et al., 2004; Lampson et al., 2004). Purified kinetochores attach to microtubules more strongly when the linkage is placed under tension (Akiyoshi et al., 2010). This effect does not depend on Ipl1, suggesting that there are at least two ways in which tension regulates kinetochore–microtubule interactions. Two defects activate the spindle checkpoint: microtubule depolymerization (Li and Murray, 1991; Hoyt et al., 1991) and chromosomes that are not under tension (Li and Nicklas, 1995; Stern and Murray, 2001; Biggins and Murray, 2001; Ditchfield et al., 2003; Hauf et al., 2003; Pinsky et al., 2003, 2006). The checkpoint arrests the cell cycle by inhibiting the anaphase-promoting complex (APC), which triggers the ubiquitinylation and destruction of securin (Pds1 in budding yeast), allowing the sister chromatids to separate from each other.
Mad1 and Mad2 are components of the spindle checkpoint. They bind to each other and are recruited to the kinetochore, where their complex is hypothesized to “template” the conversion of free Mad2 from an open to a closed conformation, allowing Mad2 to bind to and inhibit Cdc20, an APC cofactor that is required for the metaphase-to-anaphase transition (for review see Musacchio and Salmon, 2007). We explored the consequences of removing one copy of MAD2 in diploid budding yeast. MAD2/mad2Δ heterozygotes respond normally to microtubule depolymerization, but they have an increased rate of chromosome loss and fail to respond to chromosomes that are not under tension.
RESULTS
Reduced Mad2 expression increases chromosome loss
Mutations that inactivate the spindle checkpoint cause increased chromosome loss. We measured the chromosome loss frequency of MAD2/MAD2, MAD2/mad2Δ, and mad2Δ/mad2Δ diploids using a colony color assay (Hieter et al., 1985; Spencer et al., 1990). The strains carry a nonessential chromosome fragment carrying an ochre-suppressing tRNA gene (SUP11). The host strains carry the ade2-101 ochre mutation; loss of the chromosome fragment leads to accumulation of a red pigment. Cells that lose the chromosome fragment during their initial division produce a sectored colony with one red and one white half.
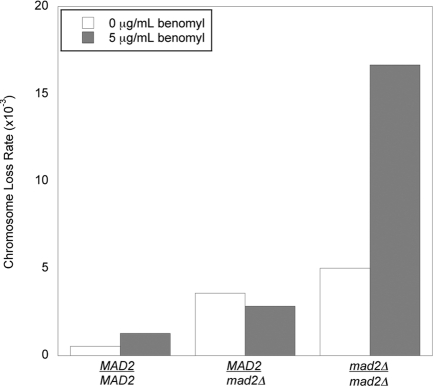
We examined the effect of two variables on the rate of chromosome loss: the level of Mad2 and the presence or absence of a low dose (5 μg/ml) of benomyl, an inhibitor of microtubule polymerization (Figure 1 and Table 1). In the absence of benomyl, the rate of chromosome loss was significantly increased in both the MAD2/mad2Δ and mad2Δ/mad2Δ diploids relative to the MAD2/MAD2 control (p < 0.0001, χ2 test; Table 2). The addition of benomyl led to a statistically significant increase in the chromosome loss rate of MAD2/MAD2 (p = 0.001) and mad2Δ/mad2Δ (p < 0.0001) cells. However, in MAD2/mad2Δ cells, adding benomyl did not significantly increase the chromosome loss rate (p = 0.2). Because microtubule depolymerization creates kinetochores that are not bound to microtubules, this result suggests that MAD2/mad2Δ cells respond normally to unattached kinetochores.
FIGURE 1:
Chromosome loss rates in MAD2/MAD2, MAD2/mad2Δ, and mad2Δ/mad2Δ diploids. Diploid cells contained a genetically marked chromosome fragment whose loss rates was measured using a colony color-sectoring assay. Measurements were made on plates in the absence (white) or presence (shaded) of 5 μg/ml benomyl.
TABLE 1:
Measured chromosome loss rates.
| 0 μg benomyl | 5 μg benomyl | |||||
|---|---|---|---|---|---|---|
| Strain | Sectored | Total | Rate (×10−3) | Sectored | Total | Rate (×10−3) |
| MAD2/MAD2 | 24 | 45,336 | 0.529 | 29 | 22,774 | 1.273 |
| MAD2/mad2Δ | 129 | 36,321 | 3.552 | 43 | 15,159 | 2.837 |
| mad2Δ/mad2Δ | 44 | 8,671 | 5.074 | 106 | 6,363 | 16.659 |
| MAD1/mad1ΔMAD2/mad2Δ | 23 | 28,149 | 0.817 | |||
TABLE 2:
Comparing chromosome loss rates by the χ2 test. p values are shown.
| MAD2/mad2Δ | mad2Δ/mad2Δ | MAD2/MAD2 +benomyl | MAD2/mad2Δ +benomyl | mad2Δ/mad2Δ +benomyl | MAD1/mad1Δ MAD2/mad2Δ | ||
|---|---|---|---|---|---|---|---|
| Rate ×10−3 | 3.552 | 5.074 | 1.273 | 2.837 | 16.659 | 0.817 | |
| MAD2/MAD2 | 0.529 | <0.0001 | <0.0001 | 0.001 | <0.0001 | <0.0001 | 0.134 |
| MAD2/mad2Δ | 3.552 | 0.0404 | <0.0001 | 0.2015 | <0.0001 | <0.0001 | |
| mad2Δ/mad2Δ | 5.074 | <0.0001 | 0.0061 | <0.0001 | <0.0001 | ||
| MAD2/MAD2+benomyl | 1.273 | 0.0006 | <0.0001 | 0.1093 | |||
| MAD2/mad2Δ +benomyl | 2.837 | <0.0001 | <0.0001 | ||||
| mad2Δ/mad2Δ +benomyl | 16.659 | <0.0001 |
MAD2/mad2Δ heterozygotes respond to microtubule depolymerization
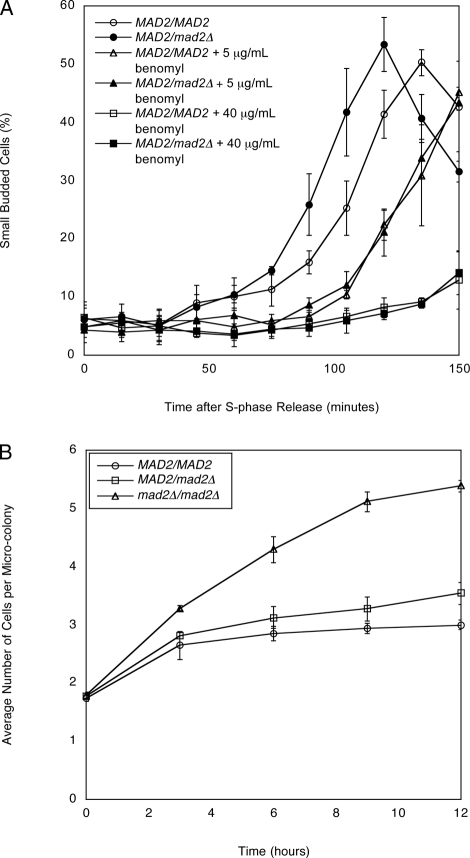
To directly determine whether MAD2/mad2Δ diploids respond to microtubule depolymerization, we compared the ability of benomyl to arrest MAD2/MAD2 and MAD2/mad2Δ cells in mitosis. We arrested cells in S phase, released them into different concentrations of benomyl, and measured their passage through mitosis by detecting the bud they formed as they entered the subsequent cell cycle. To ensure that any delay we measured occurred at the metaphase–anaphase transition rather than at the transition between anaphase and G1 (mitotic exit), we performed these experiments in diploid bub2Δ/bub2Δ mutant cells, which lack the spindle positioning checkpoint (Li, 1999; Bardin et al., 2000; Bloecher et al., 2000; Pereira et al., 2000). MAD2/MAD2 bub2Δ/bub2Δ diploids rebud after release from S-phase arrest, with the fraction of rebudded cells peaking at 135 min (Figure 2A). Cells heterozygous for MAD2 consistently progress through the cell cycle even faster, with the fraction of rebudded cells peaking at 120 min. This faster cell cycle progression is reminiscent of the observed acceleration of anaphase in haploid mad2Δ cells compared with their MAD2 counterparts (Li, 1999; Shonn et al., 2000). A low benomyl level (5 μg/ml) makes cells rebud more slowly, and the kinetics of rebudding are identical for MAD2/MAD2 and MAD2/mad2Δ cells. A higher benomyl level (40 μg/ml) severely delays rebudding in both cell types. These results show that MAD2/mad2Δ cells respond normally to kinetochores that are not attached to microtubules.
