Rap1 stabilizes cell–cell junctions by directly binding to KRIT1, displacing it from microtubules and enabling localization at the junctions.
Abstract
Activation of Rap1 small GTPases stabilizes cell–cell junctions, and this activity requires Krev Interaction Trapped gene 1 (KRIT1). Loss of KRIT1 disrupts cardiovascular development and causes autosomal dominant familial cerebral cavernous malformations. Here we report that native KRIT1 protein binds the effector loop of Rap1A but not H-Ras in a GTP-dependent manner, establishing that it is an authentic Rap1-specific effector. By modeling the KRIT1–Rap1 interface we designed a well-folded KRIT1 mutant that exhibited a ∼40-fold-reduced affinity for Rap1A and maintained other KRIT1-binding functions. Direct binding of KRIT1 to Rap1 stabilized endothelial cell–cell junctions in vitro and was required for cardiovascular development in vivo. Mechanistically, Rap1 binding released KRIT1 from microtubules, enabling it to locate to cell–cell junctions, where it suppressed Rho kinase signaling and stabilized the junctions. These studies establish that the direct physical interaction of Rap1 with KRIT1 enables the translocation of microtubule-sequestered KRIT1 to junctions, thereby supporting junctional integrity and cardiovascular development.
INTRODUCTION
Rap1 small GTPase regulates both cell–cell and cell–matrix adhesions and numerous other biological functions (Bos, 2005). As is true of other Ras-family GTPases, Rap1 cycles between GTP-bound and GDP-bound forms. In the GTP-bound form, Rap1 binds to downstream effectors, and these effectors are responsible for its biological activities. Many Rap1 effectors are shared with other Ras-family small GTPases; however, the effects of Rap1 on cell adhesion appear distinct from those of other Ras-family members (Kinbara et al., 2003; Bos, 2005). Hence there is a continuing effort to identify Rap1-specific effectors that regulate adhesion.
Krev Interaction Trapped gene 1 (KRIT1) was identified as a Rap1-binding protein in a yeast two-hybrid screen; however, the GTP dependence of the interaction was not evaluated (Serebriiskii et al., 1997). KRIT1 protein is the product of the KRIT1 gene (Laberge-le Couteulx et al., 1999; Sahoo et al., 1999). Loss-of-function mutations in this gene are responsible for ∼40% of cases of human autosomal dominant familial cerebral cavernous malformations (CCMs). Furthermore, KRIT1 is required for normal blood vessel and heart development in vertebrates, since mouse homozygous null mutations of the Krit1 gene are embryonic lethal due to vascular defects (Whitehead et al., 2004), and zebrafish embryos lacking krit1 have dilated hearts (Mably et al., 2006). KRIT1 has four ankyrin repeats and a C-terminal band 4.1/ezrin/radixin/moesin (FERM) domain (Serebriiskii et al., 1997; Sahoo et al., 2001). KRIT1 FERM domain binds to heart of glass (HEG1; Kleaveland et al., 2009), a transmembrane protein that is genetically linked to KRIT1 (Mably et al., 2006). The N-terminus of KRIT1 contains three NPxY(F) motifs, certain of which bind to the phosphotyrosine-binding domain protein ICAP1α (Zawistowski et al., 2002) or CCM2 (Zawistowski et al., 2005). As noted, Rap1A binds to a fragment of KRIT1; both Rap1 and KRIT1 are regulators of endothelial junctional integrity (Cullere et al., 2005; Fukuhara et al., 2005; Kooistra et al., 2005; Wittchen et al., 2005; Glading et al., 2007). Thus it is possible that Rap1 binding to KRIT1 might be responsible for stabilization of those junctions.
In the present study, we found that KRIT1 is an authentic Rap1-specific effector that binds strongly to the effector loop of active Rap1A [Rap1A(G12V)] but not to an inactive Rap1A(S17N) or to an active variant of H-Ras. We modeled the binding interface between Rap1A and KRIT1 to create a structure-based point mutant (R452E) with markedly reduced affinity for Rap1. KRIT1(R452E) failed to support endothelial cell–cell junction stabilization in vitro or cardiovascular development in Danio rerio. KRIT1(R452E) did not localize to cell–cell junctions because it was sequestered on microtubules; however, it bound to both HEG1 and CCM2. Thus Rap1 binding to the F1 subdomain of the KRT1 FERM domain frees KRIT1 from microtubules, enabling KRIT1 to localize to cell–cell junctions and to stabilize those junctions.
RESULTS
KRIT1 is an authentic Rap1-specific effector
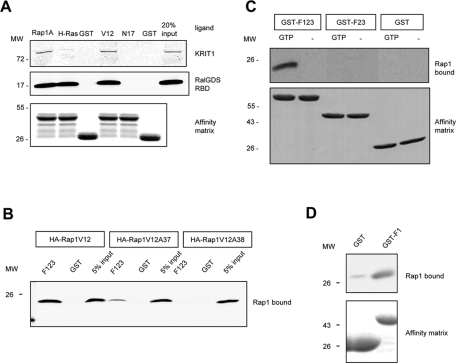
A fragment of KRIT1 was identified as a binding partner for Rap1 small GTPase in a yeast two-hybrid screen (Serebriiskii et al., 1997), and that analysis suggested significantly less interaction between this protein and H-Ras, a closely related small GTPase. However, others have failed to see an interaction with full-length KRIT1 (Zhang et al., 2001; Francalanci et al., 2009). To test whether KRIT1 is indeed a Rap1-specific effector, we examined the interaction of recombinant human full-length KRIT1 with these small GTPases by affinity chromatography. Rap1A, but not H-Ras, bound strongly to KRIT1 (Figure 1A), whereas both small GTPases bound equally well to RalGDS, a known common effector (Spaargaren and Bischoff, 1994). To assess the GTP dependence of this interaction, similar affinity chromatography was performed with either constitutively GTP-bound Rap1A (G12V) or dominant-negative Rap1A (S17N). Only the active Rap1 bound to KRIT1 (Figure 1A). Thus full-length native KRIT1 is both Rap1 specific and GTP dependent in its interactions with the Ras subfamily of small GTPases, consistent with function as a Rap1-specific effector.
FIGURE 1:
KRIT1 is a Rap1-specific effector protein, and its interaction with Rap1 requires the F1 subregion of the FERM domain and the switch I domain of Rap1. (A) Recombinant bead-bound GST-Rap1A, GST-H-Ras, and GST were used to isolate recombinant KRIT1 and RalGDS from cell lysates. KRIT1 binds to Rap1A but not H-Ras or GST, whereas RalGDS binds to both Rap and Ras proteins (left). KRIT1 preferentially binds active Rap1A (GST-Rap1V12) and does not bind to an inactive Rap1 variant (GST-Rap1N17) or to GST only (right). GST blot is shown as loading control. Blots are representative; n = 3. Blots were cropped, and intervening lanes were removed using Adobe Photoshop. (B) KRIT1–Rap1A interaction involves residues within the switch 1 domain of Rap1A. Recombinant Rap1A (V12) from HEK293 cell lysate binds to GST-F123 (left). Single-alanine mutation on a switch 1 domain residue D38 (Rap1V12A38) does not bind to GST-F123 (right). Another switch 1 domain mutant (Rap1V12A37) dramatically reduces the binding to GST-F123 (middle). Blots are representative of three experiments. (C) KRIT1 binding to Rap1A is GTP dependent. Recombinant Rap1A from cell lysate binds to the KRIT1 FERM domain (GST-F123) only in the presence of GTP (10 mM). Binding of Rap1A to GST-KRIT1 FERM is lost on deletion of the F1 region, as binding to truncated FERM domain protein (GST-F23) is not observed (top). Expression of GST constructs is shown at bottom; blots are representative of three experiments. (D) F1 subregion of KRIT1 FERM domain is sufficient for Rap1A binding. Top, recombinant Rap1A (V12) binds to GST-F1 (right) but not GST alone (left). Bottom, equal loading of both GST fusion proteins as judged by SDS–PAGE and Coomassie Blue staining. Blots are representative of three experiments.