FIGURE 2:
MAD2/mad2Δ cells have a mitotic delay in response to microtubule depolymerization. (A) Exponentially dividing MAD2/MAD2 bub2Δ/bub2Δ cells and MAD2/mad2Δ bub2Δ/bub2Δ cells were arrested in S phase with hydroxyurea. Cells were released into fresh media containing 0, 5, or 40 μg/ml benomyl. The percentage of small, budded cells was determined by light microscopy at the indicated time points. Error bars represent the SD of three separate trials. (B) Exponentially dividing MAD2/MAD2, MAD2/mad2Δ, and mad2Δ/mad2Δ (all bub2Δ/bub2Δ) cells were diluted in rich media (YPD) made with 0.6% low–melting point agar and 60 μg/ml benomyl. The number of cells per microcolony was counted under a light microscope. Error bars represent the SD of three separate trials.
We also performed a microcolony assay, which monitors the ability of cells to divide under conditions that prevent spindle formation. MAD2/MAD2, MAD2/mad2Δ, and mad2Δ/mad2Δ cells (all bub2Δ/bub2Δ) were placed on rich media containing 60 μg/ml benomyl, which completely depolymerizes microtubules (Figure 2B). For all three strains, the average number of cells per microcolony increased in the first 3 h because cells that had completed mitosis but not yet budded were able to bud, replicate their DNA, and arrest at the next mitosis. After 3 h, mad2Δ/mad2Δ cells kept dividing, but MAD2/MAD2 and MAD2/mad2Δ cells did not, confirming that MAD2/mad2Δ cells respond normally to kinetochores that are not attached to microtubules.
MAD2/mad2Δ heterozygotes fail to respond to short linear chromosomes
The behavior of MAD2/mad2Δ cells puzzled us: they respond normally to microtubule depolymerization, but they lose chromosomes frequently. One explanation is that they respond to chromosomes that are not attached to microtubules but cannot respond to chromosomes that are attached to microtubules but are not under tension.
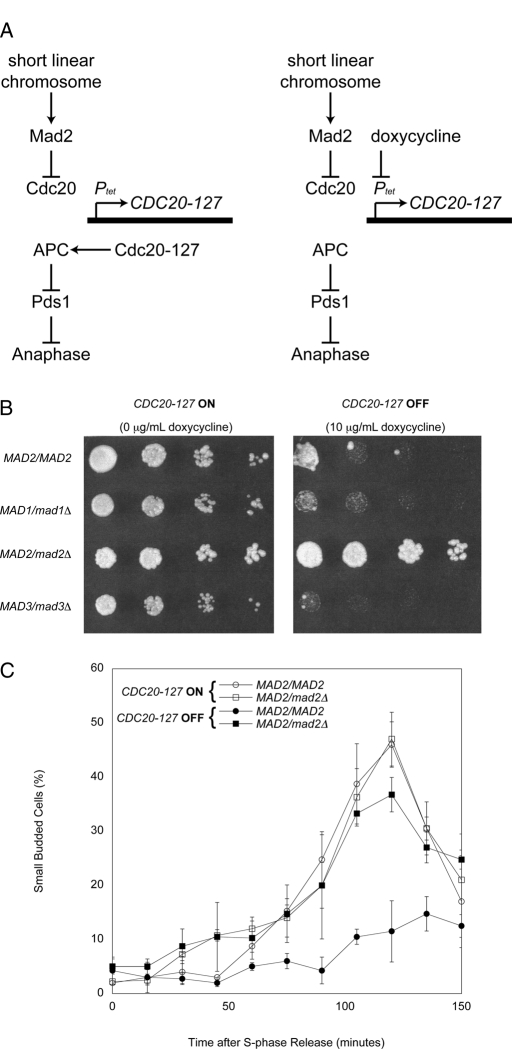
To test this possibility, we examined the response of MAD2/mad2Δ cells to chromosomes that are not under tension. Short linear chromosomes activate the spindle checkpoint because their sister chromatids separate prematurely; once this happens, they lose tension at their kinetochores (Wells and Murray, 1996). In the presence of the CDC28-VF mutation, which slows the exit from mitosis (Rudner et al., 2000), this mitotic delay is strong enough to block cell proliferation. To allow these strains to proliferate, we expressed CDC20-127, a dominant allele of CDC20 that the spindle checkpoint cannot inhibit (Hwang et al., 1998). In strains where this allele is expressed from a tetracycline-regulated promoter (Ptet), the response to the spindle checkpoint can be manipulated (Figure 3A). We used diploid strains that contain short linear chromosomes and are homozygous for Ptet-CDC20-127 and CDC28-VF. When these strains are grown in the absence of doxycycline the checkpoint is turned off: CDC20-127 is expressed, the APC is resistant to signals from the checkpoint, and cells proliferate normally. Adding doxycycline represses CDC20-127 and allows the short linear chromosomes to activate the checkpoint by inhibiting the endogenous Cdc20 and arresting cells in mitosis, thus preventing cell proliferation.
FIGURE 3:
MAD2/mad2Δ cells do not arrest in response to short linear chromosomes. (A) A diagram of the effects of short linear chromosomes on the spindle checkpoint. CDC20-127 is a dominant allele of CDC20 that the spindle checkpoint cannot inhibit; in strains in which this allele is expressed from a tetracycline-regulated promoter (Ptet), the response to the spindle checkpoint can be manipulated. In the absence of doxycycline (left) the checkpoint is turned off: CDC20-127 is expressed, activation of the APC is resistant to signals from the checkpoint, and cells execute anaphase and proliferate normally. In contrast, in the presence of doxycycline (right) the checkpoint is turned on: expression of CDC20-127 is repressed, allowing the short linear chromosomes to activate the checkpoint and cause an arrest in mitosis that prevents cell proliferation. (B) Short linear chromosome–dependent activation of the spindle checkpoint. The indicated strains were tested for their ability to proliferate in the absence (CDC20-127 expressed; left) or presence (CDC20-127 repressed; right) of the spindle checkpoint. Serial fourfold dilutions of all strains were spotted onto plates containing 0 μg/ml doxycycline (left) or 10 μg/ml doxycycline (right) and grown for 2 d. (C) MAD2/mad2Δ heterozygotes do not have a mitotic delay in response to loss of tension. Exponentially dividing cultures of MAD2/MAD2 and MAD2/mad2Δ cells containing Ptet-CDC20-127, CDC28-VF, and short linear chromosomes were arrested in S phase with hydroxyurea. Cells were released into fresh media and treated with 10 μg/ml doxycycline to repress expression of Ptet-CDC20-127, allowing short linear chromosomes to activate the spindle checkpoint. The percentage of small, budded cells after release was determined by light microscopy at the indicated time points. Error bars represent the SD of three separate trials.
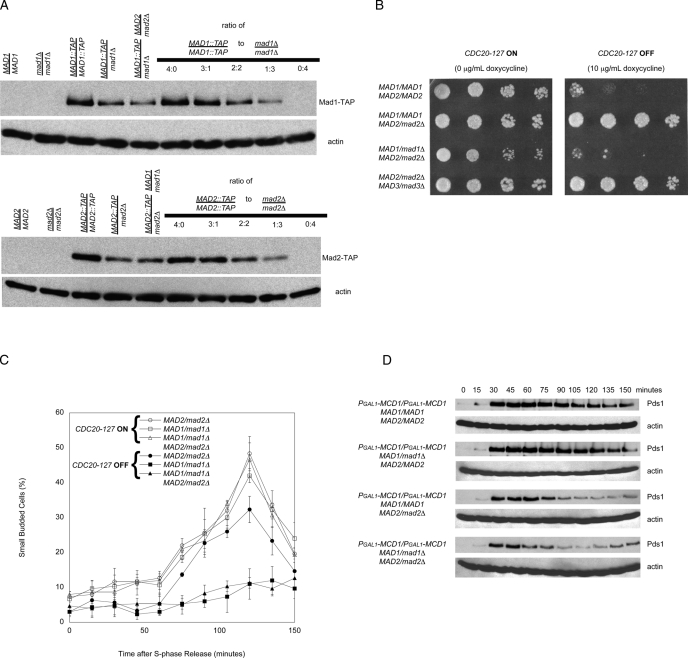
We compared the ability of short linear chromosomes to arrest MAD2/MAD2 and MAD2/mad2Δ cells. Strains were spotted on plates in the absence (checkpoint off) or presence of 10 μg/ml of doxycycline (checkpoint on). MAD2/MAD2 cells arrested on doxycycline plates, but MAD2/mad2Δ cells proliferated (Figure 3B). Among the MAD genes, the effect of heterozygosity is specific for MAD2: MAD1/mad1Δ and MAD3/mad3Δ strains did not proliferate on doxycycline plates.
We monitored the effect of short linear chromosomes on the passage through a single cell cycle. We arrested cells in S phase, released them from the arrest, and followed their progress through mitosis and into the next cell cycle, as detected by rebudding. In the absence of doxycycline, the rebudding of MAD2/MAD2 cells peaked at 120 min, but the presence of doxycycline strongly delayed rebudding (Figure 3C). In contrast, the rebudding of MAD2/mad2Δ was only slightly reduced by the presence of doxycycline, showing that halving the dose of MAD2 greatly reduces the ability of kinetochores that are not under tension to activate the spindle checkpoint.
The loss of sister chromatid cohesion does not arrest MAD2/mad2Δ heterozygotes
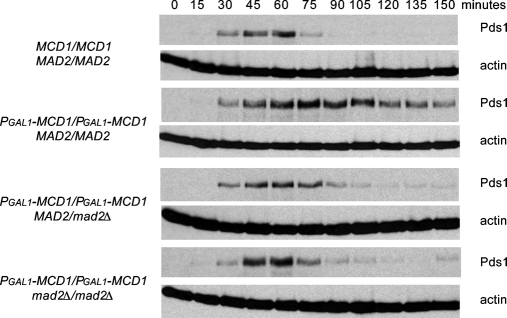
We used a second method to remove tension from the linkage between kinetochores and microtubules. Abolishing the linkage between chromosomes reduces tension at their kinetochores (Li and Nicklas, 1995), and this manipulation activates the spindle checkpoint in meiotic (Shonn et al., 2000) and mitotic (Biggins and Murray, 2001; Stern and Murray, 2001; King et al., 2007) yeast cells. To remove tension from all chromosomes, we regulated the expression of the cohesin subunit Mcd1 (also known as Scc1) by fusing its gene to the galactose-inducible GAL1 promoter (PGAL1-MCD1). We measured cell cycle progression by following the disappearance of epitope-tagged securin (Pds1–18xMyc), which marks the activation of the APC and normally leads to sister chromatid separation. When Mcd1 was expressed, the sister chromatids of MAD2/MAD2 cells were held together by cohesin until anaphase, and the cells degraded Pds1 promptly (Figure 4). When it was not, there was no cohesion, and a delay in the onset of Pds1 destruction was observed. In contrast, both MAD2/mad2Δ and mad2Δ/mad2Δ showed little or no response to a loss of tension on mitotic chromosomes: both cells initiated Pds1 destruction at the same time as cells that had normal cohesin. Our experiments show that the MAD2 heterozygotes have different responses to the two signals that activate the spindle checkpoint: MAD2/mad2Δ cells respond normally to an antimicrotubule drug, but they are defective in their ability to respond to chromosomes that are not under tension.
FIGURE 4:
MAD2/mad2Δ heterozygotes do not arrest in response to a loss of tension on all mitotic chromosomes. We measured cell cycle progression by Western blotting after release from a G1 arrest in the presence or absence of the MCD1, a component of sister chromatid cohesion. Cells were grown to mid-log phase and arrested in G1 with α-factor, and released into glucose to shut off expression of PGAL1-MCD1. Western blots against Myc (top) or actin (bottom) were performed (n = 3). Pds1-18xMyc serves as a marker for metaphase, and a decrease in its level indicates progression into anaphase. Wild-type diploids progress through the cell cycle without delay. In the absence of sister chromatid cohesion, wild-type cells display an extended arrest. However, MAD2/mad2Δ and mad2Δ/mad2Δ cells do not arrest in response to a loss of tension on all mitotic chromosomes.