A common region of Ras GTPases serves as a major binding site for most effectors, leading us to examine the role of this region in interaction with KRIT1. The crystal structure of Rap1A/c-Raf Ras-binding domain (RBD) complex (Nassar et al., 1995) reveals that the major interaction is mediated by the β2 strand of Rap1A and the β2 strand and the preceding loop from the c-Raf RBD. The Rap1A β2 strand is within the so-called switch 1 region containing five amino acids (E37, D38, S39, Y40, and R41). To test whether KRIT1–Rap1 interaction involves the Rap1 switch 1 region, we made alanine mutations at each of those five residues on the constitutively active Rap1A (G12V) background and expressed each of them in HEK293 cells to assess their binding affinities to the KRIT1 FERM domain. Two mutants, Rap1A (G12V, E37A) and Rap1A (G12V, D38A), showed reduced binding to the KRIT1 FERM domain (Figure 1B). Thus, on the basis of involvement of the Rap1A switch 1 region, GTP dependence of binding, and the binding of native full-length KRIT1 to Rap1A but not H-Ras, we conclude that KRIT1 is an authentic Rap1-specific effector.
Mapping the Rap1-binding interface in KRIT1
To analyze the functional importance of Rap1 binding to KRIT1, we mapped the KRIT1 region that binds to Rap1. Affinity chromatography showed that the KRIT1 FERM domain binds to Rap1A (Figure 1C), in agreement with a previous yeast two-hybrid analysis (Serebriiskii et al., 1997). As with the interaction of full-length KRIT1, the binding of the FERM domain (F123) to Rap1A was GTP dependent (Figure 1B). Domain analyses (http://smart.embl-heidelberg.de/, http://scansite.mit.edu/motifscan_seq.phtml) suggested a potential Ras association domain in the predicted F1 subregion of the KRIT1 FERM domain. The importance of this region was confirmed by the failure of Rap1A to bind to the F23 fragment of KRIT1, which lacks the F1 subregion (Figure 1C). We also expressed the isolated F1 subregion and found that F1 alone was sufficient for Rap1A binding (Figure 1D). Thus the FERM domain of KRIT1 binds to Rap1 in a GTP-dependent manner, and the F1 subregion mediates this interaction.
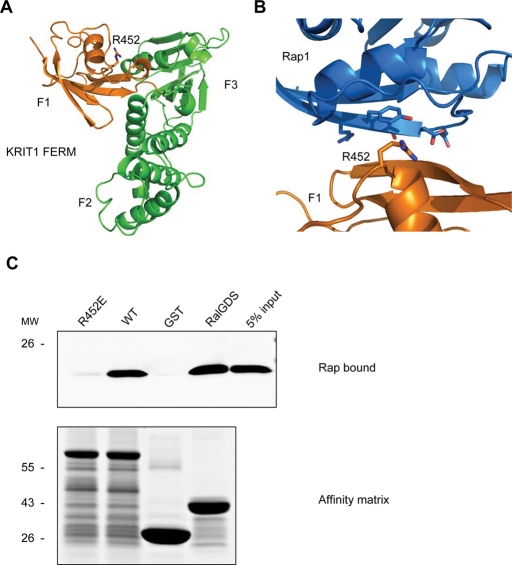
Having mapped the Rap1 binding site to the F1 subdomain, we built a homology model of the KRIT1 FERM domain using the moesin FERM domain structure (PDB 1EF1) as a template. In this model (Figure 2A), the predicted F1 subdomain (orange) contained a five-stranded β sheet interrupted by two α helices and structurally resembled the c-Raf RBD (Nassar et al., 1995). The model was subjected to conjugate energy minimization in a water-bound environment in the presence of Rap1 (Figure 2B). The model had good structure-packing quality (Z score of 0.83), indicating that the predicted structure is compatible with the sequence of the KRIT1 FERM domain. The major predicted binding interface between Rap1 (blue) and KRIT1 (orange) involved the β2 strand from Rap1 switch 1 domain, in agreement with our previous results (Figure 1B). In KRIT1 F1 subdomain, the β2 strand and the preceding loop are in close contact with Rap1A, and Arg452 of KRIT1 is proximal to Rap1A Glu-37 and Asp-38 residues. We had implicated these acidic Rap1A residues in the capacity of Rap1A to bind to KRIT1 (Figure 1B), lending additional credence to the model. KRIT1 Arg-452 corresponded to c-Raf Arg-89, a residue critical for Rap1 binding (Nassar et al., 1995).
FIGURE 2:
KRIT1 FERM domain homologous modeling reveals R452 residue mediating KRIT1–Rap1 interaction. (A) Homology model of KRIT1 FERM domain. Residues 410–736 from KRIT1 were modeled using the moesin crystal structure as the template. The modeled structure is formed by three subdomains that correspond to the F1 (orange), F2, and F3 (both in green) subregions of the FERM fold. F1 subregion structurally resembles the c-Raf RBD in a complex with Rap1. An arginine residue (R-452), whose corresponding residue in c-Raf (R-89) mediates the c-Raf–Rap1 binding, is also conserved in the KRIT1 F1 subregion. (B) A closer look at the modeled binding interface between KRIT1 F1 subregion (orange) and Rap1 (blue). The R452 residue in KRIT1 and switch 1 domain residues in Rap1 are shown in black. (C) The R452 residue is crucial for KRIT1–Rap1 binding. GST-F123 WT pulls down recombinant Rap1V12 from HEK293 cell lysate (second lane). The GST-F123(R452E) mutant form, which changes a basic residue to an acidic residue, does not bind to Rap1V12 (left lane). Bottom, the equal loading of both GST fusion proteins as judged by SDS–PAGE and Coomassie Blue staining. Blots are representative of five experiments.
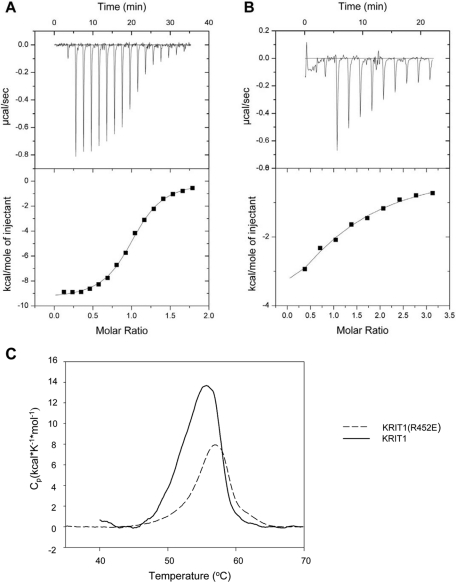
On the basis of the homology model described, we created a KRIT1(R452E) mutant and tested its interaction with Rap1. Affinity chromatography (Figure 2C) revealed that the FERM domain of KRIT1(R452E) exhibited a markedly reduced affinity for Rap1A. We also quantified the binding affinity by isothermal titration calorimetry (ITC) (Figure 3, A and B). The KRIT1 FERM domain bound to Rap1 with Kd = 1.8 ± 0.25 μM; however, the KRIT1(R452E) FERM bound with a ∼37-fold-lower affinity (Kd = 67 ± 18.7 μM; Table 1). It is significant that the R452E mutation did not affect the folding of the KRIT1 FERM domain, since it exhibited a sharp melting point (57.0 ± 0.1°C vs. 55.7 ± 0.1°C for wild-type FERM domain) in differential scanning calorimetry (Figure 3C). Thus the KRIT1(R452E) mutant remains well folded but binds to Rap1 with dramatically reduced affinity.
FIGURE 3:
Calorimetric characterization of KRIT1 FERM-domain binding to Rap1B bound to GMP-PNP, a GTP analogue. (A) Calorimetric titration of 400 μM Rap1B, out of the syringe, into 45 μM wild-type KRIT1 FERM domain in the sample cell. (B) Titration of 1.2 mM Rap1B into 45 μM KRIT1(R452E) FERM domain mutant protein. (C) KRIT1(R452E) FERM mutant does not disrupt protein folding. Differential scanning calorimetry results of wild-type and R452E FERM proteins both exhibit similar narrowly defined melting points, indicating that they are well folded.
TABLE 1:
Thermodynamic parameters for binding of KRIT1 FERM domain, wild type and R452E mutant, to Rap1B bound to GMP-PNP.
| n | Kd (μM) | ΔHº (kcal/mol) | ΔGº (kcal/mol) | TΔSº (kcal/mol) | |
|---|---|---|---|---|---|
| Wild type | 1.0 | 1.8 ± 0.25 | −9.5 | −7.9 | −1.6 |
| R452E | 1.0 | 67 ± 18.7 | −7.9 | −5.7 | −2.2 |
Kd = 1/Ka; ΔGº = –RT ln Ka ; ΔG = ΔH – TΔS.
The KRIT1–Rap1 interaction stabilizes endothelial cell–cell junctions
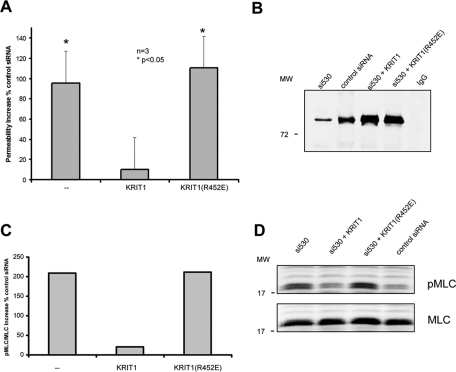
Rap1 acts to stabilize cell–cell junctions through several potential effectors (Bos, 2005), and we proposed KRIT1 as one such effector (Glading et al., 2007). Having created a folded KRIT1 point mutant with markedly impaired Rap1 binding, we used this mutant to test the role of KRIT1–Rap1 interaction in the stabilization of endothelial cell–cell junctions. Small interfering RNA (siRNA)–mediated depletion of KRIT1 led to disruption of cell junctions as measured by a twofold increase in monolayer permeability; cotransfection of KRIT1 reversed the increased permeability. In sharp contrast, cotransfection of KRIT1(R452E) failed to reverse the siRNA-induced permeability increase (Figure 4A), even though it was expressed at similar abundance to wild-type KRIT1 (Figure 4B).
FIGURE 4:
KRIT1–Rap1 binding regulates HuVEC permeability. (A) Knockdown of KRIT1 expression by KRIT1 siRNA causes an approximately twofold increase in permeability (left). Reconstitution of recombinant WT KRIT1 reverses increased permeability (center). Recombinant KRIT1(R452E) mutant does not reverse increased permeability (right). Data shown are mean increased percentage over control siRNA in permeability ± SEM; n = 3. *p < 0,05 compared with control siRNA. (B) A representative blot of KRIT1 depletion and reexpression of recombinant proteins. More than 80% knockdown of KRIT1 is observed by immunoprecipitation and blotting. Reconstituted wild-type and RE (arginine 452 to glutamic acid) mutant KRIT1 proteins are expressed at equivalent levels. (C, D) Knockdown of KRIT1 expression causes an approximately threefold increase in pMLC/total MCL ratio (left). Reconstitution of recombinant WT KRIT1 reverses MLC phosphorylation increase (center). Recombinant KRIT1(R452E) mutant does not reverse increased pMLC level (right). Data shown are mean increased percentage over control siRNA; n = 2. (D) Immunoblots of pMLC (top) and total MLC (bottom) in HuVECs.
KRIT1 suppresses the activity of RhoA and its effector, Rho kinase (ROCK), thereby stabilizing endothelial cell–cell junctions (Stockton et al., 2010). We therefore examined the capacity of KRIT1(R452E) to inhibit phosphorylation of myosin light chain (MLC), a major substrate of ROCK. Depletion of KRIT1 led to increased phosphorylated MLC (pMLC), an index of ROCK activity (Figure 4C). This increased pMLC was reversed by reconstitution of KRIT1 but not KRIT1(R452E). Reconstituted wild-type and mutant KRIT1 proteins were equally abundant in all transfections (unpublished data). Thus Rap1 binding to KRIT1 stabilizes endothelial cell–cell junctions by inhibiting the RhoA/ROCK signaling pathway.
KRIT1 requires Rap1 binding to support cardiovascular development
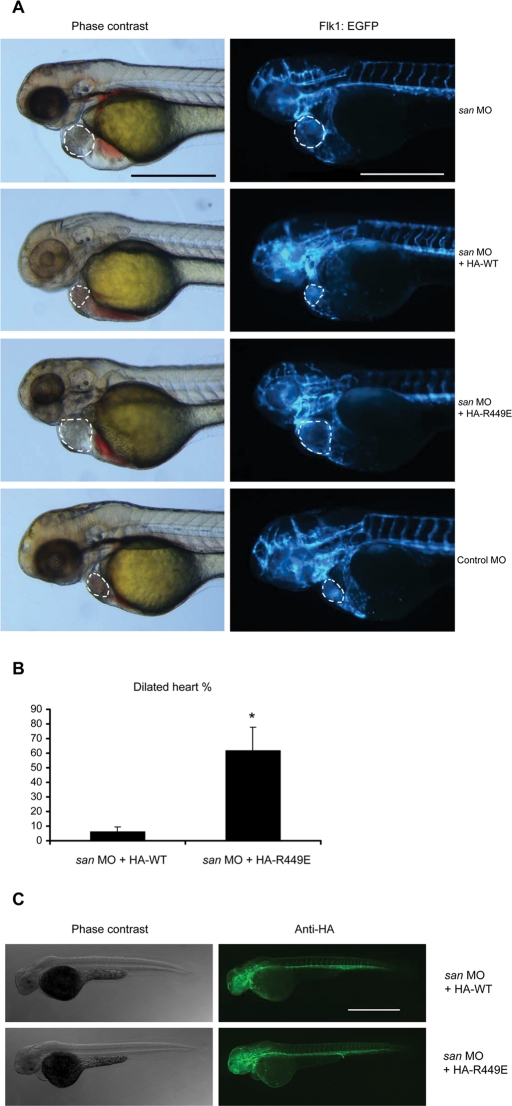
The foregoing experiments established the importance of Rap1 binding for KRIT1 function in stabilizing endothelial cell–cell junctions in vitro. In addition to its effects regulating vascular permeability, KRIT1 is important in vertebrate cardiovascular development. Zebrafish embryos lacking krit1 (also known as santa) exhibit a dilated heart phenotype (Mably et al., 2006), and zebrafish krit1 Arg-449 corresponds to human KRIT1 Arg-452. Injection of an antisense morpholino oligonucleotide targeting splice donor site at exon 14 of krit1 (san MO) (Mably et al., 2006) resulted in cardiac dilation in greater than 95% of san MO–injected fish at 48 h postfertilization (hpf). Authentic dilation was verified by identifying the endocardium in the fluorescence images of flk1:EGFP zebrafish (Figure 5A). Coinjection of wild-type krit1 cRNA together with san MO resulted in less than 5% of fish with dilated hearts (Figure 5B). In sharp contrast, 60% of the san MO + krit1(R449E) cRNA–injected fish exhibited dilated hearts. Both wild-type and R449E zebrafish krit1 cRNAs contained N-terminal hemagglutinin (HA) tag, and similar protein expression levels were confirmed by immunofluorescence microscopy (Figure 5C). In addition, both wild-type and mutant proteins were well expressed in HEK293 cells (Supplemental Figure S3). Thus the direct interaction between Rap1 and KRIT1 is required for KRIT1 function in cardiovascular development.