Reducing Mad1 expression allows MAD2/mad2Δ cells to respond to short linear chromosomes
Because the relative levels of Mad1 and Mad2 affect the spindle checkpoint (Chung and Chen, 2002; Sironi et al., 2002), we hypothesized that decreasing Mad1 expression would allow MAD2/mad2Δ cells to respond to chromosomes that were not under tension. We confirmed that Mad1 and Mad2 levels in diploid cells reflected gene dose: MAD1/mad1Δ and MAD2/mad2Δ heterozygotes express half as much of the corresponding proteins as wild-type cells (Figure 5A). We tested whether reducing the level of Mad1 affected the response of MAD2/mad2Δ diploids to two perturbations that affect tension at kinetochores: short linear chromosomes and the absence of cohesin. As a control, we generated a MAD2/mad2Δ MAD3/mad3Δ double heterozygote.
FIGURE 5:
MAD1/mad1Δ MAD2/mad2Δ cells arrest in response to loss of tension and loss of attachment. (A) Western blots to detect the relative levels of Mad1-TAP (top) or Mad2-TAP (bottom) epitope-tagged strains that are homozygous diploids, heterozygous diploids, or double heterozygous diploids. Both Mad1 and Mad2 proteins levels appear to be lowered by approximately one-half the amount in the heterozygous diploids and double heterozygous diploids relative to the homozygous diploid in comparison to mixed ratios of homozygous TAP-tagged diploids and homozygous nulls (n = 3). (B) The indicated strains were tested for their ability to proliferate in the absence (CDC20-127 expressed; left) or presence (CDC20-127 repressed; right) of the spindle checkpoint. Serial fourfold dilutions of all strains were spotted onto plates containing 0 μg/ml doxycycline (left) or 10 μg/ml doxycycline (right) and grown for 2 d. (C) MAD1/mad1Δ MAD2/mad2Δ double heterozygotes have a mitotic delay in response to loss of tension on chromosomes. Error bars represent the SD of three separate trials. (D) MAD1/mad1Δ MAD2/mad2Δ double heterozygotes do not arrest in response to a loss of tension on all mitotic chromosomes. We measured cell cycle progression by Western blotting after release from a G1 arrest in the absence of the MCD1. Western blots against Myc (top) or actin (bottom) were performed (n = 3). In the absence of sister chromatid cohesion, wild-type and MAD1/mad1Δ cells display an extended arrest. MAD1/mad1Δ MAD2/mad2Δ diploid cells do not arrest in response to a loss of cohesion on all mitotic chromosomes.
The dose of Mad1 affected the response to short linear chromosomes. Two strains that had reduced levels of Mad2 and normal levels of Mad1 (a MAD2/mad2Δ single heterozygote and a MAD2/mad2Δ MAD3/mad3Δ double heterozygote) failed to respond to the short linear chromosomes. In contrast, the response of the MAD1/mad1Δ MAD2/mad2Δ double heterozygote resembled that of wild-type MAD2/MAD2 cells. This conclusion held for both long-term proliferation on plates (Figure 5B) and passage through mitosis in a single cell cycle (Figure 5C).
The dose of Mad1 did not affect the response to loss of cohesin. We measured the response of the MAD1/mad1Δ MAD2/mad2Δ double heterozygote to the loss of tension produced by repressing Mcd1. Like wild-type cells, MAD1/mad1Δ cells arrested in mitosis in response to the loss of cohesin (Figure 5D). However, MAD1/mad1Δ MAD2/mad2Δ double heterozygotes showed no cell cycle arrest. We discuss the difference between the responses to short linear chromosomes and cohesin depletion in what follows.
MAD1 heterozygosity suppresses MAD2/mad2Δ haploinsufficiency for chromosome loss
Reducing Mad2 expression increases chromosome loss (Figure 1). To determine whether a corresponding reduction in Mad1 expression can suppress this phenotype, we measured chromosome loss in MAD1/mad1Δ MAD2/mad2Δ cells (Table 1). The rate of chromosome loss in MAD1/mad1Δ MAD2/mad2Δ cells is not significantly different from the loss rate in wild-type MAD1/MAD1 MAD2/MAD2 cells (p = 0.13, χ2 test, Table 2), but is significantly lower than that of MAD2/mad2Δ cells (p < 0.0001, χ2 test).
DISCUSSION
Halving the dose of Mad2 in budding yeast increases chromosome loss and keeps cells from responding to chromosomes that are not under tension but has no effect on their response to microtubule depolymerization. The response to short linear chromosomes depends on the Mad2:Mad1 ratio, supporting the hypothesis that Mad1 can both activate and inhibit Mad2 (Chung and Chen, 2002; Sironi et al., 2002).
MAD2 haploinsufficiency in response to loss of tension
MAD2/mad2Δ heterozygotes fail to arrest in response to short linear chromosomes or removal of a cohesin subunit. Among the MAD genes, haploinsufficiency is specific to MAD2: cells heterozygous for MAD1 or MAD3 respond normally to short linear chromosomes, and cells heterozygous for MAD1 respond normally to cohesin depletion.
The two perturbations we used to remove tension from chromosomes are different. Short linear chromosomes have a wide distribution of copy numbers, with an average of ∼10 per cell (Murray and Szostak, 1983). This distribution means that we cannot exclude the possibility that the spindle checkpoint is activated normally in the small fraction of MAD2/mad2Δ cells that contain many short linear chromosomes (Figures 3C and 5C). This reservation does not apply to cells that lack cohesin: every diploid cell that enters mitosis contains 64 single chromatids that attach to one of the spindle poles.
Halving the amount of Mad2 prevents cells from responding to either defect, but cells differ in their response to also removing one copy of MAD1 to restore the normal Mad2:Mad1 ratio. Doubly heterozygous MAD1/mad1Δ MAD2/mad2Δ cells recover the ability to arrest in response to short linear chromosomes, but they still fail to arrest in response to cohesin depletion, even though none of the chromosomes are under tension. These results seem surprising: removing tension from every chromosome fails to activate the checkpoint in cells that have lower expression of both Mad2 and Mad1, but creating a smaller number of single chromatids succeeds in arresting cells. The discrepancy suggests that cohesin may play a role in the checkpoint's response to chromosomes that are not under tension.
Our work is consistent with previous demonstrations that the Mad2:Mad1 ratio affects the spindle checkpoint. Lowering the Mad2:Mad1 ratio increases chromosome loss in budding yeast (Warren et al., 2002), inactivates the checkpoint in Xenopus egg extracts (Chen et al., 1998), and produces aberrant mitoses and tumors in mice (Michel et al., 2001). Increasing the Mad2:Mad1 ratio activates the checkpoint in the absence of spindle defects in frogs (Chen et al., 1996) and fission yeast (He et al., 1997).