FIGURE 5:
KRIT1–Rap1 interaction is required for normal zebrafish cardiovascular development in vivo. (A) Flk1:EGFP transgenic krit1-morphant (san MO) embryo showed enlarged heart phenotype. Coinjecting the cRNA encoding HA-tagged wild-type (WT) zebrafish krit1 protein reduced the heart size of injected fish, whereas fish coinjected with cRNA encoding HA-tagged Krit1(R449E) (RE, homologous to human R452E mutant) still exhibited a dilated heart phenotype. Dilated heart phenotype was scored by cardiac dilation in living fish in combination with a slower heart rate. Authentic dilation was verified by identifying the endocardium in the fluorescence images, which is indicated by the dotted lines. All microscopic images were taken at 48 hpf. Scale bars, 500 μm. (B) Bar graphs showing effects of Krit1 RE mutant on zebrafish cardiovascular development. Data are expressed as number of embryos with dilated heart phenotype divided by total number of embryos used per experiment times 100%, mean ± SD. Asterisk indicates p < 0.05 compared to san MO + HA-WT cRNA group. Data are from three independent experiments. Total number of animals used: 85 in san MO + HA-WT group, 120 in san MO + HA-RE group. (C) Lateral views of fixed animals at 48 hpf. Right, immunofluorescence images revealing that both HA-WT and HA-RE proteins were expressed at similar levels and throughout the entire fish. Scale bar, 500 μm.
Mechanisms of Rap1 regulation of KRIT1
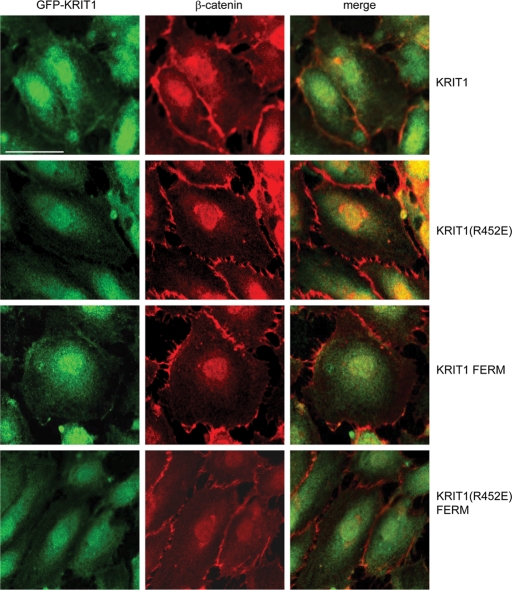
The foregoing experiments established that Rap1 interaction with KRIT1 is required for KRIT1 function. In endothelial cells, KRIT1 shows three different subcellular localizations: endothelial cell–cell junctions, cytoplasm, and nucleus (Glading et al., 2007). In dermal microvascular endothelial cells, KRIT1(R452E) and KRIT1(R452E) FERM domain failed to localize to cell–cell junctions; however, both were present in the cytoplasm and the nucleus (Figure 6). β-Catenin staining was used as a bona fide endothelial cell–cell junction marker. Our results reveal that KRIT1 localization to cell–cell junctions is controlled by direct binding of the FERM domain to Rap1.
FIGURE 6:
Rap1 binding determines KRIT1 localization to endothelial cell–cell junctions. GFP-fused KRIT1 full-length protein (top) and FERM domain (third row from top) colocalize with β-catenin (red) in HuVEC cell–cell junctions. GFP-fused KRIT1(R452E) (second row from top) and KRIT1(R452E) FERM (bottom) mutants do not localize to HuVEC cell–cell junctions, whereas β-catenin (red) localizes to junctions. Confocal images are representative; n = 3. Bar, 50 μm.
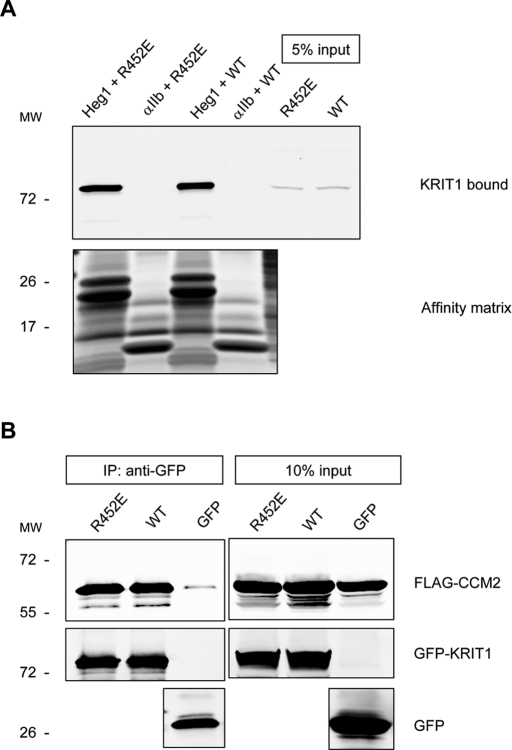
KRIT1 has several binding partners that regulate its subcellular localization, including CCM2 (OSM) (Zawistowski et al., 2005; Stockton et al., 2010) and heart of glass (HEG1) (Kleaveland et al., 2009). The HEG1 cytoplasmic domain bound KRIT1(R452E) to the same extent as wild-type KRIT1 (Figure 7A). Similarly, KRIT1(R452E) interacted with CCM2 (Figure 7B). Furthermore, the KRIT1 FERM domain lacks the CCM2-binding site (Zhang et al., 2007) and is sufficient to target KRIT1 to cell–cell junctions (Glading et al., 2007). The isolated KRIT1(R452E) mutation failed to target KRIT1 to cell–cell junctions, thus establishing that Rap1 binding is required to physically target the KRIT1 FERM domain to cell–cell junctions without influencing other known binding partners.
FIGURE 7:
Rap1 binding does not affect KRIT1 interactions with HEG1 or CCM2. (A) HEG1 cytoplasmic tail model protein binds to recombinant KRIT1 WT and R452E mutant proteins from HEK293 cell lysates. Integrin αIIb tail model protein does not bind to KRIT1. Bottom, the equal loading of tail proteins as judged by SDS–PAGE and Coomassie Blue staining. Blots are representative of three experiments. (B) Both GFP-fused KRIT1 WT and GFP-fused KRIT1(R452E) are associated with CCM2 at equivalent levels as assessed by coimmunoprecipitation and immunoblotting. Blots are representative of three experiments.
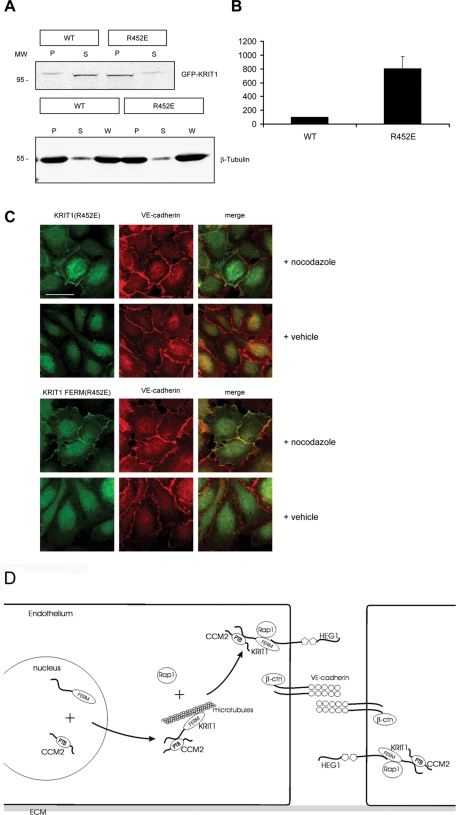
KRIT1 associates with microtubules, and transfection of activated Rap1 can reduce this association (Gunel et al., 2002; Beraud-Dufour et al., 2007). To directly test the importance of KRIT1–Rap1 physical interaction in limiting KRIT1 association with microtubules, we expressed green fluorescent protein (GFP)–fused KRIT1 or KRIT1(R452E) and examined interaction with microtubules. The Rap1-binding–deficient mutant KRIT1(R452E) exhibited an approximately eightfold increase in association with microtubules compared with the wild-type protein (Figure 8, A and B). When the microtubules were depolymerized by nocodazole treatment, neither KRIT1 nor KRIT1(R452E) was sedimented (Supplemental Figure S2). Thus the physical association between KRIT1 and microtubules is inhibited by the direct KRIT1–Rap1 interaction.