The “template” model for the checkpoint (Musacchio and Salmon, 2007) rationalizes these results. In this model, Mad1 plays two roles: it forms a complex with Mad2, which interacts with and activates free Mad2 molecules, and it contains a peptide that is similar to and competes with the Mad2-binding peptide in Cdc20. In vitro, Mad1 and Cdc20 compete for binding to Mad2 (Sironi et al., 2002). The opposing roles of Mad1 imply that cells need the right Mad2:Mad1 ratio. Without Mad1, they cannot convert enough Mad2 into the closed conformation to bind to and inhibit Cdc20, but if they contain too much Mad1, most of the Mad2 binds to Mad1 and there is not enough Mad2–Cdc20 binding to rapidly arrest the cell cycle.
Implications for understanding the spindle checkpoint
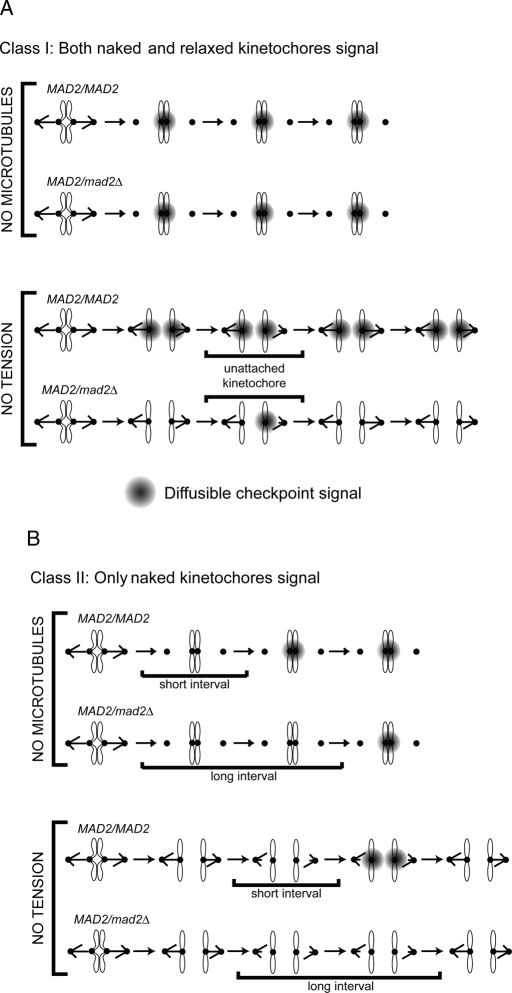
Reducing the dose of Mad2 affects the response to chromosomes that are not under tension but has no effect on the response to microtubule depolymerization. For clarity, we refer to kinetochores that are not bound to microtubules as naked and those that are bound to microtubules but not under tension as relaxed. What accounts for the different response to the two types of kinetochores, especially since relaxed kinetochores release their microtubules (Biggins et al., 1999; Tanaka et al., 2002; Hauf et al., 2003; Dewar et al., 2004; Lampson et al., 2004), making them resemble the naked kinetochores in cells whose microtubules have been depolymerized?
We argue that the response of the checkpoint depends on kinetochore dynamics, and these dynamics depend on whether cells have microtubules. In a cell without microtubules, naked kinetochores are continuously free of microtubules. In a cell with microtubules, although a relaxed kinetochore lets go of microtubules, it must rebind them in order to reorient; thus these kinetochores go through cycles of attachment and detachment until they establish tension. These cycles have been observed in meiotic insect cells (Li and Nicklas, 1995). Different authors have reached different conclusions about whether (Pinsky et al., 2006) or not (King et al., 2007) chromosomes must detach from microtubules to arrest yeast cells in mitosis. Thus we propose two classes of explanations, which make different assumptions about whether relaxed kinetochores can send a signal that inhibits the APC and keeps cells from starting anaphase.
The first class (Figure 6A) depends on three assumptions: 1) both naked and relaxed kinetochores can send the signal that inhibits the APC, 2) reducing the Mad2:Mad1 level abolishes signaling from relaxed kinetochores but has no effect on signaling from naked kinetochores, and 3) relaxed kinetochores detach from microtubules much more slowly than they reattach to them. As a result, even when all the chromosomes lack tension, the vast majority of kinetochores are always attached to microtubules. As long as this cell has the normal Mad2:Mad1 ratio, the vast majority of the signal that inhibits the APC comes from relaxed kinetochores, and there is little or no signaling from naked kinetochores. When the Mad2:Mad1 ratio is reduced, there is no signaling from relaxed kinetochores, and there are enough periods when all the kinetochores are attached to allow APC activation and sister chromatid segregation.
FIGURE 6:
Two classes of models for the spindle checkpoint defect in MAD2/mad2Δ cells. (A) Class I. Two types of kinetochores can generate a diffusible signal (shown in gray) that can inhibit the APC: kinetochores that are attached to microtubules but are not under tension (relaxed), and kinetochores that are not attached (naked). Both wild-type MAD2/MAD2 and heterozygous MAD2/mad2Δ cells can respond to loss of attachment induced by microtubule depolymerization (top). The relaxed kinetochores can signal in cells that have the normal Mad2:Mad1 ratio but cannot signal in MAD2/mad2Δ heterozygotes, whereas the naked kinetochores signal in either cell type. In addition, even though relaxed kinetochores release microtubules, reattachment is faster than release, so most kinetochores remain attached to microtubules. In this scenario, reducing the tension on all the kinetochores cannot arrest a MAD2/mad2Δ cell (bottom). (B) class II. Only kinetochores that are not attached to microtubules (naked) can generate a signal that inhibits the APC, and this signal is generated more slowly in MAD2/mad2Δ cells. Both wild-type MAD2/MAD2 and heterozygous MAD2/mad2Δ cells can respond to loss of attachment induced by microtubule depolymerization (top). In MAD2/mad2Δ cells, the reduced Mad2:Mad1 ratio increases the lag before naked kinetochores begin to generate a signal that inhibits the APC and arrests the cell cycle. Without microtubules, the kinetochores cannot reattach, and they eventually generate a signal that arrests the cell cycle (top). However, in the absence of tension when relaxed kinetochores detach from microtubules there is a race between generating the signal that inhibits the APC and reattaching to microtubules (bottom). In MAD2/MAD2 cells, we propose that naked kinetochores can generate a signal that can arrest the cell cycle before they reattach to a microtubule, but the naked kinetochores in MAD2/mad2Δ cells reattach to a microtubule before they can generate a signal sufficient to halt the cell cycle (bottom).
The second class of explanation argues that only naked kinetochores can signal to the APC. The two hypotheses in this class claim that the reduced Mad2:Mad1 ratio affects different steps in the spindle checkpoint. The first proposes that reducing the Mad2:Mad1 ratio slows the release of microtubules from relaxed kinetochores. Although cells lacking Mad2 show slower reorientation of mono-oriented chromosomes in meiosis (Shonn et al., 2003), this is a modest effect, and we were unable to detect it in mitotic cells (B. Stern and A.W.M, unpublished data).