FIGURE 8:
KRIT1–Rap1 interaction enables KRIT1 release from microtubules. (A) Top, KRIT1(R452E) (right two lanes) cosedimented with microtubules eightfold more than KRIT1 wild type (left two lanes). Bottom, immunoblotting of β-tubulin to show that the microtubule sedimentation is equivalent. P, pellet; S, supernatant; W, whole-cell lysate. (B) Quantification of KRIT1 cosedimented with microtubules. KRIT1 in the pellet was normalized to 100%. n = 3. (C) When microtubules were depolymerized by nocodazole, GFP-fused KRIT1(R452E) and KRIT1 FERM(R452E) localized to endothelial cell membranes and colocalized with junction marker VE-cadherin (red). Confocal images are representative; n = 3. Bar, 50 μm. (D) Mechanism of Rap1-mediated KRIT1 junctional localization. In endothelial cells, CCM2 binding enables KRIT1 egress from the nucleus. In the cytoplasm, KRIT1 is associated with microtubules. Rap1 binds KRIT1 and releases KRIT1 from microtubules. KRIT1 binds HEG1 to localize to endothelial cell–cell junctions, where it associates with junctional proteins, including β-catenin and VE-cadherin. Endothelial junction–localized KRIT1 controls junctional stability by suppressing RhoA activation. ECM, extracellular matrix.
To learn whether the association between KRIT1 and microtubules limits its membrane localization and thus targeting to endothelial cell–cell junctions, we expressed either GFP-fused KRIT1 or KRIT1(R452E) in dermal microvascular endothelial cells. Wild-type KRIT1 but not the KRIT1(R452E) mutant localized to the plasma membrane at endothelial junctions. The KRIT1(R452E) mutant was absent from the membrane and colocalized with microtubules (Supplemental Figure S1). When the microtubules were depolymerized by nocodazole, the KRIT1(R452E) mutant and the KRIT1(R452E) FERM mutant localized to the membrane and partially colocalized with VE-cadherin (Figure 8C). It is significant that nocodazole treatment is known to activate the RhoA/ROCK pathway, thus causing junctional disruption (Verin et al., 2001); hence we could not test whether microtubule depolymerization enabled KRIT1(R452E) to reverse the effect of KRIT1 silencing on junctional stability. Thus Rap1 binding displaces KRIT1 from microtubules, enabling it to translocate to the membrane and to cell–cell junctions.
DISCUSSION
Rap1 GTPase promotes cell–matrix adhesion and cell–cell junction formation and maintenance. A large number of potential effectors have been identified that might mediate these actions (Bos, 2005); however, decisive proof that Rap1 interaction with these effectors regulates adhesion is lacking. We previously showed that KRIT1, a fragment of which binds to activated Rap1A, stabilizes cell–cell junctions in endothelial (Glading et al., 2007) and epithelial (Glading and Ginsberg, 2010) cells. Here we report that KRIT1 is an authentic Rap1-specific effector, in that native KRIT1 binds to the switch 1 region of Rap1A in a GTP-dependent manner. By modeling the KRIT1–Rap1 interface, we developed a point mutant of KRIT1 that remains folded and interacts with other KRIT1-binding partners but exhibits an ∼40-fold-reduced affinity for Rap1A. By use of this mutant, we prove that the KRIT1–Rap1 interaction stabilizes endothelial cell–cell junctions in vitro and is required for cardiovascular development in vivo. We report that Rap1 binding enables KRIT1 function by freeing KRIT1 from the microtubule cytoskeleton, enabling targeting of KRIT1 to cell–cell junctions. Our data establish that direct physical interaction of Rap1 and its effector KRIT1 is required for KRIT1 junctional localization and function in endothelial cells and in cardiovascular development.
KRIT1 is an authentic Rap1-specific effector based on four lines of evidence. First, full-length KRIT1 protein binds to activated Rap1A but not activated H-Ras, indicating that KRIT1 specifically binds Rap1 among the Ras family of small GTPases. Second, full-length KRIT1 and the KRIT1 FERM domain bind to GTP-bound Rap1A (V12) but not the nucleotide-free form of Rap1A (N17), indicating that KRIT1 binds to Rap1 in a GTP-dependent manner. Third, point mutations on two residues of the Rap1A switch 1 region (Glu37 and Asp38) disrupt KRIT1 binding, indicating that KRIT1 interacts with the Rap1A effector–binding domain, which is a hallmark of small GTPase–effector interactions (Herrmann and Nassar, 1996). Fourth, the F1 domain of KRIT1 resembles a Ras-binding domain, and we show that it interacts with Rap1 in a manner analogous to the Rap1–cRaf interaction. Most important, mutation of the Rap1-binding site in KRIT1 blocks KRIT1 function, establishing the biological relevance of the direct KRIT1–Rap1 interaction. It is particularly notable that among the many Rap1-binding proteins known, KRIT1 appears to be unusual in its tight binding to Rap1 but not to Ras (Raaijmakers and Bos, 2009). Within cells, many Ras-like GTPases may be active. The remarkable Rap1 specificity and affinity of KRIT1 suggest that at modest levels of Rap1 activation, KRIT1 will be preferentially engaged rather than other effectors such as RIAM or afadin. This preferential engagement provides a plausible explanation for the capacity of KRIT1, a relatively trace protein (Glading and Ginsberg, 2010), to play a critical role in the preservation of cell–cell junctions (Glading et al., 2007).
KRIT1–Rap1 physical interaction is important for KRIT1 function both in vitro and in vivo because 1) KRIT1 (R452E) mutant did not reduce endothelial monolayer permeability and MLC phosphorylation in endothelial cells, and 2) zebrafish KRIT1(R449E) mutant did not support cardiovascular development. Both CCM2 and HEG1 bind to KRIT1, and their interactions are important for KRIT1 function (Zawistowski et al., 2005; Kleaveland et al., 2009; Stockton et al., 2010). KRIT1(R452E) mutant interacts with both HEG1 and CCM2 to a similar extent as wild-type KRIT1 does. This indicates that Rap1 binding does not affect KRIT1 interaction with CCM2 and HEG1 per se. KRIT1(R452E) mutant colocalizes and cosediments with tubulin and does not localize to cell–cell junctions. Depolymerization of microtubules enables KRIT1(R452E) to reach the plasma membrane. Thus Rap1 controls KRIT1 localization to cell–cell junctions by facilitating the release of KRIT1 from microtubules.
In endothelial cells, KRIT1 shows three different subcellular localizations: endothelial cell–cell junctions, cytoplasm, and nucleus (Glading et al., 2007). Our data indicate that junctional-localized KRIT1 is required for its functions in junctional stabilization and in development. That said, the potential functions of cytoplasmic or nuclear KRIT1 remain open questions. Nevertheless, it is clear from the present work that the regulation of the subcellular localization of KRIT1 is important in the integrity of cell–cell junctions. In particular, Figure 8D illustrates that KRIT1 binds to CCM2 and thereby exits the nucleus (Zawistowski et al., 2005; Francalanci et al., 2009); in the cytosol, KRIT1 associates with and is sequestered by microtubules. It is possible that KRIT1 is transported along microtubules toward cell–cell junctions, thereby contributing to the stabilization of the junctions (Verin et al., 2001; Tozer et al., 2005). Activation of Rap1 at cell–cell junctions (Hogan et al., 2004; Sakurai et al., 2006) leads to its binding to KRIT1, thereby displacing KRIT1 from microtubules and increasing its affinity for phosphatidylinositol 4,5-P2 (Beraud-Dufour et al., 2007). Activated Rap1 may thus facilitate KRIT1 localization to the junctional membrane, where it suppresses RhoA activity to stabilize the junction. In endothelial cells this pathway functions to stabilize vascular development and integrity; however, it is also functional in epithelial cells. Indeed, in these cells the KRIT1–Rap1 pathway stabilizes cell–cell junctions, thereby inhibiting the canonical Wnt signaling pathway and tumorigenesis triggered by that pathway (Glading and Ginsberg, 2010). Thus the present studies define a mechanistic basis whereby Rap1 signaling can regulate cell junctional integrity in multiple physiological and pathological contexts.
MATERIALS AND METHODS
Cell culture and transfection
Human embryonic kidney (HEK) 293 cells were maintained in DMEM (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (FBS) (Sigma-Aldrich, St. Louis, MO), 1% nonessential amino acids, 1% l-glutamine, and 1% penicillin and streptomycin (all from Invitrogen, Carlsbad, CA). HEK293 cells were transfected using Lipofectamine Plus (Invitrogen) according to the manufacturer's protocol. Human umbilical vein endothelial cells (HuVECs; Lonza, Basel, Switzerland) and human dermal microvascular endothelial cells (HMVECs; ScienCell, San Diego, CA) were maintained in EGM-2 supplemented with bullet kit (Lonza) or Endothelial Medium (ScienCell) supplemented with 10% FBS, antibiotic–antimycotic solution, and endothelial cell growth substances (Invitrogen). HuVECs and HMVECs were transfected either by AMAXA nucleoporation (Lonza) or by using FuGENE 6 (Roche, Indianapolis, IN) according to the manufacturer's protocol. U2OS cells were maintained in McCoy's 5A medium (Cellgro) supplemented with 10% FBS (Sigma-Aldrich), 1% nonessential amino acids, 1% l-glutamine, and 1% penicillin and streptomycin (all from Invitrogen). U2OS cells were transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol.
Antibodies, cDNAs, siRNA, and reagents
Monoclonal anti-HA and monoclonal anti–FLAG M2 antibodies (both from Sigma-Aldrich) were used for immunoblotting at 1:4000. Monoclonal anti-KRIT1 (15B2) and polyclonal anti-KRIT1 (6832) antibodies were previously described (Glading et al., 2007). Mouse anti–KRIT1 15B2 antibody was used for immunoprecipitation. Rabbit anti–KRIT1 6832 antibody was used for immunoblotting at 1:1000. Polyclonal anti-pMLC antibody (Cell Signaling, Danvers, MA) was used for immunoblotting at 1:1000. Polyclonal anti-GFP antibody (Clontech, Mountain View, CA) was used for immunoprecipitation. Monoclonal anti-GFP antibody (Clontech) was used for immunoblotting at 1:5000. Monoclonal anti–β-tubulin antibody (BD Biosciences, San Jose, CA) was used for immunoblotting at 1:2000. Monoclonal anti–β-actin antibody (Sigma-Aldrich) was used for immunoblotting at 1:4000.
Human KRIT1 siRNA, cDNAs encoding HA- and GFP-tagged full-length KRIT1, and KRIT1 FERM domain were previously described (Glading et al., 2007). Human KRIT1 R452E mutant was generated by site-directed mutagenesis using the QuikChange SD mutagenesis kit (Agilent, Santa Clara, CA). The mutant was then subcloned into pGEX2TK (GE Healthcare, Piscataway, NJ), pcDNA3.1 (Invitrogen), and pEGFP-C1 (Clontech) vectors for expressing GST-, HA-, and GFP-tagged proteins, respectively. Zebrafish krit1 cDNA was a gift from B. Weinstein (National Institutes of Health, Bethesda, MD) and subcloned into pcDNA3.1 vector (Invitrogen) with an N-terminal HA tag. Zebrafish krit1 (R449E) mutant was generated using the QuikChange SD mutagenesis kit. pRK5-CCM2 encoding FLAG-tagged human CCM2 has been previously described and was the generous gift of Doug Marchuk (Duke University, Durham, NC) (Zawistowski et al., 2005). The recombinant human HEG1 cytoplasmic tail model protein affinity matrix was previously described (Lim et al., 2007; Kleaveland et al., 2009). Mammalian expression constructs for HA-Rap1A-G12V (Rap1V12) and HA-Rap1GAP (RapGAP) have been described (Han et al., 2006). HA-Rap1A-G12V E37A, D38A, S39A, Y40A, and R41A mutants were generated using the QuikChange SD mutagenesis kit. pGEX-H-Ras construct was a gift from D. Broek (University of Southern California, Los Angeles, CA).
Homology modeling of KRIT1 FERM domain–Rap1 interaction
We used a DeepView Swiss-PdbViewer (Swiss Institute of Bioinformatics, Lausanne, Switzerland) (Guex and Peitsch, 1997) to align the KRIT1 FERM and moesin FERM sequences and to build an initial model of KRIT1 FERM domain. We then sent the alignment to I-TASSER server for protein structure and function prediction (Zhang, 2008, 2009; Roy et al., 2010) and built a revised model of the KRIT1 FERM domain. We replaced the c-Raf RBD from the Rap1/c-Raf RBD complex crystal structure (PDB 1C1Y) with our KRIT1 FERM model and adjusted the spatial restraints with Rap1 by using Coot (Emsley et al., 2010). This Rap1/KRIT1 FERM complex model was then completely immersed in a water sphere, and the whole system was subjected to energy minimization by molecular dynamics simulation using NAMD program (Phillips et al., 2005). The simulation continued for 5 ns until the total energy reached the minimum and the RMSD was stabilized. The quality of the model was assessed by the program QMEAN (Benkert et al., 2009). The final model was rendered for publication in PyMOL (Schrödinger, New York, NY).
In vitro protein interaction assay
Bacterial expression plasmids encoding glutathione S-transferase (GST)–KRIT1 FERM, partial KRIT1 FERM domains (F23, F1), R452E mutant, GST–RalGDS RBD, or GST vector were expressed in BL21 (DE3) (Novagen, Madison, WI), and recombinant proteins were purified on glutathione-Sepharose beads according to manufacturer's instructions (GE Healthcare). HEG1 and αIIb intracellular tail model proteins were expressed and purified as previously described (Pfaff et al., 1998). To perform pulldown assays using cell lysates, HEK293 cells were transfected with indicated cDNAs. At 24 h after transfection, cells were lysed in cold lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.5% NP-40, and 5 mM MgCl2) plus protease and phosphatase inhibitor cocktails (Roche). A total of 10 μg of immobilized bead-bound proteins was added to 350 μg of clarified cell lysates. Reactions were kept at 4°C overnight. After washing the beads with lysis buffer, samples were fractionated on 4–20% SDS–PAGE gel (Invitrogen). Bound proteins were analyzed by immunoblotting or Coomassie Blue staining.
Immunoprecipitation
HEK293 cells were transfected with indicated cDNAs and incubated for 24 h. Cells were scraped on ice in lysis buffer plus protease and phosphatase inhibitor cocktails (Roche). A total of 2 μg of monoclonal anti-GFP antibody was added to 350 μg of cell lysates and incubated at 4°C overnight. Protein G-Sepharose (Invitrogen) was added to the reaction mixture and further incubated for 4 h at 4°C. After three washes with lysis buffer, beads were mixed with sample buffer and subjected to SDS–PAGE. Immunoprecipitated proteins were detected by immunoblotting.
Isothermal titration calorimetry and differential scanning calorimetry
Proteins were dialyzed into the ITC buffer (20 mM sodium phosphate, pH 6.5, 150 mM NaCl, 3 mM MgCl2, and 0.1 mM guanylimidodiphosphate [GMP-PNP]). The thermodynamic parameters are determined using an isothermal titration calorimeter iTC200 (MicroCal, Piscataway, NJ) at 25°C in 20 mM sodium phosphate (pH 6.5), 150 mM NaCl, 3 mM MgCl2, and 0.1 mM GMP-PNP. For binding, 0.4 or 1.2 mM Rap1B bound to GMP-PNP was titrated from the syringe into the sample cell containing 45 μM KRIT1 FERM domain wild-type or R452E mutant protein, respectively. Titration was carried out by injecting volumes of 2.5 μl into the sample cell, where the time between injections was 2 min. Further data evaluation was done using the MicroCal Origin program.
Differential scanning calorimetry experiments were performed at a scanning rate of 1 K/min under 3.0 atm of pressure using N-DSC II differential scanning calorimeter (Calorimetry Sciences Corp, Provo, UT). Differential scanning calorimetry samples contained phosphate-buffered saline (PBS) (pH 7.4) and 0.8 mg/ml KRIT1 F123 or KRIT1F123(R452E).