The second hypothesis proposes a short lag before naked kinetochores start signaling to the checkpoint and that this delay increases as the Mad2:Mad1 ratio falls (Figure 6B). In cells treated with microtubule poisons, there are few or no microtubules, so naked kinetochores stay naked and will eventually inhibit the APC and halt cell cycle progression even though the reduced Mad2:Mad1 ratio causes a longer lag before they start signaling (Figure 6B, top). At the normal Mad2:Mad1 ratio, relaxed kinetochores let go of their microtubules in response to loss of tension, and the resulting naked kinetochores start signaling to the APC well before they capture another microtubule (Figure 6B, bottom). Thus chromosomes start to signal before they reattach to the spindle, and cells arrest in response to relaxed kinetochores. Reducing the Mad2:Mad1 ratio increases the lag before the naked kinetochore can start signaling. After a relaxed kinetochore lets go of its microtubule, it reattaches to another microtubule before it can begin to generate the signal that inhibits the APC and cells cannot arrest.
Implications for genome stability
The majority of tumors show chromosome instability—the loss and gain of chromosomes and chromosome fragments (Lengauer et al., 1997). Inactivating one copy of MAD2 in diploid yeast increases the rate of chromosome loss sevenfold. In mice, Mad1 and Mad2 are essential, most likely because the chromosome loss rate seen in homozygous mutant cells is too high to maintain viability (Michel et al., 2001; Iwanaga et al., 2007). In contrast, inactivation of only one allele of Mad1 or Mad2 allows the checkpoint to keep cells alive and increases the rate of chromosome loss that contributes to tumor formation and evolution (Michel et al., 2001; Iwanaga et al., 2007). Thus the mammalian Mad1 and Mad2 genes may be examples of a class of essential genes that make widespread contributions to cancer development: haploinsufficient tumor suppressor genes in which a single hit will lead to genetic instability.
MATERIALS AND METHODS
Yeast strains and methods
Strains used in this study are listed in Table 3. All strains are derivatives of W303 (ura3-1 leu2-3, 112 his3-11, 15 trp1-1 ade2-101 can1-100) or the S288c derivative BY4741 (MATa ura3Δ0 leu2Δ0 his3Δ1 met15Δ0) (Open Biosystems, Thermo Biosystems, Huntsville, AL). Strains were constructed using standard genetic techniques, and media were prepared according to established recipes (Sherman et al., 1974). All media contain 2% wt/vol glucose as a carbon source, unless otherwise indicated. To activate the spindle checkpoint, cells were exposed to methyl 1-(butylcarbamoyl)-2-benzimidazolecarbamate (benomyl) (Sigma-Aldrich, St. Louis, MO). To measure cell cycle arrest in mitosis, cells growing exponentially at 30°C were arrested in S phase with 10 mg/ml hydroxyurea (Sigma-Aldrich) and then released into fresh media. The percentage of small, budded cells due to passage through mitosis and into the next cell cycle was determined by light microscopy.
TABLE 3:
Strains used in this study.
| Strain | MAT | Genotype | Source |
|---|---|---|---|
| EBY54 | MATa/MATα | Ptet-CDC20-127@TRP1/Ptet-CDC20-127@TRP1 CDC28-VF::HIS3/CDC28-VF::HIS3 A241p1::LEU2 mad1Δ::HIS3/MAD1 mad2Δ::Kanr/MAD2 | This study |
| EBY63 | MATa/MATα | Ptet-CDC20-127@TRP1/Ptet-CDC20-127@TRP1 CDC28-VF::HIS3/CDC28-VF::HIS3 A241p1::LEU2 mad2Δ::HIS3/MAD2 mad3Δ/MAD3 | This study |
| EBY69 | MATa/MATα | bub2Δ::LEU2/bub2Δ::LEU2 | This study |
| EBY70 | MATa/MATα | bub2Δ::LEU2/bub2Δ::LEU2 mad2Δ::URA3/MAD2 | This study |
| EBY73 | MATa/MATα | mad2Δ::Kanr/MAD2 SUP11::URA3/SUP11::URA3 | This study |
| EBY74 | MATa/MATα | SUP11::URA3/SUP11::URA3 | This study |
| EBY75 | MATa/MATα | mad2Δ::Kanr/mad2Δ::LEU2 SUP11::URA3/SUP11::URA3 | This study |
| EBY76 | MATa/MATα | mad2Δ::Kanr/MAD2 mad1Δ::HIS3/MAD1 SUP11::URA3/SUP11::URA3 | This study |
| MAS381 | MATa/MATα | bub2Δ::LEU2/bub2Δ::LEU2 mad2Δ::URA3/mad2Δ::URA3 | This study |
| RDY473 | MATa/MATα | Ptet-CDC20-127@TRP1/Ptet-CDC20-127@TRP1 CDC28-VF::HIS3/CDC28-VF::HIS3 A241p1::LEU2 | This study |
| RDY353 | MATa/MATα | Ptet-CDC20-127@TRP1/Ptet-CDC20-127@TRP1 CDC28-VF::HIS3/CDC28-VF::HIS3 A241p1::LEU2 mad1Δ::HIS3/MAD1 | This study |
| RDY354 | MATa/MATα | Ptet-CDC20-127@TRP1/Ptet-CDC20-127@TRP1 CDC28-VF::HIS3/CDC28-VF::HIS3 A241p1::LEU2 mad2Δ::Kanr/MAD2 | This study |
| RDY355 | MATa/MATα | Ptet-CDC20-127@TRP1/Ptet-CDC20-127@TRP1 CDC28-VF::HIS3/CDC28-VF::HIS3 A241p1::LEU2 mad3Δ/MAD3 | This study |
| SCSY78 | MATa/MATa | PDS1::18xMYC::LEU2/PDS1::18xMYC::LEU2 | This study |
| SCSY215 | MATa/MATαΔ::URA3 | PDS1::18xMYC::LEU2/PDS1::18xMYC::LEU2 Kanr::PGAL1–3xHA::MCD1/Kanr::PGAL1–3xHA::MCD1 | This study |
| SCSY302 | MATa/MATα | his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/met15Δ0 ura3Δ0/ura3Δ0 | BY4743, Open Biosystems |
| SCSY345 | MATa/MATαΔ::URA3 | PDS1::18xMYC::LEU2/PDS1::18xMYC::LEU2 Kanr::PGAL1–3xHA::MCD1/Kanr::PGAL1–3xHA::MCD1 MAD2/mad2Δ::HPH | This study |
| SCSY348 | MATa/MATαΔ::URA3 | PDS1::18xMYC::LEU2/PDS1::18xMYC::LEU2 Kanr::PGAL1–3xHA::MCD1/Kanr::PGAL1–3xHA::MCD1 mad2Δ::HPH/mad2Δ::HPH | This study |
| SCSY414 | MATa/MATα | MAD1::TAP::HIS3/mad1Δ::URA3 | This study |
| SCSY415 | MATa/MATα | mad1Δ::HIS3/mad1Δ::URA3 | This study |
| SCSY416 | MATa/MATα | MAD2::TAP::HIS3/mad2Δ::Kanr | This study |
| SCS430 | MATa/MATαΔ::URA3 | PDS1::18xMYC::LEU2/PDS1::18xMYC::LEU2 Kanr::PGAL1–3xHA::MCD1/Kanr::PGAL1–3xHA::MCD1 MAD1/mad1Δ::HIS3 | This study |
| SCSY434 | MATa/MATα | mad2Δ::URA3/mad2Δ::Kanr | This study |
| SCSY436 | MATa/MATα | MAD1::TAP::HIS3/MAD1::TAP::HIS3 | This study |
| SCSY438 | MATa/MATα | MAD2::TAP::HIS3/MAD2::TAP::HIS3 | This study |
| SCSY446 | MATa/MATα | MAD1/mad1Δ::HIS3 MAD2::TAP::HIS3/mad2Δ::Kanr | This study |
| SCSY451 | MATa/MATα | MAD1::TAP::HIS3/mad1Δ::HIS3 MAD2/mad2Δ::Kanr | This study |
| SCSY456 | MATa/MATαΔ::URA3 | PDS1::18xMYC::LEU2/PDS1::18xMYC::LEU2 Kanr::PGAL1–3xHA::MCD1/Kanr::PGAL1–3xHA::MCD1 MAD2/mad2Δ::HPH MAD1/mad1Δ::HIS3 | This study |
Chromosome loss measurements
Chromosome loss was measured by a colony color assay (Hieter et al., 1985; Spencer et al., 1990). Strains containing a nonessential, SUP11-marked chromosome fragment were grown to saturation in selective media and plated on complete synthetic medium (CSM) with a limiting adenine concentration (6 μg/ml) to increase the accumulation of red pigment in ade2-101 cells that had lost SUP11. The chromosome loss frequency was calculated by dividing the number of half-sectored colonies by the total number of colonies whose initiating cell contained the chromosome fragment.
Microcolony assays
Cultures of mid-log-phase cells were adjusted to a density of 107 cells/ml, sonicated briefly to dissociate cells that had divided but had failed to separate, diluted to 105 cells/ml in rich media (yeast extract and peptone [YPD]) made with 0.6% low–melting point agar and 60 μg/ml benomyl, spotted on glass slides, and incubated at 23°C. The number of cells per microcolony was counted under a light microscope every 3 h for 12 h, and the average number of cells per microcolony was calculated for each time point. Every visible cell was counted; cells with buds were counted as a two-cell microcolony.
Short linear chromosomes
The test strains contained a CDC28 allele (CDC28-VF) that prevents adaptation to prolonged activation of the spindle checkpoint (Rudner et al., 2000) and a checkpoint-resistant allele of CDC20 (CDC20-127) under the control of a tetracycline-inducible promoter (Ptet). Diploid cells homozygous for CDC28-VF and Ptet-CDC20-127 and containing an 11-kb short linear chromosome (Indjeian et al., 2005) were grown in CSM without leucine to maintain the short linear chromosomes. To visualize cell growth on solid media, serial fourfold dilutions of saturated cultures were spotted on solid plates (CSM without leucine) either without or with 10 μg/ml doxycycline (to repress expression of CDC20-127) and grown for 2 d at 30°C.
Cell cycle analysis
For experiments with short linear chromosomes, exponentially dividing cells containing Ptet-CDC20-127, CDC28-VF, and short linear chromosomes were arrested in S phase with 10 mg/ml hydroxyurea. Cells were released into fresh media with or without 10 μg/ml doxycycline, which represses expression of Ptet-CDC20-127, allowing short linear chromosomes to activate the spindle checkpoint. Small buds arise after cells pass through mitosis, and their number was determined by light microscopy.
We used strains that conditionally expressed the cohesin gene, MCD1, from the GAL1 promoter to investigate the response of cells to the absence of sister chromatid cohesion. Diploid cells were made responsive to α-factor either by overexpressing the HO endonuclease to convert the MATa/MATα diploids to homozygous MATa/MATa diploids or by knocking out the MATα locus using a plasmid marked with URA3 (SCSB184). Cells were grown overnight at 30°C to mid-log phase in fresh rich media (yeast extract and peptone [YEP]) with 2% galactose (wt/vol). A total of 2 ml of culture was added to 13 ml of YEP with 2% galactose (wt/vol) containing 10 μg/ml α-factor (Bio-Synthesis, Lewisville, TX). Cells were incubated for 2 h at 30°C and then harvested by centrifugation in a table-top centrifuge at top speed for 3 min at room temperature. Cells were resuspended in YEP with 2% glucose (wt/vol) (YPD) with 10 μg/ml α-factor and incubated for 1 h at 30°C. G1 arrest, as indicated by shmoo formation, was confirmed by light microscopy. Cells were washed twice to remove α-factor, resuspended in 15 ml of YPD, and placed at 30°C. One-milliliter samples were collected every 15 min, and the cells were pelleted by centrifugation for 1 min at room temperature. The supernatant was removed. Cell pellets were frozen and stored at −20°C. During the time course, 10 μg/ml α-factor was added after 50 min to prevent cells from progressing into the next S phase.
Cell pellets were lysed using a NaOH/β-mercaptoethanol–based protocol (Yaffe and Schatz, 1984). Protein samples were loaded onto and separated in 10% SDS–PAGE gels. Proteins were transferred overnight to nitrocellulose (Whatman, Piscataway, NJ). Western blotting for a Myc epitope–tagged cell cycle marker (Pds1) was performed using anti-MYC 9E10 antibodies (Roche Applied Science, Indianapolis, IN) at a 1:500 dilution, and a loading control was detected with antiactin antibodies (Abcam, Cambridge, MA) used at a 1:1000 dilution. The secondary antibody, horseradish peroxidase–linked sheep anti-mouse (Amersham, GE Healthcare Bio-Sciences, Piscataway, NJ), was used at a 1:2000 dilution. The secondary antibody was detected by enhanced chemiluminescence (Amersham), which exposed BioMax MS film (Eastman Kodak, Rochester, NY).
Measuring the level of checkpoint proteins
Cells were grown overnight at 30°C to mid-log phase, and cell density was measured by Coulter counting (Beckman-Coulter, Brea, CA). Cells were diluted into fresh YPD to yield equal cell densities for different strains and grown for an additional 6 h at 30°C. The cell density was measured by Coulter counting. Normalized volumes of cells were pelleted by centrifugation for 1 min at room temperature. The supernatant was removed. Cell pellets were frozen and stored at −20°C. The cells were lysed and prepared for Western blotting as described earlier. Calibration samples were produced by mixing extracts of diploid epitope-tagged strains with the corresponding homozygous null at several different ratios.
Acknowledgments
This work was supported by funds from the National Institutes of Health GM043987 (A.W.M.) and GM067509 (S.C.S.) and by the Chang Gung Memorial Hospital CMRP-D190211 (S.C.S.). R.K.D. was supported by a Howard Hughes Medical Institute postdoctoral grant for physician-scientists. We thank D. Pellman for plasmids.