Protein expression and purification for ITC and differential scanning calorimetry
The cDNA encoding human KRIT1 residues 417–736 was synthesized by PCR using a human KRIT1 cDNA as template and cloned into the expression vector pLEICS-07 (Protex, Leicester, United Kingdom). Constructs were expressed in Escherichia coli BL21 Star (DE3) (Invitrogen) cultured in LB media. Recombinant His-tagged polypeptides were purified by nickel-affinity chromatography following standard procedures. The His tag was removed by cleavage with AcTEV protease (Invitrogen), and the protein was further purified by cation-exchange chromatography. The protein concentration was assessed using the A280 extinction coefficient of 45,090 M−1 before dialysis into buffers. Human Rap1 isoform Rap1B (residues 1–167) was cloned into pTAC vector and expressed in the E. coli strain CK600K (Tucker et al., 1986). Cultures were grown at 37°C to A600 of 0.8, when they were induced with 200 μM IPTG and then cultured at 18°C overnight. Rap1B protein was purified by ion exchange, followed by Superdex-75 gel filtration. Nucleotide exchange was achieved as previously described (Gorzalczany et al., 2000), and the final sample was dialyzed into ITC buffer. The protein concentration was assessed using the CB protein assay (EMD, San Diego, CA).
In vitro endothelial permeability assay
Human umbilical vein endothelial cells were detached and transfected with indicated DNA and/or siRNA using an Amaxa (Lonza) nucleoporation device, then plated onto fibronectin-coated 6-mm Transwell filters and grown for 48 h. In some experiments, cells were plated into filters and grown to confluence, then transfected using FuGENE 6. Upper and lower chamber medium was replaced with phenol-free DMEM. Fluorescein isothiocyanate (FITC)–labeled 40-kDa dextran (2 μg/ml) was added to upper chambers for 2 h, and then FITC was quantified in lower chamber medium by fluorimetry. Raw fluorescence units were normalized as percentage of control treatment.
Immunofluorescence
Human umbilical vein endothelial cells were grown to ∼80% confluence on fibronectin-coated glass coverslips, then transfected using FuGENE 6 reagent and the indicated KRIT1 cDNA. Transfected cells were grown for 24 h and then treated for 2 h at 37°C with 5 μM nocodazole or vehicle, then for 60 min at 37°C with 2 μM taxol or vehicle (Invitrogen). Cells were fixed with 3.7% formaldehyde for 60 min at 37°C, permeabilized for 7 min with 0.15% Triton X-100, and blocked for 1 h with 10% normal goat serum (Invitrogen) in Tris-buffered saline, pH 7.4, with 0.01% Triton X-100 (TBST). Cells were probed with mouse anti–β-catenin antibody (BD Biosciences), anti–VE-cadherin (Cell Signaling), or mouse antitubulin (Sigma-Aldrich) in TBST and with rabbit anti-GFP (Clontech) (all at 1:1000) overnight in a humidified chamber. Coverslips were washed with TBST and then incubated with indicated Alexa 568–labeled anti–mouse immunoglobulin (IgG) and anti–rabbit IgG Alexa 488 (Invitrogen) at 1:1000 overnight at 4°C. Coverslips were washed in TBST and PBS (pH 7.4) and then mounted using Prolong Gold (Invitrogen). Imaging was performed by a microscope (Leica LR23) with a charge-coupled device camera (model 2.2.1; Diagnostic Instruments, Sterling Heights, MI) or a confocal microscopy system (Leica TCS SP2 AOBS with DMRE microscope and HCX PL APO 63×/1.32 oil objective). Images were acquired for each experiment using the same gamma setting for all samples and processed using Photoshop CS II software (Adobe, San Jose, CA). Images shown are representative of results from at least three independent experiments.
Zebrafish studies
We maintained and bred antibody wild-type zebrafish and Tg(flk1:EGFP) mutant zebrafish. We injected antisense morpholino oligonucleotides (Gene-Tools, Eugene, OR) that target splice junctions of krit1 (Mably et al., 2006) or standard nontargeting control morpholino oligonucleotides (Gene-Tools) into the yolks of one-cell-stage embryos at a dose of 1 ng. To rescue the enlarged heart phenotype caused by the krit1 morpholino, we coinjected the one-cell-stage embryos with 1 ng of the krit1 morpholino and 400 pg of cRNA encoding HA-tagged krit1 or krit1 (R449E). We used the mMessage mMachine T7 ultra kit (Ambion, Austin, TX) to prepare cRNAs. We mounted the 48-hpf live embryos laterally in 1.2% agarose and acquired images using a fluorescence imaging system (Leica AF6000 with M205 FA stereomicroscope and Leica DFC310 FX digital color camera).
Paraformaldehyde-fixed (4%) embryos were kept in methanol and rehydrated in incubation buffer containing 1× PBS, 0.5% Triton X-100, 5% bovine serum albumin, and 2% goat serum. The primary antibody, rabbit anti-HA tag (Clontech), was used at 1:200 dilution. The secondary antibody, goat anti–rabbit IgG (H+L) Alexa Fluor 488 (Invitrogen), was used at 1:500 dilution. Stained embryos were mounted in 50% glycerol images acquired as described earlier. Images were acquired for each experiment using the same gamma setting for all examples and processed using Photoshop CS II software.
Microtubule sedimentation assay
U2OS cells were transfected with plasmids and treated with either vehicle or nocodazole as indicated. The microtubule fraction was prepared using a cytoskeleton isolation and assay kit (Cytoskeleton, Denver, CO). After centrifugation, the pellets were redissolved in cold water on ice for 1 h. To assess GFP-KRIT1 cosedimentation, polyclonal anti-GFP antibody (Clontech) was used for immunoprecipitation at 4°C overnight. Protein G-Sepharose was added to the reaction mixture and further incubated for 4 h at 4°C. After three washings with lysis buffer, beads were mixed with sample buffer and subjected to SDS–PAGE. Proteins were detected by immunoblotting with monoclonal anti-GFP antibody. The presence of tubulin was assessed by immunoblotting with monoclonal anti–β-tubulin antibody (BD Biosciences).
Statistics
We calculated p values by using an unpaired two-tailed Student's t test.
Supplementary Material
Acknowledgments
The authors are grateful to I. L. Barsukov (University of Liverpool, Liverpool, United Kingdom) for providing access to the ITC facility and help with the ITC experiments and B. T. Goult (Leicester University, Leicester, United Kingdom) for technical advice and assistance. J.J.L. was supported by an American Heart Association postdoctoral fellowship. These studies were supported by grants from the National Institutes of Health to M.H.G.
Abbreviations used:
- CCM
cerebral cavernous malformations
- FERM
band 4.1/ezrin/radixin/moesin
- hpf
hours postfertilization
- ITC
isothermal titration calorimetry
- KRIT1
Krev Interaction Trapped gene 1
- RBD
Ras-binding domain
- RE
arginine 452 to glutamic acid
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-02-0157) on June 1, 2011.