Abbreviations used:
- APC
anaphase-promoting complex
- CSM
complete synthetic media
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-01-0029) on May 18, 2011.
REFERENCES
- Akiyoshi B, Sarangapani KK, Powers AF, Nelson CR, Reichow SL, Arellano-Santoyo H, Gonen T, Ranish JA, Asbury CL, Biggins S. Tension directly stabilizes reconstituted kinetochore-microtubule attachments. Nature. 2010;468:576–579. doi: 10.1038/nature09594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A, Venturi GM, Tatchell K. Anaphase spindle position is monitored by the BUB2 checkpoint. Nat Cell Biol. 2000;2:556–558. doi: 10.1038/35019601. [DOI] [PubMed] [Google Scholar]
- Chen RH, Shevchenko A, Mann M, Murray AW. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J Cell Biol. 1998;143:283–295. doi: 10.1083/jcb.143.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen RH, Waters JC, Salmon ED, Murray AW. Association of spindle assembly checkpoint component XMAD2 with unattached kinetochores. Science. 1996;274:242–246. doi: 10.1126/science.274.5285.242. [DOI] [PubMed] [Google Scholar]
- Chung E, Chen RH. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2. Mol Biol Cell. 2002;13:1501–1511. doi: 10.1091/mbc.02-01-0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar H, Tanaka K, Nasmyth K, Tanaka TU. Tension between two kinetochore suffices for bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T, Mortlock A, Keen N, Taylor SS. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauf S, Cole RW, LaTerra S, Zimmer C, Schnapp G, Walter R, Heckel A, van Meel J, Rieder CL, Peters JM. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Patterson TE, Sazer S. The Schizosaccharomyces pombe spindle checkpoint protein mad2p blocks anaphase and genetically interacts with the anaphase-promoting complex. Proc Natl Acad Sci USA. 1997;94:7965–7970. doi: 10.1073/pnas.94.15.7965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieter P, Mann C, Snyder M, Davis RW. Mitotic stability of yeast chromosomes: a colony color assay that measures nondisjunction and chromosome loss. Cell. 1985;40:381–392. doi: 10.1016/0092-8674(85)90152-7. [DOI] [PubMed] [Google Scholar]
- Hoyt MA, Totis L, Roberts BT. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991;66:507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Hwang LH, Lau LF, Smith DL, Mistrot CA, Hardwick KG, Hwang ES, Amon A, Murray AW. Budding yeast Cdc20: a target of the spindle checkpoint. Science. 1998;279:1041–1044. doi: 10.1126/science.279.5353.1041. [DOI] [PubMed] [Google Scholar]
- Indjeian VB, Stern BM, Murray AW. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Iwanaga Y, Chi YH, Miyazato A, Sheleg S, Haller K, Peloponese JM, Jr, Li Y, Ward JM, Benezra R, Jeang KT. Heterozygous deletion of mitotic arrest-deficient protein 1 (MAD1) increases the incidence of tumors in mice. Cancer Res. 2007;67:160–166. doi: 10.1158/0008-5472.CAN-06-3326. [DOI] [PubMed] [Google Scholar]
- King EM, Rachidi N, Morrice N, Hardwick KG, Stark MJ. Ipl1p-dependent phosphorylation of Mad3p is required for the spindle checkpoint response to lack of tension at kinetochores. Genes Dev. 2007;21:1163–1168. doi: 10.1101/gad.431507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson MA, Renduchitala K, Khodjakov A, Kapoor TM. Correcting improper chromosome-spindle attachments during cell division. Nat Cell Biol. 2004;6:232–237. doi: 10.1038/ncb1102. [DOI] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Li R. Bifurcation of the mitotic checkpoint pathway in budding yeast. Proc Natl Acad Sci USA. 1999;96:4989–4994. doi: 10.1073/pnas.96.9.4989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Murray AW. Feedback control of mitosis in budding yeast. Cell. 1991;66:519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Li X, Nicklas RB. Mitotic forces control a cell-cycle checkpoint. Nature. 1995;373:630–632. doi: 10.1038/373630a0. [DOI] [PubMed] [Google Scholar]
- Michel LS, Liberal V, Chatterjee A, Kirchwegger R, Pasche B, Gerald W, Dobles M, Sorger PK, Murty VV, Benezra R. MAD2 Haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells. Nature. 2001;409:355–359. doi: 10.1038/35053094. [DOI] [PubMed] [Google Scholar]
- Murray AW, Szostak JW. Construction of artificial chromosomes in yeast. Nature. 1983;305:189–193. doi: 10.1038/305189a0. [DOI] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Pereira G, Höfken T, Grindlay J, Manson C, Schiebel E. The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol Cell. 2000;6:1–10. [PubMed] [Google Scholar]
- Pinsky BA, Biggins S. The spindle checkpoint: tension versus attachment. Trends Cell Biol. 2005;15:486–493. doi: 10.1016/j.tcb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kung C, Shokat KM, Biggins S. The Ipl1-Aurora protein kinase activates the spindle checkpoint by creating unattached kinetochores. Nat Cell Biol. 2006;8:78–83. doi: 10.1038/ncb1341. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- Rudner AD, Hardwick KG, Murray AW. Cdc28 activates exit from mitosis in budding yeast. J Cell Biol. 2000;149:1361–1376. doi: 10.1083/jcb.149.7.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink G, Lawrence C. Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press; 1974. Methods in Yeast Genetics. [Google Scholar]
- Shonn MA, McCarroll R, Murray AW. Requirement of the spindle checkpoint for proper chromosome segregation in budding yeast meiosis. Science. 2000;289:300–303. doi: 10.1126/science.289.5477.300. [DOI] [PubMed] [Google Scholar]
- Shonn MA, Murray AL, Murray AW. Spindle checkpoint component Mad2 contributes to biorientation of homologous chromosomes. Curr Biol. 2003;13:1979–1984. doi: 10.1016/j.cub.2003.10.057. [DOI] [PubMed] [Google Scholar]
- Sironi L, Mapelli M, Knapp S, De Antoni A, Jeang KT, Musacchio A. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a “safety belt” binding mechanism for the spindle checkpoint. EMBO J. 2002;21:2496–2506. doi: 10.1093/emboj/21.10.2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F, Gerring SL, Connelly C, Hieter P. Mitotic chromosome transmission fidelity mutants in Saccharomyces cerevisiae. Genetics. 1990;124:237–249. doi: 10.1093/genetics/124.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern BM, Murray AW. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Warren CD, Brady DM, Johnston RC, Hanna JS, Hardwick KG, Spencer FA. Distinct chromosome segregation roles for spindle checkpoint proteins. Mol Biol Cell. 2002;13:3029–3041. doi: 10.1091/mbc.E02-04-0203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells WA, Murray AW. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996;133:75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe MP, Schatz G. Two nuclear mutations that block mitochondrial protein import in yeast. Proc Natl Acad Sci USA. 1984;81:4819–4823. doi: 10.1073/pnas.81.15.4819. [DOI] [PMC free article] [PubMed] [Google Scholar]