REFERENCES
- Benkert P, Kunzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud-Dufour S, Gautier R, Albiges-Rizo C, Chardin P, Faurobert E. Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. FEBS J. 2007;274:5518–5532. doi: 10.1111/j.1742-4658.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bos JL. Linking Rap to cell adhesion. Curr Opin Cell Biol. 2005;17:123–128. doi: 10.1016/j.ceb.2005.02.009. [DOI] [PubMed] [Google Scholar]
- Cullere X, Shaw SK, Andersson L, Hirahashi J, Luscinskas FW, Mayadas TN. Regulation of vascular endothelial barrier function by Epac, a cAMP-activated exchange factor for Rap GTPase. Blood. 2005;105:1950–1955. doi: 10.1182/blood-2004-05-1987. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francalanci F, et al. Structural and functional differences between KRIT1A and KRIT1B isoforms: a framework for understanding CCM pathogenesis. Exp Cell Res. 2009;315:285–303. doi: 10.1016/j.yexcr.2008.10.006. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Sakurai A, Sano H, Yamagishi A, Somekawa S, Takakura N, Saito Y, Kangawa K, Mochizuki N. Cyclic AMP potentiates vascular endothelial cadherin-mediated cell-cell contact to enhance endothelial barrier function through an Epac-Rap1 signaling pathway. Mol Cell Biol. 2005;25:136–146. doi: 10.1128/MCB.25.1.136-146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading A, Han J, Stockton RA, Ginsberg MH. KRIT-1/CCM1 is a Rap1 effector that regulates endothelial cell–cell junctions. J Cell Biol. 2007;179:247–254. doi: 10.1083/jcb.200705175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glading AJ, Ginsberg MH. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis Model Mech. 2010;3:73–83. doi: 10.1242/dmm.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorzalczany Y, Sigal N, Itan M, Lotan O, Pick E. Targeting of Rac1 to the phagocyte membrane is sufficient for the induction of NADPH oxidase assembly. J Biol Chem. 2000;275:40073–40081. doi: 10.1074/jbc.M006013200. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Gunel M, Laurans MS, Shin D, DiLuna ML, Voorhees J, Choate K, Nelson-Williams C, Lifton RP. KRIT1, a gene mutated in cerebral cavernous malformation, encodes a microtubule-associated protein. Proc Natl Acad Sci USA. 2002;99:10677–10682. doi: 10.1073/pnas.122354499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, et al. Reconstructing and deconstructing agonist-induced activation of integrin alphaIIbbeta3. Curr Biol. 2006;16:1796–1806. doi: 10.1016/j.cub.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Herrmann C, Nassar N. Ras and its effectors. Prog Biophys Mol Biol. 1996;66:1–41. doi: 10.1016/s0079-6107(96)00015-6. [DOI] [PubMed] [Google Scholar]
- Hogan C, Serpente N, Cogram P, Hosking CR, Bialucha CU, Feller SM, Braga VM, Birchmeier W, Fujita Y. Rap1 regulates the formation of E-cadherin-based cell-cell contacts. Mol Cell Biol. 2004;24:6690–6700. doi: 10.1128/MCB.24.15.6690-6700.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinbara K, Goldfinger LE, Hansen M, Chou FL, Ginsberg MH. Ras GTPases: integrins’ friends or foes? Nat Rev Mol Cell Biol. 2003;4:767–776. doi: 10.1038/nrm1229. [DOI] [PubMed] [Google Scholar]
- Kleaveland B, et al. Regulation of cardiovascular development and integrity by the heart of glass-cerebral cavernous malformation protein pathway. Nat Med. 2009;15:169–176. doi: 10.1038/nm.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooistra MR, Corada M, Dejana E, Bos JL. Epac1 regulates integrity of endothelial cell junctions through VE-cadherin. FEBS Lett. 2005;579:4966–4972. doi: 10.1016/j.febslet.2005.07.080. [DOI] [PubMed] [Google Scholar]
- Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF, Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- Lim CJ, Han J, Yousefi N, Ma Y, Amieux PS, McKnight GS, Taylor SS, Ginsberg MH. Alpha4 integrins are type I cAMP-dependent protein kinase-anchoring proteins. Nat Cell Biol. 2007;9:415–421. doi: 10.1038/ncb1561. [DOI] [PubMed] [Google Scholar]
- Mably JD, Chuang LP, Serluca FC, Mohideen MA, Chen JN, Fishman MC. santa and valentine pattern concentric growth of cardiac myocardium in the zebrafish. Development. 2006;133:3139–3146. doi: 10.1242/dev.02469. [DOI] [PubMed] [Google Scholar]
- Nassar N, Horn G, Herrmann C, Scherer A, McCormick F, Wittinghofer A. The 2.2 A crystal structure of the Ras-binding domain of the serine/threonine kinase c-Raf1 in complex with Rap1A and a GTP analogue. Nature. 1995;375:554–560. doi: 10.1038/375554a0. [DOI] [PubMed] [Google Scholar]
- Pfaff M, Liu S, Erle DJ, Ginsberg MH. Integrin beta cytoplasmic domains differentially bind to cytoskeletal proteins. J Biol Chem. 1998;273:6104–6109. doi: 10.1074/jbc.273.11.6104. [DOI] [PubMed] [Google Scholar]
- Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J Comput Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raaijmakers JH, Bos JL. Specificity in Ras and Rap signaling. J Biol Chem. 2009;284:10995–10999. doi: 10.1074/jbc.R800061200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, et al. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum Mol Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- Sahoo T, et al. Computational and experimental analyses reveal previously undetected coding exons of the KRIT1 (CCM1) gene. Genomics. 2001;71:123–126. doi: 10.1006/geno.2000.6426. [DOI] [PubMed] [Google Scholar]
- Sakurai A, Fukuhara S, Yamagishi A, Sako K, Kamioka Y, Masuda M, Nakaoka Y, Mochizuki N. MAGI-1 is required for Rap1 activation upon cell-cell contact and for enhancement of vascular endothelial cadherin-mediated cell adhesion. Mol Biol Cell. 2006;17:966–976. doi: 10.1091/mbc.E05-07-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21–22. Oncogene. 1997;15:1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- Spaargaren M, Bischoff JR. Identification of the guanine nucleotide dissociation stimulator for Ral as a putative effector molecule of R-ras, H-ras, K-ras, and Rap. Proc Natl Acad Sci USA. 1994;91:12609–12613. doi: 10.1073/pnas.91.26.12609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207:881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tozer GM, Kanthou C, Baguley BC. Disrupting tumour blood vessels. Nat Rev Cancer. 2005;5:423–435. doi: 10.1038/nrc1628. [DOI] [PubMed] [Google Scholar]
- Tucker J, Sczakiel G, Feuerstein J, John J, Goody RS, Wittinghofer A. Expression of p21 proteins in Escherichia coli and stereochemistry of the nucleotide-binding site. EMBO J. 1986;5:1351–1358. doi: 10.1002/j.1460-2075.1986.tb04366.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verin AD, Birukova A, Wang P, Liu F, Becker P, Birukov K, Garcia JG. Microtubule disassembly increases endothelial cell barrier dysfunction: role of MLC phosphorylation. Am J Physiol Lung Cell Mol Physiol. 2001;281:L565–L574. doi: 10.1152/ajplung.2001.281.3.L565. [DOI] [PubMed] [Google Scholar]
- Whitehead KJ, Plummer NW, Adams JA, Marchuk DA, Li DY. Ccm1 is required for arterial morphogenesis: implications for the etiology of human cavernous malformations. Development. 2004;131:1437–1448. doi: 10.1242/dev.01036. [DOI] [PubMed] [Google Scholar]
- Wittchen ES, van Buul JD, Burridge K, Worthylake RA. Trading spaces: Rap, Rac, and Rho as architects of transendothelial migration. Curr Opin Hematol. 2005;12:14–21. doi: 10.1097/01.moh.0000147892.83713.a7. [DOI] [PubMed] [Google Scholar]
- Zawistowski JS, Serebriiskii IG, Lee MF, Golemis EA, Marchuk DA. KRIT1 association with the integrin-binding protein ICAP-1: a new direction in the elucidation of cerebral cavernous malformations (CCM1) pathogenesis. Hum Mol Genet. 2002;11:389–396. doi: 10.1093/hmg/11.4.389. [DOI] [PubMed] [Google Scholar]
- Zawistowski JS, Stalheim L, Uhlik MT, Abell AN, Ancrile BB, Johnson GL, Marchuk DA. CCM1 and CCM2 protein interactions in cell signaling: implications for cerebral cavernous malformations pathogenesis. Hum Mol Genet. 2005;14:2521–2531. doi: 10.1093/hmg/ddi256. [DOI] [PubMed] [Google Scholar]
- Zhang J, Clatterbuck RE, Rigamonti D, Chang DD, Dietz HC. Interaction between krit1 and icap1alpha infers perturbation of integrin beta1-mediated angiogenesis in the pathogenesis of cerebral cavernous malformation. Hum Mol Genet. 2001;10:2953–2960. doi: 10.1093/hmg/10.25.2953. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rigamonti D, Dietz HC, Clatterbuck RE. Interaction between krit1 and malcavernin: implications for the pathogenesis of cerebral cavernous malformations. Neurosurgery. 2007;60:353–359. doi: 10.1227/01.NEU.0000249268.11074.83. discussion 359. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.